Abstract
Breast cancer is one of the most common malignant diseases in women. Triple‐negative breast cancer (TNBC) shows higher aggressiveness and recurrence rates than other subtypes, and there are no effective targets or tailored treatments for TNBC patients. Thus, finding effective prognostic markers for TNBC could help clinicians in their ability to care for their patients. We used tissue microarrays (TMAs) to detect microRNA‐493 (miR‐493) expression in breast cancer samples. A miRCURY LNA detection probe specific for miR‐493 was used in in situ hybridization assays. Staining results were reviewed by two independent pathologists and classified as high or low expression of miR‐493. Kaplan–Meier survival plots and multivariate Cox analysis were carried out to clarify the relationship between miR‐493 and survival. In the Kaplan–Meier analysis, patients with high miR‐493 expression had better disease‐free survival than patients with low miR‐493 expression. After adjusting for common clinicopathological factors in breast cancer, the expression level of miR‐493 was still a significant prognostic factor in breast cancer. Further subtype analysis revealed that miR‐493 expression levels were only significantly prognostic in TNBC patients. These results were validated in the Molecular Taxonomy of Breast Cancer International Consortium database for overall survival. We proved the prognostic role of miR‐493 in TNBC by using one of the largest breast cancer TMAs available and validated it in a large public RNA sequencing database.
Keywords: breast cancer, DFS, microRNA 493, prognostic factor, triple‐negative
1. INTRODUCTION
Breast cancer is one of the most common malignant diseases in women, and is responsible for the second highest cancer‐specific death rates.1, 2, 3, 4, 5 According to the American Cancer Society Surveillance Information Services, Surveillance and Health Services’ report, there will be 252 710 new breast cancer cases, which account for 30% of all new cancer cases diagnosed in US women in 2017; and about 40 610 patients will die of breast cancer, accounting for 14% of all cancer‐related deaths.1 The 5‐year survival rate of breast cancer can be as high as 90%,1 and many patients will experience disease recurrence or metastasis. Once this occurs, oncologists have little to offer in the way of treatment.6, 7, 8, 9 Therefore, finding useful approaches or markers for predicting breast cancer recurrence can allow treatment in the adjuvant setting to be tailored to these patients, which can be effective in decreasing relapse and cancer‐related deaths.10, 11, 12 This kind of strategy might be more significant in triple‐negative breast cancer (TNBC; estrogen receptor‐negative, progesterone receptor‐negative, and human epidermal growth factor receptor 2 [HER2]‐negative), which usually shows higher aggressiveness and recurrence rates and is widely accepted as a heterogeneous disease. For TNBC patients, there are no effective targets or tailored treatments. The standard adjuvant treatment for TNBC is chemotherapy, the doses and regimens of which can be modified according to the patient's risk of recurrence. Thus, finding effective prognostic markers for TNBC might help clinicians in their ability to take better care of their patients.11, 12
MicroRNA (miRNA) are small non‐coding elements, usually 21‐25 nt in length, which regulate expression of various genes and some important cellular processes.13, 14 The role of miRNAs in breast cancer has been widely studied.15, 16, 17, 18 There are oncogenic miRNAs, like miR‐21,15, 16 miR‐155,17, 18 miR‐10b,19, 20 and miR27a,21, 22 which are frequently upregulated in breast cancer and influence tumor cell survival, promote proliferation and metastasis, and inhibit cell death. There are also tumor‐suppressive miRNAs, like the let‐7 family,23, 24 the miR‐200 family,25, 26 miR‐145,27 and miR‐205,28, 29 which are significantly downregulated in breast cancer and can silence different target genes, and negatively regulate epithelial–mesenchymal transition and tumor development.
MicroRNA‐493 has been explored in cancer in previous studies. According to these studies, the roles of miR‐493 in cancer are mainly suppressive. In lung cancer, Gu et al30 found that miR‐493 was markedly reduced in pulmonary carcinomas and that it could directly target E2F1, which in turn decreased growth, invasion, and metastasis of lung cancer cells. Another study by Cui et al31 showed that overexpression of miR‐493 suppressed cell proliferation and the cell cycle in human melanoma cells. Furthermore, miR‐493 was a new tumor suppressor miRNA in bladder cancer and inhibited cell motility through downregulation of RhoC and FZD4.32 Moreover, the suppressor role of miR‐493 has also been reported in colon cancer metastasis. Sakai et al33 observed that miR‐493 inhibited MAPK kinase 7 expression by targeting the binding site at the 3′‐UTR of the mkk7 gene, which resulted in marked suppression of liver metastasis of colon cancer cells. Also, it was shown in the Okamoto et al34 study that in a subset of colon cancer, upregulation of miR‐493 during carcinogenesis prevented liver metastasis through the induction of cell death of metastasized cells.
Several studies have shown that miRNAs can be prognostic factors in breast cancer.13, 35, 36 In the present study, we focused on miR‐493, and have used our center's well‐developed tissue microarray (TMA) to explore its prognostic value in breast cancer, especially in TNBC.
2. MATERIALS AND METHODS
2.1. Study cohort
We used our prepared TMAs, which have been described previously,35, 36, 37, 38 to detect miR‐493 expression in breast cancer samples. A total of 382 breast cancer patients were included in the present study, with their miR‐493 expression levels available. All analyzed patients were diagnosed with stage I‐III breast cancer from August 2001 to November 2007 at Fudan University Shanghai Cancer Center (FUSCC; Shanghai, China), and confirmed using pathology. Clinical and pathological factors were collected by using the FUSCC inpatient system, and survival data was retrieved through the FUSCC outpatient system or by telephone. Disease‐free survival (DFS) events were recorded with any of the following situations: the first recurrence of invasive disease at a local, regional, or distant site, contralateral breast cancer, and death from any cause. Patients without DFS events were censored at the last follow‐up. The median DFS and overall survival (OS) times for the cohorts were 90.28 and 94.73 months, respectively. The independent ethics committee/institutional review board of FUSCC (Shanghai Cancer Center Ethics Committee) approved the present study. All patients gave their written informed consent.
2.2. In situ hybridization of miR‐493
All staining was undertaken on the well‐developed TMAs. The detailed method used in the development of the TMAs has been described previously.35, 36, 37, 38 A miRCURY LNA detection probe specific for miR‐493 was purchased from Exiqon (Vedbaek, Denmark). The probe, 5′‐CCTGGCACACAGTAGACCTTCA‐3′, was 5′‐digoxigenin‐ and 3′‐digoxigenin‐labeled. For in situ hybridization, we used Enhanced Sensitive ISH Detection I (POD) kits from Boster (Wuhan, China). All experiments were undertaken according to the manufacturer's protocol and described previously. Briefly, TMAs were rewarmed at 65°C for 4 hours, deparaffinized in xylene, and sequentially hydrated in gradient ethanol solutions (three times in 100%, once in 95%, once in 85%, and once in 75%). After this procedure, TMAs were washed with PBS three times and incubated with 3% hydrogen peroxide for 10 minutes at room temperature to block endogenous peroxidase activity. Then TMAs were washed with 0.1% DEPC‐H2O for 5 minutes, and then incubated with pepsin diluted 10‐fold by citrate at 37°C for 20 minutes to expose the nucleic acid fraction of RNA. After the digestion procedure, TMAs were washed with PBS three times for 5 minutes each and with 0.1% DEPC‐H2O once for 5 minutes. After incubation with prehybridization solution for 3 hours at 37°C, the TMAs were incubated with 200 μL miRNA probe (20 nmol/L) that had been preheated for 10 minutes at 80°C and quickly transferred to an ice/water mixture for 5 minutes, in a hybridization box at 60°C overnight. The next day, TMAs were subjected to a stringent washing procedure with 2× saline sodium citrate (SSC; preheated at 37°C, two washes, 5 minutes each), 0.5× SSC (one wash, 15 minutes), and 0.2× SSC (one wash, 15 minutes). After a 30‐minute wash in blocking solution, TMAs were sequentially incubated with biotinylated digoxin (60 minutes), streptavidin–biotin complex (20 minutes), and peroxidase (20 minutes) with a 5‐minute wash in 0.5 mol/L PBS between each. The results were visualized after staining with 3, 3‐diaminobenzidine and counterstained with Gill hematoxylin. For all experiments, a U6 probe was used as a positive control.
Two experienced pathologists reviewed the in situ hybridization staining independently, using the staining index (SI), which incorporates intensity and percentage of positive cells. The strength of the staining was scored as: 0, no staining; 1, weak; 2, moderate; and 3, strong. The percentage of cells stained was scored as: 0, no staining; 1, <10%; 2, 10%‐50%; and 3, >50% tumor cells. If there was a disagreement between the two pathologists, a third pathologist would be consulted. The SI was derived by multiplying the two results mentioned above. Samples with SIs >4 were considered as miR‐493 high expression. Examples of high and low expression stains are shown in Figure 1(A).
Figure 1.

MicroRNA‐493 (miR‐493) expression is correlated with breast cancer survival. A, Examples of high and low expression of miR‐493 staining in samples at ×10 and ×40 magnification. B, Kaplan–Meier plot of miR‐493 in relation to disease‐free survival
2.3. MicroRNA‐493 expression in METABRIC database
The Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) is an integrated genomic/transcriptomic database of breast cancers.39 In the following research, Lánczky et al40developed a tool, miRpower, which helps to easily access the miRNA expression data in METABRIC, and they provided a website for public use (http://kmplot.com/analysis/index.php?p=service&cancer=breast_mirna). All miRNA expression data in the METABRIC database are based on the microarray platform. To validate our finding of the prognostic effect of miR‐493 in breast cancer, we downloaded miR‐493 expression and responding clinical data from the METABRIC database using miRpower. The cut‐off of miR‐493 expression value, classifying its expression as high or low, was retrieved by using the auto select best cut‐off function in miRpower in the whole available METABRIC breast cancer cohort, and applied it to subtype analyses.
2.4. Statistical analysis
The distribution of miR‐493 expression in relation with different clinicopathological factors was evaluated using Pearson's chi‐square‐test. Kaplan–Meier survival plots were used to assess the impact of miR‐493 expression on DFS, with log–rank test results reported. Multivariate Cox analysis was carried out to clarify the relationship between miR‐493 and survival. All statistical analyses were undertaken using SPSS software (version 22; IBM, Armonk, NY, USA). All P‐values were two‐sided unless stated otherwise and only P‐values <.05 were considered statistically significant.
3. RESULTS
Detailed information of miR‐493 expression with different clinicopathological factors is listed in Table 1. Among 384 patients, 116 patients (30.1%) were classified as having high miR‐493 expression. There was no statistically significant difference in miR‐493 expression among any of the clinicopathological factors studied (Table 1).
Table 1.
Clinicopathological characteristics of microRNA (miR)‐493 levels in the Fudan University Shanghai Cancer Center cohort of patients with triple‐negative breast cancer
| miR‐493 expression level, n (%) | P‐value, χ2‐test | ||
|---|---|---|---|
| Low | High | ||
| Age, years | .353 | ||
| <40 | 28 (60.9) | 18 (39.1) | ‐ |
| 41‐60 | 197 (70.4) | 83 (29.6) | ‐ |
| >61 | 41 (73.2) | 15 (26.8) | ‐ |
| Menopause | .069 | ||
| No | 129 (65.5) | 68 (34.5) | ‐ |
| Yes | 137 (74.1) | 48 (25.9) | ‐ |
| Grade | .328 | ||
| II | 136 (66.3) | 69 (33.7) | ‐ |
| III | 74 (71.8) | 29 (28.2) | ‐ |
| Tumor size | .746 | ||
| T1 | 125 (69.8) | 54 (30.2) | ‐ |
| T2 | 123 (69.1) | 55 (30.9) | ‐ |
| T3 | 14 (77.8) | 4 (22.2) | ‐ |
| Lymph nodes | .236 | ||
| Negative | 155 (72.1) | 60 (27.9) | ‐ |
| Positive | 111 (66.5) | 56 (33.5) | ‐ |
| ER | .216 | ||
| Negative | 147 (67.1) | 72 (32.9) | ‐ |
| Positive | 119 (73.0) | 44 (27.0) | ‐ |
| PR | .417 | ||
| Negative | 181 (71.0) | 74 (29.0) | ‐ |
| Positive | 85 (66.9) | 42 (33.1) | ‐ |
| HER2 | .707 | ||
| Negative | 150 (70.4) | 63 (29.6) | ‐ |
| Positive | 116 (68.6) | 53 (31.4) | ‐ |
‐, not applicable; ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; PR, progesterone receptor.
In the Kaplan–Meier analysis, patients with high miR‐493 expression had better DFS than patients with low miR‐493 expression (Figure 1B; log–rank, P = .008). Although there was no distribution difference of miR‐493 expression level with the studied clinicopathological factors, we still undertook multivariate analysis using a Cox proportional hazards regression model to rule out potential confounders. As shown in Table 2, after adjusting for the common clinicopathological factors in breast cancer, the expression level of miR‐493 was still a significant prognostic factor in breast cancer. MicroRNA‐493 high‐expression patients have less risk of disease recurrence compared with low‐expression patients (HR = 0.51; 95% CI, 0.28‐0.92; P = .026). Other significant factors were lymph node status and HER2 status (Table 2).
Table 2.
Multivariate survival analyses of factors associated with disease‐free survival in the the Fudan University Shanghai Cancer Center cohort of patients with triple‐negative breast cancer
| HR (95% CI) | P‐value | |
|---|---|---|
| Age, years | ||
| <40 | Reference | ‐ |
| 41‐60 | 0.53 (0.25‐1.13) | .101 |
| >61 | 0.27 (0.09‐0.80) | .019 |
| Menopause | ||
| No | Reference | ‐ |
| Yes | 1.37 (0.79‐2.36) | .260 |
| Grade | ||
| II | Reference | ‐ |
| III | 0.86 (0.49‐1.48) | .577 |
| Tumor size | ||
| T1 | Reference | ‐ |
| T2 | 1.22 (0.72‐2.04) | .462 |
| T3 | 2.16 (0.72‐6.45) | .167 |
| Lymph nodes | ||
| Negative | Reference | ‐ |
| Positive | 2.31 (1.40‐3.81) | .001 |
| ER | ||
| Negative | Reference | ‐ |
| Positive | 0.92 (0.43‐2.00) | .840 |
| PR | ||
| Negative | Reference | ‐ |
| Positive | 0.44 (0.19‐1.00) | .051 |
| HER2 | ||
| Negative | Reference | ‐ |
| Positive | 0.52 (0.28‐0.92) | .018 |
| Chemotherapy | ||
| No | Reference | ‐ |
| Yes | 0.33 (0.13‐0.82) | .017 |
| Unknown | 0.81 (0.23‐2.84) | .744 |
| miR‐493 status | ||
| Low expression | Reference | ‐ |
| High expression | 0.51 (0.28‐0.92) | .026 |
‐, not applicable; CI, confidence interval; ER, estrogen receptor; FUSCC, Fudan University Shanghai Cancer Center; HER2, human epidermal growth factor receptor 2; HR, hazard ratio; miR, microRNA; PR, progesterone receptor.
To further explore miR‐493 prognostic value in breast cancer, we carried out Kaplan–Meier analyses in different breast cancer subtypes. Interestingly, miR‐493 expression level was only significantly related with DFS in TNBC patients (Figure 2D, P = .031), but not in luminal A, luminal B, or HER2‐positive groups (Figure 2A‐C; P = .397, .326, and .159, respectively). This prognostic value difference was not caused by distribution imbalance of miR‐493 expression among the different subtypes (Figure 2; P = .535).
Figure 2.
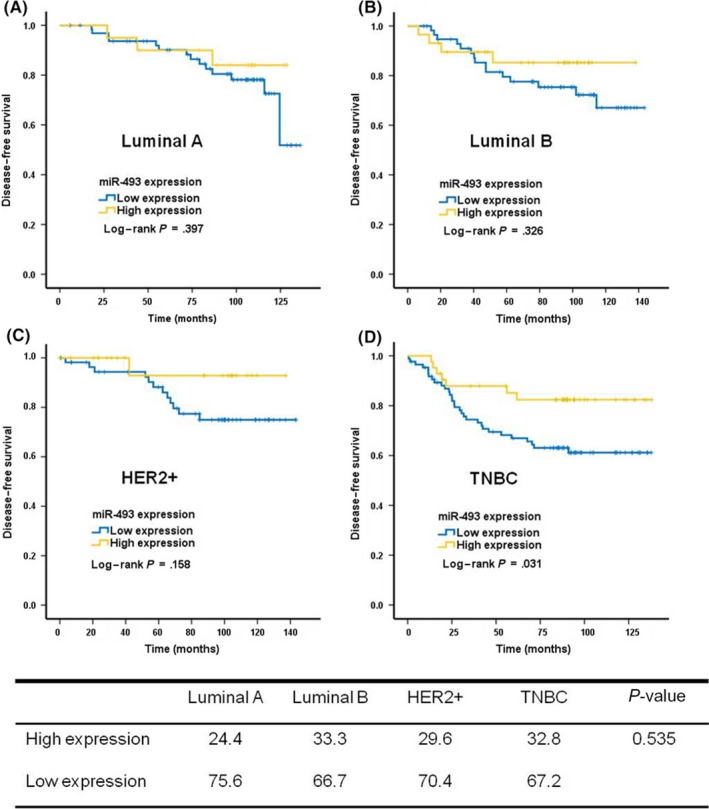
Prognostic value of microRNA‐493 (miR‐493) in breast cancer varies among different subtypes. A, Luminal A. B, Luminal B. C, Human epidermal growth factor receptor 2 (HER2)+. D, Triple‐negative breast cancer (TNBC). The table shows miR‐493 expression in different subtypes
To validate our finding of the prognostic effect of miR‐493 in breast cancer, we downloaded miR‐493 expression data from the METABRIC database using miRpower. In the whole breast cancer cohort, the difference in expression level of miR‐493, the cut‐off for which was determined by miRpower automatically, was not significantly related with OS in patients (Figure 3A; P = .162). In detailed subtype analyses, we found the same prognostic value of miR‐493 in TNBC patients; that is, patients with high miR‐493 expression level would have better OS than those with low‐level expression (Figure 3B; P = .048). In the HER2‐positive group, high expression of miR‐493 was also correlated with better OS (Figure 3E; P = .05); however, only 80 patients were available for this subtype analysis. As before, miR‐493 expression was not prognostic in luminal A or luminal B patients (Figure 3C,D; P = .127 and .242, respectively).
Figure 3.

Validation of microRNA‐493 (miR‐493) prognostic value in the Molecular Taxonomy of Breast Cancer International Consortium database of breast cancers. A, All breast cancers in the database. B, Triple‐negative breast cancer (TNBC) subtype. C, Luminal A subtype. D, Luminal B subtype. E, Human epidermal growth factor receptor 2 (HER2)+ subtype
4. DISCUSSION
In the present study, we found that miR‐493 could be a prognostic factor in breast cancer, especially in the TNBC subtype. Patients with high miR‐493 expression had better survival outcomes compared with patients with low expression. Further analysis in the different subtypes showed that the prognostic value of miR‐493 was mainly significant in TNBC patients, but not in luminal A or luminal B patients. More samples are needed to clarify the prognostic role of miR‐493 for HER2‐positive patients.
In our study, we found that miR‐493 could be a prognostic marker for breast cancer, especially in TNBC. Patients with miR‐493 high expression had better survival, which was in line with the results shown in studies of other cancer types discussed above. The prognostic value of miR‐493 was also validated in the METABRIC database. In breast cancer, few studies have reported investigating the role of miR‐493. In a study by Zhao et al,41 the authors showed that miR493‐5p was differentially expressed between breast cancer tissues, matched adjacent tissue samples, and could be a novel suppressor of invasiveness and tumorigenicity of breast cancer cells through targeting of fucosyltransferase IV. Another study reported that miR‐493 might be a prognostic factor in breast cancer. Using a miRNA expression profiling technique, Gasparini et al42 identified a four‐miRNA signature that included miR‐155, miR‐493, miR‐30e, and miR‐27a, which allowed subdivision of TNBCs into high‐ and low‐risk groups. They proved that this subclassification was a powerful diagnostic and prognostic biomarker in TNBCs. Taken together with our study, these results proved that miR‐493 could be an important tumor suppressor and prognostic marker in breast cancer.
Our study had some limitations. First, this was a retrospective analysis in a relatively small patient cohort. More samples will be needed to further validate the prognostic value of miR‐493. Second, determining miR‐493 expression levels using TMAs in breast cancer might be subjective, even though we used two independent pathologists who were blinded to patients’ survival outcomes and consulted a third pathologist to resolve any discord. Use of the METABRIC database in the validation of the results might help in attenuating these limitations.
Taken together, by using one of the largest breast cancer TMAs available in conjunction with a large public RNA sequencing database, we have shown that miR‐493 can be used as a prognostic marker in TNBC. Clarifying the mechanism by which miR‐493 plays a tumor‐suppressor role in breast cancer will require additional functional studies.
CONFLICT OF INTEREST
The authors have no conflict of interest.
ACKNOWLEDGMENTS
This research was supported by grants from the National Natural Science Foundation of China (81202082) and the Shanghai Cancer Center Funds (YJYQ201402).
Yao L, Liu Y, Cao Z, et al. MicroRNA‐493 is a prognostic factor in triple‐negative breast cancer. Cancer Sci. 2018;109:2294–2301. https://doi.org/10.1111/cas.13644
Yao, Liu and Cao equally contributed to this work.
REFERENCES
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7‐30. [DOI] [PubMed] [Google Scholar]
- 2. DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin. 2014;64:52‐62. [DOI] [PubMed] [Google Scholar]
- 3. DeSantis C, Siegel R, Bandi P, Jemal A. Breast cancer statistics, 2011. CA Cancer J Clin. 2011;61:409‐418. [DOI] [PubMed] [Google Scholar]
- 4. DeSantis CE, Fedewa SA, Goding Sauer A, Kramer JL, Smith RA, Jemal A. Breast cancer statistics, 2015: convergence of incidence rates between black and white women. CA Cancer J Clin. 2016;66:31‐42. [DOI] [PubMed] [Google Scholar]
- 5. Ward EM, DeSantis CE, Lin CC, et al. Cancer statistics: breast cancer in situ. CA Cancer J Clin. 2015;65:481‐495. [DOI] [PubMed] [Google Scholar]
- 6. de la Mare JA, Contu L, Hunter MC, et al. Breast cancer: current developments in molecular approaches to diagnosis and treatment. Recent Pat Anti‐Cancer Drug Discovery. 2014;9:153‐175. [DOI] [PubMed] [Google Scholar]
- 7. Matsen CB, Neumayer LA. Breast cancer: a review for the general surgeon. JAMA Surg. 2013;148:971‐979. [DOI] [PubMed] [Google Scholar]
- 8. Murphy CG, Dickler MN. The role of CDK4/6 inhibition in breast cancer. Oncologist. 2015;20:483‐490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gradishar WJ, Anderson BO, Balassanian R, et al. NCCN guidelines insights: breast cancer, version 1.2017. J Natl Compr Canc Netw. 2017;15:433‐451. [DOI] [PubMed] [Google Scholar]
- 10. Jiang YZ, Liu YR, Xu XE, et al. Transcriptome analysis of triple‐negative breast cancer reveals an integrated mRNA‐lncRNA signature with predictive and prognostic value. Can Res. 2016;76:2105‐2114. [DOI] [PubMed] [Google Scholar]
- 11. Liu YR, Jiang YZ, Xu XE, Hu X, Yu KD, Shao ZM. Comprehensive transcriptome profiling reveals multigene signatures in triple‐negative breast cancer. Clin Cancer Res. 2016;22:1653‐1662. [DOI] [PubMed] [Google Scholar]
- 12. Liu YR, Jiang YZ, Xu XE, et al. Comprehensive transcriptome analysis identifies novel molecular subtypes and subtype‐specific RNAs of triple‐negative breast cancer. Breast Cancer Res. 2016;18:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gyparaki MT, Basdra EK, Papavassiliou AG. MicroRNAs as regulatory elements in triple negative breast cancer. Cancer Lett. 2014;354:1‐4. [DOI] [PubMed] [Google Scholar]
- 14. Zaleska K. miRNA – Therapeutic tool in breast cancer? Where are we now? Rep Pract Oncol Radiother. 2015;20:79‐86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR‐21 in breast cancer cells. J Biol Chem. 2008;283:1026‐1033. [DOI] [PubMed] [Google Scholar]
- 16. Qi J, Wang J, Katayama H, Sen S, Liu SM. Circulating microRNAs (cmiRNAs) as novel potential biomarkers for hepatocellular carcinoma. Neoplasma. 2013;60:135‐142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kong W, He L, Coppola M, et al. MicroRNA‐155 regulates cell survival, growth, and chemosensitivity by targeting FOXO3a in breast cancer. J Biol Chem. 2010;285:17869‐17879. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18. Jiang S, Zhang HW, Lu MH, et al. MicroRNA‐155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Can Res. 2010;70:3119‐3127. [DOI] [PubMed] [Google Scholar]
- 19. Haque I, Banerjee S, Mehta S, et al. Cysteine rich 61‐connective tissue growth factor‐nephroblastoma‐overexpressed 5 (CCN5)/Wnt‐1‐induced signaling protein‐2(WISP‐2) regulates microRNA‐10b via hypoxia‐inducible factor‐1α‐TWIST signaling networks in human breast cancer cells. J Biol Chem. 2011;286:43475‐43485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu Y, Zhao J, Zhang PY, et al. MicroRNA‐10b targets E‐cadherin and modulates breast cancer metastasis. Med Sci Monit 2012;18:Br299‐Br308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tang W, Yu F, Yao H, et al. miR‐27a regulates endothelial differentiation of breast cancer stem like cells. Oncogene. 2014;33:2629‐2638. [DOI] [PubMed] [Google Scholar]
- 22. Guttilla IK, White BA. Coordinate regulation of FOXO1 by miR‐27a, miR‐96, and miR‐182 in breast cancer cells. J Biol Chem. 2009;284:23204‐23216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hu X, Guo J, Zheng L, et al. The heterochronic microRNA let‐7 inhibits cell motility by regulating the genes in the actin cytoskeleton pathway in breast cancer. Mol Cancer Res. 2013;11:240‐250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nimmo RA, Slack FJ. An elegant miRror: microRNAs in stem cells, developmental timing and cancer. Chromosoma. 2009;118:405‐418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li X, Roslan S, Johnstone CN, et al. MiR‐200 can repress breast cancer metastasis through ZEB1‐independent but moesin‐dependent pathways. Oncogene. 2014;33:4077‐4088. [DOI] [PubMed] [Google Scholar]
- 26. Lim YY, Wright JA, Attema JL, et al. Epigenetic modulation of the miR‐200 family is associated with transition to a breast cancer stem‐cell‐like state. J Cell Sci. 2013;126:2256‐2266. [DOI] [PubMed] [Google Scholar]
- 27. Sachdeva M, Mo YY. MicroRNA‐145 suppresses cell invasion and metastasis by directly targeting mucin 1. Can Res. 2010;70:378‐387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang S, Huang J, Lyu H, et al. Functional cooperation of miR‐125a, miR‐125b, and miR‐205 in entinostat‐induced downregulation of erbB2/erbB3 and apoptosis in breast cancer cells. Cell Death Dis. 2013;4:e556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chao CH, Chang CC, Wu MJ, et al. MicroRNA‐205 signaling regulates mammary stem cell fate and tumorigenesis. J Clin Investig. 2014;124:3093‐3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gu Y, Cheng Y, Song Y, et al. MicroRNA‐493 suppresses tumor growth, invasion and metastasis of lung cancer by regulating E2F1. PLoS ONE. 2014;9:e102602. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31. Cui A, Jin Z, Gao Z, et al. Downregulation of miR‐493 promoted melanoma proliferation by suppressing IRS4 expression. Tumour Biol. 2017;39:1010428317701640. [DOI] [PubMed] [Google Scholar]
- 32. Ueno K, Hirata H, Majid S, et al. Tumor suppressor microRNA‐493 decreases cell motility and migration ability in human bladder cancer cells by downregulating RhoC and FZD4. Mol Cancer Ther. 2012;11:244‐253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sakai H, Sato A, Aihara Y, et al. MKK7 mediates miR‐493‐dependent suppression of liver metastasis of colon cancer cells. Cancer Sci. 2014;105:425‐430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Okamoto K, Ishiguro T, Midorikawa Y, et al. miR‐493 induction during carcinogenesis blocks metastatic settlement of colon cancer cells in liver. EMBO J. 2012;31:1752‐1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cao ZG, Huang YN, Yao L, et al. Positive expression of miR‐361‐5p indicates better prognosis for breast cancer patients. J Thorac Dis. 2016;8:1772‐1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cao ZG, Li JJ, Yao L, et al. High expression of microRNA‐454 is associated with poor prognosis in triple‐negative breast cancer. Oncotarget. 2016;7:64900‐64909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ye FG, Song CG, Cao ZG, et al. Cytidine deaminase axis modulated by miR‐484 differentially regulates cell proliferation and chemoresistance in breast cancer. Can Res. 2015;75:1504‐1515. [DOI] [PubMed] [Google Scholar]
- 38. Chen L, Yang L, Qiao F, et al. High levels of nucleolar spindle‐associated protein and reduced levels of BRCA1 expression predict poor prognosis in triple‐negative breast cancer. PLoS ONE. 2015;10:e0140572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Curtis C, Shah SP, Chin SF, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346‐352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lanczky A, Nagy A, Bottai G, et al. miRpower: a web‐tool to validate survival‐associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat. 2016;160:439‐446. [DOI] [PubMed] [Google Scholar]
- 41. Zhao L, Feng X, Song X, et al. miR‐493‐5p attenuates the invasiveness and tumorigenicity in human breast cancer by targeting FUT4. Oncol Rep. 2016;36:1007‐1015. [DOI] [PubMed] [Google Scholar]
- 42. Gasparini P, Cascione L, Fassan M, et al. MicroRNA expression profiling identifies a four microRNA signature as a novel diagnostic and prognostic biomarker in triple negative breast cancers. Oncotarget. 2014; 5: 1174‐1184. [DOI] [PMC free article] [PubMed] [Google Scholar]


