Abstract
Reprogramming of the chromatin landscape is a critical component to the transcriptional response in breast cancer. Effects of sex hormones such as estrogens and progesterone have been well described to have a critical impact on breast cancer proliferation. However, the complex network of the chromatin landscape, enhancer regions, and mode of function of steroid receptors (SRs) and other transcription factors (TFs), is an intricate web of signaling and functional processes that is still largely misunderstood at the mechanistic level. In this review, we describe what is currently known about the dynamic interplay between TFs with chromatin and the reprogramming of enhancer elements. Emphasis has been placed on characterizing the different modes of action of TFs in regulating enhancer activity, specifically, how different SRs target enhancer regions and reprogram chromatin in breast cancer cells. In addition, we discuss current techniques employed to study enhancer function at a genome-wide level. Further, we have noted recent advances in live cell imaging technology. These single cell approaches enable the coupling of population based assays with real-time studies to address many unsolved questions about SRs and chromatin dynamics in breast cancer.
Keywords: breast cancer, steroid receptor, estrogen receptor, enhancer, chromatin, chromatin dynamics, ChIP-seq, single-molecule
INTRODUCTION
Transcriptional regulation is one of the most important biological processes in organisms, contributing to every aspect of health and disease. The regulation of gene expression is a complex process involving a multitude of proteins, including transcription factors (TFs), cofactors and RNA polymerases. These proteins bind and interact with specific regulatory elements termed enhancers on chromatin. Here the transcriptional regulation activity is influenced through a number of different processes (Long, et al. 2016; Voss and Hager 2014). The enhancer landscape and the accessibility of chromatin at enhancers is constantly changing during cellular development and differentiation (Chronis, et al. 2017). Interestingly, the enhancer landscape in all developed cells appears to represent subsets of a general population of enhancers identified in embryonic stem cells (Stergachis, et al. 2013). In many cases, TFs reprogram the chromatin landscape by altering the chromatin accessibility, thereby influencing the activity and recruitment of other TFs, cofactors, and RNA polymerases. The cofactors recruited by TFs can possess a variety of enzymatic properties, such as histone-modifying and ATP-dependent chromatin remodeling activities. These events have a crucial role in regulating chromatin accessibility (Long et al. 2016; Murakami, et al. 2017; Perissi and Rosenfeld 2005; Yi, et al. 2017). Reprogramming ensures accurate regulatory TF binding and subsequent regulation of cell-type specificity. Abnormal alterations in the activity of TFs can aberrantly program the chromatin landscape leading to many different disease states (Smith and Shilatifard 2014). The most drastic examples of abnormal alterations of TF binding and chromatin accessibility occurs in cancers (Denny, et al. 2016; Qu, et al. 2017). Breast cancer is no exception, with alterations occurring in chromatin accessibility, TF action, and regulation (D’Antonio, et al. 2017; Jeselsohn, et al. 2015; Rheinbay, et al. 2017; Toy, et al. 2017). Furthermore, the development of mammary epithelial cells to breast carcinoma can create novel enhancers unique to the progression of this cancer subtype (Stergachis et al. 2013). It has been shown that deletion of single nucleotide polymorphisms (SNPs) containing the enhancer that up-regulates MYC in intestinal cancer results in decrease of MYC expression and resistance to tumorigenesis (Sur, et al. 2012). This highlights the importance of enhancers in cancers (Sur and Taipale 2016). Furthermore, this mechanism is not exclusive to cancer, as SNPs at nuclear receptor regulatory regions can influence metabolic disease risk and subsequent effectiveness of therapeutics (Soccio, et al. 2015). In addition to SNPs, a large number of genomic alterations and mutation occur in cancers (Beroukhim, et al. 2010; Bignell, et al. 2010; Rheinbay et al. 2017). For example, a mutational hotspot at the FoxA1 promoter region leads to increased protein expression that can influence the action of other TFs in breast cancer (Rheinbay et al. 2017). In addition, FoxA1 can drive enhancer reprogramming during development of pancreatic cancer (Roe, et al. 2017). In the last year, it has also been recognized that not only direct DNA binding TFs, but also recruited cofactors can drive chromatin reprogramming. This has prominently been described with chromatin remodeling complexes whose action can drastically change the chromatin landscape in various cancers (Boulay, et al. 2017; Kadoch, et al. 2017; Pulice and Kadoch 2016). In addition, many other cofactors, such as histone modification writers, readers, and erasers are misregulated in cancers (Dawson 2017; Kim and Roberts 2016; Lonard and O’Malley 2016). Due to these observations, there is a growing interest to develop therapeutics targeting cofactors rather than TFs (Bennett and Licht 2017; Illendula, et al. 2015; Lasko, et al. 2017; Ribich, et al. 2017; Song, et al. 2016).
The discovery and history of nuclear receptors, including SRs, has been extensively reviewed by others (Chawla, et al. 2001; Lazar 2017; Mangelsdorf, et al. 1995). Here, we will focus on the reprogramming of the breast cancer chromatin landscape through steroid receptors (SRs). As the female sex hormone estrogen plays a predominate role in breast cancer growth, emphasis will be placed on the estrogen receptor (ER) and its cooperative action with other TFs. Furthermore, we will review the current knowledge of how TFs bind and interact with chromatin to reprogram the chromatin landscape.
BREAST CANCER
Breast cancer is the leading cause of cancer related death in women. One in 8 women will develop the disease in their lifetime in the United States alone. In addition, there will be 230,000 new cases diagnosed and approximately 40,000 women succumbing to the disease each year (Siegel, et al. 2011). The epithelial cells that line the lobules or ducts are the predominant site for breast cancer initiation. These first detectable lesions are neoplastic growths confined within individual ducts, considered pre-invasive and termed in situ carcinoma. Invasive carcinoma is the next stage in breast cancer development. Here the cells breach the basement membrane and invade the surrounding breast stromal tissue (Roses 1999). The last stage in the progression of the disease is metastasis of the invasive cells. During this stage, the cells can migrate from the primary tumor site, via the blood stream or lymphatic system, where they transplant into the lymph nodes or other organs. It is well documented that 17β-estradiol (E2), an active metabolite of estrogen, is required for the development, growth, and homeostatic maintenance of normal and malignant breast tissue. Historically, it has been determined that the removal of the ovaries suppressed the growth of breast cancer (Beatson 1896; Mauvais-Jarvis, et al. 1990; Wittliff 1984). This was first reported in 1896, when a bilateral oophorectomy was performed in a premenopausal patient, resulting in a complete remission of the disease (Beatson 1896). Many years later the estrogen receptor (ER) was discovered (Jensen and DeSombre 1973) and the association to the removal of the ovaries and tumor remission could be attributed to the dependence of E2 on breast cancer growth. This discovery was quickly followed by the first cloning of ER (Walter, et al. 1985) and then isolation of a complementary DNA clone from translated mRNA of ER. This isolation was from the MCF-7 human breast cancer cells, which manifest functional expression of the protein (Greene, et al. 1986).
The steroid receptors (SRs) that mediate the effects of steroid hormones, such as E2, are involved in the progression and prognosis of hormone associated cancers. The estrogen receptor (ER) is expressed in approximately 50–88% of all breast cancers, with the progesterone receptor (PR) expressed in 45–82% (McGuire 1978; Rosa, et al. 2008). Primary diagnosis of breast cancer is subtyped by ER, PR, and the human epidermal growth factor 2 (HER2) expression to determine current treatment approaches. Given the known role of ER in breast cancer development, it is primarily utilized as a therapeutic target in the clinic. PR is a well described E2 regulated gene, and its dependence on ER signaling is utilized solely as a marker of a functional ER (Creighton, et al. 2009). At present, the standard of care for patients with ER positive breast cancers is to inhibit the receptors functionality. This is achieved by active competition of ER with antagonists such as tamoxifen. Alternatively, the use of aromatase inhibitors results in ER signaling inhibition by blocking the catalytic processes of estrogen production (Arpino, et al. 2009). Generally, these treatments are effective short term; however, 30–50% of ER positive tumors display resistance to these therapies and generally all metastatic ER positive breast cancer acquire resistance (Arpino et al. 2009; Mouridsen, et al. 2003). Two major trials, the Women’s Health Initiative (WHI) and the Million Women Study, investigated the effects of estrogen and progestin on breast cancer incidence. Both studies looked at the use of hormone replacement therapy (HRT) (i.e. estrogen only) and combined hormone replacement therapy (cHRT) (i.e. estrogen and progestin in combination). The WHI concluded that women on cHRT had an increased risk of invasive breast cancer compared to placebo treated women, with an incidence of 0.38% and 0.3% respectively (Rossouw, et al. 2002). Similar findings were found in the Million Women Study, concluding that women on HRT or cHRT were at a higher risk of developing breast cancer compared to women that had not used, or were not currently using either of these therapies (Beral 2003). However, these results have remained controversial over the years. Recently there has been the suggestion that HRT may have no effect on breast cancer incidence in younger women and could provide a level of protection in older women (Santen 2014). The level of proposed protection in older women has been suggested to be a result of estrogen-induced apoptosis in breast tumors (Jordan 2015; Santen 2014) This finding contrasts historic and current data and is an avenue of importance that needs to be further explored.
For the SR family, the androgen receptor (AR) and the glucocorticoid receptor (GR) each have a multitude of functions in human biology and disease progression. In addition to ER and PR, AR and GR have also been implicated in breast cancer progression. Specifically, AR is found to be expressed in up to 85% of primary breast tumors (Honma, et al. 2012; Qi, et al. 2012). Tumors expressing AR/ER/PR present with a better prognosis (Garreau, et al. 2006) compared to AR positive ER/PR negative tumors (Lin Fde, et al. 2012). Further, the patients with AR/ER/PR positive tumors have smaller tumor size, lower Ki-67 expression (marker of proliferation), and better disease-free survival compared to AR negative ER/PR positive tumor patients (Hu, et al. 2011; Qi et al. 2012). More recently the role of GR in mediating breast cancer development has begun to immerge. It appears the effect of GR on breast cancer is dependent on the expression of ER. Specifically, there has been an association with chemo-resistance and a short disease free survival period in triple negative breast cancer (breast cancer which lacks expression of ER, PR, and HER2 growth factor) (Pan, et al. 2011). However, cancers that express ER, PR, and have high GR expression, demonstrate an increased overall disease free survival (Pan et al. 2011). It is important to note that the overall levels of circulating endogenous ligand of the SRs change throughout the female monthly menstrual cycle. In addition, levels of estrogen are markedly decreased once a female goes through menopause. It is becoming clear that the role of SR signaling in breast cancer progression is complex and likely involves a crosstalk between multiple TFs. To fully understand the complexity behind these SR signaling pathways we need to increase our knowledge of the genetic elements that they bind, and the modes of action by which ER, PR, AR and GR collaborate at these enhancer elements. Advances in our understanding of the genomic responses of these factors will undoubtedly lead to improved patient therapies.
TECHNIQUES USED TO STUDY CHROMATIN DYNAMICS
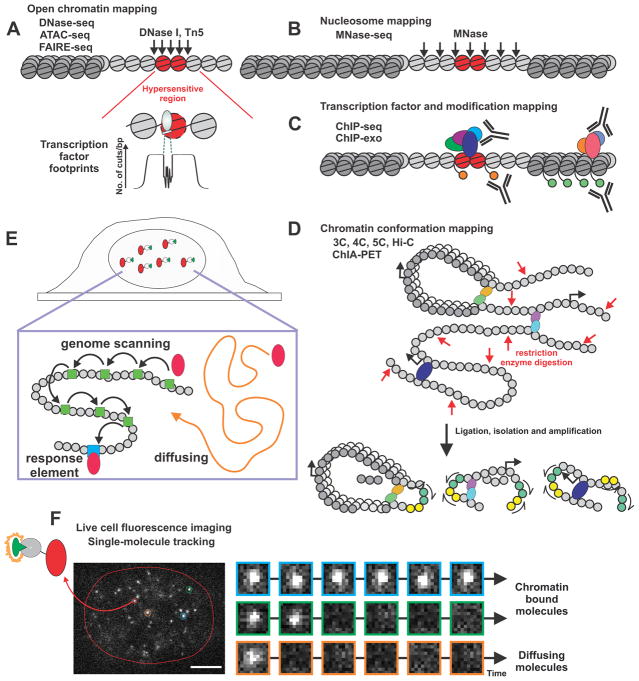
The chromatin landscape predominately defines the genomic response of TFs, with the accessibility of enhancer elements effecting the gene transcription response of a given stimulus. Enhancers serve as the critical regulatory elements of the cells; characterization their functional activity states can be achieved by multiple population based assays (Figure 1). In the last 10 years these techniques have become available to almost all scientists (Soon, et al. 2013). These methods can be used to characterize a wide-range of chromatin landscape properties including chromatin accessibility, nucleosome mapping, TF occupancy and long range chromatin conformations (Maston, et al. 2012). For transcriptional regulation of an enhancer region, the chromatin landscape must be accessible to the TF attempting to elucidate its response.
Figure 1.
Illustration of current techniques utilized to study enhancer elements through population based assays and single molecule approaches. (A) Mapping of open chromatin accessibility via DNase- ATAC- or FAIRE-seq. Nuclease such as DNase I or transposase such as Tn5 can target hypersensitive region (black arrows). The hypersensitive region is marked in red with the ability to detect the TF footprint at this region. The protection by the TF in the footprint is usually counted as number of cuts per bp (No. of cuts/bp). (B) Mapping of nucleosome positioning using MNase-seq. Micrococcal nuclease (MNase) preferentially cuts DNA strand between nucleosomes (black arrows) enabling chromatin structure mapping. (C) Mapping of the genomic location of TF (colored ovals) and chromatin modifications (small orange or green circles) by ChIP-seq and ChIP-exo. The TF or histone modification of interest is targeted by an antibody located at open or closed regions of chromatin. (D) The capture of chromatin interactions via 3C, 4C, 5C, Hi-C and ChIP-PET. Illustrations demonstrates the restriction enzyme digestion, ligation, isolation, and amplification interaction events. Depending on the technique used different degree of interaction can be captured and measured. (E) Capture of single-molecules bound to a fluorescent tag demonstrating TF (red oval) diffusing in the nucleus of the cell (orange arrow), scanning the genome (green square), and binding to a response element (RE) (blue square). (F) Live cell fluorescent imaging of a TF with post-translational labeling tag upon activation via a fluorescent ligand can be tracked in the nucleus and a residence time of the bound molecules can be determined. Orange represents a diffusing molecule, green a fast bound molecule and blue a TF binding at a response element (RE) termed slow bound. White scale bar 5 μm.
Open chromatin mapping
The most widely utilized assay for measuring the DNA accessibility is the DNase hypersensitivity assay followed by sequencing (DNase-seq) (Figure 1A) (Boyle, et al. 2008). This technique was based on the initial concept that nucleases can preferentially cut chromatin at regions with disrupted nucleosome structure [DNase I hypersensitive sites (DHSs)] (Keene, et al. 1981; McGhee, et al. 1981). Mapping DHSs in breast cancer can be used to identify chromatin sites that influence cancer development (D’Antonio et al. 2017). Formaldehyde assisted isolation of regulatory elements (FAIRE)-seq is an alternative technique whereby chromatin is cross-linked using formaldehyde, sonicated, and then phenol-chloroform extracted. The fragments in the aqueous phase are considered to derive from accessible regions and are sequenced (Gaulton, et al. 2010). In general, FAIRE-seq and DNase-seq can identify the same open chromatin sites (Song, et al. 2011) however, there are also unique regions identified by both techniques. More recently, an assay for transposase-accessible chromatin (ATAC)-seq has been developed. This alternative technique assays for accessible chromatin using the hyperactive Tn5 transposase (Buenrostro, et al. 2013). Here, Tn5 will cut and insert sequencing adapters to open chromatin sites, which enables direct deep sequencing and mapping of genome-wide chromatin accessibility from the sample.
ATAC-seq is gaining favor over DNase- and FAIRE-seq, due to the small amount of starting material required and the much faster time to effect the assay. It can also be used for frozen as well as fresh tissue samples (Corces, et al. 2017). The limits of DNase-seq and ATAC-seq have been extensively explored. Both these assays have been performed on single-cells (Buenrostro, et al. 2015; Cooper, et al. 2017); in addition to measuring chromatin accessibility, DNase-seq and ATAC-seq data can be utilized to measure TF footprints (Baek, et al. 2017; Buenrostro et al. 2013). The short region within an accessible enhancer sequence that is bound by a TF is sometimes protected from enzymatic attack. This level of protection provides a TF footprint (Galas and Schmitz 1978; He, et al. 2014; Neph, et al. 2012; Sung, et al. 2016). If the DNA cut fragments are sequenced with enough depth the number of cuts per base pair (no. of cuts/bp) can be determined. The footprint can be quantified by an aggregation cut count plot across the motif and motif flanking regions (Baek and Sung 2016) (Figure 1A [bottom]). These TF footprints have been described as a signature of TF binding to the protected site from a single experiment (Siersbaek, et al. 2014; Stergachis, et al. 2014). However, we and others have recently demonstrated that many TFs lack a detectable footprint (Baek et al. 2017; He et al. 2014; Sung et al. 2016; Sung, et al. 2014). The “absence” of these footprints is likely related to the rapid exchange observed for many transcription factors on chromatin in living cells (see below; Live Cell Imaging).
Nucleosome mapping
Characterization of chromatin accessibility is valuable tool assessing the status of active chromatin landscape. However, the techniques mentioned above do not provide adequate information on the structure of chromatin, specifically, the positions of nucleosomes. This can be achieved with genome-wide nucleosome mapping by digestion of the chromatin with micrococcal nuclease (MNase) (Schones, et al. 2008). This nuclease preferentially attacks linker regions between nucleosomes (Figure 1B). Thus, sequencing the DNA fragments insensitive to MNase digestion produces a map of nucleosome positions, or the lack thereof. Traditional mapping of nucleosome positions around promoters has been used to determine gene activity, as active promoters tend to be nucleosome depleted. For TF binding events, nucleosome content can be used to evaluate whether factors can bind to nucleosomal DNA (Ballare, et al. 2012; Iwafuchi-Doi, et al. 2016). This approach has been used as a rationale to determine whether TFs act as pioneer factors (further details below). Although MNase-seq is a powerful technique to map nucleosome positions, there are potential drawbacks. MNase digestion has a sequence preference, with digestion occurring at A/T rich regions over G/C rich regions. Furthermore, due to exonuclease activity of the MNase, nucleosome position can be altered due to concentration of MNase utilized in the experiments (Chereji, et al. 2017; Lai and Pugh 2017; Voong, et al. 2017). To overcome this issue, MNase-seq experiments are often performed at differing concentrations of enzymes. This facilitates a more accurate evaluation of nucleosome positions (Iwafuchi-Doi et al. 2016). In addition to MNase-seq, several other techniques, such as ATAC-seq and RICC-seq (using ionizing radiation) have the capabilities to map nucleosome positions (Buenrostro et al. 2013; Risca, et al. 2017). Furthermore, an increased resolution of nucleosome positions can be achieved using chemical cleavage mapping of nucleosome centers (Voong, et al. 2016; Voong et al. 2017).
Transcription factor and modification mapping
One of the most highly used techniques to study TF binding to chromatin to date is chromatin immunoprecipitation (ChIP) (Orlando and Paro 1993; Solomon, et al. 1988). This technique (Figure 1C) uses crosslinking (via formaldehyde), DNA fragmentation (sonication), and immunoprecipitation to capture all the occupying sites for a given protein, such as TF, cofactor or histone modification (Massie and Mills 2008). Before the wide availability of deep sequencing, chromosome-wide mapping of TF action, such as ER, was mapped using the ChIP-on-chip (ChIP coupled with tiling array) technique (Carroll, et al. 2005). Soon after the discovery of this technique ChIP was coupled with deep sequencing (ChIP-seq) (Johnson, et al. 2007; Robertson, et al. 2007), allowing the genome-wide mapping of TFs (including ER) binding events (Welboren, et al. 2009). Due to the ease of accessibility to deep sequencing platforms, ChIP-seq has become the standard technique to characterize the genome-wide occurrence of a given protein (Furey 2012). However, some of the binding events mapped by ChIP-seq can arise from non-specific enrichment, creating a phenomenon termed “Phantom peaks” (Jain, et al. 2015). Thus, several controls are needed to discriminate between a real binding and a false peak. As a result, a substantial number of modifications to the technique have been introduced, improving and expanding the output of the data. The usage of exonuclease digestion with bound protein of interest protecting the binding site (ChIP-exo) increases the resolution to a single nucleotide level (Rhee and Pugh 2011). Furthermore, ChIP-seq can be performed on single-cells (Rotem, et al. 2015), and coupling mass spectrometry to ChIP enables the detection of interactomes at the chromatin level (Mohammed, et al. 2013; Rafiee, et al. 2016). The ChIP-seq technique is by no means restricted to cell lines and fresh tissue samples, as ChIP-seq can also be performed from fixed clinical samples (Cejas, et al. 2016) and core needle biopsy samples (Zwart, et al. 2013). Thus, recent developments in sequencing techniques now enable researchers to perform both open chromatin techniques (Corces et al. 2017; Jin, et al. 2015), as well as TF binding mapping from clinical samples (Cejas et al. 2016), as well as older cataloged material.
Chromatin conformation mapping
After the realization that many TFs binding events are located at far distances from promoters, several methods to characterize long-range chromosomal interactions were developed (Bernstein, et al. 2012; Davies, et al. 2017; Thurman, et al. 2012). Most of these techniques are based on the digestion and ligation of interacting sites, termed chromosome conformation capture (3C) (Figure 1D) (Davies et al. 2017; Dekker, et al. 2002). However, newer techniques are emerging that allow the study of genome-wide interaction of these sites (Beagrie, et al. 2017) such as genome-wide 3C, called Hi-C, (Lieberman-Aiden, et al. 2009) and chromatin interaction analysis by paired-end tag sequencing (ChIA-PET) (Fullwood, et al. 2009). While these techniques are similar, Hi-C is used to map all DNA-DNA interactions while ChIA-PET uses ChIP to pre-select specific TF interacting sites. Initially, Hi-C was capable of 1 megabase resolution (Lieberman-Aiden et al. 2009), while ChIA-PET could identify individual interactions on a kilobase resolution due to antibody selection (Li, et al. 2010). However, improvements in Hi-C technique (in situ Hi-C) have enabled detection of interactions in 1 kilobase resolution (Rao, et al. 2014). This detailed resolution was achieved by performing the DNA-DNA proximity ligation in intact nucleic and deeply sequencing the data. Higher resolution in Hi-C can also be increased by focusing on certain interaction such as promoters via capture enriched Hi-C (Javierre, et al. 2016; Schoenfelder, et al. 2015). Both Hi-C and ChIA-PET techniques have been used to study chromatin architecture in breast cancer. Hi-C data clearly shows that interactions differ between mammary epithelial and breast cancer cells (Barutcu, et al. 2015). Furthermore, ER (Fullwood et al. 2009) and RNA polymerase II (Li, et al. 2012) ChIA-PET in breast cancer cells indicates that they are anchored to promoters through long-range interactions.
Live cell fluorescence imaging
All the above-mentioned assays are valuable tools to characterize the action of TFs. However, they suffer from two major drawbacks; the assays average signals across populations of heterogeneous cells and rely on dead cells. While biochemical and population-based assays suggest that TFs are assembled in a well-ordered manner to chromatin, live cell fluorescent imaging has indicated a more stochastic assembly (Coulon, et al. 2013; Stasevich and McNally 2011). This difference arises due to the temporal resolution of biochemical and population-based assays, which cannot resolve the dynamic binding of TFs that occurs in live cells (Hager, et al. 2009). In the past fifteen years, several fluorescent microscopy techniques, such as fluorescent recovery after photobleaching (FRAP) and fluorescent correlation spectroscopy (FCS), have been used to resolve the dynamic action of TFs (Mueller, et al. 2013). Many early FRAP experiments indicated that TF binding to chromatin occurs in the range of seconds (McNally, et al. 2000; Stenoien, et al. 2001). Furthermore, the rapid exchange of TFs with chromatin can influence transcriptional output (Karpova, et al. 2008; Stavreva, et al. 2004). Although FRAP and FCS can be used to resolve milli-sec to min range dynamic processes, both techniques are restricted to molecular populations in single cells. Hence, FRAP data will mostly represent diffusing TFs rather than bound ones. Recent advancements in imaging technologies (Chen, et al. 2014; Gebhardt, et al. 2013; Tokunaga, et al. 2008), protein tags (Gautier, et al. 2008; Los, et al. 2008) and fluorescent dyes (Grimm, et al. 2015), have enabled direct measurement of TF action at the single-molecule level (Mazza, et al. 2012). This technique, known as single-molecule tracking (SMT) or single-particle tracking (SPT), utilizes bright and stable fluorophores, and EMCCD cameras to resolve fluorescent signals originating from single fluorophores (Presman, et al. 2017). Several recent reviews have extensively covered the details and challenges of SMT (Liu, et al. 2015; Liu and Tjian 2018; Manzo and Garcia-Parajo 2015; Vera, et al. 2016; von Diezmann, et al. 2017). Single-molecules in general can be divided into two modes, bound and unbound states (Figure 1E) (Paakinaho, et al. 2017). It has been suggested that unbound molecules that can be captured on a single-frame but not tracked represent diffusing molecules (Figure 1F). These diffusing molecules can be classified into several types of diffusion (Izeddin, et al. 2014; Mazza et al. 2012).
Many investigators in the field have adopted an empirical method to describe the dynamics of bound molecules. This method involves fitting the dwell time data to alternate exponential distributions, and choosing the model that provides the best fit (Ball, et al. 2016; Chen et al. 2014; Hansen, et al. 2017; Kieffer-Kwon, et al. 2017; Kilic, et al. 2015; Loffreda, et al. 2017; Morisaki, et al. 2014; Schmidt, et al. 2016; Sugo, et al. 2015; Zhen, et al. 2016). The most frequent models currently invoked argue for 2 or 3 component distributions for the bound fraction. In a two component version, fast bound molecules are proposed to represent non-specific binding. The TF scans the genome attempting to find its specific binding sites remaining bound only for short period of time (Figure 1E–F) (Chen et al. 2014; Elf, et al. 2007; Paakinaho et al. 2017). In contrast, slow bound molecules remain bound for longer periods (5–15 sec) and are proposed to represent TF binding to specific response element (Figure 1E–F). In support of this interpretation, mutating the DNA-binding domain of a TF drastically reduces or abolishes the slow bound population of single-molecules (Chen et al. 2014; Morisaki et al. 2014; Paakinaho et al. 2017; Sugo et al. 2015). Work in breast cancer cell lines has indicated that chromatin binding of SRs is a very dynamic process (Swinstead, et al. 2016a). Interestingly, the pioneer factor FoxA1 (to be described below) also displays rapid dynamics in breast cancer cells. Essentially all TFs studied so far show dynamic action at the single-molecule level in live cells (Chen et al. 2014; Goldstein, et al. 2017a; Mazza et al. 2012; Morisaki et al. 2014; Paakinaho et al. 2017; Sugo et al. 2015; Teves, et al. 2016; Zhen et al. 2016), while structural proteins such as CTCF show much slower binding dynamics (Hansen et al. 2017). It should be emphasized that current interpretations for single molecule tracking are not based on rigorous thermodynamic models. It is likely that more accurate descriptions of real time TF/chromatin interactions will emerge. However, current results clearly demonstrate that TF action in general is a very rapid and dynamic process. Eventually population-based and live cell fluorescent microscopy perspectives should be resolved within a single comprehensive model (Paakinaho et al. 2017). We are already beginning to see this combination with the development of synthetic techniques such as ATAC-see (Chen, et al. 2016).
ENHANCER REPROGRAMMING MODES OF TRANSCRIPTION FACTORS
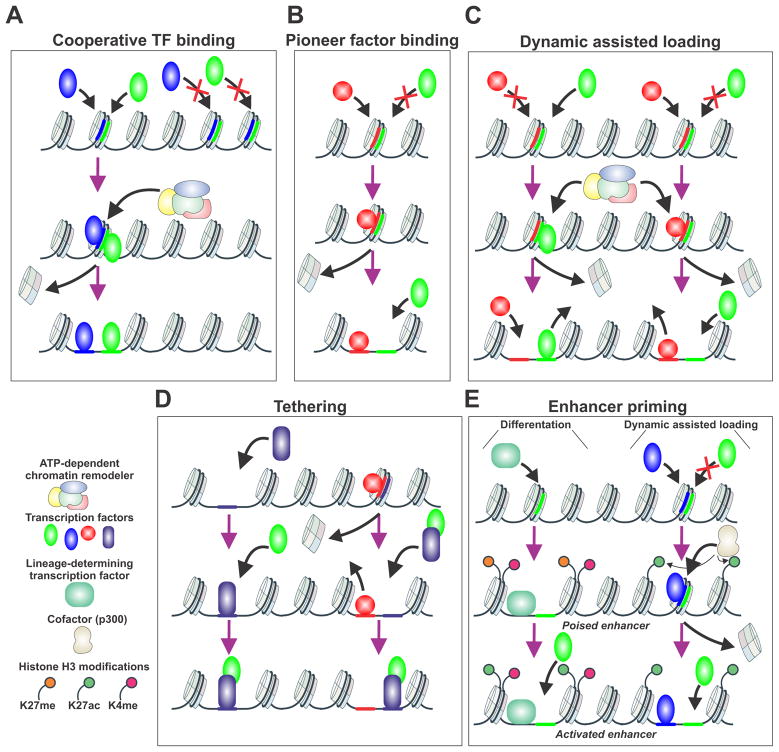
Enhancers are short stretches of regulatory elements generally located distal to promoters. During transcriptional regulation TFs reprogram enhancers by altering chromatin accessibility thereby influencing the recruitment of other factors including RNA polymerases. Thus, the initial enhancer reprogramming determines the transcriptional outcomes (Schaffner 2015; Smith and Shilatifard 2014). To reprogram enhancers and regulate transcription, TFs must be able to access enhancer regions on chromatin. Current models envisage TFs binding and enhancer reprogramming through five major modes (Figure 2) (Long et al. 2016; Spitz and Furlong 2012; Voss and Hager 2014).
Figure 2.
Illustration of enhancer reprogramming models. (A) Cooperative TF binding model. The binding region of interest is inaccessible until two TF concomitantly binding, recruit chromatin remodeling factors resulting in an accessible region. (B) Pioneer factor binding. Pioneer factors such as FoxA1 bind to closed regions of chromatin, expel the nucleosomes, allowing binding of a secondary factor in the absence of ATP-dependent processes. (C) Dynamic Assisted Loading. The initiating factor binds to a closed chromatin region, upon recruitment of chromatin remodeling factors the secondary factor can binding to a region previously deemed inaccessible. This is usually a bimodal switch between two TF depending on the chromatin landscape and the enhancer region. (D) The tethering model. One TF binds to a chromatin region with the secondary TF recruited upon binding or initially tether to the first TF. (E) Enhancer priming. This mechanism is functional in differentiation or dynamic assisted loading. The bound lineage-determining TF can alter the chromatin landscape by changing enhancer accessibility and histone modifications. Conversely in assisted loading the initiating factor binding actives enhancer region by increasing active histone modifications inducing the recruitment of the secondary factor to these sites. In both cases, poised enhancer state exists between the inactive and active enhancer states.
1. Cooperative transcription factor binding
It has been generally proposed that TFs require cooperative action of two of more factors to gain access to binding sites in closed chromatin regions (Figure 2A), through direct or indirect interactions (Long et al. 2016; Spitz and Furlong 2012). These events are thought to be ATP-dependent, requiring the recruitment of chromatin remodeling complexes enabling an alteration to chromatin accessibility. The direct interaction model relies on a physical contact or simultaneous binding between the cooperative TFs. Furthermore, TF pairs can have closely proximal recognition motifs on DNA influencing TF concomitant binding (Jolma, et al. 2015; Morgunova and Taipale 2017). In the case of indirect interaction, cooperativity occurs without apparent physical contact but with TFs binding in close proximity to each other. For some TFs, the indirect interaction mode can be classified as pioneer action (to be described). It is likely that both direct and indirect cooperative action can occur simultaneously. For example, it was recently described that during reprogramming of somatic cells to pluripotency (Takahashi and Yamanaka 2006), Oct4, Sox2, and Klf4 interact cooperatively to reprogram enhancers for pluripotency (Chronis et al. 2017). However, this cooperative action can function in the pioneer factor mode to induce the binding of somatic TFs. Further, during T cell activation, two pairs of TFs can cooperatively act to reprogram a different set of enhancers (Bevington, et al. 2016). In this case, enhancer priming (to be described) is regulated by the cooperative action of ETS-1 and RUNX1, while activation of inducible enhancer is regulated by AP-1 and NFAT. Interestingly, integration of several genomic and interactome datasets can identify several layers of cooperative TF action in liver (Dubois-Chevalier, et al. 2017) and during adipogenesis (Siersbaek et al. 2014). Finally, cooperative TF binding most likely influences enhancer activity since it is seemingly dependent on the other TFs in the immediate vicinity rather than an actual TF binding event (Grossman, et al. 2017).
2. Pioneer factor binding
It has been postulated that most TFs act in a cooperative manner. However, pioneer factors have been described as a small class of unique TFs that can penetrate chromatin on their own and assist the binding of non-pioneer proteins to the chromatin (Figure 2B) (Drouin 2014; Zaret and Carroll 2011). This group includes Oct4, Sox2, Klf4 (Soufi, et al. 2015), factors that reprogram somatic cells to pluripotency, and GATA and FoxA family members (Cirillo, et al. 2002). In addition, Pax7 can act as pioneer factor in pituitary melanotrope cells (Mayran, et al. 2018). Their capability to bind and remodel nucleosomes is the major characteristic of a pioneer factor (Zaret and Mango 2016). In the case of Oct4, Sox2 and Klf4, these factors can recognize their motifs on the surface of nucleosomes (Soufi et al. 2015), and increase the binding of other non-pioneer factors. In comparison, FoxA also has capability to bind to core histones (Cirillo et al. 2002); however, the structure of FoxA’s DNA-binding domain resembles that of linker histone H1 (Zaret and Carroll 2011). This suggests that FoxA can efficiently displace H1 from the chromatin thus maintaining accessible chromatin (Iwafuchi-Doi et al. 2016). Pax7 can bind to heterochromatin regions and slowly promote chromatin opening providing epigenetic memory (Mayran et al. 2018). Most interesting is the proposal that the action of pioneer factors occurs in an ATP-independent fashion (Cirillo et al. 2002). However recent discoveries suggest, at least for Oct4 and GATA pioneer factors, that ATP-dependent chromatin remodeling complexes are required for efficient pioneer factor function during enhancer reprograming (King and Klose 2017; Swinstead, et al. 2016b; Takaku, et al. 2016). Interestingly, the pioneer activity of FoxA2, Oct4 and GATA4 is not increased by ectopic expression of the factor (Donaghey, et al. 2018). However, the activity of FoxA2 is increased when coexpressed together with GATA4, suggesting that pioneer factors might operate in cooperative manner.
Pioneer factors, particularly FoxA1, have been shown to be exceptionally important for SR recruitment (Lupien and Brown 2009; Zaret and Carroll 2011). A major fraction of chromatin binding for both ER and AR depends on the pioneer activity of FoxA1 (Hurtado, et al. 2010; Lupien, et al. 2008; Robinson, et al. 2011), with GATA3 also playing a pioneer role for ER in breast cancer (Theodorou, et al. 2012). In the case of GR, AP-1 functions as a pioneer factor for a significant fraction of the receptor binding events (Biddie, et al. 2011; John, et al. 2011). While it has been suggested that SRs themselves can function as pioneer factors, the mechanism behind the SR “pioneer activity” appears to be more complex than the classically described process (Sahu, et al. 2011; Swinstead et al. 2016a).
3. Dynamic Assisted loading
Based on in vitro experiments it is expected that TFs that bind to the same DNA sites would in fact compete for binding. However, it was shown in vivo that two SRs failed to compete for the same binding site; rather one receptor assisted the binding of the secondary receptor (Voss, et al. 2011). These findings led to the proposal of a new model, termed “dynamic assisted loading.” In this model, one TF binds to a closed chromatin site, induces chromatin remodeling though ATP dependent processes, and thereby assists the binding of the second TF to the site (Figure 2C). It is further suggested that competition does not occur at the assisted loading sites in vivo due to the rapid interaction of TFs with chromatin in live cells (Paakinaho et al. 2017; Swinstead et al. 2016a). Furthermore, some TFs have been shown to be mobile during chromatin remodeling reactions (Li, et al. 2015; Nagaich, et al. 2004). In this scenario, the initiating TF inducing chromatin remodeling is actively displaced by the remodeler before the binding of the assisted factor. The assisted loading model differs from the cooperative and pioneer model in several distinct ways (Swinstead et al. 2016b). In comparison to the cooperative model, the two factors do not physically interact. In addition, assisted loading can be a symmetric event, with one factor acting as an initiator at some sites, while the other acts as initiator at different sites. This bimodal symmetry highlights the difference of assisted loading from the pioneer model. Furthermore, ATP-dependent chromatin remodeling factors are crucial for assisted loading, while the classically described pioneer model suggests ATP-independent action.
Although assisted loading occurs in symmetric manner, it also occurs in an enhancer-specific manner. This is a major mechanism of enhancer reprograming for SRs (Goldstein et al. 2017a; Grontved, et al. 2013; Swinstead et al. 2016a) and other nuclear receptors (Madsen, et al. 2014; Soccio et al. 2015) in various cellular contexts. For example, GR will assist the binding of ER at a subset of enhancers while ER will assist the binding of GR at another subset of enhancers (Miranda, et al. 2013). In addition to nuclear receptors, other TFs have been described to also operate through the assisted loading mechanism (Goldstein, et al. 2017b; Zhu, et al. 2015).
4. Tethering
An alternative model for TF function at enhancers involves a tethering event, whereby one TF can access an assessable site by physically tethering to another TF (Figure 2D). Genome-wide ChIP-seq studies alone are insufficient to determine if a binding event is a classical direct binding event or a tethering paradigm. Consequently, the DNA that is immunoprecipitated by the antibody of interest will represent sites of direct binding events, as well as sites of protein-protein interaction. DNA binding motif identification is frequently used to distinguish between these modes of action.
Classically, the anti-inflammatory action of GR is thought to be the consequence of GR tethering to pro-inflammatory factors thereby inhibiting their action (Cain and Cidlowski 2017; Petta, et al. 2016). However, recent results suggest that tethering might not be as prominent a mode of action as was previously believed (Oh, et al. 2017; Uhlenhaut, et al. 2012). Furthermore, GR can tether to other TFs without inflammatory stimuli to influence their action (Langlais, et al. 2012). In the case of classical ER binding events, the receptor binds to an estrogen response element (ERE); however, there are a number of identified sites missing the canonical ERE, suggesting a tethering phenomenon. Specifically, in MBA-MD-231 cells breast cancer cells transfected with wild type ER or DBD mutant ER incapable of binding to EREs, there is a clear difference between the transcription profiles of the two receptors (Stender, et al. 2010). This segregation was used to identify direct ER binding or tethering events. As tethered sites were enriched for the RUNX motif in the absence of an ERE, it was concluded that ER tethers RUNX to mediate DNA-independent gene regulation (Stender et al. 2010). It is important to note that the tethering event is largely different from the dynamic assisted loading model. Specifically, during dynamic assisted loading there is a presence of binding response elements of both TFs (initiating and secondary) (Swinstead et al. 2016a). However, there can be collaboration between the tethering and the dynamic assisted loading model. At a population of ER sites that are assisted by GR, AP-1 is tethered to ER facilitating the binding of ER at a number of assisted loading sites (Miranda et al. 2013). However, because formaldehyde crosslinks both DNA-protein and protein-protein interactions, the tethering mode can only be proposed but not confirmed by ChIP assays. Alternative techniques should be used, such as expression of DBD mutant TFs (Langlais et al. 2012; Stender et al. 2010). In addition, the improved resolution obtained by ChIP-exo can be used to distinguish direct DNA binding and tethering events (Starick, et al. 2015). More sophisticated techniques are being harnessed to address these issues. Previously, UV-laser cross-linking has been used in vitro (Nagaich et al. 2004), to study direct DNA binding of TFs. Interestingly, very recently it has been shown that UV-laser cross-linking can be coupled to ChIP (Steube, et al. 2017) to resolve tethering events from direct DNA binding. Future development of this technique will further help to distinguish protein-protein from protein-DNA interactions.
5. Enhancer priming
The selection and function of TF binding events can prime the enhancer for a resultant transcriptional response (Figure 2E). Enhancer priming is prevalent in differentiation. Specifically, a TF can bind, altering a poised enhancer to an activated enhancer through histone H3 modifications. This results in a recruitment of a secondary factor (Heinz, et al. 2015). This has been most clearly shown in the differentiation of hematopoietic cells, where lineage-determining factors, such as PU.1, prime enhancers for macrophage or B cell differentiation (Heinz, et al. 2010). Eventually these primed enhancers will serve as a platform for signal-dependent TF binding driving differentiation to a specific direction. Furthermore, H3K4 methylation and enhancer transcription seem especially important for enhancer priming (Heinz et al. 2010; Kaikkonen, et al. 2013). Another example of enhancer priming is illustrated in the phenomenon of T cell memory acquisition. Here, activation of T cells induce NCAT and AP-1 binding, resulting in a number of new DHS site and subsequent recruitment of ETS-1 and RUNX1. Interestingly, the DHSs remained stable long after T cell activation, maintaining open chromatin regions at active enhancer regions (Bevington et al. 2016). The dynamic assisted loading model can be extended further with evidence for enhancer priming. The dynamic crosstalk between two factors through this interaction results in alteration of H3K27ac and an associated recruitment of P300 (Goldstein et al. 2017b).
ENHANCER REPROGRAMMING BY STEROID RECEPTORS IN BREAST CANCER
Specifically relevant to breast cancer, a number of investigators are beginning to examine the mechanistic processes of SR recruitment to enhancer elements. Many of the enhancer reprogramming modes are utilized by SRs in breast cancer. These studies have explored the different modes of action a SR can have on the chromatin landscape and the consequential output for gene regulation and transcription profiles. Cooperative action among SRs is beginning to appear as a major mode of enhancer reprogramming in breast cancer cells (see next section for details). However, other pathways can cooperatively reprogram enhancers with SRs. Growth factors, independently or in cooperation with ER, can reprogram the enhancer landscape influencing the ER cistrome in breast cancer cells (Lupien, et al. 2010). Interestingly, in non-tumorigenic mammary cells, growth factors and GR can act in a cooperative and antagonistic manners to reprogram enhancers and gene regulation (Enuka, et al. 2017). In addition to growth factors, inflammatory pathways can reprogram the ER enhancer landscape potentially influencing clinical outcome of breast cancer (Franco, et al. 2015), or endocrine resistance (Stender, et al. 2017). Thus, it is expected that other signaling pathways will cooperatively influence SR enhancer reprogramming. In addition, mutations in ER (ESR1) arising from endocrine resistance, can reprogram ER binding (Jeselsohn, et al. 2018; Martin, et al. 2017; Toy et al. 2017), influencing receptor action on chromatin. In the case of tethering, Carroll and colleagues reported that activated PR could reprogram the ER enhancer landscape, contributing to an inhibition of breast cancer tumor growth under the dual treatment conditions. These newly acquired ER sites under the dual activation of both receptors was proposed to be through a tethering event, whereby PR and ER physically interact with cofactors and FoxA1 at binding sites. It was proposed that there is a lack of a classical ERE at these unique sites (Mohammed, et al. 2015). Myers and colleagues have also suggested that cell type specific ER and GR binding events that lack a strong canonical response element represent tethering events (Gertz, et al. 2013). Interestingly, this model also suggests that these SR tethered enhancers are primed by other TFs. These results imply that in some cases SRs can have only a minor effect on enhancer reprogramming as they bind to already accessible chromatin. This is in line with the pioneer factor model, where SRs binding is dictated by other TFs (Biddie et al. 2011; Hurtado et al. 2010; John et al. 2011; Theodorou et al. 2012). As indicated above, the main property of a pioneer factor is the capability to bind histones at closed chromatin sites (Drouin 2014; Zaret and Carroll 2011).
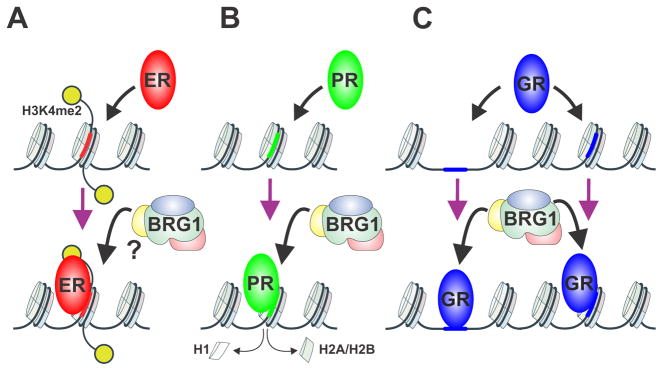
In this vein, SRs can target nucleosomes in breast cancer cells transforming them to bona fide pioneer factors. It has been suggested that the vast majority of ER and PR binding regions are largely nucleosome rich (Ballare et al. 2012; He, et al. 2012) (Figure 3A–B). ER binding in breast cancer cells is largely marked by H3K4me2 implying nucleosome rich binding regions (He et al. 2012) (Figure 3A). However, whether ER binds to nucleosomes without a H3K4me2 mark or how chromatin remodelers influence these processes is unknown. In the case for PR, Beato and colleagues described PR binding events at PRE enhancer regions rich with nucleosomes. The majority of sites are DNase I hypersensitive and hormone activation results in displacement of H1 and H2A/H2B dimers (Ballare et al. 2012) (Figure 3B). Further it has been described that Brg1 is largely involved in the resultant gene transcriptional response at enhancer regions (Ceballos-Chavez, et al. 2015). Thus, two important factors for breast cancer development, ER and PR, can both act as pioneer factors, and at least ER can regulate the binding of other factors by this pioneer activity (Swinstead et al. 2016a). For GR, initial genome-wide studies indicated frequent binding at sites with pre-existing DNase hypersensitivity. This accessibility is often interpreted as areas lacking nucleosomes (John et al. 2011) (Figure 3C). However, we have recently completed high resolution nucleosome positioning studies, and discovered that many DHS elements retain modified nucleosomes. This has led to a refined view of the chromatin structures present at responsive GR enhancers (Johnson, et al. 2018). In this view, they can be located either in pre-existing nucleosome depleted regions or within a nucleosome (Figure 3C). The nucleosomal depleted GR enhancers are already marked with Brg1, a chromatin remodeling factor, and flanked by H2A.Z. However, GR sites that are rich with nucleosomes can be segregated further into i) DNase I hypersensitive sites associated with Brg1, or ii) sites insensitive to DNase I and lacking Brg1, suggestive of a true GR pioneer function by dynamic assisted loading (Johnson et al. 2018) (Figure 3C). Thus, although GR action differs from that of ER and PR, it can also act as a pioneer factor targeting nucleosomal chromatin sites. The association of Brg1 and SR transcriptional responses has been well characterized (Swinstead et al. 2016b). However, an in-depth understanding of how these different enhancer region states relate specifically to SR binding and transcriptional response in breast cancer remains incomplete.
Figure 3.
Nucleosomal enhancer reprogramming by steroid receptors in breast cancer. (A) ER is bound to regions largely marked by H3K4me2 and rich in nucleosomes. ER potentially recruits chromatin remodelers to increase chromatin accessibility. (B) PR binds to nucleosome rich regions, recruits the Brg1 chromatin remodeler, resulting in a hypersensitive site and displacement of H1 and H2A/H2B dimers. (C) GR can bind to nucleosome depleted sites or sites enriched with a nucleosome. The nucleosomal depleted GR enhancers are marked with Brg1. GR sites that are rich with nucleosomes are suggestive of dynamic assisted loading or the pioneer model.
COLLABORATIVE CROSSTALK OF STEROID RECEPTOR BINDING EVENTS
Classically ER and PR binding events in breast cancer have been studied as single receptor binding events. It is becoming apparent that i) SRs collaborate with each other by the various mechanisms described above, and ii) there is an increasing appreciation for the importance of AR and GR signaling in breast cancer. The role of activated PR and the collaboration with ER signaling, and consequences for breast cancer growth is becoming well documented (Daniel, et al. 2015; Finlay-Schultz, et al. 2017; Hegde, et al. 2016; Mohammed et al. 2015; Singhal, et al. 2016). PR influences ER genomic recruitment (Mohammed et al. 2015; Singhal et al. 2016) potentially influencing decisions on breast cancer therapies. Interestingly, ER/PR crosstalk can be defined by PR isoforms, wherein PR-A inhibits while PR-B redistributes chromatin binding of ER (Singhal, et al. 2018). In addition, recently it was shown that PR can decrease the expression of proteins needed for translation, such as tRNAs in breast cancer (Finlay-Schultz et al. 2017). This decrease of tRNAs will restrict the translation of ER-regulated genes related for breast cancer growth. This highlights the importance of looking at these factors in a common setting. In addition, AR has been described to facilitate ER binding at a number of loci with enzalutamide, an AR antagonist, attenuating the response (D’Amato, et al. 2016). Furthermore, AR can collaborate with ER enhancing its transcriptional activity in aromatase inhibited breast cancer cells (Rechoum, et al. 2014). Lastly, the role of GR and ER crosstalk is emerging in the field, with several studies suggesting GR can induce or repress a number of ER binding events (Miranda et al. 2013; Yang, et al. 2017). Post-translational modifications of GR, such as those mediated by small ubiquitin-related modifier (SUMO; SUMOylation) is seemingly important in the repression of ER (Yang et al. 2017). However, this repression was only shown for a few SR-regulated loci. On a genome-wide level GR SUMOylation fine-tunes GR chromatin occupancy (Paakinaho, et al. 2014), suggesting that SUMOylation can have an even wider effect on SR crosstalk. Conversely, activated ER can result in enrichment of GR at proximal promoter regions, with increased GR chromatin association at ER, FOX, and AP-1 binding response regions (West, et al. 2016). The profiling of the SR landscape in human male breast cancer tumors indicate extensive overlap between ER, PR, AR, and GR binding (Severson, et al. 2018). This suggests that the interplay of SRs is also important in primary patient samples. Finally, analysis of nuclear receptor networks in breast cancer cells has revealed not only SRs interactions, but also complex interactions among nuclear receptors and other TFs related to breast cancer growth (Kittler, et al. 2013).
While many studies have started to uncover the concomitant crosstalk of SRs in breast cancer biology, what is still unclear are the mechanisms associated with the collaboration. Whether these events occur through the pioneer factor model, dynamic assisted loading, tethering, or direct binding events associated with other cofactors is still poorly understood at the level of molecular mechanism.
CONCLUSION
To date the clear majority of studies investigating TF signaling in breast cancer have focused on individual SR binding events. This is not a representative capture of the biologically important functions in the human body. Above we have provided an in-depth description of the dual collaboration of TFs on enhancer regulation. Lacking in the field of SR biology is a comprehensive understanding of the collaborative crosstalk of multiple SRs and other TFs at any given time, and the underlying mechanisms associated with these events. In addition, as many SRs in breast cancer cells can act as pioneer factors, existing models should be refined. It is becoming more apparent that there is no certain set of pioneer factors, but rather multiple TFs that in certain settings possess pioneering activities.
As we more fully understand the different types of SR binding events in breast cancer and underlying chromatin landscape, we can assess the direct effects on gene transcriptional profiles. These events are critical to driving breast cancer progression and proliferation. This information will assist in the understanding of the complex and in-depth systems associated with TF biology in breast cancer and the effects that the chromatin landscape has on these events.
Acknowledgments
This work was supported by the Intramural Research Program of the National Institutes of Health (NIH), the National Cancer Institute (NCI), the Center for Cancer Research (CCR). E.E.S. was supported by the Office of the Assistant Secretary of Defense for Health Affairs, through the Breast Cancer Research Program under Award No. W81XWH-17-1-0067. Opinions, interpretations, conclusions and recommendations are those of the author and are not necessarily endorsed by the Department of Defense. V.P. was supported by the University of Eastern Finland strategic funding and the Sigrid Jusélius Foundation.
Footnotes
DECLARATION OF INTEREST
The authors have no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
References
- Arpino G, De Angelis C, Giuliano M, Giordano A, Falato C, De Laurentiis M, De Placido S. Molecular mechanism and clinical implications of endocrine therapy resistance in breast cancer. Oncology. 2009;77(Suppl 1):23–37. doi: 10.1159/000258493. [DOI] [PubMed] [Google Scholar]
- Baek S, Goldstein I, Hager GL. Bivariate Genomic Footprinting Detects Changes in Transcription Factor Activity. Cell Rep. 2017;19:1710–1722. doi: 10.1016/j.celrep.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek S, Sung MH. Genome-Scale Analysis of Cell-Specific Regulatory Codes Using Nuclear Enzymes. Methods Mol Biol. 2016;1418:225–240. doi: 10.1007/978-1-4939-3578-9_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball DA, Mehta GD, Salomon-Kent R, Mazza D, Morisaki T, Mueller F, McNally JG, Karpova TS. Single molecule tracking of Ace1p in Saccharomyces cerevisiae defines a characteristic residence time for non-specific interactions of transcription factors with chromatin. Nucleic Acids Res. 2016;44:e160. doi: 10.1093/nar/gkw744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballare C, Castellano G, Gaveglia L, Althammer S, Gonzalez-Vallinas J, Eyras E, Le DF, Zaurin R, Soronellas D, Vicent GP, et al. Nucleosome-driven transcription factor binding and gene regulation. Mol Cell. 2012;49:67–79. doi: 10.1016/j.molcel.2012.10.019. [DOI] [PubMed] [Google Scholar]
- Barutcu AR, Lajoie BR, McCord RP, Tye CE, Hong D, Messier TL, Browne G, van Wijnen AJ, Lian JB, Stein JL, et al. Chromatin interaction analysis reveals changes in small chromosome and telomere clustering between epithelial and breast cancer cells. Genome Biol. 2015;16:214. doi: 10.1186/s13059-015-0768-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beagrie RA, Scialdone A, Schueler M, Kraemer DC, Chotalia M, Xie SQ, Barbieri M, de Santiago I, Lavitas LM, Branco MR, et al. Complex multi-enhancer contacts captured by genome architecture mapping. Nature. 2017 doi: 10.1038/nature21411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatson GT. On the treatment of inoperable cases of carcinoma of the mamma: suggestions for a new method of treatment, with illustrative cases. The Lancet. 1896;148:162–165. [PMC free article] [PubMed] [Google Scholar]
- Bennett RL, Licht JD. Targeting Epigenetics in Cancer. Annu Rev Pharmacol Toxicol. 2017 doi: 10.1146/annurev-pharmtox-010716-105106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beral V. Breast cancer and hormone-replacement therapy in the Million Women Study. Lancet. 2003;362:419–427. doi: 10.1016/s0140-6736(03)14065-2. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevington SL, Cauchy P, Piper J, Bertrand E, Lalli N, Jarvis RC, Gilding LN, Ott S, Bonifer C, Cockerill PN. Inducible chromatin priming is associated with the establishment of immunological memory in T cells. EMBO J. 2016 doi: 10.15252/embj.201592534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddie SC, John S, Sabo PJ, Thurman RE, Johnson TA, Schiltz RL, Miranda TB, Sung MH, Trump S, Lightman SL, et al. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol Cell. 2011;43:145–155. doi: 10.1016/j.molcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignell GR, Greenman CD, Davies H, Butler AP, Edkins S, Andrews JM, Buck G, Chen L, Beare D, Latimer C, et al. Signatures of mutation and selection in the cancer genome. Nature. 2010;463:893–898. doi: 10.1038/nature08768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulay G, Sandoval GJ, Riggi N, Iyer S, Buisson R, Naigles B, Awad ME, Rengarajan S, Volorio A, McBride MJ, et al. Cancer-Specific Retargeting of BAF Complexes by a Prion-like Domain. Cell. 2017;171:163–178. e119. doi: 10.1016/j.cell.2017.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, Greenleaf WJ. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015 doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain DW, Cidlowski JA. Immune regulation by glucocorticoids. Nat Rev Immunol. 2017;17:233–247. doi: 10.1038/nri.2017.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Ceballos-Chavez M, Subtil-Rodriguez A, Giannopoulou EG, Soronellas D, Vazquez-Chavez E, Vicent GP, Elemento O, Beato M, Reyes JC. The Chromatin Remodeler CHD8 Is Required for Activation of Progesterone Receptor-Dependent Enhancers. PLoS Genet. 2015;11:e1005174. doi: 10.1371/journal.pgen.1005174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejas P, Li L, O’Neill NK, Duarte M, Rao P, Bowden M, Zhou CW, Mendiola M, Burgos E, Feliu J, et al. Chromatin immunoprecipitation from fixed clinical tissues reveals tumor-specific enhancer profiles. Nat Med. 2016;22:685–691. doi: 10.1038/nm.4085. [DOI] [PubMed] [Google Scholar]
- Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: opening the X-files. Science. 2001;294:1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- Chen J, Zhang Z, Li L, Chen BC, Revyakin A, Hajj B, Legant W, Dahan M, Lionnet T, Betzig E, et al. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell. 2014;156:1274–1285. doi: 10.1016/j.cell.2014.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Shen Y, Draper W, Buenrostro JD, Litzenburger U, Cho SW, Satpathy AT, Carter AC, Ghosh RP, East-Seletsky A, et al. ATAC-see reveals the accessible genome by transposase-mediated imaging and sequencing. Nat Methods. 2016;13:1013–1020. doi: 10.1038/nmeth.4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chereji RV, Ocampo J, Clark DJ. MNase-Sensitive Complexes in Yeast: Nucleosomes and Non-histone Barriers. Mol Cell. 2017;65:565–577. e563. doi: 10.1016/j.molcel.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chronis C, Fiziev P, Papp B, Butz S, Bonora G, Sabri S, Ernst J, Plath K. Cooperative Binding of Transcription Factors Orchestrates Reprogramming. Cell. 2017;168:442–459. e420. doi: 10.1016/j.cell.2016.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- Cooper J, Ding Y, Song J, Zhao K. Genome-wide mapping of DNase I hypersensitive sites in rare cell populations using single-cell DNase sequencing. Nat Protoc. 2017;12:2342–2354. doi: 10.1038/nprot.2017.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, Satpathy AT, Rubin AJ, Montine KS, Wu B, et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods. 2017;14:959–962. doi: 10.1038/nmeth.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulon A, Chow CC, Singer RH, Larson DR. Eukaryotic transcriptional dynamics: from single molecules to cell populations. Nat Rev Genet. 2013;14:572–584. doi: 10.1038/nrg3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton CJ, Kent Osborne C, van de Vijver MJ, Foekens JA, Klijn JG, Horlings HM, Nuyten D, Wang Y, Zhang Y, Chamness GC, et al. Molecular profiles of progesterone receptor loss in human breast tumors. Breast Cancer Res Treat. 2009;114:287–299. doi: 10.1007/s10549-008-0017-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amato NC, Gordon MA, Babbs B, Spoelstra NS, Carson Butterfield KT, Torkko KC, Phan VT, Barton VN, Rogers TJ, Sartorius CA, et al. Cooperative Dynamics of AR and ER Activity in Breast Cancer. Mol Cancer Res. 2016;14:1054–1067. doi: 10.1158/1541-7786.MCR-16-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Antonio M, Weghorn D, D’Antonio-Chronowska A, Coulet F, Olson KM, DeBoever C, Drees F, Arias A, Alakus H, Richardson AL, et al. Identifying DNase I hypersensitive sites as driver distal regulatory elements in breast cancer. Nat Commun. 2017;8:436. doi: 10.1038/s41467-017-00100-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel AR, Gaviglio AL, Knutson TP, Ostrander JH, D’Assoro AB, Ravindranathan P, Peng Y, Raj GV, Yee D, Lange CA. Progesterone receptor-B enhances estrogen responsiveness of breast cancer cells via scaffolding PELP1- and estrogen receptor-containing transcription complexes. Oncogene. 2015;34:506–515. doi: 10.1038/onc.2013.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JO, Oudelaar AM, Higgs DR, Hughes JR. How best to identify chromosomal interactions: a comparison of approaches. Nat Methods. 2017;14:125–134. doi: 10.1038/nmeth.4146. [DOI] [PubMed] [Google Scholar]
- Dawson MA. The cancer epigenome: Concepts, challenges, and therapeutic opportunities. Science. 2017;355:1147–1152. doi: 10.1126/science.aam7304. [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Denny SK, Yang D, Chuang CH, Brady JJ, Lim JS, Gruner BM, Chiou SH, Schep AN, Baral J, Hamard C, et al. Nfib Promotes Metastasis through a Widespread Increase in Chromatin Accessibility. Cell. 2016;166:328–342. doi: 10.1016/j.cell.2016.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaghey J, Thakurela S, Charlton J, Chen JS, Smith ZD, Gu H, Pop R, Clement K, Stamenova EK, Karnik R, et al. Genetic determinants and epigenetic effects of pioneer-factor occupancy. Nat Genet. 2018 doi: 10.1038/s41588-017-0034-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouin J. Minireview: pioneer transcription factors in cell fate specification. Mol Endocrinol. 2014;28:989–998. doi: 10.1210/me.2014-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois-Chevalier J, Dubois V, Dehondt H, Mazrooei P, Mazuy C, Serandour AA, Gheeraert C, Guillaume P, Bauge E, Derudas B, et al. The logic of transcriptional regulator recruitment architecture at cis-regulatory modules controlling liver functions. Genome Res. 2017;27:985–996. doi: 10.1101/gr.217075.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elf J, Li GW, Xie XS. Probing transcription factor dynamics at the single-molecule level in a living cell. Science. 2007;316:1191–1194. doi: 10.1126/science.1141967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enuka Y, Feldman ME, Chowdhury A, Srivastava S, Lindzen M, Sas-Chen A, Massart R, Cheishvili D, Suderman MJ, Zaltsman Y, et al. Epigenetic mechanisms underlie the crosstalk between growth factors and a steroid hormone. Nucleic Acids Res. 2017;45:12681–12699. doi: 10.1093/nar/gkx865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay-Schultz J, Gillen AE, Brechbuhl HM, Ivie JJ, Matthews SB, Jacobsen BM, Bentley DL, Kabos P, Sartorius CA. Breast Cancer Suppression by Progesterone Receptors Is Mediated by Their Modulation of Estrogen Receptors and RNA Polymerase III. Cancer Res. 2017;77:4934–4946. doi: 10.1158/0008-5472.CAN-16-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco HL, Nagari A, Kraus WL. TNFalpha Signaling Exposes Latent Estrogen Receptor Binding Sites to Alter the Breast Cancer Cell Transcriptome. Mol Cell. 2015;58:21–34. doi: 10.1016/j.molcel.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furey TS. ChIP-seq and beyond: new and improved methodologies to detect and characterize protein-DNA interactions. Nat Rev Genet. 2012;13:840–852. doi: 10.1038/nrg3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas DJ, Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978;5:3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garreau JR, Muller P, Pommier R, Pommier S. Transgenic introduction of androgen receptor into estrogen-receptor-, progesterone-receptor-, and androgen-receptor-negative breast cancer cells renders them responsive to hormonal manipulation. Am J Surg. 2006;191:576–580. doi: 10.1016/j.amjsurg.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Gaulton K, Nammo T, Pasquali L, Simon J, Giresi PG, Fogarty M, Panhuis T, Mieczkowski P, Secchi A, Bosco D, et al. A map of open chromatin in human pancreatic islets. Nat Genet. 2010;42:255–259. doi: 10.1038/ng.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier A, Juillerat A, Heinis C, Correa IR, Jr, Kindermann M, Beaufils F, Johnsson K. An engineered protein tag for multiprotein labeling in living cells. Chem Biol. 2008;15:128–136. doi: 10.1016/j.chembiol.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Gebhardt JC, Suter DM, Roy R, Zhao ZW, Chapman AR, Basu S, Maniatis T, Xie XS. Single-molecule imaging of transcription factor binding to DNA in live mammalian cells. Nat Methods. 2013;10:421–426. doi: 10.1038/nmeth.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz J, Savic D, Varley KE, Partridge EC, Safi A, Jain P, Cooper GM, Reddy TE, Crawford GE, Myers RM. Distinct properties of cell-type-specific and shared transcription factor binding sites. Mol Cell. 2013;52:25–36. doi: 10.1016/j.molcel.2013.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein I, Baek S, Presman DM, Pakkinaho V, Swinstead EE, Hager GL. Transcription factor assisted loading and enhancer dynamics dictate the hepatic fasting response. Genome Res. 2017a;27:427–439. doi: 10.1101/gr.212175.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein I, Paakinaho V, Baek S, Sung MH, Hager GL. Synergistic gene expression during the acute phase response is characterized by transcription factor assisted loading. Nat Commun. 2017b;1:1849. doi: 10.1038/s41467-017-02055-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene GL, Gilna P, Waterfield M, Baker A, Hort Y, Shine J. Sequence and expression of human estrogen receptor complementary DNA. Science. 1986;231:1150–1154. doi: 10.1126/science.3753802. [DOI] [PubMed] [Google Scholar]
- Grimm JB, English BP, Chen J, Slaughter JP, Zhang Z, Revyakin A, Patel R, Macklin JJ, Normanno D, Singer RH, et al. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat Methods. 2015;12:244–250. doi: 10.1038/nmeth.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grontved L, John S, Baek S, Liu Y, Buckley JR, Vinson C, Aguilera G, Hager GL. C/EBP maintains chromatin accessibility in liver and facilitates glucocorticoid receptor recruitment to steroid response elements. EMBO J. 2013;32:1568–1583. doi: 10.1038/emboj.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman SR, Zhang X, Wang L, Engreitz J, Melnikov A, Rogov P, Tewhey R, Isakova A, Deplancke B, Bernstein BE, et al. Systematic dissection of genomic features determining transcription factor binding and enhancer function. Proc Natl Acad Sci U S A. 2017;114:E1291–E1300. doi: 10.1073/pnas.1621150114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager GL, McNally JG, Misteli T. Transcription dynamics. Mol Cell. 2009;35:741–753. doi: 10.1016/j.molcel.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AS, Pustova I, Cattoglio C, Tjian R, Darzacq X. CTCF and cohesin regulate chromatin loop stability with distinct dynamics. Elife. 2017:6. doi: 10.7554/eLife.25776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He HH, Meyer CA, Chen MW, Jordan VC, Brown M, Liu XS. Differential DNase I hypersensitivity reveals factor-dependent chromatin dynamics. Genome Res. 2012;22:1015–1025. doi: 10.1101/gr.133280.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He HH, Meyer CA, Hu SS, Chen MW, Zang C, Liu Y, Rao PK, Fei T, Xu H, Long H, et al. Refined DNase-seq protocol and data analysis reveals intrinsic bias in transcription factor footprint identification. Nat Methods. 2014;11:73–78. doi: 10.1038/nmeth.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde SM, Kumar MN, Kavya K, Kumar KM, Nagesh R, Patil RH, Babu RL, Ramesh GT, Sharma SC. Interplay of nuclear receptors (ER, PR, and GR) and their steroid hormones in MCF-7 cells. Mol Cell Biochem. 2016;422:109–120. doi: 10.1007/s11010-016-2810-2. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Romanoski CE, Benner C, Glass CK. The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol. 2015;16:144–154. doi: 10.1038/nrm3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma N, Horii R, Iwase T, Saji S, Younes M, Ito Y, Akiyama F. Clinical importance of androgen receptor in breast cancer patients treated with adjuvant tamoxifen monotherapy. Breast Cancer. 2012;20:323–330. doi: 10.1007/s12282-012-0337-2. [DOI] [PubMed] [Google Scholar]
- Hu R, Dawood S, Holmes MD, Collins LC, Schnitt SJ, Cole K, Marotti JD, Hankinson SE, Colditz GA, Tamimi RM. Androgen receptor expression and breast cancer survival in postmenopausal women. Clin Cancer Res. 2011;17:1867–1874. doi: 10.1158/1078-0432.CCR-10-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet. 2010;43:27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illendula A, Pulikkan JA, Zong H, Grembecka J, Xue L, Sen S, Zhou Y, Boulton A, Kuntimaddi A, Gao Y, et al. A small-molecule inhibitor of the aberrant transcription factor CBF+Ý-SMMHC delays leukemia in mice. Science. 2015;347:779–784. doi: 10.1126/science.aaa0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi-Doi M, Donahue G, Kakumanu A, Watts JA, Mahony S, Pugh BF, Lee D, Kaestner KH, Zaret KS. The Pioneer Transcription Factor FoxA Maintains an Accessible Nucleosome Configuration at Enhancers for Tissue-Specific Gene Activation. Mol Cell. 2016;62:79–91. doi: 10.1016/j.molcel.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izeddin I, Recamier V, Bosanac L, Cisse II, Boudarene L, Dugast-Darzacq C, Proux F, Benichou O, Voituriez R, Bensaude O, et al. Single-molecule tracking in live cells reveals distinct target-search strategies of transcription factors in the nucleus. Elife. 2014;3:e02230. doi: 10.7554/eLife.02230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain D, Baldi S, Zabel A, Straub T, Becker PB. Active promoters give rise to false positive ‘Phantom Peaks’ in ChIP-seq experiments. Nucleic Acids Res. 2015;43:6959–6968. doi: 10.1093/nar/gkv637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javierre BM, Burren OS, Wilder SP, Kreuzhuber R, Hill SM, Sewitz S, Cairns J, Wingett SW, Varnai C, Thiecke MJ, et al. Lineage-Specific Genome Architecture Links Enhancers and Non-coding Disease Variants to Target Gene Promoters. Cell. 2016;167:1369–1384. e1319. doi: 10.1016/j.cell.2016.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen EV, DeSombre ER. Estrogen-receptor interaction. Science. 1973;182:126–134. doi: 10.1126/science.182.4108.126. [DOI] [PubMed] [Google Scholar]
- Jeselsohn R, Bergholz JS, Pun M, Cornwell M, Liu W, Nardone A, Xiao T, Li W, Qiu X, Buchwalter G, et al. Allele-Specific Chromatin Recruitment and Therapeutic Vulnerabilities of ESR1 Activating Mutations. Cancer Cell. 2018;33:173–186. e175. doi: 10.1016/j.ccell.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeselsohn R, Buchwalter G, De Angelis C, Brown M, Schiff R. ESR1 mutations-a mechanism for acquired endocrine resistance in breast cancer. Nat Rev Clin Oncol. 2015;12:573–583. doi: 10.1038/nrclinonc.2015.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Tang Q, Wan M, Cui K, Zhang Y, Ren G, Ni B, Sklar J, Przytycka TM, Childs R, et al. Genome-wide detection of DNase I hypersensitive sites in single cells and FFPE tissue samples. Nature. 2015;528:142–146. doi: 10.1038/nature15740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Johnson TA, Chereji RV, Stavreva DA, Morris SA, Hager GL, Clark DJ. Conventional and pioneer modes of glucocorticoid receptor interaction with enhancer chromatin in vivo. Nucleic Acids Res. 2018;46:203–214. doi: 10.1093/nar/gkx1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature. 2015;527:384–388. doi: 10.1038/nature15518. [DOI] [PubMed] [Google Scholar]
- Jordan VC. The new biology of estrogen-induced apoptosis applied to treat and prevent breast cancer. Endocr Relat Cancer. 2015;22:R1–31. doi: 10.1530/ERC-14-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C, Williams RT, Calarco JP, Miller EL, Weber CM, Braun SM, Pulice JL, Chory EJ, Crabtree GR. Dynamics of BAF-Polycomb complex opposition on heterochromatin in normal and oncogenic states. Nat Genet. 2017;49:213–222. doi: 10.1038/ng.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD, Chun HB, Tough DF, Prinjha RK, Benner C, et al. Remodeling of the Enhancer Landscape during Macrophage Activation Is Coupled to Enhancer Transcription. Mol Cell. 2013;51:310–325. doi: 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpova TS, Kim MJ, Spriet C, Nalley K, Stasevich TJ, Kherrouche Z, Heliot L, McNally JG. Concurrent fast and slow cycling of a transcriptional activator at an endogenous promoter. Science. 2008;319:466–469. doi: 10.1126/science.1150559. [DOI] [PubMed] [Google Scholar]
- Keene MA, Corces V, Lowenhaupt K, Elgin SC. DNase I hypersensitive sites in Drosophila chromatin occur at the 5′ ends of regions of transcription. Proc Natl Acad Sci U S A. 1981;78:143–146. doi: 10.1073/pnas.78.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer-Kwon K-R, Nimura K, Rao SSP, Xu J, Jung S, Pekowska A, Dose M, Stevens E, Mathe E, Dong P, et al. Myc regulates chromatin decompaction and nuclear architecture during B cell activation. Molecular Cell. 2017;67:566–578. e510. doi: 10.1016/j.molcel.2017.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilic S, Bachmann AL, Bryan LC, Fierz B. Multivalency governs HP1alpha association dynamics with the silent chromatin state. Nat Commun. 2015;6:7313. doi: 10.1038/ncomms8313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Roberts CW. Targeting EZH2 in cancer. Nat Med. 2016;22:128–134. doi: 10.1038/nm.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King HW, Klose RJ. The pioneer factor OCT4 requires the chromatin remodeller BRG1 to support gene regulatory element function in mouse embryonic stem cells. Elife. 2017:6. doi: 10.7554/eLife.22631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler R, Zhou J, Hua S, Ma L, Liu Y, Pendleton E, Cheng C, Gerstein M, White KP. A comprehensive nuclear receptor network for breast cancer cells. Cell Rep. 2013;3:538–551. doi: 10.1016/j.celrep.2013.01.004. [DOI] [PubMed] [Google Scholar]
- Lai WKM, Pugh BF. Understanding nucleosome dynamics and their links to gene expression and DNA replication. Nat Rev Mol Cell Biol. 2017;18:548–562. doi: 10.1038/nrm.2017.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlais D, Couture C, Balsalobre A, Drouin J. The Stat3/GR Interaction Code: Predictive Value of Direct/Indirect DNA Recruitment for Transcription Outcome. Mol Cell. 2012;47:38–49. doi: 10.1016/j.molcel.2012.04.021. [DOI] [PubMed] [Google Scholar]
- Lasko LM, Jakob CG, Edalji RP, Qiu W, Montgomery D, Digiammarino EL, Hansen TM, Risi RM, Frey R, Manaves V, et al. Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature. 2017;550:128–132. doi: 10.1038/nature24028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar MA. Maturing of the nuclear receptor family. J Clin Invest. 2017;127:1123–1125. doi: 10.1172/JCI92949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Fullwood MJ, Xu H, Mulawadi FH, Velkov S, Vega V, Ariyaratne PN, Mohamed YB, Ooi HS, Tennakoon C, et al. ChIA-PET tool for comprehensive chromatin interaction analysis with paired-end tag sequencing. Genome Biol. 2010;11:R22. doi: 10.1186/gb-2010-11-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Hada A, Sen P, Olufemi L, Hall MA, Smith BY, Forth S, McKnight JN, Patel A, Bowman GD, et al. Dynamic regulation of transcription factors by nucleosome remodeling. Elife. 2015;4:e06249. doi: 10.7554/eLife.06249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lin FM, Pincerato KM, Bacchi CE, Baracat EC, Carvalho FM. Coordinated expression of oestrogen and androgen receptors in HER2-positive breast carcinomas: impact on proliferative activity. J Clin Pathol. 2012;65:64–68. doi: 10.1136/jclinpath-2011-200318. [DOI] [PubMed] [Google Scholar]
- Liu Z, Lavis LD, Betzig E. Imaging Live-Cell Dynamics and Structure at the Single-Molecule Level. Mol Cell. 2015;58:644–659. doi: 10.1016/j.molcel.2015.02.033. [DOI] [PubMed] [Google Scholar]
- Liu Z, Tjian R. Visualizing transcription factor dynamics in living cells. J Cell Biol. 2018 doi: 10.1083/jcb.201710038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffreda A, Jacchetti E, Antunes S, Rainone P, Daniele T, Morisaki T, Bianchi ME, Tacchetti C, Mazza D. Live-cell p53 single-molecule binding is modulated by C-terminal acetylation and correlates with transcriptional activity. Nat Commun. 2017;8:313. doi: 10.1038/s41467-017-00398-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonard DM, O’Malley BW. Molecular Pathways: Targeting Steroid Receptor Coactivators in Cancer. Clin Cancer Res. 2016;22:5403–5407. doi: 10.1158/1078-0432.CCR-15-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long HK, Prescott SL, Wysocka J. Ever-Changing Landscapes: Transcriptional Enhancers in Development and Evolution. Cell. 2016;167:1170–1187. doi: 10.1016/j.cell.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, Wood MG, Learish R, Ohana RF, Urh M, et al. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem Biol. 2008;3:373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- Lupien M, Brown M. Cistromics of hormone-dependent cancer. Endocr Relat Cancer. 2009;16:381–389. doi: 10.1677/ERC-09-0038. [DOI] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien M, Meyer CA, Bailey ST, Eeckhoute J, Cook J, Westerling T, Zhang X, Carroll JS, Rhodes DR, Liu XS, et al. Growth factor stimulation induces a distinct ER(alpha) cistrome underlying breast cancer endocrine resistance. Genes Dev. 2010;24:2219–2227. doi: 10.1101/gad.1944810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen MS, Siersbaek R, Boergesen M, Nielsen R, Mandrup S. Peroxisome Proliferator-Activated Receptor gamma and C/EBPalpha Synergistically Activate Key Metabolic Adipocyte Genes by Assisted Loading. Mol Cell Biol. 2014;34:939–954. doi: 10.1128/MCB.01344-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzo C, Garcia-Parajo MF. A review of progress in single particle tracking: from methods to biophysical insights. Rep Prog Phys. 2015;78:124601. doi: 10.1088/0034-4885/78/12/124601. [DOI] [PubMed] [Google Scholar]
- Martin LA, Ribas R, Simigdala N, Schuster E, Pancholi S, Tenev T, Gellert P, Buluwela L, Harrod A, Thornhill A, et al. Discovery of naturally occurring ESR1 mutations in breast cancer cell lines modelling endocrine resistance. Nat Commun. 2017;8:1865. doi: 10.1038/s41467-017-01864-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massie CE, Mills IG. ChIPping away at gene regulation. EMBO Rep. 2008;9:337–343. doi: 10.1038/embor.2008.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maston GA, Landt SG, Snyder M, Green MR. Characterization of enhancer function from genome-wide analyses. Annu Rev Genomics Hum Genet. 2012;13:29–57. doi: 10.1146/annurev-genom-090711-163723. [DOI] [PubMed] [Google Scholar]
- Mauvais-Jarvis P, Kuttenn F, Malet C, Gompel A. Normal breast cells in culture. Effect of estrogens, progestins, and antiestrogens. Ann N Y Acad Sci. 1990;595:117–129. doi: 10.1111/j.1749-6632.1990.tb34287.x. [DOI] [PubMed] [Google Scholar]
- Mayran A, Khetchoumian K, Hariri F, Pastinen T, Gauthier Y, Balsalobre A, Drouin J. Pioneer factor Pax7 deploys a stable enhancer repertoire for specification of cell fate. Nat Genet. 2018;50:259–269. doi: 10.1038/s41588-017-0035-2. [DOI] [PubMed] [Google Scholar]
- Mazza D, Abernathy A, Golob N, Morisaki T, McNally JG. A benchmark for chromatin binding measurements in live cells. Nucleic Acids Res. 2012;40:e119. doi: 10.1093/nar/gks701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee JD, Wood WI, Dolan M, Engel JD, Felsenfeld G. A 200 base pair region at the 5′ end of the chicken adult beta-globin gene is accessible to nuclease digestion. Cell. 1981;27:45–55. doi: 10.1016/0092-8674(81)90359-7. [DOI] [PubMed] [Google Scholar]
- McGuire WL. Hormone receptors: their role in predicting prognosis and response to endocrine therapy. Semin Oncol. 1978;5:428–433. [PubMed] [Google Scholar]
- McNally JG, Mueller WG, Walker D, Wolford RG, Hager GL. The glucocorticoid receptor: Rapid exchange with regulatory sites in living cells. Science. 2000;287:1262–1265. doi: 10.1126/science.287.5456.1262. [DOI] [PubMed] [Google Scholar]
- Miranda TB, Voss TC, Sung MH, Baek S, John S, Hawkins M, Grontved L, Schiltz RL, Hager GL. Reprogramming of the chromatin landscape: interplay of the estrogen and glucocorticoid receptors at the genomic level. Cancer Res. 2013;73:5130–5139. doi: 10.1158/0008-5472.CAN-13-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed H, D’Santos C, Serandour AA, Ali HR, Brown GD, Atkins A, Rueda OM, Holmes KA, Theodorou V, Robinson JL, et al. Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep. 2013;3:342–349. doi: 10.1016/j.celrep.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed H, Russell IA, Stark R, Rueda OM, Hickey TE, Tarulli GA, Serandour AA, Birrell SN, Bruna A, Saadi A, et al. Progesterone receptor modulates ERalpha action in breast cancer. Nature. 2015;523:313–317. doi: 10.1038/nature14583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgunova E, Taipale J. Structural perspective of cooperative transcription factor binding. Curr Opin Struct Biol. 2017;47:1–8. doi: 10.1016/j.sbi.2017.03.006. [DOI] [PubMed] [Google Scholar]
- Morisaki T, Muller WG, Golob N, Mazza D, McNally JG. Single-molecule analysis of transcription factor binding at transcription sites in live cells. Nat Commun. 2014;5:4456. doi: 10.1038/ncomms5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouridsen H, Gershanovich M, Sun Y, Perez-Carrion R, Boni C, Monnier A, Apffelstaedt J, Smith R, Sleeboom HP, Jaenicke F, et al. Phase III study of letrozole versus tamoxifen as first-line therapy of advanced breast cancer in postmenopausal women: analysis of survival and update of efficacy from the International Letrozole Breast Cancer Group. J Clin Oncol. 2003;21:2101–2109. doi: 10.1200/JCO.2003.04.194. [DOI] [PubMed] [Google Scholar]
- Mueller F, Stasevich TJ, Mazza D, McNally JG. Quantifying transcription factor kinetics: at work or at play? Crit Rev Biochem Mol Biol. 2013;48:492–514. doi: 10.3109/10409238.2013.833891. [DOI] [PubMed] [Google Scholar]
- Murakami S, Nagari A, Kraus WL. Dynamic assembly and activation of estrogen receptor alpha enhancers through coregulator switching. Genes Dev. 2017;31:1535–1548. doi: 10.1101/gad.302182.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaich AK, Walker DA, Wolford RG, Hager GL. Rapid periodic binding and displacement of the glucocorticoid receptor during chromatin remodeling. Mol Cell. 2004;14:163–174. doi: 10.1016/s1097-2765(04)00178-9. [DOI] [PubMed] [Google Scholar]
- Neph S, Vierstra J, Stergachis AB, Reynolds AP, Haugen E, Vernot B, Thurman RE, John S, Sandstrom R, Johnson AK, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh KS, Patel H, Gottschalk RA, Lee WS, Baek S, Fraser IDC, Hager GL, Sung MH. Anti-inflammatory chromatinscape suggests alternative mechanisms of glucocorticoid receptor action. Immunity. 2017;47:298–309. e295. doi: 10.1016/j.immuni.2017.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando V, Paro R. Mapping Polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin. Cell. 1993;75:1187–1198. doi: 10.1016/0092-8674(93)90328-n. [DOI] [PubMed] [Google Scholar]
- Paakinaho V, Kaikkonen S, Makkonen H, Benes V, Palvimo JJ. SUMOylation regulates the chromatin occupancy and anti-proliferative gene programs of glucocorticoid receptor. Nucleic Acids Res. 2014;42:1575–1592. doi: 10.1093/nar/gkt1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paakinaho V, Presman DM, Ball DA, Johnson TA, Schiltz RL, Levitt P, Mazza D, Morisaki T, Karpova TS, Hager GL. Single-molecule analysis of steroid receptor and cofactor action in living cells. Nat Commun. 2017;8:15896. doi: 10.1038/ncomms15896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Kocherginsky M, Conzen SD. Activation of the glucocorticoid receptor is associated with poor prognosis in estrogen receptor-negative breast cancer. Cancer Res. 2011;71:6360–6370. doi: 10.1158/0008-5472.CAN-11-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissi V, Rosenfeld MG. Controlling nuclear receptors: the circular logic of cofactor cycles. Nat Rev Mol Cell Biol. 2005;6:542–554. doi: 10.1038/nrm1680. [DOI] [PubMed] [Google Scholar]
- Petta I, Dejager L, Ballegeer M, Lievens S, Tavernier J, De Bosscher K, Libert C. The interactome of the glucocorticoid receptor and its influence on the actions of glucocorticoids in combatting inflammatory and infectious diseases. Microbiol Mol Biol Rev. 2016;80:495–522. doi: 10.1128/MMBR.00064-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presman DM, Ball DA, Paakinaho V, Grimm JB, Lavis LD, Karpova TA, Hager GL. Quantifying transcription factor dynamics at the single-molecule level in live cells. Methods. 2017;123:76–88. doi: 10.1016/j.ymeth.2017.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulice JL, Kadoch C. Composition and Function of Mammalian SWI/SNF Chromatin Remodeling Complexes in Human Disease. Cold Spring Harb Symp Quant Biol. 2016;81:53–60. doi: 10.1101/sqb.2016.81.031021. [DOI] [PubMed] [Google Scholar]
- Qi JP, Yang YL, Zhu H, Wang J, Jia Y, Liu N, Song YJ, Zan LK, Zhang X, Zhou M, et al. Expression of the androgen receptor and its correlation with molecular subtypes in 980 chinese breast cancer patients. Breast Cancer (Auckl) 2012;6:1–8. doi: 10.4137/BCBCR.S8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu K, Zaba LC, Satpathy AT, Giresi PG, Li R, Jin Y, Armstrong R, Jin C, Schmitt N, Rahbar Z, et al. Chromatin Accessibility Landscape of Cutaneous T Cell Lymphoma and Dynamic Response to HDAC Inhibitors. Cancer Cell. 2017;32:27–41. e24. doi: 10.1016/j.ccell.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafiee MR, Girardot C, Sigismondo G, Krijgsveld J. Expanding the Circuitry of Pluripotency by Selective Isolation of Chromatin-Associated Proteins. Mol Cell. 2016;64:624–635. doi: 10.1016/j.molcel.2016.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014:159. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechoum Y, Rovito D, Iacopetta D, Barone I, Ando S, Weigel NL, O’Malley BW, Brown PH, Fuqua SA. AR collaborates with ERalpha in aromatase inhibitor-resistant breast cancer. Breast Cancer Res Treat. 2014;147:473–485. doi: 10.1007/s10549-014-3082-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011;147:1408–1419. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheinbay E, Parasuraman P, Grimsby J, Tiao G, Engreitz JM, Kim J, Lawrence MS, Taylor-Weiner A, Rodriguez-Cuevas S, Rosenberg M, et al. Recurrent and functional regulatory mutations in breast cancer. Nature. 2017;547:55–60. doi: 10.1038/nature22992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribich S, Harvey D, Copeland RA. Drug Discovery and Chemical Biology of Cancer Epigenetics. Cell Chem Biol. 2017;24:1120–1147. doi: 10.1016/j.chembiol.2017.08.020. [DOI] [PubMed] [Google Scholar]
- Risca VI, Denny SK, Straight AF, Greenleaf WJ. Variable chromatin structure revealed by in situ spatially correlated DNA cleavage mapping. Nature. 2017;541:237–241. doi: 10.1038/nature20781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- Robinson JL, MacArthur S, Ross-Innes CS, Tilley WD, Neal DE, Mills IG, Carroll JS. Androgen receptor driven transcription in molecular apocrine breast cancer is mediated by FoxA1. EMBO J. 2011;30:3019–3027. doi: 10.1038/emboj.2011.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe JS, Hwang CI, Somerville TDD, Milazzo JP, Lee EJ, Da Silva B, Maiorino L, Tiriac H, Young CM, Miyabayashi K, et al. Enhancer Reprogramming Promotes Pancreatic Cancer Metastasis. Cell. 2017;170:875–888. e820. doi: 10.1016/j.cell.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa FE, Caldeira JR, Felipes J, Bertonha FB, Quevedo FC, Domingues MA, Moraes Neto FA, Rogatto SR. Evaluation of estrogen receptor alpha and beta and progesterone receptor expression and correlation with clinicopathologic factors and proliferative marker Ki-67 in breast cancers. Hum Pathol. 2008;39:720–730. doi: 10.1016/j.humpath.2007.09.019. [DOI] [PubMed] [Google Scholar]
- Roses DF. Breast cancer. Churchill Livingstone; 1999. [Google Scholar]
- Rossouw JE, Anderson GL, Prentice RL, LaCroix AZ, Kooperberg C, Stefanick ML, Jackson RD, Beresford SA, Howard BV, Johnson KC, et al. Risks and benefits of estrogen plus progestin in healthy postmenopausal women: principal results From the Women’s Health Initiative randomized controlled trial. JAMA. 2002;288:321–333. doi: 10.1001/jama.288.3.321. [DOI] [PubMed] [Google Scholar]
- Rotem A, Ram O, Shoresh N, Sperling RA, Goren A, Weitz DA, Bernstein BE. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat Biotechnol. 2015;33:1165–1172. doi: 10.1038/nbt.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu B, Laakso M, Ovaska K, Mirtti T, Lundin J, Rannikko A, Sankila A, Turunen JP, Lundin M, Konsti J, et al. Dual role of FoxA1 in androgen receptor binding to chromatin, androgen signalling and prostate cancer. EMBO J. 2011;30:3962–3976. doi: 10.1038/emboj.2011.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santen RJ. Menopausal hormone therapy and breast cancer. J Steroid Biochem Mol Biol. 2014;142:52–61. doi: 10.1016/j.jsbmb.2013.06.010. [DOI] [PubMed] [Google Scholar]
- Schaffner W. Enhancers, enhancers - from their discovery to today’s universe of transcription enhancers. Biol Chem. 2015;396:311–327. doi: 10.1515/hsz-2014-0303. [DOI] [PubMed] [Google Scholar]
- Schmidt JC, Zaug AJ, Cech TR. Live Cell Imaging Reveals the Dynamics of Telomerase Recruitment to Telomeres. Cell. 2016;166:1188–1197. doi: 10.1016/j.cell.2016.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S, Furlan-Magaril M, Mifsud B, Tavares-Cadete F, Sugar R, Javierre BM, Nagano T, Katsman Y, Sakthidevi M, Wingett SW, et al. The pluripotent regulatory circuitry connecting promoters to their long-range interacting elements. Genome Res. 2015;25:582–597. doi: 10.1101/gr.185272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson TM, Kim Y, Joosten SEP, Schuurman K, van der Groep P, Moelans CB, Ter Hoeve ND, Manson QF, Martens JW, van Deurzen CHM, et al. Characterizing steroid hormone receptor chromatin binding landscapes in male and female breast cancer. Nat Commun. 2018;9:482. doi: 10.1038/s41467-018-02856-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–236. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- Siersbaek R, Baek S, Rabiee A, Nielsen R, Traynor S, Clark N, Sandelin A, Jensen ON, Sung MH, Hager GL, et al. Molecular Architecture of Transcription Factor Hotspots in Early Adipogenesis. Cell Rep. 2014;7:1434–1442. doi: 10.1016/j.celrep.2014.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal H, Greene ME, Tarulli G, Zarnke AL, Bourgo RJ, Laine M, Chang YF, Ma S, Dembo AG, Raj GV, et al. Genomic agonism and phenotypic antagonism between estrogen and progesterone receptors in breast cancer. Sci Adv. 2016;2:e1501924. doi: 10.1126/sciadv.1501924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal H, Greene ME, Zarnke AL, Laine M, Al Abosy R, Chang YF, Dembo AG, Schoenfelt K, Vadhi R, Qiu X, et al. Progesterone receptor isoforms, agonists and antagonists differentially reprogram estrogen signaling. Oncotarget. 2018;9:4282–4300. doi: 10.18632/oncotarget.21378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Shilatifard A. Enhancer biology and enhanceropathies. Nat Struct Mol Biol. 2014;21:210–219. doi: 10.1038/nsmb.2784. [DOI] [PubMed] [Google Scholar]
- Soccio RE, Chen ER, Rajapurkar SR, Safabakhsh P, Marinis JM, Dispirito JR, Emmett MJ, Briggs ER, Fang B, Everett LJ, et al. Genetic Variation Determines PPARgamma Function and Anti-diabetic Drug Response In Vivo. Cell. 2015;162:33–44. doi: 10.1016/j.cell.2015.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon MJ, larsen pl, Varshavsky A. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell. 1988;53:937–947. doi: 10.1016/s0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- Song L, Zhang Z, Grasfeder LL, Boyle AP, Giresi PG, Lee BK, Sheffield NC, Graf S, Huss M, Keefe D, et al. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 2011:21. doi: 10.1101/gr.121541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Chen J, Zhao M, Zhang C, Yu Y, Lonard DM, Chow DC, Palzkill T, Xu J, O’Malley BW, et al. Development of potent small-molecule inhibitors to drug the undruggable steroid receptor coactivator-3. Proc Natl Acad Sci U S A. 2016;113:4970–4975. doi: 10.1073/pnas.1604274113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soon WW, Hariharan M, Snyder MP. High-throughput sequencing for biology and medicine. Mol Syst Biol. 2013;9:640. doi: 10.1038/msb.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Garcia MF, Jaroszewicz A, Osman N, Pellegrini M, Zaret KS. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell. 2015;161:555–568. doi: 10.1016/j.cell.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Furlong EE. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- Starick SR, Ibn-Salem J, Jurk M, Hernandez C, Love MI, Chung HR, Vingron M, Thomas-Chollier M, Meijsing SH. ChIP-exo signal associated with DNA-binding motifs provide insights into the genomic binding of the glucocorticoid receptor and cooperating transcription factors. Genome Res. 2015 doi: 10.1101/gr.185157.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasevich TJ, McNally JG. Assembly of the transcription machinery: ordered and stable, random and dynamic, or both? Chromosoma. 2011;120:533–545. doi: 10.1007/s00412-011-0340-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavreva DA, Muller WG, Hager GL, Smith CL, McNally JG. Rapid glucocorticoid receptor exchange at a promoter is coupled to transcription and regulated by chaperones and proteasomes. Mol Cell Biol. 2004;24:2682–2697. doi: 10.1128/MCB.24.7.2682-2697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stender JD, Kim K, Charn TH, Komm B, Chang KC, Kraus WL, Benner C, Glass CK, Katzenellenbogen BS. Genome-wide analysis of estrogen receptor alpha DNA binding and tethering mechanisms identifies Runx1 as a novel tethering factor in receptor-mediated transcriptional activation. Mol Cell Biol. 2010;30:3943–3955. doi: 10.1128/MCB.00118-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stender JD, Nwachukwu JC, Kastrati I, Kim Y, Strid T, Yakir M, Srinivasan S, Nowak J, Izard T, Rangarajan ES, et al. Structural and Molecular Mechanisms of Cytokine-Mediated Endocrine Resistance in Human Breast Cancer Cells. Mol Cell. 2017;65:1122–1135. e1125. doi: 10.1016/j.molcel.2017.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenoien DL, Patel K, Mancini MG, Dutertre M, Smith CL, O’Malley BW, Mancini MA. FRAP reveals that mobility of oestrogen receptor-alpha is ligand- and proteasome-dependent. Nat Cell Biol. 2001;3:15–23. doi: 10.1038/35050515. [DOI] [PubMed] [Google Scholar]
- Stergachis AB, Neph S, Reynolds A, Humbert R, Miller B, Paige SL, Vernot B, Cheng JB, Thurman RE, Sandstrom R, et al. Developmental fate and cellular maturity encoded in human regulatory DNA landscapes. Cell. 2013;154:888–903. doi: 10.1016/j.cell.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergachis AB, Neph S, Sandstrom R, Haugen E, Reynolds AP, Zhang M, Byron R, Canfield T, Stelhing-Sun S, Lee K, et al. Conservation of trans-acting circuitry during mammalian regulatory evolution. Nature. 2014;515:365–370. doi: 10.1038/nature13972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steube A, Schenk T, Tretyakov A, Saluz HP. High-intensity UV laser ChIP-seq for the study of protein-DNA interactions in living cells. Nat Commun. 2017;8:1303. doi: 10.1038/s41467-017-01251-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugo N, Morimatsu M, Arai Y, Kousoku Y, Ohkuni A, Nomura T, Yanagida T, Yamamoto N. Single-Molecule Imaging Reveals Dynamics of CREB Transcription Factor Bound to Its Target Sequence. Sci Rep. 2015;5:10662. doi: 10.1038/srep10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung M-H, Baek S, Hager GL. Genome-wide footprinting: ready for prime time? Nat Methods. 2016;13:222–228. doi: 10.1038/nmeth.3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung MH, Guertin MJ, Baek S, Hager GL. DNase footprint signatures are dictated by factor dynamics and DNA sequence. Mol Cell. 2014;56:275–285. doi: 10.1016/j.molcel.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sur I, Taipale J. The role of enhancers in cancer. Nat Rev Cancer. 2016;16:483–493. doi: 10.1038/nrc.2016.62. [DOI] [PubMed] [Google Scholar]
- Sur IK, Hallikas O, Vaharautio A, Yan J, Turunen M, Enge M, Taipale M, Karhu A, Aaltonen LA, Taipale J. Mice lacking a Myc enhancer that includes human SNP rs6983267 are resistant to intestinal tumors. Science. 2012;338:1360–1363. doi: 10.1126/science.1228606. [DOI] [PubMed] [Google Scholar]
- Swinstead EE, Miranda TB, Paakinaho V, Baek S, Goldstein I, Hawkins M, Karpova TS, Ball DA, Mazza D, Lavis LD, et al. Steroid Receptors Reprogram FoxA1 Occupancy through Dynamic Chromatin Transitions. Cell. 2016a;165:593–605. doi: 10.1016/j.cell.2016.02.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinstead EE, Paakinaho V, Presman DM, Hager GL. Pioneer factors and ATP-dependent chromatin remodeling factors interact dynamically: A new perspective: Multiple transcription factors can effect chromatin pioneer functions through dynamic interactions with ATP-dependent chromatin remodeling factors. Bioessays. 2016b;38:1150–1157. doi: 10.1002/bies.201600137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takaku M, Grimm SA, Shimbo T, Perera L, Menafra R, Stunnenberg HG, Archer TK, Machida S, Kurumizaka H, Wade PA. GATA3-dependent cellular reprogramming requires activation-domain dependent recruitment of a chromatin remodeler. Genome Biol. 2016;17:36. doi: 10.1186/s13059-016-0897-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teves SS, An L, Hansen AS, Xie L, Darzacq X, Tjian R. A dynamic mode of mitotic bookmarking by transcription factors. Elife. 2016:5. doi: 10.7554/eLife.22280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodorou V, Stark R, Menon S, Carroll JS. GATA3 acts upstream of FOXA1 in mediating ESR1 binding by shaping enhancer accessibility. Genome Res. 2012 doi: 10.1101/gr.139469.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga M, Imamoto N, Sakata-Sogawa K. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat Methods. 2008;5:159–161. doi: 10.1038/nmeth1171. [DOI] [PubMed] [Google Scholar]
- Toy W, Weir H, Razavi P, Lawson M, Goeppert AU, Mazzola AM, Smith A, Wilson J, Morrow C, Wong WL, et al. Activating ESR1 Mutations Differentially Affect the Efficacy of ER Antagonists. Cancer Discov. 2017;7:277–287. doi: 10.1158/2159-8290.CD-15-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlenhaut NH, Barish GD, Yu RT, Downes M, Karunasiri M, Liddle C, Schwalie P, Hubner N, Evans RM. Insights into Negative Regulation by the Glucocorticoid Receptor from Genome-wide Profiling of Inflammatory Cistromes. Mol Cell. 2012;49:158–171. doi: 10.1016/j.molcel.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera M, Biswas J, Senecal A, Singer RH, Park HY. Single-Cell and Single-Molecule Analysis of Gene Expression Regulation. Annu Rev Genet. 2016;50:267–291. doi: 10.1146/annurev-genet-120215-034854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Diezmann A, Shechtman Y, Moerner WE. Three-Dimensional Localization of Single Molecules for Super-Resolution Imaging and Single-Particle Tracking. Chem Rev. 2017;117:7244–7275. doi: 10.1021/acs.chemrev.6b00629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voong LN, Xi L, Sebeson AC, Xiong B, Wang JP, Wang X. Insights into Nucleosome Organization in Mouse Embryonic Stem Cells through Chemical Mapping. Cell. 2016;167:1555–1570. e1515. doi: 10.1016/j.cell.2016.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voong LN, Xi L, Wang JP, Wang X. Genome-wide Mapping of the Nucleosome Landscape by Micrococcal Nuclease and Chemical Mapping. Trends Genet. 2017;33:495–507. doi: 10.1016/j.tig.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss TC, Hager GL. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat Rev Genet. 2014;15:69–81. doi: 10.1038/nrg3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss TC, Schiltz RL, Sung MH, Yen PM, Stamatoyannopoulos JA, Biddie SC, Johnson TA, Miranda TB, John S, Hager GL. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell. 2011;146:544–554. doi: 10.1016/j.cell.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Green S, Greene G, Krust A, Bornert JM, Jeltsch JM, Staub A, Jensen E, Scrace G, Waterfield M, et al. Cloning of the human estrogen receptor cDNA. Proc Natl Acad Sci USA. 1985;82:7889–7893. doi: 10.1073/pnas.82.23.7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welboren WJ, van Driel MA, Janssen-Megens EM, van Heeringen SJ, Sweep FC, Span PN, Stunnenberg HG. ChIP-Seq of ERalpha and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 2009;28:1418–1428. doi: 10.1038/emboj.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West DC, Pan D, Tonsing-Carter EY, Hernandez KM, Pierce CF, Styke SC, Bowie KR, Garcia TI, Kocherginsky M, Conzen SD. GR and ER co-activation alters the expression of differentiation genes and associates with improved ER+ breast cancer outcome. Mol Cancer Res. 2016 doi: 10.1158/1541-7786.MCR-15-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittliff JL. Steroid-hormone receptors in breast cancer. Cancer. 1984;53:630–643. doi: 10.1002/1097-0142(19840201)53:3+<630::aid-cncr2820531308>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Yang F, Ma Q, Liu Z, Li W, Tan Y, Jin C, Ma W, Hu Y, Shen J, Ohgi KA, et al. Glucocorticoid Receptor:MegaTrans Switching Mediates the Repression of an ERalpha-Regulated Transcriptional Program. Mol Cell. 2017;66:321–331. e326. doi: 10.1016/j.molcel.2017.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi P, Wang Z, Feng Q, Chou CK, Pintilie GD, Shen H, Foulds CE, Fan G, Serysheva I, Ludtke SJ, et al. Structural and Functional Impacts of ER Coactivator Sequential Recruitment. Mol Cell. 2017;67:733–743. e734. doi: 10.1016/j.molcel.2017.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Mango SE. Pioneer transcription factors, chromatin dynamics, and cell fate control. Curr Opin Genet Dev. 2016;37:76–81. doi: 10.1016/j.gde.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen CY, Tatavosian R, Huynh TN, Duc HN, Das R, Kokotovic M, Grimm JB, Lavis LD, Lee J, Mejia FJ, et al. Live-cell single-molecule tracking reveals co-recognition of H3K27me3 and DNA targets polycomb Cbx7-PRC1 to chromatin. Elife. 2016:5. doi: 10.7554/eLife.17667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Gates LA, Stashi E, Dasgupta S, Gonzales N, Dean A, Dacso CC, York B, O’Malley BW. Coactivator-Dependent Oscillation of Chromatin Accessibility Dictates Circadian Gene Amplitude via REV-ERB Loading. Mol Cell. 2015;60:769–783. doi: 10.1016/j.molcel.2015.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart W, Koornstra R, Wesseling J, Rutgers E, Linn S, Carroll JS. A carrier-assisted ChIP-seq method for estrogen receptor-chromatin interactions from breast cancer core needle biopsy samples. BMC Genomics. 2013;14:232. doi: 10.1186/1471-2164-14-232. [DOI] [PMC free article] [PubMed] [Google Scholar]