Summary
The H3K27 lysine-specific demethylase UTX (or KDM6A) is targeted by loss-of-function mutations in multiple cancers. Here, we demonstrate that UTX suppresses myeloid leukemogenesis through noncatalytic functions, a property shared with its catalytically inactive Y-chromosome paralog, UTY (or KDM6C). In keeping with this, we demonstrate concomitant loss/mutation of UTX and UTY in multiple human cancers. Mechanistically, global genomic profiling showed only minor changes in H3K27Me3, but significant and bidirectional alterations in H3K27Ac and chromatin accessibility; a predominant loss of H3K4Me1 modifications; alterations in ETS and GATA-factor binding; and altered gene expression after Utx loss. By integrating proteomic and genomic analyses, we link these changes to UTX regulation of ATP-dependent chromatin remodeling, coordination of the COMPASS complex and enhanced pioneering activity of ETS factors during evolution to AML. Collectively, our findings reveal a dual role for UTX in suppressing acute myeloid leukaemia via repression of oncogenic ETS and upregulation of tumor-suppressive GATA programs.
Introduction
Enzymatic modifications of histones play a central role in the control of gene expression to orchestrate diverse biological processes1. The JmjC-domain-containing protein ubiquitously transcribed tetratricopeptide repeat, X-linked (UTX), which demethylates dimethylated as well as trimethylated histone H3 Lys27 (H3K27Me3)2, is a frequent target of somatic loss-of-function mutations in multiple cancer types3–8 including leukemia9–12. Reintroduction of intact UTX into mutation-bearing cancer cells leads to significant transcriptional changes and a decrease in proliferation7, in keeping with UTX’s role as a tumor suppressor. However the mechanism whereby UTX suppresses malignancy are poorly studied. Mechanistic insights into the tumor-suppressive function of UTX have come from studies of T-cell acute lymphoblastic leukaemia (T-ALL), in which an absence of UTX catalytic function is crucial for T-ALL initiation and maintenance9. Importantly, UTX mutations in T-ALL are almost exclusively found in males, reflecting the fact that the gene is X linked and escapes X inactivation13, such that females (but not males) with single-allele loss-of-function UTX mutations retain UTX expression14. Interestingly, in T-ALL mutations are concentrated in the catalytic JmjC domain, whereas this bias is not seen in other cancers where the mutations are spread throughout the gene15, thus raising the possibility of different tumor-suppressive mechanisms. Of potential relevance, UTY, the Y-chromosome homologue of UTX, has markedly lower demethylase activity owning to point substitutions affecting substrate binding16. In contrast, in common with UTX, UTY contains an intact tetratricopeptide-repeat region involved in protein-protein interactions and mediates demethylase-independent functions17. Tantalizingly, deletion of UTY is seen more frequently than expected in cancer cell lines with mutations in UTX, than in those without such mutations7, raising the possibility of a functional redundancy between the two paralogs.
Using myeloid malignancies as an exemplar, we investigate the role of Utx loss in oncogenesis and its interaction with Uty in mice following haematopoietic-specific loss of Utx. Our findings reveal that UTX prevents leukemogenesis through coordinate repression of pro-oncogenic E-twenty-six (ETS) and maintenance of tumor-suppressive GATA transcriptional programs. These functions are mediated by differential effects on genome-wide acetylated H3 Lys27 (H3K27Ac), monomethylated H3Lys4 (H3K4Me1) and chromatin accessibility, and their functional consequences were found to be rescued by both UTY and enzymatically dead UTX, confirming their independence from demethylase activity.
Results
Homozygous loss of Utx induces spontaneous leukaemia in a mouse model
To investigate the function of UTX in the hematopoietic system, we generated conditional-Utx-knock-out mice, in which exon 3 of Utx is flanked with loxP sites (Utxf/f)18 (Fig.1a). Utxf/f mice were crossed into the inducible Mx1-Cre line, thus enabling efficient Utx recombination in hematopoietic stem and progenitor cells (HSPC) after polyinosinic-polycytidylic acid (pIpC), which activates the Mx1 promoter. Utxf/f;Mx1-Cre mice treated with pIpC (hereafter denoted Utx-/- mice) showed loss of Utx mRNA and protein (Fig.1b,c), as compared with levels in pIpC-treated Utxf/f mice (hereafter denoted Utx+/+ or wild type). Female Utx-/-, Utx+/- and Utx+/+ mice were then allowed to aged and were monitored for leukemia development for up to 22 months. Utx-/- mice demonstrated significantly lower survival than did Utx+/- and Utx+/+ female mice (Fig.1d). At necropsy, Utx-/- mice had a significantly higher spleen weight (Fig.1e) and a predominance of myeloid cells in the spleen and bone marrow (BM) (Fig.1f). Blood leukocyte counts (WBC) showed a variable rise, and both platelet (PLT) and hemoglobin (HGB) levels were lower than those in Utx+/+ (Supplementary Fig. 1 a-c). Histological examination showed that most of the mice (63%) developed acute myeloid leukaemia (AML) (Fig.1g-h). In contrast, no Utx+/- or Utx+/+ mice developed AML. Splenocytes from leukaemic Utx-/- mice propagated the disease in secondary recipients (Fig.1i), thus verifying their full leukemogenic potential. Exome sequencing of seven Utx-/- AMLs showed no recurrently mutated genes, with the exception of Skint11 (two of seven samples), and only occasional copy number alterations (Supplementary Fig. 1 d-f). To more accurately mimic human disease we expressed the AML1-ETO fusion gene, which commonly co-occurs with UTX mutations10,19,20 in Utx-/- HSPCs, and observed significantly reduced survival of recipient mice (Supplementary Fig. 1 g-i).
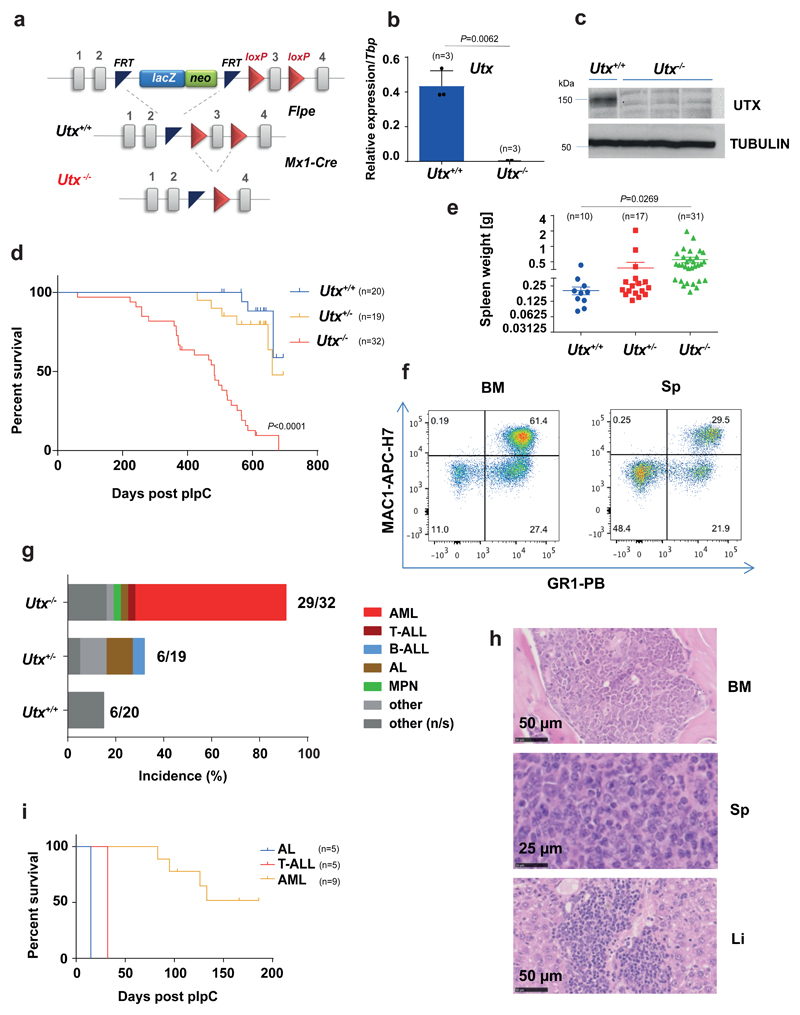
Figure 1. Utx-/- mice develop acute myeloid leukaemia.
(a) Structure of the Utx conditional allele. (b) qRT-PCR for exons 2-3 of Utx confirms Utx loss in Utx-/- HSPCs. The mean ± s.e.m is shown; n, number of mice per genotype; P by two-sided t-test, t=10.93, df=63. (c) Immunoblot showing loss of UTX protein in Utx-/- BM. Results of one representative experiment are shown (n=3 experiments). Uncropped images are shown in Supplementary Fig. 12. MW, molecular weight; α-tubulin, loading control (d) Kaplan-Meier survival curves for female Utx-/- (median 483 days), Utx+/- (median 661 days) and Utx+/+(median survival not reached) mice; n, number of mice per genotype; P by Log-rank (Mantel-Cox) test, df=2. (e) Spleen weights of Utx-/-, Utx+/- and Utx+/+mice; mean ± s.e.m. is shown; n, number of mice per genotype; P by one-way analysis of variance (ANOVA) with Bonferroni correction, t=2.554, df=55. (f) Characteristic MAC1/GR1 fluorescence activated cell sorting (FACS) analysis of BM and spleen cells from one diseased Utx-/- mice (similar result was observed in n=12). (g) Histopathological diagnoses of moribund mice of the indicated genotypes. The number of mice with a cancer diagnosis and the total analysed is indicated for each genotype. B-ALL: B-cell ALL; MPN: myeloproliferative neoplasm; other: non-hematological tumors; other (n/s), nonspecified. (h) Characteristic histology from one mouse with AML is shown (similar results were observed in n=20 mice). Sp, spleen; Li, liver. (i) Kaplan-Meier survival curves of mice transplanted with Utx-/- leukemias: AL (n=5), T-ALL (n=5), and two AML (n=9).
Deregulation of HSPC number, function and differentiation after Utx loss
Our findings suggested that Utx loss confers a pre-leukaemic state on HSPCs, with transformation reliant on additional mutations. To characterize this preleukemic phase, we analyzed mice early after Utx deletion (4-5 weeks post-pIpC). As reported previously21, Utx-/- mice demonstrated splenic enlargement (Fig.2a-b). We next examined the effect of UTX on hematopoietic differentiation and composition of the HSPC compartment, likely to harbor the initial target cell for transformation. Utx-/- mice demonstrated a significant expansion of HSPC progenitors (lineage negative, Lin-) (Fig.2c), a raised long-term and short-term hematopoietic stem cell (LT-HSC and ST-HSC, respectively) frequency (Fig.2d and Supplementary Fig. 2a), an increase in the granulocyte–monocyte progenitor (GMP) and common myeloid progenitor (CMP), and a decrease in the megakaryocyte-erythroid progenitor (MEP) compartments (Fig.2e,f and Supplementary Fig. 2b), as well as a significant decrease in common lymphoid progenitors (CLPs)(Fig.2g and Supplementary Fig. 2c). To assess HSPC function, we performed serial replating assays, observing enhanced self-renewal and proliferative potential of Utx-/- progenitors (Fig.2h and Supplementary Fig.3a). Regarding mature-cell numbers in BM, spleen and blood, only peripheral-blood thrombocytopenia was noted at 5 weeks after pIpC (Supplementary Fig.3b-d). However, at later time points (36 weeks post pIpC) in otherwise healthy animals, there was also an increase in peripheral-blood MAC1+ myeloid cells and total WBC, and a decrease in B-cells in Utx-/- compared with Utx+/+ mice (Supplementary Fig. 3e-g). Collectively, these results demonstrate that biallelic loss of Utx leads to dramatic and progressive alterations in the composition, function and differentiation of HSPCs and their progeny, including enhanced self-renewal, myeloid expansion and a block in lymphoid and erythroid/megakaryocytic differentiation (Fig.2i).
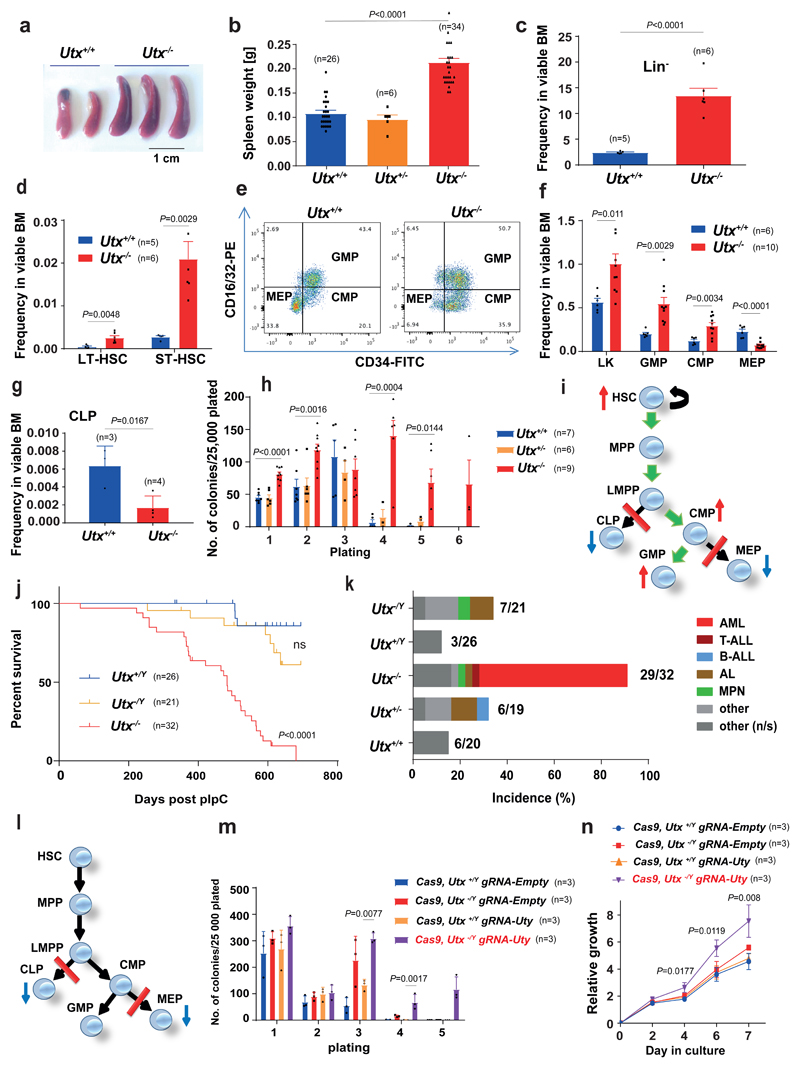
Figure 2. Utx loss expands hematopoietic stem/progenitor cells and imparts a myeloid bias, characteristics rescued by Uty.
a–d, Preleukemic spleens (a), spleen weights (t = 10.93, df = 63) (b) and frequency of Lin− cells (HSPCs) (t = 6.908, df = 9) (c), LT-HSCs (t = 3.712, df = 9) and ST-HSCs (d) in BM from Utx+/+ and Utx−/− mice (t = 4.049, df = 9). e, Representative flow cytometry profiles of CMP, GMP and MEP (similar results observed for 5 other mice). f, Quantification of Lin−Sca1−c-Kit+ (LK) (t = 2.927, df = 14), CMP (t = 3.518, df = 14), GMP (t = 3.608, df = 14) and MEP (t = 6.181, df = 15) from Utx+/+ and Utx−/− mice. g, CLP frequency in BM from Utx+/+ and Utx−/− mice (t = 3.534, df = 5). h, Serial replating of BM-derived colonies from Utx+/+, Utx+/− and Utx−/− (for Utx+/+ versus Utx−/− in plating: 1, t = 7.164; df = 19; 2, t = 3.991, df = 19; 3, t = 0.7332, df = 13; 4, t = 5.489, df = 11; 5, t = 3.292, df = 11) i, Schematic summary of progenitor differentiation in Utx−/−. Green arrows, preferential differentiation; red lines, differentiation block. MPP, multipotent progenitors; LMPP, lymphoid primed multipotent progenitors. j, Kaplan–Meier survival curves of Utx−/Y and Utx+/Y compared with Utx−/−; P = nonsignificant between Utx−/Y and Utx+/Y by log-rank (Mantel–Cox) test, df = 1. k, Histopathological diagnoses of moribund mice. Numbers of mice with a cancer diagnosis and total number analyzed are indicated for each genotype. l, Schematic summary of progenitor differentiation in Utx−/Y. m,n, Serial replating (m) and proliferation (n) of Cas9-expressing HSPCs after Uty editing. Cells were isolated from n = 3 mice per genotype; the mean ± s.e.m. is shown; P by one-way ANOVA with Bonferroni correction (P shown for Cas9, Utx−/Y gRNA-Uty versus Cas9, Utx+/Y gRNA-Uty, in m for plating: 3 (t = 4.313, df = 8), 4 (t = 5.522, df = 8); in n for culture days: 4 (t = 3.176, df = 8); 6, (t = 3.994, df = 8); 7, (t = 4.537, df = 7). In c, d, f and g, the mean ± s.e.m. is shown; n, number of mice; P by two-sided t test. In b and h, mean ± s.e.m. is shown; P by one-way ANOVA with Bonferroni correction.
UTY also suppresses leukemia induction and rescues the UTX-deficient preleukemic phenotypes
To define a possible role for UTY in suppressing leukemogenesis, we also monitored hemizygous (Utx-/Y, lacking Utx but expressing Uty) and control (Utx+/Y) male mice over the same time period. Remarkably, and in stark contrast to Utx-/- females, Utx-/Y males showed no difference in survival or hematological phenotype as compared with those of Utx+/Y mice (Fig.2j). In particular, we observed no differences in spleen and liver weights, WBC counts, or platelet and hemoglobin levels (Supplementary Fig. 3h-l). Moreover, no Utx-/Y mice developed AML, indicating that UTY also suppresses myeloid leukemogenesis (Fig.2k). Similarly, the presence of UTY in hemizygous males was sufficient to abrogate the abnormalities in preleukemic HSPCs, apart from decreases in the MEP and CLP compartments (Fig.2l and Supplementary Fig. 4a-i). Importantly, CRISPR-Cas9-mediated knockout of Uty in Utx-/Y mice increased HSPC self-renewal, thus recapitulating the phenotype of Utx-/- female mice (Fig.2m, n).
Because the only significant difference between UTY and UTX proteins is the lack of catalytic activity in the former, these findings suggested that catalytic activity is dispensable for their tumor suppressor functions. To further test this hypothesis, we identified an AML cell line, MONO-MAC6, with deletion of both UTX and UTY. Lentiviral expression of UTX, UTY or a catalytically-dead UTX mutant (UTX-MT2)22 in MONO-MAC6 confirmed this hypothesis: all constructs significantly suppressed proliferation in vitro (Fig.3a-c). In xenotransplantation assays, MONO-MAC6 expressing UTX, UTX-MT2 or UTY, compared to FLAG-expressing control cells, demonstrated slower growth and significantly longer recipient survival (Fig.3d-f). Together, these studies demonstrate that the tumor-suppressor functions of UTX do not require its catalytic activity and are shared with its catalytically inactive paralog UTY.
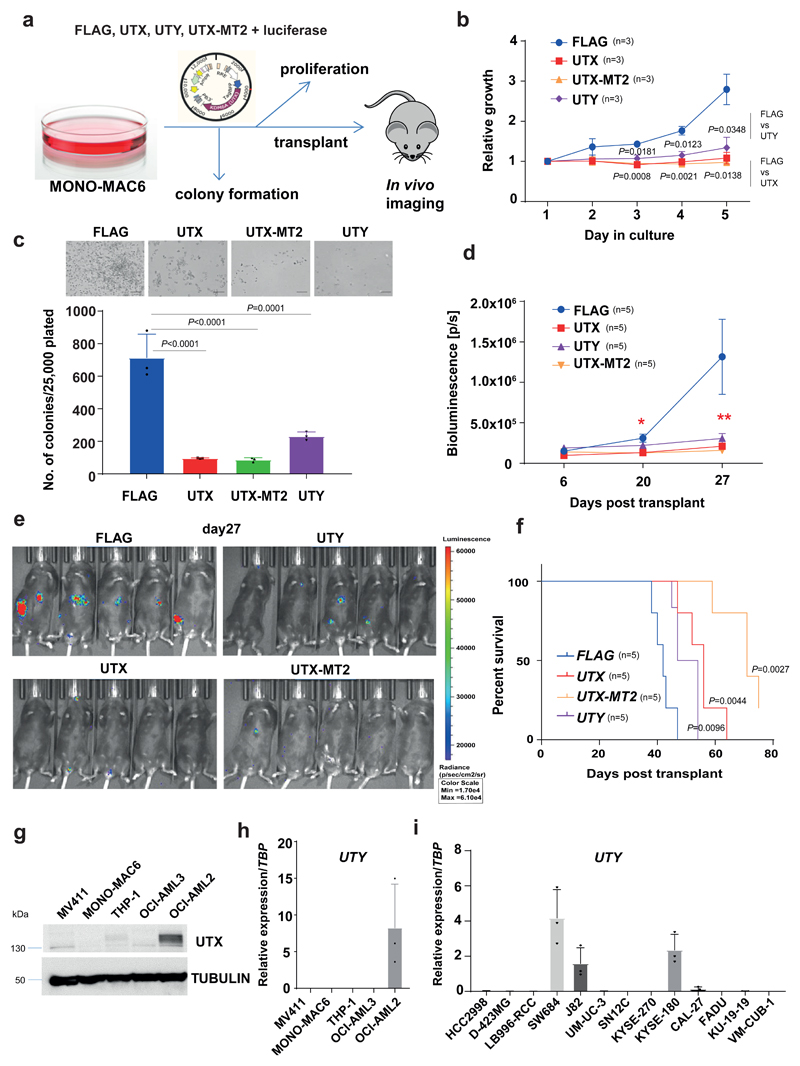
Figure 3. H3K27 demethylase activity is not required for tumor suppression by UTX.
a, Experimental approach for study of UTX-mutant MONO-MAC6 cells after expression of FLAG, UTX, UTY and UTX-MT2. b, UTX, UTY and UTX-MT2, as compared to FLAG, decreased MONO-MAC6 proliferation; mean ± s.e.m. is shown; n, number of independent cultures; P by one-way ANOVA with Bonferroni correction. Day 3 (compared to FLAG): UTX (t = 5,633; df = 8), UTY (t = 3,95; df = 8); day 4: UTX (t = 6.866; df = 8), UTY (t = 5.444; df = 8); day 5: UTX (t = 4.976; df = 8), UTY (t = 4.216; df = 8). c, Colony formation in semisolid medium. Top, similar result observed in n = 3 cultures; bottom, colony quantification. The mean ± s.e.m. is shown; n = 3 independent cultures; P by one-way ANOVA with Bonferroni correction; compared to FLAG: for UTX t = 10.19, df = 8; UTX-MT2 t = 10.36, df = 8; UTY t = 7.955; df = 8. d,e, In vivo growth after transplantation into immunocompromised mice. Cells were also transduced with luciferase-expressing vector, and mice were imaged on days 6, 20 and 27 after transplantation. d, Quantified bioluminescence, shown as mean ± s.e.m.; P by one-way ANOVA with Bonferroni correction: **P (FLAG versus UTX) = 0.0131, t = 3.317, df = 16; P (FLAG versus UTY) = 0.0242, t = 3.025, df = 16; P (FLAG versus UTX-MT2) = 0.0095, t = 3.469, df = 16; *P (FLAG versus UTX) = 0.0013, t = 4.408, df = 16; P (FLAG versus UTY) = 0.1284, t = 2.2, df = 16; for P (FLAG versus UTX-MT2) = 0.0010, t = 4.524; df = 16. e, Bioluminescence imaging of mice. f, Kaplan–Meier survival curves of transplanted mice; n, number of mice, P by log-rank (Mantel–Cox) test, reported versus FLAG, df = 1. g, Immunoblot analysis of UTX in AML lines; similar results were observed in n = 3 experiments. Uncropped images are shown in Supplementary Fig. 12. α-tubulin, loading control. h,i, qRT–PCR for UTY in AML (h) and in nonhematopoietic cancer cell lines with UTX mutations (i). In h and i, the mean ± s.e.m. is shown; n = 3 independent cell cultures.
Concomitant loss of both UTX and UTY tumor suppression occurs in multiple human cancer types
Our findings indicate that UTY can suppress myeloid leukemogenesis; however, unlike T-ALL14, UTX-mutated AML cases show no sex bias. We therefore analysed the status of UTY in human male AML cell lines carrying UTX mutations, identified through the COSMIC database (Fig.3g). We confirmed loss of UTY expression in all four studied male AML lines with UTX mutations/deletions (Fig.3h). Analysis of exome sequencing data from COSMIC showed that all lines contained a UTY microdeletion. Systematically extending our analysis, we identified an additional seven male hematopoietic cell line with UTX mutations, of which four lines had UTY microdeletions (Supplementary Table 1). Strikingly, in informative solid organ cancers, 20/25 (80%) UTX-mutant male cell lines also demonstrated UTY microdeletion/mutation (Supplementary Table 1), and we confirmed loss of UTY expression in a subset (10/13; Fig.3i).
Integrated genome-scale analysis identifies altered enhancer function as a mediator of leukemogenesis following Utx loss
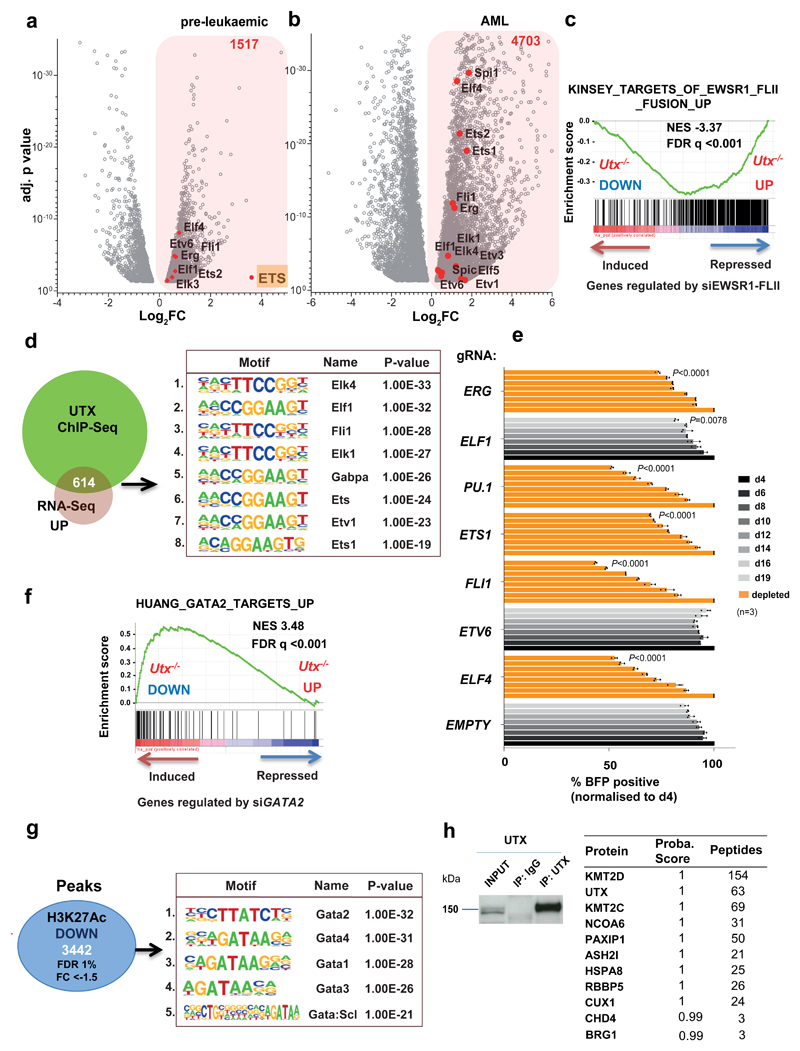
To determine the molecular basis of UTX-mediated leukemia suppression, we performed integrated genome-scale analyses, RNA-Seq, chromatin immunoprecipitation (ChIP)-Seq and assay for transposase-accessible chromatin using sequencing (ATAC-Seq) in HSPCs from age-matched pre-leukemic Utx-/-, Utx-/Y and Utx+/+ mice. As anticipated, the number of differentially expressed genes in comparison to controls was significantly greater for Utx-/- than Utx-/Y (4497 vs 673 genes, P < 0.05; Supplementary Table 2-4). Because loss of their single Utx allele did not lead to AML in hemizygous Utx-/Y males, we removed these 673 genes from subsequent analysis. Focusing on mRNAs differentially expressed with a log2 fold change (FC) above 0.5 or below -0.5, we identified 2686 genes (Fig. 4a-b). Interestingly, and somewhat counterintuitive to the perception of UTX as solely a transcriptional activator, similar numbers of genes were upregulated (1517, 57%) and downregulated (1169, 43%) after Utx loss. Importantly, although additional genes were also differentially expressed upon evolution to frank AML in Utx-/- mice, substantial components of the preleukemic transcriptional programs were retained (Fig. 4c and Supplementary Table 5). Using ChIP-Seq in wild-type mice, we documented 8304 UTX-binding sites, corresponding to 6734 genes (Supplementary Table 6), most of which were found at the promoter or within the gene body (Fig. 4d). Correlation with gene expression demonstrated that 581/1169 (50%) of downregulated and 614/1517 (40%) of upregulated gene loci were bound by UTX (Supplementary Table 7), thus suggesting that approximately half of deregulated genes are direct UTX targets (Fig. 4e) and supporting the view that UTX can be a transcriptional activator or repressor.
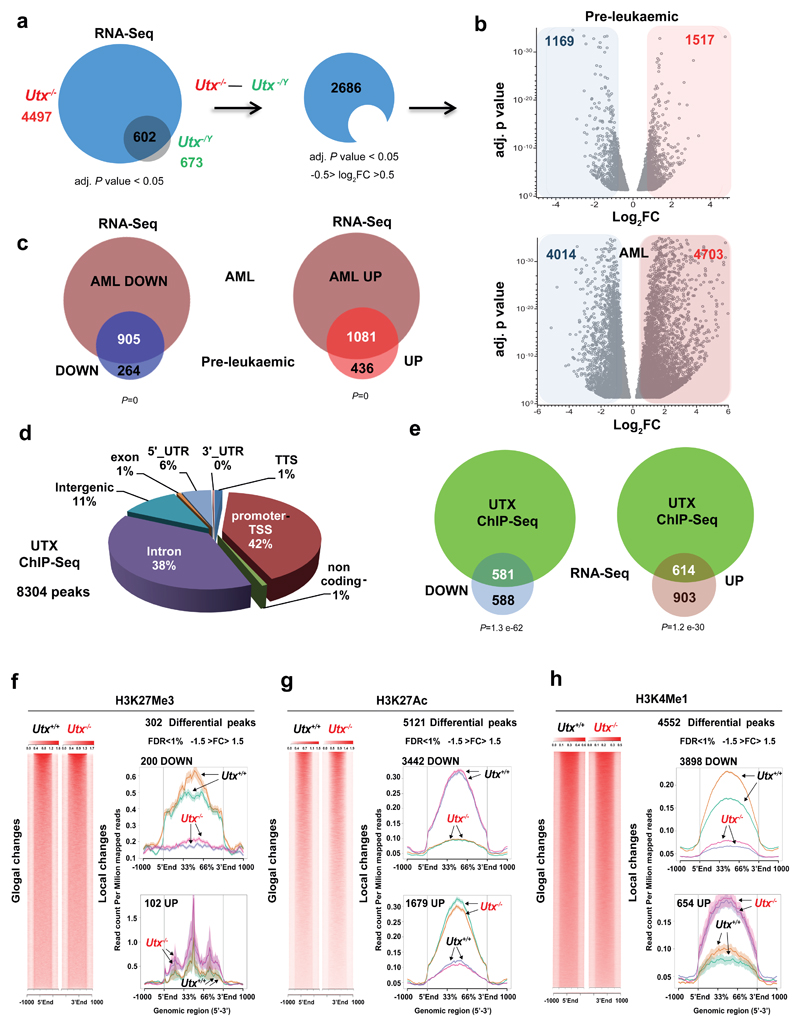
Figure 4. Utx loss drives both up and down regulation of gene expression primarily through effects on H3K27 acetylation.
a, Preleukemic gene expression changes in HSPCs from Utx−/− female (n = 2 mice) and Utx−/Y male (n = 2 mice) compared with sex-matched wild-type controls (n = 2 mice); genes with adjusted (adj.) P < 0.05 are shown. Subtraction of genes differentially expressed in males versus females defines a differential transcriptional program of interest; log2 fold change (FC) (–0.5 > log2 FC > 0.5). b, Volcano plot of log2 FC (–0.5 > log2 FC > 0.5) and adjusted P < 0.05 (only transcripts with P values between 0.05 and 1 × 10−38 are shown) for genes differentially expressed in the preleukemic and AML setting. In a and b, P values were generated with a negative binominal generalized linear model (DESeq2). c, Overlap between differentially expressed genes in Utx−/− preleukemic HSPCs (n = 2 mice) and AMLs (n = 3 mice), each compared to Utx+/+ HSPCs (n = 2 mice); P by hypergeometric test. Down, decreased genes or peaks; up, increased genes or peaks. d, Distribution of UTX ChIP–seq peaks in annotated regions of the genome. e, Highly significant enrichment of UTX-bound genes among those differentially expressed in preleukemic Utx−/− HSPCs; P by hypergeometric test. f–h, H3K27me3 (f), H3K27ac (g) and H3K4me1 (h) density plots (left) and average read counts (right) across all (global changes) or differentially modified regions (local changes). H3K27me3 signal density shows that only 302 genomic regions were differential modified, in contrast to similar plots revealing 5,120 differential modifications for H3K27ac and 4,552 differential modifications for H3K4me1 in Utx−/− versus Utx+/+ HSPCs. Arrows show each replicate (mouse) per genotype. False discovery rate (FDR) was calculated with the DiffBind tool; n = 2 mice per genotype. Plots are peak centered, scaled and ±1 kb for each locus. Shaded region in the line graphs in f–h indicate standard errors.
In keeping with our finding that H3 Lys27-demethylase activity is redundant for tumor suppression, only 302 differentially modified H3K27Me3 peaks were observed between Utx-/- and Utx+/+ HSPC. Further corroborating this notion, the majority (200/302, 67%) also showed decreased rather than increased modification (Fig. 4f and Supplementary Table 8). In marked contrast, the coordinated acetylation at the same lysine residue, H3 Lys27, was markedly altered in its distribution in Utx-/- compared with Utx+/+ mice. We observed 5121 regions with differential H3K27Ac in either direction (corresponding to 2916 gene loci), including 3442 peaks (2054 gene loci) significantly decreased and 1679 (953 gene loci) increased, after UTX loss (Fig.4g and Supplementary Tables 9 and 10). Comparing these putative enhancer regions to a recently published promoter-based capture-HiC dataset in the hematopoietic stem and progenitor cell line HPC7 (ref.23), we observed that 23% of upregulated and 32% of downregulated regions interacted with promoters (Supplementary Fig. 5a-c), thus suggesting substantial enhancer remodeling on UTX loss. The observed changes were locus specific, because global levels of H3K27Ac and H3K27Me3 were similar between Utx+/+ and Utx-/- BM (Supplementary Fig. 5d). To define direct co-occurrence on chromatin, we overlapped UTX peaks and differential H3K27Ac regions. However, we found only a modest co-occurrence, with only 282/5121 regions (6%) in common (Supplementary Fig. 5e). UTX-peaks and regions with differential H3K27Ac were then annotated to their associated/adjacent genes, defining larger genomic areas for comparison. In contrast to the limited peak-to-peak co-occurrence, when we compared whole gene loci showing differential H3K27Ac and UTX binding, we identified a highly significant overlap of 1396/2916 regions (48%, Supplementary Fig. 5f and Supplementary Table 11), thus suggesting that UTX indirectly regulates acetylation of regions adjacent to its binding. To further address the impact of UTX on enhancer function, we performed ChIP-Seq for the canonical enhancer-defining mark H3K4Me1 in preleukaemic Utx-/- and Utx+/+ HSPCs. Of particular interest, H3K4Me1 is deposited by KMT2C/D, components of the COMPASS complex that are known UTX interaction partners24. We observed 4552 differentially modified H3K4Me1 peaks (Fig. 4h and Supplementary Table 12); most of which (3898) were downregulated in Utx-/- mice. Differentially downregulated H3K4Me1 regions highly correlated with peaks that also lost H3K27Ac (1589 common peaks, 46% overlap). UTX did not bind directly at these sites, thus suggesting an indirect role of UTX and the COMPASS complex in early enhancer specification. Together, these data confirm that H3K27 demethylase activity is dispensable for tumor suppression, identify UTX as both a transcriptional activator and repressor and suggest that UTX/COMPASS-mediated indirect regulation of enhancers is important for transformation.
Loss of Utx activates an oncogenic ETS transcriptional program during leukemia development
As our genomic data demonstrated both loss and gain of activating chromatin marks and up- and downregulation of gene expression upon Utx loss, we speculated that both processes were necessary for leukemia development, but may be mediated by different mechanisms. We therefore analysed up and down regulated gene programs separately. Notably, several ETS transcription factors, including Elf4, Etv6, Erg, Fli1, Ets2, Spi1 and Elk3 were upregulated immediately after Utx loss in the preleukaemic phase (Fig.5a). Importantly, overexpression of these ETS factors persisted in Utx-/- AML (Fig.5b). Gene-set enrichment analysis (GSEA) also demonstrated a significant correlation between upregulated pre-leukaemic transcriptional programs and genes repressed after knockdown of the oncogenic ETS-factor fusion EWSR1-FLI1 (Fig.5c). In addition, motif analysis of 614 UTX-bound and upregulated genes demonstrated a significant enrichment in ETS binding motifs (Fig.5d and Supplementary Table 30). To directly link an ETS program to leukemogenesis, we mapped global chromatin occupancy of the ETS factor SPI1 (PU.1) in Utx-/- and Utx+/+ HSPCs. We observed 8329 enriched and 5869 depleted PU.1 peaks in Utx-/- (Supplementary Table 13). Many genes encoding ETS factors overexpressed in Utx-/- and directly bound by UTX in wild-type cells, including Ets2 and Fli1 showed higher PU.1 promoter occupancy in the absence of UTX (Supplementary Fig. 5g). These data suggest enhanced ETS factor binding and transcriptional auto-regulation of specific ETS genes in the absence of Utx. On a global scale however, UTX binding did not colocalize with PU.1 chromatin occupancy (only 115 peaks/14198, 0.8%), thus suggesting that the profound general redistribution of PU.1 binding may relate more to its overexpression. ETS factors are known to recruit histone acetyltransferases25, and consequently increase H3K27Ac deposition. Therefore, we next asked whether the changes in differential H3K27Ac and H3K4Me1 observed in Utx-/- might correlate with UTX-dependent redistribution of PU.1 binding. Indeed, we observed that 51% of gained H3K4Me1 (335/654) and 30% of gained H3K27Ac (470/1679) peaks also demonstrated increased PU.1 binding (Supplementary Fig. 6a, b). These direct binding data provide a mechanistic explanation for the gain of enhancer marks after Utx loss.
Figure 5. Utx loss activates an oncogenic ETS transcriptional program while suppressing a GATA program.
a,b, Volcano plots of differentially expressed genes in preleukemic (n = 2 mice) (a) and AML Utx−/− (n = 3 mice) (b) compared with wild-type controls (n = 2 mice), revealing overexpression of multiple ETS factors (red dots). FC (–0.5 < log2 FC > 0.5) and adjusted P < 0.05 (only transcripts with P values between 0.05 and 1 × 10−38 are shown in the graph); P values were generated in DESeq2. c, GSEA plot, showing significant overlap with a known ETS oncogenic program driven by the EWSR1–FLI1 fusion. The ‘si’ prefix denotes short interfering RNA. The Kinsey data are from the GSEA database (URLs). d, Motif analysis of UTX ChIP–seq peaks that overlap with overexpressed genes; number indicates motif rank. e, MONO-MAC6 proliferation after editing of the indicated gene. The BFP-positive fraction was compared with the nontransduced population and normalized to day 4 for each gRNA. The mean ± s.d. is shown; n, number of independent cell cultures; P by one-way ANOVA with Bonferroni correction; P shown for day 19 compared with control gRNA (empty) for: ELF4 (t = 32.32), ETV6 (t = 10.03), FLI1 (t = 41.1), ETS1 (t = 15.85), SPI1 (t = 33.56), ELF1 (t = 3.967) and ERG (t = 12.35); df = 16. f, GSEA plot showing enrichment of genes differentially expressed in Utx−/− HSPCs with a published dataset of GATA2 targets. g, Motif analysis of 3,442 downregulated H3K27ac peaks (FDR < 1%, FC < –1.5) identified in Utx−/− HSPCs; number indicates motif rank. The Huang data are from the GSEA database (URLs). h, Selected proteins identified by mass spectrometry after immunoprecipitation of endogenous UTX from mouse myeloid cells (416B) (n = 2 independent cell cultures). Motif and statistical analysis in d and g were determined in HOMER software (Methods and Supplementary Table 30).
To investigate the functional importance of deregulated ETS factors, we used CRISPR-Cas9 genome editing to ablate several of these factors in Cas9-expressing, UTX/UTY mutant, MONO-MAC6 cells. Importantly, we observed substantial growth suppression after editing of the same ETS factors overexpressed immediately after Utx loss: FLI1, ERG, SPI1 (PU.1), ETS1, and ELF4 (Fig.5e). These data indicate that ETS factors drive leukemia induction and maintenance in the context of UTX/UTY loss.
Utx loss affects BRG1 (SMARCA4)-dependent chromatin remodeling, thereby repressing a tumor-suppressive GATA program during leukemia development
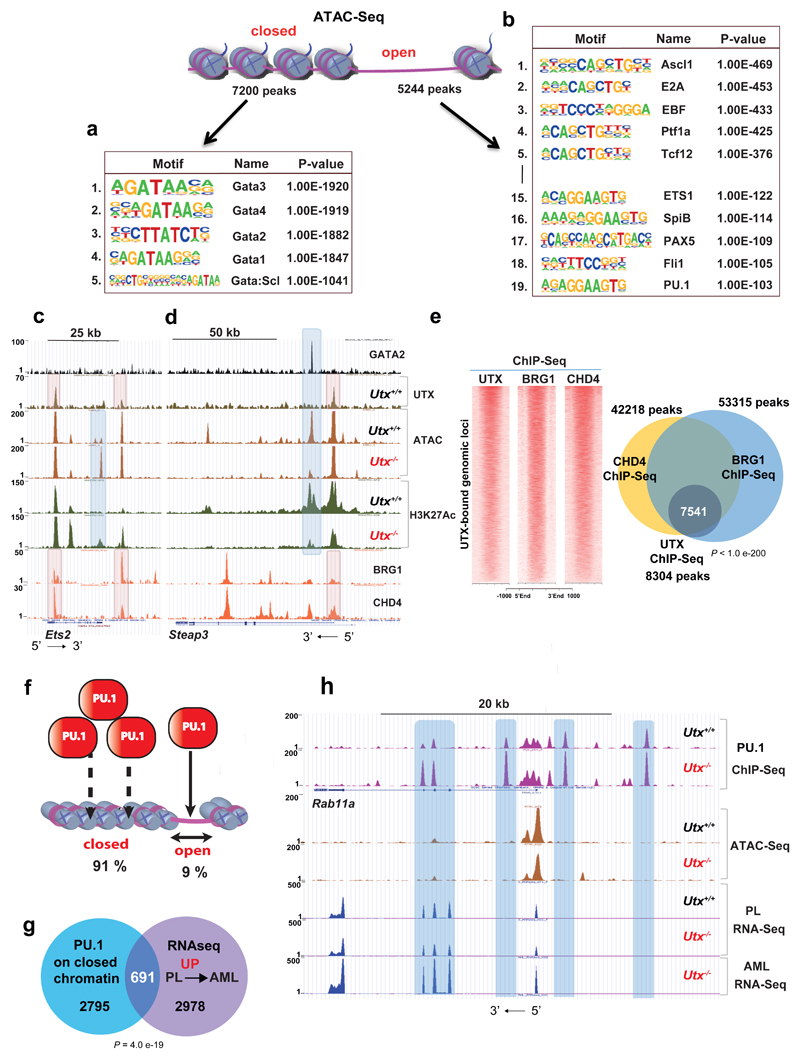
GSEA of the downregulated gene-expression program demonstrated enrichment in GATA2 targets (Fig.5f). Furthermore, motif analysis at the 3442 regions with significantly decreased H3K27Ac after Utx loss demonstrated a striking and almost exclusive enrichment in GATA-binding motifs (Fig.5g and Supplementary Table 30). Similarly to the ETS motifs, loss of H3K27Ac did not directly overlap with UTX binding (Supplementary Fig. 6c,d) but occurred nearby and probably affected the same genes (Supplementary Fig. 6e and Supplementary Table 14). To better understand the indirect effects3 of UTX on acetylation changes at GATA and ETS sites, we performed pulldown of endogenous UTX in the mouse myeloid cell line 416B and subsequent mass spectrometric analysis. We did not observe UTX interaction with ETS factors or GATA2, but we did identify multiple known UTX interactors, including KMT2C/D and other COMPASS members (Fig.5h and Supplementary Tables 15 and 16). Interestingly, our proteomic analysis also demonstrated lower-level interactions between UTX and the ATP-dependent chromatin-remodeling-complex members SMARCA4 (BRG1) and CHD4, which we further verified (Supplementary Fig.6f). Speculating that changes in acetylation and GATA binding occurred through alterations in chromatin accessibility, we performed ATAC-Seq analysis in preleukaemic Utx-/- HSPCs and Utx+/+ controls. Strikingly, we observed significant and bidirectional changes in chromatin accessibility, including 7200 sites where chromatin accessibility decreased and 5244 sites that became accessible upon Utx loss (Supplementary Tables 17 and 18). Loss of chromatin accessibility correlated strongly with decreased H3K27Ac (2274/3442 peaks, 73%) and H3K4Me1 deposition (2264/3898, 58%) (Supplementary Fig. 7a,d) and decreased gene expression (Supplementary Fig.7b and Supplementary Table 19). Furthermore, analysis of closed chromatin sites showed a striking enrichment in GATA motifs (Fig.6a and Supplementary Fig.7a,b). Comparisons of GATA2 peaks in HPC-7 (ref.26) with differential chromatin accessibility in Utx-/- HSPC demonstrated direct correlation only with closed chromatin regions (15%, 409/2796 peaks), because only 0.7% (19/2796 peaks) overlapped with open chromatin. As before, only limited overlap was found between closed chromatin and UTX binding (Supplementary Fig.7c). We next asked if UTX binds with BRG1 and CHD4 on chromatin. Importantly, we observed a highly significant overlap, 91% (7541/8304 peaks) of UTX sites with binding of both BRG1 and CHD4 from published ChIP-Seq27 (Fig.6c-e). To validate co-occupancy we employed CRISPR-Cas9 genome editing of Utx in 416B cells targeting Utx exon-3, recapitulating our mouse model. We observed significantly lower chromatin binding for BRG1 and CHD4 at exemplar loci (Aff1 and Lrrc8c) in the absence of UTX, in keeping with a role for UTX in the recruitment of complexes containing these proteins (Supplementary Fig. 7e-g).
Figure 6. UTX interacts with chromatin modifiers to maintain chromatin accessibility.
a,b, Motif analysis of ATAC–seq closed (a) and open (b) peaks, revealing dramatic enrichment in GATA motifs in the former and ETS, among other motifs, in the latter. Number indicates motif rank. Motif and statistical analysis was performed in HOMER software (Supplementary Table 30). c,d, Genomic snapshot of GATA2, UTX, ATAC–seq and H3K27ac ChIP–seq in Utx+/+ and Utx−/− HSPCs at the Ets2 (c) and Steap3 loci (d). Colocalization of GATA2 binding with dynamically closed chromatin and loss of H3K27ac after UTX loss were found without evidence of GATA2-UTX cobinding. In contrast, binding of the chromatin remodelers SMARCA4 and CHD4 directly colocalized with UTX binding (lower two tracks). At the Ets2 locus, newly accessible chromatin was also seen after UTX loss, again at regions not directly bound by UTX or chromatin remodelers. e, Density plots of UTX, SMARCA4 and CHD4 ChIP–seq on UTX-bound genomic loci; Venn diagram shows overlap among all UTX, SMARCA4 and CHD4 ChIP–seq peaks; P by Fisher's exact test for ChIP–seq: UTX versus SMARCA4/CHD4. f, Schematic representation of PU.1 occupancy occurring mostly on closed chromatin. g, Overlap of genes associated with enhanced PU.1 binding (in Utx−/−) on closed chromatin with gene expression changes from preleukemia (PL) to AML. For PU.1 ChIP–seq, n = 3 mice; ATAC–seq, n = 3 mice; PL RNA-seq, n = 2 mice; AML RNA-seq, n = 3 mice. P by hypergeometric test. h, Genomic snapshot demonstrating an enhanced PU.1 occupancy in Utx−/− HSPCs that occurs on closed chromatin at the Rab11a locus and a correlation with PL and AML RNA-seq. Rab11a expression increased only after progression to AML.
To further address molecular mechanism of UTX-dependent chromatin remodeling, we analysed the activity of BRG1 and CHD4 in Utx-/- and Utx+/+ HSPCs. We crossed Utxf/f;Mx1-Cre mice with Cas9-expressing mice28 and induced Utx deletion. Five weeks post gene deletion we isolated HSPCs and used CRISPR-Cas9 genome editing to target Smarca4 and Chd4, using an empty guide RNA (gRNA) construct as control. We then analyzed chromatin accessibility through ATAC-Seq, hypothesizing that loss of Smarca4 and/or Chd4 would at least partially mimic the effect of UTX loss. We found 1150 sites with significantly decreased accessibility after Smarca4 editing and only 16 sites with increased accessibility, thus suggesting that SMARCA4 is mainly involved in the opening or maintenance of specific open chromatin loci (Supplementary Table 20). We then overlapped these sites with regions that were differentially lost between Utx+/+ versus Utx-/- cells, using the same culture conditions (2871 peaks; Supplementary Table 21). We observed that 21% (244/1150) of regions closed in Smarca4-edited cells overlapped with sites that closed after Utx deletion (Supplementary Fig. 8a-b). Of note, there was no further alteration of accessibility at these 244 sites when Smarca4 was edited in Utx-/- cells (Supplementary Table 22). This suggests a degree of functional redundancy between UTX and SMARCA4 loss for chromatin accessibility. Performing motif analysis of these 244 sites, we again observed high enrichment for GATA motifs (Supplementary Fig. 8a-b). Similar analysis for CHD4 demonstrated no significant overlap upon Utx loss (Supplementary Fig.8c and Supplementary Table 23). These findings, along with our proteomic data, suggest that UTX interacts with SMARCA4 and subsequently maintains chromatin accessibility at GATA-bound regions.
Utx loss allows chromatin accessibility to other transcription factors and facilitates the pioneering function of ETS factors during AML evolution
For sites newly accessible by ATAC-Seq following Utx loss, the converse was seen, with a correlation between these sites and increased H3K27Ac (766/1679 peaks, 45%; Supplementary Fig.9a), H3K4Me1 (438/654 peaks, 67%; Supplementary Fig.9b) and gene expression close to the accessible sites (389/1517, 25%; Supplementary Fig.9c and Supplementary Table 24). Furthermore, motif analysis of newly accessible sites showed enrichment for a number of transcription factors including ASCL1, E2A, EBF, PTF1a, TCF12, in addition to ETS factors (Fig.6b). Three transcription factors represented in the top five motifs (ASCL1, EBF, PTF1A) were not expressed in MONO-MAC6. Assessing the functional relevance of the two remaining expressed TFs, TCF3 (E2A) and TCF12 (HEB), we utilized CRISPR-Cas9 editing in MONO-MAC6 and observed a significant decrease in cell growth upon knockout of either TCF3 or TCF12 (Supplementary Fig.9d) These data suggest that the transcriptional activity of TCF3 and TCF12 maintain AML growth in the absence of UTX. In assessing ETS sites, PU.1-binding sites gained in the absence of UTX only minimally overlapped with open chromatin at the preleukemic stage (758/8329 peaks, 9%). However, of significant interest, although the remaining 91% (7592 peaks, linked to 3450 genes) occurred in ATAC inaccessible chromatin in the preleukemic stage and had no effect on gene expression after Utx loss, we documented that the expression of 691 of these linked genes (20%) was upregulated in the later transition to AML (Fig.6f-h and Supplementary Table 25 and 26). These data suggest that pioneering function of redistributed ETS TFs “primes” a later leukemogenic transcriptional program for upregulation during AML evolution.
Discussion
The mechanism of tumor suppression by UTX has been previously been thought to be dependent on its demethylase catalytic function, a notion supported by data from T-ALL9,14. However, during the evolution of AML, we demonstrated that the demethylase function of UTX is redundant for tumor suppression. Noncatalytic functions of UTX have previously been demonstrated in embryonic development22,29,30 and in mammary luminal lineage development31. UTX catalytic activity has been reported to upregulate expression of the master transcription factor PLZF and to modulate superenhancer accessibility in invariant natural killer T cells32. Additionally, UTX function was recently linked to enhancer activity and gene activation via coordination of COMPASS mediated H3K4monomethylation and CREBBP/p300-mediated H3K27Ac deposition33, as also demonstrate here. However, no role for these functions in tumor suppression has been demonstrated until now. Furthermore, our study confirms and defines the molecular basis for the frequent co-occurrence of UTX and UTY mutation/loss6,7, identifying UTY as a tumor suppressor in its own right and highlighting non-catalytic functions of UTX/UTY as the dominant mediators of tumor suppression.
Contrary to the prevailing perception that UTX is solely a transcriptional activator17, our study identifies that it also functions as a repressor. We demonstrate that its loss leads to significant alterations in chromatin accessibility, bidirectional alterations in H3K27Ac, a predominant loss of H3K4Me1 and coordinated changes in gene expression that impart proleukemic properties on HSPCs and that are maintained during evolution to AML (Supplementary Fig.11). In particular, UTX loss upregulates a transcriptional program driven by the ETS family of pioneer transcription factors34–36. ETS factors are known to be oncogenic37,38. Overexpression of a single ETS factor, ERG, able to generate AML in mice39, and ERG expression levels are one of the strongest prognostic factors in human AML40. Novel binding events of the exemplar ETS factor PU.1, lead to an increase in chromatin accessibility, activation of enhancer modifications and, via its pioneering activity, gene activation occurring at later time points during leukemia evolution. Furthermore, loss of UTX also downregulates a program of GATA-driven genes through loss of chromatin accessibility and local H3K27 acetylation. GATA factors are also critical regulators of hematopoiesis and leukemia41, and germline and somatic loss-of-function mutations of GATA2 described in AML42,43. Together, our data demonstrate that UTX loss coordinates a “double-hit” mechanism reminiscent of genomic inversion that removes a critical enhancer and consequently downregulates GATA2 expression and relocates it, thereby driving oncogenic expression of the transcription factor EVI1 (ref.44).
Our proteomic data also suggest that noncatalytic tumor suppressive functions operate through protein-protein interactions with tetratricopeptide repeats, the other major protein domain of UTX/UTY17. We demonstrated protein interactions between endogenous UTX and the KMT2C/D-containing COMPASS complex and the ATP-dependent chromatin remodeling factors SMARCA4 and CHD4 in myeloid cells. The interaction between UTX and SMARCA4 was previously demonstrated in T lymphocytes45 and during cardiac development46. Loss of Utx led to a marked decrease in the deposition of the canonical early enhancer mark H3K4Me1. Notably, mutations of KMT2D are responsible for most (60-80%) cases of Kabuki syndrome, with UTX mutations causing ~ 10% of cases, thus highlighting the mechanistic links between the two proteins. Furthermore, we could demonstrate co-occupancy of UTX, SMARCA4 and CHD4 (ref.27) at specific genomic loci associated with alterations in chromatin accessibility after UTX loss (Fig. 6c-e). Furthermore, we functionally demonstrated that SMARCA4 loss at least in part recapitulated the phenotype of loss of chromatin accessibility after Utx deletion. Recent studies have demonstrated that H3K4Me1 is required for binding of the SMARCA4-containing BAF complex to chromatin and enhances BAF chromatin-remodeling activity47. Our own data corroborate and extend this model, further defining a role for UTX in linking H3K4Me1 deposition with chromatin remodeling via SMARCA4. Altogether, these mechanistic data demonstrate that loss of UTX leads to an upregulated activity of ETS transcription factors with both immediate and later pioneering effects, thus facilitating chromatin accessibility, as well as loss of coordination of COMPASS-mediated H3K4Me1 enhancer specification and SMARCA4-mediated chromatin accessibility. Together, these responses lead to alter patterns of gene expression that induce and maintain leukemia (Supplementary Fig. 11).
Our findings identify UTX as a complex transcriptional regulator capable of activating or repressing transcription, through effects on pioneering transcription factors, enhancer function and chromatin accessibility. These results have obvious implications regarding the role of UTX in tumor suppression and other critical cellular processes. Finally, our framing of UTY as a bona fide tumor-suppressor gene establishes a pathogenic role of Y-chromosome-specific genes in carcinogenesis, and sheds new light on the role of Y-chromosome loss in diverse cancer types and on the importnace of age-related clonal hematopoiesis associated with “loss of Y” in otherwise healthy men48–50.
Methods
Mice
The in vivo experiments were performed under project license PPL 80/2564 issued by the United Kingdom Home Office, in accordance with the Animal Scientific Procedures Act 1986. The Utx mouse model, C57Bl6, was developed at the Sanger Institute. Utxf/f mice were crossed with Flpe mice and then with Mx1-Cre mice. Cre expression was induced by intraperitoneal injection of 5- to 6-week-old mice with pIpC (Sigma P1530, 400 µg/mouse; five doses over a period of 10 d). All preleukemic experiments were performed 4–6 weeks after pIpC injection. Cas9-expressing mice were as reported previously28.
Cell lines
293FT (Invitrogen) were cultured in DMEM (Invitrogen) supplemented with 10% FBS. 416B cells were cultured in RPMI1640 (Invitrogen), 10% FBS. SN12C, KU-19-19, KYSE-180 and HCC2998 were cultured in RPMI (Invitrogen) supplemented with 10% FBS (Invitrogen); J82, UM-UC-3 and FADU were cultured in EMEM (Invitrogen), 10% FBS; CAL-27 and VM-CUB-1 were cultured in DMEM (Invitrogen), 10% FBS. SW684 was cultured in L15, 10% FBS. LB996-RCC was cultured in IMDM (Invitrogen), 10% FBS (Invitrogen). D-423MG was cultured in Gibco Zinc Option (Invitrogen), 10% FBS; KYSE-270 in RPMI and Ham’s F12, 2% FBS. Each medium was supplemented with 1% penicillin–streptomycin–glutamine (PSG, Invitrogen). AML cell lines: MV4-11, MONO-MAC6 and THP1 were cultured in RPMI1640 (Invitrogen) supplemented with 10% FBS and 1% PSG. OCI-AML2 and OCI-AML3 were cultured in alpha-MEM (Lonza), 20% FBS and 1% PSG. All cancer cell lines were obtained from the Sanger Institute Cancer Cell Collection.
cDNA synthesis, PCR and qRT-PCR
cDNA synthesis was performed with qScript cDNA SuperMix (Quanta Biosciences) according to the manufacturer’s instructions. PCR was performed with REDTaq ReadyMix PCR Reaction Mix (Sigma) according to the manufacturer's instructions. qRT–PCR was performed with TaqMan Fast Universal PCR Master Mix (Thermo Fisher Scientific) and a Universal Probe Library system (Roche). The TBP housekeeping gene was used for data normalization. qRT–PCR primer sequences are presented in Supplementary Table 27.
Protein extraction, immunoblotting and coimmunoprecipitation
The cells were lysed in whole-cell lysis buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl, 0.1% NP-40 and 1 mM EDTA) supplemented with 1 mM DTT, protease inhibitors (Sigma) and phosphatase inhibitors (Sigma). Protein concentrations were assessed with Bradford assays (Bio-Rad), and an equal amount of protein was loaded per track. Before loading, the samples were supplemented with SDS–PAGE sample buffer, and DTT was added to each sample. 10–40 µg of protein was separated on a 10% SDS–PAGE gel and blotted onto polyvinylidene difluoride membranes (Millipore). The following primary antibodies were used: anti-UTX (Bethyl, A302-374A), anti-UTX (GeneTex, GTX121246) and anti-IgG (Santa Cruz Biotechnology, sc-2027); anti-α-tubulin (Sigma, T6074) or anti-ACTIN (Santa Cruz Biotechnology, sc-1616) was used as a loading control. The secondary antibodies used in the study were as follows: HRP-linked donkey anti-rabbit (GE Healthcare UK, NA934) and ECL HRP-linked anti-mouse (Santa Cruz Biotechnology, sc-2005). Visualization was performed with LumiGLO Chemiluminescent Substrate (KPL, 54-61-00). Coimmunoprecipitation was performed on the extracts isolated with cell lysis buffer (described above). 2–6 μg of antibody was bound to 20 µl of Dynabeads Protein G (Thermo Fisher Scientific) and incubated with 500–1,000 µg of freshly extracted protein for 1.5 h at 4 °C with rotation. Immunoprecipitates were washed four times with IP wash buffer (10 mM Tris-HCl, pH 8, 150 mM NaCl, 0.1% NP-40 and 1 mM EDTA) supplemented with protease inhibitor (Sigma). Immunoprecipitation samples with beads were then resuspended in 1× NuPAGE LDS Sample Buffer (Thermo Fisher Scientific) supplemented with NuPAGE Sample Reducing Agent (Thermo Fisher Scientific). The following antibodieswere used for immunoprecipitation; anti-CHD4 (Abcam, ab72418), anti-BRG1 (Santa Cruz Biotechnology, sc-10768) anti-UTX (Bethyl, A302-374A) and IgG (Santa Cruz Biotechnology, sc-2027).
Histological analysis of mouse tissue
The tissues were fixed in 10% formaldehyde and subsequently paraffin embedded. Bones were decalcified using 0.38 M EDTA pH=7. Tissue sections (4 µm) were stained with Hematoxylin and Eosin (Thermo Fisher Scientific). Histology assessment was performed using the Bethesda criteria for mouse hematological tumors51,52.
Blood-count analysis
Blood count measurement was performed on a VetabC analyzer (Horiba ABX).
Isolation of mouse hematopoietic progenitors
Freshly isolated bone marrow was suspended in erythrocyte lysis solution (BD PharmLyse, BD Bioscience), followed by magnetic bead selection of Lin- cells, using the Lineage Cell Depletion Kit (Miltenyi Biotec, cat. no. 130-090-858) according to the manufacturer’s instructions. c-KIT+ progenitors were selected with mouse CD117 MicroBeads, (Miltenyi Biotec, #130-091-224) according to the manufacturer’s instructions.
Culture of mouse hematopoietic progenitors
Primary mouse cells were cultured in X-VIVO 20 media (Lonza) supplemented with 5% serum (Stem Cell Technologies), 10ng/ml IL3 (Peprotech), 10ng/ml IL6 (Peprotech) 50ng ml/ml of SCF (Peprotech) and 1% penicillin-streptomycin-glutamine (Gibco).
Flow cytometry analysis
BM cells were incubated in erythrocyte lysis buffer (0.85% NH4Cl; Sigma) and blocked with anti-mouse CD16/32 (BD Pharmigen, 553142) and 10% mouse serum (Sigma M5905) for LSK and CLP staining or 10% mouse serum alone for LK staining. LSK, MPP, LMPP and LT/ST-HSC flow cytometry staining was performed with antibodies against CD4 (BioLegend, 100514), CD5 (BioLegend, 100610), CD8a (BioLegend, 100710), CD11b (BioLegend, 101210), B220 (BioLegend, 103210), TER-119 (BioLegend, 116210), GR-1 (BioLegend, 108410), SCA-1 (BioLegend, 122520), CD117 (eBioscience, 47-1171), CD48 (BioLegend, 103411), CD150 (BioLegend, 115913), CD34 (BD Pharmigen, 553733) and FLT3 (eBioscience, 12-1351). GMP, MEP and CMP staining was performed with the following biotin-conjugated lineage markers: MAC1, GR1, CD3, B220, TER119 (BD Pharmigen, 559971), IL7Ra (BioLegend, 121103) and streptavidin (BioLegend, 405206) alongside CD34 (BD Pharmigen, 553733), CD16/32 (BD Pharmigen, 553145), c-KIT (BioLegend, 105812) and SCA-1 (BioLegend, 122520). For the detection of the CLP population, cells were stained for Flt3 (eBioscience, 12-1351), IL7Ra (BioLegend, 135008) and the lineage biotin-conjugated markers MAC1, GR1, CD3, B220 and TER119 (BD Pharmigen, 559971), as well as NK (LSBio, LS-C62548) c-KIT (BioLegend, 105812) and SCA-1 (BioLegend, 122520). Differentiated BM, spleen and peripheral blood cells were stained with CD45 (BD Pharmigen, 563891), CD11b (BD Pharmigen, 557657), B220 (BioLegend, 103210), GR1 (BD Pharmigen, 560603), c-KIT (BioLegend, 105812), TER119 (BD Pharmigen, 557915) and CD3e (eBioscience, 12-0031-82). LT-HSCs were defined as Lin–c-KIT+SCA1+FLT3–CD48–CD150+CD34–. ST-HSCs were defined as Lin–c-KIT+SCA1+FLT3–CD48–CD150+CD34+. MPPs were defined as Lin–c-KIT+SCA1+FLT3+. LMPPs were defined as Lin–c-KIT+SCA1+FLT3hi. CLPs were defined as Lin–FLT3hiIL7Ra+c-KITloSCA-1lo. GMPs were defined as Lin–IL7Ra–c-KIT+SCA1–CD34+CD16/32+. CMPs were defined as Lin–IL7Ra–c-KIT+SCA1–CD34+CD16/32–. MEPs were defined as Lin–IL7Ra–c-KIT+SCA1–CD34–CD16/32–. Flow cytometry analysis was performed with an LSRFortessa instrument (BD), and data were analyzed in FlowJo software.
Serial replating assays
For re-plating assays 50,000 bone marrow cells were plated in two wells of 6-well-plate of M3434 methylcellulose (Stem Cell Technologies). The colonies were counted 7 days later and further 30,000 cells re-seeded and re-counted after a week until no colonies were observed.
Proliferation assays
104 cells /well were plated onto 96-well plates and assayed daily for growth using CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay (Promega) according to manufactures’ instructions.
Plasmids and cloning
Plasmids for expression of FLAG-tagged versions of UTX, UTY, UTX-MT2 were purchased from Addgene (pCS2-UTX-F 24168, pCS2-UTY-F 17439 and pCS2-UTX-F-MT2 40619). The lentiviral UTX, UTY, UTX-MT2 and FLAG expression vectors were constructed in pKLV-puro as follows. First, the lentiviral backbone vector, pKLV-U6(Flip)gRNA(BbsI)-PGKpuro2ABFP53 was digested with BbsI and KpnI to remove the U6gRNA(BbsI)-PGKpuro2ABFP cassette. Gibson cloning (NEB) was performed to clone the PCR products for the EF1α promoter, UTX/UTY/UTX-MT2/FLAG cDNA and a puromycin (Puro)-resistance gene with the primers in Supplementary Table 28. The EF1α promoter was PCR-amplified from a pLVX-EF1α-IRES-ZsGreen1 construct (Clontech, 631982). The Puro-resistance gene was amplified from the pKLV-U6(Flip)gRNA(BbsI)-PGKpuro2ABFP construct. Gibson cloning was performed according to the manufacturer's specifications. Firefly luciferase–expressing plasmid (EF1α-GFP-T2A-Luciferase) was obtained from System Biosciences (BLIV503-MC-1-SBI). AML–ETO9a plasmid was as previously described54. For CRISPR–Cas9 experiments, gRNAs were cloned into a BbsI-digested pKLV2-U6gRNA(BbsI)PGKpuro2ABFP backbone28. Sequences of gRNAs used in the study are provided in Supplementary Table 29.
Lentiviral-vector production and transduction
Lentiviruses were produced in HEK293 cells using ViraPower Lentiviral Expression System (Invitrogen) according to manufacturer's instructions. Viral supernatant was concentrated by centrifugation at 6000g, 16h, at 4 °C. The cells were transduced by spinoculation (60 min, 800 g, 32 °C) in culture medium supplemented with 4µg/ml of polybrene (Millipore) and further incubated overnight at 37 °C. The following day, the transduced cells underwent selection on puromycin, (1.5ug/ml, Sigma) for 3 d.
Transplantation and in vivo imaging and quantification
MONO-MAC6 cells were transduced with lentiviral vectors for expression of UTX, UTY, UTX-MT2 or FLAG control, and puromycin selected for 4 d. Then 0.8 × 106 cells were transplanted via intravenous tail injection into immunocompromised recipient mice (Il2rg−/−; Rag2−/−). Five mice were injected per group. For bioluminescence examination, mice were injected with D-luciferin (BioVision; 3 mg/20 g intraperitoneally) and subsequently anesthetized with isoflurane. Bioluminescence was quantified with an In vivo Imaging System IVIS Lumina II (Caliper), with Living Image version 4.3.1 software (PerkinElmer) according to the manufacturer’s instructions.
AML-ETO9a transplants
c-KIT+ positive cells from the BM of Utx+/+ and Utx-/- mice and transduced them with lentiviral vectors expressing the AML-ETO9a fusion. Then 1×106 cells were subsequently transplanted into lethally irradiated syngeneic recipient mice (n=9 or 10 per group).
Secondary transplant of mouse Utx-/- leukemias
1×106 splenocytes were injected into sublethally irradiated recipient mice: acute leukaemia unclassified (n=5 mice), T-ALL (n=5 mice), and two AML (n=9 mice).
RNA extraction and RNA-Seq analysis
RNA was extracted from HSPC/Lin– BM cells with an Arcturus Picopure RNA Isolation Kit (Thermo Fisher Scientific) according to the manufacturer's instructions. RNA from mouse primary AML samples was extracted with TRIzol reagent (Thermo Fisher Scientific). RNA from human cell lines was extracted with an RNeasy kit (Qiagen) according to the manufacturer's instructions. An RNA-seq library was generated with a TruSeq Stranded mRNA Sample Prep Kit (Illumina) and sequenced on the Illumina HiSeq2000 v4 platform with 75-bp paired-end sequencing. RNA-seq reads for each library were mapped with TopHat version 2.0.13 against the mouse genome, build GRCm38.68, downloaded via the Ensembl BioMart data-mining tool. HiSeq libraries were aligned with the following options: --keep-fasta-order --no-sort-bam -r 100 -p 12 --library-type fr-firststrand --no-coverage-search --microexon-search --transcriptome-index = GRCm38.known, where GRCm38.known is a transcriptome index file in GTF format prepared beforehand in an initial single TopHat run without input reads. Raw counts for each gene in the Genome Reference Consortium genome assembly (GRCm38.68) were obtained with Bioconductor package GenomicAlignments version 1.2.2 with mode ‘Union’. Differential expression analysis was carried out with these counts in the Bioconductor package DESeq2, version 1.6.3, with a BH independent filtering method with an FDR of 1%; both packages were used according to their vignettes. Fold changes in expression were also calculated in DESeq2 after correction for differences in library sizes.
ChIP-Seq and ChIP-qPCR
ChIP–seq experiments were performed on primary HSPC cells. For histone ChIP–seq, cells were fixed in 1% formaldehyde (FA, Thermo Fisher Scientific, 28906) for 5 min at room temperature. The reaction was stopped by addition of glycine (0.125 M, Sigma), and the cells were washed in ice-cold PBS. The cells were then processed with an iDeal ChIP–seq Kit for Histones (Diagenode) with antibodies to the following proteins: H3K4me1 (Diagenode, pAb-194-050), H3K27me3 (Abcam, Ab6002) and H3K27ac (Diagenode, pAb-196-050). For PU.1 ChIP–seq, cells were fixed with 1% FA for 10 min at room temperature and processed with an iDeal ChIP–seq Kit for Transcription Factors (Diagenode) according to the manufacturer's instructions, with anti-PU.1 antibody (Santa Cruz Biotechnology, sc-352x). ChIP–seq for UTX was performed in HSPCs. The cells were cross-linked with 2 mM disuccinimidyl glutarate (Sigma) for 30 min at RT, and this was followed by a second cross-linking with 1% FA for 30 min at 4 °C. The cells were then processed with an iDeal ChIP–seq Kit for Transcription Factors, with anti-UTX antibody (Bethyl, A302-374A). Primer sequences used for ChIP–qPCR are listed in Supplementary Table 27.
For SMARCA4, CHD4 and UTX ChIP–qPCR, 416B cells were cross-linked with 2 mM disuccinimidyl glutarate (Sigma) for 30 min at RT, and this was followed by a second cross-linking with 1% FA for 10 min at RT. Cross-linking was stopped by addition of 125 mM glycine. Cells were resuspended in ChIP Lysis Buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl, pH 8, and protease inhibitors) and sonicated in a Bioruptor Pico instrument (Diagenode) for ten cycles. Sonicated chromatin was diluted 1:10 in modified RIPA buffer (1% Triton X-100, 0.1% deoxycholate, 90 mM NaCl, 10 mM Tris-HCl, pH 8, and EDTA-free protease inhibitors) and incubated overnight with 3 μg of anti-SMARCA4 (Santa Cruz Biotechnology, sc-10768x) or anti-CHD4 (Abcam, ab72418). Next, protein A/G (50% A 50% G) Dynabeads (Invitrogen) were added to the chromatin and incubated 2 h at 4 °C, and magnetic separation was then performed. Beads were subsequently washed twice with mixed-micelle buffer (150 mM NaCl, 0.2% SDS, 20 mM Tris-Cl, pH 8.0, 5 mM EDTA, 5.2% sucrose and 1% Triton X-100), high-salt buffer (250 mM NaCl, 5 mM Tris-Cl, pH 8.0, 0.5 mM EDTA, 0.05% sodium deoxycholate, 25 mM HEPES, pH 8.0, and 0.5% Triton X-100) and LiCl buffer (250 mM LiCl, 10 mM Tris-Cl, pH 8.0, 10 mM EDTA, 0.5% NP-40 and 0.5% sodium deoxycholate), then once with elution buffer (1% SDS and 100 mM NaHCO3). Beads were then resuspended in elution buffer supplemented with DNase-free RNase (Roche, 11119915001). Cross-linking was reverted by incubation at 37 °C for 30 min, then at 65 °C overnight. DNA was purified with a Chip-DNA purification kit (Zymo).
Chip-Seq, motif analysis, and GSEA data visualization
Adaptor sequences were trimmed for all paired-end reads and mapped against the mm10 reference genome with Bowtie2 (ref. 55). All the samples were processed independently, and uniquely mapped reads were retained. Peaks were called with SICER56 with W200 and G600 for broad peaks and W200 and G200 for narrow peaks. Peak calling was performed for each replicate individually. Motif analyses and peaks were annotated with HOMER35. Detailed output of the HOMER motif analyses is included in Supplementary Table 30. Peaks in intergenic regions were assigned to genes if they were within a 100-kb window from the TSS. Differential binding analysis was performed with DiffBind57 by grouping replicates together. Overlapping peak analysis was performed with intersect in bedtools58. The statistical analysis of the overlapping peaks was performed with Fisher's exact test in bedtools. Each of the ChIP–seq experiments, with the exception of the PU.1 ChIP–seq, was performed in biological duplicate. PU.1 ChIP–seq was performed in biological triplicate. GSEA tools were obtained from The Broad Institute59. ChIP–seq, RNA-seq and ATAC–seq data were visualized in UCSC Genome Browser60. Venn diagrams were generated with the BioVenn web application61. All graphs were generated in GraphPad Prism unless otherwise specified.
Promoter-enhancer interaction analysis
Promoter-associated interaction matrix of multipotent hematopoietic progenitor cell line 7 (HPC-7) was generated in a previous study using Promoter Capture Hi-C (PCHi-C) method and data were analysed with CHiCAGO package23. Genome coordinates from the interaction matrix were converted from mm9 to mm10. In total, 54,339 regions that form significant interactions (CHiCAGO score ≥ 5) with promoter baits were defined as promoter-interacting regions (PIRs). Differential H3K27Ac peaks in Utx-/- versus Utx+/+ were grouped as increased (UP) or decreased (DOWN) peaks and intersected with PIRs using bedtools. Interactions at specific gene loci were visualized in WashU Epigenome Browser.
ATAC
The ATAC-Seq method was used based on the established protocol62 with modifications63. Briefly, 200,000 cells were washed in 0.3 mL of ice-cold Dulbecco’s phosphate buffered saline without calcium and magnesium. This was followed by centrifugation at 300g for 3 minutes before resuspending in 400 µL of freshly-made ice-cold sucrose buffer (10 mM Tris-Cl pH 7.5, 3 mM CaCl2, 2 mM MgCl2 and 0.32 M sucrose) and incubated on ice for 12 minutes. 10% Triton X-100 was added to a final concentration of 0.5 % and the cells were vortexed briefly before incubating on ice for a further 6 minutes to access nuclei. The nuclei were briefly vortexed again before another centrifugation at 300g for 3 minutes at 4 °C. The sucrose/triton lysis buffer was removed before immediately resuspending the nuclei pellet in 50 µL of Nextera tagmentation master mix, comprising 25 µL 2x Tagment DNA buffer, 20 µL nuclease-free water and 5 µL Tagment DNA Enzyme 1 (Illumina FC-121-1030). The tagmentation reaction mixture was immediately transferred to a 1.5 mL low-bind microfuge tube and incubated at 37 °C for 30 minutes. The tagmentation reaction was stopped by the addition of 500 µL Buffer PB (Qiagen). The tagmented chromatin was purified using the MinElute PCR purification kit (Qiagen 28004), according to the manufacturer’s instructions, eluting in 10 µL of buffer EB (Qiagen). 10 µL of the tagmented chromatin was mixed with 2.5 µL Nextera PCR primer cocktail and 7.5 µL Nextera PCR mastermix (Illumina FC-121-1030) in a 0.2 mL low-bind PCR tube. 2.5 µL of an i5 primer and 2.5 µL of an i7 primer (Illumina FC-121-1011) were added per PCR, totaling 25 µL. PCR amplification was performed as follows: 72 °C for 3 minutes and 98 °C for 30 seconds, followed by 12 cycles of 98 °C for 10 seconds, 63 °C for 30 seconds and 72 °C for 3 minutes. Libraries were size-selected on a 1 % agarose TAE gel, collecting library fragments from 120 bp to 1 kb. Gel slices were extracted with the MinElute Gel Extraction kit (Qiagen 28604), eluting in 20 µL of Elution Buffer. Samples were further purified using Agencourt AMPure XP magnetic beads (Beckman Coulter A63880) at a ratio of 1.2 AMPure beads :1 PCR sample (v/v), according the manufacturer’s instructions, eluting in 20 µL of Buffer EB (Qiagen). Before sequencing, each ATAC-seq library was assessed on an Agilent 2100 Bioanalyzer using a High Sensitivity DNA chip (Agilent Technologies 5067-4626).
ATAC-Sequencing analysis
Similar to ChIP-Seq analyses all the adapter sequenced of ATAC-Seq paired end reads were trimmed and mapped against mm10 reference genome using Bowtie2. All the samples were processed independently and uniquely mapped reads were retained. Peaks were called with MACS2 (ref.64) with –nomodel and –nolambda parameters. Peak calling was performed individually for each of the replicate. Differential binding analysis was performed using DiffBind57 by groping replicates together. Overlapping peak analysis were performed using intersected from bedtools58. The statistical analysis of the overlapping peaks were performed using fisher’s exact test from bedtools. ATAC-Seq in HSPC was performed in biological triplicates, ATAC-Seq experiments for remodelers were performed in biological duplicates.
GEO accession codes for publicly available datasets
Data were downloaded from the GEO database under accession codes GSM552234 (GATA2); GSM1296403 and GSM1296404 (CHD4); and GSM1296402 (SMARCA4).
Preparation of IP samples for MS
107 416B cells were lysed in the whole cell lysis buffer (50 mM Tris-HCl pH=8, 150 mM NaCl, 0.1% NP-40, 1 mM EDTA), supplemented with 1 mM DTT, protease inhibitors (Sigma), and phosphatase inhibitors (Sigma). Cell were homogenized and lysate cleared by centrifugation. UTX immunoprecipitation was performed in the whole cell lysis buffer with 16 ug of antibody bound to 100ul of Dynabeads Protein G (Thermo Fisher Scientific) and incubated for 1,5h at 4°C with rotation. IP was washed five times with IP wash buffer (10 mM Tris-HCl pH=8, 150 mM NaCl, 0.1% NP-40, 1 mM EDTA), supplemented with protease inhibitor (Sigma). UTX immunoprecipitates were eluted by boiling in 1x LDS loading buffer, reduced with 5 mM TCEP, alkylated with 10 mM iodoacetamide and electrophoresed in Novex NuPAGE Bis-Tris 4%–12% gels (Life Technologies). Gels were stained with colloidal Coomassie (Sigma). Whole lanes were cut in slices and samples processed for MS analysis as described previously65.
Sample preparation and LC-MS/MS analysis
The affinity purified material was electrophoresed in 4-12% Bis-Tris NuPAGE gels (Life Technologies). Gels were fixed and stained with Coomassie as previously described65. The whole gel lanes were excised into five sections and IgG bands were discarded. Gel pieces were digested with trypsin and peptides extracted as previously described65. The peptides were re-dissolved in 0.5% formic acid and analysed with on-line nano liquid chromatography (Ultimate 3000 RSLCnano System) tandem mass spectrometry on an LTQ Orbitrap Velos mass spectrometer. The sample was desalted on a PepMap C18 nano-trap (100 µm i.d. x 20 mm, 100Å, 5µm), then separated on a PepMap RSLC C18 column (75 µm i.d. x 250 mm, 100 Å, 2 µm) in a linear gradient of 4-32% CH3CN/0.1% formic acid in 90 mins. The HPLC, columns and mass spectrometer were all from Thermo Fisher Scientific. The Orbitrap mass spectrometer was operated in the standard “top 15” data-dependent acquisition mode while the preview mode was disabled. The MS full scan was set at m/z 380 – 1600 with the resolution at 30,000 at m/z 400 and AGC at 1x106 with a maximum injection time at 200 msec. The siloxane ion at 445.120030 was used as lock mass. The 15 most abundant multiply-charged precursor ions (z ≥ 2), with a minimal signal above 3000 counts, were dynamically selected for CID (Collision Induced Dissociation) fragmentation in the ion trap, which had the AGC set at 5000 with the maximum injection time at 100 msec. The precursor isolation width was set at 2 Da. The normalized collision energy for CID MS/MS was set at 35%. The dynamic exclusion duration time for the selected ions for MS/MS was set for 60 sec with ±10 ppm exclusion mass width.
MS data analysis
Raw MS files were processed in Proteome Discoverer v 1.4 (Thermo Fisher Scientific). Database searches were performed in Mascot (v 2.5, Matrix Science) with the mouse Swiss-Prot database (December 2015, 16,942 sequences) supplemented with an in-house contaminant database. The search parameters were as follows: trypsin digestion, two missed cleavages, 10-p.p.m. mass tolerance for precursor ions, 0.5-Da mass tolerance for fragment ions, variable modifications of carbamidomethyl (C), N-acetylation (protein), formyl (N-terminal), oxidation (M), deamidated (NQ) and pyro-glu (N-terminal Q). Database search results were further processed with Percolator66–68 in Proteome Discoverer. Protein identification required at least one high-confidence peptide (FDR <1% according to q value) and a minimum Mascot protein score of 20. Protein lists for bait and control experiments (two and three replicates, respectively) were compared in SAINTexpress with default settings69. External protein contaminants (such as albumin, casein and keratin) were removed from the protein identification list before further data analysis. Prey with a SAINT probability score ≥ 0.99 is reported in the final high-confidence interactors list. The complement protein C1qc was manually removed from the list because, despite having a SAINT probability of 1, it was detected with a very similar number of peptides in both bait and control samples (Supplementary Tables 15 and 16).
Exome sequencing
DNA from 7 Utx-/- AML cases and matched normal DNA extracted from tail tips before pIpC-mediated Utx deletion, was extracted using DNeasy blood & tissue kit (Qiagen) according to the manufacturer’s instructions. The extracted DNA was quantified (using Invitrogen’s dsDNA Quant-IT PicoGreen), followed by normalizing each sample to 4.17ng/ul in 120ul in preparation for library creation. The first step of library preparation involved shearing the DNA into fragments of 150bp (using the Covaris LC220 and Agilent Bravo automated workstation for liquid handling) followed by library creation and PCR using unique index tags and adaptors (Agilent’s SureSelectXT Automated Library Prep and Capture Kits and MJ Tetrad). The amplified libraries were then purified (using Agencourt AMPure XP and a Beckman Coulter Biomek NX96 for liquid handling) and eluted in nuclease-free water followed by another round of quantification (using the Caliper GX). The quantified, size-selected libraries were then diluted down to an appropriate concentration for introduction into the exome capture stage. Exome pulldown (or hybridisation) was performed using Mouse-All Exon RNA-baits (designed by Agilent, supplier ID: S0276129) for 23 hours at 65°C. Eight uniquely indexed samples were baited and captured in a single pool as part of the agreed multiplexing strategy. The pulldown was then purified and eluted using streptavidin-coated Dynal beads ready to be amplified using PCR (MJ Tetrad). The PCR was then further purified using Agencourt AMPure XP (and Beckman Coulter Biomek NX96 for liquid handling), followed by quantification of the amplified pulldown product using the Agilent Bioanalyser. Samples were exome sequenced as paired-end 75bp inserts using Illumina HiSeq v4 flow-cell chemistry.
Exome data analysis
Somatic variants- point mutations and indels - were called using the Caveman70 and Pindel71,72 pipelines respectively, and filtered for artefacts (including sufficient tumor fraction, strand bias, presence of tumor allele in the normal, presence of tumor allele in panel of normal samples also sequenced at Sanger). Genome-wide copy number variation was called using the Control Freek73 software package. This packages accepts paired tumor/ normal sequence files. We noticed that in a few of our samples that the sequenced paired normal displayed an artifactual noisiness in sequencing depth and opted to use a single ‘quiescent’ normal (MD5280a) as a constant comparator for all samples. Control Freek was run using these parameters: step=1000000, window=5000000, breakpointtype=4, breakpointthreshold=1.2, readcountthreshold=50. The copy-number plots show the normalised bam depth-ratio as produced by ControlFreek (black points), as well as a mark (red points) for regions marked with CopyNumber = 2. Copy-number variation looking for a focal change at exon 3 of Utx was run (as before) with an unmatched normal - MD5280a - as well as the following parameters: step=250, window=500, breakpointthreshold=0.6, breakpointtype=4, readcountthreshold=50.
Statistical analysis
All statistical analyses were performed using two-sided Student’s T test or one-way ANOVA as specified in figure legends. Error bars represent the standard error of the mean (s.e.m.) or the standard deviation (s.d.). P values ≤ 0.05 were considered statistically significant. Representative data/images were replicated in at least three independent experiments as specified in the relevant figure legend. Hypergeometric distribution was calculated with the online tool, GeneProf. The number of independent experiments used to generate statistically significant data is defined in the relevant figure legends.
Supplementary Material
Acknowledgements
This study was primarily funded by a joint Bloodwise Program Grant (17006) to B.J.P.H. and G.S.V. Work in the laboratory of B.J.P.H. is also funded by an ERC consolidator award (grant 647685 COMAL), a Cancer Research UK program award, the Medical Research Council, (MRC) the Wellcome Trust (WT) and the Cambridge NIHR BRC. We acknowledge the WT/MRC Center grant (097922/Z/11/Z) and support from WT strategic award 100140. G.S.V. is funded by a Cancer Research UK Senior Cancer Research Fellowship (C22324/A23015). The laboratory of G.S.V. is also supported by the Kay Kendall Leukemia Fund and core funding from the Sanger Institute (WT098051).
Footnotes
Author Contributions
M.G., G.S.V. and B.J.P.H. conceived the study, designed the experiments and prepared the manuscript. M.G. conducted most of the experiments. E. Meduri performed ChIP–seq, ATAC–seq and motif analysis. A.J.K. performed ATAC–seq experiments. M.P. and M.G. prepared samples for mass spectrometry; M.P., L.Y. and J.S.C. conducted mass spectrometry and related data analysis. E. Metzakopian designed and generated pKLV-puro vectors. V.I. and D.A. performed exome analysis. H.Y. performed promoter–enhancer interaction analysis. N.P. and I.V. performed experimental and computational analysis. G.C., M.M., M.D., O.D., K.T., D.A.G., E.D.B. and J.C. performed cell culture and mouse experiments. R.B. performed analysis of RNA-seq data. P.A.N., B.G. and L.B. provided genomic data and expertise. S.K. helped with vector generation. All authors reviewed and agreed with the final submission.
Competing Financial Interests
Authors states no competing financial interests.
URLs
TopHat version 2.0.13, http://ccb.jhu.edu/software/tophat/; BioMart data-mining tool, http://www.ensembl.org/info/data/biomart/; GenomicAlignments version 1.2.2, http://bioconductor.org/packages/3.0/bioc/html/GenomicAlignments.html with mode = ‘Union’; DESeq2 version 1.6.3, http://bioconductor.org/packages/3.0/bioc/html/DESeq2.html; UCSC Genome Browser, http://genome.ucsc.edu/; BioVenn tool, http://www.biovenn.nl/; hypergeometric test, https://www.geneprof.org/GeneProf/tools/hypergeometric.jsp/; ENA, http://www.ebi.ac.uk/ena/; Gene Expression Omnibus (GEO), https://www.ncbi.nlm.nih.gov/geo/; ProteomeXchange, http://www.proteomexchange.org/; processed RNA-seq data, ftp://ngs.sanger.ac.uk/production/casm/2018/Gozdecka_et_al_NatGenet/UTX_FPKM_RNAseq/; Kinsey targets of EWSR1-FLI1, http://software.broadinstitute.org/gsea/msigdb/geneset_page.jsp?geneSetName=KINSEY_TARGETS_OF_EWSR1_FLII_FUSION_UP; Huang GATA2 Targets, http://software.broadinstitute.org/gsea/msigdb/geneset_page.jsp?geneSetName=HUANG_GATA2_TARGETS_DN.
Data availability.
All sequencing and proteomic raw data have been deposited in public databases. There are no restrictions to data access. The ChIP–seq and ATAC–seq data have been deposited in the GEO database under accession numbers GSE86490 and GSE101307. The exome sequencing data have been deposited in the ENA database under accession number ERP017908. MS data are available via the ProteomeXchange database under identifier PXD005011. A description of raw and processed RNA-seq files is provided in Supplementary Table 31.
References
- 1.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 2.Agger K, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 3.Dalgliesh GL, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey P, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 5.Gui Y, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nature genetics. 2011;43:875–878. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robinson G, et al. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488:43–48. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Haaften G, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nature genetics. 2009;41:521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huether R, et al. The landscape of somatic mutations in epigenetic regulators across 1,000 paediatric cancer genomes. Nat Commun. 2014;5:3630. doi: 10.1038/ncomms4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ntziachristos P, et al. Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature. 2014;514:513–517. doi: 10.1038/nature13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cancer Genome Atlas Research, N. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. The New England journal of medicine. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jankowska AM, et al. Mutational spectrum analysis of chronic myelomonocytic leukemia includes genes associated with epigenetic regulation: UTX, EZH2, and DNMT3A. Blood. 2011;118:3932–3941. doi: 10.1182/blood-2010-10-311019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wouters BJ, Delwel R. Epigenetics and approaches to targeted epigenetic therapy in acute myeloid leukemia. Blood. 2016;127:42–52. doi: 10.1182/blood-2015-07-604512. [DOI] [PubMed] [Google Scholar]
- 13.Greenfield A, et al. The UTX gene escapes X inactivation in mice and humans. Hum Mol Genet. 1998;7:737–742. doi: 10.1093/hmg/7.4.737. [DOI] [PubMed] [Google Scholar]
- 14.Van der Meulen J, et al. The H3K27me3 demethylase UTX is a gender-specific tumor suppressor in T-cell acute lymphoblastic leukemia. Blood. 2015;125:13–21. doi: 10.1182/blood-2014-05-577270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arcipowski KM, Martinez CA, Ntziachristos P. Histone demethylases in physiology and cancer: a tale of two enzymes, JMJD3 and UTX. Curr Opin Genet Dev. 2016;36:59–67. doi: 10.1016/j.gde.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walport LJ, et al. Human UTY(KDM6C) is a male-specific N-methyl lysyl demethylase. J Biol Chem. 2014;289:18302–18313. doi: 10.1074/jbc.M114.555052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van der Meulen J, Speleman F, Van Vlierberghe P. The H3K27me3 demethylase UTX in normal development and disease. Epigenetics. 2014;9:658–668. doi: 10.4161/epi.28298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skarnes WC, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faber ZJ, et al. The genomic landscape of core-binding factor acute myeloid leukemias. Nature genetics. 2016;48:1551–1556. doi: 10.1038/ng.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papaemmanuil E, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. The New England journal of medicine. 2016;374:2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thieme S, et al. The histone demethylase UTX regulates stem cell migration and hematopoiesis. Blood. 2013;121:2462–2473. doi: 10.1182/blood-2012-08-452003. [DOI] [PubMed] [Google Scholar]
- 22.Wang C, et al. UTX regulates mesoderm differentiation of embryonic stem cells independent of H3K27 demethylase activity. Proc Natl Acad Sci U S A. 2012;109:15324–15329. doi: 10.1073/pnas.1204166109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson NK, et al. Integrated genome-scale analysis of the transcriptional regulatory landscape in a blood stem/progenitor cell model. Blood. 2016;127:e12–23. doi: 10.1182/blood-2015-10-677393. [DOI] [PubMed] [Google Scholar]
- 24.Cho YW, et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. The Journal of biological chemistry. 2007;282:20395–20406. doi: 10.1074/jbc.M701574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foulds CE, Nelson ML, Blaszczak AG, Graves BJ. Ras/mitogen-activated protein kinase signaling activates Ets-1 and Ets-2 by CBP/p300 recruitment. Molecular and cellular biology. 2004;24:10954–10964. doi: 10.1128/MCB.24.24.10954-10964.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson NK, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell stem cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 27.Morris SA, et al. Overlapping chromatin-remodeling systems collaborate genome wide at dynamic chromatin transitions. Nat Struct Mol Biol. 2014;21:73–81. doi: 10.1038/nsmb.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tzelepis K, et al. A CRISPR Dropout Screen Identifies Genetic Vulnerabilities and Therapeutic Targets in Acute Myeloid Leukemia. Cell reports. 2016;17:1193–1205. doi: 10.1016/j.celrep.2016.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shpargel KB, Starmer J, Yee D, Pohlers M, Magnuson T. KDM6 demethylase independent loss of histone H3 lysine 27 trimethylation during early embryonic development. PLoS Genet. 2014;10:e1004507. doi: 10.1371/journal.pgen.1004507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morales Torres C, Laugesen A, Helin K. Utx is required for proper induction of ectoderm and mesoderm during differentiation of embryonic stem cells. PloS one. 2013;8:e60020. doi: 10.1371/journal.pone.0060020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoo KH, et al. Histone Demethylase KDM6A Controls the Mammary Luminal Lineage through Enzyme-Independent Mechanisms. Molecular and cellular biology. 2016;36:2108–2120. doi: 10.1128/MCB.00089-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beyaz S, et al. The histone demethylase UTX regulates the lineage-specific epigenetic program of invariant natural killer T cells. Nature immunology. 2017;18:184–195. doi: 10.1038/ni.3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang SP, et al. A UTX-MLL4-p300 Transcriptional Regulatory Network Coordinately Shapes Active Enhancer Landscapes for Eliciting Transcription. Molecular cell. 2017;67:308–321 e306. doi: 10.1016/j.molcel.2017.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zaret KS, Mango SE. Pioneer transcription factors, chromatin dynamics, and cell fate control. Curr Opin Genet Dev. 2016;37:76–81. doi: 10.1016/j.gde.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ciau-Uitz A, Wang L, Patient R, Liu F. ETS transcription factors in hematopoietic stem cell development. Blood Cells Mol Dis. 2013;51:248–255. doi: 10.1016/j.bcmd.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 37.May WA, et al. Ewing sarcoma 11;22 translocation produces a chimeric transcription factor that requires the DNA-binding domain encoded by FLI1 for transformation. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:5752–5756. doi: 10.1073/pnas.90.12.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 39.Goldberg L, et al. Genome-scale expression and transcription factor binding profiles reveal therapeutic targets in transgenic ERG myeloid leukemia. Blood. 2013;122:2694–2703. doi: 10.1182/blood-2013-01-477133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marcucci G, et al. Overexpression of the ETS-related gene, ERG, predicts a worse outcome in acute myeloid leukemia with normal karyotype: a Cancer and Leukemia Group B study. J Clin Oncol. 2005;23:9234–9242. doi: 10.1200/JCO.2005.03.6137. [DOI] [PubMed] [Google Scholar]
- 41.Gao J, Chen YH, Peterson LC. GATA family transcriptional factors: emerging suspects in hematologic disorders. Exp Hematol Oncol. 2015;4:28. doi: 10.1186/s40164-015-0024-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hahn CN, et al. Heritable GATA2 mutations associated with familial myelodysplastic syndrome and acute myeloid leukemia. Nature genetics. 2011;43:1012–1017. doi: 10.1038/ng.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greif PA, et al. GATA2 zinc finger 1 mutations associated with biallelic CEBPA mutations define a unique genetic entity of acute myeloid leukemia. Blood. 2012;120:395–403. doi: 10.1182/blood-2012-01-403220. [DOI] [PubMed] [Google Scholar]
- 44.Groschel S, et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell. 2014;157:369–381. doi: 10.1016/j.cell.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 45.Miller SA, Mohn SE, Weinmann AS. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Molecular cell. 2010;40:594–605. doi: 10.1016/j.molcel.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee S, Lee JW, Lee SK. UTX, a histone H3-lysine 27 demethylase, acts as a critical switch to activate the cardiac developmental program. Developmental cell. 2012;22:25–37. doi: 10.1016/j.devcel.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Local A, et al. Identification of H3K4me1-associated proteins at mammalian enhancers. Nature genetics. 2018;50:73–82. doi: 10.1038/s41588-017-0015-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Forsberg LA, et al. Mosaic loss of chromosome Y in peripheral blood is associated with shorter survival and higher risk of cancer. Nat Genet. 2014;46:624–628. doi: 10.1038/ng.2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dumanski JP, et al. Mutagenesis. Smoking is associated with mosaic loss of chromosome Y. Science. 2015;347:81–83. doi: 10.1126/science.1262092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou W, et al. Mosaic loss of chromosome Y is associated with common variation near TCL1A. Nat Genet. 2016;48:563–568. doi: 10.1038/ng.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kogan SC, et al. Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice. Blood. 2002;100:238–245. doi: 10.1182/blood.v100.1.238. [DOI] [PubMed] [Google Scholar]
- 52.Morse HC, 3rd, et al. Bethesda proposals for classification of lymphoid neoplasms in mice. Blood. 2002;100:246–258. doi: 10.1182/blood.v100.1.246. [DOI] [PubMed] [Google Scholar]
- 53.Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera Mdel C, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nature biotechnology. 2014;32:267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- 54.Yan M, et al. A previously unidentified alternatively spliced isoform of t(8;21) transcript promotes leukemogenesis. Nat Med. 2006;12:945–949. doi: 10.1038/nm1443. [DOI] [PubMed] [Google Scholar]
- 55.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu S, Grullon S, Ge K, Peng W. Spatial clustering for identification of ChIP-enriched regions (SICER) to map regions of histone methylation patterns in embryonic stem cells. Methods in molecular biology. 2014;1150:97–111. doi: 10.1007/978-1-4939-0512-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ross-Innes CS, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kent WJ, et al. The human genome browser at UCSC. Genome research. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hulsen T, de Vlieg J, Alkema W. BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC genomics. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kumasaka N, Knights AJ, Gaffney DJ. Fine-mapping cellular QTLs with RASQUAL and ATAC-seq. Nature genetics. 2016;48:206–213. doi: 10.1038/ng.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pardo M, et al. An expanded Oct4 interaction network: implications for stem cell biology, development, and disease. Cell stem cell. 2010;6:382–395. doi: 10.1016/j.stem.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kall L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nature methods. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 67.Brosch M, Yu L, Hubbard T, Choudhary J. Accurate and sensitive peptide identification with Mascot Percolator. Journal of proteome research. 2009;8:3176–3181. doi: 10.1021/pr800982s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Spivak M, Weston J, Bottou L, Kall L, Noble WS. Improvements to the percolator algorithm for Peptide identification from shotgun proteomics data sets. Journal of proteome research. 2009;8:3737–3745. doi: 10.1021/pr801109k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Teo G, et al. SAINTexpress: improvements and additional features in Significance Analysis of INTeractome software. Journal of proteomics. 2014;100:37–43. doi: 10.1016/j.jprot.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jones D, et al. cgpCaVEManWrapper: Simple Execution of CaVEMan in Order to Detect Somatic Single Nucleotide Variants in NGS Data. Current protocols in bioinformatics. 2016;56:15 10 11–15 10 18. doi: 10.1002/cpbi.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Raine KM, et al. cgpPindel: Identifying Somatically Acquired Insertion and Deletion Events from Paired End Sequencing. Current protocols in bioinformatics. 2015;52:15 17 11–12. doi: 10.1002/0471250953.bi1507s52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boeva V, et al. Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics. 2012;28:423–425. doi: 10.1093/bioinformatics/btr670. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.