Abstract
Carbohydrates, or glycans, are as integral to biology as nucleic acids and proteins. In immunology, glycans are well known to drive diverse functions ranging from glycosaminoglycan-mediated chemokine presentation and selectin-dependent leukocyte trafficking, to the discrimination of self and non-self through the recognition of sialic acids by Siglec receptors. In recent years, a number of key immunological discoveries are now driving a renewed and burgeoning appreciation for the importance of glycans. In this review, we highlight these findings which collectively help to define and refine our knowledge of the function and impact of glycans within the immune response.
A renewed focus on glycosylation as an immune modulator
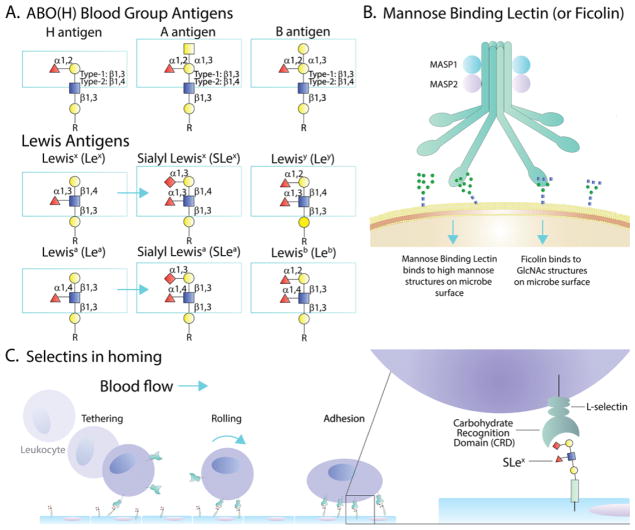
The study of how glycans impact the immune system has a long and storied history. For example, the human ABO(H) and Lewis blood groups (Fig 1A), the lectin pathway of complement (Fig 1B), and leukocyte trafficking mediated by selectin-glycan interactions (Fig 1C) are well established pillars of modern immunology which cannot be omitted from even a cursory treatise of the immune response. Yet, the depth to which glycans are important to the immune system is only recently becoming widely recognized, fueled in large part by more approachable glycobiology-oriented technologies that have been developed over the last decade (Box 1). The impact of glycans in clinical settings and novel therapeutics is equally impressive, and highlights the major implications surrounding the translation of glycomics. For example, IgG glycosylation impacts the function and production of biologic drugs, and HIV glycan recognition can be induced during the development of broadly neutralizing antibodies through vaccination. On the receptor side, the galectins, β-galactoside binding proteins, have been implicated in cancer immunosurveillance, while the Siglecs (sialic acid-binding Ig-like lectins) play a role in the differentiation of self and non-self through sialic acid recognition. The underlying findings in some of these areas are the focus of this review, but it should be noted that many other worthy findings have been omitted simply due to thematic focus and space limitations, not because of importance.
Figure 1.
Examples of Glycan Function in the Immune System. (A) The ABO(H) and some of the Lewis blood group antigens are shown, illustrating their glycan structures. The Lewis antigens are also known to be sulfated in multiple patterns not shown. (B) The complement pathway is broken into the Classical, Alternative, and Lectin pathways. Shown is a very simplified schematic of where glycan binding by Mannose Binding Lectin (MBL) and Ficolin drives the lectin pathway of activation and cellular cytotoxicity. (C) Selectins are C-type Lectins which drive leukocyte trafficking to sites of infection and/or inflammation. The process of tethering, rolling, and ultimately adhesion to the endothelium is predominantly dependent upon sialyl-LewisX binding by selectins. Symbols: fucose, red triangles; galactose, yellow circles; N-acetylglucosamine, blue squares; N-acetylgalactosamine, yellow squares; sialic acid, red diamonds.
Box 1. Glycobiology Tools.
The unapproachable nature of some common glycobiology analytical tools has traditionally limited the growth of the field. Glycan-specific mass spectrometry (MS), for example, is extremely powerful, but mostly rests in the hands of a limited number of experts and is not commonly performed in the average proteomics core at most institutions. Despite this limitation, one area of growth which may begin to bridge this gap has been the combined use of genetic tools with MS technologies. For example, analyzing the O-GalNAc glycome using cells expressing truncated O-GalNAc glycans simplifies the analysis and enables a discovery-oriented strategy for identifying sites of O-GalNAc glycosylation [63].
Since the creation of the Consortium for Functional Glycomics (CFG; http://www.functionalglycomics.org/) about 15 years ago, tool and resource development has been a high priority to lower the barrier of entry into the glycosciences. One key development was the creation of glycan arrays by the CFG and now numerous other groups in which hundreds of chemically-defined glycan structures derived from both mammalian and microbial systems are immobilized on an array and then interrogated for glycan binding properties of biological samples/proteins. This enables the rapid analysis of glycan binding specificity with high precision.
Another tool that has gained traction is the widespread use of plant lectins. While these have been used for many years, glycan arrays have clarified their binding specificities which facilitates the interpretation of their binding. Importantly, many well-characterized plant lectins are commercially available as fluorescent and biotinylated conjugates, making them suitable for nearly any application in which antibodies are now used. This includes Western blots, immunohistochemistry and imaging, flow cytometry, protein purification/enrichment, ELISA, and others, thereby enabling a glycomic measurement within familiar techniques for essentially all biomedical laboratories. The success of this approach is also fueling the development of other glycan binding probes as reagents in these platforms.
Important developments in glycan analysis also include the expanded use of hydrophilic interaction liquid chromatography (HILIC) for the quantification of glycan structures. In the past, HPLC (high-performance liquid chromatography)-based glycan analyses required monitoring anion-exchange column elution at very high pH with a pulsed amperometric detector, which was fairly uncommon. Modern HPLC utilizes HILIC applications with fluorescence as a monitoring system, which is far more ubiquitous in research labs.
Finally, there has been a major push to develop informatic platforms for glycobiology. While many of these remain under development, there has been some important progress made. For example, platforms to interrogate mass spectrometry data now exist [64], while databases are being created to house large-scale glycomics data [65–71] with improved structural annotation [72–75]. Although the ultimate goal is to integrate glycomics with genomics and proteomics within the NCBI, the availability of glycomic informatics is dramatically improved over years past.
At the heart of many of these pathways is protein glycosylation, which can take multiple forms that include asparagine (N-linked) glycosylation, O-GalNAc (mucin-type) glycans, O-linked GlcNAc, O-linked fucose, O-linked mannose, and O-linked glucose (Box 2)[1]. Not surprisingly, these post-translational modifications strongly impact and/or directly participate in a multitude of cellular and extracellular functions [2, 3]. Consider the impact of a phosphorylation event on a protein, which adds a single negative charge and a mere 80 Da in molecular mass, and then compare this to the impact of one complex N-glycan, which can occupy the same physical space as an Ig domain, add four or more negative charges, and easily add more than 3500 Da. Moreover, glycosylation is a template-independent process in that the composition of any glycan is not dictated or copied from an information source within the cell. Instead, glycan structure is influenced by the metabolic state of the cell, availability of nucleotide-sugar donors, expression patterns of glycosidases, glycosyltransferases, epimerases, nucleotide-sugar transporters and others in the endoplasmic reticulum and Golgi apparatus, and many other factors [1]. Without a template, cell-specific ‘programming’ of homogeneous protein glycosylation is not possible, though significant trends within the heterogeneity of the glycome are observed as a function of disease and environment, and these differences direct major changes in cellular and molecular properties. It is equally important to recognize that the possible variability in glycan structure and composition is immense considering the many theoretical glycosidic linkages (α vs. β, and between nearly any hydroxyl of one monosaccharide and any carbon on another) and variations within the monosaccharides themselves (which can be acetylated, phosphorylated, sulfated, etc. at one or more positions). However, rest assured that the in vivo reality of glycan structure is much less complex than theoretically possible, likely reflecting both the specificity of the enzymes responsible for building glycans and conservation to preserve biological function.
Box 2. Protein Glycosylation.
Protein glycosylation can be split into three classes: co-translational N-linked glycosylation, post-translational O-linked glycosylation, and the O-GlcNAc modification. N-linked glycosylation originates from transfer of a large precursor oligosaccharide from a membrane-embedded dolichol phosphate lipid to the asparagine of a nascent protein while being synthesized in the rough ER. The glycan(s) assists in protein folding by mediating interactions with ER chaperones like calnexin and calreticulin, thereby serving as a quality control checkpoint. Upon proper folding, the glycans are trimmed to “high mannose” structures prior to trafficking to the Golgi apparatus, where they undergo further trimming and then rebuilding through the combined action of various glycosyltransferases. This yields both hybrid and complex-type N-glycans which are common at the plasma membrane and on secreted glycoproteins, including IgG and essentially every surface protein on a cell.
O-linked glycosylation is a broad category comprising various modifications named for the glycosidic linkage of oxygen on threonine or serine residues, and includes O-GalNAc, O-fucose, and O-mannose glycans. Unlike N-glycosylation, all of these are built progressively from a single initiating sugar post-translationally. O-GalNAc (or mucin type) glycans are initiated by a family of 20 homologous galactosyltransferases (GALNTs) with overlapping activities and various degrees of site specificity. After adding the initial O-GalNAc, it is assembled into one of 8 core structures which are further elaborated into complex structures through various glycosyltransferases in the Golgi [1]. O-GalNAc glycans are best known as mucin-like glycans, as they comprise most of the dry weight of mucin proteins and function in that context through their ability to retain water, creating a viscous barrier between the microbial world and the mammalian host.
O-mannose, like O-GalNAc, can be elaborated from an initial linkage into several core structures through the activity of a single initiating heterodimeric O-mannosyltransferase in the ER and a series of dedicated enzymes in the Golgi apparatus. Elaborated O-mannose glycans act as binding sites on α-dystroglycan for extracellular matrix proteins and are implicated in many congenital muscular dystrophies [76]. The EGF repeats of Notch, a receptor family that plays an integral role in development, are highly glycosylated with a variety of interesting structures including O-fucose and O-glucose, both of which modulate interactions and signaling. Moreover, the Notch O-fucose can be further modified by GlcNAc through the action of Fringe enzymes which are known for their modulation of the Notch pathway. Indeed, it is clear that these modifications go beyond Notch and play many important roles in developmental biology [77].
The O-GlcNAc modification is uniquely found in the reducing environment of the cytosol and is extremely dynamic compared to other glycan modifications. O-GlcNAcylation is an ancient modification found in multicellular organisms whereby GlcNAc is rapidly added or removed by a pair of conserved enzymes: O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA). These enzymes have broad specificity, acting as holoenzymes to refine their activity on any of thousands identified targets in humans [78]. This modification acts as a signaling modality, interacting with protein O-phosphorylation through complex allosteric effects and direct competition, and acting as the signal itself. O-GlcNAc is sensor of cellular condition, feeding back information about metabolism and stress to decisions about changes in cell cycle progression, protein degradation, and gene expression [79].
Here, we review recent discoveries that link glycan composition with the manipulation of protein-carbohydrate and protein-protein interactions in the immune system. In so doing, we show that glycobiology is revolutionizing our understanding of the proteome in much the same way that epigenetics has revolutionized our understanding of the genome – and in no other field has this been clearer than immunology.
The Role of Siglecs in Immune Regulation
Siglecs are comprised of a family of 16 receptors (Box 3) in humans, most of which are expressed on cells of hematopoietic lineages [4] and bind to glycans containing terminal sialic acids in a linkage-sensitive fashion. Their ligands, sialic acids, are negatively charged, 9 carbon monosaccharides that hold a strategic spatial location as the terminal sugar residue on glycan structures, and are attached through α2,3, α2,6 and α2,8 glycosidic linkages to the underlying carbohydrate [1]. These properties give sialic acids and Siglecs diverse biological roles, in particular in the regulation of immune responses, many of which are mediated by one or more immunoreceptor tyrosine-based inhibitory motifs (ITIMs) that associate with phosphatases like Shp1 to dampen immune signals [5].
Box 3. Siglecs.
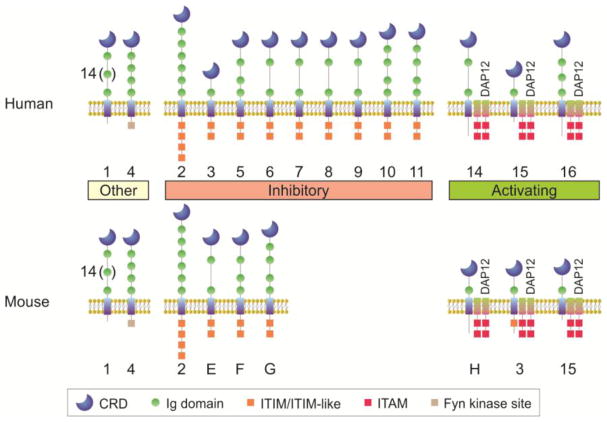
Sialic binding immunoglobulin-like lectins (Siglecs) are a subfamily of the I-type lectin family. I-type lectins are glycan binding proteins which contain repeated immunoglobulin-like domains, making them a part of the immunoglobulin superfamily (hence “I-type”). There are 16 known human Siglecs and 9 mouse Siglecs, and all of these receptors have the ability to bind sialic acid. They diverge in domain structure, sialic acid linkage preference, expression pattern, and function (Figure 1).
With the exception of Siglec-4 and Siglec-6, all of the Siglec family members are found in cells from the hematopoietic lineage. Some Siglecs show highly selective expression patterns, such as CD22/Siglec-2 on human and murine B cells and Siglec-F on murine eosinophils and alveolar macrophages. Most Siglecs contain immunoreceptor tyrosine-based inhibitory motifs (ITIMs), imbuing them with the ability to down-regulate signaling cascades and limit immune responses through the recruitment of phosphatases like Shp1. These receptors give Siglecs their reputation as regulatory receptors that recognize “self” in the form of terminal sialic acids, a sugar found predominantly in mammals. There are several “activating” Siglecs, however, and these contain basic residues in their transmembrane domain. These residues allow for the association with DAP12, a signaling molecule containing immunoreceptor tyrosine-based activation motifs (ITAMs) that promote signaling and immune responses. Finally, Siglec-1 is not known to be directly involved with signaling, but rather is thought to function primarily in cell-cell adhesion. Thus, Siglecs are a family of surface receptors which recognize and bind sialic acid-containing glycans in the host and on microbes, and provide an array of signaling outcomes that drives the immune response.
Figure I. The Human and Mouse Siglec Family.
Siglec nomenclature is based on the human Siglecs, numbered 1 to 16 (Siglec-1, Siglec-2, etc.). The Siglecs conserved across humans and mice retain the number designation (Siglec-1, Siglec-2, Siglec-3, Siglec-4, and Siglec-15), although key differences in the domains of Siglec-3 renders it inhibitory in humans and activating in mice. Mouse Siglecs without a conserved counterpart in humans are given letter designations (Siglec-E, Siglec-F, Siglec-G, Siglec-H). Most Siglecs fall into an inhibitory category, carrying one or more ITIM sequences, while others are activating receptors in which their transmembrane domain contains a charged residue that leads to association with DAP12, which carries ITAMs. Siglec-1 has no defined ability to transmit signals, and contains an unusual number of Ig domains (16 in total), while Siglec-4 contains a cytoplasmic domain with a Fyn kinase phosphorylation site. Finally, several Siglecs are known by other names. Siglec-1 is also called sialoadhesin or CD169. Siglec-2 is also called CD22. Siglec-3 is also called CD33.
One theme derived from the Siglec literature is that immune evasion by both cancer and a variety of pathogens can be promoted through hijacking the homeostatic nature of Siglec signaling (Fig 2A). For example, recent findings reveal that a cancer-specific mucin-1 (MUC1) glycoform interacts with Siglec-9 [6]. The resulting signaling induces macrophages to polarize into tumor-associated macrophage (TAM)-like phenotypes, which are known for their ability to modulate PD-1/PD-L1 expression resulting in the ability to downregulate T cell activity and tumor clearance [7]. Simply put, tumors can promote glycomic remodeling leading to a suppressive immune microenvironment mediated by Siglec receptors as a means of immune evasion.
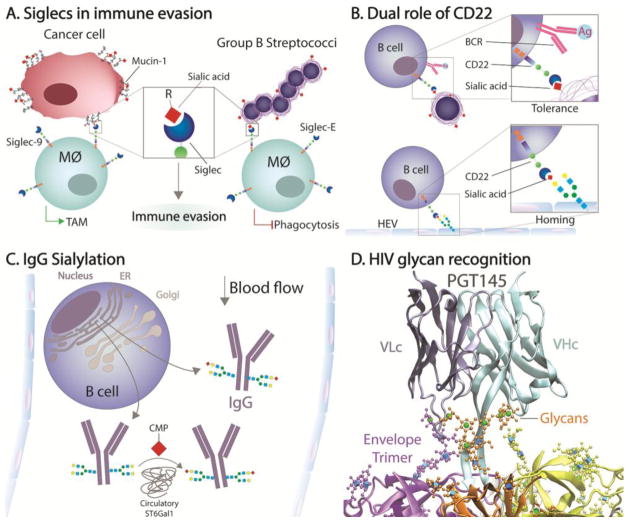
Figure 2.
Highlighted Glycan-Influenced Pathways. (A) Recognition of MUC1 by Siglec-9 or GBS capsule by Siglec-E on macrophages leads to the differentiation into tumor-associated inhibitory macrophages (TAM) and reduces phagocytosis, respectively, leading to immune evasion. (B) CD22 is a Siglec that leads to B cell tolerance when ligated in trans while the B cell receptor (BCR) is also engaged, but can also function to promote adherence to high endothelial venules (HEV). (C) The sialyltransferase ST6Gal1 can sialylate Fc-localized IgG glycans outside of the B cell, within the blood stream, but the degree to which B cells also sialylate IgG in the Golgi apparatus prior to secretion remains debatable. (D) The binding of HIV broadly neutralizing antibody PGT145 requires both protein-protein and glycan-protein contacts with high mannose glycans present on the HIV envelope trimer (coordinates from PDB# 5v8l). Green spheres are mannose residues and blue cubes are N-acetylglucosamine (GlcNAc) residues. The image was created using VMD software with a GLYCAM plugin for glycan visualization and rendered in POV-Ray.
Likewise, although most bacteria cannot produce sialic acid, some microbes have evolved the ability to either produce sialic acids themselves [8], or to appropriate them from host glycans through the action of trans-sialidases [9]. The idea is that the microbe becomes able to masquerade as “sialylated self” and evade host defenses [10]. In 2014, the presence of sialic acids in the capsule of group B streptococcus (GBS) was found to selectively bind Siglec-E [11] (murine-specific Siglecs are given letter designations rather than number designations as in humans; Box 3), which is primarily expressed by monocytes and other myeloid cells [11]. In mice lacking Siglec-E, GBS infection resulted in an increased pro-inflammatory cytokine release, enhanced phagocytosis and more potent bactericidal activity. Without Siglec-E, both NF-κB and MAPK-mediated signaling was increased, culminating in reduced GBS invasion into the CNS. However, at high multiplicity of infection, the lack of Siglec-E resulted in a cytokine storm and increased mortality in mice, indicating that its absence led to impaired host immunomodulation. In a related study, Siglec-E ablation was also found to enhance dendritic cell responses to a wide spectrum of microbial toll-like receptor (TLR) ligands [12]. In the context of T cells, the interplay between Siglec E and TLR signaling was driven by direct binding between the receptors, presumably through cis-interactions between the Siglec and sialic acid on the numerous glycans on each TLR molecule, thereby recruiting phosphatases to regulate TLR signaling under normal conditions. These findings are also consistent with data demonstrating that sialic acids induce immunologic tolerance by inhibiting T cell proliferation and inducing antigen-specific regulatory T cells (Tregs) through dendritic cell-mediated presentation of sialylated antigens [13]. Together, these findings illustrate the careful balance between inflammation and immune homeostasis established by Siglec molecules, and how this can be commandeered by both cancer and infectious diseases to evade protective immunity.
Fortunately, evolution works both ways, as encapsulated by the Red Queen hypothesis that describes the immunologic arms race that drives evolution in both the host and microbe [14]. Recent discoveries show that mammalian immune systems have also evolved responses to combat pathogens that seek to subvert immunity through Siglec signaling. For example, TLR recognition of danger signals was reported to upregulate the expression of the sialidase Neu1 at the cell surface [12], which removes terminal sialic acids from glycans [15]. Elimination of sialic acids led to the release of Siglec binding and TLR inhibition, thereby triggering a potent immune response that protected the host from endotoxemia [12].
Another mechanism to combat Siglec hijacking is selective receptor pairing. While Siglecs are representative of this phenomenon, immune regulation via receptor pairing has also been described for killer-cell immunoglobulin-like receptors (KIRs) and Ly49 on NK cells, dendritic cell immune receptor (DCIR) which is also a C-type lectin receptor, CD200R, and several other gene families [16]. Pairing-mediated regulation is based on the notion that the extracellular ligand-binding domains of paired receptors are very similar, while their intracellular machinery lead to opposite signals. For Siglecs, GBS again is an excellent example of this phenomenon, as it was recently shown that the pairing of Siglec-5 and Siglec-14 resulted in the activation of the MAPK and AKT signaling pathways in neutrophils [17]. Siglec-5 contains the typical ITIM and is generally a suppressive receptor, yet Siglec-14 actually contains an immunoreceptor tyrosine-based activation motif (ITAM) that overrides the inhibitory effect of Siglec-5 [17]. Through the paired expression of Siglec-5 and Siglec-14, the cells rapidly titrate the balance between activation and inhibition upon recognition of sialylated glycans.
Another member of the Siglec family has garnered much attention in recent years – CD22, or Siglec-2. CD22 is a typical inhibitory Siglec with an endoplasmic tail containing an ITIM, and near exclusive expression on lymphocytes, especially B cells [18]. There are several reasons CD22 holds great interest. CD22 is a key regulatory element of B cell receptor (BCR) signaling on B cells and possesses implications in the context of autoimmunity (Fig 2B)[19]. In 2013, it was found that cis-interactions at the B cell surface between CD22 and the BCR regulates calcium-dependent signaling [20]. Mutation in the ITIMs associated with CD22 led to calcium signal increase, but interestingly, mutation in the carbohydrate recognition domain (CRD) led to signal decrease. The emerging model [19–21] is that CD22 self-associates and is sequestered away from the BCR under normal conditions. This allows BCR signals to proceed unless the antigen carries sialic acids. In that case, CD22 is recruited to the BCR and eliminates the signaling in an ITIM-dependent fashion. However, elimination of sialic acid binding through mutagenesis results in loss of CD22 sequestration, allowing BCR proximity and limited signaling in an ITIM-dependent fashion.
Another reason CD22 came into focus was the discovery that it can act as a homing receptor in a selectin-like fashion (Fig 2B). It has been known for years that post-capillary high endothelial venules (HEVs) in lymph nodes and Peyer’s patches are highly efficient at recruiting lymphocytes [22]. However, it was found that the endothelium expresses high levels of the sialyltransferase ST6Gal1, which synthesizes the α2,6-linked sialic acids preferred by CD22 [23]. In the absence of CD22, both T and B cells were deficient in HEV homing [23], which is analogous to the glycan-dependent homing mediated by the well-characterized selectin family [24].
The final reason CD22 has gained attention is because it is rapidly and constitutively endocytosed and recycled from the plasma membrane, and thus acts as an efficient sialylated ligand delivery system [25]. Coupled with the B cell selectivity of expression, CD22 has become an outstanding Siglec target for drug development. Conjugation of a CD22-target reagent, such as an α2,6-sialylated glycan or an anti-CD22 antibody, to an immunotoxin enables the specific targeting of B cells and efficient endocytosis-mediated intracellular delivery of the toxin to selectively eliminate B cells in cases of acute lymphoblastic leukemia (ALL). One early example in 2007 utilized a CD22-specific antibody coupled to a cytotoxic agent called CMC-544 [26]. Much more recently in 2017, anti-CD22 antibody conjugated to RFB4 was used in a Phase I clinical trial for children with relapsed or refractory ALL [27]. The response rate was 32%, which included an overall 23% composite complete response and five patients negative for minimal residual disease. These exciting findings are mirrored in related studies targeting other Siglecs, such as the use of Siglec-1 (aka, Sialoadhesin and CD169) to deliver cargo to macrophages for the induction of iNKT cells [28], and are fueling the design of synthetic glycan ligands to target Siglecs in the clinical setting [4].
Immunoglobulin Glycosylation
In 2006, it was discovered that terminal α2,6-linked sialylation of complex N-glycans located at asparagine 297 of the human IgG heavy chain served as a functional switch [29]. The essential observation was that sialylated IgG imposed a net inhibitory effect on the immune response, thereby explaining the anti-autoimmune nature of intravenous immunoglobulin therapy [29]. Although this was not the first time that IgG glycosylation was shown in influence function [30], this and subsequent reports [31, 32] have led to immense interest in the role of glycan composition on IgG function. If altering glycosylation of IgG fundamentally impacts IgG function, then the regulation of glycosylation impacts every process driven by antibodies – from vaccination to antibody-based biologic drug development and commercial process control. Since the functional ramifications of IgG glycosylation have been reviewed very recently [33], we will instead focus on the related question of in vivo regulation of IgG glycosylation (Fig 2C), which is poorly understood.
One theory promoted by a number of groups is that antibody glycosylation is programmable and uniformly hard-wired into each clonal population of plasma cells or memory B cells. One line of evidence for this is population-based human studies. For example, in 2016, the glycosylation profiles of gp120, p24 and hemagglutinin (HA) specific IgG were compared to the total IgG glycosylation from the same individual. It was found that these specific antibodies differed from the bulk IgG in terms of total sialylation, presence of bisecting GlcNAc residues, and fucosylation [34]. Moreover, it was reported that the IgG glycoforms correlated with the inflammatory state of the patient, and that these forms had different functions in promoting natural killer (NK) cell activation. In mice, similar results have been reported in which vaccination against HA generated IgG glycoforms which were divergent from total IgG, and that the nature of the glycoforms predicted overall vaccine efficacy [35]. It was suggested that sialylation of the Fc domain drives affinity selection through the Type-II Fc receptor CD23, which is also a C-type lectin family member [36], by altering the threshold of BCR signaling and pushing towards higher affinity antibodies. The model which emerges from these data suggests that B cells are permanently programmed on an antigen-specific basis depending on the inflammatory milieu at the time of vaccination or exposure, and therefore careful design of vaccines could not only generate the desired IgG specificity, but also specific and permanent glycoform programming to optimize the desired activity.
A competing model comes from the observations that protein glycoforms are not template-driven but is sensitive to inflammation, metabolism, expression of glycosidases, glycosyltransferases, nucleotide-sugar transporters, and nucleotide-sugar synthetic pathway enzymes, availability of glucose, Golgi structure and a host of other parameters [1]. As these factors change, so does the glycome of any given cell and the glycoproteins they express [1, 37]. Another complication to the pre-programmed model is the observation that a B cell-specific knockout of ST6Gal1, the only B cell-expressed enzyme capable of placing an α2,6-linked sialic acid on IgG [38, 39], does not show differences in IgG sialylation, indicating that B cells do not always control IgG sialylation [40]. In fact, ST6Gal1 exists in an active state within plasma [40–43], having been identified as an acute phase reactant many years ago [43]. Consistent with this, murine models in which liver-expressed ST6Gal1 is missing lack sialylation on IgG molecules [42] even though the B cells express the enzyme. Finally, when mice are treated with the sialic acid precursor N-acetylmannosamine, IgG sialylation is increased [44], further showing the variable nature of IgG sialylation. Although this pathway has not yet been demonstrated in humans, it is noteworthy that ST6Gal1 is also an acute phase reactant in humans, regulated by inflammation, and is active within the plasma [45]. These data collectively demonstrate that sialylation cannot be programmed within the B cell on a clonal basis, and that the glycoforms of specific antibodies is variable depending on the environment at the time of harvest and analysis.
Beyond sialylation, the evidence for a lack of permanent glycome programming of B cells is much less direct but no less powerful. As we have reviewed previously, the glycome in general is strongly influenced by the current in vivo circumstances [2, 46]. Inflammation, cancer, and infection are all known to reversibly dysregulate the glycome in ways that sometimes promote disease and/or immune evasion. Given the fact that all IgG is antigen-specific, the mere fact that the global glycome of IgG changes during disease testifies to its malleability, supporting the notion protein glycosylation is not hard-wired into any given cell and is a product of the physiological milieu. Thus, the reported “antigen-specific” IgG glycosylation most likely reflects the glycome at the time of IgG secretion. Moreover, since the plasma half-life of human IgG is approximately three weeks, the difference between specifically induced IgG through vaccination and the broad population of pre-existing IgG would be expected. This could easily be tested using mass spectroscopy-based structural analysis of antigen-specific IgG glycans over time and as a function of inflammation or disease.
HIV Broadly Neutralizing Antibodies
Another area of glycobiology that has garnered much attention in recent years is the usability of broadly neutralizing antibodies (bnAbs) against HIV. Of particular note are the V2 apex region-specific antibodies targeting the envelop (Env) trimeric complex at the viral surface. The V2 apex site is a leucine-dense region of the Env trimer, near the point of the 3-fold axis, and is neighbored by three sites of N-linked glycosylation: N160, N156 and N173 [47]. For N160, the predominant glycoform is high mannose, whereas both N156 and N173 tend to carry sialylated hybrid N-glycans. As a whole, the HIV research community describes Env glycosylation as a ‘glycan shield’, a concept of immune evasion first proposed in 2003 [48]. This model is accurate in some ways, but insufficient in others because it implies that the primary role of HIV glycans is the prevention of immune access to the underlying Env [49]. Instead, there is evidence that the gp120 glycans serve an active role in infection through DC-SIGN interactions [50] and are key components of some HIV epitopes (reviewed next; Fig 2D), which certainly goes beyond any physical barrier attributes.
In 2014, two bnAbs were described to selectively bind to the pre-fusion conformation of gp41 [51], PGT151 and PGT152. Both of these antibodies mediate antibody-dependent cellular cytotoxicity (ADCC), but the epitope is comprised of complex tri- and tetra-antennary glycans within the Env protein. Interestingly, this was only seen in the cleaved trimers, suggesting that conformation and orientation of these glycans are critical for recognition. Another example is the CAP256 family of bnAbs. In this case, the bnAbs associate with both glycans and protein surfaces in the V2 apex region. It was also found that sialic acid-bearing glycans associate with residues within the heavy chain CDR2 loop, whereas the long CDR3 loop inserts beyond the glycans and makes contacts with the underlying protein [47]. The data lead to a model in which the CDR2 loops anchor a given antibody to the site to maintain site localization through glycan-protein interactions, while allowing for affinity maturation within the CDR3 loop to enable deeper protein-protein contact and optimal overall binding. In a similar report, the PGT145 bnAb was also found to associate with the apex region of Env (Fig 2D); however, in this case, the preference was for the high mannose N-glycans nearby in conjunction with protein-protein contacts [52].
Most recently, the antibody VRC-PG05 was revealed to bind a glycan epitope utilizing N-glycans at N262, N295 and N448, which is located at the so-called ‘silent face’ of the Env trimer [53]. Similar to PGT145, this bnAb primarily binds high mannose-type N-glycans, however, the positioning of these glycans were found to be critical for binding. Mutations in which the glycan at N448 is moved to N446 resulted in the loss of VRC-PG05 binding and evasion of neutralization [53], which indicates that the spacing and orientation of the N-glycans, in addition to their composition, is critical for the formation of the proper epitope.
These and other similar findings using bnAbs show that HIV glycans can act as the sole epitope, as shown with the previously described 2G12 bnAb [54], or as a portion of the epitope as described here [47, 51–53]. They can also serve as a means of cellular attachment and delivery [50], and are undoubtedly critical for protein folding given that N-glycosylation plays a dominant role in glycoprotein quality control within the ER [1, 2]. As a result, a better understanding of what regulates HIV glycosylation and its impact on the viral life cycle could facilitate new approaches for therapy, including HIV vaccination strategies in which the glycans themselves are included as a target.
Translational Glycomics
We have already introduced one example of the translation from glycobiology to clinical trial where an anti-CD22 delivery antibody (inotuzumab) was coupled to the cytotoxin CMC-544, trade named Besponsa, as a therapy for ALL [26], but another recent example is crizanlizumab. This is an anti-P-selectin antibody that is currently in Phase III trials for the treatment of the very painful vaso-occlusive crises common to patients with Sickle cell disease [55]. This trial comes on the heels of a successful Phase II trial and the subsequent acquisition of the originating Selexys Pharmaceuticals by Novartis for $665 million dollars in 2016 (https://relationshipscience.com/organization/selexys-pharmaceuticals-corp-195590). The scope of success, so far, for both of these glycobiology-rooted therapies demonstrates the untapped and potentially game-changing impact of the glycome on the pharmaceutical landscape.
Another area of clinical translation that has been expanding exponentially is the impact of IgG glycosylation on its function. Consider the fact that there are many current and developing antibody-based biologic drugs, with three of the top-five selling drugs (based on profit) in 2017 being antibodies. Humira, an antibody which neutralizes tumor necrosis factor α [56], is the best-selling drug in the world for several consecutive years at $18 billion in 2017 alone (The Motley Fool). Some of these drugs, like Humira, are aimed at immune suppression, while others seek to promote ADCC or other immune activating properties to, for example, kill tumor cells. Rituxan is the best example of this, and is the fourth best-selling drug in 2017 (The Motley Fool). Thus, it stands to reason that glycoengineering these biologic drugs is becoming a top priority to optimize activity and efficacy. For example, if the biologic drug aims to suppress an autoimmune disease, then sialylated glycoforms of the drug would be preferential to optimize immune inhibition [29, 31, 32]. Thus, it is clear that controlling the glycosylation of these drugs is critical, and could significantly increase the effectiveness of each drug.
Still another aspect where glycobiology is making a translational impact is the design of vaccines. The advances in our knowledge about bnAbs and their specificity of binding the HIV Env trimeric complex through protein and glycan contacts drives new approaches to elicit similar antibodies through vaccination to prevent HIV infection. The question then moves to the methods of eliciting such responses, and there are at least two new advances to mention. First, chimeric antigen receptor (CAR) T cell therapy has been reported against Tn (Thomsen-Friedenreich)-antigen-containing MUC1 [57] and was based on a Tn-MUC1 glycopeptide-specific monoclonal antibody [58]. The Tn-antigen is a truncated form of a Core 1 O-GalNAc glycan consisting of a β1,3-linked galactose on the initiating serine/threonine linked GalNAc, which has long been associated with cancer [59]. Second, novel glycan-protein conjugates are being investigated to increase antibody affinity and B cell memory beyond current vaccine conjugates like Prevnar-13. The use of viral particles carrying small and chemically-defined glycan epitopes administered with NKT-activating adjuvants are showing promise to elicit IgG with nanomolar affinity [60].
These are merely three groups of examples in which the power of the glycome is finally being harnessed for human health, but it potentially goes much deeper. The identification of congenital disorders of glycosylation is increasing [61], and novel approaches to target galectin molecules (mammalian lectins which associate with common glycan signatures on host cells) in anti-cancer therapies are ongoing and gaining a lot of attention [62]. And despite these exciting developments, there is much more to be discovered.
Concluding Remarks
It has become clear that glycosylation can and usually has a profound impact upon the structure and function of a protein, and can drive the nature of their interactions. However, the ability to target those glycans or their binding partners remains in its infancy (see Outstanding Questions). For IgG, the ability to modulate function through glycan alterations represents a paradigm shift, but very little is known about the regulation of IgG glycan composition. Siglecs are gatekeepers of immune homeostasis through the recognition of self via sialic acid binding, but despite the potential, it remains to be seen whether these molecules can be therapeutically harnessed to treat diseases characterized by unwanted inflammation. Perhaps most importantly, essentially all secreted and plasma membrane proteins are glycoproteins, including all cytokines, chemokines and their receptors. And yet, almost nothing is known about the functional impact of their corresponding glycans, or the extent to which they change depending on the originating cell or their environmental milieu. Relative to the number of current translational efforts, the glycome remains nearly completely untapped as a source of therapeutic approaches, but the tide has turned on our collective recognition for the importance of glycans in immunity, and this is beginning to change.
Outstanding Questions.
What are the in vivo controls of IgG glycosylation inside and outside of the B cell?
How can vaccination against HIV harness both glycan and protein contacts in the design of novel immunogens?
Can targeting Siglecs be useful in manipulating autoimmunity, allergies, and other inflammatory diseases?
In what ways does the glycoform of cytokines, other immune mediators and their receptors drive function and lifetime in vivo?
Highlights.
Protein glycosylation drives molecular interactions that modulate immune system signaling and function.
Sialic acid-binding (Siglec) receptors modulate cancer surveillance, host response to infection, and B cell function.
Antibody glycosylation is sensitive to the physiologic milieu, thereby driving IgG effector function.
HIV glycosylation is more than a shield against immunity, but can also directly participate in antibody-mediated recognition.
Glycoimmunology is increasingly being translated into the clinical setting, revealing the largely untapped potential of the glycome in novel therapeutics.
Acknowledgments
We would like to thank Carlos Alvarez for critical reading of this manuscript. We also acknowledge support from the National Institutes of Health through grants to BAC (GM082916, GM115234 and AI089474).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Essentials of Glycobiology. 2. Cold Spring Harbor Laboratory Press; 2009. [PubMed] [Google Scholar]
- 2.Varki A. Biological roles of glycans. Glycobiology. 2017;27(1):3–49. doi: 10.1093/glycob/cww086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Varki A, Gagneux P. Multifarious roles of sialic acids in immunity. Ann N Y Acad Sci. 2012;1253:16–36. doi: 10.1111/j.1749-6632.2012.06517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Angata T, et al. Therapeutic Targeting of Siglecs using Antibody- and Glycan-Based Approaches. Trends Pharmacol Sci. 2015;36(10):645–660. doi: 10.1016/j.tips.2015.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coxon CH, et al. ITIM receptors: more than just inhibitors of platelet activation. Blood. 2017;129(26):3407–3418. doi: 10.1182/blood-2016-12-720185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beatson R, et al. The mucin MUC1 modulates the tumor immunological microenvironment through engagement of the lectin Siglec-9. Nat Immunol. 2016;17(11):1273–1281. doi: 10.1038/ni.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santoni M, et al. Triple negative breast cancer: Key role of Tumor-Associated Macrophages in regulating the activity of anti-PD-1/PD-L1 agents. Biochim Biophys Acta. 2018;1869(1):78–84. doi: 10.1016/j.bbcan.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 8.Vimr E, Lichtensteiger C. To sialylate, or not to sialylate: that is the question. Trends Microbiol. 2002;10(6):254–7. doi: 10.1016/s0966-842x(02)02361-2. [DOI] [PubMed] [Google Scholar]
- 9.Ferrero-Garcia MA, et al. The action of Trypanosoma cruzi trans-sialidase on glycolipids and glycoproteins. Eur J Biochem. 1993;213(2):765–71. doi: 10.1111/j.1432-1033.1993.tb17818.x. [DOI] [PubMed] [Google Scholar]
- 10.Chang YC, Nizet V. The interplay between Siglecs and sialylated pathogens. Glycobiology. 2014;24(9):818–25. doi: 10.1093/glycob/cwu067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang YC, et al. Group B Streptococcus engages an inhibitory Siglec through sialic acid mimicry to blunt innate immune and inflammatory responses in vivo. PLoS Pathog. 2014;10(1):e1003846. doi: 10.1371/journal.ppat.1003846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen GY, et al. Broad and direct interaction between TLR and Siglec families of pattern recognition receptors and its regulation by Neu1. Elife. 2014;3:e04066. doi: 10.7554/eLife.04066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perdicchio M, et al. Sialic acid-modified antigens impose tolerance via inhibition of T-cell proliferation and de novo induction of regulatory T cells. Proc Natl Acad Sci U S A. 2016;113(12):3329–34. doi: 10.1073/pnas.1507706113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Varki A. Nothing in glycobiology makes sense, except in the light of evolution. Cell. 2006;126(5):841–5. doi: 10.1016/j.cell.2006.08.022. [DOI] [PubMed] [Google Scholar]
- 15.Carrillo MB, et al. Cloning and characterization of a sialidase from the murine histocompatibility-2 complex: low levels of mRNA and a single amino acid mutation are responsible for reduced sialidase activity in mice carrying the Neu1a allele. Glycobiology. 1997;7(7):975–86. doi: 10.1093/glycob/7.7.975. [DOI] [PubMed] [Google Scholar]
- 16.Akkaya M, Barclay AN. How do pathogens drive the evolution of paired receptors? Eur J Immunol. 2013;43(2):303–13. doi: 10.1002/eji.201242896. [DOI] [PubMed] [Google Scholar]
- 17.Ali SR, et al. Siglec-5 and Siglec-14 are polymorphic paired receptors that modulate neutrophil and amnion signaling responses to group B Streptococcus. J Exp Med. 2014;211(6):1231–42. doi: 10.1084/jem.20131853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Reilly MK, et al. CD22 Is a Recycling Receptor That Can Shuttle Cargo between the Cell Surface and Endosomal Compartments of B Cells. Journal of Immunology. 2011;186(3):1554–1563. doi: 10.4049/jimmunol.1003005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nitschke L. CD22 and Siglec-G regulate inhibition of B-cell signaling by sialic acid ligand binding and control B-cell tolerance. Glycobiology. 2014;24(9):807–17. doi: 10.1093/glycob/cwu066. [DOI] [PubMed] [Google Scholar]
- 20.Muller J, et al. CD22 ligand-binding and signaling domains reciprocally regulate B-cell Ca2+ signaling. Proc Natl Acad Sci U S A. 2013;110(30):12402–7. doi: 10.1073/pnas.1304888110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grewal PK, et al. ST6Gal-I restrains CD22-dependent antigen receptor endocytosis and Shp-1 recruitment in normal and pathogenic immune signaling. Mol Cell Biol. 2006;26(13):4970–81. doi: 10.1128/MCB.00308-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ager A. High Endothelial Venules and Other Blood Vessels: Critical Regulators of Lymphoid Organ Development and Function. Front Immunol. 2017;8:45. doi: 10.3389/fimmu.2017.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee M, et al. Transcriptional programs of lymphoid tissue capillary and high endothelium reveal control mechanisms for lymphocyte homing. Nat Immunol. 2014;15(10):982–95. doi: 10.1038/ni.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosen SD. Ligands for L-selectin: homing, inflammation, and beyond. Annu Rev Immunol. 2004;22:129–56. doi: 10.1146/annurev.immunol.21.090501.080131. [DOI] [PubMed] [Google Scholar]
- 25.Collins BE, et al. High-affinity ligand probes of CD22 overcome the threshold set by cis ligands to allow for binding, endocytosis, and killing of B cells. J Immunol. 2006;177(5):2994–3003. doi: 10.4049/jimmunol.177.5.2994. [DOI] [PubMed] [Google Scholar]
- 26.Dijoseph JF, et al. Therapeutic potential of CD22-specific antibody-targeted chemotherapy using inotuzumab ozogamicin (CMC-544) for the treatment of acute lymphoblastic leukemia. Leukemia. 2007;21(11):2240–5. doi: 10.1038/sj.leu.2404866. [DOI] [PubMed] [Google Scholar]
- 27.Wayne AS, et al. Phase 1 study of the anti-CD22 immunotoxin moxetumomab pasudotox for childhood acute lymphoblastic leukemia. Blood. 2017;130(14):1620–1627. doi: 10.1182/blood-2017-02-749101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawasaki N, et al. Targeted delivery of lipid antigen to macrophages via the CD169/sialoadhesin endocytic pathway induces robust invariant natural killer T cell activation. Proc Natl Acad Sci U S A. 2013;110(19):7826–31. doi: 10.1073/pnas.1219888110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaneko Y, et al. Anti-inflammatory activity of immunoglobulin G resulting from Fc sialylation. Science. 2006;313(5787):670–3. doi: 10.1126/science.1129594. [DOI] [PubMed] [Google Scholar]
- 30.Rademacher TW, et al. Agalactosyl glycoforms of IgG autoantibodies are pathogenic. Proc Natl Acad Sci U S A. 1994;91(13):6123–7. doi: 10.1073/pnas.91.13.6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anthony RM, et al. Recapitulation of IVIG anti-inflammatory activity with a recombinant IgG Fc. Science. 2008;320(5874):373–6. doi: 10.1126/science.1154315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anthony RM, et al. Intravenous gammaglobulin suppresses inflammation through a novel T(H)2 pathway. Nature. 2011;475(7354):110–U133. doi: 10.1038/nature10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jennewein MF, Alter G. The Immunoregulatory Roles of Antibody Glycosylation. Trends Immunol. 2017;38(5):358–372. doi: 10.1016/j.it.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 34.Mahan AE, et al. Antigen-Specific Antibody Glycosylation Is Regulated via Vaccination. PLoS Pathog. 2016;12(3):e1005456. doi: 10.1371/journal.ppat.1005456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang TT, et al. Anti-HA Glycoforms Drive B Cell Affinity Selection and Determine Influenza Vaccine Efficacy. Cell. 2015;162(1):160–9. doi: 10.1016/j.cell.2015.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bettler B, et al. Binding site for IgE of the human lymphocyte low-affinity Fc epsilon receptor (Fc epsilon RII/CD23) is confined to the domain homologous with animal lectins. Proc Natl Acad Sci U S A. 1989;86(18):7118–22. doi: 10.1073/pnas.86.18.7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–64. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 38.Hennet T, et al. Immune regulation by the ST6Gal sialyltransferase. Proc Natl Acad Sci U S A. 1998;95(8):4504–9. doi: 10.1073/pnas.95.8.4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barb AW, et al. Branch-specific sialylation of IgG-Fc glycans by ST6Gal-I. Biochemistry. 2009;48(41):9705–7. doi: 10.1021/bi901430h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones MB, et al. B-cell-independent sialylation of IgG. Proc Natl Acad Sci U S A. 2016;113(26):7207–12. doi: 10.1073/pnas.1523968113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones MB, et al. Role for hepatic and circulatory ST6Gal-1 sialyltransferase in regulating myelopoiesis. J Biol Chem. 2010;285(32):25009–17. doi: 10.1074/jbc.M110.104406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones MB, et al. Anti-inflammatory IgG production requires functional P1 promoter in beta-galactoside alpha2,6-sialyltransferase 1 (ST6Gal-1) gene. J Biol Chem. 2012;287(19):15365–70. doi: 10.1074/jbc.M112.345710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaplan HA, et al. Studies on the effect of inflammation on rat liver and serum sialyltransferase. Evidence that inflammation causes release of Gal beta 1 leads to 4GlcNAc alpha 2 leads to 6 sialyltransferase from liver. J Biol Chem. 1983;258(19):11505–11509. [PubMed] [Google Scholar]
- 44.Harre U, et al. Glycosylation of immunoglobulin G determines osteoclast differentiation and bone loss. Nat Commun. 2015;6:6651. doi: 10.1038/ncomms7651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thorne-Tjomsland G, et al. Increased levels of GALbeta1-4GLCNACalpha2-6 sialyltransferase pretransplant predict delayed graft function in kidney transplant recipients. Transplantation. 2000;69(5):806–8. doi: 10.1097/00007890-200003150-00022. [DOI] [PubMed] [Google Scholar]
- 46.Kreisman LS, Cobb BA. Infection, inflammation and host carbohydrates: a Glyco-Evasion Hypothesis. Glycobiology. 2012;22(8):1019–30. doi: 10.1093/glycob/cws070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andrabi R, et al. Glycans Function as Anchors for Antibodies and Help Drive HIV Broadly Neutralizing Antibody Development. Immunity. 2017;47(3):524–537e3. doi: 10.1016/j.immuni.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wei X, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422(6929):307–12. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 49.Townsley S, et al. Conserved Role of an N-Linked Glycan on the Surface Antigen of Human Immunodeficiency Virus Type 1 Modulating Virus Sensitivity to Broadly Neutralizing Antibodies against the Receptor and Coreceptor Binding Sites. J Virol. 2016;90(2):829–41. doi: 10.1128/JVI.02321-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Geijtenbeek TBH, et al. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell. 2000;100(5):587–597. doi: 10.1016/s0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- 51.Falkowska E, et al. Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity. 2014;40(5):657–68. doi: 10.1016/j.immuni.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee JH, et al. A Broadly Neutralizing Antibody Targets the Dynamic HIV Envelope Trimer Apex via a Long, Rigidified, and Anionic beta-Hairpin Structure. Immunity. 2017;46(4):690–702. doi: 10.1016/j.immuni.2017.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou T, et al. A Neutralizing Antibody Recognizing Primarily N-Linked Glycan Targets the Silent Face of the HIV Envelope. Immunity. 2018;48(3):500–513e6. doi: 10.1016/j.immuni.2018.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scanlan CN, et al. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1-->2 mannose residues on the outer face of gp120. J Virol. 2002;76(14):7306–21. doi: 10.1128/JVI.76.14.7306-7321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ataga KI, et al. Crizanlizumab for the Prevention of Pain Crises in Sickle Cell Disease. N Engl J Med. 2017;376(5):429–439. doi: 10.1056/NEJMoa1611770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lorenz HM. Technology evaluation: adalimumab, Abbott laboratories. Curr Opin Mol Ther. 2002;4(2):185–90. [PubMed] [Google Scholar]
- 57.Posey AD, Jr, et al. Engineered CAR T Cells Targeting the Cancer-Associated Tn-Glycoform of the Membrane Mucin MUC1 Control Adenocarcinoma. Immunity. 2016;44(6):1444–54. doi: 10.1016/j.immuni.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sorensen AL, et al. Chemoenzymatically synthesized multimeric Tn/STn MUC1 glycopeptides elicit cancer-specific anti-MUC1 antibody responses and override tolerance. Glycobiology. 2006;16(2):96–107. doi: 10.1093/glycob/cwj044. [DOI] [PubMed] [Google Scholar]
- 59.Uhlenbruck G. The Thomsen-Friedenreich (TF) receptor: an old history with new mystery. Immunol Commun. 1981;10(3):251–64. doi: 10.3109/08820138109093459. [DOI] [PubMed] [Google Scholar]
- 60.Polonskaya Z, et al. T cells control the generation of nanomolar-affinity anti-glycan antibodies. J Clin Invest. 2017;127(4):1491–1504. doi: 10.1172/JCI91192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Francisco R, et al. Keeping an eye on congenital disorders of O-glycosylation: a systematic literature review. J Inherit Metab Dis. 2018 doi: 10.1002/jimd.12025. [DOI] [PubMed] [Google Scholar]
- 62.Mendez-Huergo SP, et al. Galectins: emerging regulatory checkpoints linking tumor immunity and angiogenesis. Curr Opin Immunol. 2017;45:8–15. doi: 10.1016/j.coi.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 63.Campos D, et al. Probing the O-glycoproteome of gastric cancer cell lines for biomarker discovery. Mol Cell Proteomics. 2015;14(6):1616–29. doi: 10.1074/mcp.M114.046862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsai PL, Chen SF. A Brief Review of Bioinformatics Tools for Glycosylation Analysis by Mass Spectrometry. Mass Spectrom (Tokyo) 2017;6(Spec Iss):S0064. doi: 10.5702/massspectrometry.S0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aoki-Kinoshita K, et al. GlyTouCan 1.0--The international glycan structure repository. Nucleic Acids Res. 2016;44(D1):D1237–42. doi: 10.1093/nar/gkv1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eavenson M, et al. Qrator: a web-based curation tool for glycan structures. Glycobiology. 2015;25(1):66–73. doi: 10.1093/glycob/cwu090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Al Jadda K, et al. EUROCarbDB(CCRC): a EUROCarbDB node for storing glycomics standard data. Bioinformatics. 2015;31(2):242–5. doi: 10.1093/bioinformatics/btu609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ranzinger R, et al. GlycomeDB - integration of open-access carbohydrate structure databases. BMC Bioinformatics. 2008;9:384. doi: 10.1186/1471-2105-9-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ranzinger R, York WS. GlycomeDB. Methods Mol Biol. 2015;1273:109–24. doi: 10.1007/978-1-4939-2343-4_8. [DOI] [PubMed] [Google Scholar]
- 70.Miura N, et al. Functional network in posttranslational modifications: Glyco-Net in Glycoconjugate Data Bank. Methods Mol Biol. 2015;1273:149–57. doi: 10.1007/978-1-4939-2343-4_11. [DOI] [PubMed] [Google Scholar]
- 71.Maeda M, et al. JCGGDB: Japan Consortium for Glycobiology and Glycotechnology Database. Methods Mol Biol. 2015;1273:161–79. doi: 10.1007/978-1-4939-2343-4_12. [DOI] [PubMed] [Google Scholar]
- 72.Ranzinger R, et al. GlycoRDF: an ontology to standardize glycomics data in RDF. Bioinformatics. 2015;31(6):919–25. doi: 10.1093/bioinformatics/btu732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Herget S, et al. GlycoCT-a unifying sequence format for carbohydrates. Carbohydr Res. 2008;343(12):2162–71. doi: 10.1016/j.carres.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 74.Lutteke T. Handling and conversion of carbohydrate sequence formats and monosaccharide notation. Methods Mol Biol. 2015;1273:43–54. doi: 10.1007/978-1-4939-2343-4_4. [DOI] [PubMed] [Google Scholar]
- 75.Damerell D, et al. Annotation of glycomics MS and MS/MS spectra using the GlycoWorkbench software tool. Methods Mol Biol. 2015;1273:3–15. doi: 10.1007/978-1-4939-2343-4_1. [DOI] [PubMed] [Google Scholar]
- 76.Stalnaker SH, et al. Mammalian O-mannosylation: unsolved questions of structure/function. Curr Opin Struct Biol. 2011;21(5):603–9. doi: 10.1016/j.sbi.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Takeuchi H, Haltiwanger RS. Significance of glycosylation in Notch signaling. Biochem Biophys Res Commun. 2014;453(2):235–42. doi: 10.1016/j.bbrc.2014.05.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zachara N, et al. Essentials of Glycobiology. 2015. The O-GlcNAc Modification; pp. 239–251. (rd et al. eds) [Google Scholar]
- 79.Hart GW. Three Decades of Research on O-GlcNAcylation - A Major Nutrient Sensor That Regulates Signaling, Transcription and Cellular Metabolism. Front Endocrinol (Lausanne) 2014;5:183. doi: 10.3389/fendo.2014.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]