Abstract
Cell division involves mechanical processes, such as chromosome transport and centrosome separation. Quantitative micromanipulation-based approaches have been central to dissecting the forces driving these processes. We highlight two biophysical assays that can be employed for such analyses. First, an in vitro “mini-spindle” assay is described that can be used to examine the collective mechanics of mitotic motor proteins cross-linking two microtubules. In the spindle, motor proteins (e.g., kinesin-5, kinesin-14, and dynein) can localize to overlapping microtubules that slide relative to each other, work as an ensemble, and equilibrate between cytoplasm and the microtubules. The “mini-spindle” assay can recapitulate these features and allows measurements of forces generated between adjacent microtubules and their dependence on filament orientation, sliding speed, overlap length, and motor protein density. Second, we describe a force-calibrated microneedle-based “whole-spindle” micromechanics assay. Microneedle-based micromanipulation can be a useful technique to examine cellular scale mechanics, but its use has been restricted by the difficulty in getting probes to penetrate the plasma membrane without disrupting cell physiology. As detailed here, the use of cell-free extracts prepared from metaphase-arrested Xenopus eggs can address this limitation. These micromanipulation studies also benefit from the use of frozen stocks of Xenopus egg extract. Together, these approaches can be used to decipher how micromechanics and biochemical activities ensure successful cell division.
Keywords: Spindle assembly, Microtubules, Kinesin-5, In vitro reconstitution, Xenopus egg extract, Frozen extract, Force measurement, Optical tweezers, Force-calibrated microneedles
1. INTRODUCTION
During cell division, chromosomes and centrosomes can undergo directional intracellular movement across micrometers within minutes. These dynamics are regulated, by integrating micromechanical and biochemical cues, to ensure accurate chromosome segregation (Compton, 2000; Kapoor, 2017; Walczak & Heald, 2008). Quantitative micromanipulation can be a powerful approach for dissecting force-based mechanisms underlying such cellular dynamics. A major challenge, which has restricted the use of these approaches, is that the typically large, micrometer-sized micromanipulation probes must be introduced into the cytoplasm of living cells without perturbing physiology (Dufrene et al., 2011). On the other hand, several biochemical techniques have been developed for examining cell division mechanisms in cell-free systems (e.g., Desai, Murray, Mitchison, & Walczak, 1999; Kapitein et al., 2005). With an appropriate choice of biophysical force probes, these cell-free systems allow quantitative analyses. This chapter presents two biophysical assays that can be used to examine the micromechanics of the cell division apparatus.
First, we describe a “mini-spindle” micromechanics assay, in which forces generated by mitotic motor proteins cross-linking pairs of microtubules can be measured using optical tweezers. In general, optical tweezers are used to analyze forces generated by single motor proteins (e.g., vesicle-transporting kinesin) by firmly attaching these enzymes to microbeads and tracking motion along single filaments (e.g., microtubules) (Fazal & Block, 2011). On the other hand, spindle motor proteins (e.g., kinesin-5, kinesin-14, and dynein) often cross-link overlapping microtubules, work as an ensemble to slide adjacent filaments, and equilibrate between cytoplasm and the microtubule-based structure (Kapoor, 2017). The “mini-spindle” assay recapitulates these spindle features, allowing for analysis of collective motor protein dynamics that occur within overlapping microtubules (Shimamoto, Forth, & Kapoor, 2015). In this assay, the extent of microtubule overlap, filament sliding speed, and relative filament orientation can be controlled. This assay is performed using an inverted microscope equipped with TIRF (total internal reflection fluorescence) and epifluorescence illuminators, which allow for imaging the motion and localization of individual motor proteins on microtubules.
Second, we describe a “whole-spindle” micromechanics assay, in which bipolar spindles assembled in Xenopus egg extracts can be micromanipulated using a pair of force-calibrated glass microneedles. Microneedles are particularly useful for applying and measuring forces on the order of nN (vs ~ pN using optical tweezers) and have been used to probe forces in cells. In particular, the seminal work by Nicklas employed microneedle probes to capture mitotic chromosomes in a cell and examine how forces control the metaphase to anaphase transition (Li & Nicklas, 1995; Nicklas, 1997). The grasshopper spermatocyte cells, which have particularly robust cell membranes, were well suited for these micromanipulation experiments (Zhang & Nicklas, 1999). The assay we describe here uses Xenopus egg extracts, which is a cell-free system that is devoid of the plasma membrane and the cell cortex (Desai et al., 1999). This allows us to perform measurements without perturbing these cellular boundaries (Shimamoto, Maeda, Ishiwata, Libchaber, & Kapoor, 2011). The Xenopus system also allows a range of chemical and biochemical perturbations (e.g., pharmacological inhibition, immunodepletion, and recombinant protein addition) to examine molecular mechanisms. This assay is performed using a confocal microscope equipped with two excitation lasers for imaging microtubules and chromosomes in the spindle.
2. OPTICAL TRAP-BASED “MINI-SPINDLE” ASSAY
The “mini-spindle” assay described here was used to measure forces generated within overlapping microtubules cross-linked by kinesin-5 (Shimamoto et al., 2015). Kinesin-5 is a bipolar homotetrameric motor protein, which uses pairs of dimeric motor domains to cross-link and push apart antiparallel microtubules with their minus-ends leading. Kinesin-5 also promotes the formation of parallel microtubule bundles. Examining the forces exerted by kinesin-5 within overlapping microtubules is crucial to understand cell division, as it plays essential roles in centrosome separation and bipolar spindle assembly (Compton, 2000).
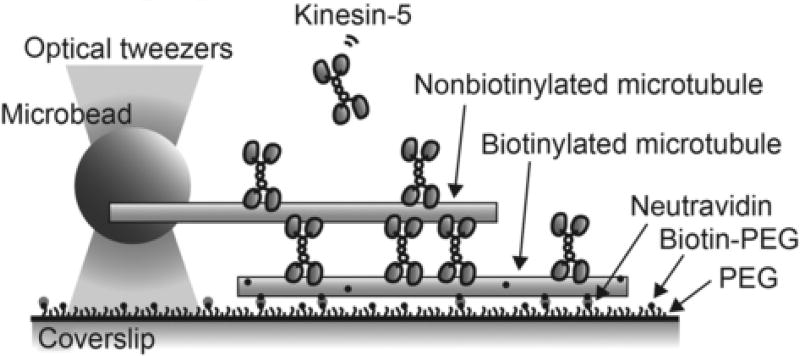
Priorto examining the force-generating functions of kinesin-5 we need to consider : (i) collective activity of multiple motors within regions of micron-sized microtubule overlap, (ii) dynamic turnover between the spindle and the cytoplasm, and (iii) spatial localization such as that near the spindle equator where many antiparallel microtubules overlap, or as observed near the spindle pole where microtubules are mainly parallel, and (iv) relative sliding of adjacent filaments, which is associated with spindle formation and polewards flux (Mitchison, 1989) in the metaphase spindle. This assay is designed to recapitulate all these features. Specifically pairs of microtubules are assembled in the presence of kinesin-5, resulting in a microtubule-based “mini-spindle” (Fig. 1). Specific labeling of one of the two filaments in each microtubule pair allows the labeled filament to be immobilized onto the bottom coverslip of a flow cell. The other filament, not attached to the surface, is captured by an optically trapped microbead. In this geometry, kinesin-5 motors slide the bead-attached filament over the surface-immobilized one, pushing the microbead away from the center of the optical trap. The magnitude of this pushing force can be determined by measuring the displacement of the microbead and the stiffness of the trap. Kinesin-5 molecules are also present in solution and can associate with the overlapping microtubules. The extent of filament overlap can be determined using fluorescence images of microtubules. Further, a C-terminal GFP tag on kinesin-5 allows analyses of the motion and localization of individual tetrameric motor protein molecules along microtubules. The assay involves preparation of labeled microtubules for surface immobilization, passivation of coverslips for minimizing nonspecific protein binding, and coating of microbeads for microtubule capture. While the assay is described for kinesin-5, it can be used for other proteins such as NuMA and PRC1 (Forth, Hsia, Shimamoto, & Kapoor, 2014), which also target to the spindle and cross-link microtubules.
Fig. 1.
“Mini-spindle” force assay. Schematic illustration showing a side view of the assay in which a pair of microtubules cross-linked by an ensemble of kinesin-5 is attached to coverslip surface and the force developed within the pair is measured by single-beam optical tweezers.
2.1. Assay Design
2.1.1. Microscope
For the measurement of forces that develop within a microtubule overlap, optical tweezers are an excellent choice over other biophysical methods such as those using magnetic tweezers or atomic force microscopy. This is because optical tweezers allow facile micromanipulation of trapped particles (e.g., latex beads) in three dimensions and are well suited for controlling the interaction geometry of microtubule pairs (e.g., filament orientation, overlap length). The use of a strong laser (power: ~ 10–100 mW), which is required for achieving a stable trap, is the major source of background noise in fluorescence imaging. However, proper choice of the laser wavelength (e.g., 1064 nm) and placement of additional optical filters (e.g., an IR-cut filter) in an observation light path can be used to eliminate most of the background associated with the trap laser, allowing for the simultaneous use of optical tweezers and single molecule fluorescence imaging.
The imaging of kinesin-5 is performed using a TIRF-based method, in which the excitation light penetrates within only a few 100 nm past the coverslip surface and thus can be used to image GFP-tagged kinesin-5 molecules proximal to the surface but not those diffusing in solution at a greater distance from the coverslip. On the other hand, microtubules are imaged using epifluorescence illumination such that the filaments located microns away from the coverslip surface, such as those captured by an optically trapped microbead, can be visualized. The flow cell is placed on a three-axis piezo stage, which allows controlling the motion of surface-immobilized microtubules relative to the optically trapped filament. Detailed construction procedures for optical tweezers and TIRF systems have been described by others (Joo & Ha, 2012; Neuman & Block, 2004; Selvin & Ha, 2007).
2.1.2. Surface chemistry
Coverslip surface in a flow cell is functionalized such that microtubules can be immobilized but nonspecific binding of other proteins is reduced. To this end, the surface is passivated by grafting a layer of polymer brush comprised of polyethylene glycol (PEG) (Selvin & Ha, 2007). Incorporation of biotinylated PEG in the polymer layer allows for specific binding of biotinylated microtubules by means of neutravidin, which has multiple biotin-binding sites. Blocking with casein minimizes nonspecific protein binding to potential surface “defects” that remain uncoated with polymer brush. Microtubules that are held by optical tweezers are not biotinylated and therefore, do not attach to the surface; the association of these microtubules with the surface-immobilized filaments depends on kinesin-5.
2.1.3. Bead–microtubule linkage
To achieve a stable attachment of microtubules with a microbead, we use ATPase-dead rigor kinesin (Rice et al., 1999). Densely decorating the microbead surface with rigor kinesin results in bead-microtubule attachments that can withstand up to ~ 50 pN force, which is greater that the force generated by tens of kinesin-5 molecules. An avidin–biotin linkage could be an alternative choice for the bead–microtubule attachment. However, in our assay design an incorporation of biotin in the bead-attached filaments would increase the risk of their binding on coverslips surface, which should be avoided for measuring the motor protein-dependent forces.
2.2. Buffers, Reagents, and Equipment
For microtubule preparation
Bovine or porcine tubulins (unlabeled, biotinylated, and X-rhodamine labeled, 5–10 mg/mL; each prepared according to protocols from Tim Mitchison's lab (https://mitchison.hms.harvard.edu/microtubules) and stored at − 80°C). We note that these tubulins are mixtures of different isotypes and can have posttranslational modifications that impact interactions with associated proteins and polymerization properties. Methods to generate recombinant tubulins have been developed and may be used to address some of these potential limitations (Minoura et al., 2013; Ti et al., 2016; Vemu et al., 2016)
BRB80: 80 mM K-PIPES (pH 6.8), 1 mM MgCl2, 1 mM EGTA
GMPCPP
DTT
Taxol
TLA-100 ultracentrifuge (Beckman)
TLA120.1 rotor (Beckman)
For coverslip passivation
Vectabond (Vector Labs, product #SP-1800)
2% Micro-90 detergent in ddH2O
0.1 M KOH
100% Ethanol
Acetone, HPLC grade
mPEG-SVA and Biotin-PEG-SVA, MW 5000 (Laysan-Bio) in 0.1 M NaHCO3
Humidified chamber (can be prepared by putting moist Kimwipes in a plastic box)
For beads coating
Rigor kinesin (K560 G234A, expressed in bacteria and purified using Ni-NTA followed by a Mono-Q anion exchange column (Rice et al., 1999), ~ 5 µM)
Carboxylated microbeads (1 µm in diameter, ~ 5 × 1010 particles/mL or ~ 100 pM, Polysciences, Inc)
Wash buffer: 0.1 M K-Hepes, pH 8.0
Dilution buffer: 1 × BRB80 plus 1 mM DTT
Storage buffer: 1 × BRB80 plus 1 mM DTT and 0.5 mg/mL α-casein
Tabletop centrifuge (Beckman Microfuge 18 or similar)
For “mini-spindle” force assay
BRB80: 80 mM K-PIPES (pH 6.8), 1 mM MgCl2, 1 mM EGTA
Casein buffer: 0.5 mg/mL α-casein in 1 × BRB80 plus 1 mM DTT
Neutravidin buffer: 0.2 mg/mL in 1 × BRB80 plus 1 mM DTT and 0.5 mg/mL α-casein
Kinesin-5-GFP (prepared according to Weinger, 2011)
ATP
Taxol
KCl
Oxygen scavenging system: 4.5 mg/mL glucose, 34 U/mL glucose oxidase, and 350 U/mL catalase
VaLaP: an equal mixing ratio of vaseline, lanolin, and paraffin
2.3. Methods
2.3.1. Preparation of microtubules
Microtubules are prepared by polymerizing a mixture of unlabeled tubulin, biotinylated tubulin, and X-rhodamine-labeled tubulin at appropriate mixing ratios and concentrations. Incorporation of biotinylated tubulin into the microtubule lattice provides stable tethering of the filaments onto biotin-PEG coverslip surface via neutravidin. In our experience, excess biotinylated tubulin (> 0.3 µM, or 10%) can interfere with microtubule formation. Therefore, the mixing ratio should be carefully optimized in order to obtain microtubules of adequate lengths for force measurement assay (e.g., 5–20 µm). X-rhodamine-labeled tubulin is also incorporated into the microtubule lattice for fluorescence imaging. Nonbiotinylated microtubules are labeled ~ twofold brighter than biotinylated microtubules for distinguishing them in imaging assays. The prepared microtubules should be stable at room temperature for ~ 1 week.
Mix unlabeled, biotinylated, and X-rhodamine-labeled tubulins on ice at appropriate ratios in BRB80 plus 1 mM DTT. A good starting point is 20:1:1 for biotinylated microtubules and 10:0:1 for nonbiotinylated microtubules. Add 0.5 mM GMPCPP to each reaction.
Incubate each tubulin mix on ice for 10 min and then spin in a TLA120.1 rotor at 90,000 rpm for 7 min at 2°C.
The clarified tubulin mix can be snap-frozen and stored at − 80°C, or proceed to polymerization.
Add BRB80 plus 1 mM DTT to each tubulin mix such that the total tubulin concentration is 0.3–0.5 mg/mL. A lower tubulin concentration typically yields longer microtubules.
Incubate the reaction for 2 h at 37°C to promote polymerization.
Dilute the reaction in BRB80 plus 1 mM DTT and 20 µM Taxol.
Spin in a TLA120.1 rotor at 90,000 rpm for 7 min at 35°C.
Resuspend pelleted microtubules in BRB80 plus 1 mM DTT and 20 µM Taxol. Store at room temperature until required.
2.3.2. Passivation of coverslips
Coverslips used in flow cell chambers are passivated by amino-silanating the surface and then grafting a mixture of PEG and biotinylated PEG onto it. The mixing ratio of PEG to biotin-PEG should be determined empirically, but a good starting point would be 80:1. If poor microtubule binding is observed, the amount of biotin-PEG can be increased. The prepared coverslips can be kept in a vacuum chamber for 6–7 days without noticeable deterioration.
Clean coverslips (18 × 18 mm square) by sonication in buffers in the following order: ddH2O, 1 N KOH, ddH2O, 2% Micro-90, and then ddH2O.
Wash the coverslips in ethanol, dried in a stream of nitrogen, and then immerse them in Vectabond reagent for 5 min for amino-silanation of the surface. Wash the coverslip surface with ddH2O.
Prepare a mixture of PEG and biotinylated PEG in 0.1 M NaHCO3 and graft it onto the amino-silanated coverslip surface. This can be done by making a sandwich of PEG suspension between two coverslips.
Following 3 h incubation in a humidified chamber, wash the coverslips with ddH2O, dried in a stream of nitrogen, and then stored in a vacuum container until required.
2.3.3. Preparation of microbeads
Microbeads used to capture microtubules in an optical trap are prepared by coating the surface of 1-µm diameter carboxylated microbeads with ATPase-dead “rigor” kinesin. The prepared beads should be kept on ice until required and used within 3–4 h.
Wash a suspension of carboxylated microbeads with wash buffer using a centrifugal spin (10,000 rpm, 5 min, at room temperature). Repeat the wash 3 ×.
Resuspend the bead pellet in wash buffer such that the bead concentration is ~ 4 pM.
Add freshly thawed rigor kinesin to the bead suspension such that the final protein concentration is ~ 5 µM (protein:beads = 106:1).
Incubate the mixture for 15 min on ice.
Remove unbound kinesins by spinning down the beads in dilution buffer at 10,000 rpm for 2 min. A prolonged spinning is not recommended as the beads become clumped.
Repeat the wash once, and then resuspend the pellet in storage buffer. Keep the bead suspension on ice until required.
2.3.4. “Mini-spindle” force assay
For the measurement of forces developed within overlapping microtubules, pairs of microtubules, each consisting of a nonbiotinylated and a biotinylated microtubule, are assembled on a PEG-coated coverslip surface. To measure the force, the leading end of a nonbiotinylated microtubule sliding over a surface-immobilized microtubule, which is driven by kinesin-5, is captured by a microbead trapped in optical tweezers.
Assemble a flow cell using a clean slide and a PEG-coated coverslip with two stripes of double-stick tape as a spacer. The channel volume should be 10–20 µL.
Perfuse buffers into the channel and incubate for defined periods in the following order: BRB80 for 1 min, 0.5 mg/mL casein buffer for 2 min, and 0.2 mg/mL neutravidin buffer for 5 min. Flush the channel with BRB80 plus 20 µM Taxol.
Perfuse biotinylated microtubules into the channel and incubate for 10 min to allow them to bind to coverslip surface. Flush unbound filaments with BRB80 plus Taxol.
Perfuse assay buffer, which consists of nonbiotinylated microtubules, rigor kinesin beads, 2 mM MgATP, 1 nM kinesin-5-GFP, 0.5 mg/mL α-casein, 20 µM Taxol, 1 mM DTT, 1 × BRB80, and oxygen scavenging system. Seal the channel using VaLaP.
Place the flow cell in the assay microscope. Brightly labeled filaments should be nonbiotinylated microtubules, which diffuse in solution and land on dimly labeled biotinylated microtubules on coverslip surface. A fraction of microtubule pairs should show directional sliding, which is the indicative of antiparallel filaments.
Capture a free diffusing microbead in optical tweezers.
While avoiding the contact with surface microtubules and those in solution, bring the captured microbead into contact with an overhanging end of the nonbiotinylated microtubule in a microtubule pair on the coverslip surface. The attachment should be near (3–5 µm) the microtubule overlap region to avoid filament buckling due to motor force, which will be developed as soon as the microbead is attached to the filament.
Let the microtubule pair develop force and reach a balance with opposing force from the trap. Once equilibrium is reached, determine the magnitude of force.
Determine the amount of microtubule overlap and the number of motor protein molecules using fluorescence images. The microtubule overlap can be defined as the higher intensity area of the microtubule pair. The motor number can be estimated based on the total intensity of GFP signal within the overlap and an average intensity of single kinesin-5-GFP measured separately.
In completing the force measurement, the bead-attached microtubule can be dissociated from the surface microtubule by pulling apart the two filaments. This can be done by moving the trapped bead relative to the coverslip surface. Micromanipulating the position and orientation of the detached filament allows for reassembling the microtubule pair with a surface microtubule in different geometry (e.g., overlap length, relative polarity). Antiparallel microtubule pairs show unidirectional force. On the other hand, parallel microtubule pairs typically exhibit bidirectional force fluctuation. The force measurement can be repeated until the bead–microtubule linkage breaks.
3. MICRONEEDLE-BASED “WHOLE-SPINDLE” ASSAY
This assay is developed for examining mechanical responses of the spindle apparatus to applied force (Shimamoto et al., 2011). The measurement relies on the use of force-calibrated microneedles, which are capable of applying and measuring forces on the order of nN and their position in three dimensions can be readily controlled (Shimamoto & Kapoor, 2012). By using two microneedles, single metaphase spindles assembled in Xenopus egg extract, a widely used cell-free system to study mitosis (Desai et al., 1999; Hannak & Heald, 2006), can be captured and micromanipulated with desired force magnitude, speed, and direction. The assay involves (i) preparation of extracts, (ii) fabrication and calibration of microneedles, and (iii) micromanipulation of spindles. Typical assays are performed using freshly prepared extract, which retains activity for only 2–8 h and thus allows for a limited number of data collection in each preparation batch. This can be circumvented by preparing frozen extracts that can retain nearly endogenous spindle assembly activity (Takagi & Shimamoto, 2017), for which the protocol is also detailed here.
3.1. Assay Design
3.1.1. Micromanipulation
The spindle assembled in Xenopus egg extract is a bipolar, micron-sized structure free-floating in the cytoplasm with continuous turnover of components (e.g., tubulin) (Desai et al., 1999; Mitchison et al., 2005). Micromanipulating this dynamic structure requires : (1) forces on the order of nN to be applied and measured in controlled magnitudes and directions, and (2) dynamics of spindle components must be retained. The primary reason of employing microneedles is that the use of two micromanipulation probes can be kept in close proximity (< 2–3 µm at minimum) such that the spindle apparatus can be captured without immobilizing it on a substrate. Other typical biophysical force measurement tools such as magnetic tweezers and atomic force microscopy do not readily incorporate this feature. Microneedles are also capable of applying and measuring forces ranged from sub-nN to µN by monitoring the movement of the precalibrated microneedle tips, which cannot be easily achieved using optical trap-based assays. Typically, optical tweezers can be used to study forces up to a few hundred pN, but formation of an optical gradient in crude extract results in an accumulation of unwanted particles (e.g., micron-sized vesicles) in the trap focus. Our setup is equipped with a pair of force-calibrated microneedles; one is stiff (> 100 nN/µm) and attached to a piezo actuator such that the tip's motion can be precisely controlled by applying voltage to the actuator. The tip of the other microneedle is much more flexible (~ 0.1–0.5 nN/µm) and acts as a force sensor. The tips of these two microneedles can be inserted near the ends of the bipolar spindle and then be used to stretch the structure along selected directions. The force-sensing microneedle tips are made compliant enough to probe forces with sub-nN precision while being rigid enough to control motion in viscous extract. The tips of microneedles and the surface of the coverslip in the experimental chamber are passivated for minimizing nonspecific binding of spindle and extract proteins. The extract is covered by mineral oil to prevent sample desiccation during the measurement.
3.1.2. Imaging
Spindle microtubules and chromosomes are imaged using a spinning-disk confocal system equipped with 488 and 561 nm excitation lasers. Confocal imaging has several advantages over epifluorescence illumination, as it allows for imaging thick samples with lower out-of-focus background and less photobleaching/toxicity. Further, the spinning-disk confocal allows imaging “snap-shots” of fast-moving objects at subsec intervals, which cannot be achieved by other confocal methods, such as laser-scanning systems (Oreopoulos, Berman, & Browne, 2014).
3.2. Buffers, Reagents, and Equipment
For spindle assembly in Xenopus egg extract
PMSG: 100 IU/mL in ddH2O
hCG: 1000 IU/mL in ddH2O
MMR: 5 mM Na-Hepes, 100 mM NaCl, 0.1 mM EDTA, 2 mM KCl, 1 mM MgCl2, 2 mM CaCl2, pH 7.85 at 25 × stock
XB: 10 mM Hepes, 100 mM KCl, 1 mM MgCl2, 0.1 mM CaCl2, 50 mM sucrose, pH 7.7
CSF-XB: 10 mM Hepes, 5 mM EGTA, 100 mM KCl, 2 mM MgCl2, 0.1 mM CaCl2, 50 mM sucrose, pH 7.7
Cysteine: 4% in XB
Protease inhibitor cocktail (100 × stock): 10 mg/mL each of leupeptin, pepstatin A, and cymostatin
Cytochalasin D (100 × stock): 10 mg/mL
Energy mix (50 × stock): 190 mM creatine phosphate, 25 mM MgATP, 5 mM EGTA, pH 7.7 adjusted by KOH
Demembranated sperm (~ 2 × 104/µL, prepared from male Xenopus laevis testis)
X-rhodamine tubulin (5–10 mg/mL)
SYTOX green DNA dye: 1 mM stock in CSF-XB
SW-55 rotor and tubes (Beckman)
Tabletop centrifuge (Eppendorf 5424R or similar)
Centrifugal filter unit (Amicon 100-kDa filter, 500 µL capacity)
Tube cooler (should allow for sample cooling at ~−1°C/min)
For microneedle preparation
Glass rod, 1 mm in diameter
Glass capillary puller
Microforge
Microgas burner
Platinum wire (for calibrated weights)
Inverted microscope with a low magnification objective (e.g., 20 ×)
For coverslip coating
Coverslips
Coverslip holder
Agarose
Alkaline ethanol solution: 5% sodium hydroxide and 80% ethanol in ddH2O
For spindle micromechanics assay
Micromanipulators (Narishige's MHW-3 or similar)
Inverted microscope with a spinning-disk confocal (Yokogawa CSU-X1 or similar) and a high-sensitivity camera (Andor Neo or similar)
Rubber plate with a central aperture, used to make an open micromanipulation chamber
Mineral oil, used to cover the sample in the chamber
3.3. Methods
3.3.1. Preparation of extracts
Cell-free extract arrested at metaphase of meiosis II is prepared from unfertilized eggs of X. laevis. The freshly prepared extract can be used for several hours for spindle assembly assays. This extract can be further processed to make frozen stocks that retain nearly endogenous spindle assembly activity. Prepared extracts are supplemented with demembranated Xenopus sperm and cycle through interphase back into metaphase to assemble bipolar spindles.
Fresh CSF extract
Prime female frogs by injection of 50 IU PMSG > 3 days and induce laying eggs by injection of 500 IU hCG ~ 16 h prior to the extract preparation.
Collect eggs in MMR.
Wash eggs in MMR and then decant to remove as much MMR as possible.
Dejelly the eggs in cysteine in a glass beaker precoated with gelatin. Keep swirling the beaker such that the treatment is homogeneous. The dejellying typically completes in 3–6 min. Remove as much cysteine as possible.
Wash the dejellied eggs in XB 3 ×. Remove as much CSF-XB as possible at each wash step.
Wash eggs in CSF-XB 3 ×.
Wash eggs in CSF-XB plus protease inhibitor cocktail (PI) 3 ×.
Transfer the eggs to clear centrifugal tubes that are prefilled with 1 mL CSF-XB plus PI and cytochalasin D.
Pack the eggs in tubes by two rounds of low speed spins (1st spin: 2000 rpm for 1 min, second spin: 2000 rpm for 1 min followed by 3000 rpm for 30 s). Aspirate off as much buffer as possible after each spin.
Spin in a SW55 swing rotor at 10,000 rpm for 15 min at 16°C.
Collect cytoplasmic extract fraction using a 16-gauge needle and a 1-mL syringe by punctuating the tube wall and syringe out the fraction.
Transfer the extract in a precooled test tube, add LPC, cytochalasin D, and energy mix, and keep on ice until required.
Frozen extract preparation capable of spindle assembly
Immediately after the CSF extract preparation described earlier, transfer 200 µL each of fresh extract to centrifugal filter devices. 200 µL per device typically yields the best result.
Spin at 17,000 × g for 10 min at 4°C in a tabletop centrifuge. This step should be completed within 30 min following the fresh extract preparation, as a prolonged incubation of extracts markedly reduces the quality of frozen stocks.
Separate the filter device from each centrifugal unit and place it upside down in a collection tube. Spin for ~ 10 s in a tabletop centrifuge (~ 2000 × g) to collect the concentrate in the tubes. Meanwhile, keep the flow-through fraction on ice.
Transfer the concentrate and flow-through fractions to separate test tubes precooled on ice. Make sure all the viscous concentrate fraction in each tube is collected.
Place the test tubes in a tube cooler, which has been precooled at 4°C.
Place the tube cooler in a − 80°C deep freezer. The frozen extract fractions can be stored at − 80°C or in a vapor phase of liquid nitrogen for several weeks to months.
Prior to its use, take out each fraction of frozen concentrate and flow-through from the freezer and let them thaw on ice for 10 min.
Transfer the entire flow-through fraction to the concentrate and mix thoroughly.
Incubate the extract on ice for > 20 min and the extract should recover its activity.
Spindle assembly
In a test tube, add demembranated sperm (~ 2 × 102 nuclei/µL at final) and calcium (0.4 mM at final) to CSF-arrested extract that was prepared freshly or recovered from the frozen stock.
Incubate for 80 min at 18°C.
Add an equal volume of CSF-arrested extract to the cycling reaction and incubate for 60 min.
For fluorescence imaging, add DNA dye (e.g., SYTOX green for 488°nm excitation) and X-rhodamine tubulin to the extract.
3.3.2. Fabrication and calibration of microneedles
Microneedles used for measuring spindle force are prepared using a burner and a microforge, by heating and pulling a glass rod of 1-mm diameter. The processed tips are typically of 1 µm in diameter and 50–200 µm in length, of which stiffness is determined by cross-calibrating each microneedle tip against a reference microneedle of a known stiffness. The reference microneedle's stiffness is calibrated by measuring the tip's bending against the gravitational force generated by thin platinum wires of known weights.
Preparation of microneedles
Heat and pull the middle of a glass rod using a capillary puller.
Place the glass rod in a microforge and contact its pulled tip with a glass bead, which is preformed on the microforge's heating filament.
Raise the temperature of the heating filament. As soon as the tip and the glass bead melt and fuse together, quickly pull the glass rod (~ 1–10 cm in ~ 100 ms) away from the bead such that the fused part is stretched to form a long and thin fiber. The speed and timing of the pulling and the filament temperature determine the microneedle tip's length and diameter. A fast pulling at high temperature typically yields a long and thin, flexible fiber.
Cut the tip of the fiber to a desired length using precision scissors.
Calibration of reference microneedles
Mount a reference microneedle horizontally on an inverted microscope.
Cut a piece of platinum wire into various lengths (~ 1–20 mm). Estimate the weight of the individual pieces based on its average mass per unit length. Use forceps to bend each piece of the wire into a horseshoe shape.
Hang the horseshoe-shaped piece of wire at the tip of the reference microneedle. Using a low magnification objective (10 × or 20 ×), measure the vertical displacement of the tip caused by the wire piece. Record the weight and the resultant displacement of the needle tip.
Repeat step 4 for several different pieces of wire.
Plot the weight (y-axis) against the displacement (x-axis). Determine the stiffness (= kREF, N/m) by calculating the slope using linear regression.
Calibration of micromanipulation microneedles
Hold the precalibrated reference microneedle and a microneedle to be calibrated, each in a micromanipulator mounted on an inverted microscope.
Bring the tips of the two microneedles into contact and push the one tip against the other. Record the bending displacements of the tips of two microneedles, and then withdraw the microneedle back to its original position. Repeat the measurement at several different equilibrium points.
Determine the stiffness of the micromanipulating microneedle (= km) according to km = kr·xr/xm, where kr is the precalibrated stiffness of the reference microneedle, and xr/xm is the relative bending displacement of the tips of the reference (xr) and micromanipulating microneedles (xm) measured at the equilibrium points in step 2. Repeat the calibration using reference microneedles of different stiffness values.
After the tip of the micromanipulation microneedle is calibrated, bend its shaft using a burner. The degree of bending should be such that it allows for spindle micromanipulation from nearly vertical angle (i.e., perpendicular to coverslip surface).
Store the force-calibrated microneedles in a dust-free box until required. Prevent mechanical shock or vibration as it may damage the calibrated tips.
3.3.3. Preparation of agarose-coated coverslips
Coverslips used in assay chambers are precoated with a thin layer of agarose in order to minimize nonspecific binding of extract and spindle proteins.
Wash coverslips in alkaline ethanol by sonication for 20 min.
Wash the coverslips 5–6 × with ddH2O and then sonicate for 5–10 min.
Repeat step 2 3 ×.
Dry the coverslips in a clean bench.
Prepare 0.1% (w/v) agarose solution in a conical tube. Heat to boil until all the agarose is dissolved. Make sure the weight of the solution is maintained during boiling and add ddH2O if necessary. Then, lower the temperature to ~ 80°C and maintain it.
Dip a cleaned coverslip into the warm agarose solution for a few seconds and then gently retrieve it from the solution. Too thick agarose layer would reduce imaging quality. Too thin layer would not prevent stickiness of proteins. The surface of a properly coated coverslip typically has a rainbow-colored reflection.
Place the coated coverslips in a rack and dry them in a clean bench for 10–20 min. The prepared coverslips can be stored in a humidified chamber at 4°C for 2–3 days.
3.3.4. “Whole-spindle” force assay
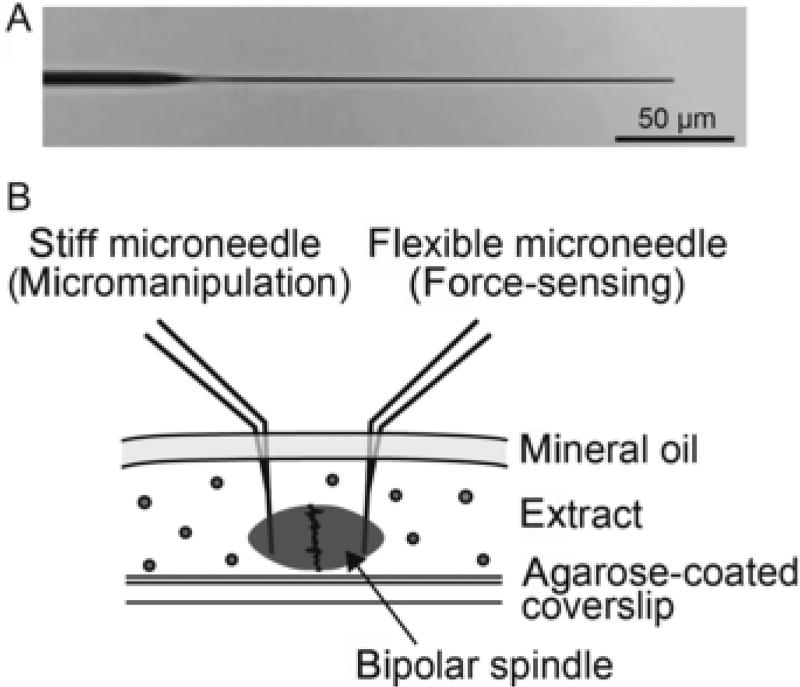
The spindle force measurement assay is performed using an inverted microscope equipped with two three-axis micromanipulators, a spinning-disk confocal unit, two excitation lasers, and a high-sensitivity camera. Preassembled single metaphase spindles in CSF-arrested egg extract can be micromanipulated using a pair of force-calibrated glass microneedles (Fig. 2).
Assemble an open experimental chamber by attaching a rubber plate (~ 10 mm central aperture) onto an agarose-coated coverslip. Vaseline or silicone can be used for achieving tight attachment between the plate and the coverslip.
Spread ~ 4 µL of metaphase-arrested extract, which contained preassembled spindles, on the coverslip surface. Cover the extract with mineral oil.
Place the chamber in an inverted microscope and sit stand for 2–3 min to settle.
Locate a bipolar spindle of typical morphology (30–40 µm in length, focused poles, and tight equatorial alignment of chromosomes) at the center of the field of view.
Move down the microneedles and insert their tips into the bipolar spindle. For spindle micromanipulation along the pole-to-pole axis, the two microneedle tips can be inserted into each half of the bipolar structure. For micromanipulation along the perpendicular axis, the tips can be inserted into one half of the spindle. The microneedle tips are brought down to above ~ 1 µm from coverslip surface to prevent frictional drag during the measurement.
Move one microneedle tip against the other to stretch or compress the spindle. This can be done by manually rotating the micromanipulator's knob or applying a control voltage to the piezo actuator, which can be attached to the shaft of the manipulating microneedle.
Monitor the motion of the force-sensing microneedle tip to estimate the amount of force developed within the structure. This can be done by multiplying the displacement of the tip from its equilibrium by the tip's precalibrated stiffness. Deformation can be estimated based on spindle microtubule images acquired using the confocal system.
Fig. 2.
“Whole-spindle” force assay. (A) Bright-field image showing the tip of a force-calibrated microneedle. The tip's diameter is ~ 1 µm. (B) Schematic illustrating showing a side view of the assay in which a single bipolar spindle assembled in Xenopus cell-free egg extract is micromanipulated using a pair of force-calibrated microneedles.
Acknowledgments
We thank Dr. Jun Takagi for development of the frozen extract protocol, Dr. Scott Forth for optical trap construction, JSPS KAKENHI 16748896 (to Y.S.) and NIH GM65933 (to T.M.K.) for support.
References
- 1.Compton DA. Spindle assembly in animal cells. Annual Review of Biochemistry. 2000;69:95–114. doi: 10.1146/annurev.biochem.69.1.95. [DOI] [PubMed] [Google Scholar]
- 2.Desai A, Murray A, Mitchison TJ, Walczak CE. The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods in Cell Biology. 1999;61:385–412. doi: 10.1016/s0091-679x(08)61991-3. [DOI] [PubMed] [Google Scholar]
- 3.Dufrene YF, Evans E, Engel A, Helenius J, Gaub HE, Muller DJ. Five challenges to bringing single-molecule force spectroscopy into living cells. Nature Methods. 2011;8:123–127. doi: 10.1038/nmeth0211-123. [DOI] [PubMed] [Google Scholar]
- 4.Fazal FM, Block SM. Optical tweezers study life under tension. Nature Photonics. 2011;5:318–321. doi: 10.1038/nphoton.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forth S, Hsia KC, Shimamoto Y, Kapoor TM. Asymmetric friction of non-motor MAPs can lead to their directional motion in active microtubule networks. Cell. 2014;157:420–432. doi: 10.1016/j.cell.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hannak E, Heald R. Investigating mitotic spindle assembly and function in vitro using Xenopus laevis egg extracts. Nature Protocols. 2006;1:2305–2314. doi: 10.1038/nprot.2006.396. [DOI] [PubMed] [Google Scholar]
- 7.Joo C, Ha T. Single-molecule FRET with total internal reflection microscopy. Cold Spring Harbor Protocols. 2012;2012 doi: 10.1101/pdb.top072058. [DOI] [PubMed] [Google Scholar]
- 8.Kapitein LC, Peterman EJ, Kwok BH, Kim JH, Kapoor TM, Schmidt CF. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435:114–118. doi: 10.1038/nature03503. [DOI] [PubMed] [Google Scholar]
- 9.Kapoor TM. Metaphase spindle assembly. Biology (Basel) 2017;6:8. doi: 10.3390/biology6010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X, Nicklas RB. Mitotic forces control a cell-cycle checkpoint. Nature. 1995;373:630–632. doi: 10.1038/373630a0. [DOI] [PubMed] [Google Scholar]
- 11.Minoura I, Hachikubo Y, Yamakita Y, Takazaki H, Ayukawa R, Uchimura S, et al. Overexpression, purification, and functional analysis of recombinant human tubulin dimer. FEBS Letters. 2013;587(21):3450–3455. doi: 10.1016/j.febslet.2013.08.032. [DOI] [PubMed] [Google Scholar]
- 12.Mitchison TJ. Polewards microtubule flux in the mitotic spindle: Evidence from photoactivation of fluorescence. The Journal of Cell Biology. 1989;109:637–652. doi: 10.1083/jcb.109.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitchison TJ, Maddox P, Gaetz J, Groen A, Shirasu M, Desai A, et al. Roles of polymerization dynamics, opposed motors, and a tensile element in governing the length of Xenopus extract meiotic spindles. Molecular Biology of the Cell. 2005;16:3064–3076. doi: 10.1091/mbc.E05-02-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neuman KC, Block SM. Optical trapping. The Review of Scientific Instruments. 2004;75:2787–2809. doi: 10.1063/1.1785844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicklas RB. How cells get the right chromosomes. Science. 1997;275:632–637. doi: 10.1126/science.275.5300.632. [DOI] [PubMed] [Google Scholar]
- 16.Oreopoulos J, Berman R, Browne M. Spinning-disk confocal microscopy: Present technology and future trends. Methods in Cell Biology. 2014;123:153–175. doi: 10.1016/B978-0-12-420138-5.00009-4. [DOI] [PubMed] [Google Scholar]
- 17.Rice S, Lin AW, Safer D, Hart CL, Naber N, Carragher BO, et al. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- 18.Selvin P, Ha T. Single-molecule techniques: A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. [Google Scholar]
- 19.Shimamoto Y, Forth S, Kapoor TM. Measuring pushing and braking forces generated by ensembles of Kinesin-5 crosslinking two microtubules. Developmental Cell. 2015;34:669–681. doi: 10.1016/j.devcel.2015.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimamoto Y, Kapoor TM. Microneedle-based analysis of the micromechanics of the metaphase spindle assembled in Xenopus laevis egg extracts. Nature Protocols. 2012;7:959–969. doi: 10.1038/nprot.2012.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimamoto Y, Maeda YT, Ishiwata S, Libchaber AJ, Kapoor TM. Insights into the micromechanical properties of the metaphase spindle. Cell. 2011;145:1062–1074. doi: 10.1016/j.cell.2011.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takagi J, Shimamoto Y. High-quality frozen extracts of Xenopus laevis eggs reveal size-dependent control of metaphase spindle micromechanics. Molecular Biology of the Cell. 2017;28:2170–2177. doi: 10.1091/mbc.E17-03-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ti S-C, Pamula MC, Howes SC, Duellberg C, Cade NI, Kleiner RE, et al. Mutations in human tubulin proximal to the kinesin-binding site alter dynamic instability at microtubule plus- and minus-ends. Developmental Cell. 2016;37:72–84. doi: 10.1016/j.devcel.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vemu A, Atherton J, Spector JO, Szyk A, Moores CA, Roll-Mecak A. Structure and dynamics of single-isoform recombinant neuronal human tubulin. The Journal of Biological Chemistry. 2016;291(25):12907–12915. doi: 10.1074/jbc.C116.731133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walczak CE, Heald R. Mechanisms of mitotic spindle assembly and function. International Review of Cytology. 2008;265:111–158. doi: 10.1016/S0074-7696(07)65003-7. [DOI] [PubMed] [Google Scholar]
- 26.Weinger JS, Qiu M, Yang G, Kapoor TM. A nonmotor microtubule binding site in kinesin-5 is required for filament crosslinking and sliding. Current Biology: CB. 2011;21:154–160. doi: 10.1016/j.cub.2010.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang D, Nicklas RB. Micromanipulation of chromosomes and spindles in insect spermatocytes. Methods in Cell Biology. 1999;61:209–218. doi: 10.1016/s0091-679x(08)61982-2. [DOI] [PubMed] [Google Scholar]