Abstract
Background and objective
Nasopharyngeal carcinoma (NPC) is a common head and neck malignancy. Despite recent advances in treatment, the prognosis, particularly for those at the advanced stages, remains poor. Moreover, the underlying genetic and molecular events have remained obscure so far. Recently, increasing evidence has demonstrated that long noncoding RNAs (lncRNAs) could act as either oncogenes or tumor suppressor genes in various cancers depending on their targets. And some lncRNAs have been shown to be aberrantly expressed in NPC. In this meta-analysis, we try to elucidate the possible role of lncRNAs and their expression on prognosis in NPC.
Methods
We searched the databases of PubMed, Embase, and Web of Science for relevant articles ranging from January 2000 to December 2017. Pooled hazard ratios (HRs) and 95% confidence intervals (CIs) were used to evaluate the prognostic value of lncRNAs in NPC. Odds ratios (ORs) were used to assess the association between lncRNAs and clinicopathological characteristics.
Results
A total of 14 eligible publications including 14 on prognosis and eight on clinicopathological characteristics were identified. Our results demonstrated that the high expression of lncRNAs was related to poor overall survival (OS; HR =1.55; 95% CI =1.01, 2.40; P=0.05) and disease-free survival (DFS; HR =1.83; 95% CI =1.07, 3.13; P=0.03) of NPC. Moreover, the expression of lncRNAs was correlated with male gender (OR =1.42; 95% CI =1.05, 1.91; P=0.02), lymph node status (OR =2.20; 95% CI =1.29, 3.73; P=0.004), and tumor node metastasis (TNM) clinical stage (OR =2.55; 95% CI =1.12, 5.78; P=0.03).
Conclusion
This meta-analysis shows that the level of expression of lncRNAs may be a potential prognostic indicator in NPC.
Keywords: long noncoding RNAs, nasopharyngeal carcinoma, prognosis, overall survival, meta-analysis
Introduction
Nasopharyngeal carcinoma (NPC) is a distinctive type of head and neck cancer and originates from nasopharyngeal epithelial cells. Although rarely occurring worldwide, the incidence and mortality rates of this tumor are remarkably high among Southeast Asia and Southern China.1,2 The distinct geographical distribution highlights the significance of several etiologic factors in NPC tumorigenesis, including Epstein–Barr virus (EBV) infection, genetic predisposition, and intake of preserved food. The World Health Organization (WHO) recognizes the following three histological patterns: type I, keratinizing squamous-cell carcinoma; type II, differentiated nonkeratinizing carcinoma; and type III, undifferentiated carcinoma; the nonkeratinizing subtypes constitute most cases.3 However, the understanding of the development of NPC is still unclear. Local recurrence and metastasis to cervical lymph nodes are the main cause of mortality in NPC. During the last decades, the outcome of the treatment has improved considerably. Concurrent chemoradiotherapy has become the choice of treatment for advanced NPC.4
Patients with NPC often do not show specific symptoms in the early stage, and when first diagnosed, most of them have stepped into advanced stage. So far, the clinical tumor node metastasis (TNM) staging system is the most commonly used predictor of prognosis for NPC patients; what is troubling, the prognosis often varies even at the same stage. Therefore, more specific and sensitive prognostic indicators need to be discovered and applied for the early diagnosis and individualized treatment for NPC patients.
Previous studies have found that long noncoding RNAs (lncRNAs) are extensively transcribed from genomes. They have been shown to play a role in carcinogenesis and cancer progression by modulating the expression of many oncogenes or tumor suppressor genes.5–7 However, there are limited studies on lncRNAs in human NPC and the expression and function of lncRNAs in NPC remain uncovered. lncRNAs, >200 nucleotides (nts), with limited capacity of protein coding,8,9 were considered transcriptional “noise” or “junk”. Now, increasing evidence has demonstrated that these lncRNAs are junk no more, and in contrast, these lncRNAs have important biological functions in transcriptional regulation, epigenetic gene regulation, and disease.8–12 Due to their essential role at every level of gene expression, lncRNAs have attracted major attention as molecules with structural and functional roles in various physiological processes, including development, differentiation, and metabolism.13,14 Nevertheless, functional roles of the majority of these molecules remain to be identified.
To date, several studies have shown that a number of lncRNAs are involved in the development and progression of NPC, including HOTAIR, MALAT1, NEAT1, HNF1A-AS, AFAP1-AS1, and LINC00312.15–20 He et al21 summarized the different types of associated lncRNAs and their functional mechanisms in the development of NPC, and lncRNA was considered as a novel biomarker for the clinical diagnosis and treatment of NPC. Many studies had already evaluated the role of lncRNAs in NPC for prognosis; however, the small number of studies, limited number of lncRNAs, and paucity of multivariate analyses are the limitations; hence, this meta-analysis aims to comprehensively assess the value of lncRNAs both in the prognosis and clinical outcomes for patients with NPC.
Methods
Publication search strategy
PubMed, Embase, and Web of Science were searched for relevant literature published from January 2000 to December 2017. We mainly focused on “lncRNA”, “nasopharyngeal”, and “carcinoma”, and the specific search strategy was as follows: “long non-coding RNA”, “lncRNA”, “lincRNA”, “long intergenic non-coding RNA”, “long untranslated RNA” and “NPC”, “nasopharyngeal carcinoma”, “nasopharyngeal neoplasm”, “nasopharyngeal cancer”. Studies explored the expression level of lncRNAs in different data set were considered to be different studies.
Inclusion and exclusion criteria
In this meta-analysis, the inclusion criteria for the eligible studies were as follows: 1) diagnosed with NPC; 2) analyzed the association between lncRNAs and NPC; 3) prognostic values such as overall survival (OS), disease-free survival (DFS), and recurrence-free survival (RFS) were investigated; 4) usable and sufficient published data were provided to calculate hazard ratios (HRs) and 95% confidence intervals (CIs); and 5) more complete and updated studies and data are preferred. Exclusion criteria were as follows: 1) no usable or insufficient data; 2) case reports, reviews, letters, and conference abstracts; and 3) animal experiments and Chinese literature.
Data extraction
Three of us (HuanHuan Guo, Shuo Huang, and Shuang Li) extracted the provided data including author, publication year, lncRNAs and their biotypes, methods, case number, outcomes, cut-off value, and follow-up months. The HRs and 95% CIs for survival analysis were obtained from the articles if available, and for studies that did not provide OS, DFS, or RFS directly, we also digitized and extracted the data from the given Kaplan–Meier survival curves using the Engauge Digitizer version 4.1.22
Statistical analysis
The relation between lncRNAs and prognosis in NPC was evaluated by the calculated HRs and 95% CIs. HR >1 implied a worse survival for the group with increased lncRNAs expression. Conversely, HR <1 implied a better survival for the group with increased lncRNAs expression. Meanwhile, the association between lncRNAs and clinicopathological characteristics (including gender, histological classification, tumor classification, lymph node status, metastasis, and TNM clinical stage) were assessed by odds ratios (ORs) and 95% CIs. RevMan 5.2 software (RevMan; Cochrane Collaboration, Copenhagen, Denmark) was used to perform this meta-analysis, and the heterogeneity within studies was evaluated by Cochrane’s Q and I2 tests.23 When heterogeneity was observed (I2≤50% and P>0.1), the fixed-effect model was applied, otherwise, only the random-effect model was applied to calculate the pooled HRs or ORs. We evaluated the sensitivity and publication bias of the included articles by the Stata12.0 Software (StataCorp LP, College Station, TX, USA), and publication bias was evaluated by Begg’s rank correlation method and Egger’s weighted regression method. P<0.05 was considered statistically significant.
Results
Study characteristics
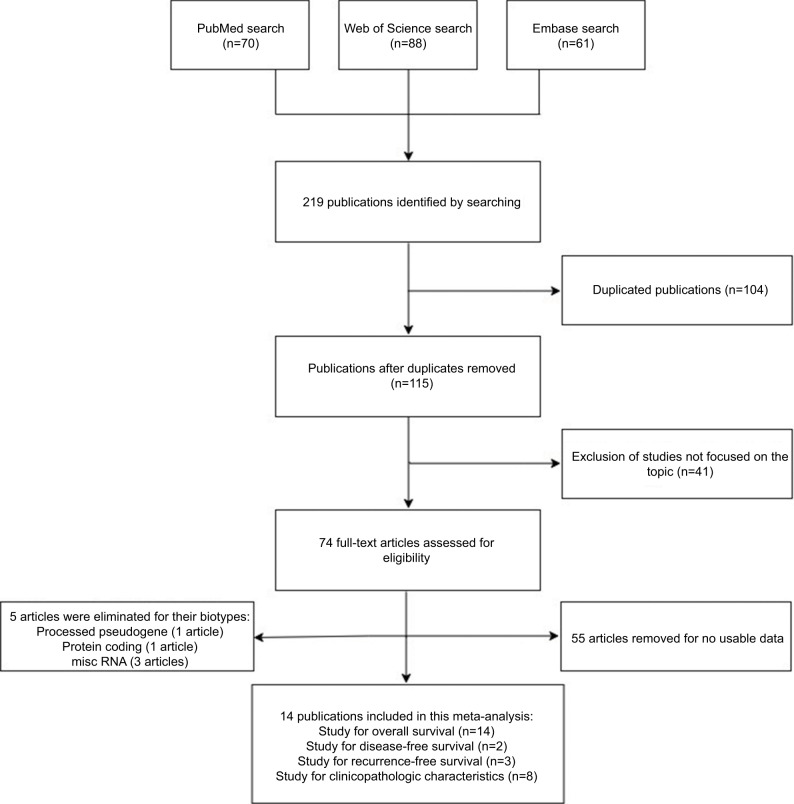
We searched 219 publications in the databases including 70 in PubMed, 61 in Embase, and 88 in Web of Science, and the flowchart of the literature review is shown in Figure 1. A total of 104 duplicated publications were removed, then 41 articles were excluded after screening the titles and abstracts, and 55 articles were removed for no usable data. After reviewing the studies, five articles were eliminated for their biotypes (Table S1). As a result, 14 articles were included in this systematic review and meta-analysis.16–19,24–33
Figure 1.
Flowchart of the search and selection of studies for the meta-analysis.
The main characteristics of the included 14 studies are summarized in Table 1, and the biotypes of different lncRNAs according to the Ensembl are shown later. Eight studies analyzed the association between the expression of lncRNAs and gender,16–19,24,25,28,29 two studies indicated that lncRNAs were related to histological classification,17,24 eight studies were on tumor classification,16–19,24,25,28,29 seven studies were about lymph node status,16–19,24,28,29 eight studies referred to the metastasis,16–19,24,25,28,29 and eight studies reported that lncRNAs were significantly correlated with TNM clinical stage (Table 2).16–19,24,25,28,29
Table 1.
Characteristics of studies included in this meta-analysis
| Author | Year | lncRNAs | Biotype (Ensembl)/length (bp) | Method | Case number (high/low) | Outcome | Cut-off | Follow-up months |
|---|---|---|---|---|---|---|---|---|
| Nie et al17 | 2013 | HOTAIR | Antisense RNA/2421 | qRT-PCR and ISH | 91/69 | OS, DFS, and RFS | SI =6 | Median of 69 |
| Bo et al24 | 2015 | AFAP1-AS1 | Antisense RNA/6795 | qRT-PCR and ISH | 23/55 | OS and RFS | 1.5-Fold | 120 |
| Sun et al25 | 2015 | LET | Sense intronic/2283 | qRT-PCR | 34/34 | OS and RFS | Median | 100 |
| Jin et al18 | 2016 | MALAT1 | lincRNA/8708 | qRT-PCR and ISH | 66/65 | OS | SI =6 | 60 |
| Lu et al19 | 2016 | NEAT1 | lincRNA/22743 | qRT-PCR and ISH | 66/65 | OS | SI =6 | 60 |
| Song et al26 | 2016 | XIST | lincRNA/19275 | qRT-PCR | 76/32 | OS | 2.31-Fold | 140 |
| Song and Yin27 | 2016 | EWSAT1 | lincRNA/2498 | qRT-PCR | 76/32 | OS | 2.36-Fold | 140 |
| Zou et al28 | 2016 | ANRIL | Antisense RNA/3835 | qRT-PCR | 44/44 | OS and DFS | Median | 100 |
| Jiang and Liu29 | 2017 | HULC | lincRNA/556 | qRT-PCR | 78/42 | OS | 2.5-Fold | 50 |
| Liu et al31 | 2017 | PCAT7 | Retained intron/1164 | qRT-PCR | 38/12 | OS | NA | 140 |
| Su et al30 | 2017 | CASC9 | lincRNA/1164 | qRT-PCR | 45/45 | OS | 2.5-Fold | 60 |
| Tang et al16 | 2017 | AFAP1-AS1 | Antisense RNA/6795 | qRT-PCR and ISH | 68/28 | OS | NA | 120 |
| Wang et al32 | 2017 | NEAT1 | lincRNA/22743 | qRT-PCR and AM | 39/31 | OS | 2-Fold | 50 |
| Kong et al33 | 2018 | LINC00460 | lincRNA/739 | qRT-PCR | 25/25 | OS | Median | 140 |
Abbreviations: AM, affymetrix microarray; DFS, disease-free survival; ISH, in situ hybridization; lincRNA, long intergenic noncoding RNA; lncRNAs, long noncoding RNAs; NA, not available; NPC, nasopharyngeal carcinoma; OS, overall survival; qRT-PCR, quantitative real time polymerase chain reaction; RFS, recurrence-free survival; SI, staining index.
Table 2.
Correlation between high expression of lncRNAs and clinicopathological characteristics of patients with NPC
| Characteristics | Studies | Case number | Pooled OR (95% CI) | P | Heterogeneity
|
|
|---|---|---|---|---|---|---|
| I2 (%) | P | |||||
| Gender (male/female) | 8 | 470 | 1.42 (1.05, 1.91) | 0.02 | 0 | 0.54 |
| Histological classification (WHO type II/III) | 2 | 106 | 0.91 (0.39, 2.13) | 0.82 | NA | NA |
| T classification (T1–T2/T3–T4) | 8 | 455 | 1.33 (0.69, 2.56) | 0.39 | 80 | <0.00001 |
| N classification (N0–N1/N2–N3) | 7 | 447 | 2.20 (1.29, 3.73) | 0.004 | 67 | 0.005 |
| Metastasis (no/yes) | 8 | 499 | 1.41 (0.69, 2.87) | 0.34 | 74 | 0.0004 |
| TNM clinical stage (I–II/III–IV) | 8 | 497 | 2.55 (1.12, 5.78) | 0.03 | 85 | <0.00001 |
Abbreviations: CI, confidence interval; lncRNAs, long noncoding RNAs; NA, not applicable; NPC, nasopharyngeal carcinoma; OR, odds ratio; TNM, tumor node metastasis; WHO, World Health Organization.
Prognosis
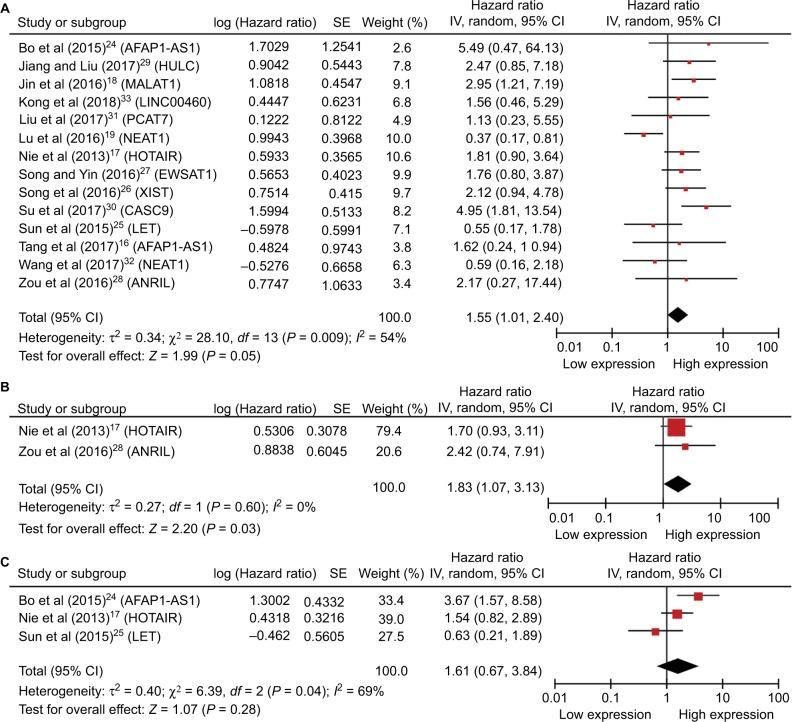
Among the included studies, 14 studies performed the correlation between lncRNAs and OS.16–19,24–33 HRs and 95% CIs were extracted or calculated by the provided data in these studies. The expression of lncRNAs was related to OS in NPC (HR =1.55; 95% CI =1.01, 2.40; P=0.05, random-effect) (Figure 2A). From the forest plot, we found that the high level of AFAP1-AS1, HULC, MALAT1, LINC00460, PCAT7, HOTAIR, EWSAT1, XIST, CASC9, and ANRIL was correlated with poor prognosis, whereas the low level of NEAT1 and LET was correlated with poor prognosis in NPC.
Figure 2.
(A) Forest plot of studies evaluating HRs of lncRNAs’ expression and the overall survival in NPC. (B) Forest plot of studies evaluating HRs of lncRNAs’ expression and the disease-free survival in NPC. (C) Forest plot of studies evaluating HRs of lncRNAs’ expression and the recurrence-free survival in NPC. The point estimate is bounded by a 95% CI, and the perpendicular line represents no increased risk for the outcome.
Abbreviations: CI, confidence interval; HRs, hazard ratios; lncRNAs, long noncoding RNAs; NPC, nasopharyngeal carcinoma.
Only two studies explored that the level of lncRNAs was associated with DFS in patients with NPC.17,28 The elevated level of HOTAIR and ANRIL indicated a relatively poor prognosis. And the enrolled studies showed the correlation between the increased expression of lncRNAs and DFS in NPC (HR =1.83; 95% CI =1.07, 3.13; P=0.03, fixed-effect) (Figure 2B).
And three studies showed that lncRNAs’ expression was associated with the RFS in NPC (HR =1.61; 95% CI =0.67, 3.84; P=0.28, random-effect) (Figure 2C).17,24,25 The high expression of AFAP1-AS1 and HOTAIR predicted a shorter RFS time, while the high expression of LET demonstrated a better outcome.
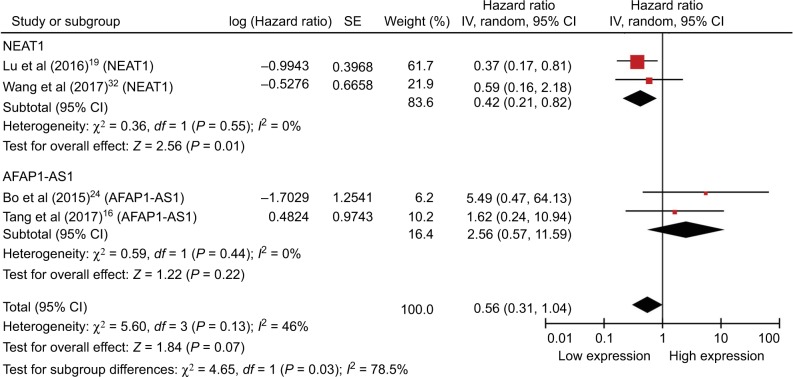
With only one or two studies included in this meta-analysis, with maybe the exception of NEAT1 (Figure 3), the analysis may be considered inadequate and this may be of limited practicability and should be elaborated prudently; furthermore, comprehensive and larger sample size studies are required to be validated.
Figure 3.
Forest plot showing the combined HR from studies for the association between high NEAT1 and AFAP1-AS1 levels and overall survival.
Abbreviations: CI, confidence interval; HR, hazard ratio.
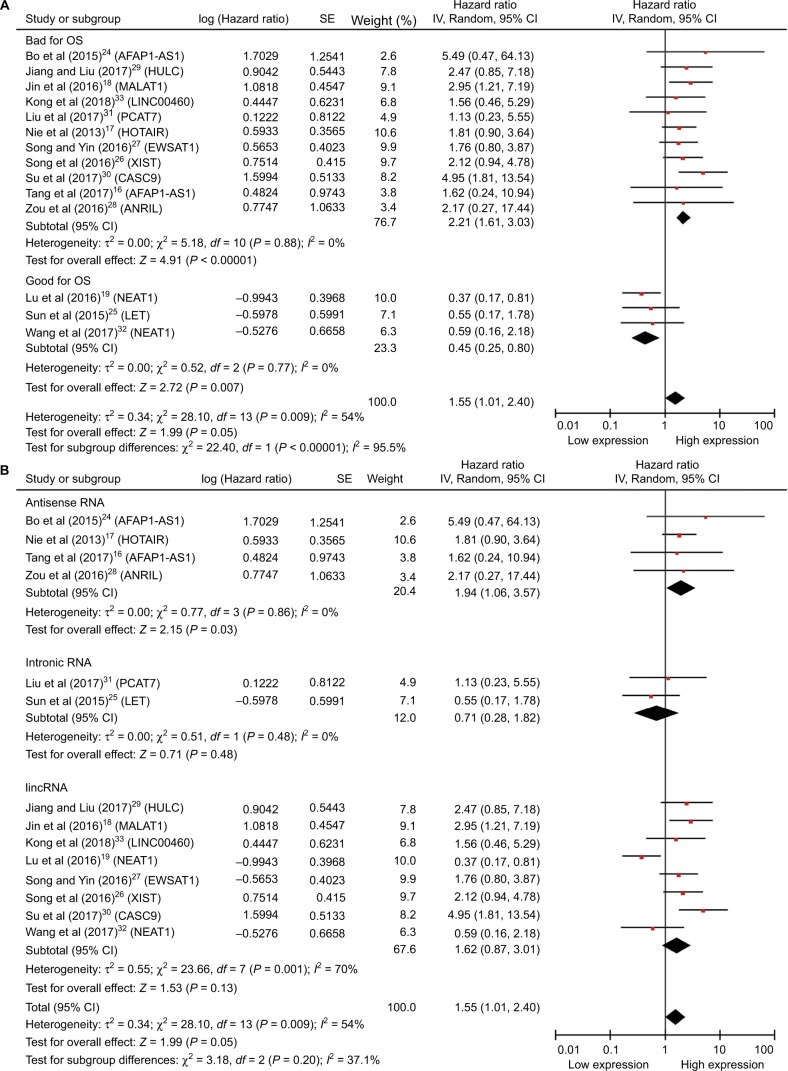
Due to the expression of lncRNAs that varies in NPC and the high expression as shown earlier, some may be associated with improved survival whereas others may definitely be correlated with reduced survival; thus, we performed the subgroup analyses (Figure 4). The upregulated lncRNAs were divided into two groups according to helpful for prognosis and harmful to prognosis (Figure 4A). And we also divided the included studies into three subgroups based on the biotypes of lncRNAs in the Ensembl database (Figure 4B). As shown in the forest plots, the heterogeneity has been reduced to some extent in several subgroups, but the total heterogeneity still cannot be ignored. Due to the small sample sizes of the included studies, we did not perform meta-regression.
Figure 4.
Forest plots of studies evaluating HRs of upregulated lncRNAs and the OS of NPC patients.
Notes: (A) Subgroup outcome and (B) subgroup biotype.
Abbreviations: CI, confidence interval; HRs, hazard ratios; lincRNA, long intergenic noncoding RNA; lncRNAs, long noncoding RNAs; OS, overall survival; NPC, nasopharyngeal carcinoma.
Correlation between the expression of lncRNAs and clinicopathological characteristics in NPC
Table 2 summarizes the association between the expression levels of lncRNAs and clinicopathological characteristics of NPC patients. ORs >1 implied that elevated expression of lncRNAs might be more susceptible to the characteristic. And the analysis indicated that the increased expression of lncRNAs was correlated with gender (OR =1.42; 95% CI =1.05, 1.91; P=0.02, fixed-effect), lymph node status (OR =2.20; 95% CI =1.29, 3.73; P=0.004, random-effect), and TNM clinical stage (OR =2.55; 95% CI =1.12, 5.78; P=0.03, random-effect) (Figure S1). Unfortunately, there was no correlation with histological classification (OR =0.91; 95% CI =0.39, 2.13; P=0.82, fixed-effect), tumor classification (OR =1.33; 95% CI =0.69, 2.56; P=0.39, random-effect), and metastasis (OR =1.41; 95% CI =0.69, 2.87; P=0.34, random-effect) (Figure S2). As shown earlier, a significant heterogeneity was observed among tumor classification (I2=80%), lymph node status (I2=67%), metastasis (I2=74%), and TNM clinical stage (I2=85%). We suspected that the main causes of the significant heterogeneity in this analysis were the different cut-off definitions of the expression and the different roles of the lncRNAs in different studies. And we did not perform subgroup analysis due to the limited number of the enrolled studies, and further studies should be conducted to verify this conclusion.
Publication bias and sensitivity analysis
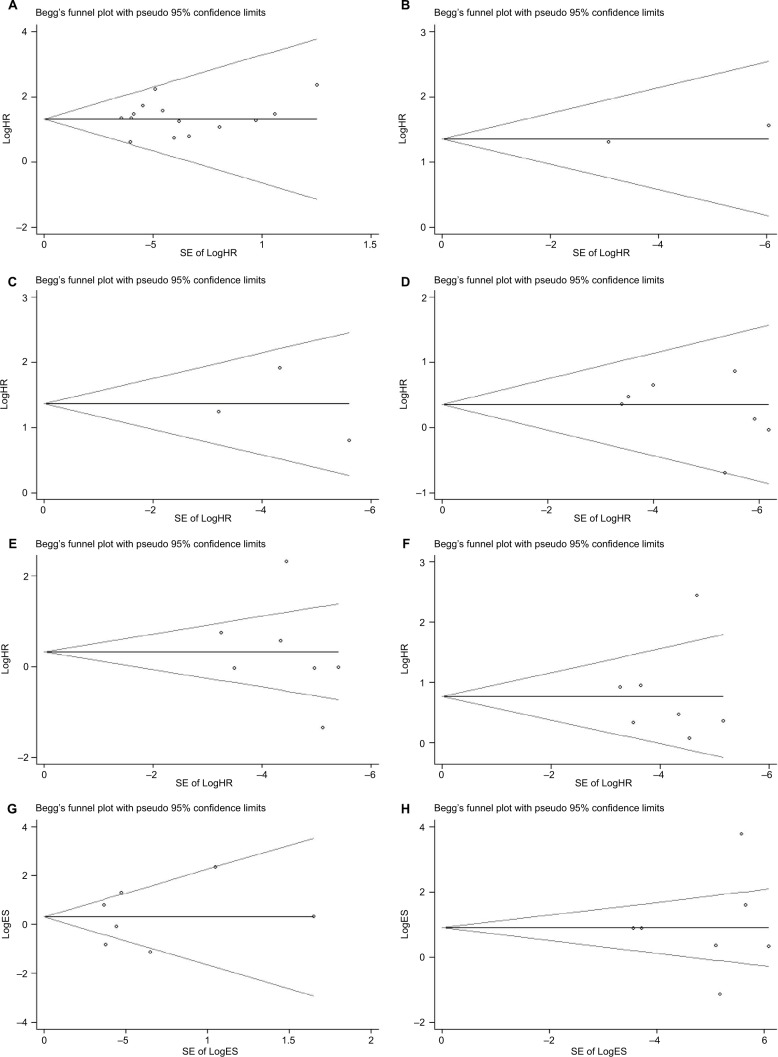
Begg’s funnel plot and Egger’s test were performed to assess the potential publication bias in the available literature. The shape of funnel plots did not reveal any evidence of funnel plot asymmetry (Figure 5) and all the values of P>0.05. Egger’s test also showed that there was no statistical significance for the evaluation of publication bias (Figure S3; OS: P=0.732, RFS: P=0.900; gender: P=0.294, tumor classification: P=0.679, lymph node status: P=0.878, metastasis: P=0.811, and TNM stage: P=0.826).
Figure 5.
Begg’s test for publication bias.
Notes: (A) Overall survival, (B) disease-free survival, (C) recurrence-free survival, (D) gender, (E) tumor classification, (F) lymph node status, (G) metastasis, and (H) TNM stage.
Abbreviations: HR, hazard ratio; TNM, tumor node metastasis; ES, effect size; SE, standard error.
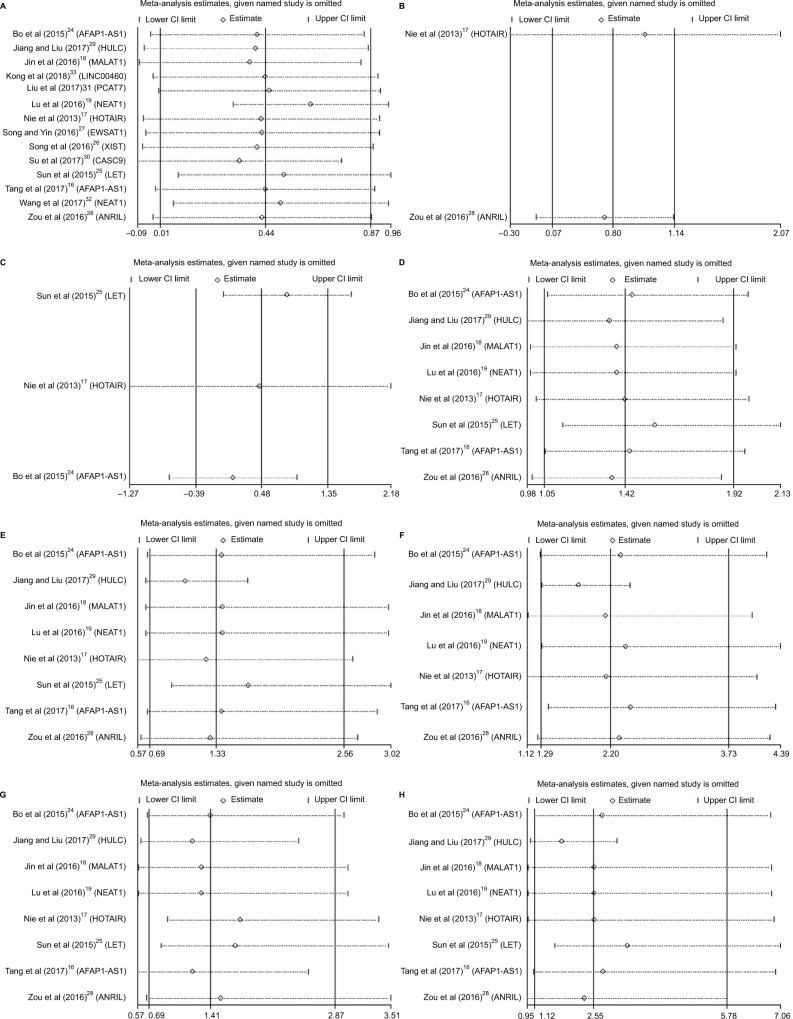
Sensitivity analysis, after removing one study at a time, was performed to evaluate the stability of the result. We found little change in the estimated results (Figure 6), indicating that our results were statistically robust.
Figure 6.
Sensitivity analyses of the studies.
Notes: (A) Overall survival, (B) disease-free survival, (C) recurrence-free survival, (D) gender, (E) tumor classification, (F) lymph node status, (G) metastasis, and (H) TNM stage.
Abbreviations: CI, confidence interval; TNM, tumor node metastasis.
Discussion
In the patients with NPC, many factors including stage of disease, nodal involvement, distant metastasis, histopathologic type, tumor volume, age, and parapharyngeal extension have been evaluated as potential prognostic indicators.3,34 In recent years, the searches for novel prognostic factors that more reliably predict the biological behavior of the tumors have focused on the role of various molecular biomarkers at the genetic levels,35 including lncRNAs.36 In this meta-analysis, we comprehensively assessed the associations of aberrantly expressed lncRNAs with the prognosis and clinicopathological parameters in NPC and we demonstrated that the higher expression of lncRNAs was significantly associated with the worse OS and DFS.
Recently, increasing evidence suggests that the dysregulation of lncRNAs was involved in cancer, and alterations in lncRNA expression and their mutations promote tumorigenesis and metastasis.5,6,37,38 Many cancer-associated lncRNAs have been identified and characterized, and these lncRNAs help further understand the molecular mechanism of cancer.13,39 In addition, most of these cancer-associated lncRNAs could be effective prognostic biomarkers and even therapeutic targets.39–41 HOTAIR is transcribed from the antisense strand of homeobox C (HOXC) gene locus in chromosome 12, and it coordinates with chromatin-modifying enzymes and regulates gene silencing. It is a scaffolding lncRNA, silences genes via interaction with PRC2 and LSD1, and aids protein degradation via interaction with E3 ubiquitin ligases; the knockdown of HOTAIR reduces tumor invasiveness and disrupts epithelial mesenchymal transition. And it is one of the well-studied lncRNAs that is overexpressed in various cancers including breast, pancreatic, colorectal, hepatocellular, gastrointestinal, and non-small-cell lung carcinomas.42–44 Furthermore, the aberrantly upregulated expression in NPC correlates with clinical stage progression and contributes to the malignant character of NPC cells through involvement in diverse cellular processes, including migration, invasion, and proliferation, and also indicates a poorer prognosis for DFS and OS. AFAP1-AS1 promoted cancer cell metastasis via regulation of actin filament integrity. And its knockdown significantly inhibited the NPC cell migration and invasive capability. The study indicated that the AFAP1-AS1 expression was upregulated in NPC and associated with NPC metastasis and poor prognosis.24 H19 affected the expression of enhancer of zeste homolog 2 (EZH2) to promote cell invasion by suppressing the activity of miR-630. Furthermore, H19 inhibited E-cadherin expression and promoted the cell invasion of NPC cells via the miR-630/EZH2 pathway.45 MALAT1 was originally found to be overexpressed in primary non-small-cell lung cancers,46,47 and it undergoes posttranscriptional processing to produce a short RNA (cytoplasmic MALAT1-associated small cytoplasmic RNA [mascRNA]) and a long MALAT1 transcript that are localized to nuclear speckles and influence the level of phosphorylated splicing-associated serine arginine (SR) proteins. And it is also overexpressed in other cancers including bladder carcinoma, breast cancer, prostate cancer, and ovarian cancer and is a potential biomarker and therapeutic target. HULC, an oncogenic lncRNA, which was first identified in hepatocellular carcinoma (HCC), acts as a miRNA sponge and sequesters miR-372, and its knockdown inhibits cell proliferation and increases chemosensitivity. HULC promoter possesses a binding site for transcription factor cAMP response element binding (CREB), and its expression is potentially regulated by CREB phosphorylation. It is highly expressed in NPC patients and correlated with a poor prognosis in cancer patients. Overexpressed HULC promotes NPC cell growth, while downregulated HULC activated p53 and induced the increased expression of p21, which finally caused cell cycle arrest and cell apoptosis.29 Prostate cancer-associated transcript 7 (PCAT7), a novel lncRNA, was found to be overexpressed and associated with good prognosis in NPC, which might contribute to the tumor progression by functioning as a competitive endogenous RNA (ceRNA) to sponge miR-134-5p and regulating miR-134-5p/ELF2 signal pathway.31 FOXCUT, which is located upstream of forkhead box C1 (FOXC1), was upregulated in clinical NPC tissues and cultured NPC cell lines, and the high levels of FOXCUT expression were correlated with lymph node metastasis and distant metastasis.48
There are also some reports suggesting that some lncRNAs may serve as antitumor factors in NPC and could predict a good prognosis, such as MEG3,49 LINC0086,50 LINC00312,20 LOC401317,51 LncRNA-LET,25 and NEAT1.19,32 LINC00312, also called NPC-associated gene 7 (NAG7), could inhibit proliferation and induce apoptosis in NPC cells but also stimulate NPC cell invasion. And positive expression of LINC00312 was associated with good prognosis in NPC patients with no lymph node metastasis and was associated with poor prognosis in NPC patients with lymph node metastasis.20 Further studies indicated that LOC401317 is directly regulated by p53 through a p53-binding site adjacent to its potential promoter and that LOC401317’s overexpres-sion inhibits HNE2 cell proliferation in vitro and in vivo by inducing cell cycle arrest and apoptosis. And these results suggest that LOC401317 exerts antitumor effects in HNE2 NPC cells.51 LncRNA-LET was transcriptionally repressed by EZH2-mediated H3K27 histone methylation on the LET promoter and significantly downregulated in NPC, and its decreased level is significantly related to advanced clinical stage, larger tumor size, increased lymph node tumor burden, and poor survival in NPC patients.25
Resistance to radiotherapy and chemotherapy is the primary cause of NPC patients’ death. In the current studies, some lncRNAs played a critical functional role in chemo-resistance or radioresistance. LincRNA-ROR was highly associated with the proliferation, metastasis, and apoptosis of NPC.52 And the enrichment of lincRNA-ROR was associated with chemoresistance.53 Further investigation found that NEAT1 upregulated ZEB1 expression by negatively regulating miR-204 expression and regulated radioresistance by modulating epithelial mesenchymal transition phenotype in NPC.19 Furthermore, MALAT1 regulated cancer stem cell activity and radioresistance by modulating miR-1/slug axis.18 Moreover, knockdown of ANRIL represses tumorigenicity and enhances cisplatin (DDP)-induced cytotoxicity via regulating microRNA let-7a in NPC cells.54 Upregulated CCAT1 results in significantly enhancing paclitaxel resistance in nasopharyngeal cancer cells. And lncRNA CCAT1 regulates the sensitivity of paclitaxel in NPC cells via miR-181a/CPEB2 axis.55 Together, the current study provide a molecular basis for a comprehensive understanding of, and exploring new therapies for, NPC.35
At present, a number of studies have shown that the expression of lncRNAs was correlated with clinicopathological characteristics in NPC: histological type, tumor size, TNM clinical stage, and some others. In this meta-analysis, we also found that the lncRNAs were related to the male, lymph node status, and TNM clinical stage. There are some limitations in this meta-analysis. First, our results were based on unadjusted estimates, while the lack of information (such as age and family history) for the date analysis may cause serious confounding bias. Second, because of incomplete raw data or publication limitations (the enrolled studies are only English and all from China), some relevant studies could not be included in our analysis. Third, the number of published studies was not sufficiently large for a comprehensive analysis and some studies with small size may not have enough statistical power to explore the real association.
Taking these observations into consideration, the novel molecular mechanisms by which the lncRNAs regulate carcinogenesis and metastasis are expected to be elucidated. And they will developed to be new clinical prognostic biomarker as well as new therapeutic target for NPC. Nevertheless, discovering novel lncRNAs, identifying their function and association with various cancer subtypes, and developing novel lncRNA-based strategies for diagnosis and targeted therapies appear very promising, bring a new paradigm in cancer research, and may emerge as a major therapeutic strategy for the treatment of cancer in the near future.
Supplementary materials
Forest plot of studies evaluating ORs of lncRNAs’ expression and the clinicopathological characteristics of NPC patients. (A) Gender; (B) lymph node status; and (C) TNM clinical stages.
Abbreviation: CI, confidence interval.
Forest plot of studies evaluating ORs of lncRNAs’ expression and the clinicopathological characteristics of NPC patients. (A) histological classification; (B) tumor classification; and (C) metastasis.
Abbreviations: CI, confidence interval; WHO, World Health Organization; M-H, Mantel Haenszel test.
Egger’s test for publication bias.
Notes: (A) Overall survival; (B) disease-free survival; (C) recurrence-free survival; (D) gender; (E) tumor classification; (F) lymph node status; (G) metastasis; (H) TNM stage.
Table S1.
Characteristics of studies (included and excluded) in this meta-analysis
| Author | Year | lncRNAs | Biotype (Ensembl)/length (bp) | Method | Case number (high/low) | Outcome | Cut-off | Follow-up months |
|---|---|---|---|---|---|---|---|---|
| Nie et al1 | 2013 | HOTAIR | Antisense RNA/2421 | qRT-PCR and ISH | 91/69 | OS, DFS, and RFS | SI =6 | Median of 69 |
| Zhang et al9 | 2013 | LINC00312 | Misc RNA/2887 | qRT-PCR and ISH | 169/160 | OS and DFS | NA | 100 |
| Bo et al2 | 2015 | AFAP1-AS1 | Antisense RNA/6795 | qRT-PCR and ISH | 23/55 | OS and RFS | 1.5-Fold | 120 |
| Sun et al3 | 2015 | LET | Sense intronic/2283 | qRT-PCR | 34/34 | OS and RFS | Median | 100 |
| Zhang et al19 | 2015 | LOC84740 | NA | qRT-PCR | 23/82 | DFS | Fold-change | 48 |
| Zhang et al19 | 2015 | ENST00000498296 | Processed pseudogene/909 | qRT-PCR | 64/42 | DFS | Fold-change | 48 |
| Zhang et al19 | 2015 | AL359062 | NA/1762 | qRT-PCR | 58/48 | DFS | Fold-change | 48 |
| Zhang et al19 | 2015 | ENST00000438550 | Processed pseudogene/371 | qRT-PCR | 81/25 | DFS | Fold-change | 48 |
| Jin et al4 | 2016 | MALAT1 | lincRNA/8708 | qRT-PCR and ISH | 66/65 | OS | SI =6 | 60 |
| Lu et al5 | 2016 | NEAT1 | lincRNA/22743 | qRT-PCR and ISH | 66/65 | OS | SI =6 | 60 |
| Song et al10 | 2016 | XIST | lincRNA/19275 | qRT-PCR | 76/32 | OS | 2.31-Fold | 140 |
| Song and Yin11 | 2016 | EWSAT1 | lincRNA/2498 | qRT-PCR | 76/32 | OS | 2.36-Fold | 140 |
| Zou et al6 | 2016 | ANRIL | Antisense RNA/3835 | qRT-PCR | 44/44 | OS and DFS | Median | 100 |
| Guo et al12 | 2017 | LINC0086 | Protein coding/5230 | qRT-PCR and ISH | 44/68 | OS | NA | 80 |
| Jiang and Liu7 | 2017 | HULC | lincRNA/556 | qRT-PCR | 78/42 | OS | 2.5-Fold | 50 |
| Liu et al13 | 2017 | PCAT7 | Retained intron/1164 | qRT-PCR | 38/12 | OS | NA | 140 |
| Su et al14 | 2017 | CASC9 | lincRNA/1164 | qRT-PCR | 45/45 | OS | 2.5-Fold | 60 |
| Sun et al17 | 2017 | LOC100129148 | Misc RNA/461 | qRT-PCR | 39/43 | OS | Mean | 80 |
| Tang et al8 | 2017 | AFAP1-AS1 | Antisense RNA/6795 | qRT-PCR and ISH | 68/28 | OS | NA | 120 |
| Wang et al15 | 2017 | NEAT1 | lincRNA/22743 | qRT-PCR and AM | 39/31 | OS | 2-Fold | 50 |
| Yang et al18 | 2017 | LINC01420 | Misc RNA/694 | qRT-PCR and ISH | 65/45 | OS | 1.2-Fold | 125 |
| Kong et al16 | 2018 | LINC00460 | lincRNA/739 | qRT-PCR | 25/25 | OS | Median | 140 |
Note: Bold fonts represent the included studies.
Abbreviations: AM, affymetrix microarray; DFS, disease-free survival; ISH, in situ hybridization; lincRNA, long intergenic noncoding RNA; lncRNAs, long noncoding RNAs; NA, not available; NPC, nasopharyngeal carcinoma; OS, overall survival; qRT-PCR, quantitative real time polymerase chain reaction; RFS, recurrence-free survival; SI, staining index.
References
- 1.Nie Y, Liu X, Qu S, Song E, Zou H, Gong C. Long non-coding RNA HOTAIR is an independent prognostic marker for nasopharyngeal carcinoma progression and survival. Cancer Sci. 2013;104(4):458–464. doi: 10.1111/cas.12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bo H, Gong Z, Zhang W, et al. Upregulated long non-coding RNA AFAP1-AS1 expression is associated with progression and poor prognosis of nasopharyngeal carcinoma. Oncotarget. 2015;6(24):20404–20418. doi: 10.18632/oncotarget.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun Q, Liu H, Li L, et al. Long noncoding RNA-LET, which is repressed by EZH2, inhibits cell proliferation and induces apoptosis of nasopharyngeal carcinoma cell. Med Oncol. 2015;32(9):226. doi: 10.1007/s12032-015-0673-0. [DOI] [PubMed] [Google Scholar]
- 4.Jin C, Yan B, Lu Q, Lin Y, Ma L. The role of MALAT1/miR-1/slug axis on radioresistance in nasopharyngeal carcinoma. Tumour Biol. 2016;37(3):4025–4033. doi: 10.1007/s13277-015-4227-z. [DOI] [PubMed] [Google Scholar]
- 5.Lu Y, Li T, Wei G, et al. The long non-coding RNA NEAT1 regulates epithelial to mesenchymal transition and radioresistance in through miR-204/ZEB1 axis in nasopharyngeal carcinoma. Tumour Biol. 2016;37(9):11733–11741. doi: 10.1007/s13277-015-4773-4. [DOI] [PubMed] [Google Scholar]
- 6.Zou ZW, Ma C, Medoro L, et al. LncRNA ANRIL is up-regulated in nasopharyngeal carcinoma and promotes the cancer progression via increasing proliferation, reprograming cell glucose metabolism and inducing side-population stem-like cancer cells. Oncotarget. 2016;7(38):61741–61754. doi: 10.18632/oncotarget.11437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang X, Liu W. Long noncoding RNA highly upregulated in liver cancer activates p53-p21 pathway and promotes nasopharyngeal carcinoma cell growth. DNA Cell Biol. 2017;36(7):596–602. doi: 10.1089/dna.2017.3686. [DOI] [PubMed] [Google Scholar]
- 8.Tang Y, He Y, Shi L, et al. Co-expression of AFAP1-AS1 and PD-1 predicts poor prognosis in nasopharyngeal carcinoma. Oncotarget. 2017;8(24):39001–39011. doi: 10.18632/oncotarget.16545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang W, Huang C, Gong Z, et al. Expression of LINC00312, a long intergenic non-coding RNA, is negatively correlated with tumor size but positively correlated with lymph node metastasis in nasopharyngeal carcinoma. J Mol Histol. 2013;44(5):545–554. doi: 10.1007/s10735-013-9503-x. [DOI] [PubMed] [Google Scholar]
- 10.Song P, Ye LF, Zhang C, Peng T, Zhou XH. Long non-coding RNA XIST exerts oncogenic functions in human nasopharyngeal carcinoma by targeting miR-34a-5p. Gene. 2016;592(1):8–14. doi: 10.1016/j.gene.2016.07.055. [DOI] [PubMed] [Google Scholar]
- 11.Song P, Yin SC. Long non-coding RNA EWSAT1 promotes human nasopharyngeal carcinoma cell growth in vitro by targeting miR-326/-330-5p. Aging (Albany NY) 2016;8(11):2948–2960. doi: 10.18632/aging.101103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo J, Ma J, Zhao G, Li G, Fu Y, Luo Y. Long non-coding RNA LINC0086 functions as a tumor suppressor in nasopharyngeal carcinoma by targeting miR-214. Oncol Res. 2017;25(7):1189–1197. doi: 10.3727/096504017X14865126670075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, Tao Z, Qu J, Zhou X, Zhang C. Long non-coding RNA PCAT7 regulates ELF2 signaling through inhibition of miR-134-5p in nasopharyngeal carcinoma. Biochem Biophys Res Commun. 2017;491(2):374–381. doi: 10.1016/j.bbrc.2017.07.093. [DOI] [PubMed] [Google Scholar]
- 14.Su X, Li G, Liu W. The long noncoding RNA cancer susceptibility candidate 9 promotes nasopharyngeal carcinogenesis via stabilizing HIF1alpha. DNA Cell Biol. 2017;36(5):394–400. doi: 10.1089/dna.2016.3615. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Wang C, Chen C, et al. Long non-coding RNA NEAT1 regulates epithelial membrane protein 2 expression to repress nasopharyngeal carcinoma migration and irradiation-resistance through miR-101-3p as a competing endogenous RNA mechanism. Oncotarget. 2017;8(41):70156–70171. doi: 10.18632/oncotarget.19596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kong YG, Cui M, Chen SM, Xu Y, Xu Y, Tao ZZ. LncRNA-LINC00460 facilitates nasopharyngeal carcinoma tumorigenesis through sponging miR-149-5p to up-regulate IL6. Gene. 2018;639:77–84. doi: 10.1016/j.gene.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 17.Sun KY, Peng T, Chen Z, Song P, Zhou XH. Long non-coding RNA LOC100129148 functions as an oncogene in human nasopharyngeal carcinoma by targeting miR-539-5p. Aging. 2017;9:999–1011. doi: 10.18632/aging.101205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang L, Tang Y, He Y, et al. High Expression of LINC01420 indicates an unfavorable prognosis and modulates cell migration and invasion in nasopharyngeal carcinoma. J Cancer. 2017;8:97–103. doi: 10.7150/jca.16819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang W, Wang L, Zheng F, et al. Long noncoding RNA expression signatures of metastatic nasopharyngeal carcinoma and their prognostic value. Biomed Res Int. 2015;2015:618924. doi: 10.1155/2015/618924. [DOI] [PMC free article] [PubMed] [Google Scholar]
Footnotes
Author contributions
All authors contributed toward data analysis, drafting and critically revising the paper and agree to be accountable for all aspects of the work. All the authors approved the final paper.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Chang ET, Adami HO. The enigmatic epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol Biomarkers Prev. 2006;15(10):1765–1777. doi: 10.1158/1055-9965.EPI-06-0353. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Brennan B. Nasopharyngeal carcinoma. Orphanet J Rare Dis. 2006;1:23. doi: 10.1186/1750-1172-1-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chua MLK, Wee JTS, Hui EP, Chan ATC. Nasopharyngeal carcinoma. Lancet. 2016;387(10022):1012–1024. doi: 10.1016/S0140-6736(15)00055-0. [DOI] [PubMed] [Google Scholar]
- 5.Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31(43):4577–4587. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gong Z, Zhang S, Zhang W, et al. Long non-coding RNAs in cancer. Sci China Life Sci. 2012;55(12):1120–1124. doi: 10.1007/s11427-012-4413-9. [DOI] [PubMed] [Google Scholar]
- 7.Wu J, Hann SS. Functions and roles of long-non-coding RNAs in human nasopharyngeal carcinoma. Cell Physiol Biochem. 2018;45(3):1191–1204. doi: 10.1159/000487451. [DOI] [PubMed] [Google Scholar]
- 8.Sun M, Kraus WL. From discovery to function: the expanding roles of long noncoding RNAs in physiology and disease. Endocr Rev. 2015;36(1):25–64. doi: 10.1210/er.2014-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 11.Huarte M, Guttman M, Feldser D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loewer S, Cabili MN, Guttman M, et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42(12):1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wen X, Tang X, Li Y, et al. Microarray expression profiling of long non-coding RNAs involved in nasopharyngeal carcinoma metastasis. Int J Mol Sci. 2016;17(11):1956. doi: 10.3390/ijms17111956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhuang K, Wu Q, Jin CS, Yuan HJ, Cheng JZ. Long non-coding RNA HNF1A-AS is upregulated and promotes cell proliferation and metastasis in nasopharyngeal carcinoma. Cancer Biomark. 2016;16(2):291–300. doi: 10.3233/CBM-150567. [DOI] [PubMed] [Google Scholar]
- 16.Tang Y, He Y, Shi L, et al. Co-expression of AFAP1-AS1 and PD-1 predicts poor prognosis in nasopharyngeal carcinoma. Oncotarget. 2017;8(24):39001–39011. doi: 10.18632/oncotarget.16545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nie Y, Liu X, Qu S, Song E, Zou H, Gong C. Long non-coding RNA HOTAIR is an independent prognostic marker for nasopharyngeal carcinoma progression and survival. Cancer Sci. 2013;104(4):458–464. doi: 10.1111/cas.12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin C, Yan B, Lu Q, Lin Y, Ma L. The role of MALAT1/miR-1/slug axis on radioresistance in nasopharyngeal carcinoma. Tumour Biol. 2016;37(3):4025–4033. doi: 10.1007/s13277-015-4227-z. [DOI] [PubMed] [Google Scholar]
- 19.Lu Y, Li T, Wei G, et al. The long non-coding RNA NEAT1 regulates epithelial to mesenchymal transition and radioresistance in through miR-204/ZEB1 axis in nasopharyngeal carcinoma. Tumour Biol. 2016;37(9):11733–11741. doi: 10.1007/s13277-015-4773-4. [DOI] [PubMed] [Google Scholar]
- 20.Zhang W, Huang C, Gong Z, et al. Expression of LINC00312, a long intergenic non-coding RNA, is negatively correlated with tumor size but positively correlated with lymph node metastasis in nasopharyngeal carcinoma. J Mol Histol. 2013;44(5):545–554. doi: 10.1007/s10735-013-9503-x. [DOI] [PubMed] [Google Scholar]
- 21.He R, Hu Z, Wang Q, et al. The role of long non-coding RNAs in nasopharyngeal carcinoma: as systemic review. Oncotarget. 2017;8(9):16075–16083. doi: 10.18632/oncotarget.14211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 24.Bo H, Gong Z, Zhang W, et al. Upregulated long non-coding RNA AFAP1-AS1 expression is associated with progression and poor prognosis of nasopharyngeal carcinoma. Oncotarget. 2015;6(24):20404–20418. doi: 10.18632/oncotarget.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun Q, Liu H, Li L, et al. Long noncoding RNA-LET, which is repressed by EZH2, inhibits cell proliferation and induces apoptosis of nasopharyngeal carcinoma cell. Med Oncol. 2015;32(9):226. doi: 10.1007/s12032-015-0673-0. [DOI] [PubMed] [Google Scholar]
- 26.Song P, Ye LF, Zhang C, Peng T, Zhou XH. Long non-coding RNA XIST exerts oncogenic functions in human nasopharyngeal carcinoma by targeting miR-34a-5p. Gene. 2016;592(1):8–14. doi: 10.1016/j.gene.2016.07.055. [DOI] [PubMed] [Google Scholar]
- 27.Song P, Yin SC. Long non-coding RNA EWSAT1 promotes human nasopharyngeal carcinoma cell growth in vitro by targeting miR-326/-330-5p. Aging (Albany NY) 2016;8(11):2948–2960. doi: 10.18632/aging.101103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zou ZW, Ma C, Medoro L, et al. LncRNA ANRIL is up-regulated in nasopharyngeal carcinoma and promotes the cancer progression via increasing proliferation, reprograming cell glucose metabolism and inducing side-population stem-like cancer cells. Oncotarget. 2016;7(38):61741–61754. doi: 10.18632/oncotarget.11437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang X, Liu W. Long noncoding RNA highly upregulated in liver cancer activates p53-p21 pathway and promotes nasopharyngeal carcinoma cell growth. DNA Cell Biol. 2017;36(7):596–602. doi: 10.1089/dna.2017.3686. [DOI] [PubMed] [Google Scholar]
- 30.Su X, Li G, Liu W. The long noncoding RNA cancer susceptibility candidate 9 promotes nasopharyngeal carcinogenesis via stabilizing HIF1alpha. DNA Cell Biol. 2017;36(5):394–400. doi: 10.1089/dna.2016.3615. [DOI] [PubMed] [Google Scholar]
- 31.Liu Y, Tao Z, Qu J, Zhou X, Zhang C. Long non-coding RNA PCAT7 regulates ELF2 signaling through inhibition of miR-134-5p in nasopharyngeal carcinoma. Biochem Biophys Res Commun. 2017;491(2):374–381. doi: 10.1016/j.bbrc.2017.07.093. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y, Wang C, Chen C, et al. Long non-coding RNA NEAT1 regulates epithelial membrane protein 2 expression to repress nasopharyngeal carcinoma migration and irradiation-resistance through miR-101-3p as a competing endogenous RNA mechanism. Oncotarget. 2017;8(41):70156–70171. doi: 10.18632/oncotarget.19596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kong YG, Cui M, Chen SM, Xu Y, Xu Y, Tao ZZ. LncRNA-LINC00460 facilitates nasopharyngeal carcinoma tumorigenesis through sponging miR-149-5p to up-regulate IL6. Gene. 2018;639:77–84. doi: 10.1016/j.gene.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 34.Wei WI, Sham JS. Nasopharyngeal carcinoma. Lancet. 2005;365(9476):2041–2054. doi: 10.1016/S0140-6736(05)66698-6. [DOI] [PubMed] [Google Scholar]
- 35.Lin DC, Meng X, Hazawa M, et al. The genomic landscape of nasopharyngeal carcinoma. Nat Genet. 2014;46(8):866–871. doi: 10.1038/ng.3006. [DOI] [PubMed] [Google Scholar]
- 36.Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J Pathol. 2010;220(2):126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 37.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xing Z, Lin A, Li C, et al. lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell. 2014;159(5):1110–1125. doi: 10.1016/j.cell.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang G, Chen H, Liu J. The long noncoding RNA LINC01207 promotes proliferation of lung adenocarcinoma. Am J Cancer Res. 2015;5(10):3162–3173. [PMC free article] [PubMed] [Google Scholar]
- 41.Ma Y, Yang Y, Wang F, et al. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/beta-catenin signalling pathway via suppression of activator protein 2alpha. Gut. 2016;65(9):1494–1504. doi: 10.1136/gutjnl-2014-308392. [DOI] [PubMed] [Google Scholar]
- 42.Kim K, Jutooru I, Chadalapaka G, et al. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32(13):1616–1625. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakagawa T, Endo H, Yokoyama M, et al. Large noncoding RNA HOTAIR enhances aggressive biological behavior and is associated with short disease-free survival in human non-small cell lung cancer. Biochem Biophys Res Commun. 2013;436(2):319–324. doi: 10.1016/j.bbrc.2013.05.101. [DOI] [PubMed] [Google Scholar]
- 44.Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X, Lin Y, Yang X, Wu X, He X. Long noncoding RNA H19 regulates EZH2 expression by interacting with miR-630 and promotes cell invasion in nasopharyngeal carcinoma. Biochem Biophys Res Commun. 2016;473(4):913–919. doi: 10.1016/j.bbrc.2016.03.150. [DOI] [PubMed] [Google Scholar]
- 46.Gutschner T, Hammerle M, Eissmann M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73(3):1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ji P, Diederichs S, Wang W, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22(39):8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 48.Xu YZ, Chen FF, Zhang Y, et al. The long noncoding RNA FOXCUT promotes proliferation and migration by targeting FOXC1 in nasopharyngeal carcinoma. Tumour Biol. 2017;39 doi: 10.1177/1010428317706054. 1010428317706054. [DOI] [PubMed] [Google Scholar]
- 49.Chak WP, Lung RW, Tong JH, et al. Downregulation of long non-coding RNA MEG3 in nasopharyngeal carcinoma. Mol Carcinog. 2017;56(3):1041–1054. doi: 10.1002/mc.22569. [DOI] [PubMed] [Google Scholar]
- 50.Guo J, Ma J, Zhao G, Li G, Fu Y, Luo Y. Long non-coding RNA LINC0086 functions as a tumor suppressor in nasopharyngeal carcinoma by targeting miR-214. Oncol Res. 2017;25(7):1189–1197. doi: 10.3727/096504017X14865126670075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gong Z, Zhang S, Zeng Z, et al. LOC401317, a p53-regulated long non-coding RNA, inhibits cell proliferation and induces apoptosis in the nasopharyngeal carcinoma cell line HNE2. PLoS One. 2014;9(11):e110674. doi: 10.1371/journal.pone.0110674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pan Y, Li C, Chen J, et al. The emerging roles of long noncoding RNA ROR (lincRNA-ROR) and its possible mechanisms in human cancers. Cell Physiol Biochem. 2016;40(1–2):219–229. doi: 10.1159/000452539. [DOI] [PubMed] [Google Scholar]
- 53.Li L, Gu M, You B, et al. Long non-coding RNA ROR promotes proliferation, migration and chemoresistance of nasopharyngeal carcinoma. Cancer Sci. 2016;107(9):1215–1222. doi: 10.1111/cas.12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y, Cheng N, Luo J. Downregulation of lncRNA ANRIL represses tumorigenicity and enhances cisplatin-induced cytotoxicity via regulating microRNA let-7a in nasopharyngeal carcinoma. J Biochem Mol Toxicol. 2017;31(7):e21904. doi: 10.1002/jbt.21904. [DOI] [PubMed] [Google Scholar]
- 55.Wang Q, Zhang W, Hao S. LncRNA CCAT1 modulates the sensitivity of paclitaxel in nasopharynx cancers cells via miR-181a/CPEB2 axis. Cell Cycle. 2017;16(8):795–801. doi: 10.1080/15384101.2017.1301334. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Forest plot of studies evaluating ORs of lncRNAs’ expression and the clinicopathological characteristics of NPC patients. (A) Gender; (B) lymph node status; and (C) TNM clinical stages.
Abbreviation: CI, confidence interval.
Forest plot of studies evaluating ORs of lncRNAs’ expression and the clinicopathological characteristics of NPC patients. (A) histological classification; (B) tumor classification; and (C) metastasis.
Abbreviations: CI, confidence interval; WHO, World Health Organization; M-H, Mantel Haenszel test.
Egger’s test for publication bias.
Notes: (A) Overall survival; (B) disease-free survival; (C) recurrence-free survival; (D) gender; (E) tumor classification; (F) lymph node status; (G) metastasis; (H) TNM stage.
Table S1.
Characteristics of studies (included and excluded) in this meta-analysis
| Author | Year | lncRNAs | Biotype (Ensembl)/length (bp) | Method | Case number (high/low) | Outcome | Cut-off | Follow-up months |
|---|---|---|---|---|---|---|---|---|
| Nie et al1 | 2013 | HOTAIR | Antisense RNA/2421 | qRT-PCR and ISH | 91/69 | OS, DFS, and RFS | SI =6 | Median of 69 |
| Zhang et al9 | 2013 | LINC00312 | Misc RNA/2887 | qRT-PCR and ISH | 169/160 | OS and DFS | NA | 100 |
| Bo et al2 | 2015 | AFAP1-AS1 | Antisense RNA/6795 | qRT-PCR and ISH | 23/55 | OS and RFS | 1.5-Fold | 120 |
| Sun et al3 | 2015 | LET | Sense intronic/2283 | qRT-PCR | 34/34 | OS and RFS | Median | 100 |
| Zhang et al19 | 2015 | LOC84740 | NA | qRT-PCR | 23/82 | DFS | Fold-change | 48 |
| Zhang et al19 | 2015 | ENST00000498296 | Processed pseudogene/909 | qRT-PCR | 64/42 | DFS | Fold-change | 48 |
| Zhang et al19 | 2015 | AL359062 | NA/1762 | qRT-PCR | 58/48 | DFS | Fold-change | 48 |
| Zhang et al19 | 2015 | ENST00000438550 | Processed pseudogene/371 | qRT-PCR | 81/25 | DFS | Fold-change | 48 |
| Jin et al4 | 2016 | MALAT1 | lincRNA/8708 | qRT-PCR and ISH | 66/65 | OS | SI =6 | 60 |
| Lu et al5 | 2016 | NEAT1 | lincRNA/22743 | qRT-PCR and ISH | 66/65 | OS | SI =6 | 60 |
| Song et al10 | 2016 | XIST | lincRNA/19275 | qRT-PCR | 76/32 | OS | 2.31-Fold | 140 |
| Song and Yin11 | 2016 | EWSAT1 | lincRNA/2498 | qRT-PCR | 76/32 | OS | 2.36-Fold | 140 |
| Zou et al6 | 2016 | ANRIL | Antisense RNA/3835 | qRT-PCR | 44/44 | OS and DFS | Median | 100 |
| Guo et al12 | 2017 | LINC0086 | Protein coding/5230 | qRT-PCR and ISH | 44/68 | OS | NA | 80 |
| Jiang and Liu7 | 2017 | HULC | lincRNA/556 | qRT-PCR | 78/42 | OS | 2.5-Fold | 50 |
| Liu et al13 | 2017 | PCAT7 | Retained intron/1164 | qRT-PCR | 38/12 | OS | NA | 140 |
| Su et al14 | 2017 | CASC9 | lincRNA/1164 | qRT-PCR | 45/45 | OS | 2.5-Fold | 60 |
| Sun et al17 | 2017 | LOC100129148 | Misc RNA/461 | qRT-PCR | 39/43 | OS | Mean | 80 |
| Tang et al8 | 2017 | AFAP1-AS1 | Antisense RNA/6795 | qRT-PCR and ISH | 68/28 | OS | NA | 120 |
| Wang et al15 | 2017 | NEAT1 | lincRNA/22743 | qRT-PCR and AM | 39/31 | OS | 2-Fold | 50 |
| Yang et al18 | 2017 | LINC01420 | Misc RNA/694 | qRT-PCR and ISH | 65/45 | OS | 1.2-Fold | 125 |
| Kong et al16 | 2018 | LINC00460 | lincRNA/739 | qRT-PCR | 25/25 | OS | Median | 140 |
Note: Bold fonts represent the included studies.
Abbreviations: AM, affymetrix microarray; DFS, disease-free survival; ISH, in situ hybridization; lincRNA, long intergenic noncoding RNA; lncRNAs, long noncoding RNAs; NA, not available; NPC, nasopharyngeal carcinoma; OS, overall survival; qRT-PCR, quantitative real time polymerase chain reaction; RFS, recurrence-free survival; SI, staining index.