Abstract
The myeloma bone marrow microenvironment promotes proliferation of malignant plasma cells and resistance to therapy. Activation of JAK/STAT signaling is thought to be a central component of these microenvironment-induced phenotypes. In a prior drug repurposing screen, we identified tofacitinib, a pan-JAK inhibitor Food and Drug Administration (FDA) approved for rheumatoid arthritis, as an agent that may reverse the tumor-stimulating effects of bone marrow mesenchymal stromal cells. Herein, we validated in vitro, in stromal-responsive human myeloma cell lines, and in vivo, in orthotopic disseminated xenograft models of myeloma, that tofacitinib showed efficacy in myeloma models. Furthermore, tofacitinib strongly synergized with venetoclax in coculture with bone marrow stromal cells but not in monoculture. Surprisingly, we found that ruxolitinib, an FDA approved agent targeting JAK1 and JAK2, did not lead to the same anti-myeloma effects. Combination with a novel irreversible JAK3-selective inhibitor also did not enhance ruxolitinib effects. Transcriptome analysis and unbiased phosphoproteomics revealed that bone marrow stromal cells stimulate a JAK/STAT-mediated proliferative program in myeloma cells, and tofacitinib reversed the large majority of these pro-growth signals. Taken together, our results suggest that tofacitinib reverses the growth-promoting effects of the tumor microenvironment. As tofacitinib is already FDA approved, these results can be rapidly translated into potential clinical benefits for myeloma patients.
Introduction
Multiple myeloma (MM) is the second most common hematologic malignancy in the United States of America and still has no known cure. Years of research have revealed that a major driver of malignant plasma cell proliferation, as well as therapeutic resistance, is signaling to the tumor cells from the bone marrow (BM) microenvironment.1–3 Cell types within the BM that influence myeloma cells include mesenchymal stromal cells, osteoblasts, osteoclasts, and multiple classes of immune cells.1–3 Overcoming the growth-promoting phenotype of the BM microenvironment is thought to be a promising therapeutic strategy in MM.
One approach to identifying new therapeutic agents for many diseases is drug repurposing. In this context, a large library of drugs, all of which are either Food and Drug Administration (FDA)-approved, or at the minimum shown to be safe in humans, is screened against the biological system of interest.4,5 The premise behind these screens is that small molecules, initially designed for one indication, may actually have beneficial effects across other diseases. In fact, the use of thalidomide in MM is one of the most impactful examples of successful drug repurposing. If new indications are found for already existing drugs, clinical development times and associated costs can be drastically reduced, thus accelerating the potential benefits to patients.6,7 To identify agents which may reverse the tumor-promoting effects of the MM BM microenvironment, we recently reported a repurposing screen of 2684 compounds against three MM cell lines, either grown alone (monoculture) or in coculture with MM patient-derived BM mesenchymal stromal cells.8 From that screen, we identified tofacitinib citrate, an FDA-approved small molecule for the treatment of rheumatoid arthritis (RA), as an agent which may reverse stromal-induced growth proliferation of malignant plasma cells.
Tofacitinib citrate is a potent inhibitor of all four members of the Janus kinase (JAK) family, with preferential inhibition of JAK1 and JAK3 over JAK2 and TYK2 in cellular assays.9 JAK signaling, mediated by downstream STAT transcription factors, is necessary for lymphocyte stimulation in response to encountered antigens.10 Therefore, JAK inhibition holds promise for the treatment of autoimmune diseases like RA.11 In parallel, the JAKs have gained interest as therapeutic targets in MM as they mediate signaling via interleukin-6 (IL-6). IL-6 is secreted by many cell types within the BM microenvironment, as well as by malignant plasma cells themselves, and it is thought that proliferation of malignant plasma cells within the human BM is dependent on this cytokine.12 This dependence on IL-6 was underscored by the recent development of a patient-derived xenograft model of MM, where primary plasma cell growth only occurred in immunocompromised mouse BM after knock-in of human IL-6.13
In fact, a number of groups have variously targeted JAK1/JAK2,14,15 JAK2,16–18 or all four JAKs19 with reported preclinical therapeutic efficacy in MM. However, per the registry at clinicaltrials.gov, none of these experimental JAK inhibitors have entered into MM clinical trials. Therefore, all of these agents are very far from use in MM patients, if they ever become available. Herein, we demonstrate that the already FDA-approved agent tofacitinib has robust preclinical activity in MM models. We further use ribonucleic acid sequencing (RNA-seq) and unbiased mass spectrometry-based phosphoproteomics to delineate pro-proliferative signals from the BM stroma and show that they are largely reversed by tofacitinib treatment. Furthermore, we find that an alternate repurposing candidate, the FDA-approved JAK1/2 inhibitor ruxolitinib, surprisingly does not show the same anti-myeloma properties. Therefore, our results support the rapid repurposing of tofacitinib as an anti-myeloma therapeutic to reverse the pro-growth effects of the BM microenvironment and potentiate the effects of existing myeloma therapies.
Methods
Cell culture conditions
All cell lines were authenticated by DNA genotyping at ATCC. All cells, including patient BM mononuclear cells, were maintained in complete media with roswell park memorial institute-1640 (RPMI-1640; Gibco) supplemented with 10% fetal bone serum (FBS; Gemini), 1% penicillin-streptomycin University of California San Francisco (UCSF), and 2 mM L-Glutamine (UCSF) with 5% CO2. INA-6 media was supplemented with 50 ng/mL recombinant human IL-6 (ProSpec). Additional details are provided in the Online Supplementary Methods.
MM and bone marrow stromal cells (BMSC) coculture and viability testing
Cocultures were seeded into 384 well plates (Corning) with the Multidrop Combi (Thermo Scientific). 800 stromal cells were seeded and incubated overnight. 17 hours later, 700 myeloma cells were added on top of the stromal cells. On the third day, 24 hours after the addition of myeloma cells, cocultures were treated with tofacitinib (LC Laboratories), ruxolitinib (Selleck Chemicals), JAK3i,20 or IL-6 blocking antibody (R&D Systems). For drug combination studies, on the fourth day, melphalan (Sigma Aldrich), carfilzomib (Selleck Chemicals), or venetoclax (Selleck Chemicals) were additionally added to cocultures. On the fifth day, myeloma cell viability was detected with the addition of luciferin (Gold Biotechnology) and read for luminescence on Glomax Explorer plate reader (Promega) as previously described.21 For monoculture studies cell viability was measured using CellTiter-Glo reagent (Promega). All measurements were performed in quadruplicate. All viability data are reported as normalized to dimethyl sulfoxide (DMSO)-treated cell line in monoculture.
RNA-seq
For coculture RNA-seq, 5×106 MM.1S cells were grown with 3×106 HS5 cells for 24 hours. CD138+ enrichment to >95% was verified by flow cytometry for mCherry expression (Online Supplementary Figure S1A,B). MM.1S harvested from coculture, as well as MM.1S and HS5 grown in monoculture, were then processed for RNA-seq as previously described.22 Significantly upregulated transcripts were identified by DESeq23 and bioinformatic analysis was performed using Enrichr.24 Raw sequencing data are available at the Gene Expression Omnibus (GEO) repository (Accession number GSE99293). Additional details are provided in the Online Supplementary Methods.
Western Blot Analysis
Described in the Online Supplementary Methods.
Liquid chromatography–tandem mass spectrometry phosphoproteomics
For coculture experiments, 5×106 HS5 cells were seeded into a T75 flask. Seventeen hours later, cultures were washed with phosphate buffered saline (PBS), before the addition of 107 MM.1S mC/Luc cells. Twenty-four hours later, cocultures were treated with 1 μM tofacitinib for 1.5 hours and 24 hours. MM.1S cells in suspension were harvested by aspiration, centrifuged, washed with PBS, and flash-frozen prior to analysis. For untreated MM.1S monoculture or HS5 monoculture experiments, 107 cells were used. For sample preparation, frozen cell pellets were lysed in 8 M urea. 1 mg of total protein was then reduced in tris(2-car-boxyethyl)phosphine (TCEP) and free cysteines alkylated with iodoacetamide. Proteins were then digested at room temperature for 18 hours with trypsin. Peptides were desalted, lyophilized, and enriched for phosphopeptides using immobilized-metal affinity column (IMAC) with Fe-NTA loaded beads.25 Phosphopeptides were analyzed on a Thermo Q-Exactive Plus mass spectrometer coupled to a Dionex Ultimate 3000 NanoRSLC liquid chromatography instrument with 3.5 hour linear gradient. Raw proteomic data files are available at the ProteomXchange PRIDE repository (Accession number PXD006581). Additional details are provided in the Online Supplementary Methods.
Xenograft mouse model
NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were obtained from the Jackson laboratory. 106 MM.1S mC/Luc cells, stably expressing luciferase, were transplanted via tail vein injection into each mouse. Tumor burden was assessed through weekly bioluminescent imaging, beginning 13 days after implantation and on the same day as treatment initiation. Mice were treated for four weeks with vehicle or tofacitinib as indicated (five mice/arm.) Tofacitinib was formulated in 50% DMSO, 10% (polyethylene glycol 400) PEG 400, and 40% water and administered at 21.5 mg/kg daily by continuous subcutaneous infusion. All mouse studies were performed according to UCSF Institutional Animal Care and Use Committee-approved protocols.
Patient samples
De-identified primary MM BM samples were obtained from the UCSF Hematologic Malignancy Tissue Bank in accordance with the UCSF Committee on Human Research-approved protocols and the Declaration of Helsinki. BM mononuclear cells were isolated by density gradient centrifugation Histopaque-1077 (Sigma Aldrich), then adjusted to 2 × 105/well in a 96 well plate. Primary cells were stimulated with 50 ng/ml recombinant human IL-6 (ProsPec) for 17 hours before treatment with tofacitinib for 24 hours. Cells were then stained with Alexa-Fluor 647 mouse anti-human CD138 antibody (BD Pharmingen) and SYTOX Green (Thermo) and analyzed on a CytoFLEX instrument (BD).
Results
Tofacitinib targets the BM microenvironment and reverses BMSC-mediated growth promotion
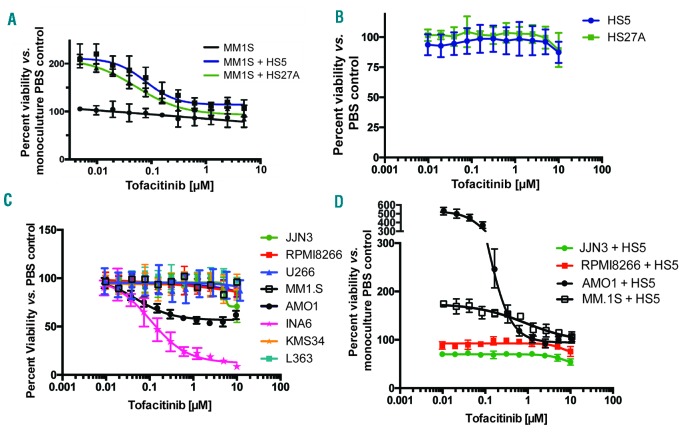
To initially validate findings from our drug repurposing screen, we cocultured the human MM cell line MM.1S, which was included in the screen,8 with the immortalized BMSC lines HS5 and HS27A (Figure 1A) and low-passage stromal cells derived from primary myeloma patient BM (Online Supplementary Figure S1C). MM.1S cell numbers strongly increased compared to monoculture growth after 24 hours, confirming stromal-induced proliferative signaling in this cell line. Tofacitinib treatment reduced MM.1S cell numbers in a dose-dependent manner, such that at >1 μM tofacitinib, MM.1S cell numbers in coculture return to approximately monoculture levels (Figure 1A). Tofacitinib has no effect on MM.1S cell viability alone nor on stromal cells alone (Figure 1B). We further studied the effect of tofacitinib on several other MM cell lines. In monoculture we found that tofacitinib only demonstrates strong anti-MM activity in the IL-6 dependent cell line INA-6, with limited effect on the AMO-1 cell line and minimal to no effect on the other MM cell lines (Figure 1C). We further evaluated four myeloma cell lines (MM.1S, RPMI-8226, JJN-3, AMO-1) in which luciferase was stably expressed, allowing for the distinction of MM cell viability versus stromal cell viability in coculture.21 Only the stromal-responsive cell lines exhibit any sensitivity to tofacitinib treatment (Figure 1D). Taken together, these results suggest that tofacitinib selectively targets the growth-promoting interaction between MM cells and the stromal microenvironment known to occur in patients.
Figure 1.
Tofacitinib inhibits stromal cell-mediated proliferation in MM cells. A. Tofacitinib has no effect vs. MM.1S MM cells in monoculture, but instead reverses proliferation induced by BMSC lines HS5 and HS27A. B. Tofacitinib has no viability effect vs. BMSC C. Tofacitinib has limited or minimal effects vs. most MM cell lines in monoculture, except the IL-6 dependent line INA-6. D. In stromal cell coculture, tofacitinib does not have anti-MM effects vs. JJN-3 and RPMI-8226 cell lines, which do not proliferate in response to stroma, but shows strong reversal of proliferation in both MM.1S and AMO1 lines. All error bars represent +/− S.D. from CellTiter-Glo assay performed in quadruplicate in 384-well plates. PBS: phosphate buffered saline.
BMSC-mediated plasma cell proliferation is through a mechanism partially dependent on IL-6
We next focused on the MM.1S cell line as it showed the unique phenotype of responsiveness to tofacitinib only in the context of stromal stimulation. To further characterize the nature of pro-growth signaling, we performed RNA-seq on MM.1S cells grown alone or in coculture with HS5 stromal cells. We first noticed that the most significantly upregulated transcript in MM.1S in the coculture setting was SOCS3, part of a well-characterized negative feedback mechanism strongly induced by JAK-STAT activation10 (Figure 2A). We further examined all 67 transcripts significantly upregulated in MM.1S in the coculture vs. monoculture setting (P< 0.005 per DESeq tool;23 listed in Online Supplementary Dataset S1). Using the Enrichr tool,24 ChIP-X enrichment analysis (ChEA) of chromatin immunoprecipitation sequence (ChIP-seq) datasets26 found the most significant enrichment of STAT3-binding sites at the promoter of these upregulated transcripts, among all transcription factors (Figure 2B). Furthermore, Protein ANalysis THrough Evolutionary Relationships (Panther)27 pathway analysis found the only two significantly enriched pathways to be related to JAK/STAT signaling and interleukin signaling (Figure 2C). Taken together, these RNA-seq findings suggest that factors secreted from stromal cells mediate proliferation by activating JAK/STAT signaling, with STAT3 playing a central role.28 Notably, similar activation of SOCS3 and related JAK/STAT genes were previously found after exposure to IL-6 in the INA-6 cell line.29
Figure 2.
Stromal-induced signatures in MM.1S identified by transcriptome analysis. A. Examples of significantly upregulated genes in MM.1S cells cocultured with HS5 stromal cells in comparison to MM.1S grown in monoculture. B. ChEA analysis of 67 significantly upregulated transcripts from untreated MM.1S in HS5 coculture vs. monoculture (P<0.005 based on DESeq analysis) demonstrates a significant enrichment of STAT3 transcription-factor binding sites based on ChIP-seq data. C. Panther pathway analysis of this gene list demonstrates significant upregulation of interleukin signaling and JAK-STAT signaling. D. Recombinant cytokines known to be secreted from HS5 stromal cells30 were tested for their ability to promote MM.1S proliferation. Up to the maximum achievable concentration, neither IL-6 alone nor a combination of all tested cytokines could recapitulate the growth promotion induced by BMSC. E. An IL-6 neutralizing antibody could partially reverse stromal-induced proliferation of MM.1S. All error bars represent +/− S.D. from CellTiter-Glo assay performed in quadruplicate in 384-well plates. PBS: phosphate buffered saline; RNA-seq: ribonucleic acid sequencing; ChIP-seq: chromatin immunoprecipitation sequencing.
However, using recombinant cytokines, even at high concentrations of IL-6, we did not observe the same degree of growth promotion as coculture with stromal cells (Figure 2D). A recent study identified other cytokines besides IL-6 highly secreted from HS5 stromal cells.30 We tested four of these: macrophage inflammatory protein 3A (MIP-3A), interleukin-8 (IL-8), granulocyte colony stimulating factor (G-CSF), granulocyte-macrophage colony stimulating factor (GM-CSF), and none showed any growth promotion (Figure 2D). Furthermore, an IL-6 blocking antibody could only partially reverse stromal cell effects at the highest achievable concentration (Figure 2E). These findings support the role of IL-6 in this system, but also indicate that other factors likely play a role in MM growth promotion.
Tofacitinib inhibits JAK/STAT signaling
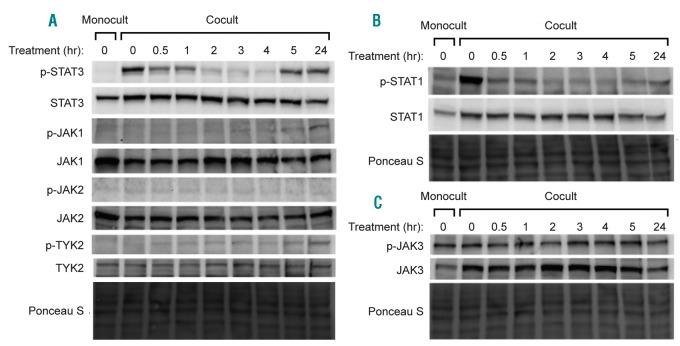
Given our results above, we chose to further evaluate downstream effects of tofacitinib inhibition of the JAK/STAT pathway. Two of the primary downstream mediators of IL-6 receptor and JAK activation are thought to be pro-proliferation signaling by STAT3 and inhibitory signaling by STAT1.28 We found that STAT3 and STAT1 phosphorylation in MM.1S dramatically increases when in coculture with HS5 (Figure 3A,B). 1 μM tofacitinib inhibits STAT3 phosphorylation in MM.1S cells in coculture almost to monoculture level by two hours of treatment. In addition, phosphorylation of JAK1, JAK2, and TYK2, which can also activate STAT3, were studied. We found evidence of a “rebound” effect by 24 hours of treatment, mediated by well-characterized feedback mechanisms,10 serving to phosphorylate the JAKs and subsequently re-activate STAT3. Despite this rebound of STAT3 activation, however, MM growth continues to be inhibited based on the dose-response results of tofacitinib. In INA-6 cells we found similar effects of decreased STAT3 phosphorylation after tofacitinib treatment and a rebound at 24 hours (Online Supplementary Figure S2A).
Figure 3.
Tofacitinib inhibits JAK/STAT signaling in MM cells in a time-dependent manner. A. Western blotting demonstrates a marked increase in STAT3 phosphorylation in untreated coculture vs. monoculture in MM.1S cells. After treatment with 1 μM tofacitinib there is a rapid decrease in STAT3 phosphorylation with rebound by 24 hours. B. STAT1 phosphorylation is also increased in response to coculture and rapidly reversed by tofacitinib treatment. C. While coculture increases JAK3 protein expression, there does not seem to be any significant change in signaling via JAK3 after tofacitinib based on phosphorylation status. Western blots are representative of assays performed in biological duplicate. Monocult: monoculture; Cocult: coculture.
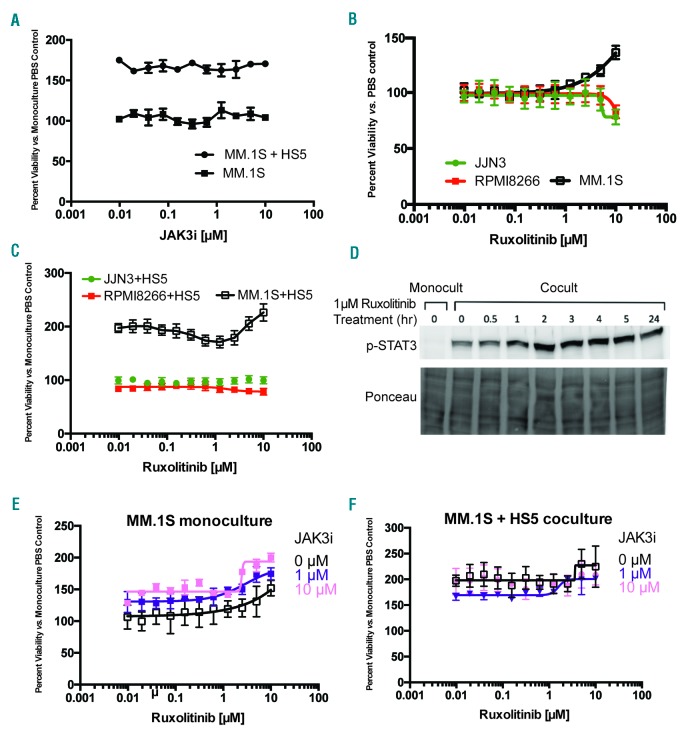
As tofacitinib is known to potently inhibit JAK3, of additional interest was an increase in JAK3 expression in MM.1S in coculture versus monoculture, found both by RNA-seq (Figure 2A) and Western blot (Figure 3C). However, tofacitinib did not lead to any significant decrease in JAK3 phosphorylation (Figure 3C). We also evaluated signaling through JAK3’s primary pro-proliferative downstream mediator STAT5.31 We found no evidence of STAT5 phosphorylation in either monoculture or coculture (Online Supplementary Figure S2B). These results suggest that JAK3/STAT5 signaling is less central to stroma-supported MM growth. We further confirmed this result using JAK3i, a newly-described, highly-specific, irreversible JAK3 inhibitor.31 JAK3i had no effect on MM.1S in mono- or coculture (Figure 4A), nor did it show any synergy with carfilzomib treatment (Online Supplementary Figure S2C,D).
Figure 4.
Ruxolitinib demonstrates less anti-MM activity than tofacitinib. A. A highly selective, irreversible inhibitor of JAK3, JAK3i, does not have any effects on MM.1S either in monoculture or in coculture with HS5. B. The JAK1/2 inhibitor ruxolitinib has minimal anti-MM effects in monoculture vs. three MM cell lines, and in fact appears to promote growth of MM.1S at higher concentrations. C. A similar phenomenon is noted in HS5 coculture. D. Ruxolitinib does not inhibit, but in fact increases signaling via STAT3 in MM.1S grown in HS5 coculture. E.-F. Combination of ruxolitinib with JAK3i, to achieve simultaneous JAK1/2/3 inhibition, does not recapitulate effects of tofacitinib in MM.1S. All error bars represent +/− S.D. from CellTiter-Glo assay performed in quadruplicate in 384-well plates. Monocult: monoculture; Cocult: coculture; PBS: phosphate buffered saline.
Taken together, these results suggest a mechanism whereby stromal cell-induced MM proliferation is mediated through STAT3 transcriptional effects. These signaling pathways are inhibited by tofacitinib, ultimately leading to the reversal of the proliferation phenotype.
Ruxolinitib has less anti-MM activity than tofacitinib
Given that our results above suggest a more important role for JAK1 and/or JAK2 in stromal-induced MM proliferation than JAK3, we turned to an alternate candidate for drug repurposing, ruxolinitib. This agent is FDA-approved for use in myeloproliferative neoplasms and has much higher affinity for JAK1 and JAK2 over JAK3 or TYK2.32 Ruxolitinib had minimal effects versus RPMI-8226 or JJN-3 cells, and, surprisingly, treatment of MM.1S actually showed a promotion of growth at higher concentrations, both in the monoculture and coculture settings (Figure 4B,C). Western blotting demonstrated that 1 μM ruxolitinib was unable to inhibit STAT3 activation in MM.1S in coculture (Figure 4D). In fact, STAT3 phosphorylation increased after two hours of treatment, consistent with the pro-proliferative effect seen in Figure 4B,C. We found that combining ruxolitinib with JAK3i was also insufficient to recapitulate tofacitinib’s effects in mono- or coculture (Figure 4E,F). Taken together, these findings suggest that ruxolitinib is unable to inhibit pro-proliferative STAT3 signaling in MM.1S cells, thereby supporting tofacitinib as having greater potential as a repurposed anti-MM therapy.
Unbiased phosphoproteomics demonstrates that tofacitinib broadly reverses pro-growth signaling induced by bone marrow stroma
Our targeted investigations above specifically focused on the JAK/STAT pathway. To further elucidate the mechanism of tofacitinib in this system, as well as gain a broader view of stromal-induced proliferation and signaling in MM cells, we pursued unbiased mass spectrometry (MS)-based phosphoproteomics. We studied four samples, performed in biological replicate: 1) MM.1S cells in monoculture, MM.1S cells in coculture, either 2) untreated (DMSO control), 3) treated with 1 μM tofacitinib for 1.5 hours, or 4) treated with 1 μM tofacitinib for 24 hours. We harvested MM.1S cells in suspension, enriched for phosphorylated peptides using immobilized metal chromatography, and analyzed by liquid chromatography-tandem mass spectrometry (LC/MS-MS) with peptide quantification performed using MaxQuant.33
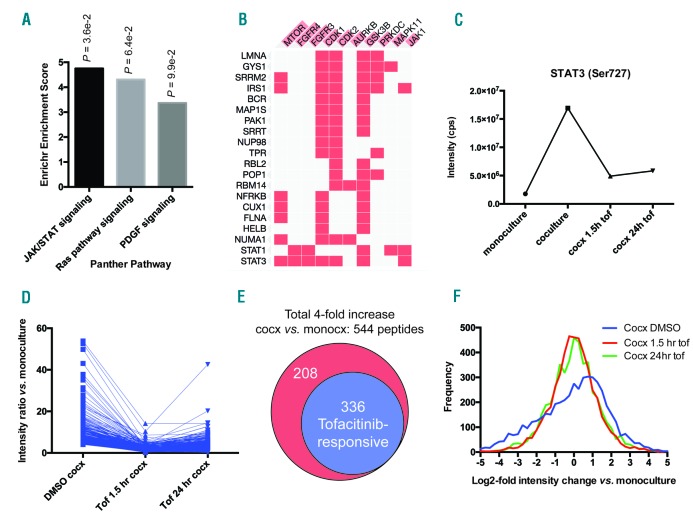
In total, 4862 phosphopeptides had intensity data in all four samples and were used for further analysis (listed in Online Supplementary Dataset S2). >99% of these sites are serine and threonine phosphorylation events, consistent with other phosphoproteomic studies using this enrichment method.34 We first evaluated for phosphosites with >4-fold intensity increases in untreated coculture vs. monoculture. Using our RNA-seq data as a proxy for protein-level changes, we verified these phosphosite changes were largely driven by changes in signaling and not protein abundance (Online Supplementary Figure S3A). Panther pathway analysis revealed the only significantly enriched pathway among the 544 upregulated phosphosites to be JAK/STAT signaling (Figure 5A). However, based on kinase enrichment analysis,35 we found enriched signatures not only of JAK1 substrates, but also of other kinases driving proliferation and the cell cycle, such as mTOR, CDK1, and CDK2 (Figure 5B). These findings demonstrate that unbiased phosphoproteomics can uncover broad signaling effects of the BM microenvironment even downstream of JAK/STAT.
Figure 5.
Unbiased phosphoproteomics reveals that tofacitinib broadly reverses pro-growth signaling from stroma to MM cells. A. Analysis of 4862 phosphorylation sites quantified by LC-MS/MS on biological replicate samples revealed 544 phosphosites to be upregulated 4-fold in untreated MM.1S in coculture with HS5 vs. monoculture. Of these upregulated phosphopeptides, Panther pathway analysis showed JAK/STAT signaling to be the only significantly enriched pathway (P=0.036). B. Kinase enrichment analysis demonstrated that many of the upregulated phosphopeptides derive from known substrates of other kinases related to proliferation, such as mTOR, CDK1, and CDK2, as well as JAK1. Phosphorylated protein substrates are on the left and enriched kinases across the top. Length of red bar in kinase name is indicative of strength of enrichment. C. Quantitative phosphoproteomic intensity of the JAK-responsive phosphosite Ser727 on STAT3, both in untreated coculture vs. monoculture, and after tofacitinib treatment, is very similar to the pattern found by Western blotting for the other known JAK responsive STAT3 phosphosite Tyr707 (Figure 3A), serving to validate this proteomics approach. D. Dynamics of all phosphosites found to be responsive to tofacitinib based on the criteria: 1) increased 4-fold in coculture vs. monoculture, 2) decreased at least-2 fold from untreated coculture after 1.5 hours of 1 μM tofacitinib treatment, and 3) phosphosite intensity remains below the untreated coculture level after 24 hours of tofacitinib treatment. E. Of 544 upregulated phosphopeptides in coculture, 336 (62%) were defined as being tofacitinib-responsive using the criteria in D. F. Examining the global phosphosite intensity across all quantified phosphopeptides demonstrates a general increase in phosphorylation of MM.1S proteins in untreated coculture with HS5 (“Cocx DMSO”; note shift of distribution maximum to log2-fold change vs. monoculture of ~1) which is then broadly reversed by tofacitinib treatment. Tof: tofacitinib; monocx: monoculture; cocx: coculture.
Next, for validation of the effects of tofacitinib treatment, we first examined Ser727 on STAT3 (Figure 5C), a known JAK-responsive phosphosite.28 Our quantitative MS results were remarkably in line with Western blotting for another JAK-responsive phosphosite on STAT3, Tyr707 (Figure 3A), with a very large increase in both phosphosites in untreated coculture compared to baseline, a >2-fold decrease in phosphorylation after short-term tofacitinib treatment, and a rebound in phosphorylation at 24 hours.
This finding both serves to validate our phosphoproteomic data as well as help us define a signature of tofacitinib-responsive phosphosites. Remarkably, 336 of the 544 up-regulated phosphosites in untreated coculture vs. monoculture (62%) met the same criteria of being tofacitinib-responsive (Figure 5D,E). Furthermore, examination across all measured phosphosites demonstrated that while phosphorylation is broadly increased in the coculture setting, noted as a general shift toward positive phosphosite intensities in the untreated sample, treatment with tofacitinib largely reverses this finding, recreating a normal distribution around the intensity values found in monoculture (Figure 5F). Taken together, these findings demonstrate that tofacitinib broadly reverses the signaling pathways driving stromal-induced proliferation in MM cells, both at the level of direct JAK targets as well as downstream proliferative signals, informing our mechanistic understanding of this treatment beyond targeted Western blots alone.
As a comparator, we also performed unbiased phosphoproteomics of HS5 stromal cells after 1.5 hours tofacitinib treatment. We found many fewer significantly changed phosphosites than identically-treated MM.1S in HS5 coculture (Online Supplementary Figure S3B,C), consistent with the lack of viability change after tofacitinib against HS5 alone (Figure 1B).
Phosphoproteomics does not reveal specific off-target activity for tofacitinib
Another advantage of unbiased phosphoproteomics is potentially detecting additional off-target effects mediating tofacitinib response. We filtered for peptides that appeared unaffected by stromal-induced signaling (less than +/− 50% intensity change in untreated coculture vs. monoculture) that decreased in intensity >4-fold after 1.5 hours tofacitinib treatment (Figure 6A). We identified only 54 peptides that fit this filter, and neither Panther nor kinase enrichment analysis identified any enriched signatures (data not shown). Given the small number of peptides and lack of any biological signatures, it appears most likely these 54 peptides are background noise in the data and do not reveal any specific off-target effects of tofacitinib.
Figure 6.
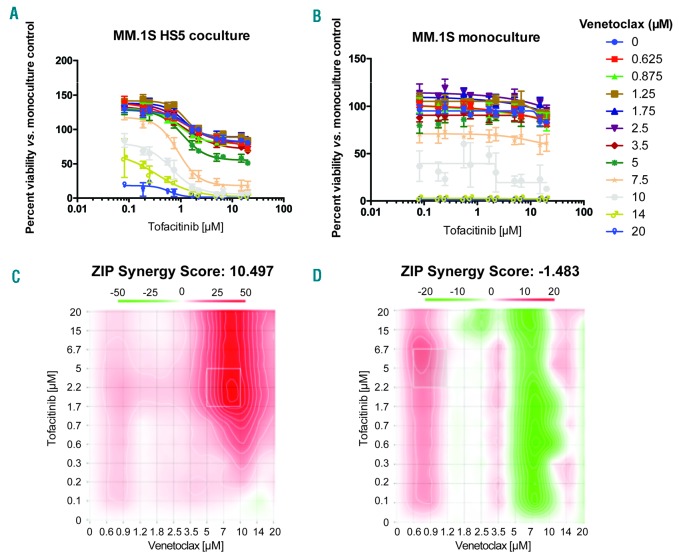
Tofacitinib shows synergy with venetoclax only in the coculture setting. A-B. Treatment was performed sequentially with 24 hours of tofacitinib followed by 24 hours of venetoclax at specified doses. C-D. For combination matrices, the interaction landscapes are shown in 2D plots. The ZIP method is used to calculate synergy across the landscape (red = positive score, synergistic; green = negative score, antagonistic) as well as to calculate an overall synergy score δ, the difference in percentage inhibition compared with the expected additive compound effect. The coculture results demonstrate synergy across all combinations with the strongest synergy at high doses of venetoclax, as well as an overall very high synergy score of ~10.5 across the landscape. The monoculture results demonstrate mild antagonism at high venetoclax doses and overall antagonistic score of ~−1.5. Note different scale bars on each 2D landscape output from ZIP. Error bars represent +/− S.D. from CellTiter-Glo assay performed in quadruplicate in 384-well plates. ZIP: zero-inflated Poisson regression.
To further investigate the possibility of any off-target effects of tofacitinib, we downloaded available data from the LINCS KINOMEscan database and plotted versus the human kinase phylogenetic tree (Online Supplementary Figure S4). These results demonstrate that, at least in cell-free assays, tofacitinib shows much greater specificity for JAK-family kinases compared to ruxolitinib. Together, these findings suggest that tofacitinib’s effects in this coculture model appear to be due to on-target activity. However, these results do not rule out off-target effects of tofacitinib not detected by these analyses herein.
Tofacitinib synergizes strongly with venetoclax only in the coculture setting
To begin to identify potential rational combinations with myeloma therapies, we first studied tofacitinib in combination with carfilzomib in MM.1S cells. Based on zero-inflated Poisson regression (ZIP) model scoring,30,36 we found very mild synergy with this proteasome inhibitor in both monoculture or HS5 coculture (Online Supplementary Figure S5). Subsequently, inspired by recent findings in acute myeloid leukemia (AML) primary samples,30 we tested the combination of tofacitinib and the Bcl-2 inhibitor venetoclax, a promising investigational agent in MM.37 Intriguingly, similar to the findings in AML, we found strong synergy of these two agents only in the coculture context but not in monoculture (Figure 6).
Tofacitinib has anti-MM activity in the BM microenvironment in vivo
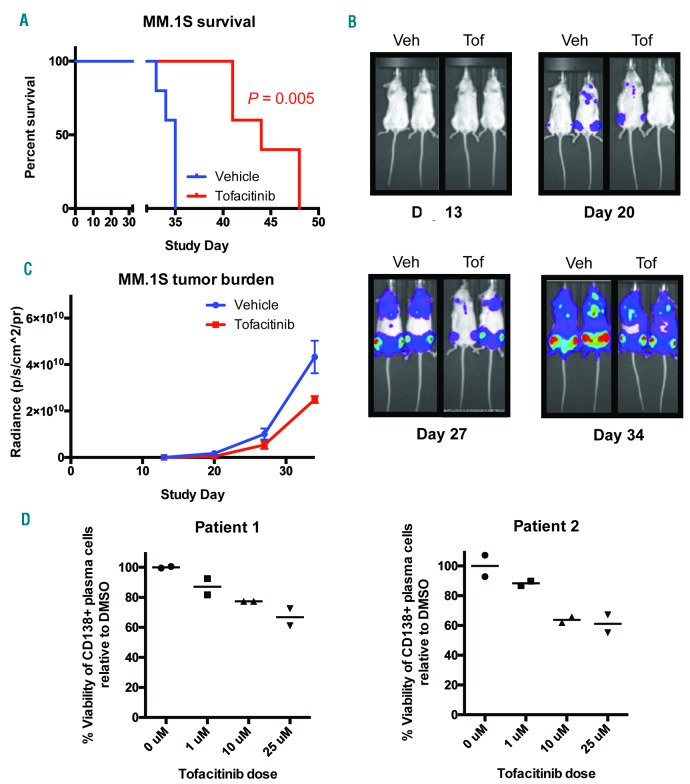
Toward the goal of repurposing tofacitinib as an anti-MM therapy in patients, we next examined the efficacy of tofacitinib in vivo. For this orthotopic disseminated xenograft model, we used a luciferase-labeled MM.1S cell line, which specifically homes to the murine BM after intravenous implantation in NOD scid γ (NSG) mice. Treatment was initiated after two weeks of tumor growth and continued for four weeks at ~2/3 of the maximal tolerated dose of tofacitinib (21.5 mg/kg/day by subcutaneous infusion).38 Encouragingly, we found significantly increased murine survival in this cell line model (Figure 7A) as well as decreased tumor burden based on bioluminescent imaging quantification (Figure 7B,C).
Figure 7.
Tofacitinib has anti-MM activity in vivo and versus primary samples ex vivo. Luciferase labeled-MM.1S MM cells were implanted intravenously into NSG mice and tumors allowed to grow for 13 days. Mice were randomized (n=5 mice per arm) and drug dosing was then begun for four weeks. Tofacitinib dose = 21.5 mg/kg/day by subcutaneous pump. A. Tofacitinib significantly increased survival of NSG mice in these aggressive mouse models of MM by log-rank test. B. Example bioluminescent images from MM.1S study showing prominent localization of tumor cells to hind limb BM. C. Bioluminescence imaging quantification of tumor burden. Error bars represent +/− S.D. D. Primary CD138+ plasma cells from two patients show modest response to 24 hour tofacitinib treatment when cultured ex vivo (with addition of 50 ng/mL IL-6 to the media) and measured by flow cytometry (n = 2 technical replicates). Veh: vehicle; Tof: tofacitinib; DMSO: dimethyl sulfoxide.
We further tested tofacitinib versus two primary MM BM samples treated ex vivo with the co-addition of 50 ng/mL IL-6. We found modest viability effects against malignant plasma cells (Figure 7D and Online Supplementary Figure S6). This result appears consistent with our in vitro and phosphoproteomic results, which suggest that plasma cell proliferation is necessary for tofacitinib to have significant effects. Primary MM plasma cells isolated ex vivo in 2D culture are known to minimally proliferate even in the presence of cytokines or stromal stimulation.39 Therefore, these results may not fully reflect the potential therapeutic efficacy of tofacitinib in MM patients, where plasma cells are constantly proliferating within the BM.
Discussion
Given the importance of the BM microenvironment for MM pathogenesis, the JAK/STAT signaling axis has generated significant interest as a therapeutic target in MM. JAK inhibition has already been validated in a number of preclinical studies as a way to target this pathway.14–19 However, the studied compounds are not yet available clinically, and may never be. Herein, following the results of a large-scale repurposing screen, we validated tofacitinib as a potential therapy that can be rapidly translated into MM patients.
Using a combination of mechanistic pharmacology and unbiased mass spectrometry-based phosphoproteomics, we found that tofacitinib appears to reverse stromal-induced proliferation of MM plasma cells by inhibiting JAK/STAT signaling. Our results also support the use of unbiased phosphoproteomics both in kinase inhibitor evaluation and more broadly in MM biology. While others have shown that IL-6 can lead to increased signaling through STAT3,40 our results suggest that additional factors derived from BMSC can lead to plasma cell proliferation. While we were unable to identify these factors, we did rule out a number of cytokines highly secreted by HS5 cells.30 Furthermore, given that murine IL-6 does not cross-react with the human IL-6 receptor,41 our in vivo results also suggest that other factors in the murine marrow microenvironment can stimulate JAK-STAT signaling, which is then reversed by tofacitinib. Candidates include leukemia inhibitory factor (LIF), which cross-reacts between mouse and humans,41,42 or the complex between murine IL-6 and soluble murine IL-6 receptor, both of which can stimulate JAK/STAT signaling in human cells.43
Surprisingly, in our studies, we found that the FDA-approved JAK1/2 inhibitor ruxolitinib did not lead to the same anti-MM effects as tofacitinib. We do note that ruxolitinib was previously evaluated in a small trial of 13 MM patients in combination with dexamethasone (clinicaltrials.gov Identifier: 00639002), and no significant anti-MM effects were noted in this small study. Our in vitro results presented herein may provide a partial explanation for the lack of ruxolitinib efficacy in that trial. Another recent study also found minimal anti-MM cell line effects of ruxolitinib unless used at extremely high doses and exposure times.44 However, it remains mechanistically unclear why ruxolitinib is unable to block JAK-STAT signaling in these myeloma models.
We note that our studies are undoubtedly limited in that they are performed in MM cell lines. While we primarily focused our analysis on stromal-responsive lines, which appear to better mimic the malignant plasma cell phenotype found in MM patients, focused clinical trials in MM will be necessary at this point to truly evaluate whether tofacitinib has anti-MM effects.
Toward this goal, the value of drug repurposing becomes readily apparent. Tofacitinib can be quickly moved into Phase I/II studies in MM as the tolerated doses and adverse event profiles of this drug are well-characterized.11 Intriguingly, a patient population with both MM and RA could readily serve as the basis of a multi-center trial. Alternatively, a patient population with early-stage disease, perhaps smoldering myeloma, may be the optimal setting for clinical use, when plasma cells may be most dependent on microenvironmental cues. Our findings of strong synergy between tofacitinib and venetoclax in the context of the BM microenvironment may be relevant given the exciting progress of venetoclax in the clinic.37 This finding may also reflect a general vulnerability specific to the BM niche, given the effectiveness of this combination both in MM and AML.30 In conclusion, tofacitinib is a promising agent to reverse the tumor-proliferative effects of the BM microenvironment that can be rapidly repurposed to benefit MM patients.
Supplementary Material
Acknowledgments
We thank Drs. Jeffrey Wolf, Tom Martin, Nina Shah, and Cammie Edwards for discussions, advice, and insight. We thank the staff of the UCSF Helen Diller Family Cancer Center Preclinical Therapeutic Core facility for completion of murine studies. We thank Dr. Diego Acosta-Alvear for providing luciferase-labeled MM cell lines.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/103/7/1218
Funding
This work was supported by the UCSF Stephen and Nancy Grand Multiple Myeloma Translational Initiative and the Myeloma Research Fund of the Silicon Valley Community Foundation (to BTA and APW), an NCI Cancer Center Support Grant (P30 CA082103) (to BCH), and an NCI Clinical Scientist Development Award (K08 CA184116), a Dale F. Frey Breakthrough Award from the Damon Runyon Cancer Research Foundation (DFS 14-15), and an American Cancer Society Individual Research Award (IRG-97-150-13) (to APW).
References
- 1.Manier S, Kawano Y, Bianchi G, Roccaro AM, Ghobrial IM. Cell autonomous and microenvironmental regulation of tumor progression in precursor states of multiple myeloma. Curr Opin Hematol. 2016;23(4):426–433. [DOI] [PubMed] [Google Scholar]
- 2.Kuehl WM, Bergsagel PL. Molecular pathogenesis of multiple myeloma and its pre-malignant precursor. J Clin Invest. 2012; 122(10):3456–3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bianchi G, Munshi NC. Pathogenesis beyond the cancer clone(s) in multiple myeloma. Blood. 2015;125(20):3049–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta SC, Sung B, Prasad S, Webb LJ, Aggarwal BB. Cancer drug discovery by repurposing: teaching new tricks to old dogs. Trends Pharmacol Sci. 2013; 34(9):508–517. [DOI] [PubMed] [Google Scholar]
- 5.Corsello SM, Bittker JA, Liu Z, et al. The Drug Repurposing Hub: a next-generation drug library and information resource. Nat Med. 2017;23(4):405–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shim JS, Liu JO. Recent advances in drug repositioning for the discovery of new anticancer drugs. Int J Biol Sci. 2014;10(7):654–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nosengo N. Can you teach old drugs new tricks? Nature. 2016;534(7607):314–316. [DOI] [PubMed] [Google Scholar]
- 8.Murnane M, Dhimolea E, Li R, et al. Defining primary marrow microenvironment-induced synthetic lethality and resistance for 2,684 approved drugs across molecularly distinct forms of multiple myeloma. Blood. 2015;126:503. [Google Scholar]
- 9.Meyer DM, Jesson MI, Li X, et al. Anti- inflammatory activity and neutrophil reductions mediated by the JAK1/JAK3 inhibitor, CP-690,550, in rat adjuvant-induced arthritis. J Inflamm. 2010;7:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shuai K, Liu B. Regulation of JAK-STAT signalling in the immune system. Nat Rev Immunol. 2003;3(11):900–911. [DOI] [PubMed] [Google Scholar]
- 11.Fleischmann R, Kremer J, Cush J, et al. Placebo-controlled trial of tofacitinib monotherapy in rheumatoid arthritis. N Engl J Med. 2012;367(6):495–507. [DOI] [PubMed] [Google Scholar]
- 12.Klein B, Zhang XG, Lu ZY, Bataille R. Interleukin-6 in human multiple myeloma. Blood. 1995;85(4):863–872. [PubMed] [Google Scholar]
- 13.Das R, Strowig T, Verma R, et al. Microenvironment-dependent growth of preneoplastic and malignant plasma cells in humanized mice. Nat Med. 2016; 22(11):1351–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monaghan KA, Khong T, Burns CJ, Spencer A. The novel JAK inhibitor CYT387 suppresses multiple signalling pathways, prevents proliferation and induces apoptosis in phenotypically diverse myeloma cells. Leukemia. 2011;25(12):1891–1899. [DOI] [PubMed] [Google Scholar]
- 15.Li J, Favata M, Kelley JA, et al. INCB16562, a JAK1/2 selective inhibitor, is efficacious against multiple myeloma cells and reverses the protective effects of cytokine and stromal cell support. Neoplasia. 2010; 12(1):28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Vos J, Jourdan M, Tarte K, Jasmin C, Klein B. JAK2 tyrosine kinase inhibitor tyrphostin AG490 downregulates the mitogen-activated protein kinase (MAPK) and signal transducer and activator of transcription (STAT) pathways and induces apoptosis in myeloma cells. Br J Hematol. 2000;109(4):823–828. [DOI] [PubMed] [Google Scholar]
- 17.Ramakrishnan V, Kimlinger T, Haug J, et al. TG101209, a novel JAK2 inhibitor, has significant in vitro activity in multiple myeloma and displays preferential cytotoxicity for CD45+ myeloma cells. Am J Hematol. 2010;85(9):675–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scuto A, Krejci P, Popplewell L, et al. The novel JAK inhibitor AZD1480 blocks STAT3 and FGFR3 signaling, resulting in suppression of human myeloma cell growth and survival. Leukemia. 2011; 25(3):538–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burger R, Le Gouill S, Tai YT, et al. Janus kinase inhibitor INCB20 has antiproliferative and apoptotic effects on human myeloma cells in vitro and in vivo. Mol Cancer Therap. 2009;8(1):26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith EJ, Olson K, Haber LJ, et al. A novel, native-format bispecific antibody triggering T-cell killing of B-cells is robustly active in mouse tumor models and cynomolgus monkeys. Sci Rep. 2015;5:17943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McMillin DW, Delmore J, Weisberg E, et al. Tumor cell-specific bioluminescence platform to identify stroma-induced changes to anticancer drug activity. Nat Med. 2010; 16(4):483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiita AP, Ziv E, Wiita PJ, et al. Global cellular response to chemotherapy-induced apoptosis. eLife. 2013;2:e01236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuleshov MV, Jones MR, Rouillard AD, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucl Acids Res. 2016;44(W1):W90–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fila J, Honys D. Enrichment techniques employed in phosphoproteomics. Amino acids. 2012;43(3):1025–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lachmann A, Xu H, Krishnan J, Berger SI, Mazloom AR, Ma’ayan A. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26(19):2438–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mi H, Huang X, Muruganujan A, et al. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucl Acids Res. 2017; 45(D1):D183–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nelson EA, Walker SR, Frank DA. Jak/STAT signaling in the pathogenesis and treatment of multiple myeloma. In: Anderson KC, ed. Advances in biology and therapy of multiple myeloma: Volume 1: Basic Science. New York: Springer, 2013: 117–138. [Google Scholar]
- 29.Brocke-Heidrich K, Kretzschmar AK, Pfeifer G, et al. Interleukin-6-dependent gene expression profiles in multiple myeloma INA-6 cells reveal a Bcl-2 family-independent survival pathway closely associated with Stat3 activation. Blood. 2004; 103(1):242–251. [DOI] [PubMed] [Google Scholar]
- 30.Karjalainen R, Pemovska T, Popa M, et al. JAK1/2 and BCL2 inhibitors synergize to counteract bone marrow stromal cell-induced protection of AML. Blood. 2017; 130(6):789–802. [DOI] [PubMed] [Google Scholar]
- 31.Smith GA, Uchida K, Weiss A, Taunton J. Essential biphasic role for JAK3 catalytic activity in IL-2 receptor signaling. Nat Chem Biol. 2016;12(5):373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quintas-Cardama A, Vaddi K, Liu P, et al. Preclinical characterization of the selective JAK1/2 inhibitor INCB018424: therapeutic implications for the treatment of myeloproliferative neoplasms. Blood. 2010; 115(15):3109–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–1372. [DOI] [PubMed] [Google Scholar]
- 34.Leitner A. Enrichment strategies in phosphoproteomics. Meth Mol Biol. 2016; 1355:105–121. [DOI] [PubMed] [Google Scholar]
- 35.Lachmann A, Ma’ayan A. KEA: kinase enrichment analysis. Bioinformatics. 2009; 25(5):684–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yadav B, Wennerberg K, Aittokallio T, Tang J. Searching for drug synergy in complex dose-response landscapes using an interaction potency model. Comp Struct Biotechnol J. 2015;13:504–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moreau P, Chanan-Khan A, Roberts AW, et al. Promising efficacy and acceptable safety of venetoclax plus bortezomib and dexamethasone in relapsed/refractory MM. Blood. 2017;130(22):2392–2400. [DOI] [PubMed] [Google Scholar]
- 38.Yokoyama S, Perera PY, Terawaki S, et al. Janus kinase inhibitor tofacitinib shows potent efficacy in a mouse model of autoimmune lymphoproliferative syndrome (ALPS). J Clin Immunol. 2015; 35(7):661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zlei M, Egert S, Wider D, Ihorst G, Wasch R, Engelhardt M. Characterization of in vitro growth of multiple myeloma cells. Exp Hemaol. 2007;35(10):1550–1561. [DOI] [PubMed] [Google Scholar]
- 40.Shain KH, Yarde DN, Meads MB, et al. Beta1 integrin adhesion enhances IL-6-mediated STAT3 signaling in myeloma cells: implications for microenvironment influence on tumor survival and proliferation. Cancer Res. 2009;69(3):1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burger R, Gunther A, Klausz K, et al. Due to interleukin-6 type cytokine redundancy only glycoprotein 130 receptor blockade efficiently inhibits myeloma growth. Haematologica. 2017;102(2):381–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burger R, Guenther A, Bakker F, et al. Gp130 and ras mediated signaling in human plasma cell line INA-6: a cytokine-regulated tumor model for plasmacytoma. Hematol J. 2001;2(1):42–53. [DOI] [PubMed] [Google Scholar]
- 43.Tenhumberg S, Waetzig GH, Chalaris A, et al. Structure-guided optimization of the interleukin-6 trans-signaling antagonist sgp130. J Biol Chem. 2008;283(40):27200–27207. [DOI] [PubMed] [Google Scholar]
- 44.de Oliveira MB, Fook-Alves VL, Eugenio AIP, et al. Anti-myeloma effects of ruxolitinib combined with bortezomib and lenalidomide: A rationale for JAK/STAT pathway inhibition in myeloma patients. Cancer Lett. 2017;403:206–215. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.