Abstract
Background
Ovarian cancer is a common type of malignant neoplasm. Its prognosis is poor because the disease is not well understood. Abnormal lipometabolism in peroxisomes is involved in tumor progression and hydroxysteroid dehydrogenase-like 2 (HSDL2), localized in peroxisomes, might be a regulatory factor in lipometabolism. However, the role of HSDL2 in ovarian cancer progression remains unknown.
Material/Methods
HSDL2 expression was detected by qPCR and immunohistochemistry in ovarian tumor samples and qPCR in human ovarian cancer cell lines. Cell proliferation was measured by Celigo and MTT assay. Cell cycle distribution and apoptosis were determined using flow cytometry. Giemsa staining was used for analyzing colony formation. Cell motility was performed using Transwell migration and invasion assays. Tumorigenesis in nude mice was also detected.
Results
HSDL2 expression was upregulated in human ovarian cancer samples and in 3 human ovarian cancer cell lines: SKOV3, HO8910, and OVCAR-3. Higher expression of HSDL2 in ovarian tumor samples was associated with more progressed tumors (P=0.03) and lymphatic metastases (P=0.03). HSDL2 down-regulation by lentiviral-mediated HSDL2 knockdown suppressed cell proliferation, colony formation, and cell motility, while it promoted cell apoptosis and resulted in cell cycle arrest at the G0/G1 phase in human ovarian cancer cell lines OVCAR-3 and SKOV3. HSDL2 knockdown also inhibited tumorigenesis in mouse models.
Conclusions
This study shows that HSDL2 upregulation is associated with ovarian cancer progression. HSDL2 knockdown inhibited cell proliferation, colony formation, motility, and tumorigenesis. It induced apoptosis and cell cycle arrest and might therefore serve as a potential target for ovarian cancer therapy.
MeSH Keywords: Apoptosis; Cell Proliferation; Ovarian Neoplasms; RNA, Small Interfering; Cell Migration Assays; Neoplasm Invasiveness
Background
Ovarian cancer, the most common lethal gynecologic malignancy, is characterized by a high rate of mortality among females with gynecological malignancies worldwide [1] and is the fourth leading cause of death from malignant disease [2]. As a consequence of being so difficult to detect at an early stage [3], patients with ovarian carcinoma are frequently diagnosed in an advanced stage, contributing to a relatively high mortality rate and poor survival [4]. The 5-year survival rate of patients with ovarian cancan has improved significantly with optimal surgical resection and standard chemotherapy [5], but the unpredictable resistance to further chemotherapeutic treatment results in the poor clinical outcomes of ovarian cancer patients, with an approximate 5-year survival rate of only 30% [6]. Hence, there is an urgent need to develop new diagnostics, treatments, and prevention methods of this disease based on an in-depth understanding of molecular tumorigenesis of ovarian cancer [7,8].
The protein superfamily of short-chain dehydrogenases/reductases (SDRs) is capable of catalyzing the oxidation and reduction of various substrates, including steroids and fatty acids, and more than 70 members have been identified in humans [9]. Several SDRs exhibit considerable multifunctionality, such as peroxisomal 17β-hydroxysteroid dehydrogenase type 4 (HSD17B4), which is responsible for catalyzing the oxidation of very long-chain fatty acids and sex steroids [10,11]. Previous studies have shown that SDR dysfunction plays a role in several diseases, such as Alzheimer’s disease [12,13], obesity [14], and cancer [15]. However, the functions of many SDRs are still unclear.
Human hydroxysteroid dehydrogenase-like 2, a characterized SDR gene, is located at the chromosome 9q32 loci and encodes the HSDL2 protein [16]. The protein consists of an N-terminal SDR domain, a C-terminal sterol carrier protein 2 (SCP2) domain, and a peroxisomal targeting signal (ARL). It is ubiquitously expressed in human tissues, with particularly high levels in the liver, kidneys, prostate, testes, and ovaries [16]. HSDL2 localizes in peroxisomes in human, mouse, and rat cells [17–19] and may be a key regulatory factor in lipometabolism [20]. Recent evidence shows that abnormal HSDL2 expression is involved human glioma progression [20]; however, the role of HSDL2 in ovarian cancer remains unclear.
In this study, the levels of HSDL2 mRNA expression in human ovarian cancer tissues and cell lines SKOV3, HO8910, and OVCAR-3 were investigated. A lentiviral-mediated small hairpin (shRNA) technology was then used to knock down HSDL2 expression to explore its effect on the biological functions of ovarian cancer cells. Knockdown of HSDL2 significantly inhibits cell proliferation and viability, colony forming ability, motility, and tumor growth in vivo, and promotes cell apoptosis in OVCAR-3 and SKOV3 cells. Taken together, these results show the HSDL2 gene is highly expressed in human ovarian cancer tissues and cells and plays a key role in cell proliferation, apoptosis, colony formation, and tumorigenesis of human malignant ovarian cancer cells. Targeted inhibition of HSDL2 appears to be a novel potential cancer treatment for ovarian cancer.
Material and methods
Patients and tissue samples
Ovarian tumor and non-tumor samples were collected from 74 inpatients in the Department of Gynecology and Obstetrics, Yijishan Hospital of Wannan Medical College (Wuhu, China) from Jan 2009 to Sep 2012. Consent was acquired from all subjects and the experimental protocols were approved by the Ethics Committee of Yijishan Hospital of Wannan Medical College. The ages of the patients at diagnosis ranged from 20 to 79 years old, with a median age of 55 years old. The clinicopathological characteristics, including age at diagnosis, pathological stage, tumor size, lymphatic metastasis, and distant metastasis, are listed in Table 1. All the patients were clinically staged using the criteria of the International Federation of Gynecology and Obstetrics (FIGO) [21]. The survival information was also recorded. Overall survival (OS) was evaluated from the first surgical date to the date of the last follow-up (Aug 2017) or the date of death.
Table 1.
Relationship between HSDL2 expression and clinicopathological characteristics of patients with ovarian cancer.
| Variable | HSDL2 expression | P-value | |
|---|---|---|---|
| Low | High | ||
| Age (year) | |||
| ≤60 | 25 | 15 | 0.85 |
| >60 | 20 | 14 | |
| Tumor size (cm) | |||
| ≤4.0 | 25 | 13 | 0.36 |
| >4.0 | 20 | 16 | |
| Pathological grade | |||
| Low | 10 | 20 | 0.00 |
| High | 35 | 7 | |
| TNM stage | |||
| I/II | 33 | 19 | 0.03 |
| III | 12 | 10 | |
| Lymph node metastasis | |||
| Nx/N0 | 36 | 17 | 0.03 |
| N1/N2/N3 | 9 | 12 | |
| Distant metastasis | |||
| M0 | 42 | 27 | 0.85 |
| M1 | 3 | 2 | |
Immunohistochemistry
Human ovarian tissues were fixed for 24 h in 10% phosphate-buffered neutral formaldehyde, and then embedded in paraffin. The serial slides (5 μm) of ovarian tissues were sliced and processed for immunohistochemical staining with primary anti-HSDL2 antibody (cat. no. ab181174, Abcam, Shanghai, China). Immunoreactivity of HSDL2 was quantified using ImageJ 1.45s freeware (National Institutes of Health, Rockville, MD, USA). The scores of 0, 1, 2, 3, or 4 were designated according to the percentage of positive cells at 0%, 1-25%, 26-50%, 51-75%, or 76-100%, respectively [22]. The staining intensity scores were defined as 0 (−), 1 (+), 2 (++), and 3 (+++). High and low expression of HSDL2 was defined as ≥2 and <2, respectively, according to the calculated product of the positive cells and intensity score.
Cell culture
Human epithelial ovarian cancer cell lines SKOV3, HO8910, and OVCAR-3, human normal ovarian epithelial cell line IOSE80, and human embryonic kidney (HEK) cell line 293T were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All cells were cultured with RPMI-1640 medium (Gibco, Scotland, UK) supplemented with 10% fetal bovine serum (FBS, Sangon Biotech, Shanghai, China), 100 U/mL penicillin/streptomycin (Sangon Biotech), and 2% L-glutamine (Sangon Biotech) at 37°C and 5% CO2 in a humidified atmosphere.
Construction of HSDL2 shRNA lentivirus
The shRNA-specific targeting sequences (5′-CCA GAA GCA GTT AGC AAG AAA-3′ and 5′-GTC TAC TAT GCT GTT TCC TTG-3′) for the HSDL2 gene and a scrambled shRNA sequence (5′-TTC TCC GAA CGT GTC ACG T-3′) used as negative control were designed and synthesized by Forevergen (Guangzhou, China). The HSDL2 or scrambled hairpin oligonucleotides were cloned into the LV-008-GFP lentiviral vector (Forevergen) with restriction endonuclease AgeI and EcoRI (NEB, Ipswich, MA, USA) and named as shHSDL2-1, shHSDL2-2, and shCtrl, respectively. The lentiviral vector system (Forevergen) was used for producing HSDL2 or scrambled shRNA. Briefly, shHSDL2 or shCtrl plasmids and packaging vectors were cotransfected into 293T cells to produce the respective lentivirus. At 72 h post-transfection, the supernatants containing infection lentivirus vectors were concentrated by ultracentrifugation at 25 000 rpm for 1.5 h and resuspended in PBS. For lentiviral infection, OVCAR-3 and SKOV3 cells were cultured in 6-well plates at a density of 5×104 cells per well and infected with either shHSDL2-1, shHSDL-2, or shCtrl lentiviruses in the presence of 5 μg/mL polybrene (Forevergen). Cells were screened with 2 μg/mL puromycin for 10–15 days to produce stable infected HSDL2-knockdown and scrambled shRNA cells.
shRNA knockdown efficiency detection
Human embryonic kidney 293T cells or human ovarian cancer cells (5×104 per well) were seeded into 6-well plates and then transfected with the plasmids shHSDL2-1, shHSDL2-2, or shCtrl. Cells were incubated at 37°C in a 5% CO2 incubator for 2 days. The appropriate lentivirus particle solution (5 μL per well of virus) was then added (MOI=5). After infection for 3 days, GFP expression from the vector was detected under a fluorescence microscope (IX71, Olympus, Tokyo, Japan) to determine infection efficiency. Cells were collected when the efficiency of the infection exceeded 50% and were then stored for further analysis as described below.
Real-time quantitative PCR (qPCR)
Total RNA was extracted with Trizol reagent (Sangon Biotech, Shanghai, China) according to the manufacturer’s instructions. After quantifying the total RNA using a NanoDrop 2000 device (Thermo Fisher Scientific, Rockford, IL, USA), reverse transcription was carried out using a Takara RNA PCR kit (Takara, Dalian, China) and Oligo dT primers (Invitrogen, Shanghai, China) to obtain cDNA samples as follows. First, 1 μg of Oligo dT was mixed with 2 μg of total RNA and ddH2O to 9 μL. This was incubated at 70°C for 10 min, followed by incubation on ice-water for primer annealing. Then, 4 μL of 5× RT buffer, 2 μL of 10 mM dNTPs, 0.5 μL of RNasin, 3.5 μL of ddH2O, and 1 μL of AMV-RTase were added and incubated at 42°C for 1 h. Finally, the mixture was incubated at 70°C for 10 min to inactivate AMV-RTase. The level of HSDL2 mRNA expression was measured with qPCR using a SYBR master mixture (Takara, Dalian, China) on a Light Cycler 480 device (Roche, Switzerland). PCR reactions were performed as follows: denaturation at 95°C for 5 min, then 40 cycles of 95°C for 10 s, and 60°C for 20 s. Each sample was assessed in triplicate. The primer sequences were designed and synthesized as follows (Forevergen): GAPDH (control) forward: 5′-TGA CTT CAA CAG CGA CAC CCA-3′; GAPDH reverse: 5′-CAC CCT GTT GCT GTA GCC AAA-3′. HSDL2-1 forward: 5′-AAG CCA CTC AAG CAA TCT ATC TG-3′; HSDL2-1 reverse: 5′-GCT CTC CAT ATC CGA CAT TCC C-3′. HSDL2-2 forward: 5′-CTG CTT TAA TCC ACA CAA GGA AC-3′; HSDL2-1 reverse: 5′-TTG TAG GCT GGA TGC TAC CCA GT-3′. The level of HSDL2 mRNA expression was normalized to GAPDH and data were analyzed using the 2−ΔΔCT method [23].
Western blot analysis
Cells were lysed with a lysis buffer (cat. no. C500001, Sangon Biotech, Shanghai, China) for the total protein extraction. Protein concentration was determined using a BCA Protein Assay Kit (cat no. C503051, Sangon Biotech, Shanghai, China). Total protein (20 μg) was separated via a 10% SDS-PAGE gel and transferred to PVDF membranes (Sangon Biotech, Shanghai, China). The PVDF membranes were blocked with 1% bovine serum albumin (BSA) dissolved in TBST for 1 h at room temperature and then incubated with primary antibodies for 1 h at room temperature or overnight at 4°C. The primary antibodies used were as follows: Rabbit anti-HSDL2 (1: 500, cat. no. HPA050453, Sigma); and mouse anti-GAPDH (1: 3000, cat. no.sc-32233, Santa Cruz). After washing with TBST 3 times, goat anti-mouse IgG coupled to HRP (1: 1000, cat. no. sc-2005, Santa Cruz) as secondary antibody was added and was detected using an EasyBlot ECL kit (cat. no. C506668, Sangon Biotech, Shanghai, China).
Cell proliferation assay
Cells were seeded into the 6-well plates at 2×105 cells per well and incubated for 48 h. Cells were collected while in the logarithmic growth phase and reseeded in 96-well plates in triplicate at 2000 cells per well. Cells were then incubated at 37°C in a 5% CO2 incubator for 24 h, and cells were quantified autonomously once a day for 5 days using the Cellomics ArrayScan VTI system (Thermo Fisher Scientific, Rockford, IL, MA, USA). Cell growth curves were generated for each condition.
MTT assay
An MTT assay was also performed to analyze cell viability. Cells growing in the logarithmic phase were reseeded into 96-well plates (2000 cells per well) and maintained at 37°C in a 5% CO2 incubator. Cell viability was measured using an MTT assay each day for 5 days. Briefly, MTT solution (20 μL per well, 5 mg/mL, Dingguo Changsheng, Beijing, China) was added into the culture medium and incubated for 4 h. After discarding the culture medium, DMSO (150 μL per well) was added to dissolve the formazan crystals. The plate was shaken gently for 10 min, and the absorbance at 490/570 nm was quantified using a Tecan Infinite 200 Pro microplate reader (Tecan, Switzerland).
Cell apoptosis assay
Cell apoptosis was measured using an annexin V-APC apoptosis detection kit (cat no. 88-8007, eBioscience, San Diego, CA, USA) according to the manufacturer’s instructions. OVCAR-3 or SKOV3 cells transfected with either the lentiviral shHSDL2-1, shHSDL2-2, or shCtrl were harvested after incubation for 4 days and then resuspended using staining buffer at a final density of 1×106 cells per mL. Then, 100-μL cell suspensions were mixed with 5 μL annexin V-APC and incubated for 10 min at room temperature. Apoptotic signalling was analyzed by a FACS Calibur flow cytometry (Becton-Dickinson, San Jose, CA, USA) according to manufacturer’s guidelines.
Cell cycle analysis
ShHSDL2-1-, shHSDL2-2-, and shCtrl-transfected OVCAR-3 or SKOV3 cells (106 per well) were fixed in ice-cold 70% ethanol for 1 h. The fixed cells were then stained with a cocktail of 50 μg/mL propidium iodide (PI, cat. no. P4170, Sigma-Aldrich, Shanghai, China) dissolved in PBS buffer containing 50 μg/mL of RNase A (cat. no. EN0531, Fermentas) and 0.1% sodium citrate for 30 min at room temperature. Cells were assessed using an FACS Calibur flow cytometer (Becton-Dickinson, San Jose, CA, USA). The cell cycle was analyzed with FlowJo software (TreeStar, Ashland, OR, USA).
Colony formation assay
Cells at the logarithmic proliferation phase were reseeded at 800 cells per well into a 6-well plate with 3 wells per experimental group. Cells were incubated at 37°C and 5% CO2 until the clones could be observed directly. After discarding the culture medium, 1 mL of 4% paraformaldehyde was added to each well and the cells were incubated for 60 min at room temperature. After removing the paraformaldehyde, the clones were dyed with 1 mL per well of crystal violet (cat. no. CB0331, Sangon Biotech, Shanghai, China) for 20 min and counted using a light microscope. Each colony-forming assay was performed in triplicate.
Migration and invasion assays
Cell migration and invasion were examined by Transwell chamber assay (8 μm; Millipore, USA) according to the manufacturer’s instructions. The Transwell chamber was coated with 80 μL Matrigel (BD Biosciences, CA) and 50 μl serum-free medium was added to hydrate Matrigel glue. The suspension (200 μl 1×105 cells/ml) of transfected cell with shHSDL2-1 or shCtrl lentivirus was added to the upper chamber and media with 30% FBS were added to the bottom wells. After culturing for 24 h, cells were fixed with 4% formaldehyde for 15 min and stained with 1% crystal violet. Cell numbers were counted under an optical microscope. Cell migration was also evaluated by using the Transwell chamber assay, while the cell migration assay was not coated with Matrigel. Each experiment was repeated at least 3 times.
Tumorigenesis detection in nude mice
Tumorigenesis in nude mice was determined as previously described [24]. Briefly, female BALB/c nude mice aged 4 weeks were subcutaneously injected with shHSDL2-1- or shCtrl-transfected OVCAR-3 cells (1×105 cell per mouse) at a single site under their front legs. After injections for 5 weeks, the average tumor weight was determined by physical measurement of the excised tumor at the time of sacrifice.
Statistical analysis
In this study, at least 3 separate experiments were carried out. Data are presented as mean ± the standard deviation (SD). Statistical tests were performed using SPSS version 13.0 software for Windows (SPSS Inc, Chicago, USA). The difference between 2 groups was calculated by Wilcoxon’s rank test or the t test. Correlations between the levels of HSDL2 expression and clinicopathological characteristics were carried out using Pearson’s chi-square test. Survival analysis was determined using the log-rank test. A value of P<0.05 was indicated to be statistically significant.
Results
HSDL2 expression in human ovarian cancer samples
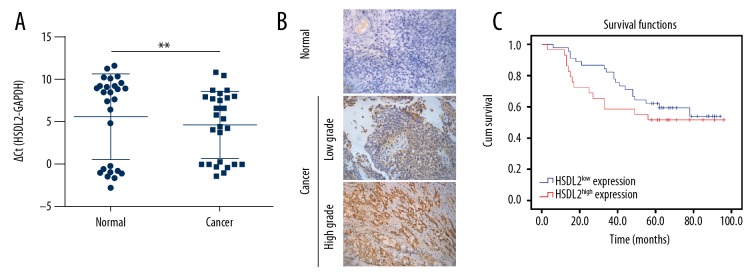
To investigate the HSDL2 expression in human ovarian cancer and matched adjacent non-tumor (referred to as normal) tissues, 27 samples were selected randomly from a total of 74 human ovarian cancer specimens to analyze the level of HSDL2 mRNA using qPCR. The results showed that the levels of HSDL2 mRNA in cancer tissues were significantly different from those in normal tissues (P<0.01) (Figure 1A). To assess the relationship between the levels of HSDL2 expression and human ovarian cancer, a total of 74 human ovarian cancer specimens with different grades and normal tissues were explored. The results revealed that the highly protein level of HSDL2 expression in ovarian tumor samples was observed compared with that in the adjacent non-tumor samples (P<0.01) (Figure 1B). However, Kaplan-Meier survival analysis suggested that the levels of HSDL2 expression in ovarian tumor samples was not correlated with that in the adjacent non-tumor samples (P value for log-rank test: 0.37) (Figure 1C).
Figure 1.
HSDL2 expression in ovarian cancer tissue samples. (A) qPCR showed that the levels of HSDL2 mRNA in cancer tissues were significantly different from those in normal tissues (P<0.01). (B) Immunohistochemical staining showed that HSDL2 was expressed at higher levels in ovarian cancer samples than in adjacent non-tumor samples. (C) Kaplan-Meier survival curve showing the correlation of overall survival with high and low expression of HSDL2 in ovarian cancer. (P=0.37).
In addition, as shown in Table 1, increased HSDL2 expression in ovarian cancer tissues was observed at a higher rate in stage III tissues compared with stage I/II tissues (P=0.03), and at a higher rate in the cases with lymphatic metastasis compared to the cases without lymphatic metastasis (P=0.03), and we also observed significant differences in the tissues with high pathological grades compared with low pathological grades (P<0.01), but expression of HSDL2 was not significantly associated with age, tumor size, or distant organ metastasis (P>0.05).
Construction of lentiviral-mediated shRNA against human HSDL2
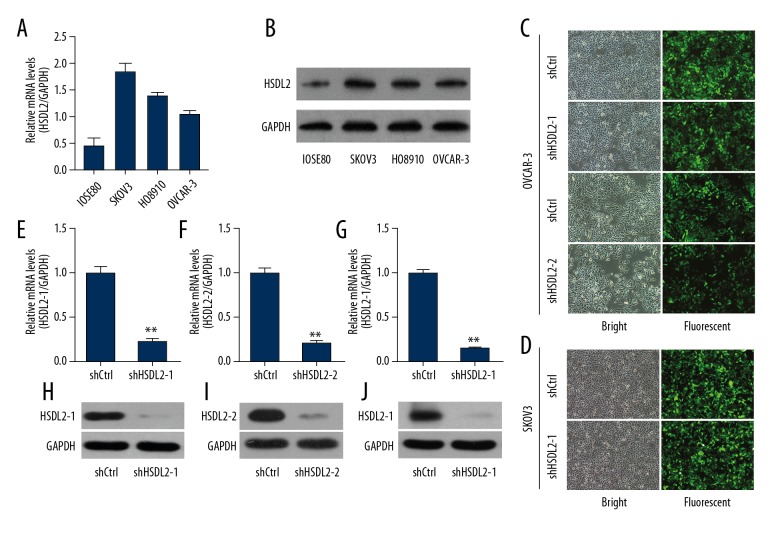
To explore the functional role of HSDL2 in ovarian cancer in vitro and in vivo, the expression of HSDL2 mRNA was initially examined in 3 human ovarian cancer cell lines – SKOV3, HO8910, and OVCAR-3 – and in the human normal ovarian epithelial cell line IOSE80. The results showed that the levels of HSDL2 mRNA in human ovarian cancer cell lines were higher compared with the human normal ovarian epithelial cell line IOSE80 (Figure 2A) and were confirmed by Western blotting (Figure 2B). Then, a lentivirus was constructed to suppress HSDL2 expression via RNAi technology for functional analysis. The results showed that GFP expression from the viral vector was observed in >90% of OVCAR-3 and SKOV3 cells at 48 h after recombinant lentivirus transfection (Figure 2C, 2D), indicating high rates of infection. qPCR analysis showed that shHSDL2, including shHSDL2-1 and shHSDL2-2, were effective in OVCAR-3 and/or SKOV3 cells, with approximately 80% knockdown efficiency observed (Figure 2E–2G). Knockdown efficiency of shHSDL2 lentivirus were then further determined in OVCAR-3 and SKOV3 cells by Western blot analysis, which showed that shHSDL2 could efficiently inhibit the protein level of HSDL2 expression (Figure 2H–2J). Taken together, these results show that a lentiviral-mediated shRNA strategy can stably suppress HSDL2 expression efficiently in human ovarian cancer cells.
Figure 2.
Efficient HSDL2 knockdown at the mRNA and protein levels in the human ovarian cancer OVCAR-3 cells using lentiviral-mediated RNAi. (A, B) HSDL2 mRNA (A) and protein (B) expression in human ovarian cancer cell lines SKOV3, HO8910, and OVCAR-3 and human normal ovarian epithelial cell line IOSE80. (C, D) Representative images of GFP expression in OVCAR-3 cells (C) and SKOV3 (D) after infection of shCtrl, shHSDL2-1, or shHSDL2-2 lentiviruses. (E–G) HSDL2 mRNA expression was analyzed by qPCR and was reduced by shHSDL2-1 or shHSDL2-2 lentiviruses compared to the shCtrl group, respectively. GAPDH was used as an internal control and the data represent the mean ±SD of 3 independent experiments. (** P<0.01). (H–J) Western blot analysis shows that expression of HSDL2 protein was inhibited by lentiviral shHSDL2-1 and shHSDL2-2 in OVCAR-3 (H, I) and lentiviral shHSDL2-1 in SKOV3 (J) cells. GAPDH was used as an internal control.
Knockdown of HSDL2 suppresses cell growth and viability significantly in ovarian cancer cells
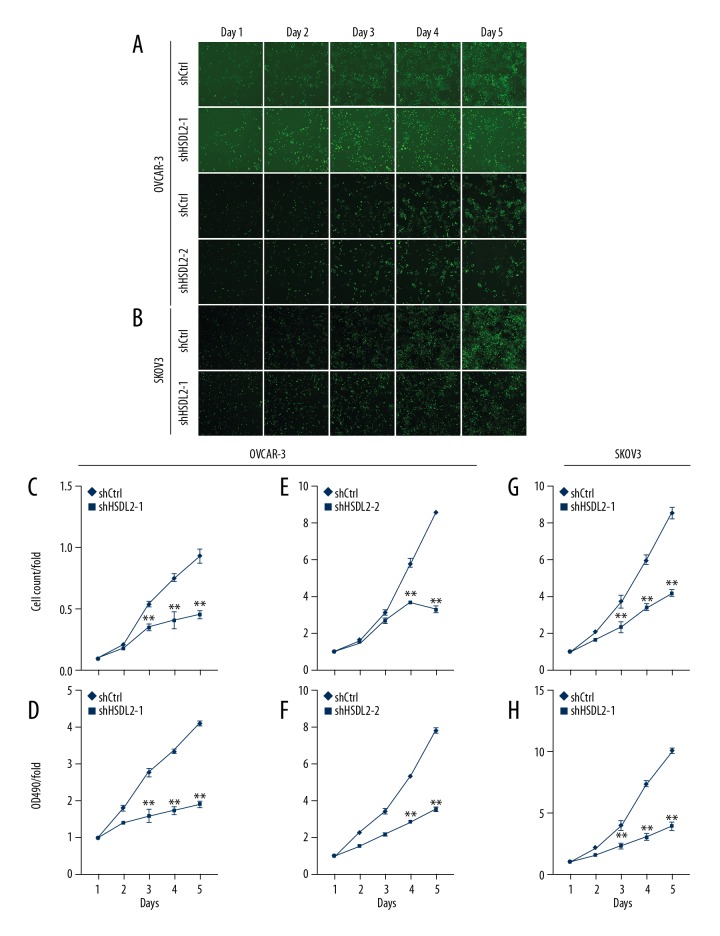
To determine the effect of HSDL2 knocking-down on human ovarian cancer cells, OVCAR-3 and SKOV3 cells were infected with lentiviral shHSDL2-1, shHSDL2-2, or shCtrl, respectively. Cells were then counted every day for 5 days (Figure 3A, 3B). After 48 h of transfection, cell growth was significantly inhibited both in OVCAR-3 and SKOV3 cells transfected with the shHSDL2-1 and shHSDL2-2 lentivirus, respectively, compared to cells transfected with the shCtrl lentivirus (P<0.01, Figure 3B, 3E, 3G). Additionally, shHSDL2-mediated inhibition of cell growth was sustained on the following 3 days.
Figure 3.
HSDL2 knockdown inhibited cell proliferation in the human ovarian cancer cell lines as assessed by the Cellomics ArrayScan VTI and MTT assays. (A, B) Representative images of OVCAR-3 (A) and SKOV3 (B) cells infected with shCtrl lentivirus (top), shHSDL2-1, or shHSDL2-2 lentivirus (bottom) at different time points. (C, E, G) Proliferation of OVCAR-3 (C, E) and SKOV3 (G) was significantly blocked when HSDL2 expression was inhibited. The results are presented as the mean ±SD of 3 separate experiments. (** P<0.01). (D, F, H) Cell proliferation was significantly inhibited as measured by the MTT assay for 5 continuous days. Cell proliferation is shown as fold change at absorbance for 5 continuous days compared to absorbance at OD490 at day 1. The results are presented as the mean ±SD of 3 separate experiments. (** P<0.01).
To further demonstrated the inhibitory effect of shHSDL2 on cell growth, an MTT assay was also carried out to monitor cell viability. As described above, OVCAR-3 and SKOV3 cells were infected with lentiviral shHSDL2-1, shHSDL2-2, or shCtrl, respectively, and the viability profile was analyzed using an MTT assay each day for 5 days. We also showed that knockdown of HSDL2 significantly decreased cell viability after 48 h of transfection with shHSDL2 lentivirus and that the decreased cell viability profiling was continued in the following 3 days, compared with those in shCtrl group (P<0.01, Figure 3D, 3F, 3H). Taken together, these results suggest that HSDL2 promotes ovarian cancer cell progression via accelerated cell growth and proliferation.
HSDL2 knockdown induced cell cycle arrest in OVCAR-3 and SKOV3 cells
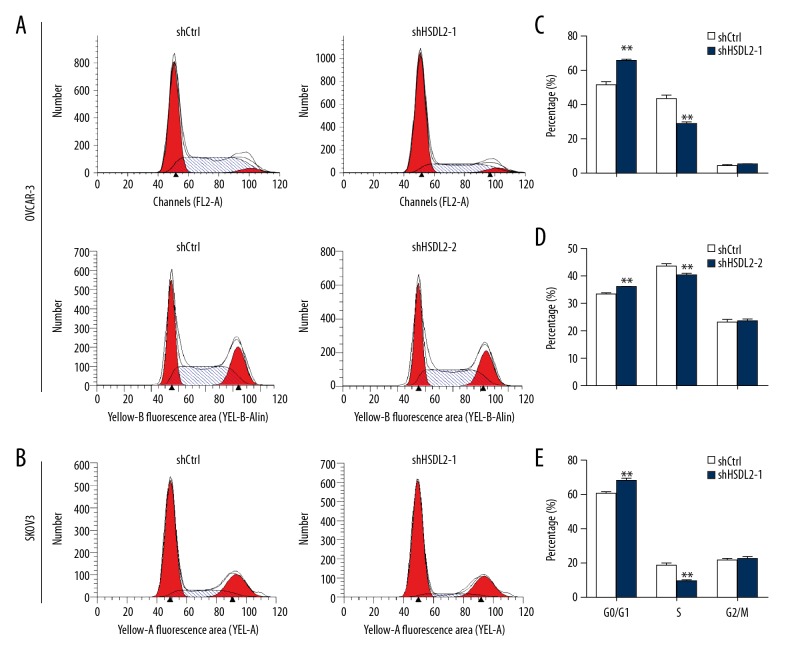
Considering that HSDL2 knockdown significantly suppressed OVCAR-3 and SKOV3 cell growth and proliferation, cell cycle analysis was then carried out to reveal the underlying mechanism of the shHSDL2 lentivirus-mediated inhibitory effect on cell proliferation. The results showed that following transfection with shHSDL2-1 lentivirus, 65.6%, 25.9%, and 5.9% of the cells were in the G0/G1, S, and G2/M phases, respectively (Figure 4A, 4C). Following transfection with shCtrl lentivirus, 37.9%, 56.1%, and 5.8% of the cells were in the G0/G1, S, and G2/M phases, respectively. Similar trends were also observed regarding cell cycle distribution in OVCAR-3 cells with shHSDL2-2 lentivirus (Figure 4A, 4D) and SKOV3 cells with shHSDL2-1 lentivirus (Figure 4B, 4E). The results suggest that suppression of shHSDL2 for HSDL2 expression arrested cells in the G0/G1 phase compared with the shCtrl group (P<0.01).
Figure 4.
HSDL2 knockdown leads to cell cycle arrest. (A, B) Representative image of cell cycle phase distribution following transfection with shHSDL2-1, shHSDL2-2, or shCtrl lentivirus. (C–E) Cell cycle phase distribution is expressed as a percentage of total cells. ** P<0.01.
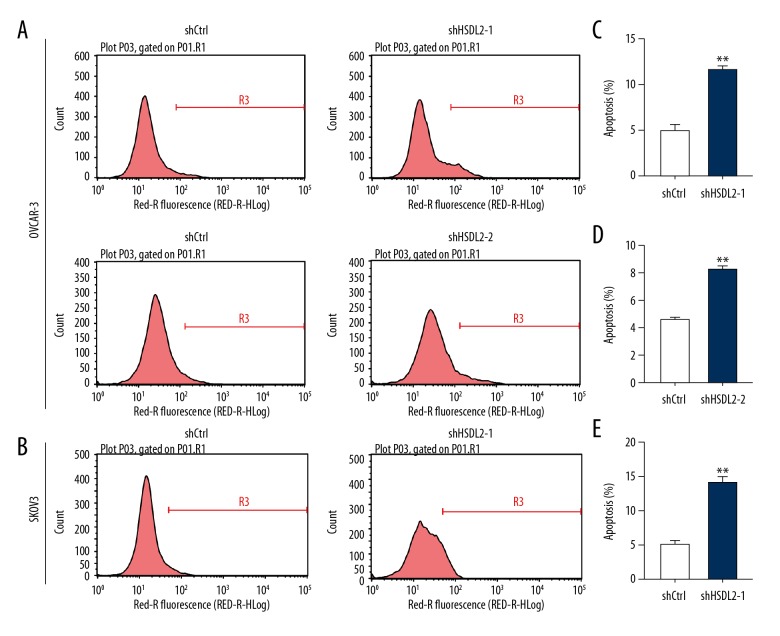
shHSDL2 induced cell apoptosis in ovarian cancer cells
To determine whether HSDL2 knockdown was involved in cell apoptosis, an annexin V-APC assay was performed and fluorescence-activated cells were sorted in OVCAR-3 and SKOV3 cells infected with lentiviral shHSDL2-1, shHSDL2-2, or shCtrl for 5 days using flow cytometry (Figure 5A, 5B). As shown in Figure 5C–5E, approximately 5.0% of apoptotic cells were infected with shCtrl lentivirus, but the apoptotic percentage was increased to 11.7% and 8.3% in OVCAR-3 cells and to 13.9% in SKOV3 cells infected with shHSDL2-1 lentivirus (Figure 5C–5E, P<0.001). The result reveal that knockdown of HSDL2 significantly induced apoptosis of human ovarian cancer cells.
Figure 5.
HSDL2 knockdown augmented cell apoptosis in human ovarian cancer lines. Representative images of cell apoptosis analysis of human ovarian cancer OVCAR-3 (A) and SKOV3 (B) cells infected with lentivirus shCtrl (left), shHSDL2-1, or shHSDL2-2 (right). Cell apoptosis in OVCAR-3 (C, D) and SKOV3 (E) cells was promoted by HSDL2 knockdown. Data shown are the mean ±SD of cell percentage in apoptosis from 3 separate experiments. (** P<0.001).
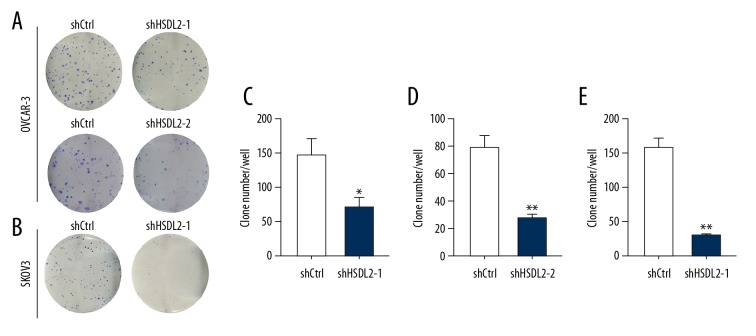
Knockdown of shHSDL2 suppressed cell colony formation
Next, the colony formation of OVCAR-3 and SKOV3 cells treated by shHSDL2 or shCtrl lentivirus was studied. Two groups of OVCAR-3 or SKOV3 cells (shHSDL2- and shCtrl-transfected cells) were grown for 14 days and allowed to form colonies. As shown in Figure 6, HSDL2 knockdown resulted in a significantly reduced number of colonies (70±13 per well in OVCAR-3 cells transfected with shHSDL2-1, Figure 6A and 6C; 79±5 per well in OVCAR-3 cells transfected with shHSDL2-2, Figure 6A and 6D; 29±1 per well in SKOV3 cells transfected with shHSDL2-1, Figure 6B and 6E) as compared with the shCtrl group (148±69 per well and 28±1 per well in OVCAR-3 cells, P<0.01, respectively; 156±8 per well in SKOV3 cells, P<0.01).
Figure 6.
HSDL2 knockdown inhibited human ovarian cancer cell colony formation. (A, B) Photomicrographs of Giemsa-stained OVCAR-3 (A) and SKOV3 (B) clones in 6-well plates at 10 days post-infection. (C–E) The average number of OVCAR-3 (C, D) and SKOV3 (E) colonies was suppressed by HSDL2 knockdown. Data shown are the mean ±SD of cell colonies from 3 separate experiments. (* P=0.017; ** P<0.01).
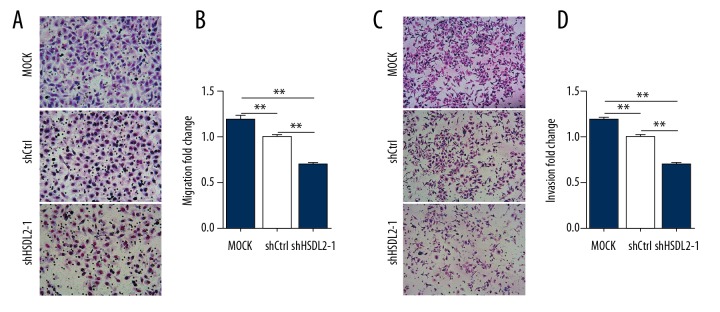
Knockdown of HSDL2 inhibited the motility of OVCAR-3 cells
To investigate the effect of HSDL2 knockdown on the migratory ability of human ovarian cancer cells, Transwell assay was carried out to validate the motility of OVCAR-3 cells transfected with shHSDL2-1 lentivirus. Compared with the shCtrl group, the relative migrated and invaded cell numbers in shHSDL2-1 group were significantly decreased (P<0.01, respectively; Figure 7A–7D), suggesting that knockdown of HSDL2 can reduce cell migration and invasion in ovarian cancer cells. This finding further emphasized the function of HSDL2 in the invasion and metastasis of ovarian cancer cells.
Figure 7.
HSDL2 knockdown inhibited the migration and invasion of human ovarian cancer OVCAR-3 cells. (A, C) Representative Transwell migration (A) and invasion (C) assay of OVCAR-3 cells transfected with shHSDL2-1 lentivirus. (B, D) Quantification of migration (B) and invasion (D) abilities of OVCAR-3 transfected with shHSDL2-1 lentivirus. (** P<0.01).
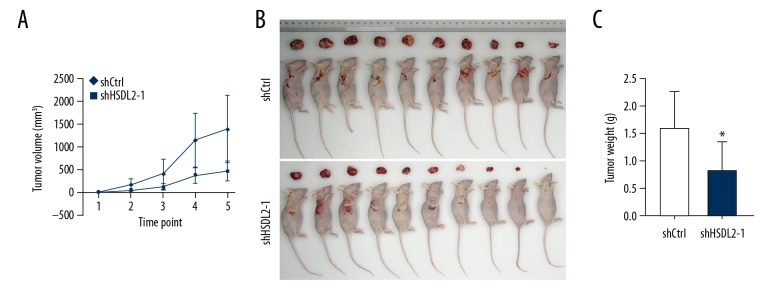
HSDL2 knockdown inhibits tumor growth in vivo
Finally, to determine the potential of shHSDL2 as a therapeutic tool for treating ovarian cancer, nude mice were subcutaneously injected with OVCAR-3 cells successfully transfected with either shHSDL2-1 or shCtrl lentivirus. Five weeks following injection, we observed that tumors grew more slowly in the shHSDL2-1 group compared with shCtrl group (Figure 8A). As expected, tumors from mice injected with the shHSDL2-1-transfected cells were smaller relative to the tumors from mice injected with the shCtrl-transfected cells (Figure 8B, 8C). The result confirmed that HSDL2 knockdown suppressed tumorigenesis in vivo. Taken together, these data indicate that targeting HSDL2 with lentiviral-shHSDL2-1 could have an inhibitory effect in vivo on ovarian cancer in which HSDL2 is overexpressed.
Figure 8.
HSDL2 knockdown repressed the growth of human ovarian cancer OVCAR-3 cells in vivo. (A) Tumors grew more slowly after injection of OVCAR-3 cells transfected with shHSDL2-1 lentivirus than those transfected with shCtrl lentivirus. (A) The sizes of OVCAR-3 implants at 5 weeks after injection. (B) Average weight of the tumors 5 weeks after cell injection, n=10. (* P<0.05).
Discussion
In the present study, results indicated that the levels of HSDL2 expression were elevated in ovarian cancer samples compared with non-tumor tissues. Additionally, higher expression of HSDL2 was observed in patients with pathological grade, advanced tumor stage, and lymphatic metastasis. HSDL2 expression did not significantly differ by age, tumor size, or distant organ metastasis. The results suggest that upregulated levels of HSDL2 expression are associated with the high risk of ovarian cancer development and requires more clinical samples to verify.
qPCR and Western blotting indicated a variable HSDL2 expression in 3 ovarian cancer cell lines – SKOV3, HO8910, and OVCAR-3 – compared with human normal ovarian epithelial cells IOSE80. OVCAR-3 and SKOV3 cell proliferation and cell colony formation were significantly reduced in shHSDL2-transfected cells, while apoptosis was significantly increased. Furthermore, HSDL2 knockdown significantly inhibited tumor growth in vivo. These results suggest that the elevation of HSDL2 expression augmented ovarian cancer cell development. Interestingly, these results show that HSDL2 knockdown arrested cell cycle at G0/G1 phase, indicating that HSDL2 promoted human ovarian cancer cell growth and colony forming ability by modulating the cell cycle. However, the potential mechanism responsible for cell cycle arrest of HSDL2-silenced OVCAR-3 and SKOV3 cells must be explored in future studies.
The encoding gene HSDL2 was initially identified in a cDNA library derived from human fetal brain tissue [16]. HSDL2 protein contains a sterol carrier protein 2 (SCP2) domain, and localizes in peroxisomes and mitochondria in humans, mice, and rats [17,19,25]. However, to date, studies related to HSDL2 function are scant, especially in cancer. Chen et al. [20] reported abnormal elevation of HSDL2 expression in human glioma tissues. HSDL2 was mainly expressed in the cytoplasm as shown by immunohistochemistry [20]. In the present study, we also found abnormally higher levels of HSDL2 mRNA in human ovarian cancer tissues, which might play a pivotal role of ovarian cancer progression.
Little is known about the role of HSDL2 in human ovarian cancer. Therefore, the functional role of HSDL2 was investigated using shHSDL2-transfected OVCAR-3 cells. Chen et al. [20] reported that knockdown of HSDL2 in 2 human glioma cell lines – U-251 and U87 MG – inhibited cell proliferation and induced cell apoptosis. Similarly, the results presented here show that HSDL2 knockdown suppressed cell proliferation and augmented cell apoptosis in OVCAR-3 and SKOV3 human ovarian cancer cells. Additionally, knockdown of HSDL2 also inhibited migration and invasion of OVCAR-3 cells. The results suggest that abnormal expression of HSDL2 is involved in human ovarian cancer progression and might be associated with the advanced tumor stage and lymphatic metastasis of ovarian cancer that we previously observed.
Conclsions
In summary, our study shows that HSDL2 expression is elevated in some ovarian cancer cell lines and tissues. Lentiviral shRNA-mediated HSDL2 knockdown suppressed cell proliferation and colony forming ability via arresting cell cycle at the G0/G1 phase and by promoting cell apoptosis. This indicates that HSDL2 plays a pivotal role in ovarian cancer progression and suggests that HSDL2 is a potential therapeutic target for ovarian cancer treatment.
Footnotes
Source of support: This work was supported by the National Natural Science Foundation of China (no. 81172790 and 81671586), the Key Project of the Natural Science Foundation of Anhui Provincial Educational Department (no. KJ2014A267), and the Academic and Technical Leaders of Wannan Medical College (no. 010202041703)
Conflicts of interest
None.
References
- 1.Reid BM, Permuth JB, Sellers TA. Epidemiology of ovarian cancer: A review. Cancer Biol Med. 2017;14(1):9–32. doi: 10.20892/j.issn.2095-3941.2016.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Bartholomeusz C, Rosen D, Wei C, et al. PEA-15 induces autophagy in human ovarian cancer cells and is associated with prolonged overall survival. Cancer Res. 2008;68(22):9302–10. doi: 10.1158/0008-5472.CAN-08-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu M, Miao Y, Qi L, et al. RNAi-mediated downregulation of FKBP14 suppresses the growth of human ovarian cancer cells. Oncol Res. 2016;23(6):267–74. doi: 10.3727/096504016X14549667333963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goff BA, Sainz de la Cuesta R, Muntz HG, et al. Clear cell carcinoma of the ovary: A distinct histologic type with poor prognosis and resistance to platinum-based chemotherapy in stage III disease. Gynecol Oncol. 1996;60(3):412–17. doi: 10.1006/gyno.1996.0065. [DOI] [PubMed] [Google Scholar]
- 6.Rustin G, van der Burg M, Griffin C, et al. Early versus delayed treatment of relapsed ovarian cancer. Lancet. 2011;377(9763):380–81. doi: 10.1016/S0140-6736(11)60126-8. [DOI] [PubMed] [Google Scholar]
- 7.Pereira D, Assis J, Gomes M, et al. Improvement of a predictive model in ovarian cancer patients submitted to platinum-based chemotherapy: Implications of a GST activity profile. Eur J Clin Pharmacol. 2016;72(5):545–53. doi: 10.1007/s00228-016-2015-3. [DOI] [PubMed] [Google Scholar]
- 8.Canevari S, Gariboldi M, Reid JF, et al. Molecular predictors of response and outcome in ovarian cancer. Crit Rev Oncol Hematol. 2006;60(1):19–37. doi: 10.1016/j.critrevonc.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 9.Kowalik D, Haller F, Adamski J, et al. In search for function of two human orphan SDR enzymes: Hydroxysteroid dehydrogenase like 2 (HSDL2) and short-chain dehydrogenase/reductase-orphan (SDR-O) J Steroid Biochem Mol Biol. 2009;117(4-5):117–24. doi: 10.1016/j.jsbmb.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Moeller G, Adamski J. Multifunctionality of human 17beta-hydroxysteroid dehydrogenases. Mol Cell Endocrinol. 2006;248(1–2):47–55. doi: 10.1016/j.mce.2005.11.031. [DOI] [PubMed] [Google Scholar]
- 11.Breitling R, Marijanovic Z, Perovic D, et al. Evolution of 17beta-HSD type 4, a multifunctional protein of beta-oxidation. Mol Cell Endocrinol. 2001;171(1–2):205–10. doi: 10.1016/s0303-7207(00)00415-9. [DOI] [PubMed] [Google Scholar]
- 12.Borger E, Aitken L, Du H, et al. Is amyloid binding alcohol dehydrogenase a drug target for treating Alzheimer’s disease? Curr Alzheimer Res. 2013;10(1):21–29. [PMC free article] [PubMed] [Google Scholar]
- 13.Borger E, Aitken L, Muirhead KE, et al. Mitochondrial beta-amyloid in Alzheimer’s disease. Biochem Soc Trans. 2011;39(4):868–73. doi: 10.1042/BST0390868. [DOI] [PubMed] [Google Scholar]
- 14.Persson B, Kallberg Y, Bray JE, et al. The SDR (short-chain dehydrogenase/reductase and related enzymes) nomenclature initiative. Chem Biol Interact. 2009;178(1–3):94–98. doi: 10.1016/j.cbi.2008.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang NS, Schultz L, Hsu LJ, et al. 17beta-Estradiol upregulates and activates WOX1/WWOXv1 and WOX2/WWOXv2 in vitro: Potential role in cancerous progression of breast and prostate to a premetastatic state in vivo. Oncogene. 2005;24(4):714–23. doi: 10.1038/sj.onc.1208124. [DOI] [PubMed] [Google Scholar]
- 16.Dai J, Xie Y, Wu Q, et al. Molecular cloning and characterization of a novel human hydroxysteroid dehydrogenase-like 2 (HSDL2) cDNA from fetal brain. Biochem Genet. 2003;41(5–6):165–74. doi: 10.1023/a:1023377627138. [DOI] [PubMed] [Google Scholar]
- 17.Gronemeyer T, Wiese S, Ofman R, et al. The proteome of human liver peroxisomes: Identification of five new peroxisomal constituents by a label-free quantitative proteomics survey. PLoS One. 2013;8(2):e57395. doi: 10.1371/journal.pone.0057395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quan S, Yang P, Cassin-Ross G, et al. Proteome analysis of peroxisomes from etiolated Arabidopsis seedlings identifies a peroxisomal protease involved in beta-oxidation and development. Plant Physiol. 2013;163(4):1518–38. doi: 10.1104/pp.113.223453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keller GA, Scallen TJ, Clarke D, et al. Subcellular localization of sterol carrier protein-2 in rat hepatocytes: Its primary localization to peroxisomes. J Cell Biol. 1989;108(4):1353–61. doi: 10.1083/jcb.108.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruokun C, Yake X, Fengdong Y, et al. Lentivirus-mediated silencing of HSDL2 suppresses cell proliferation in human gliomas. Tumour Biol. 2016;37(11):15065–77. doi: 10.1007/s13277-016-5402-6. [DOI] [PubMed] [Google Scholar]
- 21.Baak JP, Wisse-Brekelmans EC, Langley FA, et al. Morphometric data to FIGO stage and histological type and grade for prognosis of ovarian tumours. J Clin Pathol. 1986;39:1340–46. doi: 10.1136/jcp.39.12.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rui Y, Peng WJ, Wang M, et al. HIST1H3D: A promising therapeutic target for lung cancer. Int J Oncol. 2017;50(3):815–22. doi: 10.3892/ijo.2017.3856. [DOI] [PubMed] [Google Scholar]
- 23.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30(9):e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu N, Bi F, Pan Y, et al. Reversal of the malignant phenotype of gastric cancer cells by inhibition of RhoA expression and activity. Clin Cancer Res. 2004;10(18 Pt 1):6239–47. doi: 10.1158/1078-0432.CCR-04-0242. [DOI] [PubMed] [Google Scholar]
- 25.Brites P, Motley AM, Gressens P, et al. Impaired neuronal migration and endochondral ossification in Pex7 knockout mice: A model for rhizomelic chondrodysplasia punctata. Human Mol Genet. 2003;12(18):2255–67. doi: 10.1093/hmg/ddg236. [DOI] [PubMed] [Google Scholar]