Abstract
The acquisition of cellular identity is coupled to changes in the nuclear periphery and nuclear pore complexes (NPCs). Whether and how these changes determine cell fate remains unclear. We have uncovered a mechanism regulating NPC acetylation to direct cell fate after asymmetric division in budding yeast. The lysine deacetylase Hos3 associates specifically with daughter cell NPCs during mitosis to delay cell cycle entry (Start). Hos3-dependent deacetylation of nuclear basket and central channel nucleoporins establishes daughter cell-specific nuclear accumulation of the transcriptional repressor Whi5 during anaphase and perinuclear silencing of the CLN2 gene in the following G1 phase. Hos3-dependent coordination of both events restrains Start in daughter but not in mother cells. We propose that deacetylation modulates transport-dependent and -independent functions of NPCs, leading to differential cell cycle progression in mother and daughter cells. Similar mechanisms might regulate NPC functions in specific cell types and/or cell cycle stages in multicellular organisms.
Introduction
Asymmetric cell division is a conserved mechanism that generates diversity in cell populations. Asymmetric divisions are found in both unicellular organisms and metazoans, where they play a major role in stem cell self-renewal and tissue homeostasis during development 1,2. During asymmetric division, unequal partitioning of cell fate determinants between the new cells leads to their different identities.
We have investigated how the acquisition of cell identity is controlled by nuclear pore complexes (NPCs) during asymmetric cell division. NPCs are macromolecular assemblies composed of approximately 30 nucleoporins forming channels across the nuclear envelope (NE) to mediate transport between the nucleus and the cytoplasm 3–5. Nucleo-cytoplasmic transport of proteins and RNA is intimately tied to the regulation of gene expression and cell fate determination 6,7. Additionally, the NE and nucleoporins associated with the nuclear basket of NPCs can directly interact with the nuclear genome to regulate gene expression and thus affect cell differentiation [reviewed in 7–10]. In particular, the nuclear periphery is a transcriptionally repressive environment in yeast and metazoans 11–14, and gene repositioning from the nuclear interior to the periphery can result in silencing 15,16. The composition of both the NE and NPCs, and their interactions with the genome, are known to diverge during development 17–20. However, how differences in perinuclear function are established during development, and in particular during asymmetric cell divisions, remains unclear.
Budding yeast divide asymmetrically, giving rise to mother and daughter cells of different size, age, transcriptional profiles and cell cycle programs 21–23. Notably, commitment to a new division cycle is regulated asymmetrically in S. cerevisiae: daughter cells start a new cycle later than mother cells. This is due to a cell size-dependent delay that prolongs G1 until daughter cells reach a critical size 21,24 and to size-independent, daughter-specific delay of the G1/S transition 25,26. Regulatory principles controlling cell cycle entry are similar in yeast and animal cells: a transcriptional activator (SBF in yeast; E2F in mammals) drives expression of cyclin genes (CLN1/2 or cyclin E, respectively) controlling the start of S phase. SBF or E2F are inhibited in G1 by a transcriptional repressor: Whi5 in yeast, and its homolog the Rb tumour suppressor in mammals. In yeast, a key event driving the G1/S transition is the dilution of Whi5 activity by cell growth, whereby the volume increase in daughter cells during G1 lowers the concentration of Whi5 below a critical threshold 27. This allows Cyclin-dependent kinase (Cdk) complexes to inactivate Whi5, which is then evicted from the nucleus 28,29. Interestingly, the G1 concentration of Whi5 is higher in daughter cells than in mother cells 27,30. The mechanism establishing this asymmetry is not known.
Here, we reveal that cell cycle entry in budding yeast daughters is inhibited by association of the lysine deacetylase Hos3 with daughter-cell NPCs. We identify the mechanism recruiting Hos3 to NPCs during mitosis. Further, we demonstrate that Hos3-mediated NPC deacetylation establishes asymmetric segregation of the Whi5 transcriptional repressor and perinuclear silencing of the G1/S cyclin gene CLN2 in daughters, which together contribute to inhibit Start. Thus, cell-specific deacetylation of NPCs directs differences in cell identity during asymmetric division.
Results
Hos3 inhibits cell cycle entry in daughter cells
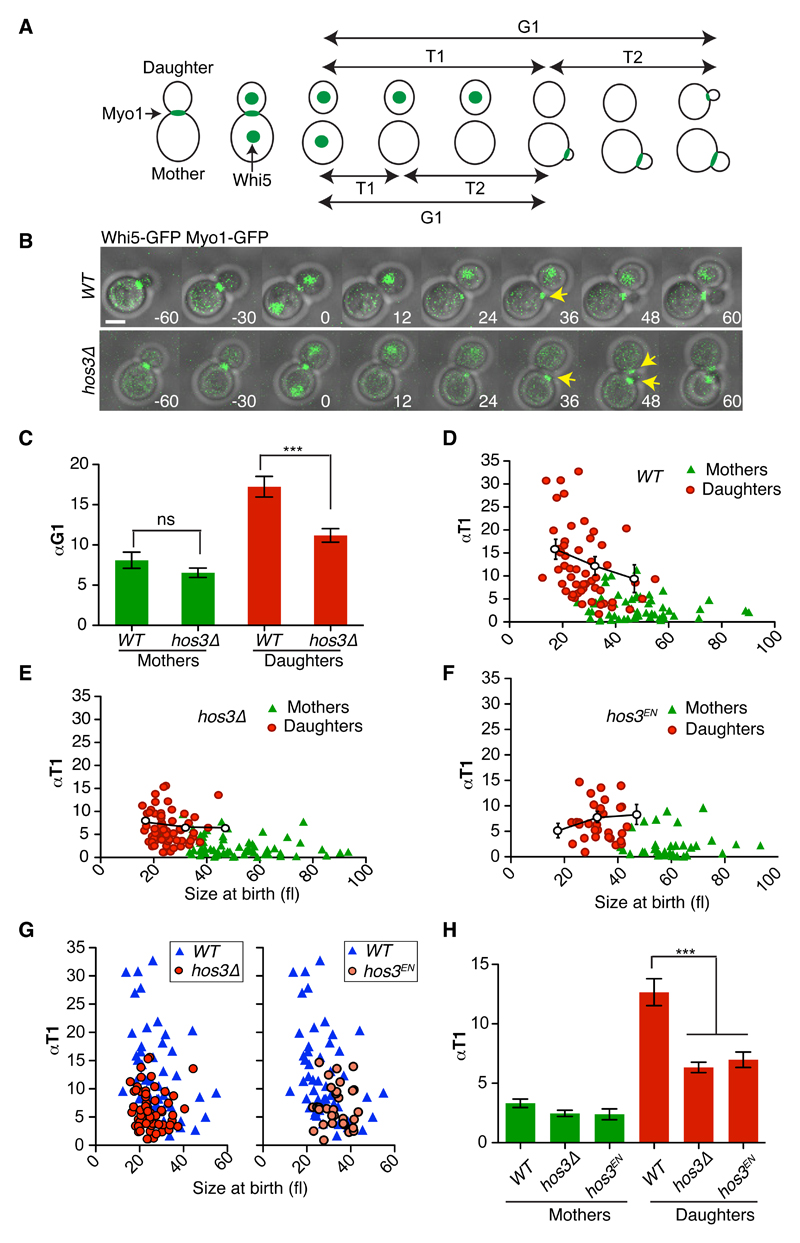
Commitment to a new division cycle in budding yeast occurs earlier in mother cells than in daughter cells. The lysine deacetylase Hos3 has been implicated in the control of G1 length and gene expression 31,32 but whether it plays a role specifically in daughter cells was not known. We therefore determined G1 duration in wild type and hos3Δ mother and daughter cells by evaluating the interval between cytokinesis and bud emergence, normalized by the rate of growth α during this period (αTG1) (Figure 1A). This revealed a shortened G1 in hos3Δ daughter cells, but not mother cells, relative to wild type (Figure 1B-C, Supplementary Figure S1A and Supplementary Table S1). Premature onset of S phase can lead to smaller cell size at Start, but this is not the case for hos3 cells 31, probably due to slightly faster growth rate of the hos3Δ mutant relative to wild type specifically during S/G2/M phases (Supplementary Figure S1B).
Figure 1. Hos3 delays Start in daughter cells.
(A) Scheme showing the separation of G1 in two distinct periods, T1 and T2, relative to Myo1 and Whi5 dynamics. The disappearance of Myo1-GFP from the bud neck marked cytokinesis; its reappearance to the new bud site marked budding. (B) Composite of phase contrast, Whi5-GFP and Myo1-GFP in wild type (WT) and hos3Δ cells. Daughter hos3Δ cells start a new bud (Myo1-GFP appearance, arrows) earlier than WT. Confocal sections spanning the entire cell were acquired at 3-minute intervals. Time is indicated in minutes; t=0 marks the last frame before cytokinesis. Scale bars, 2 μm. Experiments were repeated three times with similar results. (C-G) αTG1 values (mean and SEM) (C) and correlation between αT1 (with binned means and SEM) and cell size at the time of birth (cytokinesis) (D-G), for mother and daughter cell pairs. (H) αT1 values (mean and SEM) are shown in cells of the indicated strains. Sample size: WT (n=110 cells), hos3Δ (n=128 cells) and hos3EN (n=66 cells) pooled from three independent experiments. In panels C and H, two-sided Mann-Whitney test were used. *** denotes p < 0.0001; ns, non-significant, p > 0.05. Exact p values: 8.93x10-5 (Figure 1C, WT vs. hos3Δ), 7.20x10-7 (1H, WT vs. hos3Δ), 5.69x10-4 (1H, WT vs. hos3EN).
G1 is divided in two periods, T1 and T2, separated by nuclear export of the transcriptional repressor Whi5 (Figure 1A). Analysis of Whi5-GFP nuclear export kinetics (T1) showed that HOS3 deletion reduces αT1 in daughter cells relative to wild-type daughters (mean and standard deviation: WT: 12.6 ± 8.3, N = 55; hos3Δ: 6.3 ± 3.5, N = 64, p < 0.0001, Mann-Whitney test) (Figure 1, D-E). Shortening of G1 in hos3Δ daughter cells relative to wild type daughters was observed even for cells born at similar sizes (Figure 1G). In contrast, the scaled time between Whi5 export and budding (αT2), ascribed to molecular noise 33, was similar in hos3Δ and wild type cells (Supplementary Figure S1C). In addition, a catalytically inactive mutant of Hos3 (Hos3H196E, D231N, or Hos3EN) 34 showed advanced cell cycle entry exclusively in daughter cells in a similar manner to hos3Δ (Figure 1F-H). Therefore, Hos3 activity inhibits Start in daughter cells.
Hos3 associates with the nuclear periphery of daughter cells during anaphase
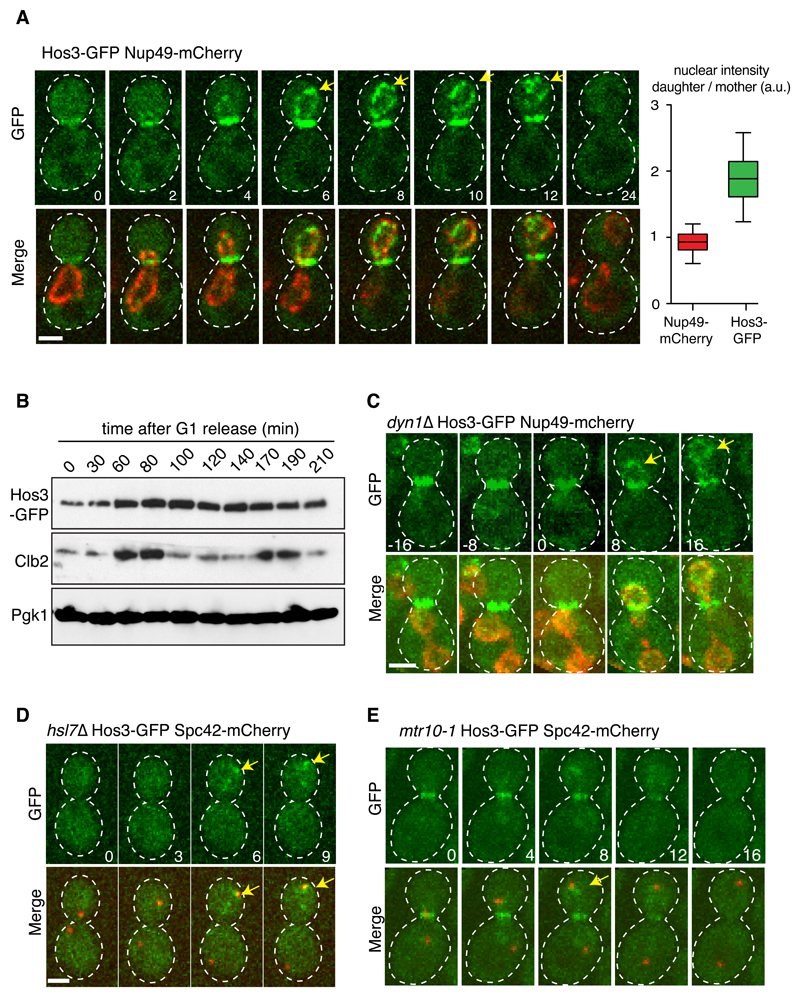
Hos3 is present in the cytoplasm during interphase and is recruited to the daughter side of the septin-based ring at the mother-bud neck during mitosis 34. Unexpectedly, time-lapse fluorescence microscopy revealed that Hos3-GFP also associates specifically with the nuclear periphery of the daughter cell during anaphase. Perinuclear Hos3 localization coincides with passage of the nucleus through the bud neck (Figure 2A, Supplementary Figure S2A, movies S1-2). Hos3 disappeared from the bud neck and nuclear periphery 1-2 minutes before cytokinesis (Supplementary Figure S2B-C and movie S3). These localization dynamics did not involve changes in total Hos3 protein levels (Figure 2B) or require Hos3 catalytic activity (Supplementary Figure S2D-E).
Figure 2. Hos3 localizes to the periphery of daughter nuclei upon nuclear migration in anaphase.
(A) Localization of Hos3-GFP (green) to the bud neck during mitosis and to the daughter nuclear periphery (Nup49-mCherry, in red) during nuclear migration into the bud. Arrows point to perinuclear Hos3-GFP. The box plot shows the ratio of nuclear intensity of GFP and mCherry signals in late anaphase cells (n = 67 cells). Boxes include 50% of data points, the line represents the median and whiskers extend to maximum and minimum values. (B) Cell extracts were prepared at the indicated times after release from a G1 block (induced with alpha factor). Hos3-GFP was detected with an anti-GFP antibody; Clb2 (B-type cyclin) serves as a cell cycle progression marker, and Pgk1(3-phosphoglycerate kinase) as loading control. This experiment was repeated two times with similar results. (C) A dyn1Δ cell showing separated anaphase nuclei (Nup49-mCherry, in red) in the mother cell. Hos3-GFP associates with the nuclear periphery only after nuclear migration across the bud neck (arrows). (D) Hos3-GFP is absent from the bud neck and nuclear periphery in the hsl7Δ mutant, but localizes to the dSPB (marked with Spc42-mCherry, red) in anaphase (arrows). (E) Hos3-GFP localizes to bud neck and spindle pole body (marked with Spc42-mCherry, arrow) in mtr10-1 cells at 30ºC, but fails to localize at the nuclear periphery. Maximal projections of whole-cell Z-stacks are shown, expect in (A) where one medial section is shown for clarity. Images were acquired at 2-4 minute intervals; time is indicated in minutes. t=0 is the last frame before the nucleus traverses the bud neck. Scale bars, 2 μm. Experiments in (C-E) were repeated three times with similar results.
We defined the determinants of Hos3 localization to the nuclear periphery of daughter cells. Firstly, this is dependent on passage of the nucleus through the bud neck. Hos3-GFP associated with nuclear protrusions extending across the bud neck and into the daughter cell, which form in cells arrested in metaphase by depletion of the APC activator Cdc20 35 (Supplementary Figure S3A). Moreover, in dyn1Δ cells that completed anaphase within the mother cell, Hos3-GFP was recruited to the perinuclear region only after migration of the anaphase nucleus into the daughter compartment (20/20 cells with delayed nuclear migration) (Figure 2C, Supplementary Figure S3B-C, and movie S4).
Secondly, recruitment of Hos3 to the nuclear periphery depends on its association with the bud neck. Septin ring perturbation in the temperature sensitive (ts) cdc12-1 mutant led to symmetric localization of Hos3 at both the bud neck and the nuclear periphery during anaphase (Supplementary Figure S3D-F). Additionally, deletion of Hsl7, which recruits Hos3 to the septin ring 34, also prevented perinuclear localization of Hos3-GFP, which localized at the anaphase daughter spindle pole body (dSPB) in hsl7Δ (71/74 cells) (Figure 2D and Supplementary Figure S3G). Interestingly, inhibition of mitotic exit in the cdc15-1 mutant retained Hos3-GFP in the bud neck and dSPB (Supplementary Figure S3H). Over-expressed Hos3-GFP also associated with the dSPB (Supplementary Figure S3I, movie S5); this may explain why perinuclear Hos3 was not detected using plasmid-borne Hos3-GFP 34.
Lastly, mutation of the importin Mtr10 impaired Hos3 recruitment to the nuclear periphery, which was instead found at the bud neck and the dSPB during anaphase (42/52 cells; Figure 2E). Thus, Hos3 is imported into the daughter nucleus during its migration across the bud neck, in a manner dependent on Mtr10. Importantly, total Hos3-GFP levels were constant in dyn1, hsl7 and mtr10 cells (Supplementary Figure S3J). The highly regulated asymmetric enrichment of Hos3 at the nuclear periphery suggests that it performs an important function in daughter cell nuclei, and might underlie the Hos3-mediated inhibition of Start in daughters.
Hos3 associates with nuclear pores in daughter cells to delay Start
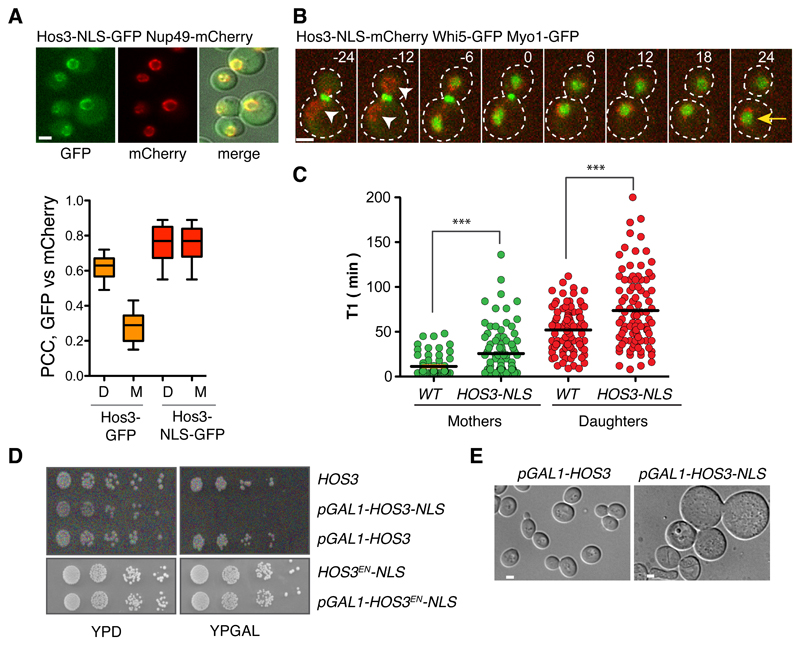
We next asked if targeting Hos3 to the mother cell nucleus prolongs G1 in these cells. Hos3-GFP fused with the nuclear localization signal of SV40 TAg 36 (Hos3-NLS-GFP) localized to the periphery of both mother and daughter cell nuclei throughout the cell cycle (Figure 3A and Supplementary Figure S4A-B), and delayed Whi5 nuclear export (T1) (Figure 3, B-C). Furthermore, over-expression of Hos3-NLS impaired growth on solid media (Figure 3D and Supplementary Figure S4C) and led to an increase in cell volume, consistent with a cell cycle delay (Figure 3E). This toxicity required the constitutive nuclear localization and the catalytic activity of Hos3, as cells grew normally when over-expressing Hos3-GFP (without the NLS) or Hos3EN-NLS-GFP (Figure 3D and Supplementary Figure S4C). Thus, perinuclear localization of active Hos3 is sufficient to inhibit Start.
Figure 3. Perinuclear Hos3 inhibits cell cycle entry.
(A) Hos3-NLS-GFP (green) localizes to the nuclear periphery (Nup49-mCherry, red) in mother and daughter cells. The graph shows the Pearson's correlation coefficient of the GFP and mCherry channels in the indicated anaphase cells. Wild type (n= 35 cells), Hos3-NLS-GFP (n= 46 cells). Boxes include 50% of data points, the line represents the median and whiskers extend to maximum and minimum values. (B) Anaphase and G1 in a cell expressing Hos3-NLS-mCherry. Arrowheads point to perinuclear Hos3-NLS in mother and daughter cells; the arrow points to persistent Whi5-GFP in the mother nucleus. Time is indicated in minutes; t=0 marks the last frame before cytokinesis. (C) Duration of pre-Start G1 phase (T1) in WHI5-GFP MYO1-GFP cells, expressing either Hos3 (WT) or Hos3-NLS-mCherry. Mother and daughter cell pairs in WT (n=177 cells), HOS3-NLS (n=191 cells) pooled from three independent experiments. Lines represent the mean. ***, p < 0.001, two-sided Mann-Whitney test. Exact p values: 1.25x10-7 (mothers), 5.67x10-4 (daughters). (D) Growth inhibition upon over-expression of Hos3-NLS from the GAL1,10 promoter. Serial dilutions of the indicated strains expressing Hos3-GFP fusions were incubated for 2 days at 30 ºC. (E) DIC images of wild type and Hos3-NLS after 2 days of growth in YP + galactose plates. Scale bars, 2 μm. All experiments in this figure were repeated three times with similar results.
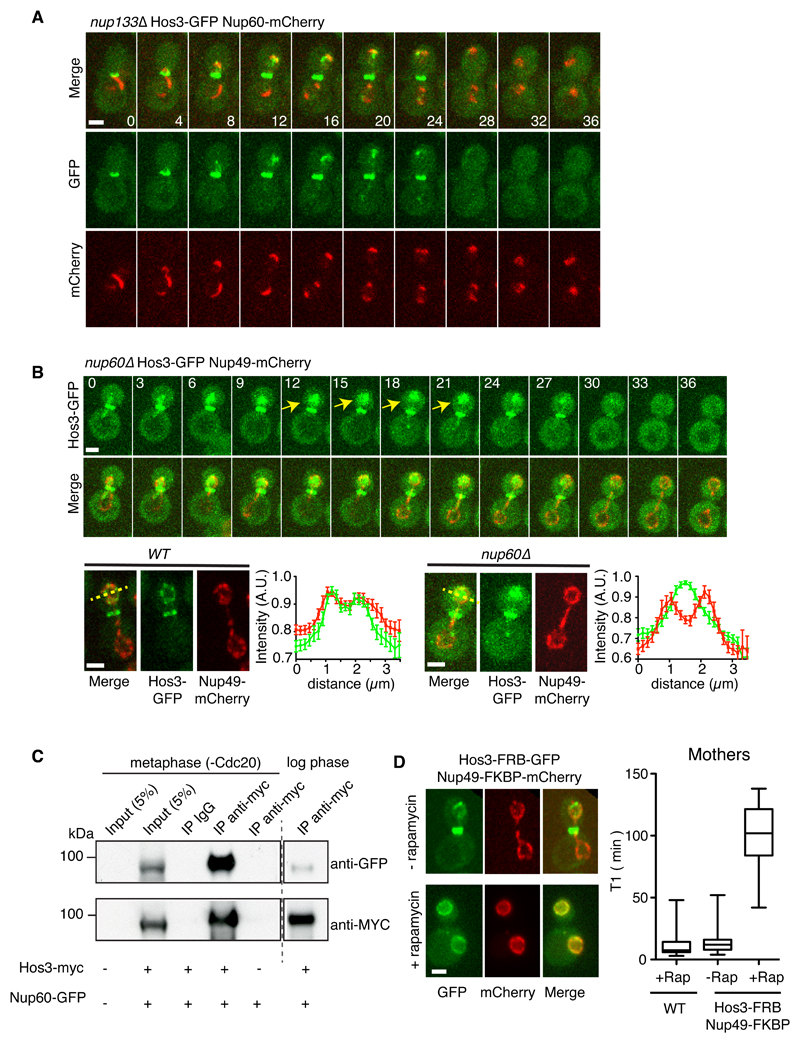
Remarkably, Hos3-NLS did not accumulate in the nucleoplasm but was instead enriched in the nuclear periphery like Hos3-GFP (see Figure 3A), and was detected by immuno-electron microscopy on the nucleoplasmic side of NPCs (Supplementary Figure S4D), suggesting that Hos3 associates with NPCs. In agreement with this, artificial NPC clustering caused by deletion of NUP133 37 led to clustering of Hos3-GFP and Hos3-GFP-NLS, which partially co-localized with a subset of pores (Figure 4A, Supplementary Figure S4E-G, and movie S6). Deletion of the nuclear pore basket component Nup60 38 prevented perinuclear localization of Hos3-NLS (Supplementary Figure S4H-I) and of Hos3-GFP, which was instead distributed in the nucleoplasm of nup60Δ anaphase daughter cells (Figure 4B and movie S7). Additionally, epitope-tagged Hos3 and Nup60 co-immuno-precipitated in logarithmically growing cultures, and their interaction was stronger in extracts prepared from metaphase-arrested cells in which Hos3 localizes to the nuclear periphery (Figure 4C and Supplementary Figure S4J). Thus, Hos3 associates with NPCs during nuclear migration into the bud. This association was sufficient to delay Whi5 export and cell cycle entry, because artificial tethering of Hos3 to NPCs with an FKBP-FRB system 39 strongly and specifically delayed Start (Figure 4D and Supplementary Figure S4K).
Figure 4. Hos3 associates with nuclear pore complexes.
(A) Hos3-GFP (green) Nup60-mCherry (red) form partially overlapping clusters during anaphase in nup133Δ cells. This experiment was repeated three times with similar results. (B) Loss of perinuclear Hos3-GFP and its distribution in the nucleoplasm during anaphase in a nup60Δ cell. The fluorescence intensity of Hos3-GFP and Nup49-mCherry (to mark the nuclear periphery) were measured across the daughter cell nucleus in wild type and nup60Δ late anaphase cells after Hos3-GFP nuclear import (yellow lines). Hos3-GFP and Nup49-mCherry profiles closely overlap in wild type whereas Hos3 is distributed throughout the nucleoplasm in nup60Δ. n=10 cells / strain, pooled from three independent experiments. Mean and SEM are shown. (C) Hos3 and Nup60 are associated in mitotic yeast extracts. Hos3-myc was immunoprecipitated in extracts prepared from metaphase-arrested cells (Cdc20 depletion) and from asynchronously growing cultures (log phase). Proteins were detected by western blot using the indicated antibodies. This experiment was repeated three times with similar results. (D) Hos3-FRB-GFP recruitment to NPCs upon rapamycin addition prolongs T1 duration in mother cells. (Left) Association of Hos3-GFP-FRB with nuclear pores (co-localization with Nup49-FKBP-RFP) was observed within 5 minutes of rapamycin addition. (Right) The experiment was repeated in the same cells, but also expressing Whi5-GFP. T1 was delayed in mother cells and was inhibited for the duration of the movie (3 hours) in daughter cells (right). WT (n=40 cells), Hos3-FRB-GFP Nup49-FKBP-mCherry, n=86 cells (-rapamycin), n= 25 cells (+rapamycin). Boxes include 50% of data points, the line represents the median and whiskers extend to maximum and minimum values. Cells were pooled from two independent experiments. In (A-B), time is indicated in minutes; t=0 marks the last frame before nuclear entry into the bud. Scale bars, 2 μm.
Hos3 is required for the enrichment of Whi5 in daughter cell nuclei
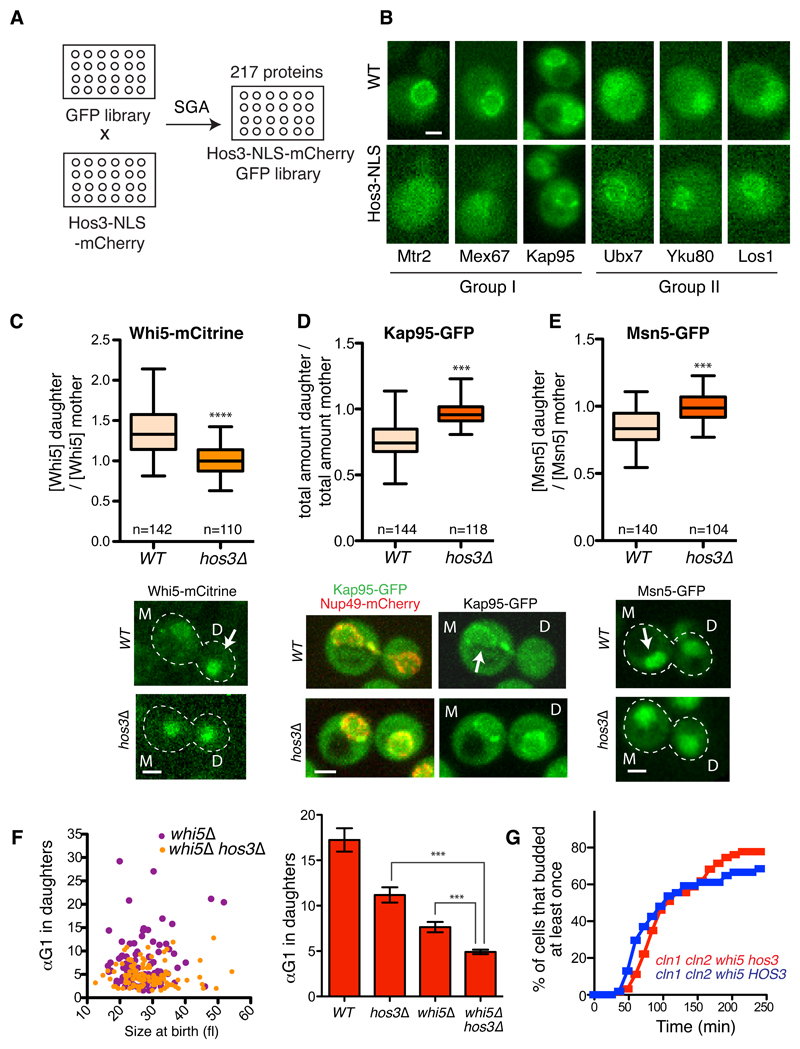
To test if perinuclear Hos3 alters interactions of proteins with NPCs, we examined the localization of 217 nuclear proteins fused to GFP, in wild type and HOS3-NLS cells (Figure 5A-B and Supplementary Table S2). 10 proteins showed altered localization in HOS3-NLS relative to wild type strains; intriguingly, the majority of these are involved in nucleo-cytoplasmic transport. The karyopherins Kap95 and Kap123, the mRNA export regulators Mtr2 and Mex67, and the TREX-2 component Sac3 showed reduced perinuclear localization in Hos3-NLS cells. In contrast, the tRNA export factor Los1, the telomeric Ku complex components Yku70 and Yku80, the UBX domain-containing Ubx7, and the karyopherin Kap114A became enriched in the nucleus or nuclear periphery in the presence of Hos3-NLS (Figure 5B and Supplementary Figure S5A). These observations raised the possibility that Hos3 regulates nucleo-cytoplasmic transport dynamics to delay Start in daughter cells.
Figure 5. Hos3 modulates localization of perinuclear factors and controls the asymmetry of Whi5 and karyopherins.
(A) Construction of a library of nuclear GFP-fusion proteins expressing Hos3 or Hos3-NLS. (B) Examples of protein localization changes induced by Hos3-NLS. Group I proteins are displaced away from the nuclear periphery, whereas group II proteins are enriched at the same location in the presence of Hos3-NLS. These experiments were repeated three times with similar results. (C-E) Hos3 affects asymmetric distribution of Whi5, Kap95 and Msn5. Nuclear fluorescence was measured in mother and daughter cells at the time of birth. Boxes include 50% of data points, the line represents the median and whiskers extend to maximum and minimum values. ***, p < 0.001, two-sided Mann-Whitney test. Exact p values: 10-15 (Whi5), 10-15 (Kap95), 2.8x10-10 (Msn5). Arrows indicate increased protein accumulation in mother or daughter cells. (F) Correlation between αTG1 and cell size at the time of birth (cytokinesis), for mother and daughter cell pairs (left); and αTG1 (mean and SEM) (right) in whi5Δ (n=178 cells) and whi5Δ hos3Δ (204 cells). WT (n=110 cells) and hos3Δ (n=128 cells) from Figure 1 are included for comparison. *** p < 0.001, two-sided Mann-Whitney test. Exact p values: 10-13 (hos3 vs hos3 whi5); 4.53x10-5 (whi5 vs hos3 whi5). Note that TG1 was used to measure Start instead of T1, since the latter is defined by Whi5 nuclear exit. Scale bars, 2 μm. (G) Frequency of budding in cells of the indicated strains, determined by time-lapse microscopy as in Figure 1. Daughter cells; cln1Δcln2Δwhi5Δ (n=63 cells), cln1Δcln2Δwhi5Δhos3Δ (n=54 cells).
The probability of entering the cell cycle is inversely proportional to Whi5 nuclear concentration, and Start is triggered when cell growth dilutes Whi5 below a critical level 27,30. Notably, Whi5 nuclear concentration is higher in daughter than in mother cells, although the reason for this asymmetry has not been elucidated. We therefore determined the nuclear concentration of Whi5 fused to mCitrine or GFP in wild type and hos3Δ cells immediately after cytokinesis. Whi5 nuclear concentration was higher in daughter than in mother cells and this asymmetry was dependent on Hos3 (Figure 5C and Supplementary Figure S5B-C). Moreover, the amount and concentration of the Kap95 importin and the Msn5 exportin, which control Whi5 nuclear levels 40,41, were higher in mother than in daughter nuclei and these asymmetries were dependent on Hos3 (Figure 5D-E and Supplementary Figure S5C-D). Deletion of Hos3 did not affect the asymmetric distribution of NPCs marked with Nup49-mCherry (Supplementary Figure S5E), which are partially retained in mother cells due to a septin-based diffusion barrier 23. These results suggest that Hos3 inhibits cell cycle entry in daughter cells through modulation of Whi5 nucleo-cytoplasmic transport in late anaphase. We propose that this leads to a higher Whi5 concentration in daughter nuclei, and consequently prolongs G1 duration of these cells.
To determine whether Hos3 regulates G1 solely through Whi5 function, we compared the time of budding for whi5Δ and hos3Δ whi5Δ cells. Deletion of WHI5 leads to advanced Start due to derepression of SBF/MBF target genes including the G1 cyclins, Cln1 and Cln2 28,29. Importantly, deletion of HOS3 advances budding in both wild type and whi5Δ cells (Figure 5F and Supplementary Figure S5F). Because hos3Δ whi5Δ cln1Δ cln2Δ cells did not advance budding relative to HOS3 whi5Δ cln1Δ cln2Δ cells (Figure 5G), Hos3 probably acts through both Whi5-dependent and Whi5-independent mechanisms to inhibit expression of G1 cyclins.
Hos3 controls CLN2 expression and subnuclear localization of the CLN2 gene locus
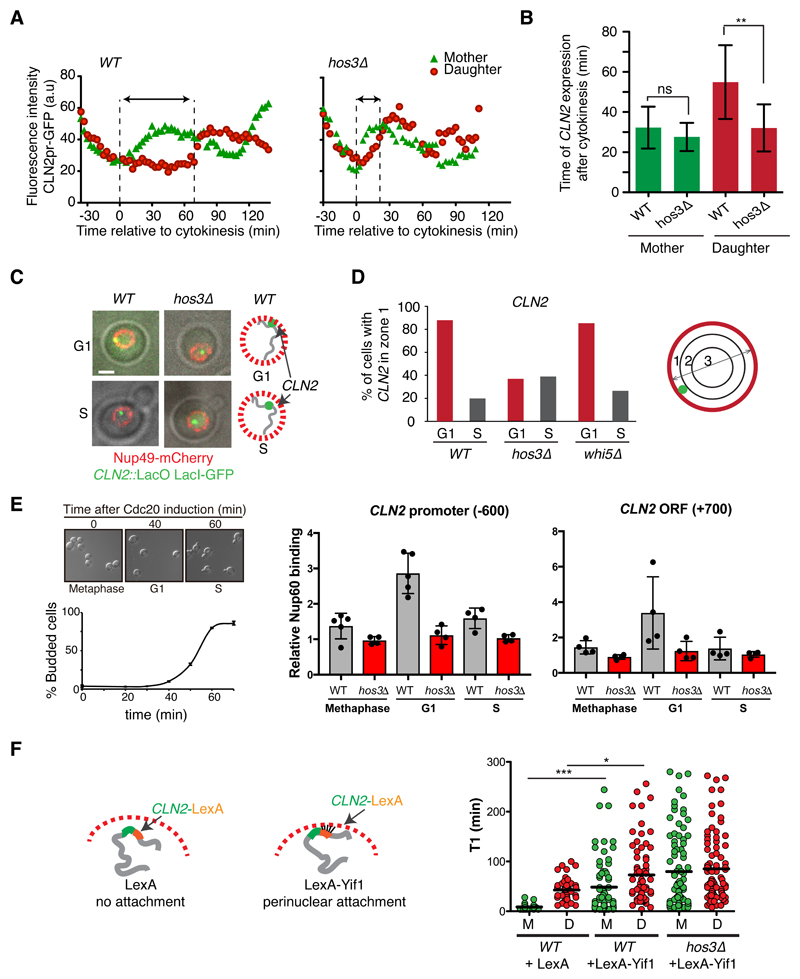
To further investigate how Hos3 delays Start, we examined the relationship between Hos3 and other regulators of cell cycle entry. Control of Start is composed of two modules, each controlled by a different G1 cyclin 33. Cln3-Cdk complexes initiate Start through inactivation of Whi5 in early G1, followed by Cln1/2-Cdk complexes, which complete Whi5 inactivation in a positive feedback loop 42–44 (Supplementary Figure S6A). Hos3 loss did not affect the Ace2 – Ash1 pathway, which delays G1 in daughters by repressing CLN3 25,26 (Supplementary Figure S6B-E). However, our data indicated that Hos3 acts through inhibition of Cln2. We analyzed CLN2 expression by time-lapse microscopy of wild type and hos3Δ cells expressing fast-folding, unstable GFP expressed from the CLN2 locus under the control of the CLN2 promoter 45. This revealed that the time of CLN2 expression relative to cytokinesis was advanced in hos3Δ daughter cells (Figure 6A-B and Supplementary Figure S7A). Hos3 was not detected at the CLN2 promoter (Supplementary Figure S7B) unlike the deacetylase Rpd3, which represses CLN2 in G1 phase 32,46. This suggests that Rpd3 and Hos3 repress CLN2 through different mechanisms.
Figure 6. Hos3-dependent perinuclear anchoring of CLN2 delays Start.
(A) Fluorescence intensity of CLN2pr-GFP in mother and daughter cells relative to cytokinesis (Myo1-mCherry disappearance; t=0) in wild type and hos3Δ. Images were acquired at 3 min intervals. The GFP signal increases shortly after cytokinesis in WT and hos3Δ mother cells. Arrows indicate the interval between cytokinesis and the half-maximum of CLN2pr-GFP intensity in daughter cells. (B) Time (mean and SD) of half-maximum CLN2pr-GFP intensity relative to cytokinesis for WT (n=20 mother-bud pairs) and hos3Δ (n=15 pairs) (**, p < 0.01; ns, non-significant; two-sided Student’s t-test). Exact p value: 2.60x10-3 (daughters). (C) Composite of phase contrast, CLN2::lacO (LacI-GFP, green) and Nup49-mCherry (red) in G1 and S-phase cells of the indicated strains. (D) Subnuclear position of CLN2 in the indicated strains and cell cycle stages, determined by mapping their localization to one of three concentric nuclear zones of equal surface in confocal optical slices. Cell numbers (n), pooled from three independent experiments with similar results: WT: 102 (G1), 92 (S). hos3Δ: 108 (G1), 108 (S). whi5Δ: 108 (G1), 82 (S). (E) (Left) Cultures of MET3:CDC20 and MET3:CDC20 hos3Δ were arrested in metaphase by adding methionine 2 mM (time 0, >95% of cells arrested). Cells were released in fresh medium lacking methionine, at 25 °C. (Right) Binding of Nup60-HA to CLN2 promoter and ORF investigated by Chromatin Inmunoprecipitation (ChIP) assays. After the release from the arrest, Nup60 binding to CLN2 loci was investigated by ChIP assays in G1 (35-40 min) and S phase (60-65 min). Values are the mean and SD derived from n=4 independent experiments; individual data points are displayed. (F) Duration of pre-Start G1 phase (T1) in cells containing LexA recognition sites within the 3’UTR of CLN2, to tether CLN2 to the nuclear periphery in the presence of LexA-Yif1 fusion protein. WT+ LexA (n=92 cell pairs), WT+LexA-Yif1 (n=136), hos3Δ+LexA-Yif1 (n=170). Lines represent the mean (* p < 0.05, *** p < 0.001, two-sided Mann-Whitney test). Exact p values: 9.14x10-10 (WT mothers, LexA vs LexA-Yif1), 4.36x10-2 (WT daughters, LexA vs LexA-Yif1).
The yeast nuclear periphery and NPCs play a role in three-dimensional genome organization and gene expression. Confocal 3D imaging showed that CLN2 labeled with the LacO/LacI-GFP system was adjacent to nuclear pores in 88% of unbudded cells and in 20% of budded cells (Figure 6C-D and Supplementary Figure S7C). Furthermore, CLN2 associated with clustered NPCs in nup133Δ mutant cells during G1 (Supplementary Figure S7D). Strikingly, the G1-specific perinuclear association of CLN2 was independent of WHI5 but required Hos3, as the fraction of G1 cells with perinuclear CLN2 was reduced to less than 40% in hos3Δ cells (Figure 6D and Supplementary Figure S7C). Moreover, the CLN2 locus associated with the nuclear basket component Nup60 specifically during G1 phase and this association was lost in hos3Δ mutants (Figure 6E). Thus, Hos3 promotes the G1-specific association of NPCs with CLN2.
CLN2 is located 60 kb away from a telomere, and Hos3 deletion led to displacement away from the nuclear periphery of subtelomeric TEL12R but not of TEL4R loci, which were perinuclear in the majority of wild type cells during both G1 and S phases (Supplementary Figure S7E). Thus, Hos3 may control the positioning of additional genes besides CLN2. However, transplanting the CLN2 locus away from telomeres did not alter its perinuclear location in G1 or advance Start (Supplementary Figure S7F), suggesting that proximity to telomeres is not required for CLN2 regulation. This is consistent with the finding that placing CLN2 in a plasmid does not alter G1 duration 47.
To determine if Hos3-dependent perinuclear association of CLN2 contributes to the role of Hos3 in Start, we evaluated ectopic recruitment of CLN2 to the nuclear periphery using the LexA-Yif1 system 48. Expression of LexA-Yif1, but not of LexA alone, increased G1 duration in both mother and daughter cells, indicating that enrichment of CLN2 in the nuclear periphery is sufficient to delay Start. This delay was not bypassed in hos3Δ cells, suggesting that Hos3 is responsible for perinuclear targeting of CLN2 but is dispensable for its repression (Figure 6F).
Daughter cell cycle control requires deacetylation of NPC components
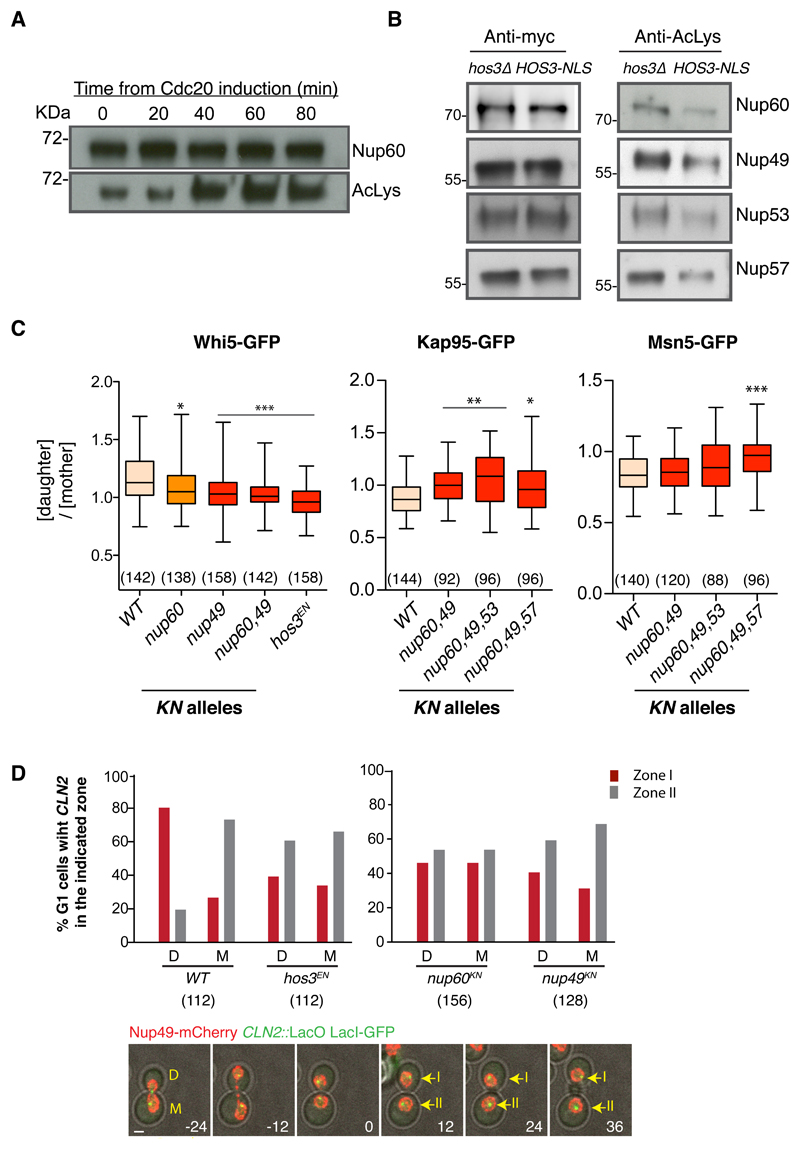
Multiple nucleoporins are acetylated in yeast, although the functional relevance of these modifications remains unclear 49. Nup60 acetylation levels are lower in late mitosis and G1 cells, and increase in S phase, mirroring Hos3 association kinetics with daughter cell NPCs (Figure 7A). Moreover, acetylation levels of Nup60 and of the central channel components Nup49, Nup53 and Nup57 were reduced in HOS3-NLS relative to hos3Δ strains (Figure 7B). To investigate if Hos3 regulates Start through deacetylation of NPC components, we generated point mutants of nucleoporins in which previously identified acetylated lysines 49 were changed to asparagine, whose biophysical properties resemble those of acetylated lysine and thus may mimic constitutive acetylation 50. Consistent with this, lysine-to-asparagine (KN) alleles of Nup60, Nup49, Nup53 and Nup57 did not perturb cell growth or Nup60 perinuclear localization, and their combination conferred resistance to the toxic effects of Hos3-NLS over-expression (Supplementary Figure S8A-C).
Figure 7. Modification of nucleoporins by Hos3 regulates Whi5 transport and CLN2 tethering.
(A) Acetylation of Nup60 in wild type cells released from a metaphase arrest. GAL1pr:CDC20 cells were arrested in glucose media for 3 h. Nup60 protein levels were assessed at the indicated times after Cdc20 induction (galactose addition) with anti-myc antibodies and acetylation levels were assayed with an antibody against acetylated lysines. (B) Acetylation of nucleoporins in Hos3 mutants. Endogenously tagged proteins were immunoprecipitated from HOS3-NLS and hos3Δ extracts and assayed as in panel (A). Experiments in A-B were repeated two times with similar results. (C) Distribution of Whi5 and its transport receptors in wild-type and acetyl-mimic (KN) mutants of the indicated nucleoporins. Nuclear fluorescence was measured in mother and daughter cells at the time of birth as in Fig. 5C. Boxes include 50% of data points, the line represents the median and whiskers extend to maximum and minimum values. The number of cells (n values) analyzed is in brackets; p < 0.05 (*), 0.01 (**) and 0.001 (***), two-sided Mann-Whitney test relative to WT. Exact p values: (Whi5) nup60 = 2.01x10-2, nup49 = 1.09x10-4, nup60 nup49 = 1.22 x10-5; hos3EN = 3.49 x10-10; (Kap95) nup60 nup49 = 1.48 x10-4; nup60 nup49 nup53 = 1.76 x10-5, nup60 nup49 nup57 = 1.58 x10-2; (Msn5) nup60 nup49 nup57 = 2.79 x10-5. (D) (Top) Subnuclear position of CLN2 (as in Fig. 6D) during the first 24 minutes following cytokinesis, determined by time-lapse imaging in daughter (D) and mother cells (M) of the indicated strains. Images were acquired at 12 min intervals (the number of cells analyzed is in brackets; cells were pooled from three independent experiments). (Bottom) GFP, mCherry and phase contrast composite images of CLN2::lacO LacI-GFP NUP49-mCherry mother (M) and daughter (D) cells. Images were acquired at 12 min intervals. Time is indicated in minutes; time 0 corresponds to cytokinesis. Maximal projections are shown. Position of CLN2 relative to the nuclear periphery (zone I or II) was determined in single confocal planes and is indicated in frames after cytokinesis.
We used time-lapse microscopy to determine whether nucleoporin KN alleles affect asymmetric Whi5 nuclear concentration and perinuclear tethering of CLN2 in daughter cells. Asymmetric partitioning of Whi5 and its nuclear transport receptors Kap95 and Msn5 was impaired in cells expressing KN alleles of multiple nucleoporins, to levels similar to cells lacking Hos3 activity (Figure 7C and Supplementary Figure S8D). Moreover, CLN2 perinuclear tethering in early G1 was defective in hos3EN, nup49KN and nup60KN daughters (Figure 7D). Thus, constitutive acetylation of putative Hos3 substrates at the NPC disrupts asymmetric partitioning of nuclear transport receptors and Whi5, and perinuclear tethering of CLN2. Interestingly, nup60Δ and nup60KN cells showed strong impairment of CLN2 tethering but relatively milder defects in Whi5 asymmetry, and the nup57KN mutation was required to disrupt asymmetry of Msn5 but not of Kap95 (Figure 7C-D and Supplementary Figure S8E). Therefore, individual nucleoporins probably make specific contributions to the establishment of asymmetries in nuclear transport and CLN2 tethering.
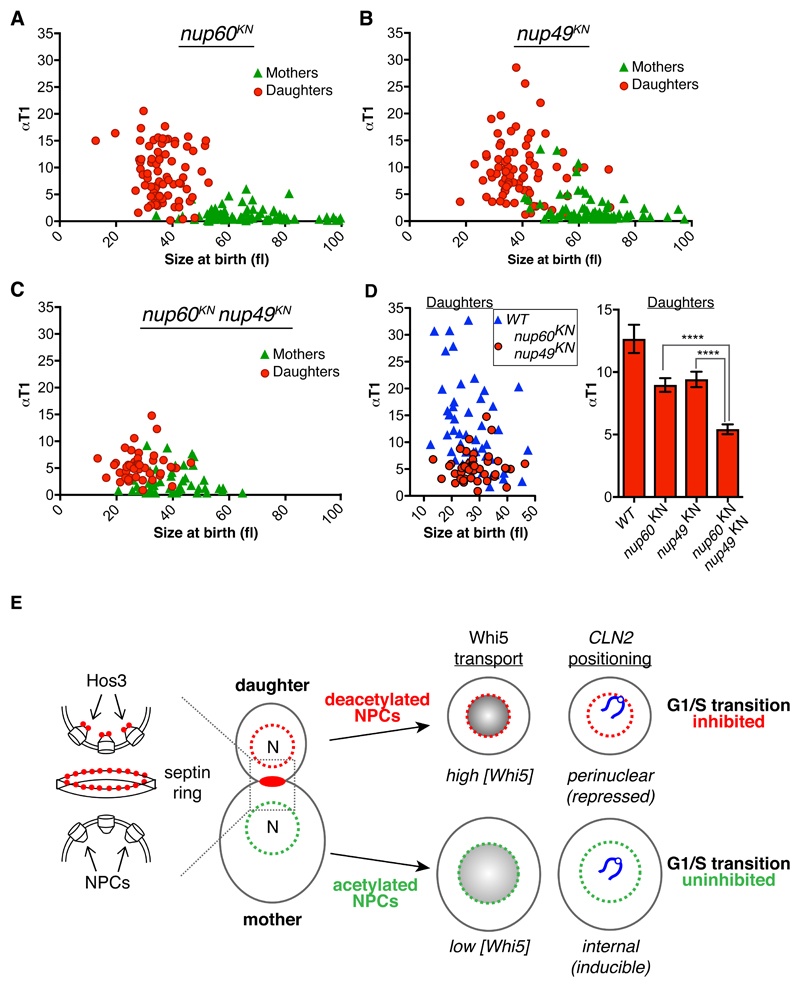
Finally, we examined progression through Start in nucleoporin mutants. Individual mutations in nucleoporins nup60KN and nup49KN slightly advanced cell cycle entry in mutant daughters relative to wild type daughters of similar size (Figure 8A-B). Deletion of NUP60 advanced S phase to a similar level as nup60KN (Supplementary Figure S8F). Remarkably, T1 duration was reduced in nup49KN nup60KN double mutant daughters to levels comparable to hos3 mutants (Figure 8C-D). We conclude that combined deacetylation of nuclear basket and central channel nucleoporins is sufficient to inhibit Start in daughter cells.
Figure 8. Modification of nucleoporins by Hos3 regulates cell cycle entry.
(A-C) Correlation between αT1 and cell size at the time of cytokinesis, for mother and daughter cell pairs in the indicated strains. Mother and daughter cell pairs in nup60KN n= 157, nup49KN n= 156, nup60KN nup49KN n= 91. (D) Correlation between αT1 and cell size at the time of cytokinesis for daughter cells of the indicated strains (left) and αT1 values (mean and SEM) for cells of size at birth 25 – 50 fl for cells shown in (A-C) (****, p < 0.00001; two-sided Mann-Whitney test) (right). Exact p values: nup60KN vs nup60KN nup49KN = 4.10x10-6; nup49KN vs nup60KN nup49KN = 2.53x10-6. (E) Model for Hos3 function in cell cycle entry. Hos3 (red circles) concentrates at the daughter side of the septin ring at bud neck prior to anaphase. Upon passage of the nucleus through the bud neck, Hos3 is imported in a Mtr10-dependent manner and associates with the nuclear basket of NPCs. Hos3 deacetylates NPCs to affect their functions in daughter cells, leading to perinuclear positioning of the silent CLN2 locus and to higher accumulation of the Whi5 transcriptional repressor in daughter nuclei, delaying Start in daughter cells.
Discussion
We have identified a mechanism that establishes differences in NPC acetylation and cell fate between mother and daughter cells in Saccharomyces cerevisiae. During mitosis, the Hos3 deacetylase associates with NPCs in the daughter cell to direct its cell cycle program. This stands in contrast to other known fate-determinant partition mechanisms, which rely on compartmentalization and polarization of the cell cortex. Because perinuclear enrichment of Hos3 required its association to the daughter side of the bud neck and nuclear migration into the daughter cell, we speculate that Hos3 association with NPCs is triggered by the close proximity between the daughter side of the septin ring and NPCs traversing the bud neck. This remains to be directly demonstrated.
Our data suggest that perinuclear Hos3 delays Start through mechanisms independent from Ace2, which represses Cln3 transcription in daughter cells 25,26, and from Rpd3, a type I HDAC that repress the CLN2 promoter 31. Instead, Hos3 acts by establishing NPC deacetylation in daughter cells, which inhibits the G1/S transition through two mechanisms. Firstly, NPC deacetylation mediates enrichment of the Start inhibitor Whi5 in daughter nuclei 27,30 and counters the accumulation of karyopherins Kap95 and Msn5 in daughters, which control nuclear levels of Whi5 40,41. This provides a mechanism by which deacetylation of NPCs can control Start in daughter cells. Secondly, NPC deacetylation confines the CLN2 gene locus to the nuclear periphery in G1 daughter cells, where it associates with the nuclear pore basket component Nup60, and which also delays Start. There is accumulating evidence that nuclear pores bind silent genomic regions or poised genes 51–54, and cell cycle regulator genes have been previously found at nuclear pores by genome-wide studies 55. Thus, Hos3 may target CLN2 to NPCs to mediate its repression, dampening the positive feedback on Cdk1 and further contributing to delaying Start in daughter cells. Interestingly, acetylation by the SAGA complex plays a major role in the recruitment of chromatin to NPCs and the nuclear periphery 56,57. Whether Hos3 counteracts SAGA to control Start in daughter cells remains to be determined.
In summary, we propose that Hos3 inhibits cell cycle entry in daughter cells through deacetylation of the nuclear basket (which predominantly modulates CLN2 tethering to NPCs) and of the central channel, which affects both CLN2 positioning and Whi5 partitioning (Figure 8E). Hos3 likely controls Whi5 nuclear levels and CLN2 gene positioning through independent mechanisms. Supporting this possibility, Whi5 is not required for CLN2 perinuclear anchoring in G1, and nup60 mutations perturb CLN2 anchoring but not Whi5 asymmetry. Hos3 may control the nucleo-cytoplasmic shuttling of other factors besides Whi5, and sub-nuclear positioning of other genes besides CLN2, which may contribute to the daughter-specific Start delay.
These data reveal that modulation of NPC functions plays a key role in the control of cell cycle progression, and establish deacetylation as a mechanism to regulate NPCs in different cell cycle stages and cell types. The segregation mechanism of Hos3 may be specific to budding yeasts, which have a specialized type of nuclear division. However, differential expression or localization of Hos3-like deacetylases might modulate NPC acetylation in other organisms in a cell cycle- or cell type-specific manner. Interestingly, the human homologues of yeast Nup60 and Nup49 (Nup153 and Nup58, respectively) are also acetylated 58. Moreover, an interplay between NPCs and tissue-specific class-II HDACs has been linked to the regulation of gene expression and positioning in cultured animal cells 59–61 and Nup153 is involved in ES cell pluripotency through gene silencing 62. Whether any of these processes is controlled by acetylation of NPCs is not known. Thus, our findings open the possibility that nucleoporin deacetylation might be a conserved mechanism to regulate NPC functions and cell fate determination.
Online Methods
Strains, cell growth
S. cerevisiae strains are derivatives of S288c except when noted (strains; Table S3). Gene deletions and insertions were generated by standard one-step PCR-based methods 63 (oligos; Table S4). An oligonucleotide encoding the SV40 NLS peptide PKKKRKVEDP was used to amplify eGFP from pYM28 or pYM27 by PCR, and yeast cells were transformed with the resulting DNA fragment to generate Hos3-NLS-GFP. Site-directed mutagenesis (QuickChange, Agilent Technologies) as used to generate the HOS3pr-HOS3(H196E, D231N)-GFP mutant in an integrative plasmid, then used to insert the mutant gene at the HIS3 locus (pRS403/NheI) in a hos3Δ background. KN mutants were generated using CRISPR/Cas9 to replace acetylated lysines [identified in 49] with asparagine, and are listed in Table S5. Positive clones were generated and verified by sequencing as described 64. At least two independent clones per strain were analyzed, with identical results.
The LacO array was inserted 87 bp downstream of CLN2 gene using a two-step PCR based method 65. GFP-LacI was integrated at HIS3 using NheI-digested pAFS135 (Straight et al., 1996). For CLN2 gene insertion and visualization at different genomic locations, the parB-mCherry/Int1 method was used 68. Int1::NAT sequence was first inserted 87 bp downstream of CLN2 gene using a PCR-based method, amplified from plasmid pFG2, pANCH1-NAT. The pCM189-ParB1-mCh::URA plasmid was than transformed in order to visualize the CLN2 gene locus. To place CLN2-INT1 at different chromosome locations, the CLN2 ORF including 1106 bp upstream promoter and downstream 400 bp UTR region was amplified using primers (Table S4); transformed in cln2Δ strain and confirmed by PCR.
The 8XLexA promoter array along with the URA3 cassette were amplified by PCR from pSH18-34 (Invitrogen) and inserted at the 3’ of CLN2 (87 bp downstream of stop codon). LexA or LexA fused to full-length Yif1 under the control of the ADH1 promoter were expressed from pRS425-based plasmids.
For G1 arrest, cells were grown in YPDA (yeast extract, peptone, dextrose, and adenine) medium to log phase, synchronized with 15 μg/ml alpha-factor (Sigma-Aldrich) for 2 h and released in fresh YPDA medium at 30°C. For induction of the GAL1,10 promoter, cells were grown overnight in YPRaf medium, washed, shifted to YPGal (yeast extract, peptone, galactose) medium for 4 hours, and then plated on YPGal plates. For arrest in metaphase, MET3pr-CDC20 cells were grown in synthetic minimal medium lacking methionine at 25°C until mid-log phase and the media was supplemented with 2 mM methionine during 2 h 30 min. For release, cells were washed twice and resuspended in minimal medium lacking methionine.
For tethering of Hos3 to nuclear pores, we used inducible dimerization of FK506-binding protein (FKBP) and FKBP-rapamycin binding (FRB) domain to tag it at C-terminus of Hos3 (Hos3-GFP-FRB; bait) and Nup49 (Nup49-RFP-FKBP; anchor) as described 39. The final strain harbors the tor1-1 mutation and lacks the endogenous FPR1 gene rendering growth insensitive to rapamycin. Cells were incubated with 25 μM rapamycin throughout the imaging experiment. Association of Hos3-GFP-FRB with nuclear pores was observed within 5 minutes of rapamycin addition.
Fluorescence microscopy
For time-lapse microscopy, cells were grown overnight in 100 ml flasks containing 25 ml minimal medium at 30°C, then diluted to O.D600 = 0.1-0.3 in fresh medium, grown for 4 hours to mid-log phase and plated in minimal synthetic medium on concanavalin A–coated (Sigma-Aldrich) Lab-Tek chambers (Thermo Fisher Scientific). Time-lapse imaging was performed using a spinning-disk confocal microscope (Revolution XD; Andor Technology) with a Plan Apochromat 100x, 1.45 NA objective equipped with a dual-mode electron-modifying charge-coupled device camera (iXon 897 E Dual Mode EM-CCD camera; Andor Technology), temperature-controlled microscopy chamber, a z-stepper and an automated stage. Images were analyzed on 2D maximum projections. Maximum projections are shown throughout, except in Fig. 2A, where one confocal section is shown for clarity. For epifluorescence microscopy, a Leica AF 6000 wide-field microscope with an Andor DU-885K-CSO-#VP camera was used.
Quantification of GFP fusion protein abundance was determined in background-subtracted 2D sum projections of whole-cell Z-stacks, with the nuclear area defined by Nup49-mCherry or by GFP fluorescence.
The position of GFP-LacI spots was mapped in confocal sections by assigning the GFP spot to one of three nuclear zones of equal area, as described 66. To map the position of GFP-LacI spots by confocal microscopy, the nuclear periphery was marked with Nup49-mCherry; 200 ms (GFP) and 250 ms (mCherry) exposure time was used to acquire 26 images (Z-step: 210 nm) to reconstruct the whole 3D nuclear volume. The distance (μm) of brightest intensity spot of GFP-LacI position from nuclear periphery was determined using ImageJ 1.48u software. Nuclear diameter was determined by measuring the nuclear long axis. Nuclear volume was divided into 3 zones as described 66. Briefly, by dividing the GFP spot to periphery distance to nuclear diameter, each spot falls into one of three consecutive zones of equal area. The most peripheral zone I has a width of 0.184 × the nuclear radius (r), zone II = 0.184 × r to 0.422 × r, and zone III has a radius of 0.422 × r. More than 100 nuclei were measured for each condition.
Analysis of G1 duration was performed essentially as described 33 except that cell size was measured on DIC stacks using ImageJ and a customized version of the BudJ3D plug-in 67 to calculate cell volume using bright-field images. Growth rates were determined by linear regression of volumetric data over time. Time-lapse series of 4 μm stacks spaced 0.2-0.3 μm were acquired every 2-4 minutes using iQ Live Cell Imaging software (Andor Technology).
For the screen in Figure 5A-B, at least 200 cells were assessed from 4 different fields for each candidate; positive hits were individually checked twice to verify the GFP localization phenotypes.
Immunolocalisation and Electron Microscopy
Cells expressing Hos3-NLS-6HA were grown until log phase, arrested in G1 with 15 μg/ml alpha-factor (Sigma) for 2 h and released in fresh YPDA medium at 30 °C. Cells were collected, filtered into a paste and cryoimmobilised by high-pressure freezing with hpm010 (ABRA Fluid). Freeze substitution was done using a EM-AFS2 device (Leica Microsystems). The freeze substitution solution used contained 0.1% uranyl acetate and 1 % water dissolved in anhydrous acetone and the samples were substituted at -90 °C for 48 h. The temperature was then increased at a rate of 5 °C/hour to -45 ºC followed by 5 h incubation at -45 °C. Samples were rinsed with acetone for three times 10 min followed by lowicryl HM20 (Polysciences) infiltration at -25 °C with 25% lowicryl in acetone 2 h, 50% for 2 h and 75% for 2h. Samples were then left in 100% lowicryl for three times 10 h before onset of polymerization. UV polymerization was applied for 48 h at -25°C and the temperature was increased to 20 °C (5 °C/h). The samples were left exposed to UV at room temperature for 48 h. Thin sections (70 nm) were cut with a Leica Ultra-cut UCT microtome and collected on Formvar-coated, palladium-copper slot grids. Immunogold labelling was done by floating grids on drops of blocking buffer consisting of 0.8% BSA and 0.1% fish skin gelatine in PBS for 30 min, followed by incubation on drops of Anti-HA antibody (Roche) diluted 1:50 in blocking buffer for 30 min, followed by incubation in rabbit anti rat antibody (Rockland) and subsequently 10 nm gold-conjugated Protein A (CMC University Medical Center Utrecht, Netherlands) for 20 min. Grids were rinsed floating on 5 drops of PBS between each incubation step. Labeling was followed by an extensive rinse of 1 h on 10 drops of PBS. Sections were then fixed in 1% glutaraldehyde in PBS for 5 min, rinsed in 10 drops of water and post-stained using uranyl acetate and lead citrate. Sections were viewed using a CM120 biotwin electron microscope (FEI) operating at 100 kV. Digital acquisitions were made with a Keen View CCD camera (Soft Imaging System).
Co-immunoprecipitation
Cells were grown to log phase or synchronized by Cdc20 depletion, lysed in bead beater with glass beads, and extracts incubated with anti-Myc antibody (05-724; Millipore) coupled to Dynabeads M-280 sheep anti-mouse IgG (11201D) to immuno-precipitate Hos3-myc. For western blotting, anti-Myc (A-14:sc-789, Santa Cruz), anti-GFP (Ref 11814460001, Roche) and anti-acetyl Lys (Cell Signaling, Ref 9681) antibodies were used. All antibodies were used at 1:500 dilution except anti-GFP, which was used at 1:1000.
Chromatin Immunoprecipitation assays
Approximately 5 x 108 cells (or 100 ml culture OD600= 0.5) were cross-linked for 15 min in formaldehyde 1% and quenched for 5 min in glycine 125 mM. Cells were washed and resuspended in 300 μL of lysis buffer (50mM HEPES-KOH pH 7.9, 40mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxicholate, 1mM PMSF, 1 mM benzamidine and Complete Mini protease inhibitor, Roche), and lysed by vortexing with glass beads for 30 min at 4 °C. Lysis buffer was supplemented to a final volume of 600 μL, and chromatin was then fragmented by sonication and the sample was centrifuged at 13,400 x g for 15 min. 20 μL from the supernatant was collected as a control of whole-cell extract (input) and the remaining was incubated with orbital rotation for 2 h at 4 °C with Dynabeads Protein G (Invitrogen) previously bound to 2 μg anti-HA (F-7) antibody (Santa Cruz Biotechnology Inc. Cat. No: SC-7392). Beads were then washed 4 times in PBS (150mM NaCl, 40mM Na2HPO4, 10mM NaH2PO4) containing 0.02% (v/v) Tween 20. Elution of bound protein was carried out twice with 40 μL of 50mM Tris-HCl pH 8.0, 10mM EDTA, 1% (w/v) SDS, by heating at 65 °C for 10 min. Cross-linking was reverted by overnight incubation at 65 °C with shaking. The eluted sample was digested for 90 min at 37 °C with 0.33 mg mL−1 proteinase K and DNA was purified with the High Pure PCR product purification kit (Roche Diagnostics). Co-immunoprecipitated DNA was analyzed in triplicate by Real Time PCR in a LightCycler® 480 Instrument II using the LightCycler® 480 SYBR® Green I Master (Roche Diagnostics). DNA analyzed are fragments from CLN2 promoter and ORF and an intergenic region as a control. Nup60 binding values indicates the specific enrichment of the investigated promoter fragments in the immunoprecipitated sample compared to the whole-cell extract (input) using the intergenic region as a control, calculated with the ΔΔCT method. Values are relative to the no tag control strain (value of 1 equivalent to no specific enrichment).
Statistical methods and reproducibility
To assess statistical significance, Student's t-test was used on normally distributed samples, and the Mann-Whitney test was used on non-Gaussian sample distributions. All experiments, including microscopy and biochemical experiments, were repeated at least twice with similar results.
Supplementary Material
Acknowledgments
We thank Martí Aldea, Jan Skotheim and all members of the Mendoza lab for discussions and sharing reagents, and Yves Barral, Lucas Carey, Ben Lehner and Life Science Editors for comments and manuscript editing. We are grateful to Frederick Cross (Rockefeller University), Josep Clotet (Universtat Internacional de Catalunya), Pedro Carvalho, Vivek Malhotra (both CRG, Barcelona), Susan Gasser (FMI, Basel), Karim Mekhail (University of Toronto) and Andrew Murray (Harvard University) for reagents, the CRG Advanced Light Microscopy Unit (Timo Zimmermann, Raquel García), and Yannick Schwab (EMBL, Heidelberg) for electron microscopy. This study was supported by the European Research Council (ERC) Starting Grant 2010-St-20091118 to M.M., the Spanish Ministry of Economy and Competitiveness, ‘Centro de Excelencia Severo Ochoa 2013-2017’, SEV-2012-0208 to the CRG, and the grant ANR-10-LABX-0030-INRT, a French State fund managed by the Agence Nationale de la Recherche under the frame program Investissements d’Avenir ANR-10-IDEX-0002-02 to the IGBMC. M.G.-A. is a recipient of a Postdoctoral Fellowship APOSTD/2017/094 from the Generalitat Valenciana.
Footnotes
Data availability: All data are available from the authors on reasonable request, and/or are included within the manuscript.
Conflict of interests statement: The authors declare no conflict of interests.
Author Contributions: Conceptualization, A.K. and M.M.; Methodology, A.K., A.D., M.G.-A., Z.S., I.M., P.S., and C.F.; Investigation, A.K., A.D., M.G.-A., Z.S., I.M., P.S., T.S. and C.F.; Writing – Original Draft, M.M.; Writing – Review & Editing, A.K., M.G.-A. and M.M.; Funding Acquisition, M.B. and M.M.; Supervision, M.M.
References
- 1.Knoblich JA. Asymmetric cell division: recent developments and their implications for tumour biology. Nat Rev Mol Cell Biol. 2010;11:849–860. doi: 10.1038/nrm3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li R. The Art of Choreographing Asymmetric Cell Division. Developmental Cell. 2013;25:439–450. doi: 10.1016/j.devcel.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 3.Terry LJ, Shows EB, Wente SR. Crossing the Nuclear Envelope: Hierarchical Regulation of Nucleocytoplasmic Transport. Science. 2007;318:1412–1416. doi: 10.1126/science.1142204. [DOI] [PubMed] [Google Scholar]
- 4.Knockenhauer KE, Schwartz TU. The Nuclear Pore Complexas a Flexible and Dynamic Gate. Cell. 2016;164:1162–1171. doi: 10.1016/j.cell.2016.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ibarra A, Hetzer MW. Nuclear pore proteins and the control of genome functions. Genes & Development. 2015;29:337–349. doi: 10.1101/gad.256495.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang J, et al. Gating pluripotency via nuclear pores. Trends in Molecular Medicine. 2013:1–7. doi: 10.1016/j.molmed.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 7.Capelson M, Doucet C, Hetzer MW. Nuclear pore complexes: guardians of the nuclear genome. Cold Spring Harb Symp Quant Biol. 2010;75:585–597. doi: 10.1101/sqb.2010.75.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat Rev Genet. 2007;8:507–517. doi: 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- 9.Zuleger N, Robson MI, Schirmer EC. The nuclear envelope as a chromatin organizer. Nucleus. 2014;2:339–349. doi: 10.4161/nucl.2.5.17846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mattout A, Cabianca DS, Gasser SM. Chromatin states and nuclear organization in development--a view from the nuclear lamina. Genome Biol. 2015;16:174. doi: 10.1186/s13059-015-0747-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andrulis ED, Neiman AM, Zappulla DC, Sternglanz R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature. 1998;394:592–595. doi: 10.1038/29100. [DOI] [PubMed] [Google Scholar]
- 12.Pickersgill H, et al. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet. 2006;38:1005–1014. doi: 10.1038/ng1852. [DOI] [PubMed] [Google Scholar]
- 13.Guelen L, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 14.Green EM, Jiang Y, Joyner R, Weis K. A negative feedback loop at the nuclear periphery regulates GAL gene expression. Mol Biol Cell. 2012;23:1367–1375. doi: 10.1091/mbc.E11-06-0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kosak ST, et al. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- 16.Zink D, et al. Transcription-dependent spatial arrangements of CFTR and adjacent genes in human cell nuclei. The Journal of Cell Biology. 2004;166:815–825. doi: 10.1083/jcb.200404107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D'Angelo MA, Gomez-Cavazos JS, Mei A, Lackner DH, Hetzer MW. A Change in Nuclear Pore Complex Composition Regulates Cell Differentiation. Developmental Cell. 2012;22:446–458. doi: 10.1016/j.devcel.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Solovei I, et al. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell. 2013;152:584–598. doi: 10.1016/j.cell.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 19.Liang Y, Franks TM, Marchetto MC, Gage FH, Hetzer MW. Dynamic association of NUP98 with the human genome. PLoS Genet. 2013;9:e1003308. doi: 10.1371/journal.pgen.1003308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korfali N, et al. The nuclear envelope proteome differs notably between tissues. Nucleus. 2014;3:552–564. doi: 10.4161/nucl.22257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartwell LH, Unger MW. Unequal division in Saccharomyces cerevisiae and its implications for the control of cell division. The Journal of Cell Biology. 1977;75:422–435. doi: 10.1083/jcb.75.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Colman-Lerner A, Chin TE, Brent R. Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell. 2001;107:739–750. doi: 10.1016/s0092-8674(01)00596-7. [DOI] [PubMed] [Google Scholar]
- 23.Shcheprova Z, Baldi S, Frei SB, Gonnet G, Barral Y. A mechanism for asymmetric segregation of age during yeast budding. Nature. 2008;454:728–734. doi: 10.1038/nature07212. [DOI] [PubMed] [Google Scholar]
- 24.Turner JJ, Ewald JC, Skotheim JM. Cell Size Control in Yeast. Current Biology. 2012;22:R350–R359. doi: 10.1016/j.cub.2012.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laabs TL, et al. ACE2 is required for daughter cell-specific G1 delay in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2003;100:10275–10280. doi: 10.1073/pnas.1833999100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Di Talia S, et al. Daughter-specific transcription factors regulate cell size control in budding yeast. PLoS Biol. 2009;7:e1000221. doi: 10.1371/journal.pbio.1000221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmoller KM, Turner JJ, Kõivomägi M, Skotheim JM. Dilution of the cell cycle inhibitor Whi5 controls budding-yeast cell size. Nature. 2015 doi: 10.1038/nature14908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Bruin RAM, Mcdonald WH, Kalashnikova TI, Yates J, Wittenberg C. Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell. 2004;117:887–898. doi: 10.1016/j.cell.2004.05.025. [DOI] [PubMed] [Google Scholar]
- 29.Costanzo M, et al. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell. 2004;117:899–913. doi: 10.1016/j.cell.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 30.Liu X, et al. Reliable cell cycle commitment in budding yeast is ensured by signal integration. Elife. 2015;4 doi: 10.7554/eLife.03977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang D, et al. Dual Regulation by Pairs of Cyclin-Dependent Protein Kinases and Histone Deacetylases Controls G1 Transcription in Budding Yeast. PLoS Biol. 2009;7:e1000188. doi: 10.1371/journal.pbio.1000188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H, Carey LB, Cai Y, Wijnen H, Futcher B. Recruitment of Cln3 Cyclin to Promoters Controls Cell Cycle Entry via Histone Deacetylase and Other Targets. PLoS Biol. 2009;7:e1000189. doi: 10.1371/journal.pbio.1000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Talia S, Skotheim JM, Bean JM, Siggia ED, Cross FR. The effects of molecular noise and size control on variability in the budding yeast cell cycle. Nature. 2007;448:947–951. doi: 10.1038/nature06072. [DOI] [PubMed] [Google Scholar]
- 34.Wang M, Collins RN. A lysine deacetylase Hos3 is targeted to the bud neck and involved in the spindle position checkpoint. Mol Biol Cell. 2014;25:2720–2734. doi: 10.1091/mbc.E13-10-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirchenbauer M, Liakopoulos D. An auxiliary, membrane-based mechanism for nuclear migration in budding yeast. Mol Biol Cell. 2013;24:1434–1443. doi: 10.1091/mbc.E12-08-0602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 37.Pemberton LF, Rout MP, Blobel G. Disruption of the nucleoporin gene NUP133 results in clustering of nuclear pore complexes. Proc Natl Acad Sci USA. 1995;92:1187–1191. doi: 10.1073/pnas.92.4.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rout MP, et al. The yeast nuclear pore complex: composition, architecture, and transport mechanism. The Journal of Cell Biology. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gallego O, et al. Detection and characterization of protein interactions in vivo by a simple live-cell imaging method. PLoS ONE. 2013;8:e62195. doi: 10.1371/journal.pone.0062195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kosugi S, Hasebe M, Tomita M, Yanagawa H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proceedings of the National Academy of Sciences. 2009;106:10171–10176. doi: 10.1073/pnas.0900604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taberner FJ, Quilis I, Igual JC. Spatial regulation of the start repressor Whi5. Cell Cycle. 2009;8:3010–3018. [PubMed] [Google Scholar]
- 42.Cross FR, Tinkelenberg AH. A potential positive feedback loop controlling CLN1 and CLN2 gene expression at the start of the yeast cell cycle. Cell. 1991;65:875–883. doi: 10.1016/0092-8674(91)90394-e. [DOI] [PubMed] [Google Scholar]
- 43.Dirick L, Nasmyth K. Positive feedback in the activation of G1 cyclins in yeast. Nature. 1991;351:754–757. doi: 10.1038/351754a0. [DOI] [PubMed] [Google Scholar]
- 44.Skotheim JM, Di Talia S, Siggia ED, Cross FR. Positive feedback of G1 cyclins ensures coherent cell cycle entry. Nature. 2008;454:291–296. doi: 10.1038/nature07118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bean JM, Siggia ED, Cross FR. Coherence and timing of cell cycle start examined at single-cell resolution. Mol Cell. 2006;21:3–14. doi: 10.1016/j.molcel.2005.10.035. [DOI] [PubMed] [Google Scholar]
- 46.Takahata S, Yu Y, Stillman DJ. The E2F functional analogue SBF recruits the Rpd3(L) HDAC, via Whi5 and Stb1, and the FACT chromatin reorganizer, to yeast G1 cyclin promoters. EMBO J. 2009;28:3378–3389. doi: 10.1038/emboj.2009.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eser U, et al. Form and function of topologically associating genomic domains in budding yeast. Proceedings of the National Academy of Sciences. 2017 doi: 10.1073/pnas.1612256114. 201612256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schober H, Ferreira H, Kalck V, Gehlen LR, Gasser SM. Yeast telomerase and the SUN domain protein Mps3 anchor telomeres and repress subtelomeric recombination. Genes & Development. 2009;23:928–938. doi: 10.1101/gad.1787509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Henriksen P, et al. Proteome-wide Analysis of Lysine Acetylation Suggests its Broad Regulatory Scope in Saccharomyces cerevisiae. Molecular & Cellular Proteomics. 2012;11:1510–1522. doi: 10.1074/mcp.M112.017251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ben-Shahar TR, et al. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science. 2008;321:563–566. doi: 10.1126/science.1157774. [DOI] [PubMed] [Google Scholar]
- 51.Casolari JM, et al. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–439. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- 52.Woolcock KJ, et al. RNAi keeps Atf1-bound stress response genes in check at nuclear pores. Genes & Development. 2012;26:683–692. doi: 10.1101/gad.186866.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van de Vosse DW, et al. A role for the nucleoporin Nup170p in chromatin structure and gene silencing. Cell. 2013;152:969–983. doi: 10.1016/j.cell.2013.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pascual-Garcia P, et al. Metazoan Nuclear Pores Provide a Scaffold for Poised Genes and Mediate Induced Enhancer-Promoter Contacts. Mol Cell. 2017;66:63–76.e6. doi: 10.1016/j.molcel.2017.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kalverda B, Pickersgill H, Shloma VV, Fornerod M. Nucleoporins Directly Stimulate Expression of Developmental and Cell-Cycle Genes Inside the Nucleoplasm. Cell. 2010;140:360–371. doi: 10.1016/j.cell.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 56.Denoth-Lippuner A, Krzyzanowski MK, Stober C, Barral Y. Role of SAGA in the asymmetric segregation of DNA circles during yeast ageing. Elife. 2014;3 doi: 10.7554/eLife.03790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dultz E, et al. Global reorganization of budding yeast chromosome conformation in different physiological conditions. The Journal of Cell Biology. 2016;212:321–334. doi: 10.1083/jcb.201507069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Choudhary C, et al. Lysine Acetylation Targets Protein Complexes and Co-Regulates Major Cellular Functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 59.Brown CR, Kennedy CJ, Delmar VA, Forbes DJ, Silver PA. Global histone acetylation induces functional genomic reorganization at mammalian nuclear pore complexes. Genes & Development. 2008;22:627–639. doi: 10.1101/gad.1632708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kehat I, Accornero F, Aronow BJ, Molkentin JD. Modulation of chromatin position and gene expression by HDAC4 interaction with nucleoporins. The Journal of Cell Biology. 2011;193:21–29. doi: 10.1083/jcb.201101046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zullo JM, et al. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell. 2012;149:1474–1487. doi: 10.1016/j.cell.2012.04.035. [DOI] [PubMed] [Google Scholar]
- 62.Jacinto FV, Benner C, Hetzer MW. The nucleoporin Nup153 regulates embryonic stem cell pluripotency through gene silencing. Genes & Development. 2015;29:1224–1238. doi: 10.1101/gad.260919.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Janke C, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 64.Laughery MF, et al. New vectors for simple and streamlined CRISPR-Cas9 genome editing in Saccharomyces cerevisiae. Yeast. 2015;32:711–720. doi: 10.1002/yea.3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rohner S, Gasser SM, Meister P. Modules for cloning-free chromatin tagging inSaccharomyces cerevisae. Yeast. 2008;25:235–239. doi: 10.1002/yea.1580. [DOI] [PubMed] [Google Scholar]
- 66.Hediger F, Neumann FR, Van Houwe G, Dubrana K, Gasser SM. Live imaging of telomeres: yKu and Sir proteins define redundant telomere-anchoring pathways in yeast. Current Biology. 2002;12:2076–2089. doi: 10.1016/s0960-9822(02)01338-6. [DOI] [PubMed] [Google Scholar]
- 67.Ferrezuelo F, et al. The critical size is set at a single-cell level by growth rate to attain homeostasis and adaptation. Nat Commun. 2012;3:1012. doi: 10.1038/ncomms2015. [DOI] [PubMed] [Google Scholar]
- 68.Saad H, et al. DNA dynamics during early double-strand break processing revealed by non-intrusive imaging of living cells. PLoS Genet. 2014;10:e1004187. doi: 10.1371/journal.pgen.1004187. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.