Abstract
Deciphering the molecular networks that discriminate organ‐confined breast cancer from metastatic breast cancer may lead to the identification of critical biomarkers for breast cancer invasion and aggressiveness. Here metabolomics, a global study of metabolites, has been applied to explore the metabolic alterations that characterize breast cancer progression. We profiled a total of 693 metabolites across 87 serum samples related to breast cancer (46 clinically localized and 41 metastatic breast cancer) and 49 normal samples. These unbiased metabolomic profiles were able to distinguish normal individuals, clinically localized and metastatic breast cancer patients. 9‐cis‐Retinoic acid, an isomer of all‐trans retinoic acid, was identified as a differential metabolite that significantly decreased during breast cancer progression to metastasis, and its levels were also reduced in urine samples from biopsy‐positive breast cancer patients relative to biopsy‐negative individuals and in invasive breast cancer cells relative to benign MCF‐10A cells. The addition of exogenous 9‐cis‐retinoic acid to MDA‐MB‐231 cells and knockdown of aldehyde dehydrogenase 1 family member A1, a regulatory enzyme for 9‐cis‐retinoic acid, remarkably impaired cell invasion and migration, presumably through preventing the key regulator cofilin from activation and inhibiting MMP2 and MMP9 expression. Taken together, our study showed the potential inhibitory role for 9‐cis‐retinoic acid in breast cancer progression by attenuating cell invasion and migration.
Keywords: 9‐cis‐retinoic acid, breast cancer, invasion, metabolomics, metastatic
1. INTRODUCTION
Breast cancer is the most frequently diagnosed and death‐related cancer in women.1 Recently, breast cancer has represented 15% of all newly diagnosed female cancers in China, the incidence of which is increasing at a rate of 3% to 4% per year.2 Cancer metastasis3 is the major cause of death for breast cancer patients, which is accompanied by complex molecular events. Deciphering the molecular networks that discriminate organ‐confined breast cancer from metastatic breast cancer may lead to the identification of critical biomarkers for breast cancer invasion and aggressiveness. Currently, however, little is known about the global metabolomic alterations that characterize breast cancer invasion and progression.
Metabolomics is a new, rapidly expanding field dedicated to extensive analysis of hundreds of metabolites in biological systems (ie, its “metabolome”),4 which is a useful complement to the characterization of several physiological and pathological conditions and offers promise as a clinical tool for disease diagnosis and prediction.5, 6 The metabolome can be considered the downstream end product of the complex interaction of genome, transcriptome and proteome, and affected by perturbations as a result of physiology, pathology and iatrogenic factors.7, 8 Discovery of regulatory enzymes and genes through differential metabolites and the known pathways will help us to gain better understanding of dysregulated metabolism in disease initiation and progression.
Breast cancer has been associated with profound alterations in metabolic systems, which have been demonstrated in many preclinical and clinical metabolomics studies of breast cancer cells, tissues, serum and urine.9, 10, 11, 12 For example, glutamate was found to successfully differentiate breast cancer patients from normal controls.13 Plasma metabolic profiles of breast cancer patients showed that a panel of 8 differential metabolites, including carnitine, lysophosphatidylcholine (20:4), proline and alanine, was identified to distinguish between subtypes of breast cancer.14 However, metabolomics biomarker and differential pathways are difficult to replicate among different studies, primarily as a result of the heterogeneity of patients and the different analytical and clinical protocols.15, 16 Moreover, limited data are available to characterize the metabolomics changes during breast cancer progression by metabolic profiling of biospecimens.
Liquid chromatography coupled with mass spectrometry (LC/MS), gas chromatography coupled with mass spectrometry (GC/MS), and NMR are the three most commonly used techniques for metabolomics studies.17, 18 LC/MS and GC/MS techniques have the advantage of higher sensitivity19 and greater separation efficiency, respectively.20 Combined use of LC/MS and GC/MS could be a better choice to profile different classes of metabolites.
In the present study, metabolomics was applied to identify the distinct metabolite signatures of patients with metastatic breast cancer and screen the metabolic markers associated with breast cancer metastasis, which could provide a theoretical basis for early diagnosis, prognosis evaluation and therapeutic target research of breast cancer. The initial training study was conducted with the use of 46 clinically localized breast cancer serum samples (LC), 41 metastatic breast cancer serum samples (MT), and 49 normal control samples (NC), which were profiled using ultra‐performance liquid chromatography‐linear ion trap mass spectrometry (UPLC‐LTQ/MS), and further validated with 50 LC and 50 MT profiled using gas chromatography‐triple quadrupole mass spectrometry (GC‐QQQ/MS). Moreover, breast cancer urine samples and invasive breast cancer cells were used to support the identified metabolic feature.
2. MATERIALS AND METHODS
2.1. Cells, reagents, and instruments
MCF‐10A, MDA‐MB‐231, MDA‐MB‐468 and MCF‐7 cells were obtained from ATCC (Shanghai, China). The cells were grown in DMEM (Invitrogen, Carlsbad, CA, USA) under 5% CO2. MDA‐MB‐231 and MCF‐7 cells were grown in charcoal‐stripped serum containing media for 24 hours, before treatment with indicated concentrations of 9‐cis‐retinoic acid (9‐cisRA; KGaA Merck, Darmstadt, Germany). Antibodies to cofilin, P‐cofilin, MMP2, MMP9, aldehyde dehydrogenase 1 family member A1 (ALDH1A1) and β‐actin were from Cell Signaling Technology, Inc. (Beverly, MA, USA).
Formic acid and acetonitrile (ACN) were obtained from Merck (KGaA Merck). All standard (l‐phenylalanine, glycerophosphocholine, chenodeoxycholic acid glycine conjugate and lysophosphatidylcholine [lysoPC] [18:0]) preparations were purchased from Sigma‐Aldrich (St Louis, MO, USA).
The ultra‐performance liquid chromatography/mass spectroscopy (UPLC/MS) portion of the platform is based on an Accela UPLC (Thermo Fisher Scientific Corporation, Waltham, MA, USA) and a LTQ‐Orbitrap XL hybrid mass spectrometer (Linear Ion Trap mass spectrometer with Orbitrap; Thermo Fisher Scientific Corporation). The GC/MS portion is based on an Agilent 7890A Series GC and a QQQ mass spectrometer (Triple Quadrupole mass spectrometer; Thermo Fisher Scientific Corporation).
2.2. Sample collection
All samples were collected from Tianjin Medical University Cancer Institute & Hospital with informed consent according to approval of the Institutional Review Board. Detailed clinical characteristics of serum samples used in the profiling phase of this study are provided in Table 1. Analogous characteristics for urine samples used in the validating phase of this study are given in Table S1. All samples were stored at −80°C until use.
Table 1.
Clinical characteristics associated with serum specimens used for metabolomic profiling
| Samples | LC group | MT group | NC group | |
|---|---|---|---|---|
| Sample no. | 46 | 41 | 49 | |
| Age (yr) | 54.1 (33‐64) | 58.3 (37‐66) | 50.5 (30‐59) | |
| Luminal B (HER2 positive and ER positive) | 23 | 11 | — | |
| Basal‐like (HER2 negative and ER negative) | 4 | 21 | — | |
| Luminal A (HER2 negative and ER positive) | 7 | 4 | — | |
| HER2‐enriched (HER2 positive and ER negative) | 12 | 5 | — | |
| TNM stage | IIA | 15 | — | — |
| IIB | 18 | — | — | |
| IIIA | 8 | — | — | |
| IIIB | 5 | — | — | |
| IV | 0 | 41 | — | |
| Metastatic sites | Liver | — | 13 | — |
| Lung | — | 15 | — | |
| Bone | — | 8 | — | |
| Soft tissue | — | 4 | — | |
| Diaphragm | — | 1 | — |
ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; LC, clinically localized breast cancer serum group; MT, metastatic breast cancer serum group; NC, normal control serum group. “—” represents the number is 0.
2.3. Sample preparation
Serum sample (100 μL each) was mixed with 400 μL methanol to precipitate proteins, and then centrifuged (4°C, 15 000 g, 30 minutes). The supernatant was filtered using a 0.22‐μm membrane and detected by LC/MS.
Serum and urine samples were centrifuged at 4°C, 15 000 g for 30 minutes, and the supernatant was extracted with 1 mL degassed isopropanol/acetonitrile/water (3/3/2) at 4°C for 5 minutes. The extracts were subsequently dried down and resuspended in 50% aqueous acetonitrile to remove most of the complex lipids. After dry evaporation, extracts were derivatized and subjected to GC/MS analysis.
As part of the quality control (QC) and system conditioning process, a pooled QC sample was prepared by mixing equal volumes (20 μL) of each sample.
2.4. Liquid chromatography coupled with mass spectrometry analysis
The chromatography system was equipped with a binary solvent delivery manager, and a sample manager. Chromatographic separation was carried out using a gradient of ACN : water (both solvents were modified by the addition of 0.1% formic acid) from 5% to 95% over a 10‐minute period, followed by 95% ACN for 4 minutes. Then, the chromatographic elution gradient was immediately reduced to 5% ACN, which was used to balance the analytical column (Hypersil GOLD C‐18; Thermo Fisher) for the final 4 minutes. The LTQ Orbitrap XL hybrid MS21 was set for continuous monitoring of positive ions, and data were collected over 15 minutes in centroid mode over the mass range 50‐1000 m/z. MS resolution was at 100 000 full‐width half‐maximum (FWHM) and the calibration standards (caffeine, Ultramark 1621) were used to assure chromatographic consistency.
2.5. Gas chromatography coupled with mass spectrometry analysis
Samples were redried under vacuum desiccation for a minimum of 24 hours prior to being derivatized under dried nitrogen using bis(trimethylsilyl)trifluoroacetamide (BSTFA). During the course of the run, temperature was ramped from 80°C to 300°C in a 35‐minute period, followed by 300°C for 8 minutes. Collision gas velocity was 2.25 mL/min for helium and 1.5 mL/min for nitrogen. QQQ mass spectrometry was set for continuous monitoring of positive ions using electron impact ionization and high resolution.
Level of target metabolite was quantified by selected ion monitoring (SIM) using isotope dilution electron‐impact ionization GC/MS, and relative area counts were obtained by manual integration of its chromatogram peaks using Xcalibur software.
2.6. Statistical analysis
MZmine 2.0 and SIMCA (version 14.1; Umetrics, Malmö, Sweden) were used for peak detection and establishing the principal component analysis (PCA) and orthogonal partial least squares discriminant analysis (OPLS‐DA) model.22, 23 Preliminary selection of differential metabolites was accomplished using the corresponding variable importance (VIP) value, coefficient plot and s‐plot. IBM SPSS Statistics for Windows, version 19.0 (IBM Corp., Armonk, NY, USA) was used for data analysis. Two‐tailed Wilcoxon rank‐sum tests were used to compare metabolite expression levels for 2‐sample tests: NC vs LC, LC vs MT. Steel‐Dwass tests were used for multiple comparisons between all groups: NC vs LC vs MT. Two‐tailed t test was used to compare pairwise differences in expression in cells, and ANOVA was used for comparisons involving multiple cells. The threshold for significance was P < .05 for all tests. Association between metabolite expression level and cell invasiveness was assessed by Pearson correlation coefficients. Hierarchical clustering was carried out on the log transformed normalized data using the MeV software package (version 4.9.0).
2.7. Metabolomics pathway analysis
Database sources including the Human Metabolome Database, KEGG and MetaboAnalyst were used for the identification of altered metabolic pathways.
2.8. RNA interference
Following the manufacturer's instructions, transient transfection was carried out by using Lipofectamine 2000 (Invitrogen) with ALDH1A1‐specific siRNA (insert CATGATTCAGTGAGTGGCAAGAAAT) and scramble sequence siRNA from Invitrogen. BLOCK‐iT Fluorescent Oligo was used to examine transfection efficiency (Invitrogen). Seventy‐two hours after transfections, cells were harvested and used for further experiments.
2.9. Reverse transcription‐PCR
Total RNA was extracted using Trizol (Invitrogen, Carlsbad, CA, USA). For the RT‐PCR, the One Step RNA PCR Kit (AMV) was used (TaKaRa, Dalian, China). The PCR procedure was done at 94°C for 1 minute, at 56°C for 1 minute and at 72°C for 1 minute with a total of 35 cycles. Specific primers for GAPDH (forward, 5′‐GCCACTAGGCGCTCACTGTTCT‐3′; reverse, 5′‐TGGGGTCGGGTCAACGCTAGG‐3′), ALDH1A1 (forward, 5′‐ GAAGGAGATAAGGAGGATGTTGA‐3′; reverse, 5′‐ CATTGACTCCATTGTCGCCAG‐3′) were from TaKaRa.
2.10. Western blotting assay
Western blotting assay was carried out as described by Sun et al.24 Briefly, cells were lysed by 1× SDS lysis buffer (Tris‐HCl, pH 6.8, 62.5 mM, 2% SDS, 10% glycerol). Equal amounts of cell lysates (20 μg per lane) were loaded onto 10% SDS‐PAGE systems and transferred onto PVDF membranes (Immobilon‐P; Millipore, Burlington, MA, USA). The membranes were probed with primary antibodies followed by HRP‐conjugated secondary antibody and visualized by using ECL reagents (Pierce, Rockford, IL, USA).
2.11. Proliferation assay
Cells were exposed to different concentrations of 9‐cisRA for 24 and 48 hours in 96‐well plates. Proliferation assay was carried out as described by the manual of CCK‐8 (CK04; Dojindo, Kumamoto, Japan).
2.12. Invasion assay
Cell invasion assay was done as previously described.25 Briefly, the upper surface of a Transwell (8‐μm pore size; Corning, Corning, NY, USA) was coated with 20 μL diluted Matrigel (BD Biosciences, Franklin Lakes, NJ, USA). Equal numbers of the indicated cells were seeded in the upper chamber of the Transwell in serum‐free medium, with FBS added to the lower chamber. After incubation for 48 hours, non‐invading cells were removed with a cotton swab, and the invaded cells were stained with crystal violet. Average invaded cell number per field of view was obtained from 5 random fields.
2.13. Scratch assay
Cells were pretreated with different concentrations of 9‐cisRA (50, 200 μmol/L) for 12 hours; thereafter, a linear scratch was lined out in the middle using a pipette tip. The cells were then incubated at 37°C, and the widths of the wounds were measured at different time points under a light microscope.
3. RESULTS
3.1. Serum metabolite profiling of breast cancer patients
Clinical characteristics of the patients with breast cancer and NC subjects are summarized in Table 1. Total ion chromatograms (TIC) of the LC group, MT group and NC group acquired by the UPLC/MS platform are shown in Figure S1A. Stability of the UPLC/MS system was adequately assessed by the analysis of QC samples during the entire experimental period.26 From the QC principal component score plot (Figure S1B) we found that the UPLC/MS system was stable throughout the analytical process.
A total of 693 metabolites were quantified from the detected spectral features of serum samples, of which 79.8% (553/693) were shared by the 3 groups (Figure 1A). Notably, there were 69 metabolites found in the LC and/or MT group but not in the NC group. These profiles are displayed as a heat map (Figure 1B) and as an NC‐based z‐score plot for each of the 693 metabolites (Figure 1C). The z‐score plots showed robust metabolic alterations in the MT group (z‐score range: −5.63 to 15.32) compared to fewer changes in LC group samples (z‐score range: −3.94 to 8.81).
Figure 1.
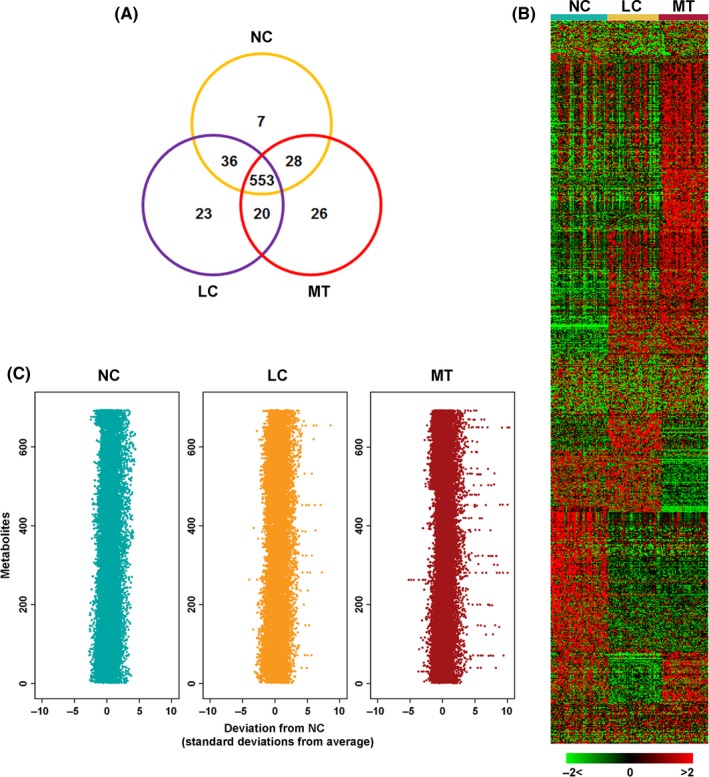
Metabolomic profiling of breast cancer. A, Venn diagram of 693 metabolites measured across 49 NC, 46 LC, and 41 MT serum samples. LC, localized breast cancer serum sample; MT, metastatic breast cancer serum sample; NC, normal control sample. B, Heat map representation of unsupervised hierarchical clustering of (A) (rows) grouped by sample type (columns). Shades of red and green represent elevation and decrease of a metabolite, respectively, relative to the median metabolite levels (see color scale). C, Z‐score plots for (A) normalized to the mean of NC samples (truncated at 10 SD for clarity)
Principal component analysis was carried out to assess the separation tendency between groups based on the 693 annotated metabolites. As shown in Figure 2A, the NC group, LC group and MT group showed a clustering tendency in the direction of the first predictive principal component (X axis), which could assess the progression of breast cancer. Further, an Orthogonal Projections to Latent Structures Discriminant Analysis (OPLS‐DA) model with 2 predictive principal components and 6 orthogonal principal components (R2X(cum) = 0.784, R2Y(cum) = 0.952, Q2(cum) = 0.867) was established using the 3 group samples. As shown in Figure 2B, the score plot of its first 2 principal components showed significant clustering tendency in the X axis direction, which also reflected the progression of breast cancer. The main focus of the present study was to discover the characteristic metabolites that significantly contributed to the clustering tendency and highly related to the progression of breast cancer. Therefore, clear discriminations were obtained by OPLS‐DA between NC and LC groups (Figure 2C), and between LC and MT groups (Figure 2F). Chance permutations at 200 times for the OPLS‐DA models produced R2‐intercept/Q2‐intercept at 0.711/−0.582 (Figure 2D) and at 0.786/−0.623 (Figure 2G), respectively, indicating that no over‐fitting was observed. Performance of the OPLS‐DA models was further tested in another group of 50 clinically localized breast cancer serum samples, 50 metastatic breast cancer serum samples and 50 normal control samples, and all test samples were correctly classified by the OPLS‐DA models.
Figure 2.
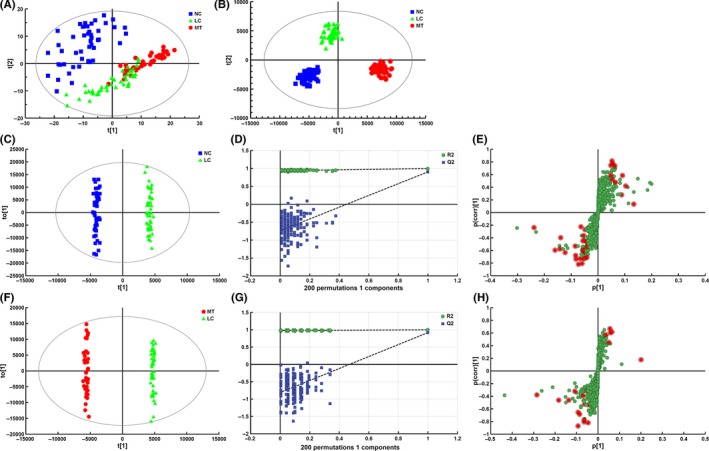
Establishment of metabolic profiles and selection of differential metabolites between NC and LC, and LC and MT. LC, localized breast cancer serum sample; MT, metastatic breast cancer serum sample; NC, normal control sample. A, Score plot of the first 2 components [t(1)/t(2)] of the serum metabolic profiling principal component analysis model. B, Score plot of metabolic profiling orthogonal partial least squares discriminant analysis (OPLS‐DA) model. OPLS‐DA score plots obtained between (C) NC vs LC, and (F) LC vs MT. Each point in the above figures represents a sample. Chance permutation at 200 times was used for the discrimination between (D) NC vs LC, and (G) LC vs MT. S‐plot of selected metabolites between (E) NC vs LC, and (H) LC vs MT (red points represent differential metabolites)
The metabolites with VIP in the project higher than 1 in OPLS‐DA models, and the metabolites with a high degree of variation (high horizontal ordinate value) and reliability (high vertical ordinate value) from the s‐plots (Figure 2E,H) were selected as differential variables between NC and LC groups, and between LC and MT groups. Additionally, the 2‐tailed Wilcoxon rank‐sum test was used to validate the significance of the difference in intensities between variables.
3.2. 9‐cisRA, a metabolic marker of breast cancer progression
A total of 57 metabolites were identified as differential variables between the LC and NC samples (P < .05). Their relative normalized quantities across samples were plotted in a heat map shown in Figure 3A. Among the perturbed metabolites, 26 were elevated in LC whereas 31 were downregulated. Figure 3B shows the relative levels of the 46 named metabolites that were differential between NC and LC groups. Similarly, 13 metabolites were found to be elevated in the MT samples compared to the LC samples whereas 8 metabolites were downregulated (P < .05) (Table S2). Figure 3C shows the levels of the 21 named metabolites altered in MT samples. A subset of 5 metabolites including l‐dihydroorotic acid, lysophosphatidic acid (LPA)(18:1(9Z)/0:0), sphinganine, LysoPC(0:0/18:0) and sphingomyelin (SM)(d18:0/16:1(9Z)) were significantly elevated upon disease progression from NC to LC to MT whereas 9‐cisRA was markedly downregulated (Table 2). These metabolites could potentially serve as biomarkers for breast cancer progression, and 9‐cisRA was the most significant differential metabolite between groups (Figure 4). Moreover, 9‐cisRA was found to be significantly lower in urine supernatants (n = 55, P = 4.354E‐17) derived from biopsy‐positive breast cancer patients as compared to negative individuals (n = 54) (Figure 5A). Overall receiver operator characteristic (ROC) curve for 9‐cisRA suggested its predictive value was modest with the area under the curve (AUC) of 0.71 (95% CI: 0.53, 0.88) for urine supernatants (Figure 5B).
Figure 3.
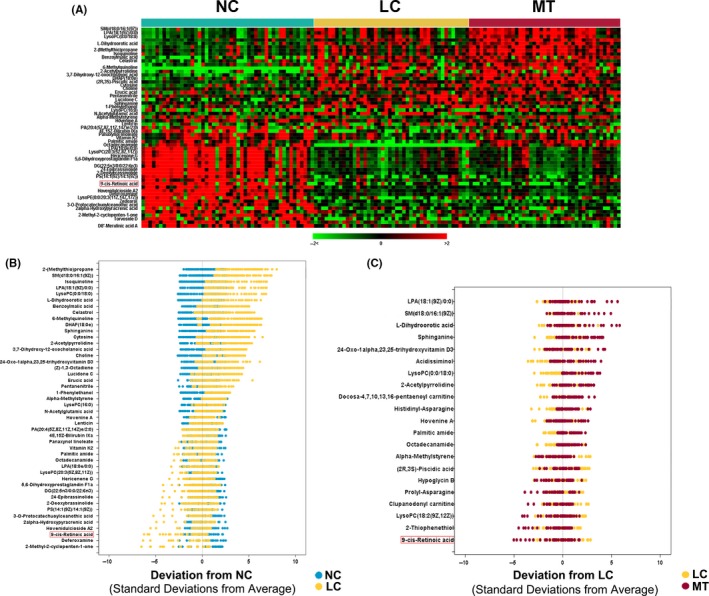
Metabolomic alterations of breast cancer progression. A, Heat map showing 57 differential metabolites in LC relative to NC samples (P < .05). B, NC‐based z‐score plot of named metabolites from (A). Each point represents 1 metabolite in 1 sample, colored by different groups (jade = NC, yellow = LC). C, As in (B) except for the comparison between MT (red) and LC (yellow), with data represented relative to the mean of LC samples. For clarity, the plots in (B) and (C) have been truncated at 10 SD above the mean of NC and LC samples, respectively. LC, localized breast cancer serum sample; MT, metastatic breast cancer serum sample; NC, normal control sample
Table 2.
Summary of potential biomarkers of breast cancer progression by UPLC/MS analysis
| m/z | RT (min) | Metabolitea | Adductb | Related pathway | Contentc | |
|---|---|---|---|---|---|---|
| LC/NC | MT/LC | |||||
| 318.241 | 7.19989 | 9‐cis‐Retinoic acid | M + NH4 [1+] | Retinol metabolism | Down* | Down** |
| 176.066 | 0.89564 | l‐Dihydroorotic acid | M + NH4 [1+] | Pyrimidine metabolism | Up* | Up** |
| 274.092 | 4.90187 | Sphinganine | M + NH4 [1+] | Sphingolipid metabolism | Up* | Up** |
| 546.353 | 8.60841 | LysoPC(0:0/18:0) | M + Na [1+] | Glycerophospholipid metabolism | Up* | Up** |
| 459.248 | 7.43676 | LPA(18:1(9Z)/0:0) | M + Na [1+] | Glycerophospholipid metabolism | Up* | Up** |
| 487.280 | 8.81888 | SM(d18:0/16:1(9Z)) | M + H [1+] | Sphingolipid metabolism | Up* | Up** |
LC, localized breast cancer serum sample; LPA, lysophosphatidic acid; lysoPC, lysophosphatidylcholine; MT, metastatic breast cancer serum sample; m/z, mass‐to‐charge ratio; NC, normal control sample; RT, retention time; SM, sphingomyelin; UPLC/MS, ultra‐performance liquid chromatography/mass spectroscopy.
Metabolites formally identified by standard comparison.
Ionospheric models of mass spectrometry cationic scanning.
Comparison of characteristic metabolites’ integral peak area in the 3 groups.
Non‐parametric test was used for comparisons among the groups (*P < .05, **P < .01).
Figure 4.
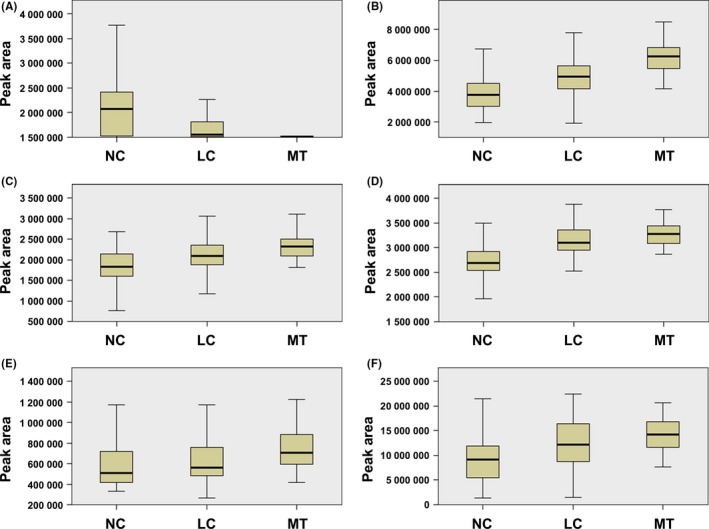
Boxplots of the differential metabolites associated with breast cancer progression. A, Boxplot showing progressive reduction of 9‐cis‐retinoic acid during progression from NC to LC to MT (NC vs LC: P = 2.349E‐5; LC vs MT: P = .00363; NC vs MT: P = 3.109E‐11). B, Boxplot showing progressive elevation of l‐dihydroorotic acid during progression from NC to LC to MT (NC vs LC: P = 2.793E‐5; LC vs MT: P = 1.571E‐6; NC vs MT: P = 1.767E‐11). C, Same as (B) but for sphingomyelin (SM)(d18:0/16:1(9Z)) (NC vs LC: P = .00488; LC vs MT: P = .01190; NC vs MT: P = 5.084E‐7). D, Same as (B) but for lysophosphatidic acid (LPA)(18:1(9Z)/0:0) (NC vs LC: P = 5.212E‐6; LC vs MT: P = 3.442E‐6; NC vs MT: P = 6.802E‐13). E, same as (B) but for sphinganine (NC vs LC: P = .04155; LC vs MT: P = .01973; NC vs MT: P = .00016). F, Same as (B) but for lysophosphatidylcholine (lysoPC)(0:0/18:0) (NC vs LC: P = .00625; LC vs MT: P = .03518; NC vs MT: P = 2.502E‐7). LC, localized breast cancer serum sample; MT, metastatic breast cancer serum sample; NC, normal control sample
Figure 5.
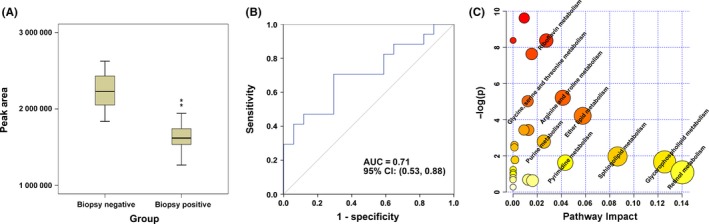
Lowered level of 9‐cis‐retinoic acid (9‐cisRA) is a metabolic feature of human breast cancer, and the disturbed metabolic pathways in breast cancer patients compared with healthy controls. A, Boxplot showing significantly lower level of 9‐cisRA in 55 biopsy‐positive breast cancer urine samples compared to 54 biopsy‐negative controls (**P = 4.354E‐17). B, Receiver operator characteristic curve for 9‐cisRA in the 109 urine supernatants. 9‐cisRA has an area under the curve (AUC) of 0.71 (95% CI: 0.53, 0.88). C, MetaboAnalyst‐generated topology map describes the impact of differential metabolites identified between breast cancer patients vs healthy controls on metabolic pathways
Using MetaboAnalyst 3.0 (http://www.metaboanalyst.ca/MetaboAnalyst/), the key metabolic pathways dysregulated in breast cancer patients were identified as retinol metabolism, glycerophospholipid metabolism, sphingolipid metabolism and pyrimidine metabolism. Among these, the retinol metabolic pathway had the most differential metabolite 9‐cisRA and showed a profound change (Figure 5C).
3.3. 9‐cis‐Retinoic acid levels correlated with breast cancer cell invasion
To determine whether 9‐cisRA decrease in breast cancer has biological relevance, we measured its endogenous levels in breast cancer cell lines MDA‐MB‐231, MDA‐MB‐468 and MCF‐7 (n = 3 each), and benign MCF‐10A breast cells (n = 3). Significantly reduced levels of the metabolite were found in breast cancer cells compared to benign MCF‐10A cells (ANOVA, P = 2.571E‐6). Additionally, 9‐cisRA levels negatively correlated with cell invasiveness (Pearson correlation = −.988, P = .0124) (Figure 6A).
Figure 6.
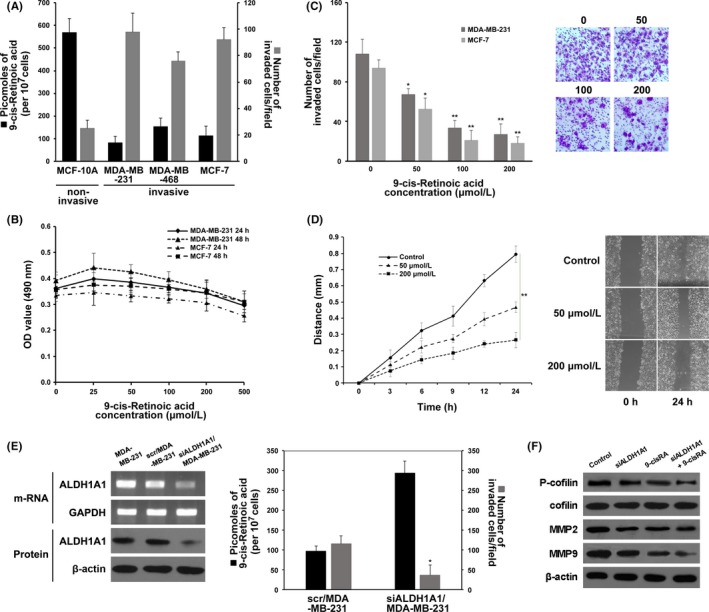
9‐cis‐Retinoic acid (9‐cisRA) levels in breast cancer cells and its association with invasion and migration. A, Decreased levels of 9‐cisRA (black bars) were found in invasive breast cancer cells compared to non‐invasive benign MCF‐10A cells. Mean ± SEM of 9‐cisRA levels (n = 3). Cell invasion (grey bars) was measured. B, Assessment of the effects of 9‐cisRA on MDA‐MB‐231 and MCF‐7 cell proliferation. C, Assessment of MDA‐MB‐231 and MCF‐7 cell invasiveness upon giving exogenous 9‐cisRA. Photographs were taken at a magnification of 200× (eg, MDA‐MB‐231). D, Assessment of MDA‐MB‐231 cell migration upon giving exogenous 9‐cisRA. E, RT‐PCR and western blotting analysis of aldehyde dehydrogenase 1 family member A1 (ALDH1A1) in MDA‐MB‐231 (left panel) and assessment of 9‐cisRA levels and cell invasiveness after knockdown of ALDH1A1 (right panel). F, Western blotting analysis of P‐cofilin, MMP2 and MMP9 from the 4 treatment groups. Each result is a representative of at least 3 independent experiments. *P < .05; **P < .01
Cell proliferation assay was then carried out to test the cytotoxic effect of exogenous 9‐cisRA against the MDA‐MB‐231 and MCF‐7 cells. As shown in Figure 6B, gradient concentrations of 9‐cisRA (25, 50, 100, and 200 μmol/L) had no significant effects on cell proliferation after 24‐ and 48‐hour treatments until the concentration of 9‐cisRA was increased to 500 μmol/L where cell proliferation was obviously inhibited.
To investigate the inhibitory effect of 9‐cisRA (50, 100, and 200 μmol/L) on the invasion and migration of cells, we carried out Matrigel invasion and scratch assays. As shown in Figure 6C, 9‐cisRA evidently prevented MDA‐MB‐231 and MCF‐7 cells from invading the Matrigel‐coated membrane in a dose‐dependent method. In the scratch assay, we found that the migration of MDA‐MB‐231 cells was also significantly reduced in a dose‐dependent method after treatment of 9‐cisRA (Figure 6D). These results suggested that 9‐cisRA was effective in preventing invasion and migration of breast cancer cells.
Furthermore, 9‐cisRA levels are downregulated by ALDH1A1, the enzyme that can convert 9‐cisRA to 9‐cis‐retinal. Knockdown of ALDH1A1 in MDA‐MB‐231 cells resulted in approximately a 3‐fold increase in intracellular 9‐cisRA levels with a significant reduction in cell invasion (t test P = .013, n = 3) compared to control cells (Figure 6E).
To explore the synergistic effect of siALDH1A1 and 9‐cisRA treatments and identify mechanisms potentially responsible for altered migratory and invasive ability of MDA‐MB‐231 cells, western blotting was carried out to quantify the expressions of various related proteins. Cofilin, an actin‐binding protein which disassembles actin filaments, is one of the major regulators of cell motility and invasion.27, 28 As shown in Figure 6F, phosphorylation of cofilin was obviously reduced in the combined siALDH1A1 + 9‐cisRA (200 μmol/L) group compared to the control, siALDH1A1 and 9‐cisRA groups. As the expression of MMP is crucial to ECM degradation and cell invasion, it is vital to determine whether MMP are involved in the inhibition of migration and invasion by siALDH1A1 and 9‐cisRA treatments. As shown in Figure 6F, MMP2 expression was moderately reduced in the siALDH1A1, 9‐cisRA and combined siALDH1A1 + 9‐cisRA groups in comparison to the control group. Also, MMP9 expression was significantly inhibited by combined siALDH1A1 + 9‐cisRA treatment compared to all other groups. Taken together, our data suggest that siALDH1A1 and 9‐cisRA coordinately inhibited breast cancer cell invasion by attenuating the phosphorylation of cofilin and the expressions of MMP2 and MMP9.
4. DISCUSSION
Metabolomics provides a direct and sensitive measurement of phenotypic changes because it reflects the downstream molecular event of the combination of all upstream genetic, transcriptomic and proteomic alterations.29 Several studies in breast cancer have investigated the metabolic profiling changes associated with cancer subtypes,14, 30 chemotherapy intervention,31 surgical treatment32 etc., whereas the metabolite profile changes and altered metabolic pathways of metastatic breast cancer have not yet been elucidated in detail. Serum metabolomic approaches, which effectively detect concurrent signals from the host and tumor, have the advantages of economic efficiency, rapidity and minimal invasiveness and may play an important role in early cancer detection32 as well as in prediction of cancer progression and metastasis.
We used combined LC/MS and GC/MS to profile serum and urine metabolites during breast cancer progression, indicating a higher degree of metabolomic alteration in serum samples from the MT group (breast cancer patients with different metastatic sites) relative to NC and LC group samples and explaining the biological variations associated with breast cancer metastasis. The current study delineated that a subset of 5 metabolites elevated upon disease progression from NC to LC to MT, whereas 9‐cisRA showed a progressive decrease from NC to LC to MT. The present data suggested that the major altered pathways in breast cancer patients compared with healthy controls included decreased retinol metabolism, glycerophospholipid metabolism and pyrimidine metabolism, and enhanced sphingolipid metabolism. As an intermediate metabolite, 9‐cisRA is involved in retinol metabolism which motivated us to examine 9‐cisRA in greater detail and further explore its possible utility in monitoring disease progression and aggressiveness.
In addition, our analysis also showed a lowered 9‐cisRA level in urine samples from biopsy‐positive breast cancer patients compared to biopsy‐negative individuals and in breast cancer cells (MDA‐MB‐231, MDA‐MB‐468 and MCF‐7) compared to MCF‐10A cells. ROC analysis using the quantitative data of 9‐cisRA in urine samples showed that 9‐cisRA (AUC = 0.71; 95% CI: 0.53, 0.88) might have the potential to distinguish breast cancer patients from healthy controls. Despite the fact that the heterogeneity within breast cancer as a result of varying gradients of metabolites and growth factors seems to complicate the metabolite profile of breast cancer, our results indicated that 9‐cisRA offered high accuracy in a heterogeneous validation set of breast cancer patients, and its potential utility will be further validated in a larger patient population.
9‐cis‐Retinoic acid, an isomer of all‐trans retinoic acid, is produced from 9‐cis‐retinal by an oxidation process. It participates in retinol metabolism and can also be converted to 9‐cis‐retinal by specific regulatory enzymes. 9‐cisRA which binds to both retinoic acid receptors (RAR) and nuclear retinoid X receptors (RXR) prevented prostate carcinogenesis in rats, reduced growth and induced apoptosis of human prostate cancer cells in a dose‐dependent method.33 9‐cisRA induced the downregulation of P‐glycoprotein, the overexpression of which develops MDR phenotype in L1210 lymphocytic leukemia cells.34 It also directly retarded the cell cycle in a concentration‐ and time‐dependent method in NCI‐H295R adrenocortical cancer cells as well as reduced tumor growth in the in vivo xenograft model.35 A number of previous studies have shown that 9‐cisRA has an anti‐tumor effect against cell proliferation but not invasion, and might represent a promising therapeutic agent for many types of cancers including mammary carcinoma,36 gastric carcinoma, pancreatic carcinoma and lung carcinoma. In the present study, we have shown that 9‐cisRA strikingly inhibited the migratory and invasive abilities of MDA‐MB‐231 and MCF‐7 cells at non‐cytotoxic concentrations in vitro. These results indicated that 9‐cisRA impaired breast cancer cell migration and invasion, and the effect was not attributed to its cytotoxicity. It was consistent with the results obtained by Flodrova et al37 that 9‐cisRA markedly inhibited the migration of MCF‐7 human breast cancer cells. ALDH1A1, an enzyme involved in retinol metabolism, could metabolize 9‐cisRA, and previous results have observed that ALDH1A1 significantly associates with tumor growth,38, 39 lymph node metastasis, and clinical stage of breast cancer patients,40 and it has been proposed as a potential target for anti‐breast cancer therapy. Our study found that knockdown of ALDH1A1 in MDA‐MB‐231 cells increased intracellular 9‐cisRA levels while notably suppressing the invasive abilities of breast cancer cells.
Furthermore, our data indicated that the synergistic effect of siALDH1A1 and 9‐cisRA against cell migration was a result of its blocking phosphorylation of the key regulator cofilin, the activity of which is pivotal for actin‐polymerization and cell migration.41 Moreover, we also found that expressions of MMP2 and MMP9, which are 2 important members of the MMP family and can degrade the ECM at tumor‐invasive fronts to overcome the ECM barrier and subsequently facilitate cancer metastasis,42, 43 were significantly inhibited attributable to the anti‐invasive effects of siALDH1A1 and 9‐cisRA treatments. Therefore, the results presented here have established at least part of the mechanism of siALDH1A1 and 9‐cisRA in preventing migration and invasion of breast cancer cells.
Taken together, we explored the metabolomic signatures during breast cancer progression. Specifically, we identified 9‐cisRA as a key metabolite that decreased most robustly in metastatic breast cancer and was detectable in the urine of patients with metastatic disease. Interestingly, 9‐cisRA, and its proximal regulatory enzyme, appear to play an inhibitory role in neoplastic progression attenuating cell invasion and migration, presumably through preventing the key regulator cofilin from activation and inhibiting the expressions of MMP2 and MMP9. Thus, 9‐cisRA and its proximal regulatory enzyme may have potential as biomarkers of breast cancer progression as well as exerting an inhibitory role in breast cancer progression. In the future, we will undertake a study of a larger prospective cohort to further validate the accuracy of our research. An evaluation of 9‐cis‐RA levels and the mechanisms of breast cancer metastasis by tissue‐targeted metabolomics as well as other systematic biological approaches could be used.
CONFLICTS OF INTEREST
Authors declare no conflicts of interest for this article.
Supporting information
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (No. 81301985), the Natural Science Foundation of Tianjin (No. 14JCQNJC14000), and the Tianjin Third Central Hospital National Natural Science Foundation Incubation Project of China (No. 2017YNR3 and No. 2017YNY2, to Jing Wu and Bingbing Liu). We also thank the participants in this study.
Wu J, Yang R, Zhang L, et al. Metabolomics research on potential role for 9‐cis‐retinoic acid in breast cancer progression. Cancer Sci. 2018;109:2315–2326. https://doi.org/10.1111/cas.13629
Jing Wu and Rui Yang contributed equally to this work.
Contributor Information
Jing Wu, Email: wujing_821013@sina.com.
ShuYe Liu, Email: lshye@163.com.
Tong Li, Email: litong3zx@sina.com.
REFERENCES
- 1. Guo C, Li X, Ye M, et al. Discriminating patients with early‐stage breast cancer from benign lesions by detection of oxidative DNA damage biomarker in urine. Oncotarget. 2017;8:53100‐53109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115‐132. [DOI] [PubMed] [Google Scholar]
- 3. Tong ZJ, Shi NY, Zhang ZJ, Yuan XD, Hong XM. Expression and prognostic value of HER‐2/neu in primary breast cancer with sentinel lymph node metastasis. Biosci Rep. 2017;37:BSR20170121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bathen TF, Geurts B, Sitter B, et al. Feasibility of MR metabolomics for immediate analysis of resection margins during breast cancer surgery. PLoS ONE. 2013;8:e61578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang Q, Xu R. MetabolitePredict: a de novo human metabolomics prediction system and its applications in rheumatoid arthritis. J Biomed Inform. 2017;71:222‐228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lee SK, Kim BK, Jung HY, et al. Metabolomics study for identification of potential biomarkers of long‐term survival in kidney transplantation recipients. Transplant Proc. 2017;49:1005‐1011. [DOI] [PubMed] [Google Scholar]
- 7. Sarver DC, Sugg KB, Disser NP, Enselman ERS, Awan TM, Mendias CL. Local cryotherapy minimally impacts the metabolome and transcriptome of human skeletal muscle. Sci Rep. 2017;7:2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang D, Zhu W, Wang Y, et al. Metabolomics approach to investigate estrogen receptor‐dependent and independent effects of o, p’‐DDT in the uterus and brain of immature mice. J Agric Food Chem. 2017;65:3609‐3616. [DOI] [PubMed] [Google Scholar]
- 9. Xie G, Zhou B, Zhao A, et al. Lowered circulating aspartate is a metabolic feature of human breast cancer. Oncotarget. 2015;6:33369‐33381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Haukaas TH, Euceda LR, Giskeødegård GF, et al. Metabolic clusters of breast cancer in relation to gene‐ and protein expression subtypes. Cancer Metab. 2016;4:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Borrego SL, Fahrmann J, Datta R, et al. Metabolic changes associated with methionine stress sensitivity in MDA‐MB‐468 breast cancer cells. Cancer Metab. 2016;4:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim KJ, Kim HJ, Park HG, et al. A MALDI‐MS‐based quantitative analytical method for endogenous estrone in human breast cancer cells. Sci Rep. 2016;6:24489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Budczies J, Pfitzner BM, Györffy B, et al. Glutamate enrichment as new diagnostic opportunity in breast cancer. Int J Cancer. 2015;136:1619‐1628. [DOI] [PubMed] [Google Scholar]
- 14. Fan Y, Zhou X, Xia TS, et al. Human plasma metabolomics for identifying differential metabolites and predicting molecular subtypes of breast cancer. Oncotarget. 2016;7:9925‐9938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Poschke I, Mao Y, Kiessling R, de Boniface J. Tumor‐dependent increase of serum amino acid levels in breast cancer patients has diagnostic potential and correlates with molecular tumor subtypes. J Transl Med. 2013;11:290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Proenza AM, Oliver J, Palou A, Roca P. Breast and lung cancer are associated with a decrease in blood cell amino acid content. J Nutr Biochem. 2003;14:133‐138. [DOI] [PubMed] [Google Scholar]
- 17. Yang X, Feng L, Zhao L, Liu X, Hassani D, Huang D. Effect of glycine nitrogen on lettuce growth under soilless culture: a metabolomics approach to identify the main changes occurred in plant primary and secondary metabolism. J Sci Food Agric. 2018;98:467‐477. [DOI] [PubMed] [Google Scholar]
- 18. Zhang P, Zhu W, Wang D, et al. A combined NMR‐ and HPLC‐MS/MS‐based metabolomics to evaluate the metabolic perturbations and subacute toxic effects of endosulfan on mice. Environ Sci Pollut Res Int. 2017;24:18870‐18880. [DOI] [PubMed] [Google Scholar]
- 19. Huang Q, Tan Y, Yin P, et al. Metabolic characterization of hepatocellular carcinoma using nontargeted tissue metabolomics. Cancer Res. 2013;73:4992‐5002. [DOI] [PubMed] [Google Scholar]
- 20. Chen T, Xie G, Wang X, et al. Serum and urine metabolite profiling reveals potential biomarkers of human hepatocellular carcinoma. Mol Cell Proteomics. 2011;10(M110):004945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Makarov A, Scigelova M. Coupling liquid chromatography to Orbitrap mass spectrometry. J Chromatogr A. 2010;1217:3938‐3945. [DOI] [PubMed] [Google Scholar]
- 22. Trygg J, Holmes E, Lundstedt T. Chemometrics in metabonomics. J Proteome Res. 2007;6:469‐479. [DOI] [PubMed] [Google Scholar]
- 23. Eriksson I, Johansson E, Kettaneh‐Wold N, Wold S. Multi‐ and Megavariate Data Analysis: Principles and Applications. Umeå, Sweden: Umetrics Academy; 2001. [Google Scholar]
- 24. Sun R, Gao P, Chen L, et al. Protein kinase C zeta is required for epidermal growth factor‐induced chemotaxis of human breast cancer cells. Cancer Res. 2005;65:1433‐1441. [DOI] [PubMed] [Google Scholar]
- 25. Wu J, Liu S, Fan Z, Zhang L, Tian Y, Yang R. A novel and selective inhibitor of PKC ζ potently inhibits human breast cancer metastasis in vitro and in mice. Tumour Biol. 2016;37:8391‐8401. [DOI] [PubMed] [Google Scholar]
- 26. Li Y, Jin Y, Yang S, et al. Strategy for comparative untargeted metabolomics reveals honey markers of different floral and geographic origins using ultrahigh‐performance liquid chromatography‐hybrid quadrupole‐orbitrap mass spectrometry. J Chromatogr A. 2017;1499:78‐89. [DOI] [PubMed] [Google Scholar]
- 27. Wang JT, Song LZ, Li LL, et al. Src controls neuronal migration by regulating the activity of FAK and cofilin. Neuroscience. 2015;292:90‐100. [DOI] [PubMed] [Google Scholar]
- 28. Wang Y, Kuramitsu Y, Kitagawa T, et al. Cofilin‐phosphatase slingshot‐1L (SSH1L) is overexpressed in pancreatic cancer (PC) and contributes to tumor cell migration. Cancer Lett. 2015;360:171‐176. [DOI] [PubMed] [Google Scholar]
- 29. Denkert C, Bucher E, Hilvo M, et al. Metabolomics of human breast cancer: new approaches for tumor typing and biomarker discovery. Genome Med. 2012;4:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cao MD, Lamichhane S, Lundgren S, et al. Metabolic characterization of triple negative breast cancer. BMC Cancer. 2014;14:941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Miller JA, Pappan K, Thompson PA, et al. Plasma metabolomic profiles of breast cancer patients after short‐term limonene intervention. Cancer Prev Res (Phila). 2015;8:86‐93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tenori L, Oakman C, Morris PG, et al. Serum metabolomic profiles evaluated after surgery may identify patients with oestrogen receptor negative early breast cancer at increased risk of disease recurrence. Results from a retrospective study. Mol Oncol. 2015;9:128‐139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Eskra JN, Kuiper JW, Walden PD, Bosland MC, Özten N. Interactive effects of 9‐cis‐retinoic acid and androgen on proliferation, differentiation, and apoptosis of LNCaP prostate cancer cells. Eur J Cancer Prev. 2017;26:71‐77. [DOI] [PubMed] [Google Scholar]
- 34. Breier A, Stetka J, Bohacova V, Macejova D, Brtko J, Sulova Z. Effect of 9‐cis retinoic acid and all‐trans retinoic acid in combination with verapamil on P‐glycoprotein expression in L1210 cells. Neoplasma. 2014;61:553‐565. [DOI] [PubMed] [Google Scholar]
- 35. Szabó DR, Baghy K, Szabó PM, et al. Antitumoral effects of 9‐cis retinoic acid in adrenocortical cancer. Cell Mol Life Sci. 2014;71:917‐932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Maeng S, Kim GJ, Choi EJ, Yang HO, Lee DS, Sohn YC. 9‐Cis‐retinoic acid induces growth inhibition in retinoid‐sensitive breast cancer and sea urchin embryonic cells via retinoid X receptor α and replication factor C3. Mol Endocrinol. 2012;26:1821‐1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Flodrova D, Benkovska D, Macejova D, et al. Proteomic analysis of changes in the protein composition of MCF‐7 human breast cancer cells induced by all‐trans retinoic acid, 9‐cis retinoic acid, and their combination. Toxicol Lett. 2015;232:226‐232. [DOI] [PubMed] [Google Scholar]
- 38. Wang J, Wang L, Ho CT, Zhang K, Liu Q, Zhao H. Garcinol from Garcinia indica downregulates cancer Stem‐like cell biomarker ALDH1A1 in nonsmall cell lung cancer A549 cells through DDIT3 activation. J Agric Food Chem. 2017;65:3675‐3683. [DOI] [PubMed] [Google Scholar]
- 39. Wang B, Chen X, Wang Z, et al. Aldehyde dehydrogenase 1A1 increases NADH levels and promotes tumor growth via glutathione/dihydrolipoic acid‐dependent NAD+ reduction. Oncotarget. 2017;8:67043‐67055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhou Y, Wang Y, Ju X, et al. Clinicopathological significance of ALDH1A1 in lung, colorectal, and breast cancers: a meta‐analysis. Biomark Med. 2015;9:777‐790. [DOI] [PubMed] [Google Scholar]
- 41. Huang TY, DerMardirossian C, Bokoch GM. Cofilin phosphatases and regulation of actin dynamics. Curr Opin Cell Biol. 2006;18:26‐31. [DOI] [PubMed] [Google Scholar]
- 42. Zhou R, Xu L, Ye M, Liao M, Du H, Chen H. Formononetin inhibits migration and invasion of MDA‐MB‐231 and 4T1 breast cancer cells by suppressing MMP‐2 and MMP‐9 through PI3K/AKT signaling pathways. Horm Metab Res. 2014;46:753‐760. [DOI] [PubMed] [Google Scholar]
- 43. Sun XF, Shao YB, Liu MG, et al. High‐concentration glucose enhances invasion in invasive ductal breast carcinoma by promoting Glut1/MMP2/MMP9 axis expression. Oncol Lett. 2017;13:2989‐2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


