Abstract
Epigenetic alterations contribute to leukemogenesis in childhood acute myeloid leukemia and therefore are of interest for potential therapeutic strategies. Herein, we performed large-scale ribonucleic acid interference screens using small hairpin ribonucleic acids in acute myeloid leukemia cells and non-transformed bone marrow cells to identify leukemia-specific dependencies. One of the target genes displaying the strongest effects on acute myeloid leukemia cell growth and less pronounced effects on nontransformed bone marrow cells, was the chromatin remodeling factor CHD4. Using ribonucleic acid interference and CRISPR-Cas9 approaches, we showed that CHD4 was essential for cell growth of leukemic cells in vitro and in vivo. Loss of function of CHD4 in acute myeloid leukemia cells caused an arrest in the G0 phase of the cell cycle as well as downregulation of MYC and its target genes involved in cell cycle progression. Importantly, we found that inhibition of CHD4 conferred anti-leukemic effects on primary childhood acute myeloid leukemia cells and prevented disease progression in a patient-derived xenograft model. Conversely, CHD4 was not required for growth of normal hematopoietic cells. Taken together, our results identified CHD4 as a potential therapeutic target in childhood acute myeloid leukemia.
Introduction
Acute myeloid leukemia (AML) is a stem cell disease, characterized by rare leukemia-initiating cells (LICs) with increased self-renewal capacity that can propagate rapidly, growing immature myeloid blast cells with limited differentiation capacity.1,2 The LICs are largely resistant to chemotherapy and therefore many patients will ultimately relapse, which accounts for the leading cause of death in AML.3
The genetic, epigenetic and transcriptomic landscape in AML differs significantly between adults and children. Many of the causative lesions identified in adult AML (e.g., IDH1 and DNMT3A mutations) are rare events in childhood AML, whereas other gene mutations are more frequent in childhood AML (e.g., MYC, IKFZ1 and EZH2).4,5 The molecular differences between adult and childhood AML also include alterations in chromosomal copy number, translocations, different micro ribonucleic acid (miRNA) and messenger (m)RNA expression levels as well as epigenetic patterns.4 For example, translocations involving the MLL gene comprise 15% to 20% of all childhood acute myeloid leukemia (AML) cases. In contrast, only around 5% of adult AML patients carry MLL-rearrangements.6 These differences manifest as dissimilar biological characteristics, clinical behavior and different response to treatment.7
The chromodomain helicase DNA-binding protein 4 (CHD4) is an adenosine triphosphate (ATP)-ase dependent chromatin remodeling factor and a component of the nucleosome remodeling and histone deacetylation (NuRD) complex and plays an important role in epigenetic transcriptional gene regulation.8 CHD4 has been linked to oncogenic processes, including control of cell cycle progression,9–12 cancer metastasis, epithelial-to-mesenchymal transition,13 and epigenetic suppression of tumor suppressor genes.13 Although the role of CHD4 in AML is largely unknown, inhibition of the chromatin remodeler has been reported to reduce AML tumor formation and sensitize AML cells to genotoxic drugs via the increased accessibility of DNA and impaired double strand break repair.14
A large number of functional screens have identified essential genes in various cancer cells,15 including AML.16–27 However, AML-specific vulnerabilities have not been studied in detail. In the study herein, we performed loss of function screens on a large scale in AML cells and non-transformed bone marrow cells (BMs) in order to identify potential AML-specific vulnerabilities. CHD4 was identified as being required for cell growth and disease progression for primary childhood AML patient samples, but not for primary blood cells. Inhibition of CHD4 resulted in a downregulation of MYC and its target genes as well as a growth arrest in the G0 phase of the cell cycle.
Methods
Cell growth assays of primary childhood AML samples
The investigation was conducted in accordance with the ethical standards and according to the Declaration of Helsinki and to national and international guidelines, and has been approved by the authors’ institutional review board. Culturing of the childhood samples was carried out as previously reported.28 MS-5 cells (DSMZ) were radiated at 80 Gy and plated at a density of 10,000 cells/well in MyeloCult media H5100 (STEMCELL Technologies Inc.) in a collagen I Cellware 96-well plate (Corning), two to three days before plating the cells. 10,000-20,000 cells suspended in MyeloCult media supplemented with recombinant human interleukin-6 (rhIL-6), recombinant human interleukin-3 (rhIL-3), recombinant human Fms-like tyrosine kinase 3/fetal liver kinase-2 (rhFl3/Flk-2) ligand, recombinant human thrombopoietin (rhTPO), recombinant human stem-cell factor (rhSCF) and recombinant human granulocyte colony-stimulating factor (rhG-CSF; STEMCELL Technologies Inc.) at a concentration of 20 ng/mL, were added to each well. The cells were maintained at normoxic conditions and effects in cell growth (LICs and leucocytes) were determined by flow cytometric analysis (see Online Supplementary Table S1 for antibodies).
Flow cytometric analysis and sorting
Flow cytometric analysis was performed with a 4-laser BD LSRFortessa. Primary childhood AML cells were harvested and incubated in anti-CD16/32 (Fc-block) antibodies against mouse (Biolegend) and human (ChromPure Mouse IgG, Jackson ImmmunoResearch) for 20 minutes on ice. Then, the cells were stained with: human CD45, CD34, CD38 and lineage antibodies (CD20, CD4, CD8, CD2, CD56, CD235b, CD3 and CD19) and incubated on ice for 20 minutes (see Online Supplementary Table S1 for antibodies). Dead cells were excluded using the Near-IR Live/Dead marker (Invitrogen). Human CD45 positive cells were analyzed by a high-throughput automated plate reader (BD LSRFortessa).
For the cell growth competition assays, cells were harvested and washed with cold phosphate-buffered saline (PBS) and thereafter stained with Near-IR Live/Dead marker in a 96-well plate in 80 μl of PBS and 2% fetal bovine serum (FBS). A high-throughput automated plate reader was used to detect the absolute number of live cells.
To determine the level of engraftment of human AML cells in transplanted NOD scid γ mice expressing human SCF, GM-CSF, and IL-3 (NSG-SGM3), BMs were isolated from the tibia and femur. The isolated BMs were incubated in mouse and human FC blocking antibodies for 20 minutes on ice. After washing, the cells were stained with human anti-CD45 on ice for 20 minutes. Then cells were incubated with Near-IR Live/Dead marker to detect live cells. Analysis was performed by FlowJo Version 9.3.3 software (Tree Star Inc.).
Results
Identification of target genes selectively required for growth of MLL-AF9 rearranged AML cells by large-scale short hairpin RNA (shRNA) screening
To identify novel target genes that are required for growth of AML cells expressing the MLL-AF9 fusion oncogene, we performed shRNA-based screens of two human AML cell lines (NOMO-1 cells derived from an adult, and THP-1 cells derived from a 1-year-old child) and a mouse AML cell line, all of which carried the MLL-AF9 translocation. Non-transformed mouse Factor-Dependent Continuous Paterson Laboratories (Factor dependent cell-Paterson [FDCP]-mix) BMs were used as control cells in the screens.29 The RNA interference (RNAi) screening systems consisted of approximately 27000 lentiviral shRNAs targeting around 5400 putative disease-associated and drug targets (Cellecta Inc.).
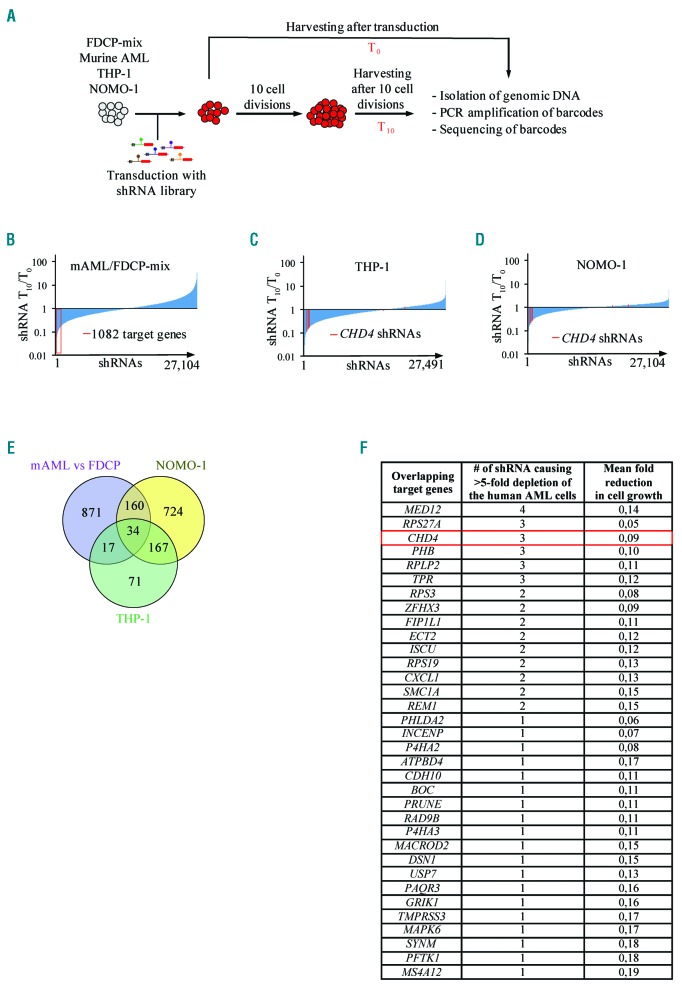
As outlined in Figure 1A, the barcoded lentiviral libraries were transduced as a pool into respective cell lines. The cells were harvested at an initial time point and after ten cell divisions. Next-generation sequencing of the polymerase chain reaction (NGS PCR) amplified barcodes from genomic DNA was followed by deconvolution and normalization of the data (Online Supplementary Table S2).
Figure 1.
Large-scale shRNA-based RNAi screens identify novel target genes required for cell growth of AML cells. A. Schematic illustration of the shRNA-based screening procedure. B. Bar chart representing ratios of barcodes from 27104 individual shRNAs after screens in murine MLL-AF9 transformed AML cells compared to murine FDCP-mix BMs, after 10 cell divisions (T10). The 1086 bar-codes with at least a five-fold lower representation in the murine MLL-AF9 AML cells compared to the murine FDCP-mix cells are highlighted in red. C and D. Bar charts representing differences in ratios of barcodes from 27491 individual shRNAs (THP-1) and 27104 (NOMO-1) after pooled loss of function screens after T10. Individual shRNAs targeting CHD4 are highlighted in red. E. Venn diagram illustrating the overlap between target genes that are five-fold more depleted in mouse AML cells compared to mouse FDCP-mix control cells, five-fold depleted target genes in THP-1 cells and NOMO-1 cells after T10. F. List of the 34 target genes overlapping in all three screens in the Venn diagram from Figure 1E together with information about the number of individual shRNAs resulting in at least a five-fold reduction in abundance of barcodes in the screens of the human AML and the mean fold reduction in abundance of barcodes in the screens after T10. FDCP: factor dependent cell-Paterson; AML: acute myeloid leukemia; PCR: polymerase chain reaction; shRNA: short hairpin RNA.
To identify target genes that were selectively required for AML cell growth, we first determined the ratio of each individual barcode in the mouse AML cells compared to the mouse FDCP-mix cells after ten cell divisions. We identified 1082 target genes with at least a fivefold higher effect in cell growth in the mouse AML cells compared to the FDCP-mix cells (Figure 1B). By comparing these 1082 target genes with the corresponding human target genes of the human THP-1 and NOMO-1 cells (Figure 1C,D), we identified 34 shRNA-target genes that overlapped with all three screens (Figure 1E). Notably, 201 of the 289 total target genes from the screens of the two human AML cell lines overlapped (P=2.4E-215, hypergeometric test), indicating a conserved functional importance of these genes in AML maintenance (Figure 1E).
Although most of the identified target genes had not been reported to have a role in AML, some have previously been linked to the disease, including MED12,30 USP7,31 FIP1L1,32 and SMC1A.33 Additionally, a significant proportion of the genes (15 of 34) were targeted by multiple shRNAs (Figure 1F). Gene ontology analysis revealed that the 34 target genes were associated with a broad range of functions and protein classes in different compartments of the cells (Online Supplementary Figure S1). Genetic alterations of the 34 target genes occurred frequently in various cancers (Online Supplementary Figure S2). In addition, TMPRSS3 (amplifications), ZFHX3 (deletions), P4HA2 (deletions) and SMC1A (missense mutations and amplifications) were found in more than 2% of AML patient samples.34 Remarkably, mRNA expression was significantly correlated with the decreased overall survival of AML for eight of the 34 target genes.35 Moreover, 17 of the target genes were found to be markedly transcriptionally upregulated in MLL-rearranged AML patient samples compared to healthy hematopoietic stem and progenitor cells (HSPCs).36 Thus, the data suggest that the screens were valid for identification of novel target genes in MLL-AF9 rearranged AML.
CHD4 is required for cell growth of leukemia cells and disease progression in vivo
In the screens, multiple shRNA vectors against CHD4 strongly inhibited expansion of the two human AML cell lines (Figure 1C,D). Moreover, three out of six shRNAs targeting CHD4 in the screens caused more pronounced reduction in cell growth in the murine AML cells compared to the FDCP-mix control cells (fold change: 0.19, 0.32, 0.38, respectively) (Online Supplementary Table S2). Both CHD4 and the NuRD complex have been reported to be required for cell growth of various types of cancer cells.9,13,14,37–39 Taken together with the fact that CHD4 is a potentially “druggable” enzyme, our studies proceeded to focus on this epigenetic factor.
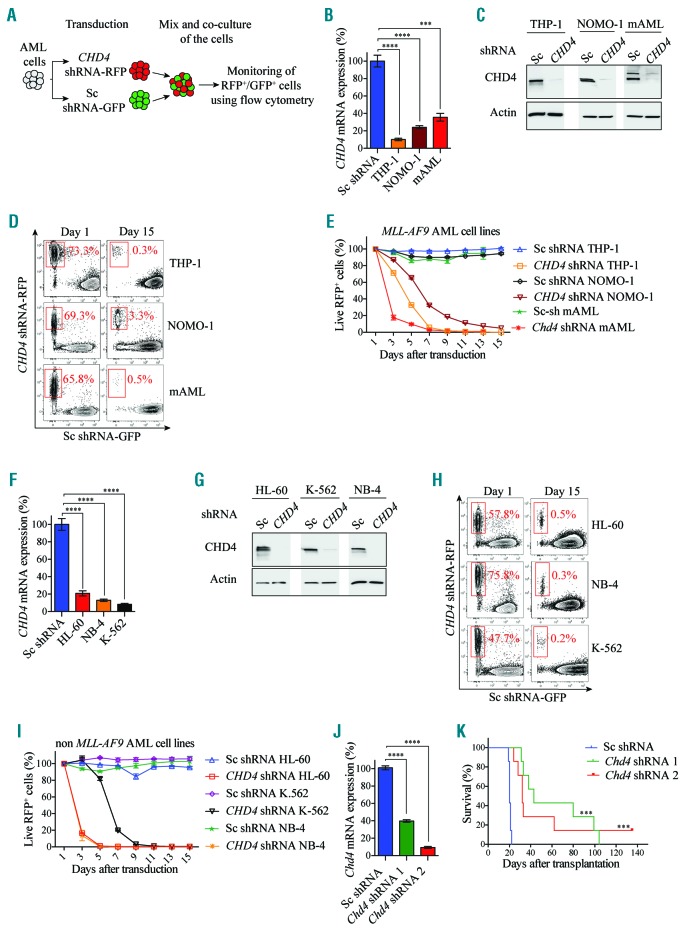
To validate the importance of CHD4 in AML, we performed a pairwise cell growth competition assay, allowing monitoring of control and CHD4 targeted cells under the same conditions (Figure 2A). shRNA-based inhibition using two different vectors resulted in efficient reduction in mRNA (Figure 2B; Online Supplementary Figure S3A) and protein levels (Figure 2C; Online Supplementary Figure S3B) of CHD4 as compared to control cells transduced with a negative control vector.
Figure 2.
CHD4 inhibition impairs cell growth of leukemic cell lines and disease progression in vivo. A. Schematic drawing of the in vitro cell growth competition assay. B and F. Bar charts represent real time PCR analysis of mRNA levels after shRNA-based knockdown of CHD4, relative to control cells transduced with vectors expressing scrambled shRNA in MLL-rearranged THP-1, NOMO-1, and murine AML cells (Figure 2B), or in non-MLL rearranged HL-60, K-562 and NB-4 cells (Figure 2F), at 72 hours post transduction. Transduced cells were used for flow cytometry and growth competition assays (Figure 2D, E, H, I). mRNA levels were normalized to UBC. The data is represented as the mean ±S.E.M., ***P<0.005, ****P<0.001 (unpaired t-test), n=3. C and G. Western blot analyses of the endogenous CHD4 and Actin levels with and without knockdown of CHD4 in the indicated cells. D and H. Representative flow cytometry plots showing percentage of live RFP+ CHD4 knockdown cells in red (CHD4 shRNA-RFP) relative to GFP+ control cells (Sc shRNA-GFP), at the indicated days after transduction. E and I. Line charts depict the percentage of live RFP+ cells normalized to 100% at the initial time point (Day one) of the indicated cells determined by flow cytometry analysis at the indicated time points. J. Bar charts represent real time PCR analysis of mRNA levels after shRNA-based knockdown of Chd4 using two independent lentiviral vectors, relative to control cells transduced with vectors expressing scrambled shRNA in mouse AML cells that were subsequently transplanted into recipient mice for survival studies (Figure 2K) at 72 hours post transduction. mRNA levels were normalized to Hprt. The data is represented as the mean ±S.E.M., ****P<0.001 (unpaired t-test), n=3. K. Kaplan-Meier survival curves of wild-type (wt) C57BL/6 mice transplanted with 200,000 of murine MLL-AF9 transformed AML cells transduced with the two individual shRNA against CHD4, or the negative control vector. Percent survival and the number of animals for each cohort (time to euthanasia of moribund animals), were plotted against time, in days. Mean time survival of animals transplanted with control vector was 20 days, whereas it was 32 and 43 days for animals transplanted with cells transduced with the two vectors against CHD4 (***P<0.005). Seven animals were used for each experiment. AML: acute myeloid leukemia; shRNA: short hairpin RNA; Sc: scramble control; RFP: red fluorescent protein; GFP: green fluorescent protein.
Longitudinal flow cytometric analysis revealed that THP-1, NOMO-1 and mouse AML cells subjected to CHD4 knockdown by two independent shRNAs were strongly inhibited in cell growth compared to control cells (Figure 2D,E; Online Supplementary Figure S3C,D).
Given the demonstrated importance in MLL-rearranged AML cells, we then investigated whether CHD4 was also required for cell growth of leukemic cells carrying differ ent types of genetic lesions. Knockdown of CHD4 with two distinct shRNAs caused a significant reduction in mRNA (Figure 2F; Online Supplementary Figure S3E) as well as protein levels (Figure 2G; Online Supplementary Figure S3F), and prevented cell growth of the HL-60 (promyelocytic leukemia cells), K562 cells (chronic myelogenous leukemia) and NB-4 (acute promyelocytic leukemia), to a similar degree as that of the MLL-AF9 rearranged AML cells (Figure 2H,I; Online Supplementary Figure S3G,H).
To investigate if CHD4 also has a role in AML disease progression, we used a murine immune competent transplantation model.40 As previously reported, wild-type (wt) mice that received transplants of MLL-AF9 transformed AML mouse cells, that were transduced with a negative control vector, resulted in pathologic and clinical manifestations of human AML within 19 to 21 days post transplantation.40 In contrast, wt recipient mice that received transplants of MLL-AF9 transformed AML cells that were transduced with two individual shRNA vectors, which mediated efficient suppression in CHD4 mRNA levels (Figure 2J), survived significantly longer than mice transplanted with control cells (Figure 2K).
CDH4 is not required for cell growth of normal hematopoietic cells
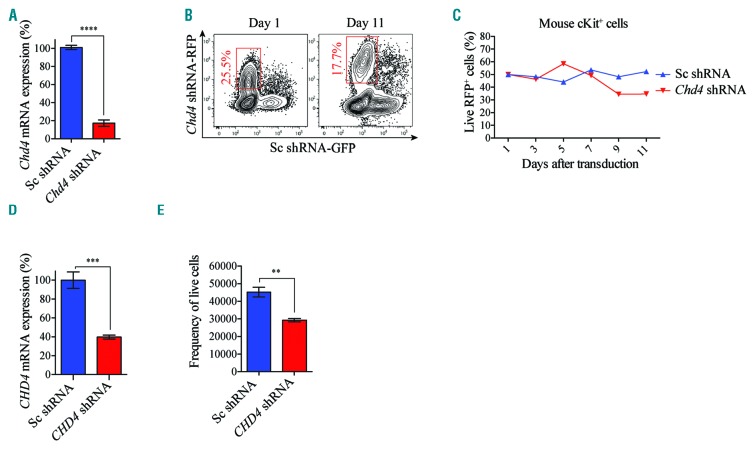
To assess the importance of Chd4 in normal hematopoietic cells, we performed shRNA-based knockdown experiments in primary mouse BMs, which resulted in efficient reduction in Chd4 mRNA levels compared to the control cells (Figure 3A). Using the cell growth assay described in Figure 2A, dynamic flow cytometric analysis displayed minor differences in cell growth between the control cells and the Chd4 depleted cells (Figure 3B,C). Similarly, CHD4 suppressed CD34 positive umbilical cord blood cells (UCBs) (Figure 3D), showed a modest reduction in the number of viable cells compared to control cells, after 14 days of culturing (Figure 3E), and as compared to the reduction in cell growth of CHD4 targeted AML cells (Figure 2; Online Supplementary Figure S3). Together, these results show that CHD4 is critical for cell growth of various types of leukemic cells and AML progression in vivo, but not for the proliferation and survival of normal primary hematopoietic cells.
Figure 3.
CHD4 inhibition causes minor effect in cell growth of normal blood cells in vitro. A. Bar charts represent real time PCR analysis of mRNA levels after shRNA-based knockdown of Chd4 relative to control cells transduced with vectors expressing scrambled shRNA, in sorted primary murine cKit+ BMs used in the competitive proliferation assays (Figure 3B,C), at 72 hours post transduction. mRNA levels were normalized to Hprt. The data is represented as the mean ±S.E.M., ****P<0.001 (unpaired t-test), n=3. B. Flow cytometry charts are representative plots of the percentage of live RFP+ Chd4 knockdown cKit+ cells (Chd4 shRNA-RFP) relative to GFP+ control cells (Sc shRNA-GFP) at the indicated time points. C. Line chart of a representative experiment of the percentage of Chd4 shRNA-RFP+ vs. Sc shRNA-GFP+ BMs propagated in suspension, normalized to 50% at the initial time point (Day one), determined by flow cytometry analysis at the indicated time points. D. Bar charts represent real time PCR analysis of mRNA levels after shRNA-based knockdown of CHD4, relative to control cells transduced with vectors expressing scrambled shRNA, in primary human CD34 enriched UCBs used in the cell growth assays (Figure 3E), 72 hours post transduction. mRNA levels were normalized to UBC. The data is represented as the mean ±S.E.M., ***P<0.005 (unpaired t-test), n=3. E. Bar chart of the total number of viable primary UCBs, transduced with shRNA against CHD4 (CHD4-shRNA), or a negative control vector (Sc-shRNA). The UCBs were propagated in suspension with supplemented cytokines and growth factors for 14 days. The number of viable UCBs was determined by flow cytometric analysis. **P<0.01 (unpaired t-test), n=3. mRNA: messenger ribonucleic acid; RFP: red fluorescent protein; GFP: green fluorescent protein; shRNA: short hairpin RNA.
Clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 mediated loss of function of CHD4 prevents cell growth of AML cells
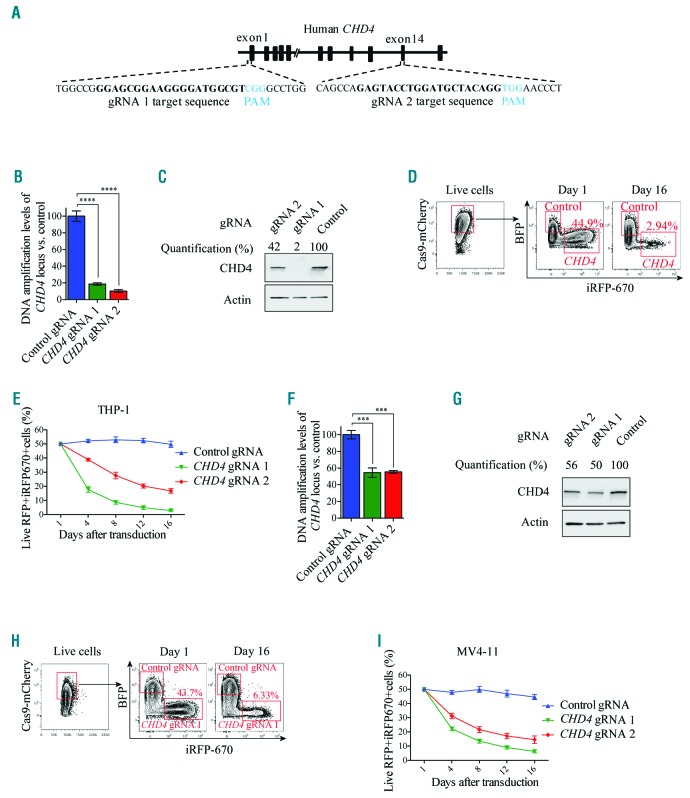
To exclude the possibility that the observed effects of the CHD4 inhibition was caused by off target effects, we took advantage of the highly specific CRISPR-Cas9 genome editing technology.41 Accordingly, we generated MV4-11 and THP-1 AML cells constitutively expressing Cas9, and transduced the cells with two individual inducible vectors expressing guide RNA (gRNA) homologous to the CHD4 locus (Figure 4A). Induction of expression of both gRNAs resulted in a significant disruption of the CHD4 coding region (Figure 4B) and reduction of protein levels (Figure 4C), in THP-1 AML cells. Although the reduction in protein levels in some experiments were relatively moderate, the specific CRISPR-Cas9 targeting of CHD4 caused a marked reduction in live THP-1 cells, confirming the essential role of CHD4 in AML cell growth (Figure 4D,E). Comparable results were obtained using the same approach with the human MV4-11 AML cells (Figure 4F-I) and the mouse MLL-AF9 AML cells (Online Supplementary Figure S4), thereby further validating that CHD4 is required for AML cell growth.
Figure 4.
CRISPR-Cas9 mediated disruption of CHD4 prevents growth of human AML cells. A. Schematic image depicting the homology of the two gRNAs used for targeting of the coding region of the human CHD4 gene. The protospacer adjacent motifs (PAM) are highlighted in blue letters and the gRNA target sequences are highlighted in bold black letters. B and F. Bar charts of real time PCR quantification of isolated genomic DNA from the targeted CHD4 gene and control locus after CRISPR-Cas9 disruption of CHD4 using two individual gRNAs, relative to the cells transduced with control vectors, in THP-1 Cas9 (Figure 4B) or MV4-11 Cas9 expressing cells (Figure 4F), at 72 hours post induction of gRNA expression. Real time PCR quantification of Cas9 targeted CHD4 loci was normalized to a control locus positioned upstream of the CHD4 gene. The transduced cells were used in the growth competition assays (Figure 4D,E,H,I). The data is represented as the mean ±S.E.M., ***P<0.005, ****P<0.001 (unpaired t-test), n=3. C and G. Western blot analyses and quantification of the endogenous CHD4 and Actin levels in the indicated cells, with and without knockdown of CHD4, at 72 hours post transduction. D and H. Representative flow cytometry charts of the percentage of live mCherry+ THP-1 (Figure 4D) or MV4-11 cells (Figure 4H) expressing Cas9, transduced with CHD4 gRNAs expressing iRF670 (CHD4-gRNA), relative to control cells transduced with empty vectors expressing BFP (control gRNA), at the indicated days after induction of gRNA expression. E and I. Line charts of the relative ratio, in percentage, of THP-1 (Figure 4E) or MV4-11 cells (Figure 4I) transduced with CHD4 gRNA-iRFP670+ knockout constructs or BFP+ control vectors, at the indicated time points using flow cytometry analysis. gRNA: guide RNA; RFP: red fluorescent protein.
CHD4 suppression impairs maintenance of primary childhood AML cells ex vivo
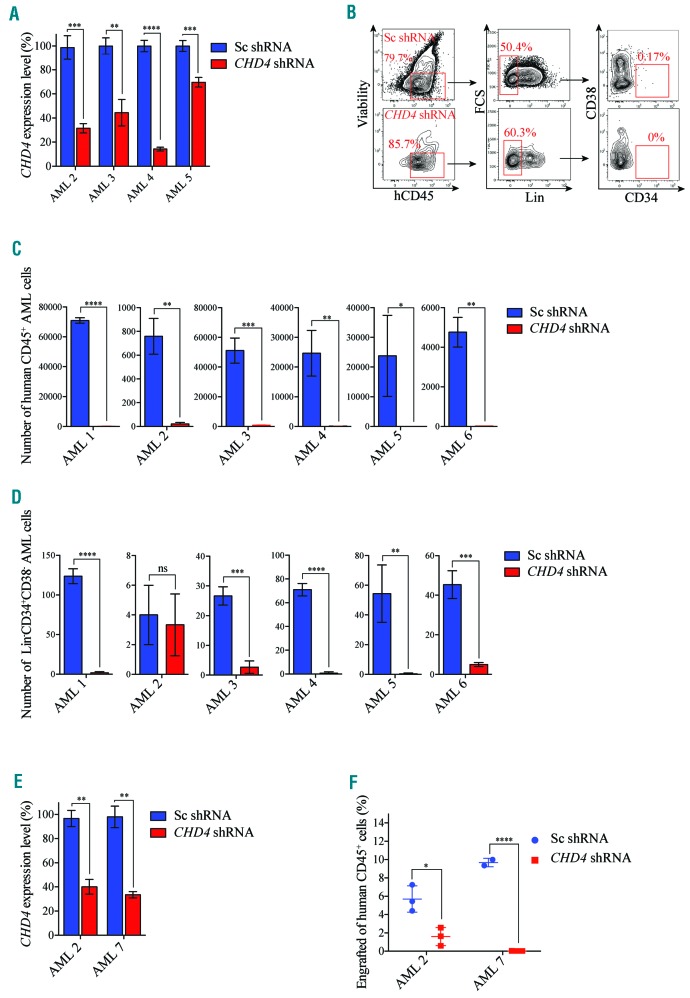
We demonstrated that CHD4 inhibition prevented growth of childhood AML cell lines (Figure 2 and Figure 4). Thus, we next investigated the importance of the inhibition of CHD4 in a cohort of childhood AML patient samples (Online Supplementary Table S3). To enable maintenance of the primary human childhood AML cells with high viability, including LICs, we utilized a stromal cell co-culture system for the cell growth assays.28 shRNA-mediated knockdown of CHD4 using two independent constructs proved to be efficient for four out of six samples from which we could extract good quality mRNA (Figure 5A; Online Supplementary Figure S5A), and indeed from one sample we had enough material for western blot analysis (Online Supplementary Figure S5B). Flow cytometric analysis showed that leukocytes (CD45+ cells) and LICs (Lin-CD34+CD38− cells) of primary childhood AML samples transduced with a negative control vector could be maintained on stromal feeder layers with high viability (80-90%) up to five weeks after transduction (Figure 5B-D; Online Supplementary Figure S5C,D).
Figure 5.
CHD4 inhibition prevents growth of patient-derived childhood AML cells ex vivo and in vivo. A. Bar charts show real time PCR analysis of mRNA levels after shRNA-based knockdown of CHD4, relative to control cells transduced with vectors expressing scrambled shRNA, of the indicated primary childhood AML samples used in the proliferation assays (Figure 5B-D), at 72 hours post transduction. mRNA levels were normalized to UBC. The data is represented as the mean ±S.E.M., **P<0.01, ***P<0.005, ****P<0.001 (unpaired t-test), n=3. B. Flow cytometry charts of expression profiles of CD34, CD38, Lin, and CD45 of cells transduced with CHD4 shRNA expressing RFP (CHD4-shRNA) or a negative control vector expressing scrambled shRNA and GFP (Control shRNA). The patient-samples were co-cultured with murine MS-5 stromal cells until the control cells reached confluence, or up to five weeks of maintenance. The number of viable CD45+ or Lin-CD34+CD38−, were determined by flow cytometric analysis. Live cell populations negative for Live/dead Near-IR dye was used for flow cytometric analysis. The numbers in each square represent the percentage of cells within each cell population. C and D. Bar chart showing the total numbers of live CD45+ (Figure C) and Lin-CD34+CD38− (Figure D) primary childhood AML cells transduced with CHD4 shRNA, or a control vector, as indicated. *P<0.05, **P<0.01, ***P<0.005, ****P<0.001 (unpaired t-test). E. Bar charts depict real time PCR analysis of mRNA levels after shRNA-based knockdown of CHD4, relative to control cells transduced with vectors expressing scrambled shRNA, of the indicated primary childhood AML samples used for the xenograft experiments (Figure F) at 72 hours post transduction. mRNA levels were normalized to UBC. The data is represented as the mean ±S.E.M., **P<0.01 (unpaired t-test), n=3. F. Scatter plot of percent of engrafted primary childhood AML cells of two childhood patient samples, transduced with shRNA against CHD4, or a negative control vector, that were transplanted into humanized NSG-SGM3 recipient mice. Each animal was transplanted by intrafemoral injection with a single cell dose of up to 300,000 unfractionated primary cells. The childhood AML BMs were harvested after eight weeks post transplantation and the level of engraftment was determined by flow cytometric analysis. Each symbol represents an individual transplanted mouse, and each sample was transplanted in triplicates. *P<0.05, ****P<0.001 (unpaired t-test). Sc: scramble control; shRNA: short hairpin RNA; ns: non-significant; AML: acute myeloid leukemia.
In contrast, shRNA-mediated targeting of CHD4 in a primary childhood sample carrying the MLL translocation (Online Supplementary Table S3, sample “AML 6”) prevented maintenance of leukocytes and LICs compared to the control sample (Figure 5C,D). We then sought to investigate if CHD4 was required for maintenance of childhood AML samples carrying alternative genetic lesions (Online Supplementary Table S3). With the exception of LICs in one patient sample (sample “AML 2”, Figure 5D), the suppression of CHD4 with two independent shRNAs prevented the maintenance of leukocytes and LICs in all of the tested non-MLL rearranged childhood AML samples compared to the control cells (Figure 5B-D; Online Supplementary Figure S5C,D). Although all non-MLL rearranged samples did not robustly expand, the suppression of maintenance was similar to that of the MLL-rearranged primary childhood AML sample (“AML 6”) (Figure 5B-D; Online Supplementary Figure S5C,D). These results show that the inhibition of CHD4 prevents growth of bulk cancer cells/leukocytes and LICs of primary childhood AML BMs with different genetic lesions.
CHD4 suppression induces anti-leukemic effects in primary childhood AML cells in vivo
To investigate whether CHD4 also was required for growth of primary childhood AML cells in vivo, we used a humanized NSG-SGM3.42 Patient sample “AML 2” and “AML 7” (Online Supplementary Table S3), transduced with CHD4 targeting shRNA vectors or control vectors (Figure 5E), were intrafemurally transplanted into recipient mice. NSG-SGM3 mice transplanted with childhood AML cells transduced with control vectors resulted in a significant level of engraftment for both transplanted patient samples (median value of 5.7% +/− 2, and 9.6% +/− 2), determined by flow cytometric analysis of CD45+ cells, eight weeks post transplantation (Figure 5F). In sharp contrast, primary childhood AML cells transduced with shRNA vectors efficiently knocking down CHD4 mRNA (Figure 5E) displayed significantly lower levels of leukemic cell engraftment compared to the control cells (median value of 1.6, 0.03, P=0.0152, P<0.001, respectively) (Figure 5F). Thus, the targeting of CHD4 inhibits cell proliferation of primary childhood AML cells and AML progression in vivo.
CHD4 controls cell cycle progression of AML cells
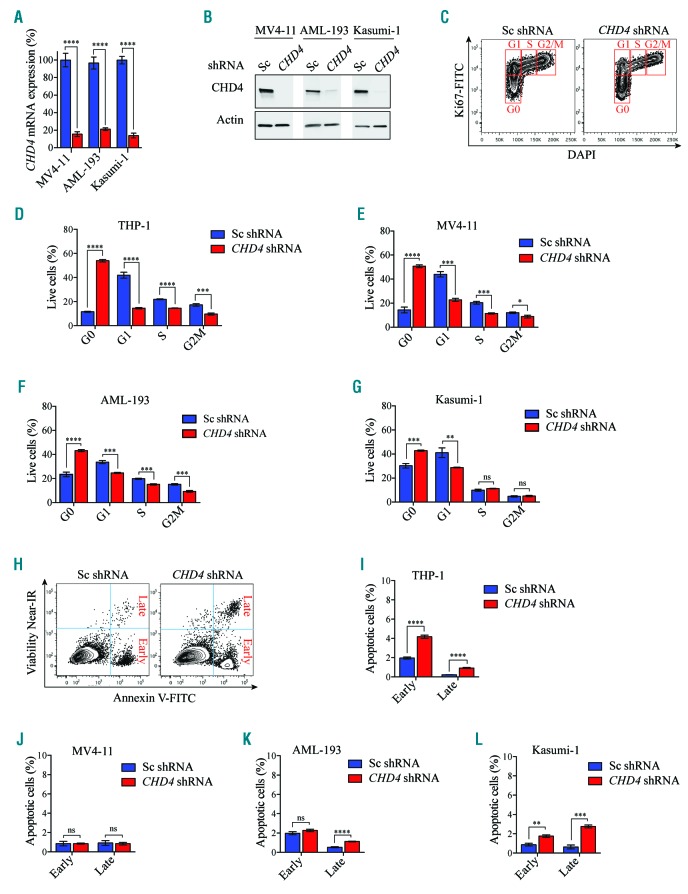
To investigate the cellular mechanisms by which CHD4 was required for AML cell growth in childhood AML, we used two MLL rearranged (THP-1 and MV4-11) and two non-MLL rearranged (AML-193 and Kasumi-1) childhood AML cell lines, and analyzed the effects in apoptosis and the cell cycle upon suppression of CHD4 expression. shRNA-mediated inhibition of CHD4 resulted in a significant reduction of mRNA (Figure 2A and Figure 6A) and protein levels (Figure 2B and Figure 6B), and a dramatic accumulation of all four cell lines in the G0 phase of the cell cycle, compared to the control cells. In addition, all cell lines displayed a decrease of cells in G1, whereas less pronounced effects was observed in the S and G2/M phase, compared to the control cells (Figure 6C-G). The robust arrest in G0 when CHD4 was inhibited (Online Supplementary Figure S3A,B; Online Supplementary Figure S6A, B), was confirmed in all four cell lines using an independent shRNA against CHD4 (Online Supplementary Figure S6C-F).
Figure 6.
CHD4 inhibition in AML cells causes an arrest in the G0 phase of the cell cycle. A. Bar charts represent real time PCR analysis of mRNA levels after shRNA-based knockdown of CHD4 in MV4-11, Kasumi-1 and AML-193 cells (knockdown of mRNA levels of CHD4 in THP-1 cells used in this assay is depicted in Figure 2A). The cells were used for apoptosis and cell cycle analysis (Figure 6C-L), relative to control samples transduced with negative control vectors expressing scrambled shRNA, at 72 hours post transduction. mRNA levels were normalized to UBC. The data is represented as the mean ±S.E.M., ****P<0.001 (unpaired t-test), n=3. B. Western blot analysis of CHD4 and Actin levels in MV4-11, AML-193 and Kasumi-1 cells, with and without knockdown of CHD4, (knockdown levels of CHD4 protein levels in the THP-1 cells used in this assay is depicted in Figure 2B). C. Representative flow cytometry charts of the THP-1 cells transduced with CHD4 shRNAs, or a negative control vector, stained with Ki-67 and DAPI, at 72 hours after transduction. Each cell cycle phase is highlighted in red. D-G. Bar graphs of flow cytometric quantification as the percentage of cells in each population of the indicated cell lines transduced with CHD4 shRNA or a control vector, at 72 hours post CHD4. The data is presented as mean ±S.E.M., ***P<0.005, ****P<0.001 (unpaired t-test), n=3. H. Representative flow cytometry charts of THP-1 cells transduced with CHD4 shRNAs, or a negative control vector, stained with near-IR and Annexin V, at 72 hours after transduction. I-L. Bar graphs illustrating flow cytometric quantification as the percentage of cells in each population of the indicated cell lines transduced with CHD4 shRNA or a control vector, at 72 hours post transduction. The data is presented as mean ±S.E.M. **P<0.01, ***P<0.005, ****P<0.001, (unpaired t-test), n=3. DAPI; 4’,6-diamidino-2-phenylindole; FITC: fluorescein isothiocyanate; ns: non-significant; Sc: scramble control; shRNA: short hairpin RNA; AML: acute myeloid leukemia; mRNA: messenger RNA.
In contrast to the strong effects in cell cycle progression, CHD4 depleted cells (Figure 2A,B and Figure 6A,B) displayed modest levels of early apoptotic cells (Annexin V+NIR−), and late apoptotic cells (Annexin V+NIR+) (Figure 6G-J). Likewise, knockdown of CHD4 using an independent shRNA (Online Supplementary Figure S3A,B and Online Supplementary Figure S6A,B) resulted in the same relatively low levels of apoptosis in all cell lines (Online Supplementary Figure S6G-J). Taken together, these data indicate that the primary cellular mechanism, whereby inhibition of CHD4 prevented cell growth of AML cells, is mediated via a growth arrest of the cells in the G0 phase of cycle progression, rather than induction of apoptosis.
CHD4 suppression induces an expression profile correlating to MYC targets
To delineate the underlying molecular mechanisms whereby CHD4 is required for the growth of AML cells, we performed RNA-Seq analysis of human THP-1 AML cells transduced with shRNAs targeting CHD4.
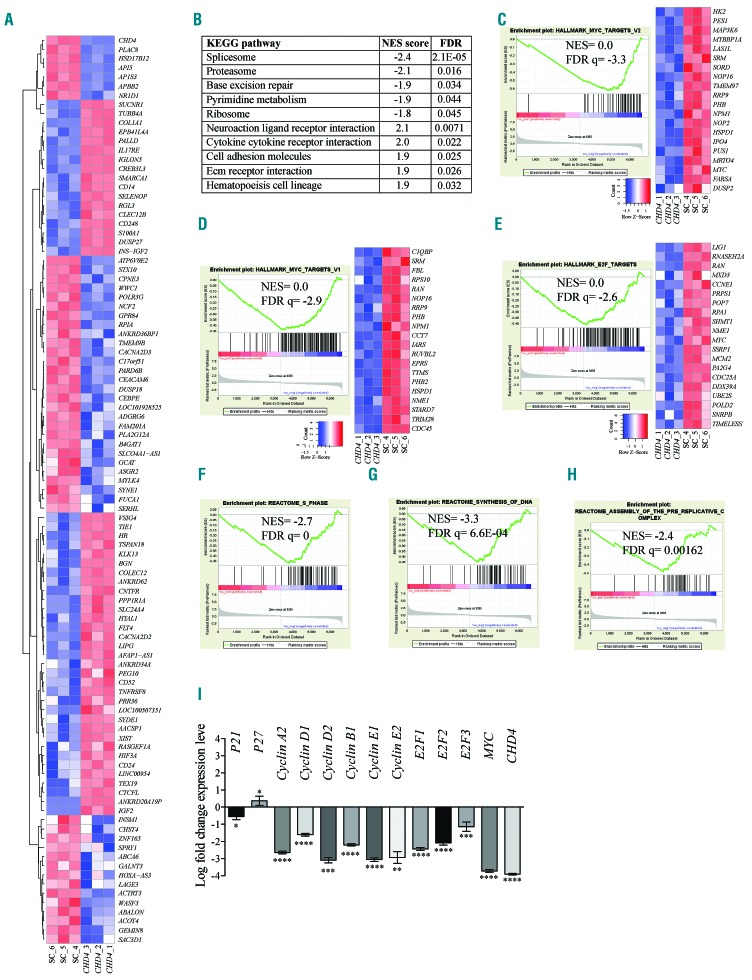
Density and boxplot analysis of the samples illustrated an even distribution of counts per million (CPM) after normalization and clustering analysis (Online Supplementary Figure S7A,B). Heatmap clustering of the top 100 upregulated or downregulated genes (Figure 7A) of the RNA-Seq data showed a high degree of reproducibility between the triplicates, and a significant differentiation between the AML CHD4 knockdown cells and the cells transduced with a scramble control.
Figure 7.
CHD4 inhibition in AML cells induces an expression profile linked to MYC and cell cycle progression. A. Heatmap of 100 genes with most significant changes in gene expression of THP-1 cells transduced with shRNA targeting CHD4, or a negative control vector, 72 hours post transduction. The data represent clustering of the individual experiments. B. List of the ten most significant KEGG gene set pathways correlating to gene expression changes resulting from shRNA-based inhibition of CHD4, relative to a negative control vector, in THP-1 cells. C-H. Enrichment plots of gene set enrichment analysis (GSEA) of mRNA gene expression profiling in response to CHD4 knockdown in THP-1 cells, demonstrating significant normalized enrichment score (NES) between MYC target V2 (Figure 7C), MYC targets V1 (Figure 7D), and E2F targets (Figure 7E), or to S phase (Figure 7F), synthesis of DNA (Figure 7G), assembly of the pre-replicative complex (Figure 7H). The bar charts represent the top ranked correlations in the predefined MSigDB: H hallmark collections (Figure 7C-E), or significant correlations to CP Reactome collection (Figure 7F-H). The heatmap on the right in C-E shows the relative level of gene expression (red = high, blue = low) of the most significant genes in the leading edge subset. I. Bar charts show real time PCR analysis of mRNA levels of genes showing changes in RNA-Seq of THP-1 cells, after shRNA-based knockdown of CHD4, relative to control cells transduced with vectors expressing scrambled shRNA, at 72 hours post transduction. mRNA levels were normalized to UBC. The data is represented as the mean ±S.E.M., *P<0.05, **P<0.01, ***P<0.005, ****P<0.001 (unpaired t-test), n=3. KEGG: Kyoto encyclopedia of genes and genomes; Sc: scramble control; FDR: false discovery rate.
Differentially expressed genes were those in which the mRNA levels were changed >1 or log2 fold change <−1; P<0.05; false discovery rate (FDR) <0.05 (Online Supplementary Table S4). Consistent with a role in gene repression,8 a majority of the genes were found to be upregulated upon knockdown of CHD4 (1011 genes were upregulated whereas 413 genes were downregulated). CHD4 and some of its previously reported target genes (e.g., IGF2, TGFB1 and PDGFA)38 were among those with the most significant changes in mRNA levels (Online Supplementary Table S4).
Gene set enrichment analysis (GSEA) was used to investigate whether the transcriptome profile generated by the inhibition of CHD4 was associated to the collection of annotated gene sets (i.e., The Molecular Signatures Database [MSigDB]).43 The Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis revealed that genes deregulated upon CHD4 suppression were enriched for gene sets such as spliceosome, proteasome and base excision repair (Figure 7B). Intriguingly, the expression changes in response to the inhibition of CHD4 compared to the Molecular Signatures Database (MSigDB) hallmark gene sets were most significantly correlated with the gene signature of two individual subgroups of MYC targets data sets (FDR Q-value=0.0 and normalized enrichment score [NES]= −3.3; FDR Q-value=0.0, NES=−2.9, respectively), and to the MYC target E2F transcription factor (E2F) and its target genes with cell cycle related functions (FDR Q-value=0.0, NES=−2.6) (Figure 7C-E). Consistent with the link to MYC and its important role in cell cycle regulation, GSEA analysis using the reactome gene sets and comparisons to changes in gene expression upon CHD4 knockdown revealed significant associations with several gene sets involving cell cycle progression, including S phase (NES=−2.7; FDR Q=0), synthesis of DNA (NES=−3.3; FDR Q=6.64E-04) and assembly of the pre-replicative complex (NES=−2.4; FDR Q=0.00162) (Figure 7F-H).
Further analysis of the RNA-Seq data revealed that inhibition of CHD4 caused decreased mRNA levels of MYC and several of its target genes involved in G1/S cell cycle transition, including cyclin D1, D2, E1, E2F1, and E2F2. Conversely, the negative cell cycle regulator p27 was shown to be upregulated. In contrast, other MYC targets with alternative roles in other stages of the cell cycle, such as cyclin A2, B1, E2, displayed less pronounced changes in mRNA levels (Online Supplementary Table S4). To validate the RNA-Seq findings, we performed an additional shRNA-based knockdown of CHD4 in THP-1 cells. qPCR analysis confirmed the RNA-Seq data, but with even more significant changes in the gene expression of MYC and an asset of its targets genes known to be involved in cell cycle progression (Figure 7I). Consistent with this, an additional RNA-Seq analysis of THP-1 cells transduced with an independent shRNA against CHD4 again showed significant correlations to MYC and E2F targets. In addition, qPCR-based validation of the RNA-Seq data confirmed the previous results in Figure 7I and the observed deregulation of MYC and its downstream targets (Online Supplementary Table S4; Online Supplementary Figure S8). Thus, inhibition of CHD4 was significantly associated with MYC targets and gene sets involved in S phase cell cycle progression.
Discussion
In this study, we performed loss of function screens on a large scale in AML cells and non-transformed BMs. We identified the epigenetic factor CHD4 as being essential for maintenance of LICs and disease progression of childhood AML, but not for normal hematopoietic cells. CHD4 inhibition in AML cells caused downregulation of MYC and its target genes and an arrest in the G0 phase of cell cycle progression.
It is of utmost importance that future treatments for AML selectively target the cancer cells without harming normal cells. Accordingly, we showed that CHD4 is required for cell growth of leukemic cells carrying various genetic lesions and for disease progression using an immune competent mouse model. However, CHD4 is not essential for primary normal murine BMs or for normal human UCBs. Our findings are supported by previous reports showing that inhibition of CHD4 is not crucial for normal hematopoietic cell growth,14 but has nonessential functions in self-renewal and lineage choice in normal hematopoiesis.44,45 Interestingly, this selectivity seems to be conserved in breast cancer.9 Indeed, CHD4 has previously been demonstrated to be required for growth of a broad range of cancer cells,9,13,37,38,46,47 including colony formation capacity of AML cells,14 implying that CHD4 may represent a cancer-specific dependency in a wider repertoire of tumors.
Most importantly, our results highlight a novel and essential role for CHD4 in maintenance of childhood AML in vitro and in vivo. The use of appropriate co-culture systems and a patient-derived xenograft mouse model for childhood AML allowed us to demonstrate that the essential role of CHD4 was consistently manifested in patient samples carrying diverse types of genetic lesions as well as the LICs. Intriguingly, the importance of CHD4 in cancer-initiating cells has also been reported in hepatocellular carcinoma47 and glioblastoma,38 indicating that the role of CHD4 in these cells that drive tumor growth may be more general than previously anticipated.
CHD4/NuRD has been shown to control cell cycle progression in a p53 dependent manner,10–12 or in a p53 independent manner,9,48 and inhibition of CHD4 was shown to cause a cell cycle arrest in G1/S.11 In the present study, inhibition of CHD4 resulted in repression of MYC and its target genes involved in cell cycle progression and consequently caused a G0 cell cycle arrest. In support of this, inhibition of MYC has been reported to cause a G0/G1 block in the cell cycle in human lymphoid and myeloid cells.49 Moreover, CHD4 has been found to directly bind to the MYC promoter in glioblastoma cells and inhibition of CHD4 resulted in a downregulation of MYC.38 In addition, MYC was also part of a set of genes suggested to have a role in colony formation in AML cells.14
Herein, we demonstrated that CHD4 is essential for maintenance of childhood AML and the LICs that are responsible for the emergence and development of the disease. Our data indicate that the importance of CHD4 in childhood AML may be mediated in part by promoting the expression of the MYC oncogene and its target genes. The cancer-specific dependency found in our studies show that CHD4 may represent a promising therapeutic target to battle childhood AML.
Supplementary Material
Acknowledgments
We thank Ying Qu for help with RNA-Seq analysis. We also thank Tim Somervaille for kindly providing the MLL-AF9 vector and Marco Herold for kindly providing the Cas9-mCherry vector. We would also like to thank NOPHO and Josefine Palle for providing the primary childhood AML cells as well as Cecilia Götherström and the National Cord Blood Bank at Karolinska University Hospital for providing the UCBs. We would like to acknowledge the MedH Core Flow Cytometry facility (Karolinska Institutet), supported by KI/SLL, for providing cell sorting services, cell analysis services, technical expertise and scientific input. We would also like to thank the Affymetrix core facility at Neo, BEA, Bioinformatics and Expression Analysis, which is supported by the board of research at the Karolinska Institute and the research committee at the Karolinska hospital.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/103/7/1169
Funding
This work was supported by the Wallenberg Foundation, The Swedish Cancer Society, the Magnus Bergvalls Foundation, the Karolinska Institutet, the Åke Wibergs Foundation, and Dr Åke Olsson Foundation for Hematological Research.
References
- 1.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3(7):730–737. [DOI] [PubMed] [Google Scholar]
- 2.Lapidot T, Sirard C, Vormoor J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367(6464):645–648. [DOI] [PubMed] [Google Scholar]
- 3.Ishikawa F, Yoshida S, Saito Y, et al. Chemotherapy-resistant human AML stem cells home to and engraft within the bone-marrow endosteal region. Nat Biotechnol. 2007;25(11):1315–1321. [DOI] [PubMed] [Google Scholar]
- 4.Farrar JE, Schuback HL, Ries RE, et al. Genomic profiling of pediatric acute myeloid leukemia reveals a changing mutational landscape from disease diagnosis to relapse. Cancer Res. 2016; 76(8):2197–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ho PA, Kutny MA, Alonzo TA, et al. Leukemic mutations in the methylation-associated genes DNMT3A and IDH2 are rare events in pediatric AML: a report from the Children’s Oncology Group. Pediatr Blood Cancer. 2011;57(2):204–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balgobind BV, Zwaan CM, Pieters R, Van den Heuvel-Eibrink MM. The heterogeneity of pediatric MLL-rearranged acute myeloid leukemia. Leukemia. 2011; 25(8):1239–1248. [DOI] [PubMed] [Google Scholar]
- 7.Creutzig U, van den Heuvel-Eibrink MM, Gibson B, et al. Diagnosis and management of acute myeloid leukemia in children and adolescents: recommendations from an international expert panel. Blood. 2012; 120(16):3187–3205. [DOI] [PubMed] [Google Scholar]
- 8.Denslow SA, Wade PA. The human Mi-2/NuRD complex and gene regulation. Oncogene. 2007;26(37):5433–5438. [DOI] [PubMed] [Google Scholar]
- 9.D’Alesio C, Punzi S, Cicalese A, et al. RNAi screens identify CHD4 as an essential gene in breast cancer growth. Oncotarget. 2016;7(49):80901–80915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo J, Su F, Chen D, Shiloh A, Gu W. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature. 2000;408(6810):377–381. [DOI] [PubMed] [Google Scholar]
- 11.O’Shaughnessy A, Hendrich B. CHD4 in the DNA-damage response and cell cycle progression: not so NuRDy now. Biochem Soc Trans. 2013;41(3):777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polo SE, Kaidi A, Baskcomb L, Galanty Y, Jackson SP. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 2010;29(18):3130–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xia L, Huang W, Bellani M, et al. CHD4 has oncogenic functions in initiating and maintaining epigenetic suppression of multiple tumor suppressor genes. Cancer Cell. 2017;31(5):653–668.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sperlazza J, Rahmani M, Beckta J, et al. Depletion of the chromatin remodeler CHD4 sensitizes AML blasts to genotoxic agents and reduces tumor formation. Blood. 2015;126(12):1462–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmidt EE, Pelz O, Buhlmann S, Kerr G, Horn T, Boutros M. GenomeRNAi: a database for cell-based and in vivo RNAi phenotypes, 2013 update. Nucleic Acids Res. 2013;41(Database issue):D1021–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuber J, Shi J, Wang E, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478(7370):524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wermke M, Camgoz A, Paszkowski-Rogacz M, et al. RNAi profiling of primary human AML cells identifies ROCK1 as a therapeutic target and nominates fasudil as an antileukemic drug. Blood. 2015; 125(24):3760–3768. [DOI] [PubMed] [Google Scholar]
- 18.Tzelepis K, Koike-Yusa H, De Braekeleer E, et al. A CRISPR dropout screen identifies genetic vulnerabilities and therapeutic targets in acute myeloid leukemia. Cell Rep. 2016;17(4):1193–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sroczynska P, Cruickshank VA, Bukowski JP, et al. shRNA screening identifies JMJD1C as being required for leukemia maintenance. Blood. 2014;123(12):1870–1882. [DOI] [PubMed] [Google Scholar]
- 20.Puram RV, Kowalczyk MS, de Boer CG, et al. Core circadian clock genes regulate leukemia stem cells in AML. Cell. 2016;165(2):303–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Porter CC, Kim J, Fosmire S, et al. Integrated genomic analyses identify WEE1 as a critical mediator of cell fate and a novel therapeutic target in acute myeloid leukemia. Leukemia. 2012;26(6):1266–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Mar BG, Zhang H, et al. The EMT regulator ZEB2 is a novel dependency of human and murine acute myeloid leukemia. Blood. 2017;129(4):497–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jude JG, Spencer GJ, Huang X, et al. A targeted knockdown screen of genes coding for phosphoinositide modulators identifies PIP4K2A as required for acute myeloid leukemia cell proliferation and survival. Oncogene. 2015;34(10):1253–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang JY, Ngai PK, Priestle JP, Joss U, Vosbeck KD, van Oostrum J. Identification of a reactive lysyl residue (Lys103) of recombinant human interleukin-1 beta. Mechanism of its reactivity and implication of its functional role in receptor binding. Biochemistry. 1992;31(11):2874–2878. [DOI] [PubMed] [Google Scholar]
- 25.Chan SM, Thomas D, Corces-Zimmerman MR, et al. Isocitrate dehydrogenase 1 and 2 mutations induce BCL-2 dependence in acute myeloid leukemia. Nat Med. 2015; 21(2):178–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carey A, Edwards DKt, Eide CA, et al. Identification of interleukin-1 by functional screening as a key mediator of cellular expansion and disease progression in acute myeloid leukemia. Cell Rep. 2017; 18(13):3204–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Banerji V, Frumm SM, Ross KN, et al. The intersection of genetic and chemical genomic screens identifies GSK-3alpha as a target in human acute myeloid leukemia. J Clin Invest. 2012;122(3):935–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Griessinger E, Anjos-Afonso F, Pizzitola I, et al. A niche-like culture system allowing the maintenance of primary human acute myeloid leukemia-initiating cells: a new tool to decipher their chemoresistance and self-renewal mechanisms. Stem Cells Transl Med. 2014;3(4):520–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spooncer E, Heyworth CM, Dunn A, Dexter TM. Self-renewal and differentiation of interleukin-3-dependent multipotent stem cells are modulated by stromal cells and serum factors. Differentiation. 1986;31(2):111–118. [DOI] [PubMed] [Google Scholar]
- 30.Bhagwat AS, Roe JS, Mok BYL, Hohmann AF, Shi J, Vakoc CR. BET Bromodomain inhibition releases the mediator complex from select cis-regulatory elements. Cell Rep. 2016;15(3):519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noguera NI, Song MS, Divona M, et al. Nucleophosmin/B26 regulates PTEN through interaction with HAUSP in acute myeloid leukemia. Leukemia. 2013; 27(5):1037–1043. [DOI] [PubMed] [Google Scholar]
- 32.Metzgeroth G, Walz C, Score J, et al. Recurrent finding of the FIP1L1-PDGFRA fusion gene in eosinophilia-associated acute myeloid leukemia and lymphoblastic T-cell lymphoma. Leukemia. 2007; 21(6):1183–1188. [DOI] [PubMed] [Google Scholar]
- 33.Thol F, Bollin R, Gehlhaar M, et al. Mutations in the cohesin complex in acute myeloid leukemia: clinical and prognostic implications. Blood. 2014;123(6):914–920. [DOI] [PubMed] [Google Scholar]
- 34.Cancer Genome Atlas Research N Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizuno H, Kitada K, Nakai K, Sarai A. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med Genomics. 2009;2:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bagger FO, Sasivarevic D, Sohi SH, et al. BloodSpot: a database of gene expression profiles and transcriptional programs for healthy and malignant haematopoiesis. Nucleic Acids Res. 2016;44(D1):D917–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bohm M, Wachtel M, Marques JG, et al. Helicase CHD4 is an epigenetic coregulator of PAX3-FOXO1 in alveolar rhabdomyosarcoma. J Clin Invest. 2016; 126(11):4237–4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chudnovsky Y, Kim D, Zheng S, et al. ZFHX4 interacts with the NuRD core member CHD4 and regulates the glioblastoma tumor-initiating cell state. Cell Rep. 2014;6(2):313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guillemette S, Serra RW, Peng M, et al. Resistance to therapy in BRCA2 mutant cells due to loss of the nucleosome remodeling factor CHD4. Genes Dev. 2015;29(5): 489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Somervaille TC, Cleary ML. PU.1 and Junb: suppressing the formation of acute myeloid leukemia stem cells. Cancer Cell. 2006; 10(6):456–457. [DOI] [PubMed] [Google Scholar]
- 41.Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Billerbeck E, Barry WT, Mu K, Dorner M, Rice CM, Ploss A. Development of human CD4+FoxP3+ regulatory T cells in human stem cell factor-, granulocyte-macrophage colony-stimulating factor-, and interleukin-3-expressing NOD-SCID IL2Rgamma(null) humanized mice. Blood. 2011;117(11): 3076–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prasad P, Lennartsson A, Ekwall K. The roles of SNF2/SWI2 nucleosome remodeling enzymes in blood cell differentiation and leukemia. Biomed Res Int. 2015; 2015:347571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoshida T, Hazan I, Zhang J, et al. The role of the chromatin remodeler Mi-2beta in hematopoietic stem cell self-renewal and multilineage differentiation. Genes Dev. 2008;22(9):1174–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mohd-Sarip A, Teeuwssen M, Bot AG, et al. DOC1-dependent recruitment of NURD reveals antagonism with SWI/SNF during epithelial-mesenchymal transition in oral cancer cells. Cell Rep. 2017;20(1):61–75. [DOI] [PubMed] [Google Scholar]
- 47.Nio K, Yamashita T, Okada H, et al. Defeating EpCAM(+) liver cancer stem cells by targeting chromatin remodeling enzyme CHD4 in human hepatocellular carcinoma. J Hepatol. 2015;63(5):1164–1172. [DOI] [PubMed] [Google Scholar]
- 48.Buddaseth S, Gottmann W, Blasczyk R, Huyton T. Dysregulation of cell cycle control caused by overexpression of the oncogene pp32r1 (ANP32C) and the Tyr>His mutant pp32r1Y140H. Biochim Biophys Acta. 2013;1833(5):1212–1221. [DOI] [PubMed] [Google Scholar]
- 49.Wickstrom EL, Bacon TA, Gonzalez A, Freeman DL, Lyman GH, Wickstrom E. Human promyelocytic leukemia HL-60 cell proliferation and c-myc protein expression are inhibited by an antisense pentadecadeoxynucleotide targeted against c-myc mRNA. Proc Natl Acad Sci USA. 1988; 85(4):1028–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.