SUMMARY
Type II cadherins are cell-cell adhesion proteins critical for tissue patterning and neuronal targeting but whose molecular binding code remains poorly understood. Here, we delineate binding preferences for type II cadherin cell-adhesive regions, revealing extensive heterophilic interactions between specific pairs, in addition to homophilic interactions. Three distinct specificity groups emerge from our analysis with members that share highly similar heterophilic binding patterns and favor binding to one another. Structures of adhesive fragments from each specificity group confirm near-identical dimer topology conserved throughout the family, allowing interface residues whose conservation corresponds to specificity preferences to be identified. We show that targeted mutation of these residues converts binding preferences between specificity groups in biophysical and co-culture assays. Our results provide a detailed understanding of the type II cadherin interaction map and a basis for defining their role in tissue patterning and for the emerging importance of their heterophilic interactions in neural connectivity.
In Brief
Type II cadherins are a family of vertebrate cell adhesion proteins expressed primarily in the CNS. Brasch et al. measure binding between adhesive fragments, revealing homophilic and extensive selective heterophilic binding with specificities that define groups of similar cadherins. Structures reveal common adhesive dimers, with residues governing cell-adhesive specificity.
INTRODUCTION
Vertebrate classical cadherins are a family of calcium-dependent cell adhesion receptors whose selective interactions are critical for morphogenesis, patterning, and maintenance of solid tissues including the CNS, in which they contribute to neural circuit assembly, axon guidance, and synapse formation and plasticity (Basu et al., 2017; Hirano and Takeichi, 2012; Redies et al., 2012; Williams et al., 2011). All are single-pass transmembrane proteins with extracellular regions composed of five successive extracellular cadherin (EC) repeats and intracellular regions containing binding sites for the adaptor proteins α-catenin, β-catenin, and p120 catenin, which link adhesion mediated by the extracellular regions to the actin cytoskeleton (Brasch et al., 2012; Hirano and Takeichi, 2012). Classical cadherins can be divided into type I cadherins, comprising E-, N-, P-, R-, and M-cadherin, and type II cadherins, which comprise a separate subfamily of thirteen members: cadherin-6 to cadherin-12, cadherin-18 to cadherin-20, cadherin-22, cadherin-24, and a divergent member, vascular endothelial (VE)-cadherin (Brasch et al., 2011). While the molecular interactions of type I cadherins have been well characterized, the larger type II cadherin subfamily is comparatively less understood.
Individual type II cadherins are differentially expressed in the CNS (Hirano and Takeichi, 2012), often with expression of distinct subsets demarcating specific subregions, as observed in the visual system (Duan et al., 2014), hippocampus (Basu et al., 2017; Bekirov et al., 2002), and spinal cord (Demireva et al., 2011; Patel et al., 2006; Price et al., 2002). In functional studies, single and double type II cadherin knockout mice show a variety of distinct non-lethal phenotypes relating to cell targeting and synaptic function in the CNS and to morphogenesis in other tissues. These phenotypes include failure of a subset of retinal ganglion cells to innervate their target neurons (Cdh6−/− mice) (Osterhout et al., 2011), reduction of high-magnitude long-term potentiation (LTP) in the hippocampus (Cdh9−/−, Cdh10−/−, Cdh6−/−, and Cdh10−/−) (Basu et al., 2017), impaired targeting of bipolar cells in the retina (Cdh8−/− and Cdh9−/−) (Duan et al., 2014), and impaired synaptic coupling in cold-sensitive sensory neurons (Cdh8−/−) (Suzuki et al., 2007), and, outside the CNS, delayed kidney development (Cdh6−/−) (Mah et al., 2000) and reduction of bone density (Cdh11−/−) (Kawaguchi et al., 2001). In addition, in vivo misexpression studies demonstrate that expression of specific complements of type II cadherins in individual neurons directs their sorting into segregated populations in the developing chicken spinal cord and mouse telencephalon (Inoue et al., 2001; Patel et al., 2006; Price et al., 2002).
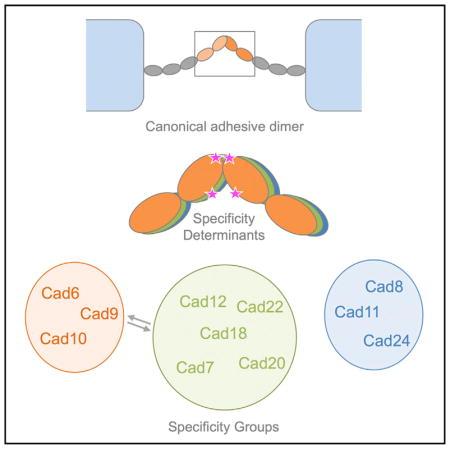
The molecular interactions of type II cadherins underlying these complex behaviors are not yet fully defined. Structural studies of cadherin-8, cadherin-11, and cadherin-20 and the divergent member VE-cadherin have revealed that type II cadherins form strand-swapped adhesive dimers between their membrane-distal EC1 domains, in which N-terminal β strands are reciprocally exchanged (Brasch et al., 2011; Patel et al., 2006). This strand exchange is anchored by docking of two conserved tryptophan residues, Trp2 and Trp4, into a hydrophobic pocket in the partner EC domain, with additional interactions contributed by a hydrophobic patch at the base of the domain (Patel et al., 2006), except in the case of VE-cadherin, which lacks these additional hydrophobic interactions (Brasch et al., 2011). Individual type II cadherins share this canonical interface but show selectivity in their binding interactions. In in vitro cell aggregation assays, type II cadherins mediate both homophilic adhesive interactions between cells expressing identical cadherins and selective heterophilic interactions between cells expressing different cadherins (Nakagawa and Takeichi, 1995; Patel et al., 2006; Shimoyama et al., 1999, 2000). Type II cadherins frequently show partially overlapping, though distinct, expression patterns in vivo, and there is evidence that both types of interaction contribute to their roles in cell sorting and targeting (Basu et al., 2017; Duan et al., 2014; Inoue et al., 2001; Osterhout et al., 2011; Patel et al., 2006; Price et al., 2002; Williams et al., 2011). Biological roles for type II cadherin heterophilic interactions have emerged from in vivo studies of cadherin-8 and cadherin-9 in the mouse retina (Duan et al., 2014) and cadherin-6, cadherin-9, and cadherin-10 in the mouse hippocampus (Basu et al., 2017). However, the precise molecular binding preferences underlying type II cadherin function remain to be fully determined.
Here, we use a comprehensive biophysical approach to quantitatively analyze homophilic and heterophilic binding interactions of type II classical cadherins. We find that heterophilic interactions among different cadherins are highly selective and are frequently preferred over homophilic interactions. Three distinct specificity groups emerge from our analysis, within which closely related cadherins preferentially interact and exhibit highly similar overall patterns of heterophilic binding preferences unique to each group. Based on these observations, we examine structural and sequence conservation of the adhesive interface between specificity groups, determine crystal structures of adhesive regions of cadherins from previously unrepresented groups, and identify critical specificity residues that can convert binding preferences between groups in biophysical and cell culture experiments.
RESULTS
Homophilic Binding Affinities of Type II Cadherin Adhesive Fragments
We used a bacterial expression system to produce soluble EC1-EC2 adhesive fragments of mouse classical type II cadherin-6, cadherin-7, cadherin-9, cadherin-10, cadherin-11, cadherin-12, cadherin-18, cadherin-20, and cadherin-22 and EC1–EC3 fragments of cadherin-8 and cadherin-24, because the latter were unstable as shorter fragments. These represent all members of the classical type II cadherin family except for cadherin-19, for which a stable protein could not be produced, and the divergent member VE-cadherin, whose characterization we reported previously (Brasch et al., 2011). Table 1 lists homophilic binding affinities determined by sedimentation equilibrium analytical ultracentrifugation (AUC) analysis of the eleven type II cadherins produced here. All formed homodimers in solution, and fitting of the data to a monomer-dimer equilibrium model yielded dissociation constant (KD) values in the low micromolar range, from 3.1 to 42.2 μM. Within this range, five cadherins show relatively tight binding affinities with KD values below 10 μM, three cadherins have intermediate affinities in the 10–30 μM range, and three have weak affinities above 30 μM. Cadherins with similar homodimerization strengths do not share the highest sequence identity. For example, cadherin-6 and cadherin-10 share the highest amino acid sequence identity (84% over EC1-EC2) but lie at each extreme of the KD range, while cadherin-6 and cadherin-22 share only 61% sequence identity yet have nearly identical homophilic affinities.
Table 1.
KD for Homodimerization of Type II Cadherin Wild-Type, Chimera, and Mutant Protein Fragments Determined by Analytical Ultracentrifugation. See also Table S2.
| Cadherin | KD (mM) | Description |
|---|---|---|
| Cadherin-6 | 3.1 ± 0.1a,b | wild-type |
| Cadherin-9 | 17.0 ± 1.1c | wild-type |
| Cadherin-10 | 42.2 ± 2.7c | wild-type |
| Cadherin-8 | 15.0 ± 0.4c | wild-type |
| Cadherin-11 | 33.8 ± 0.2c | wild-type |
| Cadherin-24 | 8.2 ± 0.3 | wild-type |
| Cadherin-7 | 32.2 ± 0.8 | wild-type |
| Cadherin-12 | 8.3 ± 1.6 | wild-type |
| Cadherin-18 | 16.8 ± 0.2 | wild-type |
| Cadherin-20 | 9.3 ± 0.6 | wild-type |
| Cadherin-22 | 3.9 ± 0.2 | wild-type |
| cad-6EC111EC2 | 5.6 ± 0.2 | chimera cad-6EC111EC2 |
| cad-11EC16EC2 | 15.6 ± 0.9 | chimera cad-11EC16EC2 |
| Cadherin-6 Y20L H97Q (LQ) | 9.63 ± 1.3 | specificity mutant |
| Cadherin-6 M3V Y20L H97Q E89P (VLQP) | 6.73 ± 0.8 | specificity mutant |
| Cadherin-11 L20Y Q97H (YH) | 11.1 ± 2.1 | specificity mutant |
| Cadherin-11 V3M L20Y Q97H P89E (MYHE) | 8.2 ± 0.8 | specificity mutant |
Errors are the SD from two or more experiments.
Previously reported in Harrison et al. (2010).
Previously reported in Brasch et al. (2011).
Heterophilic Interactions Identify Distinct Type II Cadherin Specificity Groups
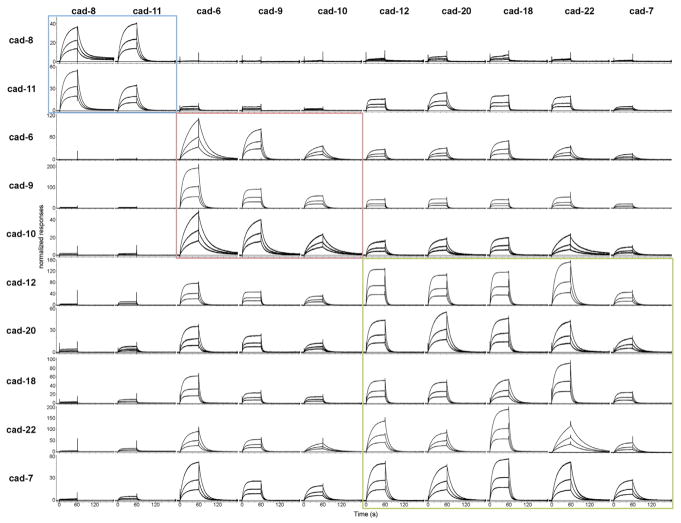
Type II cadherins have been shown to exhibit both homophilic and heterophilic binding behavior in cell aggregation studies (Nakagawa and Takeichi, 1995; Patel et al., 2006; Shimoyama et al., 1999, 2000) and in vivo (Basu et al., 2017; Duan et al., 2014; Inoue et al., 2001; Osterhout et al., 2011; Patel et al., 2006; Price et al., 2002; Williams et al., 2011). However, a comprehensive quantitative analysis of all heterophilic binding interactions in the family has not been reported. We therefore aimed to delineate the heterophilic binding behavior using surface plasmon resonance (SPR) to measure binding for all cognate pairs. In these experiments, each cadherin adhesive fragment was covalently coupled to a sensor chip surface by thiol coupling of an engineered C-terminal cysteine residue (Cys tag) to present functional EC1 domains in a favorable orientation for interaction. All Cys-tagged proteins were analyzed by AUC, confirming homophilic binding affinities are broadly similar to those of the untagged proteins (Table S1). Untagged cadherin-6 through cadherin-12, cadherin-18, cadherin-20, and cadherin-22 adhesive fragments were passed over each surface, and homophilic and heterophilic binding responses were recorded for all combinations to provide a comprehensive SPR matrix of all potential interactions (Figure 1). As expected, homophilic binding interactions were observed for each cadherin tested (Figure 1, diagonal) and dissociation rates varied up to 6-fold among cadherins (Table S2). In addition to these homophilic responses, all cadherin surfaces supported significant levels of heterophilic binding to selective subsets of cadherin family members (Figure 1, rows). Response levels for the strongest heterophilic interactions for each surface were comparable to or exceeded those of the respective homophilic interactions, suggesting functional significance. Homophilic binding was favored over all other heterophilic interactions for only two cadherins: cadherin-6 and cadherin-20 (Figures 1 and S1). Heterophilic binding affinities could not be determined from the SPR data due to competing homodimerization of surface and analyte cadherins (Katsamba et al., 2009); nevertheless, relative binding strengths could be assessed by comparing response levels over the same surface (Figure 1, rows). Based on the precise pattern of binding preferences observed for each cadherin, the type II cadherin family can be divided into three distinct specificity groups. Within these three groups, members share nearly identical binding profiles: they bind heterophilically to the same set of cadherins (Figure 1, compare rows) and show a preference for interactions within the same group (Figure 1, boxes).
Figure 1. SPR Analysis of Heterophilic Interactions of Type II Cadherins.
Profiles of type II cadherin analytes (shown in columns) binding over individual surfaces of cadherin-8 (top row), cadherin-11, cadherin-6, cadherin-9, cadherin-10, cadherin-12, cadherin-20, cadherin-18, cadherin-22, and cadherin-7 (bottom row). Analytes were tested at 12, 6, and 3 μM monomer concentrations over each surface as shown in each panel. Responses were normalized to account for the molecular weight variations of the different cadherin analytes. The normalized responses in each row (corresponding to the responses over a surface) are scaled independently, allowing quantitative comparison across rows only. Specificity groups identified based on binding preferences (see text) are boxed in blue for the cadherin-8 and cadherin-11 specificity group; in red for the cadherin-6, cadherin-9, and cadherin-10 specificity group; and in green for the cadherin-12, cadherin-18, cadherin-20, cadherin-22, and cadherin-7 specificity group. Cys-tagged cadherins were immobilized at a free monomer concentration of 60 μM, corresponding to 4,673 response units (RU) for cadherin-6, 945 RU for cadherin-7, 1,006 RU for cadherin-8, 2,174 RU for cadherin-9, 546 RU for cadherin-10, 990 RU for cadherin-11, 3,784 RU for cadherin-12, 1,112 RU for cadherin-18, 1,283 for cadherin-20, and 3,651 RU for cadherin-22. See also Figure S1 and Table S2.
Cadherin-8 and cadherin-11 comprise one such specificity group and display clear preference for heterophilic binding to each other over all other cadherins (Figure 1, top rows). The cadherin-8 surface supported heterophilic binding of cadherin-11 as the strongest observed interaction, followed by homophilic binding of the cadherin-8 analyte (Figure 1, top row). All other analytes bound at very low levels: cadherin-20 and cadherin-18 bound weakly, while cadherin-6, cadherin-9, and cadherin-10, as well as cadherin-7, cadherin-12, and cadherin-22, did not show binding above background levels. A similar binding pattern was observed for the cadherin-11 surface, with heterophilic binding of cadherin-8 comprising the strongest response, followed by homophilic binding and comparatively lower binding to all other analytes, including modest levels of binding to cadherin-12, cadherin-20, cadherin-18, and cadherin-22 (Figure 1, second row).
Cadherin-6, cadherin-9, and cadherin-10 define a second specificity group whose members share closely similar binding profiles (Figure 1). On surfaces coated with cadherin-6, cadherin-9, or cadherin-10, binding of cadherin-6 analyte showed the strongest response, followed by binding of cadherin-9 and cadherin-10. No binding of cadherin-8 and cadherin-11 was observed on any of the three surfaces, while the remaining cadherins, cadherin-12, cadherin-20, cadherin-18, cadherin-22, and cadherin-7, showed intermediate binding comparable to that of the cadherin-10 analyte.
The third specificity group comprises cadherin-12, cadherin-20, cadherin-18, cadherin-22, and cadherin-7 (Figure 1). These cadherins showed generally strong binding responses to one another (Figure 1, green box); intermediate binding to the cadherin-6, cadherin-9, and cadherin-10 specificity group; and no strong binding responses to cadherin-8 and cadherin-11. Cadherin-7 binding responses as an analyte were lower overall than those of other members in this group (Figure 1, right column), but the binding profile of cadherin-7 was close to those of cadherin-12, cadherin-18, cadherin-20, and cadherin-22 (Figure 1, bottom row).
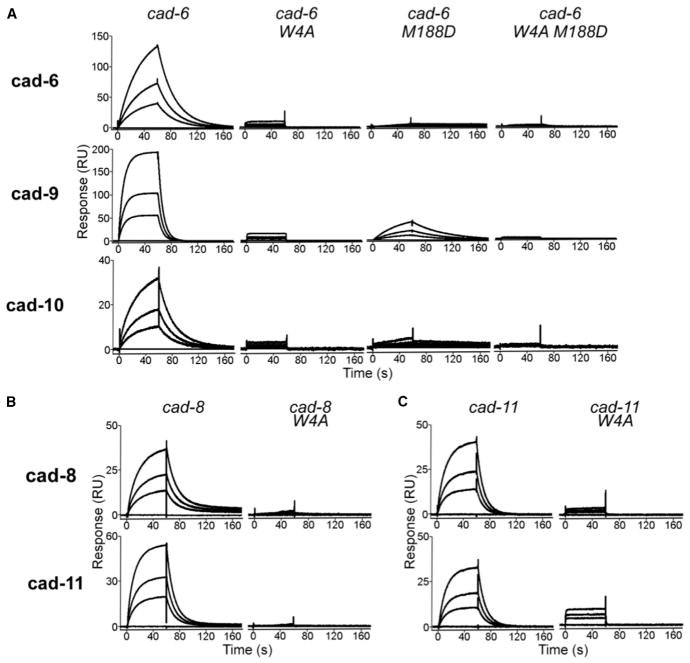
To determine whether heterophilic interactions between type II cadherins form through the same strand-swap binding mechanism shown to mediate homophilic interactions (Figure 4) (Patel et al., 2006), we tested the effects of ablating this interface in SPR (Figure 2). As expected, alanine substitution of Trp4, which anchors the strand-swap dimer, ablated homophilic binding of cadherin-6, cadherin-8, and cadherin-11 in SPR and AUC (Figure 2, W4A mutants) (Table S1) and also abolished heterophilic binding between all cognate pairs (Figure 2). In addition, mutation of the X-dimer interface (M188D) in cadherin-6, which was previously shown to be a necessary binding intermediate for homophilic interactions (Harrison et al., 2010), resulted in severely diminished heterophilic binding of cadherin-6 M188D mutant to cadherin-9 and cadherin-10 (Figure 2A). Residual heterophilic binding between cadherin-6 M188D and cadherin-9 was abolished in a double mutant, in which both strand-swap and X-dimer interfaces were mutated (Figure 2A, W4A M188D mutant). Altogether, these data confirm that homophilic and heterophilic binding interactions in the type II cadherin family form through the same adhesive interfaces.
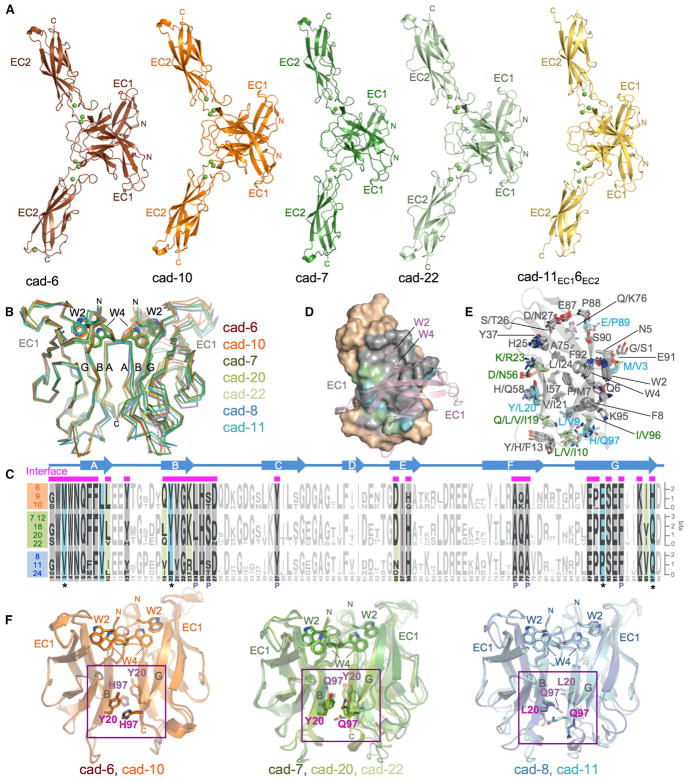
Figure 4. Crystal Structures of Type II Cadherin Homodimers and Analysis of Specificity Determinants.
(A) Ribbon representation of strand-swapped EC1-EC2 homodimer structures of cadherin-6, cadherin-10, cadherin-7, cadherin-22, and chimera cad-11EC16EC2. Three Ca2+ ions (green spheres) are coordinated by interdomain linker regions of each protomer.
(B) Superposition of EC1 homodimers reported here and in Patel et al. (2006) shown as carbon-α traces superposed over the left protomer. Docked strand-swap residues Trp2 and Trp4 are shown in stick representation.
(C) Sequence logos of aligned EC1 regions of type II cadherins from human, mouse, and chicken (Experimental Procedures) separated into specificity groups. Positions containing, or flanked by, interface residues (marked by magenta bars above alignment) are shown in bold and colored according to conservation. Cyan: residues fully conserved within but different between specificity groups. Green: residues differing between specificity groups but not fully conserved within each. Gray: positions with identical conserved consensus residues across all specificity groups. Secondary structure elements of cadherin-6 are shown above the logos. Pocket residues are indicated with P.
(D) Surface representation of EC1, with interface residues colored according to (C). Salmon: partner protomer in ribbon representation.
(E) Interface residues, colored according to (C), shown as sticks in a superposition of EC1 domain structures. The main chain ribbon is shown only for cadherin-6.
(F) Potential specificity determinants (magenta) shown as sticks on superposed EC1 domain homodimers for members of each specificity group. Trp2 and Trp4 are shown for orientation.
See also Table S3.
Figure 2. Effects of Binding Interface Mutations on Homophilic and Heterophilic Binding.
(A) SPR binding responses of wild-type cadherin-6 and its respective strand-swap mutant (W4A), X-dimer interface mutant (M188D), and a double mutant containing both mutations injected over wild-type cadherin-6 surfaces (top row), cadherin-9 surfaces (middle row), and cadherin-10 surfaces (bottom row).
(B and C) (B) Wild-type cadherin-8 and strand-swap mutant cadherin-8 W4A and (C) cadherin-11 and strand-swap mutant cadherin-11 W4A were injected over wild-type cadherin-8 surfaces (top row) and cadherin-11 surfaces (bottom row). Responses for each surface are scaled independently.
Specificity Groups Correspond to Branches of the Type II Cadherin Phylogenetic Tree
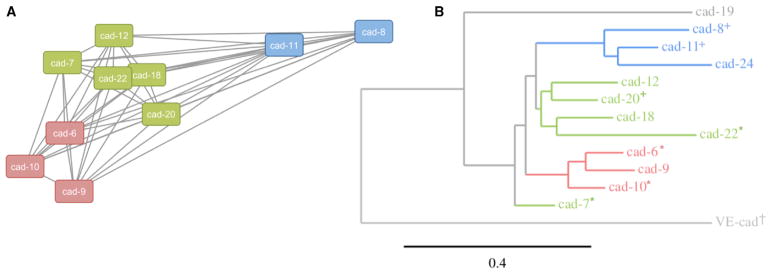
To compare the relative strengths of heterophilic interactions across the type II cadherin family, values derived from normalized responses for each interaction in our SPR matrix were used to weight a force-directed interaction network (Figure 3A). Cadherins linked closely in the network reflect strong binding interactions (e.g., cadherin-18 and cadherin-22), while cadherins linked distantly in the network reflect weak or background-level binding interactions (e.g., cadherin-8 and cadherin-9). As expected, three discrete clusters emerge from the binding network, which are consistent with the specificity groups we identified earlier based on qualitative comparison of our SPR traces (Figure 1).
Figure 3. Biophysically Identified Heterophilic Binding Specificity Groups Correspond to Branches of the Phylogenetic Tree.
(A) Force-directed binding network of type II cadherin heterophilic interactions weighted by binding responses derived from SPR experiments (Experimental Procedures). Nodes represent individual cadherins colored by specificity group; edges represent heterophilic binding interactions, with length inversely proportional to binding strength.
(B) Phylogram of the type II cadherin family computed from alignment of amino acid sequences of adhesive EC1 and EC2 domain regions using a maximum likelihood method. Branches of the phylogenetic tree are colored according to specificity group. Symbols indicate cadherins for which structures of the adhesive interface are reported in this work (asterisk), or in Patel et al. (2006) (plus sign) and Brasch et al. (2011) (dagger).
We next investigated whether cadherins belonging to the three specificity groups in SPR share other characteristics. The groups derived purely from the binding data correspond closely to the phylogenetic grouping generated from alignment of sequences of adhesive EC1 and EC2 regions (Figure 3B). This suggests that type II cadherins bind to, and share binding preferences with, family members that are closely related phylogenetically. In particular, cadherin-8 and cadherin-11 are more distantly related to the other members of the type II cadherin family (Figure 3B), which is reflected in their binding preferences. Cadherin-24, which was excluded from SPR analyses due to solubility problems (Experimental Procedures), is also likely to share binding preferences with cadherin-8 and cadherin-11, because it belongs to the same phylogenetic branch. The divergence of the cadherin-6, cadherin-9, and cadherin-10 group and the cadherin-8 and cadherin-11 group extends to the sequence of their cytoplasmic domains (Nollet et al., 2000; Sotomayor et al., 2014), suggesting the possibility of differences in downstream events upon binding.
The clustering of cadherins into specificity groups is also partially reflected in their chromosomal locations. Genes encoding the closely related cadherin-8 and cadherin-11 are located close together in mouse chromosome 8, human chromosome 16, and chicken chromosome 11 (Nollet et al., 2000). Similarly, cadherin-6, cadherin-9, and cadherin-10 are clustered on the same chromosome in mouse (chromosome 15), human (chromosome 5), and chicken (chromosome 2). Together with the phylogenetic data, these observations suggest that type II cadherins sharing similar binding preferences are likely to have diverged most recently during evolution.
Crystal Structures of Type II Cadherins Show Highly Conserved Dimer Topology across All Specificity Groups
To investigate the molecular basis of grouped specificity behavior observed in SPR, we set out to compare homodimer structures of representative type II cadherins. We have previously reported crystal structures of adhesive fragments of cadherin-8, cadherin-11, and cadherin-20 representing two members of the cadherin-8/11 group and a single member of the cadherin-7/12/18/20/22 group (Figure 3B) (Patel et al., 2006). To extend structural coverage to include multiple representatives for all specificity groups, we have now determined crystal structures of EC1-EC2 adhesive fragments of cadherin-6 and cadherin-10 belonging to the previously uncharacterized cadherin-6/9/10 group and of cadherin-7 and cadherin-22 from additional sub-branches of the cadherin-7/12/18/20/22 specificity group (Figures 3B and 4).
Crystals of cadherin-6, cadherin-7, cadherin-10, and cadherin-22 diffracted to between 1.7 and 2.7 Å resolution (Table S3), and structures were solved by molecular replacement. Each cadherin structure adopted an extended conformation rigidified by coordination of three calcium ions in the interdomain linker regions and formed strand-swapped homodimers (Figure 4A), as was observed for cadherin-8, cadherin-11, and cadherin-20 in the past (Patel et al., 2006). In each dimer, reciprocal exchange of A strands between EC1 domains is anchored by docking of Trp2 and Trp4 residues into a hydrophobic pocket of the partner molecule, bringing strands A, B, and G into intermolecular contact (Figure 4B). Further buried surface area is contributed by the BC loop of domain EC2, which packs against strands B and E of the partner EC1 domain in all four dimer structures (Figure 4A, arrows).
Superposition of these new structures with previously determined type II cadherin homodimer structures reveals that the overall dimer topology, including the angle between partner EC1 domains, is essentially identical across members of all three specificity groups (root-mean-square deviation [RMSD] < 0.9 Å between 145 and 185 aligned carbon alpha [Cα] atoms per dimer) (Figure 4B). Because of this shared topology, identical regions of the EC1 A, B, and G strands contact one another in all type II cadherin dimers. While the interface is dominated by docking of Trp2 and Trp4 into the partner hydrophobic pocket formed by residues from the B, C, F, and G strands, additional and mostly hydrophobic contacts form between paired A strands and between the B and the A or G strands, extending the dimer interface over the whole face of the domain in all structures.
Specificity Determinants in the Type II Cadherin Adhesive Interface
Structural conservation of the type II cadherin adhesive interface across all branches of the family suggests that subtype-specific differences in structurally equivalent interfacial residues could govern binding preferences. Because we observe binding within specificity groups that is favored over binding between groups, residues in the interface with group-specific conservation may be particularly important. We aligned amino acid sequences of type II cadherin EC1 domains from mouse, human, and chicken and examined sequence conservation both within and across specificity groups. Figure 4C shows a sequence logo representation calculated separately for each specificity group. Interfacial residues derived from all available crystal structures were mapped onto the sequences to identify conserved and variable regions of the interface (Figure 4C, magenta bars). Most interface residues are fully conserved, or conserved in consensus, across the type II cadherin family and comprise the core of the strand-swap interface, including residues of the exchanged A strands, the acceptor pocket, and most of the hydrophobic patch toward the base of the domain (Figures 4C–4E, gray shading). Positions of variable residue identity, in which consensus residues differ between at least two specificity groups, are restricted to ten residues at the periphery of the interface and define two distinct regions (Figures 4C–4E, highlights). First, the lower part of the interface is encircled by variable residues contributed by the base of the A strand (L/V9, L/V/I10), the base of the B strand (Q/L/V/I19, Y/L20, K/R23), the E strand (D/N56), and the base of the G strand (I/V96, H/Q97). Second, the upper periphery of the interface contains two group-specific residues, namely, M/V3 in the A strand and E/P89 at the top of the G strand.
Within the ten variable interface residues, five are not fully conserved within each respective specificity group and thus are less likely to underlie shared group binding behavior (Figures 4C–4E, green highlighting). The remaining five residues show perfect conservation within each specificity group while differing between at least two groups (Figures 4C–4E, blue highlighting), strengthening their candidacy as specificity determinants. Most strikingly, residues Y/L20 and H/Q97 form three distinct group-specific interaction pairs in the adhesive homodimer structures (Figure 4F). In cadherin-6 and cadherin-10 structures, Tyr20 and His97 are closely apposed and engage in near-parallel π-π-stacking interactions at a distance of approximately 3.3 Å (Figure 4F, left). In the cadherin-7, cadherin-20, and cadherin-22 homodimers, Tyr20 and Gln97 are also apposed and likely engage in van der Waals interactions between the amino group of Gln97 and the tyrosine ring (Burley and Petsko, 1986). In the cadherin-8/11 group, Leu20 and Gln97 are in proximity, but they do not interact closely. These subgroup-specific pairwise interactions appear to be likely to contribute to the restricted binding preferences observed in SPR. The remaining residues showing strict group-specific conservation, M/V3 and L/V9 in the A strand and E/P89 at the top of the G strand, forming the rim of the pocket, do not form such interacting pairs with each other or with any other potential specificity determining residues but instead contact fully conserved residues (Figures 4C–4E). Nonetheless, they could contribute to specificity indirectly through effects on neighboring residues or by overall effects on the shape of the interaction surface.
Targeted Mutation of Group-Specific Interface Residues Converts Binding Preferences in SPR
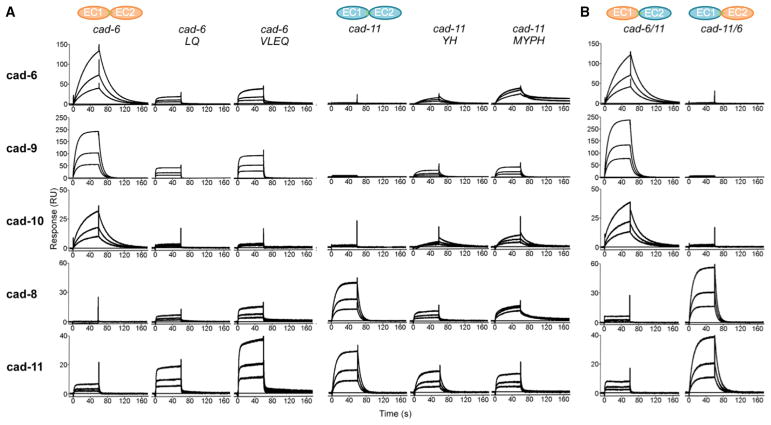
We investigated whether these group-specific residues governed binding preferences in SPR. We first introduced point mutations at positions 20 and 97, which form distinct interacting pairs at the interface in each specificity group (Figure 4F). For these experiments, we chose cadherin-6 from the cadherin-6/9/10 group and cadherin-11 from the cadherin-8/11 group, because these groups show clearly distinguishable binding preferences in SPR (Figure 1). Tyr20 and His97 in cadherin-6 were substituted with the corresponding Leu20 and Gln97 residues from cadherin-11 (cadherin-6 LQ mutant) and the reverse substitutions were made in cadherin-11 (cadherin-11 YH mutant). Binding of mutants was tested against wild-type proteins in SPR (Figure 5A). As described earlier, cadherin-6 binds selectively to cadherin-6, cadherin-9, and cadherin-10 surfaces but does not bind to cadherin-8 and cadherin-11 surfaces (Figure 5A, left). Over the same set of surfaces, cadherin-6 LQ mutant displayed decreased binding to the cadherin-6, cadherin-9, and cadherin-10 surfaces but concomitantly increased binding to cadherin-8 and cadherin-11 surfaces (Figure 5A), consistent with conversion of overall specificity. Corresponding behavior was observed for the cadherin-11 YH mutant, which compared to wild-type cadherin-11, showed dramatically decreased binding to members of the same specificity group (cadherin-8 and cadherin-11) with concomitantly increased binding to cadherin-6, cadherin-9, and cadherin-10 (Figure 5A). Our results are consistent with a decisive role for residues 20 and 97 as specificity determinants in the cadherin-6/9/10 specificity group and the cadherin-8/11 specificity group.
Figure 5. Mutational Analysis of Type II Cadherin Specificity Using SPR.
(A) SPR responses of wild-type or specificity mutants of cadherin-6 and cadherin-11 (columns) passed over wild-type cadherin-6, cadherin-9, cadherin-10, cadherin-8, and cadherin-11 surfaces (rows).
(B) Chimeric proteins cad-6EC111EC2 and cad-11EC16EC2 passed over the same surfaces reproduce the binding characteristics of the wild-type proteins with corresponding EC1 domains.
All mutant and chimeric proteins retained homophilic binding in AUC (Table 1).
Because binding responses of the cadherin-6 LQ and cadherin-11 YH mutants were lower than those of the target wild-type proteins, we tested whether conversion of additional group-specific residues enhanced changes in binding preference. We mutated the group-specifically conserved surface residues M/V3 and E/P89 in cadherin-6 LQ and cadherin-11 YH mutants to generate quadruple mutants cadherin-6 VLEQ and cadherin-11 MYPH. In SPR, cadherin-11 MYPH mutant showed a similar conversion of binding preferences to the double mutant, but with additional enhancement of responses to cadherin-6, cadherin-9, and cadherin-10 (Figure 5A). Similarly, cadherin-6 VLEQ showed enhanced binding to the opposite specificity group in comparison to the cadherin-6 LQ double mutant, though in this case binding was also increased over same specificity group surfaces (Figure 5A). Because both quadruple mutants enhanced binding to the opposite specificity group, our data suggest that M/V3 and E/P89 contribute modestly to specificity, at least in combination with residues L/Y20 and H/Q97. To interconvert all 37 non-identical EC1 residues between cadherin-6 and cadherin-11, we also prepared chimeric proteins cad-6EC111EC2 (cad-6/11) and cad-11EC16EC2 (cad-11/6). We confirmed the fidelity of these chimeric proteins in AUC (Table 1) and by structural analysis of cad-11EC16EC2 (Figure 4A, right). As expected, both formed homophilic dimers, and the structure confirmed the cad-11EC16EC2 homodimer arrangement to be nearly identical to that of wild-type cadherin-11. In SPR experiments, these chimeric proteins closely mimicked the binding behavior of the wild-type proteins from which their EC1 domains derived (Figure 5B), confirming that complete interconversion of binding behavior can be observed when sufficient residues are changed and that all specificity determinants are restricted to EC1. Thus, our data suggest that residues 20 and 97, and to a lesser extent, residues 3 and 89, are major specificity determinants with other variable interface residues in EC1, likely contributing indirectly to precise binding specificities.
Localization of Full-Length Type II Cadherins to Heterotypic Cell Contacts Depends on Specificity Determinants
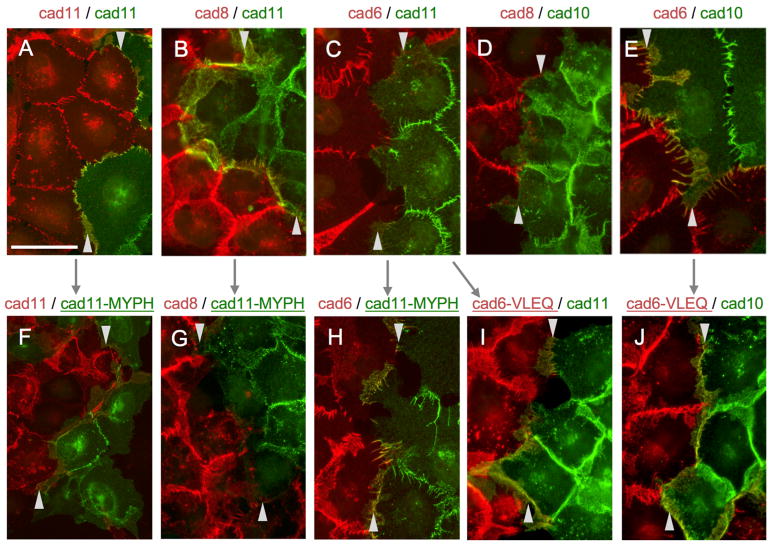
To relate our biophysical observations for adhesive fragments and their mutants to the behavior of full-length proteins at cell-cell contact sites, we examined localization of fluorescently labeled type II cadherins in transfected A431D cells (Figures 6 and S2). Full-length cadherin-8 and cadherin-11, as well as cadherin-6 and cadherin-10, representing pairs from distinct specificity groups, were labeled at their C termini with either red fluorescent mCherry, or green fluorescent Dendra2. These were transfected singly into A431D cell lines that were then co-cultured to allow formation of homotypic cell contacts between cells of the same cell line and heterotypic contacts between cells from different cell lines. When combinations of cadherins from the same specificity group, cadherin-8 and cadherin-11 or cadherin-6 and cadherin-10, were co-cultured, both cadherins co-localized at heterotypic contact sites, in addition to the respective homotypic sites, consistent with heterophilic binding observed for these pairs in SPR (Figures 6B and 6E, arrowheads). In marked contrast, co-culture of cells expressing mismatched cadherin-6 and cadherin-11 or cadherin-8 and cadherin-10 from different specificity groups produced heterotypic cell contact sites devoid of cadherins, which accumulated only at homotypic sites (Figures 6C and 6E). Lastly, a homophilic pair, cadherin-11-dendra and cadherin-11-cherry, localized equally to homotypic and heterotypic contacts (Figure 6A, arrowheads), reflecting uniform homophilic interactions. The cell-adhesive behavior of these full-length wild-type proteins thus closely mirrors binding preferences observed in biophysics.
Figure 6. Full-Length Type II Cadherin Localization at Homotypic and Heterotypic Contact Sites between Transfected A431D Cells in Co-culture.
Fluorescence images of co-cultures of A431D cells transfected with full-length type II cadherins tagged with either cherry (red) or dendra (green) in the combinations indicated.
(A–E) Wild-type cadherin-11/-11 (A), cadherin-8/-11 (B), cadherin-6/-11 (C), cadherin-8/-10 (D) and cadherin-6/-10 (E) localize to homotypic and heterotypic contact sites according to their binding preferences.
(F–J) Mutations targeting specificity determinants alter localization of mutant cadherin-11 with cadherin-11 (F), cadherin-8 (G) and cadherin-6 (H) and of mutant cadherin-6 with cadherins-11 (I) and cadherin-10 (J). Compare panels linked with arrows. Heterotypic contact sites are delimited by arrowheads at top and bottom.
Scale bar, 50 μm. See also Figure S2.
We also tested our quadruple specificity mutants in co-culture assays with full-length cadherin-6 and cadherin-11 (cadherin-6 VLEQ and cadherin-11 MYPH). Compared to wild-type protein, localization of cadherin-11 MYPH mutant to heterotypic cell-cell contacts with cadherin-8 (compare Figures 6B and 6G) or cadherin-11 (compare Figures 6A and 6F) from the same specificity group was ablated or dramatically reduced (Figure S2). At the same time, cadherin-11 MYPH mutant strikingly co-localized with cadherin-6 from the opposite specificity group (compare Figures 6C and 6H), showing a significant shift in overall binding preference (Figures 6 and S2). Similar behavior was observed for mutant cadherin-6 VLEQ, which acquired strong co-localization with cadherin-11 from the opposite specificity group (compare Figures 6C and 6I), while co-localization with cadherin-10 was concomitantly decreased (compare Figures 6E and 6J). Behavior of these mutants shows that conversion of four residues between specificity groups produces a shift of binding preferences sufficient to convert adhesive specificity between cells.
DISCUSSION
Type II cadherins represent a large family of adhesion proteins with overlapping differential expression patterns and diverse functional roles, presenting a challenge in relating their molecular and functional properties. The comprehensive matrix of binding interactions determined here reveals that all type II cadherins tested participate both in homophilic and heterophilic interactions that are of comparable strengths. Heterophilic interactions are not promiscuous: each member of the family binds to a specific subset of other members with characteristic relative response levels. These subsets and responses are shared among multiple cadherins, giving rise to three distinct specificity groups: cadherin-6, cadherin-9, and cadherin-10; cadherin-8 and cadherin-11; and cadherin-7, cadherin-12, cadherin-18, cadherin-20, and cadherin-22. Heterophilic interactions form among all members within each specificity group, and weaker heterophilic interactions form between the cadherin-6/9/10 group and the cadherin-7/12/18/20/22 group, while the cadherin-8/11 group is more isolated (Figure 1). Within groups, individual cadherins are nonetheless distinguished by widely differing homodimerization affinities (Table 1) and dissociation rates (Table S2).
We observe consistent specificity behavior between SPR experiments with purified adhesive fragments and cellular co-culture experiments with full-length proteins (Figures 1 and 6). Our results are also in agreement with previous co-culture experiments testing other pairings from the cadherin-6/9/10 specificity group (Basu et al., 2017) and with cell-cell aggregation assays using transfected human and chicken type II cadherins (Basu et al., 2017; Nakagawa and Takeichi, 1995; Patel et al., 2006; Shimoyama et al., 1999, 2000). In a broad cell aggregation study using a range of human type II cadherins, selective co-aggregation of a number of specific cadherin pairs was observed (Shimoyama et al., 2000), representing a subset of the heterophilic binding pairs we observe in SPR. Heterophilic pairs only observed in SPR were either not tested in the co-aggregation study (all interactions involving cadherin-20 and cadherin-22) (Figure 1) or did not produce detectable co-aggregation, likely due to differences in assay sensitivity and variable cadherin expression levels in transfected cells. Nonetheless, observation of closely similar specificity preferences across multiple assay systems and species supports the biological relevance of the type II cadherin adhesive interaction map determined here.
By sequence analysis, we identified potential N-linked glycosylation sites at Asn202 for cadherin-6, cadherin-9, cadherin-10, cadherin-12, cadherin-18, and cadherin-20; potential N-linked glycosylation sites at Asn127 for cadherin-8; and no sites in cadherin-7, cadherin-11, cadherin-22, and cadherin-24. Mapping the sites onto type II cadherin structures reveals Asn127 and Asn202 to be located on the B and G strands of EC1, respectively, distal from adhesive sites. Together with close correlation of binding preferences observed between SPR and cell-based experiments, this suggests that absence of glycosylation in the bacterially produced adhesive fragments is unlikely to affect interaction behavior of type II cadherins, as we have observed previously for adhesive regions of VE-cadherin (Brasch et al., 2011).
Type II family members cadherin-19, cadherin-24, and VE-cadherin were excluded from our SPR experiments. Of these, cadherin-24 is predicted to be part of the cadherin-8/11 specificity group, based on its position in the phylogenetic tree and conservation of potential specificity residues Val3, Leu20, Pro89, and Gln97 (Figures 3 and 4). Cadherin-19 cannot be assigned to one of the specificity groups identified here based on sequence information, because it contains unique residues at putative specificity sites 20 and 97 and lacks conservation of invariant residues Arg5 and Gln6 in the swapped A strand. Based on these observations, we predict that cadherin-19 is unlikely to engage in heterophilic binding with other members of the type II cadherin family, though this remains to be tested. As reported previously, the structure and interface characteristics of the VE-cadherin homodimer are divergent from, and likely incompatible with, those of other members of the type II cadherin family, consistent with its specialized biological role (Brasch et al., 2011).
Consistent with the extensive heterophilic interactions observed for type II cadherins, the structure and topology of their adhesive homodimers are nearly identical and residues mediating the core interactions of strand swapping are conserved (Figure 4). The observations that heterophilic and homophilic interactions are formed through the same interface (Figure 2) and that phylogenetically related cadherins maintain heterophilic recognition (Figure 3) suggest that divergence of a common binding mechanism gave rise to the selective heterophilic interactions we observe. In the most extreme cases, the interfaces of cadherin-8 and cadherin-11 appear to have diverged sufficiently to become incompatible with all other members of the family, particularly the cadherin-6/9/10 group (Figure 1). Variable residues are restricted to the periphery of the interface, and only a small number of these show group-specific conservation. In a previous study, we postulated that some of these variable residues could be specificity determinants (Patel et al., 2006). However, identification of specificity groups and determination of structures of multiple members of each have allowed us to identify the variable residues most likely to contribute to overall specificity preferences (Figure 4). Targeted mutations identified positions 20 and 97, a group-specific apposed pair in the strand-swapped dimer, as major determinants of the incompatibility between the cadherin-6/9/10 group and the cadherin-8/11 group (Figures 5 and 6). These same residues could in principle explain the distinct specificity of the cadherin-7/12/18/20/22 group; however, because they are partially shared with the cadherin-6/9/10 group and the cadherin-8/11 group, it is likely that other variable residues also contribute. The residues tested by mutation only partially account for differences in homophilic binding affinity and subtle differences in heterophilic binding responses of individual members in each group, which can be converted more substantially by exchange of EC1 domains in chimeric proteins (Figure 5) (Patel et al., 2006).
Binding interactions have been comprehensively determined for a limited number of other adhesion protein families within the cadherin and immunoglobulin (Ig) superfamilies. Close correspondence has been consistently observed among molecular binding affinities, behavior in cell aggregation assays, and phenotypes in vivo and has revealed a range of overall binding characteristics that differ from those observed here for type II cadherins. Like type II cadherins, clustered protocadherin and Dscams families share a canonical adhesive interface; however, they uniformly favor homophilic interactions in cell aggregation experiments, likely reflecting selection pressure for homophilic binding by their biological roles in neuronal identity and self-recognition (Goodman et al., 2016; Hattori et al., 2008; Rubinstein et al., 2017; Thu et al., 2014). Nectins and SynCAMs, families of vertebrate Ig-like proteins, are able to form both homophilic and heterophilic binding interactions through canonical interfaces as observed for type II cadherins (Fogel et al., 2007; Harrison et al., 2012; Narita et al., 2011). However, in both cases, heterophilic interactions are strongly preferred, correlating with primary biological functions in heterotypic adhesion (Takai et al., 2008). Desmosomal cadherins also show a strong preference for heterophilic binding (Harrison et al., 2016), although the biological role of this preference remains to be determined. Binding characteristics of type II cadherins are partially reminiscent of those of their close relatives: type I cadherins. These display a mixture of homophilic and specific heterophilic binding in SPR experiments (Katsamba et al., 2009; Vendome et al., 2014), but in standard cell aggregation assays, type I cadherin heterophilic pairs form only partially mixed aggregates (Katsamba et al., 2009; Shan et al., 2000) and do not fully intermix, as was observed for a number of type II cadherin pairs (Patel et al., 2006; Shan et al., 2000; Shimoyama et al., 1999, 2000). The extensive selective heterophilic binding of type II cadherins, in combination with their differential homophilic affinities (Table 1), in which neither type of interaction is dominantly preferred, may allow them to encode subtle differences in adhesiveness to drive fine sorting events within generally cohesive tissues. For heterophilic pairs that co-localize to heterotypic junctions in our co-culture assays, homophilic junctions in the same cells do not appear depleted of the respective cadherins (Figure 6). Therefore, in tissues co-expressing multiple type II cadherins, we would expect both types of interaction to contribute to adhesive identity of individual cells.
Biological roles for homophilic interactions of type II cadherin-6, cadherin-7, and cadherin-20 have been suggested by misexpression studies in the developing chicken spinal cord, chicken optic tectum, and mouse telencephalon (Inoue et al., 2001; Patel et al., 2006; Price et al., 2002; Treubert-Zimmermann et al., 2002). Experimental equalization of type II cadherin complements of normally segregated tissues led to mis-sorting or mistargeting of neurons into compartments expressing matching cadherins. Similarly, homophilic binding of cadherin-6 and cadherin-9 appear to be required for targeting of retinal ganglion cells in the mouse visual system (Osterhout et al., 2011) and for formation and differentiation of mossy fiber synapses in the hippocampus (Williams et al., 2011). While these studies do not exclude the potential involvement of heterophilic interactions, homophilic adhesion provides a parsimonious explanation for the observed phenotypes.
Biological roles for heterophilic binding of type II cadherins have emerged only recently. In mouse retina, targeting of cadherin-8-expressing type 2 bipolar cells (BC2s) and cadherin-9-expressing type 5 bipolar cells (BC5s) to distinct sublaminae of the inner plexiform layer (IPL) depended critically on their respective cadherin identities (Duan et al., 2014). However, surrounding cells of the target sublaminae do not express these cad-herins, and targeting of BC2s and BC5s expressing ectopic cadherin-8 or cadherin-9 is maintained even in cadherin knockout backgrounds, ruling out dependence on homophilic interactions (Duan et al., 2014). These data suggest heterophilic interactions of cadherin-8 and cadherin-9 with partner cadherins in the target layers, though the exact complement of type II cadherins in the IPL remains to be determined. The mouse hippocampus provides a clearer example of in vivo function driven by heterophilic interactions (Basu et al., 2017). High-magnitude LTP in the stratum oriens, where hippocampal CA3 neurons synapse with basal dendrites of CA1 neurons, depends on presynaptic cadherin-9 expressed only in CA3 neurons and on postsynaptic cadherin-6 and cadherin-10 expressed only in CA1 neurons. Knockout of either cadherin-9 alone or both cadherin-6 and cadherin-10 produces identical phenotypes, in which high-magnitude LTP is impaired (Basu et al., 2017). Because each cadherin is restricted to one side of the synapse, these findings implicate heterophilic interactions within the cadherin-6/9/10 specificity group in functioning of this neural circuit. Further functional roles for the range of selective heterophilic interactions we identify here await determination.
Specific functions of individual type II cadherins may be masked by functional redundancy partly because of their frequently overlapping expression patterns and partly because of the shared binding preferences we observe for members of each specificity group. An example of this type of redundancy was observed in the mouse hippocampal circuit described previously: single knockout of cadherin-10 alone maintained wild-type levels of high-magnitude LTP, which was impaired only when cadherin-6 and cadherin-10 were knocked out together (Basu et al., 2017). This is likely because cadherin-6 could substitute for cadherin-10 in the single knockout to bind heterophilically to cadherin-9. Surprisingly subtle phenotypes have been observed for numerous single type II cadherin knockouts (Basu et al., 2017; Duan et al., 2014; Kawaguchi et al., 2001; Mah et al., 2000; Osterhout et al., 2011; Saarimäki-Vire et al., 2011; Suzuki et al., 2007), despite their strong expression in the CNS, and these could be similarly explained by functional substitution by other cadherins belonging to the same specificity groups. Knockout of complete specificity groups should reveal additional functional roles for type II cadherins. Our analysis of specific binding patterns of type II cadherins and their segregation into distinct specificity groups provide a basis for further investigation of their biological roles.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Recombinant type II cadherin ectodomain fragments were expressed in bacteria and purified from lysate by nickel affinity chromatography, ion exchange chromatography, and gel filtration.
Biophysical Analyses
AUC experiments were performed at 25°C in a Beckman XL-A/I analytical ultracentrifuge with UV detection. For SPR, Cys-tagged proteins were captured by thiol coupling to CM4 sensor chips, and binding of analytes was assessed in a Biacore T100 biosensor at 25°C.
Structure Determination
Protein crystals were grown by vapor diffusion in hanging drops in the conditions listed in the Supplemental Experimental Procedures. X-ray diffraction data were collected from single crystals at 100 K, using a wavelength of 0.979 Å at the X4 beamlines at Brookhaven National Laboratory. Structures were solved by molecular replacement and refined using phenix (Adams et al., 2010).
Co-culture Assays
Full-length cadherins with C-terminal dendra2-Myc-or mCherry-FLAG tags in the vector pRc/CMV were transfected into human A-431D cells, and co-cultures were analyzed by fluorescence microscopy as described previously (Hong et al., 2010).
Supplementary Material
Highlights.
Mouse type II cadherins form homophilic and selective heterophilic interactions
Binding profiles are shared within groups of phylogenetically related cadherins
Interface differences in a canonical adhesive dimer govern group-wise specificity
These molecular specificities drive localization of cadherins to adhesive contacts
Acknowledgments
This work was supported by NIH grants R01 GM062270 (to L.S.), R01 AR044016 and R01 AR057992 (to S.T.), and US National Science Foundation grant MCB-1412472 (to B.H.). X-ray data were acquired at the X4A and X4C beamlines of the National Synchrotron light source, Brookhaven National Laboratory (BNL), and the beamlines are operated by the New York Structural Biology Center. We thank J. Schwanof, R. Abramowitz, and Qun Liu at BNL for support with synchrotron data collection.
Footnotes
DATA AND SOFTWARE AVAILABILITY
The accession numbers for the atomic coordinates of mouse cadherin-6 EC1-EC2, cadherin-7 EC1-EC2, cadherin-10 EC1-EC2, and cadherin-22 EC1-EC2, as well as chimera cad-11EC16EC2, reported in this paper are PDB: 6CGU, 6CGS, 6CG6, and 6CG7, and 6CGB, respectively.
Supplemental Information includes Supplemental Experimental Procedures, two figures, and three tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.04.012.
AUTHOR CONTRIBUTIONS
J.B. designed experiments, prepared proteins, crystallized proteins, and determined structures; P.S.K. designed and performed the SPR experiments; O.J.H. determined structures; G.A. performed analytical ultracentrifugation experiments; B.K. crystallized chimeric protein; A.K. crystallized cad-herin-7 protein; R.B.T., I.I., and S.T. performed all co-culture experiments; and J.B., O.J.H., P.S.K., S.T., B.H., and L.S. analyzed data and wrote the manuscript.
DECLARATION OF INTERESTS
The authors declare no competing interests.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu R, Duan X, Taylor MR, Martin EA, Muralidhar S, Wang Y, Gangi-Wellman L, Das SC, Yamagata M, West PJ, et al. Heterophilic Type II Cadherins Are Required for High-Magnitude Synaptic Potentiation in the Hippocampus. Neuron. 2017;96:160–176. doi: 10.1016/j.neuron.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekirov IH, Needleman LA, Zhang W, Benson DL. Identification and localization of multiple classic cadherins in developing rat limbic system. Neuroscience. 2002;115:213–227. doi: 10.1016/s0306-4522(02)00375-5. [DOI] [PubMed] [Google Scholar]
- Brasch J, Harrison OJ, Ahlsen G, Carnally SM, Henderson RM, Honig B, Shapiro L. Structure and binding mechanism of vascular endothelial cadherin: a divergent classical cadherin. J Mol Biol. 2011;408:57–73. doi: 10.1016/j.jmb.2011.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasch J, Harrison OJ, Honig B, Shapiro L. Thinking outside the cell: how cadherins drive adhesion. Trends Cell Biol. 2012;22:299–310. doi: 10.1016/j.tcb.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley SK, Petsko GA. Amino-aromatic interactions in proteins. FEBS Lett. 1986;203:139–143. doi: 10.1016/0014-5793(86)80730-x. [DOI] [PubMed] [Google Scholar]
- Demireva EY, Shapiro LS, Jessell TM, Zampieri N. Motor neuron position and topographic order imposed by β- and γ-catenin activities. Cell. 2011;147:641–652. doi: 10.1016/j.cell.2011.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Krishnaswamy A, De la Huerta I, Sanes JR. Type II cadherins guide assembly of a direction-selective retinal circuit. Cell. 2014;158:793–807. doi: 10.1016/j.cell.2014.06.047. [DOI] [PubMed] [Google Scholar]
- Fogel AI, Akins MR, Krupp AJ, Stagi M, Stein V, Biederer T. SynCAMs organize synapses through heterophilic adhesion. J Neurosci. 2007;27:12516–12530. doi: 10.1523/JNEUROSCI.2739-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman KM, Rubinstein R, Thu CA, Bahna F, Mannepalli S, Ahlsén G, Rittenhouse C, Maniatis T, Honig B, Shapiro L. Structural Basis of Diverse Homophilic Recognition by Clustered α- and β-Protocadherins. Neuron. 2016;90:709–723. doi: 10.1016/j.neuron.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison OJ, Bahna F, Katsamba PS, Jin X, Brasch J, Vendome J, Ahlsen G, Carroll KJ, Price SR, Honig B, Shapiro L. Two-step adhesive binding by classical cadherins. Nat Struct Mol Biol. 2010;17:348–357. doi: 10.1038/nsmb.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison OJ, Vendome J, Brasch J, Jin X, Hong S, Katsamba PS, Ahlsen G, Troyanovsky RB, Troyanovsky SM, Honig B, Shapiro L. Nectin ectodomain structures reveal a canonical adhesive interface. Nat Struct Mol Biol. 2012;19:906–915. doi: 10.1038/nsmb.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison OJ, Brasch J, Lasso G, Katsamba PS, Ahlsen G, Honig B, Shapiro L. Structural basis of adhesive binding by desmocollins and desmogleins. Proc Natl Acad Sci USA. 2016;113:7160–7165. doi: 10.1073/pnas.1606272113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori D, Millard SS, Wojtowicz WM, Zipursky SL. Dscam-mediated cell recognition regulates neural circuit formation. Annu Rev Cell Dev Biol. 2008;24:597–620. doi: 10.1146/annurev.cellbio.24.110707.175250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano S, Takeichi M. Cadherins in brain morphogenesis and wiring. Physiol Rev. 2012;92:597–634. doi: 10.1152/physrev.00014.2011. [DOI] [PubMed] [Google Scholar]
- Hong S, Troyanovsky RB, Troyanovsky SM. Spontaneous assembly and active disassembly balance adherens junction homeostasis. Proc Natl Acad Sci USA. 2010;107:3528–3533. doi: 10.1073/pnas.0911027107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Tanaka T, Takeichi M, Chisaka O, Nakamura S, Osumi N. Role of cadherins in maintaining the compartment boundary between the cortex and striatum during development. Development. 2001;128:561–569. doi: 10.1242/dev.128.4.561. [DOI] [PubMed] [Google Scholar]
- Katsamba P, Carroll K, Ahlsen G, Bahna F, Vendome J, Posy S, Rajebhosale M, Price S, Jessell TM, Ben-Shaul A, et al. Linking molecular affinity and cellular specificity in cadherin-mediated adhesion. Proc Natl Acad Sci USA. 2009;106:11594–11599. doi: 10.1073/pnas.0905349106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi J, Azuma Y, Hoshi K, Kii I, Takeshita S, Ohta T, Ozawa H, Takeichi M, Chisaka O, Kudo A. Targeted disruption of cadherin-11 leads to a reduction in bone density in calvaria and long bone metaphyses. J Bone Miner Res. 2001;16:1265–1271. doi: 10.1359/jbmr.2001.16.7.1265. [DOI] [PubMed] [Google Scholar]
- Mah SP, Saueressig H, Goulding M, Kintner C, Dressler GR. Kidney development in cadherin-6 mutants: delayed mesenchyme-to-epithelial conversion and loss of nephrons. Dev Biol. 2000;223:38–53. doi: 10.1006/dbio.2000.9738. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Takeichi M. Neural crest cell-cell adhesion controlled by sequential and subpopulation-specific expression of novel cadherins. Development. 1995;121:1321–1332. doi: 10.1242/dev.121.5.1321. [DOI] [PubMed] [Google Scholar]
- Narita H, Yamamoto Y, Suzuki M, Miyazaki N, Yoshida A, Kawai K, Iwasaki K, Nakagawa A, Takai Y, Sakisaka T. Crystal Structure of the cis-Dimer of Nectin-1: implications for the architecture of cell-cell junctions. J Biol Chem. 2011;286:12659–12669. doi: 10.1074/jbc.M110.197368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollet F, Kools P, van Roy F. Phylogenetic analysis of the cadherin superfamily allows identification of six major subfamilies besides several solitary members. J Mol Biol. 2000;299:551–572. doi: 10.1006/jmbi.2000.3777. [DOI] [PubMed] [Google Scholar]
- Osterhout JA, Josten N, Yamada J, Pan F, Wu SW, Nguyen PL, Panagiotakos G, Inoue YU, Egusa SF, Volgyi B, et al. Cadherin-6 mediates axon-target matching in a non-image-forming visual circuit. Neuron. 2011;71:632–639. doi: 10.1016/j.neuron.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SD, Ciatto C, Chen CP, Bahna F, Rajebhosale M, Arkus N, Schieren I, Jessell TM, Honig B, Price SR, Shapiro L. Type II cadherin ectodomain structures: implications for classical cadherin specificity. Cell. 2006;124:1255–1268. doi: 10.1016/j.cell.2005.12.046. [DOI] [PubMed] [Google Scholar]
- Price SR, De Marco Garcia NV, Ranscht B, Jessell TM. Regulation of motor neuron pool sorting by differential expression of type II cadherins. Cell. 2002;109:205–216. doi: 10.1016/s0092-8674(02)00695-5. [DOI] [PubMed] [Google Scholar]
- Redies C, Hertel N, Hübner CA. Cadherins and neuropsychiatric disorders. Brain Res. 2012;1470:130–144. doi: 10.1016/j.brainres.2012.06.020. [DOI] [PubMed] [Google Scholar]
- Rubinstein R, Goodman KM, Maniatis T, Shapiro L, Honig B. Structural origins of clustered protocadherin-mediated neuronal barcoding. Semin Cell Dev Biol. 2017;69:140–150. doi: 10.1016/j.semcdb.2017.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarimäki-Vire J, Alitalo A, Partanen J. Analysis of Cdh22 expression and function in the developing mouse brain. Dev Dyn. 2011;240:1989–2001. doi: 10.1002/dvdy.22686. [DOI] [PubMed] [Google Scholar]
- Shan WS, Tanaka H, Phillips GR, Arndt K, Yoshida M, Colman DR, Shapiro L. Functional cis-heterodimers of N- and R-cadherins. J Cell Biol. 2000;148:579–590. doi: 10.1083/jcb.148.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoyama Y, Takeda H, Yoshihara S, Kitajima M, Hirohashi S. Biochemical characterization and functional analysis of two type II classic cadherins, cadherin-6 and -14, and comparison with E-cadherin. J Biol Chem. 1999;274:11987–11994. doi: 10.1074/jbc.274.17.11987. [DOI] [PubMed] [Google Scholar]
- Shimoyama Y, Tsujimoto G, Kitajima M, Natori M. Identification of three human type-II classic cadherins and frequent heterophilic interactions between different subclasses of type-II classic cadherins. Biochem J. 2000;349:159–167. doi: 10.1042/0264-6021:3490159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotomayor M, Gaudet R, Corey DP. Sorting out a promiscuous superfamily: towards cadherin connectomics. Trends Cell Biol. 2014;24:524–536. doi: 10.1016/j.tcb.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki SC, Furue H, Koga K, Jiang N, Nohmi M, Shimazaki Y, Katoh-Fukui Y, Yokoyama M, Yoshimura M, Takeichi M. Cadherin-8 is required for the first relay synapses to receive functional inputs from primary sensory afferents for cold sensation. J Neurosci. 2007;27:3466–3476. doi: 10.1523/JNEUROSCI.0243-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai Y, Miyoshi J, Ikeda W, Ogita H. Nectins and nectin-like molecules: roles in contact inhibition of cell movement and proliferation. Nat Rev Mol Cell Biol. 2008;9:603–615. doi: 10.1038/nrm2457. [DOI] [PubMed] [Google Scholar]
- Thu CA, Chen WV, Rubinstein R, Chevee M, Wolcott HN, Felsovalyi KO, Tapia JC, Shapiro L, Honig B, Maniatis T. Single-cell identity generated by combinatorial homophilic interactions between α, β, and γ protocadherins. Cell. 2014;158:1045–1059. doi: 10.1016/j.cell.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treubert-Zimmermann U, Heyers D, Redies C. Targeting axons to specific fiber tracts in vivo by altering cadherin expression. J Neurosci. 2002;22:7617–7626. doi: 10.1523/JNEUROSCI.22-17-07617.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendome J, Felsovalyi K, Song H, Yang Z, Jin X, Brasch J, Harrison OJ, Ahlsen G, Bahna F, Kaczynska A, et al. Structural and energetic determinants of adhesive binding specificity in type I cadherins. Proc Natl Acad Sci USA. 2014;111:E4175–E4184. doi: 10.1073/pnas.1416737111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams ME, Wilke SA, Daggett A, Davis E, Otto S, Ravi D, Ripley B, Bushong EA, Ellisman MH, Klein G, Ghosh A. Cadherin-9 regulates synapse-specific differentiation in the developing hippocampus. Neuron. 2011;71:640–655. doi: 10.1016/j.neuron.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.