Abstract
The chaperonin GroEL is a 800 kDa nanomachine comprising two heptameric rings, each of which encloses a large cavity or folding chamber. The GroEL cycle involves ATP-dependent capping of the cavity by the cochaperone GroES to create a nanocage in which a single protein molecule can fold. We investigate how protein substrates sample the cavity prior to encapsulation by GroES using paramagnetic relaxation enhancement to detect transient, sparsely populated interactions between apo GroEL, paramagnetically labeled at several sites within the cavity, and three variants of an SH3 protein domain (the fully native wild type, a triple mutant that exchanges between a folded state and an excited folding intermediate, and a stable folding intermediate mimetic). We show that the substrate not only interacts with the hydrophobic inner rim of GroEL at the mouth of the cavity but also penetrates deep within the cavity, transiently contacting the disordered C-terminal tail, and, in the case of the folding intermediate mimetic, the base as well. Transient interactions with the C-terminal tail may facilitate substrate capture and retention prior to encapsulation.
Graphical Abstract
GroEL is a member of the chaperonins, a class of conserved protein nanomachines that facilitate protein folding and protect against misfolding and aggregation.1–3 GroEL is a barrel-shaped ~800 kDa supramolecular complex of two stacked homoheptameric rings, each of which encloses a large cavity accessible to protein substrates.4 Crystallography and cryo-electron microscopy (EM) of GroEL in a variety of states reveal a complex cycle of coupled conformational changes associated with ATP-dependent capping of the cavity by the cochaperone GroES, thereby creating a nanocage for single protein molecules to fold.5 Various studies, including cryo-EM,6,7 have shown that non-native substrates bind to the apical domain of uncapped GroEL, close to the mouth of the cavity, but following encapsulation by GroES are then located either free within the cavity7 or in contact with the cavity base and the C-terminal tail.8 The latter is 23 residues long, intrinsically disordered (i.e., invisible in X-ray electron density maps),4 and rich in Met/Gly. While the tail has been shown to have a significant impact on protein folding, it remains unclear whether the tail is actively required to facilitate substrate incorporation, as well as unfolding of non-natively folded substrates,9 or simply acts passively to ensure retention of substrate by blocking the pore at the bottom of the GroEL chamber.10
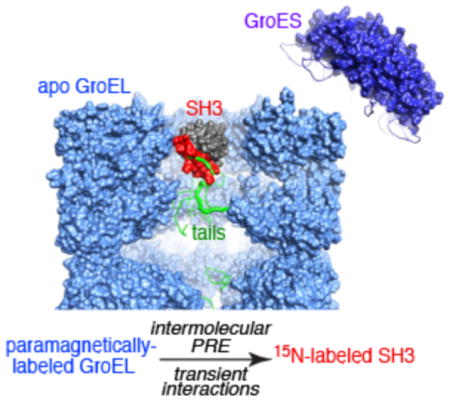
GroES encapsulation is not an absolute requirement for GroEL function.11–13 Recently, using relaxation-based methods, we showed that apo GroEL exhibits unfoldase/foldase activity, stabilizing a folding intermediate of the SH3 domain relative to both folded and unfolded states and accelerating the interconversion between folded and intermediate states.14,15 Here we elucidate the extent to which protein substrates explore the cavity of apo GroEL using intermolecular paramagnetic relaxation enhancement (PRE) measurements16 to identify transient, short-lived contacts between various paramagnetically labeled sites on GroEL and several variants of the β-rich Fyn SH3 domain as a model substrate.
In the fast and intermediate exchange regimes, intermolecular PREs in the GroEL-bound, NMR-invisible, “dark” states of the SH3 variants are transferred by exchange to the NMR visible spectrum of free SH3 where they are easily observed.16 Because the PRE is proportional to <r−6> of the distance between the paramagnetic label and the nucleus of interest, and the magnetic moment of the unpaired electron on the paramagnetic label is large, exchanging complexes with short proton–paramagnetic label contacts can be detected at populations as low as 0.5 to 1%.16 Thus the intermolecular PRE provides an exquisitely sensitive method for studying sparsely populated, transient GroEL–SH3 binding events.
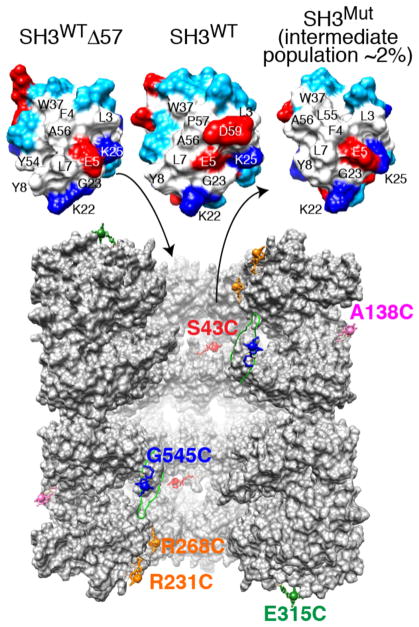
GroEL, purified to remove ~95% of protein impurities bound within the cavity,17 was paramagnetically labeled with EDTA–Mn2+16 at six separate solvent-accessible, engineered cysteine sites (see the SI for experimental details): A138C and E315C located on the outside and top, respectively, of the GroEL cylinder; R231C and R268C at the mouth of the cavity, encompassing a hydrophobic patch on the surface of the apical domain formed by helices H and I (Figure S1); G545C located in the C-terminal tail, four residues from the C-terminus; and S43C in the equatorial domain at the base of the cavity (Figure 1, bottom). Three uniformly 15N-labeled variants of the Fyn SH3 domain were employed (Figure 1, top): the native, folded wild type (SH3WT); a triple A39 V/N53P/V55L mutant (SH3Mut) that exists in dynamic equilibrium between the major folded state and a minor population of folding intermediate (~2% at 10 °C);18 and a stable mimetic of the folding intermediate, SH3WTΔ57, consisting of a three-residue C-terminal truncation of the wild-type sequence.15 The folding intermediate has the same fold as the native state, with the exception that the C-terminal β5 strand is unfolded,15,18 resulting in exposure of a large hydrophobic surface patch (Figure 1, top).
Figure 1.
Fyn SH3 variants and sites of GroEL paramagnetic labeling. Top: molecular surface of the folding intermediate mimetic SH3WTΔ57, native SH3WT, and the SH3Mut folding intermediate. Residue coloring: hydrophobic, white; hydrophilic, cyan; positively charged, dark blue; and negatively charged, red. The largest differences in chemical shift between free and GroEL-bound SH3Mut are observed for residues 2–7, 28, 37, and 50–56 that form a largely hydrophobic surface patch.14 Bottom: vertical slice through the two GroEL cavities showing five subunits for each stacked ring.4 (Note GroEL is depicted at a ~2 times smaller scale than SH3.) The locations of the solvent-accessible engineered cysteine residues used for paramagnetic labeling with EDTA–Mn2+ are indicated. Note that all 14 subunits bear a single paramagnetic label per sample. EDTA was conjugated to GroEL via a disulfide linkage to each Cys mutation to yield a (cysteaminyl–EDTA)–Cys adduct, which is then chelated to either Mn2+ (paramagnetic) or Ca2+ (diamagnetic).
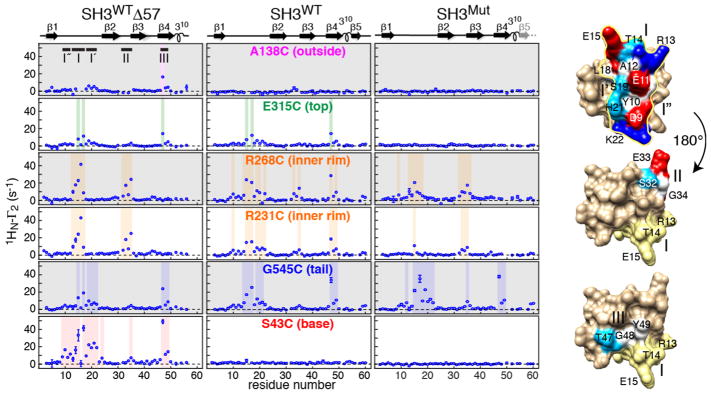
Intermolecular PRE (1HN-Γ2) profiles observed for the three SH3 variants in the presence of paramagnetically labeled GroEL are shown in Figure 2. Three major sites of intermolecular PREs are discerned (Figure 2, right): site I (13RTEDD17) with two adjacent subsidiary sites, site I′ (18LSFHK22) and site I″ (9DYEA12), located in the loop connecting strands β1 and β2; site II (32SEGD35) in the turn connecting strands β2 and β3; and site 3 (47TGY49) in strand β4. These three sites broadly coincide with sites identified by intermolecular PREs upon interaction of SH3 with three small chaperones, Spy, SurA, and Skp.19
Figure 2.
Intermolecular PRE (1HN–Γ2) profiles observed on 15N-labeled SH3WTΔ57 (left panels), SH3WT (middle panels), and SH3Mut (right panels) in the presence of GroEL paramagnetically labeled with EDTA–Mn2+ at six cysteine engineered sites. Regions with 1HN-Γ2 > 5 s−1 are shaded. The secondary structure is shown above the panels. PREs are measured using a 1H–15N correlation-based experiment21 as the difference in corresponding 1HN-R2 values between paramagnetic and diamagnetic reference samples. Data were recorded at 800 MHz and 10 °C on samples containing 100 μM 15N-labeled SH3 and 100 μM (in subunits) GroEL. The locations of sites I/I′/I″ (top), II (middle), and III (bottom) on the molecular surface of SH3WTΔ57 are shown on the right, with residues color coded as in Figure 1. In the views for sites II and III (rotated by 180° relative to the top view), site I is shown in yellow.
The surface formed by site I is contiguous with sites II and III, but the latter two are not contiguous with one another. The three sites do not overlap with the hydrophobic surface patch, located on the opposite face of the molecule, that interacts directly with the GroEL apical domain (Figure 1), as identified by chemical shift mapping derived from relaxation dispersion on a transient SH3Mut–GroEL complex.14 This is not surprising because residues buried at the GroEL–SH3 interface would be predicted to be less accessible to and hence further away from the paramagnetic labels.
Control experiments using both soluble hydroxylamine–EDTA–Mn2+ and an unrelated EDTA–Mn2+-labeled protein (maltose binding protein) show no evidence of any significant intermolecular PRE effects (Figure S2). In addition, PRE profiles are identical whether the diamagnetic control employed is EDTA–Ca2+-tagged or wild-type GroEL (Figure S3). Thus the PREs observed in the presence of paramagnetically labeled GroEL do not arise from either a solvent PRE due to random intermolecular collisions or preferential binding of the EDTA–Mn2+/Ca2+ tag to the SH3 variants.
When the paramagnetic tag is located on the outside of the GroEL cylinder (A138C), no significant PREs are observed for SH3WT or SH3Mut, and only an isolated PRE at Thr47 is seen for SH3WTΔ57 (top row Figure 2). The backbone NH of Thr47 is solvent-exposed (Figure S4), and perhaps this PRE reflects a very minor transient encounter complex. One can therefore conclude that interactions between the SH3 variants and the outside of the GroEL cylinder are minimal.
With the tag placed on the top of the GroEL cylinder (E315C), not too distant from the mouth of the cavity, a few small PREs (10–15 s−1) are apparent for a subset of residues at sites I (Glu15 and Asp17) and III (Thr47) for SH3WT and SH3WTΔ57 but not for SH3Mut (Figure 2, second row). These PREs may reflect a low population of transient encounter complexes20 as SH3 makes its way to the GroEL cavity.
When the paramagnetic tags are located on the inner rim at the mouth of the cavity (R231C and R268C), substantial PREs are observed for all three SH3 variants at sites I and II (Figure 2, third and fourth rows, and Figure 3, top) and for SH3WT at site III as well. The latter may reflect less precise sampling of the SH3WT surface as a consequence of the much shorter lifetime of the SH3WT–GroEL binary complex (40 μs versus 2–4 ms).14,15 Overall, these PRE data are consistent with the general location of non-native proteins in binary complexes with GroEL seen in low-resolution cryo-EM density maps.6,7
Figure 3.
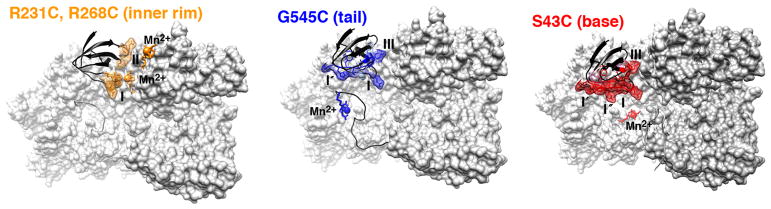
Mapping the sites of large intermolecular PREs observed for the SH3 variants originating from paramagnetic labels located at the rim (R231C and R268C), tail (G545C), and base (S43C) of the GroEL cavity. The SH3 domain is shown as a black ribbon with the regions exhibiting significant intermolecular PREs depicted as transparent surfaces. Three of seven subunits forming a single GroEL ring are depicted as a light-gray molecular surface. A cartoon of the intrinsically disordered C-terminal tail (dark gray), extending upward toward the mouth of the cavity, with the paramagnetic label at G545C, is included in the middle panel. Cys–EDTA–Mn2+ is depicted as a stick diagram with Mn2+ shown as a sphere.
The paramagnetic label at G545C, in the C-terminal tail of GroEL, makes extensive contacts with all three SH3 variants involving a contiguous surface patch comprising sites I/I′ and III (Figure 2, fifth row). Leu18 and Phe20 are located in a shallow hydrophobic pocket that may act as an attachment point for the methionines in the tail (Figure 3, middle panel). The intermolecular PREs centered around residue 35 (site II) are small, possibly because this site faces the mouth of the cavity when SH3 interacts with the cavity rim. These data provide evidence that contact with the C-terminal tail of GroEL can occur not only while SH3 is confined and diffusing within the cavity, as suggested from quantitative analysis of relaxation dispersion and lifetime line broadening data,15 but also when bound to the rim. The paramagnetic label is located at position 19 of the C-terminal tail, and the average end-to-end distance of a 19-residue random coil is predicted to be ~44 Å,22 more than sufficient to extend from the bottom to the rim of the cavity (35–40 Å), consistent with the results of molecular dynamics simulations.23
Finally, when the paramagnetic label is located at the base of the cavity (S43C, Figure 1), extensive PREs are observed for SH3WTΔ57 at sites I/I′/I″ and III, but none is observed for either SH3WT or SH3Mut (Figure 2, bottom panel), probably due to substantially reduced occupancy. This is not surprising as the populations of GroEL-bound native SH3 and SH3Mut folding intermediate are only ~0.3 and 0.7%, respectively, and the unbound population of the latter is only ~2%.14 SH3WTΔ57, on the contrary, is entirely in the folding intermediate state, and while the population of GroEL-immobilized SH3WTΔ57 is small (~1.5%), by analogy to a very closely related folding intermediate mimetic, the occupancy of a mobile, cavity-confined state, identified by relaxation dispersion, is ~20%,15 thereby significantly increasing the probability of detecting transient contacts with the base of the cavity.
In conclusion, the intermolecular PRE studies reported here show that SH3 variants sample the cavity of apo GroEL extensively and are capable of penetrating deep within the cavity prior to encapsulation by GroES. Thus in sparsely populated binary SH3–GroEL complexes, the SH3 domains not only interact transiently with the mouth of the cavity but also make contact with the disordered C-terminal tail and, in the case of the folding intermediate mimetic SH3WTΔ57, with the base of the cavity as well. The C-terminal tail itself can contact the SH3 domain when the latter is located at either the mouth or the base of the cavity, which may facilitate substrate capture and retention.
Supplementary Material
Acknowledgments
We thank J. Baber, D. S. Garrett, and J. Ying for technical support, J. Lloyd for mass spectrometry, and C. Charlier and J. Davidson for useful discussions. M.A.W. was supported in part by an Early PostdocMobility Fellowship from the Swiss National Foundation. This work was supported by the Intramural Program of the National Institute of Diabetes and Digestive and Kidney Diseases, NIH (to G.M.C.).
Footnotes
Notes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jpclett.8b01586.
Experimental details and four figures (PDF)
References
- 1.Thirumalai D, Lorimer GH. Chaperonin-Mediated Protein Folding. Annu Rev Biophys Biomol Struct. 2001;30:245–269. doi: 10.1146/annurev.biophys.30.1.245. [DOI] [PubMed] [Google Scholar]
- 2.Horwich AL, Fenton WA. Chaperonin-Mediated Protein Folding: Using a Central Cavity to Kinetically Assist Polypeptide Chain Folding. Q Rev Biophys. 2009;42:83–116. doi: 10.1017/S0033583509004764. [DOI] [PubMed] [Google Scholar]
- 3.Hartl FU, Bracher A, Hayer-Hartl M. Molecular Chaperones in Protein Folding and Proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 4.Braig K, Otwinowski Z, Hegde R, Boisvert DC, Joachimiak A, Horwich AL, Sigler PB. The Crystal Structure of the Bacterial Chaperonin GroEL at 2. 8 Å. Nature. 1994;371:578–586. doi: 10.1038/371578a0. [DOI] [PubMed] [Google Scholar]
- 5.Saibil HR, Fenton WA, Clare DK, Horwich AL. Structure and Allostery of the Chaperonin GroEL. J Mol Biol. 2013;425:1476–1487. doi: 10.1016/j.jmb.2012.11.028. [DOI] [PubMed] [Google Scholar]
- 6.Elad N, Farr GW, Clare DK, Orlova EV, Horwich AL, Saibil HR. Topologies of a Substrate Protein Bound to the Chaperonin GroEL. Mol Cell. 2007;26:415–426. doi: 10.1016/j.molcel.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clare DK, Bakkes PJ, van Heerikhuizen H, van der Vies SM, Saibil HR. Chaperonin Complex with a Newly Folded Protein Encapsulated in the Folding Chamber. Nature. 2009;457:107–110. doi: 10.1038/nature07479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen DH, Madan D, Weaver J, Lin Z, Schroder GF, Chiu W, Rye HS. Visualizing GroEL/ES in the Act of Encapsulating a Folding Protein. Cell. 2013;153:1354–1365. doi: 10.1016/j.cell.2013.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weaver J, Rye HS. The C-terminal Tails of the Bacterial Chaperonin GroEL Stimulate Protein Folding by Directly Altering the Conformation of a Substrate Protein. J Biol Chem. 2014;289:23219–23232. doi: 10.1074/jbc.M114.577205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Machida K, Kono-Okada A, Hongo K, Mizobata T, Kawata Y. Hydrophilic Residues 526KNDAAD531 in the Flexible C-terminal Region of the Chaperonin GroEL are Critical for Substrate Protein Folding Within the Central Cavity. J Biol Chem. 2008;283:6886–6896. doi: 10.1074/jbc.M708002200. [DOI] [PubMed] [Google Scholar]
- 11.Farr GW, Fenton WA, Chaudhuri TK, Clare DK, Saibil HR, Horwich AL. Folding With and Without Encapsulation by Cis- and Trans-Only GroEL-GroES Complexes. EMBO J. 2003;22:3220–3230. doi: 10.1093/emboj/cdg313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zahn R, Perrett S, Stenberg G, Fersht AR. Catalysis of Amide Proton Exchange by the Molecular Chaperones GroEL and SecB. Science. 1996;271:642–645. doi: 10.1126/science.271.5249.642. [DOI] [PubMed] [Google Scholar]
- 13.Priya S, Sharma SK, Sood V, Mattoo RU, Finka A, Azem A, De Los Rios P, Goloubinoff P. GroEL and CCT are Catalytic Unfoldases Mediating Out-of-Cage Polypeptide Refolding Without ATP. Proc Natl Acad Sci U S A. 2013;110:7199–7204. doi: 10.1073/pnas.1219867110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Libich DS, Tugarinov V, Clore GM. Intrinsic Unfoldase/Foldase Activity of the Chaperonin GroEL Directly Demonstrated Using Multinuclear Relaxation-Based NMR. Proc Natl Acad Sci U S A. 2015;112:8817–8823. doi: 10.1073/pnas.1510083112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Libich DS, Tugarinov V, Ghirlando R, Clore GM. Confinement and Stabilization of Fyn SH3 Folding Intermediate Mimetics Within the Cavity of the Chaperonin GroEL Demonstrated by Relaxation-Based NMR. Biochemistry. 2017;56:903–906. doi: 10.1021/acs.biochem.6b01237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clore GM, Iwahara J. Theory, Practice, and Applications of Paramagnetic Relaxation Enhancement for the Characterization of Transient Low-Population States of Biological Macromolecules and their Complexes. Chem Rev. 2009;109:4108–4139. doi: 10.1021/cr900033p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wälti MA, Clore GM. Disassembly/Reassembly Strategy for the Production of Highly Pure GroEL, a Tetradecameric Supra-molecular Machine, Suitable for Quantitative NMR, EPR and Mutational Studies. Protein Expression Purif. 2018;142:8–15. doi: 10.1016/j.pep.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neudecker P, Robustelli P, Cavalli A, Walsh P, Lundstrom P, Zarrine-Afsar A, Sharpe S, Vendruscolo M, Kay LE. Structure of an Intermediate State in Protein Folding and Aggregation. Science. 2012;336:362–366. doi: 10.1126/science.1214203. [DOI] [PubMed] [Google Scholar]
- 19.He L, Hiller S. Common Patterns in Chaperone Interactions With a Native Client. Angew Chem, Int Ed. 2018;57:5921–5924. doi: 10.1002/anie.201713064. [DOI] [PubMed] [Google Scholar]
- 20.Tang C, Iwahara J, Clore GM. Visualization of transient encounter complexes in protein-protein association. Nature. 2006;444:383–386. doi: 10.1038/nature05201. [DOI] [PubMed] [Google Scholar]
- 21.Iwahara J, Tang C, Clore GM. Practical Aspects of 1H Transverse Paramagnetic Relaxation Enhancement Measurements on Macromolecules. J Magn Reson. 2007;184:185–195. doi: 10.1016/j.jmr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cantor CR, Schimmel PR. Biophysical Chemistry. Vol. 3. W. H. Freeman; San Francisco: 1980. pp. 979–1018. [Google Scholar]
- 23.Dalton KM, Frydman J, Pande VS. The Dynamic Conformational Cycle of the Group I Chaperonin C-termini Revealed via Molecular Dynamics Simulation. PLoS One. 2015;10:e0117724. doi: 10.1371/journal.pone.0117724. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.