Abstract
Extracellular vesicles (EVs) are diverse, nanoscale membrane vesicles actively released by cells. Similar sized vesicles can be further classified (e.g., exosomes, microvesicles) based on their biogenesis, size and biophysical properties. Although initially thought to be cellular debris, and thus under-appreciated, EVs are now increasingly recognized as important vehicles of intercellular communication and circulating biomarkers for disease diagnoses and prognosis. Despite their clinical potential, the lack of sensitive preparatory and analytical technologies for EVs poses a barrier to clinical translation. New analytical platforms including molecular ones are thus actively being developed to address these challenges. Recent advances in the field are expected to have far-reaching impact in both basic and translational studies. This article aims to present a comprehensive and critical overview of emerging analytical technologies for EV detection, and their clinical applications.
1. INTRODUCTION
Extracellular vesicles (EVs) are increasingly being recognized as promising circulating biomarkers of disease (“liquid biopsies") and have been shown to function as a means of long range intercellular communication in the body. EVs are heterogeneous, membrane-bound phospholipid vesicles actively secreted by a variety of mammalian cells, especially by dividing cancer cells but also host cells.1–4 Initially under-appreciated as “cell dust’” and a mechanism to dispose of cellular components,5 EVs are now considered abundant and stable sources of circulating biomarkers. Biofluids can contain large quantities of EVs that shuttle various molecules from parental cells to other cells, including proteins,6–8 mRNA / miRNA9, 10 and DNA11 thus serving as cellular surrogates.12, 13 In the cancer setting, EVs are often protumorigenic while certain host cells such as sinus capsular macrophages in lymph nodes remove tumor EV’s and are thus tumor suppressive.14
EVs offer significant advantages as cancer monitors.15, 16 Although tissue biopsies remain the gold standard for molecular evaluation, their invasiveness and limited sampling challenge clinical management, especially in the face of tumor spatial heterogeneity and temporal evolution.17 EVs, on the other hand, offer a minimally invasive avenue for accessing tumor molecular information, represent collective parameters of tumor cells, and can be repeatedly sampled for longitudinal monitoring. Specifically, tumor-associated vesicles can be used as effective surrogate biomarkers to define tumor type and stage18, 19 and molecular driver mutations, as well as to monitor treatment response.20–22
EVs are relatively new targets for bioassays, and possess unique physical and biological traits: they fall in a size range (typically 50–200 nm in diameter) much smaller than cells (10–30 µm), but larger than proteins, and comprise a highly heterogenous constituency.23, 24 These properties make it difficult to define and isolate EVs. Importantly, most conventional analytical tools, optimized for other biological objects, have limited sensitivity and throughput to be practical for EV research, let alone for clinical use.25–28 Active research is thus underway to overcome these challenges and to produce EV-optimized analytical systems.
This article reviews recent advances in EV research, ranging from EV biogenesis, assay methods, and their clinical potential across various diseases. We particularly focus on describing new technical advances for EV molecular characterization, in cross-comparison with conventional assay methods. For in-depth EV biology,3, 4, 29, biogenesis30, and applications as therapeutics,31, 32 we direct the readers to recent articles in these specific topics.
2. EV BIOGENESIS AND CONTENTS
Several routes exist for EV generation, although exact mechanisms are still largely unknown. Based on their biogenesis, EVs are currently classified into three broad groups: exosomes, microvesicles and apoptotic bodies (Table 1).3, 4, 33–36 In this review, we focus on exosomes and microvesicles which come from live cells.
Table 1.
Major types of extracellular particles.
| Vesicle | Size (nm) | Density (g/mL) | Origin | Markers |
|---|---|---|---|---|
| Exosomes | 40–200 | 1.13 – 1.18 | Endosomes | Tetraspanins, Alix, TSG101 |
| Microvesicles | 200 – 2000 | 1.16 – 1.19 | Plasma membrane | Integrins, selectins, CD40 |
| Apoptotic bodies | 500 – 2000 | 1.16 – 1.28 | Plasma membrane, endoplasmic reticulum | Phosphatidylserine, genomic DNA |
| High-density lipoprotein particles | 7–13 | >1.06 | Hepatocyte | Apolipoproteins, phospholipids, cholesterol, triglycerides |
| Low-density lipoprotein particles | 21–27 | 1.02 – 1.06 | Hepatocyte | Apolipoproteins, phospholipids, cholesterol, triglycerides |
2.1. Vesicle Formation
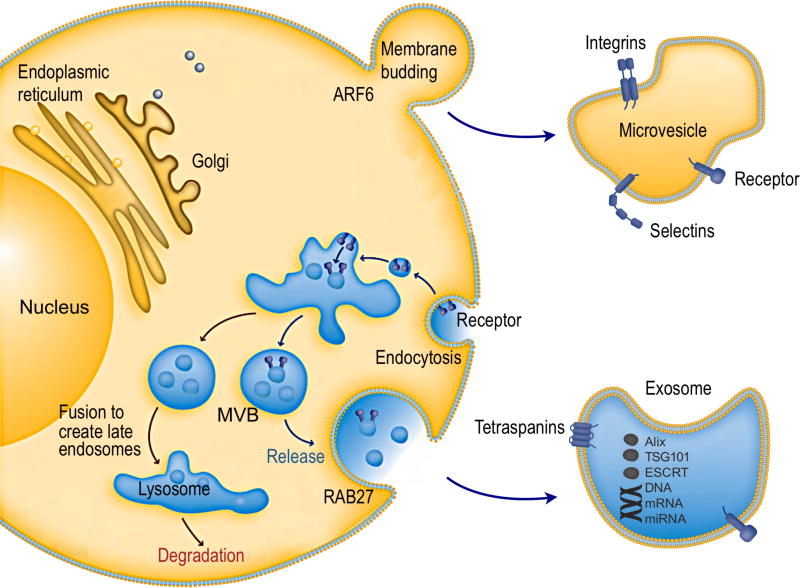
Exosomes are produced through the inward invagination of the endosomal membrane pathway.1, 37 First, the inward budding of cellular plasma membrane leads to the formation of an endosome (Figure 1).38 Small vesicles can be formed by further inward budding of the limiting membrane inside an endosome, leading to the formation of a multivesicular body (MVB), characterized by the presence of intraluminal vesicles.39 During this process, cytosolic contents, transmembrane and peripheral proteins are incorporated into the invaginating membrane.40 MVBs may then fuse with the lysosome, leading to the degradation of vesicular contents.41 Alternatively, MVBs may fuse with the plasma membrane of the cell, releasing vesicles in an exocytotic fashion to the extracellular space.4, 42 The released vesicles are considered as exosomes, small membrane-bound lipid vesicles that have a diameter ranging from 30 – 200 nm. Because of the double invagination processes, protein topology in exosomes is in the same orientation as in the plasma membrane of cells.
Figure 1. Intracellular pathways of EV biogenesis and secretion.
Cellular release of EVs occurs either through outward budding of plasma membrane (microvesicle pathway) or through the inward budding of endosomal membrane (exosome pathway). Exosomes are vesicles of endocytic origin. Following the inward invagination of the plasma membrane to form the early endosome, exosomes are formed as intraluminal vesicles due to further inward budding of the limiting membrane of endosome (now known as multivesicular body; MVB). Finally, exosomes are secreted by fusion of the MVB with the plasma membrane. Several machineries are involved regulating the cargo sorting and exocytosis of exosomes. Reprinted with permission from Ref 38. Copyright 2011 abcam.
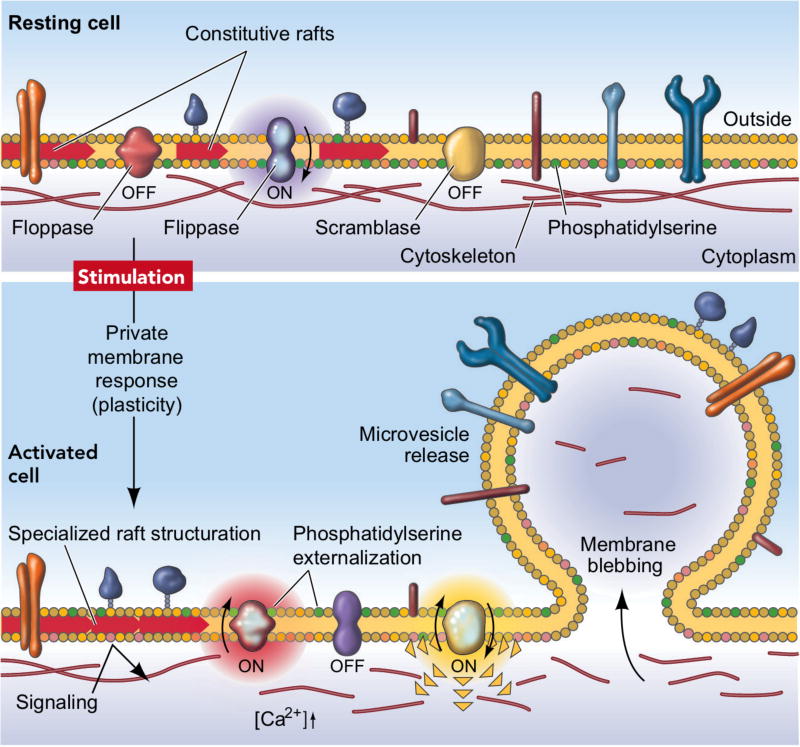
Microvesicles are derived from outward blebbing (budding) of the plasma membrane (Figure 2). The membrane bilayer maintains lipid “sideness” through asymmetric distribution of phospholipids; the outer layer is enriched with phosphatidylcholine and sphingomyelin, whereas the inner layer is predominantly formed with phosphatidylserine and phosphatidylethanolamine.43 Influx of cytosolic Ca2+, however, can disrupt this asymmetry by activating scramblase that promotes transbilyer lipid mixing. Such activation results in the redistribution of phospholipids across the membrane bilayer, promoting membrane blebbing.44 Ca2+-dependent proteolysis concomitantly degrades the membrane-associated cytoskeleton, facilitating the budding process. One reported mechanism for vesicle release is an actomyosin-based membrane abscission regulated by ARF6 activation.45 Microvesicles can also be formed through processes that parallel the virus budding from cells. Specifically, Nabhan et al. discovered that tumor susceptibility gene 101 (TSG101) interacts with a tetrapeptide PSAP motif of arrestin domain-containing protein 1 (ARRDC1). This interaction can result in the recruitment of TSG101 from the endosomes to the plasma membrane, and mediate the release of microvesicles containing TSG101, ARRDC1, and other cellular proteins.46
Figure 2. Formation of microvesicles.
The plasma membrane bilayer has an asymmetric distribution of phospholipids. The distribution is controlled by three major proteins: an inwarddirected pump, a flippase, specific for phosphatidylserine and phosphatidylethanolamine; an outward-directed pump, a floppase; and a lipid scramblase that promotes bidirectional redistribution of lipids across the bilayer. Following cell stimulation, a redistribution of lipids occurs, leading to microvesicle formation and its release. Reprinted with permission from Ref 44. Copyright 2005 Int. Union Physiol. Sci./Am. Physiol. Soc.
2.2. EV Molecular Contents
Collectively, EVs contain a trove of cellular cargos.12, 13, 47 Consistent with their biogenesis, the membrane composition of microvesicles reflects most closely the plasma membrane of the parent cells.24 In contrast, a specific subset of endosomal proteins has been identified in exosomes, suggesting a sorting mechanism during exosomal development. The endosomal sorting complex required for transport (ESCRT) has been extensively characterized for regulating and channeling specific molecules into the intraluminal vesicles of theMVBs.40, 48 The ESCRT, with it four main complexes (ESCRT 0, I, II, and III) is responsible for delivering ubiquitinated proteins for lysosomal degradation and protein recycling.49 Recent studies have shown that the depletion of specific ESCRT-family proteins can alter the protein content of exosomes and the rate of exosome release from cells.50 More interestingly, components of the ESCRT system, such as TSG101 and Alix,37 are found enriched in exosomes and thus are used as markers for exosome identification.26
The ESCRT is not the only mechanism mediating exosome formation; other ESCRT-independent processes also seem to participate, possibly in an intertwined manner, in their biogenesis and release. As such, exosomes are also enriched with molecules involved in ESCRT-independent mechanisms. For example, the tetraspanin proteins such as CD9, CD63 and CD81 have been shown to participate in endosomal vesicle trafficking.51, 52 The involvement of the Rab family of small GTPases in vesicle trafficking and fusion with the plasma membrane also suggests a role of these proteins in exosome release.53–55 In addition, sphingomyelinase has been demonstrated to be involved in vesicle release, as supported by elevated levels of ceramide in exosomes and a reduction in exosome release upon inhibition of sphigomyelinase.56
Both exosomes and microvesicles also contain nucleic acids include miRNAs, mRNAs,9, 10, DNA,11, 57 and other non-coding RNAs.58 Since the initial discovery that EVs contain RNAs,9 intense interest has been focused on using EV RNAs as diagnostic biomarkers. In a seminal work, Skog et al. found that serum exosomes of glioblastoma patients contain characteristic mutant mRNA (EGFRvIII mRNA) and miRNAs that could be used to provide diagnostic information.10 These nucleic acid discoveries led to the hypothesis that EVs can transfer genetic information between cells. Indeed, both Vakadi et al. and Skog et al. showed that EVs contain mRNA that can be transferred and translated after entering host cells.9, 10 Retrotransposons and other non-coding RNAs have also been reported in EVs.11, 58, 59 Transfer of retrotransposon sequences and miRNAs, as well as translatable mRNAs occurs via EVs.11, 58, 60 These findings and others highlight the importance of EVs as carriers and transmitters of genetic information.61,62
3. PHYSICAL CHARACTERIZATION
Microscopic methods are widely used to measure the physical features of EVs, such as vesicle size and distribution, concentration, and morphologies. This section briefly surveys these techniques, and discusses unmet needs to standardize EV characterization protocols.25, 26, 63
3.1. Microscopy based Methods
Conventional optical microscopies have a diffraction limit close to that of EV size, and are unable to generate clear images of these vesicles.64 High resolution EV images are thus produced via electron microscopy (EM) or atomic force microscopy (AFM). These methods, however, have limited throughput as specialized staining protocols and equipments are necessary.
3.1.1. Scanning Electron Microscopy
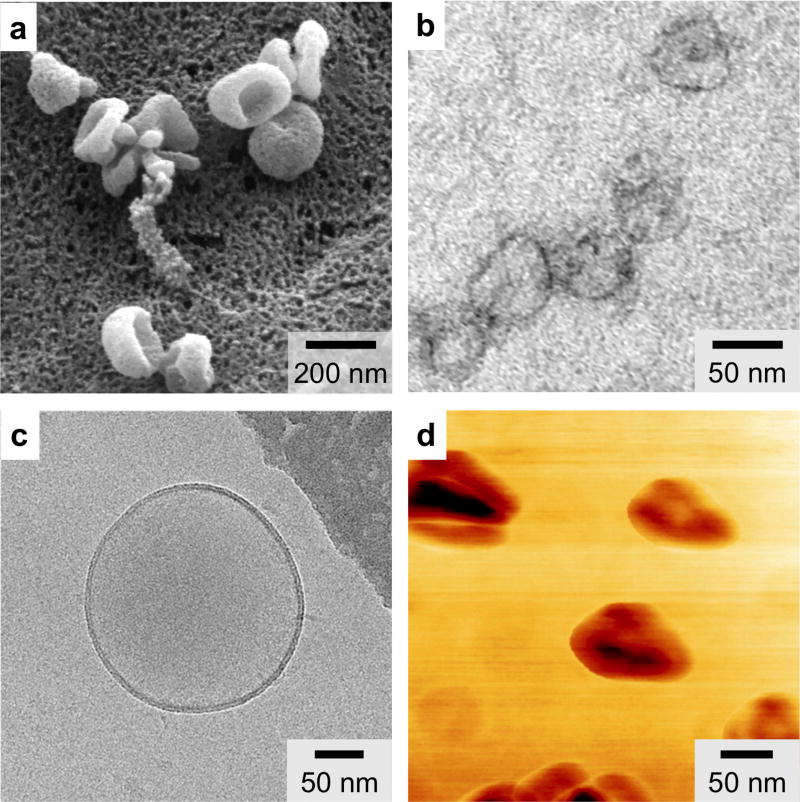
Scanning electron microscopy (SEM) is a well established and useful technique in EV research.10, 20, 65 SEM produces images of a EV sample by scanning the surface with a focused beam of electrons; the electrons interact with the atoms in the sample to produce various deducible signals that provide three-dimensional surface topography information as well as elemental composition of the sample. As the vast majority of SEM studies on EVs are performed under vacuum, the samples are typically fixed and dehydrated. Under SEM, EVs present a distorted cup-shaped morphology66 and uniform unimodal size distribution following 0.2 µm filtration (Figure 3a).20
Figure 3. Various micrographs of EVs.
(a) Scanning electron microscopy (SEM) provides three dimensional surface topology information. (b) Transmission electron microscopy (TEM) has superior image resolution and can be used with immunogold labeling to provide molecular characterization. (c) Cryo-electron microscopy (cryo-EM) enables analysis of EV morphology without extensive processing. (d) Atomic force microscopy (AFM) can provide information on both surface topology and local material properties (e.g., stiffness, adhesion). Reprinted with permission from Ref 20. Copyright 2012 Nature Publishing Group. Reprinted with permission from Ref 70. Copyright 2013 Yuana et al. Reprinted with permission from Ref 71. Copyright 2010 American Chemical Society.
3.1.2. Transmission Electron Microscopy
Transmission electron microscopy (TEM) is another popular technique for characterizing EVs.66 In TEM, a focused beam of electron is transmitted through a thin specimen to create a sample image. TEM has superior resolution, with capabilities to image <1 nm objects. In addition, heavy metal stains such as osmium tetroxide and uranyl acetate can be used to generate contrasts in lipid membrane (Figure 3b).20 Moreover, TEM can also be coupled with immunogold labeling (immuno-EM)67, 68 to provide molecular characterization.
3.1.3. Cryo-Electron Microscopy
Cyro-electron microscopy (cryo-EM) is a form of EM where samples are analyzed at very low temperature (e.g., −100 °C).69, 70 Unlike SEM or TEM that requires extensive sample fixation and staining, cryo-EM enables the analysis of EVs in frozen samples with the advantages of avoiding the effects of dehydration and chemical fixatives. For example, under cryo-EM, EVs exhibit a round morphology (Figure 3c), suggesting that the cup-shaped structures of EVs visible in other EM techniques is an artifact from dehydration.70
3.1.4. Atomic Force Microscopy
Atomic force microscopy (AFM) is another high-resolution imaging technique for EV characterization.69, 71, 72 In AFM, a mechanical cantilever is passed over a surface, with deflection indicating the presence and topography of surface structures. AFM does not require extensive sample preparation; in fact, EV samples can be adsorbed onto a mica holder and imaged after gentle drying. AFM can be operated to provide two types of information: 1) amplitude modulation detects changes in the cantilever’s vibration amplitude to inform about the surface topography, and 2) phase modulation records energy dissipation to provide information about local stiffness and adhesion properties. Under AFM, topographic AFM shows a round morphology of isolated EVs while phase images show substructures (Figure 3d), indicating variable constitutive elements (e.g., lipid, protein) making up these structures.71
3.2. Dynamic Light Scattering
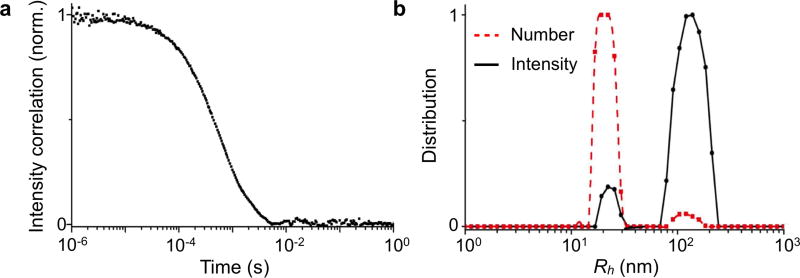
Dynamic light scattering (DLS) is an attractive technique for measuring multiple physical attributes of EVs in suspension.73–75 DLS measures the bulk scattered light from EVs when they are illuminated by a monochromatic light source. As the particles undergo Brownian motion, the scattered light from all particles interfere (constructively and destructively) and the intensity fluctuates over time. The dynamic information of the particles is derived from an autocorrelation of the intensity trace recorded during the experiment (Figure 4a).75 The fluctuation rate can be converted into the diffusivity of the particles for determining the hydrodynamic diameter (i.e., the effective particle size in fluid, Rh) through the Strokes-Einstein equation. When applied for EV analysis, the data should be carefully inspected. The original size distribution measured by DLS is intensity-weighted, proportional to Rh6, and therefore can be dominated by the presence of larger vesicles, albeit in small quantities. To spot such distortion, the number distribution, mathematically derived from the intensity distribution, should be compared as well for consistency (Figure 4b).75
Figure 4. Dynamic light scattering.
(a) Dynamic light scattering (DLS) measures bulk scattered light from EVs as the vesicles undergo continuous Brownian motion. The dynamic information of the vesicles is derived from an autocorrelation of the scattered intensity and could be used to determine vesicle size. (b) As the original size distribution measured by DLS is intensity-weighted, the data is dominated by large vesicles, even if these exist in small quantities, as the intensity is proportional to Rh6, where Rh is the effective vesicle size. Reprinted with permission from Ref 75. Copyright 2015 American Chemical Society.
3.3. Nanoparticle Tracking Analysis
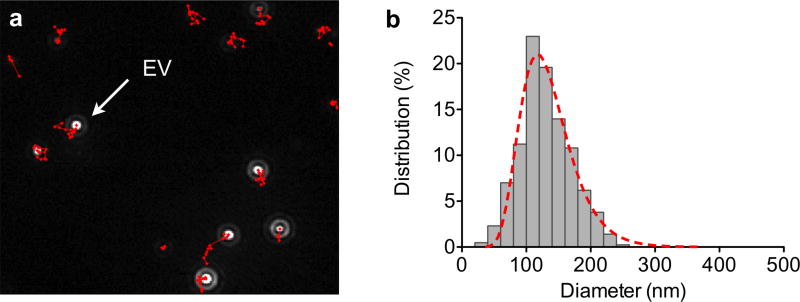
Nanoparticle tracking analysis (NTA) is an optical particle tracking method developed to determine concentration and size distribution of particles.65, 76, 77 A light beam is used to illuminate the particles in the sample. As the particles scatter light and undergo Brownian motion, a camera records the path of each particle to determine the mean velocity and diffusivity. Unlike bulk scattering measurements of DLS, NTA tracks individual particle scattering (Figure 5a).20 This information is then used to mathematically calculate the concentration (i.e., number of particles in the field of view) and size distribution (i.e., hydrodynamic diameter through the Strokes-Einstein equation, Figure 5b). For accurate quantification of concentration and size of heterogeneous populations of vesicles, the NTA procedure, however, requires accurate optimization of camera and analysis settings. Separate measurements with different settings may be needed to obtain accurate readings for EV subsets in heterogeneous mixtures.
Figure 5. Nanoparticle tracking analysis.
(a) Nanoparticle tracking analysis (NTA) tracks individual vesicle scattering over time, as they diffuse and scatter under light illumination. (b) This information is then used to mathematically determine vesicle concentration and size distribution. Reprinted with permission from Ref 20. Copyright 2012 Nature Publishing Group.
3.4. Tunable Resistive Pulse Sensing
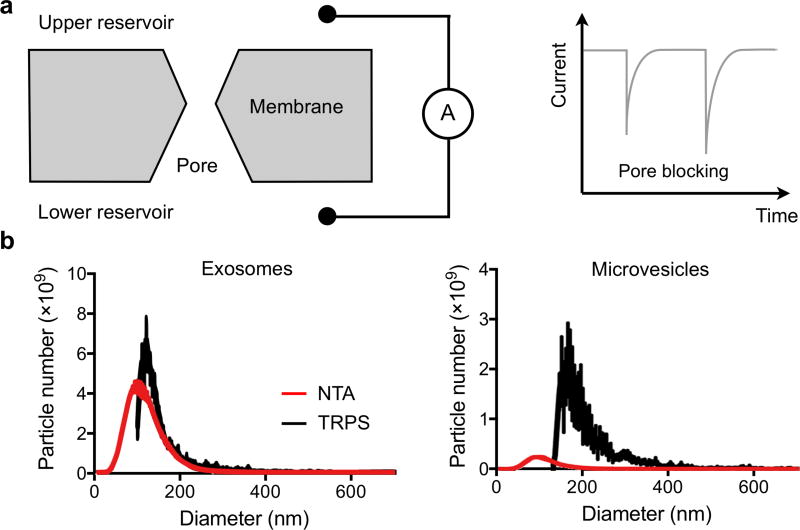
Tunable resistive pulse sensing (TRPS) can be an alternative to NTA for measuring EV concentration and size distribution.78–81 The technology is based on the Coulter principle at the nanoscale. It detects transient changes in the ionic current, generated by the transport of the vesicles through a size-tunable nanopore in a polyurethane membrane. Recently, Akers et al. compared the performance of NTA and TRPS in measuring exosomes and microvesicles derived from patient cerebrospinal fluids (Figure 6).80 The authors noted that for EVs < 150 nm in diameter, NTA consistently detected more EVs than TRPS. The reverse was true for larger EVs (>150 nm). This discrepancy indicates differences in the sensing mechanisms and suggests the need for synergistic multi-platform characterization.80
Figure 6. Tunable resistive pulse sensing (TRPS).
(a) Two fluidic reservoirs, each connected to an electrode, are separated by a membrane with a pore. The ionic current between reservoirs is then measured. When a vesicle passes through the pore, it blocks the current flow, leading to a transient current decrease. (b) Exosomes and microvesicles derived from patient cerebrospinal fluids were compared. For EVs < 150 nm in diameter, NTA consistently detected more EVs than TRPS. The reverse was true for bigger EVs (>150 nm). Reprinted with permission from Ref 80. Copyright 2016 Akers et al.
3.5. Single EV Analysis(SEA) Method
In a recent paper, a light microscopic single EV analysis (SEA) technique was described that is capable of robust, multiplexed protein biomarker measurement in individual vesicles.82 In this approach, EVs are immobilized inside a microfluidic chamber, immuno-stained, and imaged. With the vesicles immobilized on the chip surface, the achievable signal-to-noise ratio from each vesicle is generally much higher than that when the vesicles are free-floating in solution or under flow condition.
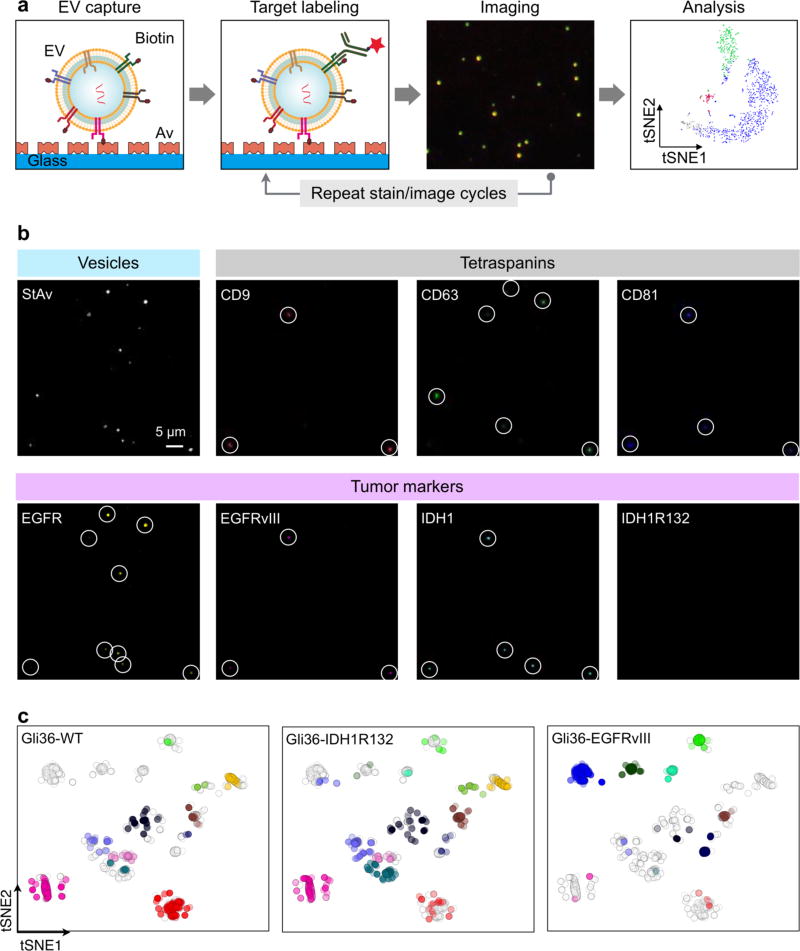
The authors further adapted the image cycling process, previously used for multiplexed cyclic cell and tissue analyses, to complement the nanoscale dimension of EVs. (Figure 7a).83, 84 Specifically, the authors tested the method using EVs derived from three isogenic glioblastoma multiforme (GBM) cell lines. EVs were stained for three protein markers at a time, and subjected to four rounds of cyclic imaging. The results showed highly heterogeneous profiles of biomarkers among EV populations (Figure 7b). Multi-dimensional data analysis, t-distributed stochastic neighbor embedding (tSNE), was then used to visualize the data; unsupervised clustering revealed the presence of potential EV subpopulations (Figure 7c). The SEA technology could be a powerful tool for studying various EV types, producing rich data sets on heterogeneity of biomarker expression, marker make-ups, and the presence of EV subpopulations.
Figure 7. Single EV analysis (SEA).
(a) EVs are biotinylated and captured on a flat surface coated with neutravidin (Av). EVs are then stained with fluorescent antibodies and imaged. Subsequently, fluorophores are quenched and the staining process is repeated for a different set of markers. (b) Example SEA image. EVs from Gli36-WT cell line were biotinylated and captured. Individual EV were detected through staining with fluorescent streptavidin StAv (top left). For molecular profiling, EVs were stained for pan-EV markers (tetraspanins; CD9, CD63, CD81) as well as tumor markers (EGFR, EGFRvIII, IDH1, IDH1R132). Spots with circles indicate individual EVs. (c) 2-dimensional tSNE mapping of individual EVs analyzed for protein markers. The original map was redrawn for EVs from a single cell line. Data from other cell lines are shaded light gray. EVs from Gli36-WT and Gli36-IDH1R132 cells lines clustered similarly, whereas EVs from Gli36-EGFRvIII cells showed distinct clusters. Reprinted with permission from Ref 82. Copyright 2017 American Chemical Society.
4. EV ENRICHMENT
EVs are heterogeneous in size, origin and molecular constituents. Aside from their intrinsic heterogeneity, they are also present in different complex biological fluids including blood,85, 86 plural effusion,87, 88 ascites,89, 90 breast milk,91, 92 saliva,93 cerebral spinal fluid,94 and urine.95 These biofluids contain varying amounts of non-vesicular macromolecular structures (e.g., RNA complexes) which could interfere with analytical results. EV isolation and enrichment is thus considered as a necessary pre-analytical requirement for biomedical investigation as well as clinical translation.25, 96 A variety of different isolation and enrichment methods have been developed; these all influence the amount, type and purity of EVs recovered. They include the “standard” ultracentrifugation, sucrose-gradient centrifugation and polymer-based precipitation. Newer fluidics systems are also being developed to enhance enrichment efficiency and to make the processes faster.
4.1. High-Throughput Bulk Methods
According to an online questionnaire drafted and distributed by ISEV in October 2015,96 bulk methods such as ultracentrifugation (81%) and density-gradient centrifugation (20%) were the most widely used primary isolation methods. Based on the principle of their separation mechanisms, these methodologies could be grouped into three major classes: density, affinity, and size. Here, we summarize the common isolation approaches with their advantages and limitations (Table 2).
Table 2.
Comparison of high-throughput bulk EV preparation methods.
| Platform | Mechanism of enrichment |
Advantages | Limitations |
|---|---|---|---|
| Ultracentrifugation | Density |
|
|
| Sucrose-gradient centrifugation | Density |
|
|
| Co-precipitation | Surface charge |
|
|
| Size-exclusion chromatography | Size and molecular weight |
|
|
| Field flow fractionation | Size and molecular weight |
|
|
4.1.1. Ultracentrifugation
Ultracentrifugation is the most commonly used conventional approach for EV isolation.96 Particles are separated with differential centrifugal forces: cellular debris are removed at a low centrifugal force (300 ×g) while a high force (100,000 ×g) is used to sediment and concentrate EVs.9, 10 Despite being the most widely used gold standard, the approach has many drawbacks such as bulky and costly instrumentation, lengthy and laborious processing, contamination with aggregated proteins and ribonuclear protein particles, and the requirement for large amount of samples. and the requirement for large amount of samples.
4.1.2. Gradient Ultracentrifugation
A more stringent form of ultracentrifugation, sucrose-gradient centrifugation helps to further fractionate different vesicular density and is typically applied to isolate exosomes,66, 97 which have been found to float at densities ranging from 1.15 to 1.19 g/mL. In this approach, a sample containing differently-sized vesicles and macromolecules is layered on the surface of a gradient whose density increases from top to bottom. During centrifugation, different molecules sediment through the gradient at different rates. Due to its resolution for further density fractionation, the approach is considered to yield EVs (specifically exosomes) at a higher purity; however, it faces many limitations associated with ultracentrifugation. Aside from sucrose gradient, newer isosmotic gradients (e.g., iodixanol gradient) are being adopted to maintain better biophysical properties of vesicles.98
4.1.3. Co-Precipitation
Recently, commercial kits (e.g., ExoQuick, Exo-Spin) that rely on polymer co-precipitation strategies have been developed for EV enrichment. These reagents typically reduce the hydration of EVs (and thus solubility) to cause precipitation. The precipitated EV products could be easily and reproducibly isolated with low centrifugal forces, bypassing the need for lengthy ultracentrifugation.99, 100 However, these kits are expensive for large-scale usage and lack specificity for EVs. The method also tends to produce heterogeneous polymeric particles. As the reagents decrease the solubility of EVs and proteins equally, the approach also co-precipitates lipoproteins and Ago-2 RNA complexes. The utility of co-precipitation is thus limited as a standalone EV isolation method.
4.1.4. Size-Exclusion Chromatography
Size exclusion chromatography separates vesicles and other molecules based on their size by filtration through a gel.101, 102 The gel consists of spherical beads which contain pores of a specific size distribution. When the sample enters the gel, small molecules diffuse into the pores while large molecules are eluted directly. Consequently, larger molecules exit the column earlier than small molecules, which makes it possible to correlate the dwell-time of molecules with its size. This separation method has been recently applied to vesicle isolation to yield purified EVs from complex biological media.102–105 Commercial columns (e.g., Sepharose, GE Healthcare; qEV, iZon) are also being developed to simplify EV isolation. These size exclusion columns contain a resin of approximately 75 nm pore size. The passage of proteins and other smaller contaminating molecules are delayed, while larger vesicles (> 75 nm) could rapidly pass through and be eluted in the void volume. Size exclusion can separate EVs from soluble proteins; several factors including media types, pore size, interactions between EVs and the media, column dimension, column packing, as well as flow rate, should be considered to improve the efficiency and resolution of the separation.
4.1.5. Field Flow Fractionation
Field flow fractionation is another separation technique where a force field is applied perpendicular to a sample flow, to enable separation based on different size and molecular weight. Recently, an asymmetric flow field flow fractionation (AF4) has been applied for EV isolation.75, 106 AF4 contains a permeable plate at the channel boundary. As the sample flows in the channel, a parabolic velocity profile is created due to the laminar flow: the fluid moves slower at the boundary than it does at the center of the flow. When the perpendicular force field is applied, analytes in the sample are driven towards the boundary. Brownian motion creates a counteracting motion such that smaller particles tend to reach an equilibrium position further away from the boundary. This type of separation spans a broad size range and could be applied to a wide variety of eluents.
4.2. New Enrichment Methods
Various new EV enrichment methods have been developed to improve the isolation efficiency and specificity form complex biological fluids. Compared to conventional methods, most of these new methods, however, have lower throughput, which should be addressed to become practical. We summarize some of the recent developments.
4.2.1. Microfluidic Filtering
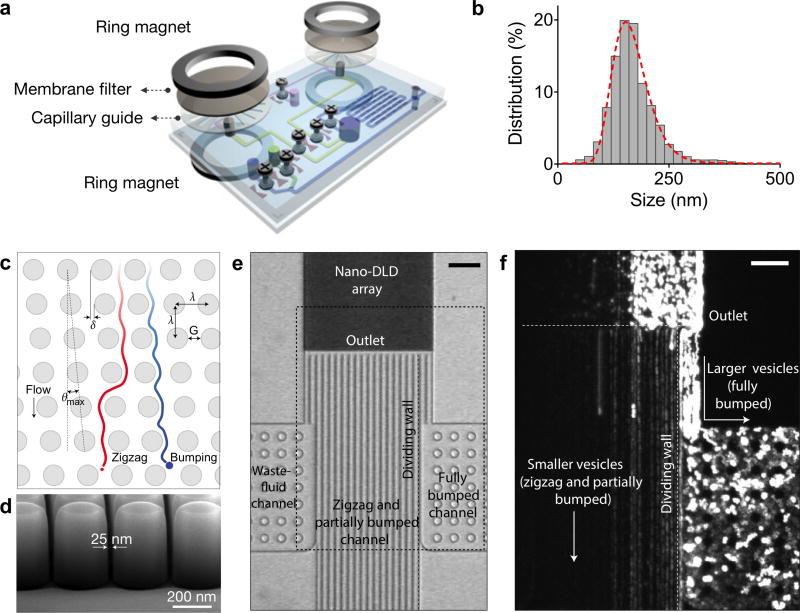
Size-based isolation represents an attractive approach to separate EVs from large cellular debris. A variety of microfluidic filter systems have been developed to isolate EVs from large cellular debris and protein aggregates, mostly based on size differentiation. For example, Rho et al. built a microfluidic device that uses membrane filters to size-selectively isolate EVs from unprocessed blood samples (Figure 8a).107 The size cutoff for the membrane filter is ~1 µm. A capillary layer, inserted underneath the membrane, is used to guide the filtered EVs towards the collection channel. The membrane filter and capillary guide are sandwiched between two ring magnets; this setup enables the filter set to be easily replaced when large volumes of samples are processed. This system can filter 300 µL of packed red blood cell samples in less than 10 min. The purified EVs revealed a single population with an average size of 167 nm (Figure 8b).
Figure 8. Microfluidic filtering methods for EV isolation and sorting.
(a) A microfluidic device that uses membrane filters to isolate EVs from unprocessed blood sample. The device consists of size-selective filters (< 1 µm) and capillary guide, and is assembled by a magnetic sandwich. (b) Filtered blood sample revealed a single EV population with an average size of 167 nm. (c) A nanoscale lateral displacement array that sorts differentially-sized vesicles through displacement trajectories. (d) The device was fabricated with advanced silicon processes to produce an pillar array with uniform gap size of 25 nm. (e) Due to the differential vesicle trajectories, larger vesicles would be displaced to the right channel (fully bumped) while small vesicles followed a zigzag path. (f) When sorted in the device, fluorescent-labelled human urine-derived EVs confirmed the differential displacement trajectories. Reprinted with permission from Ref 107. Copyright 2013 American Chemical Society. Reprinted with permission from Ref 108. Copyright 2016 Nature Publishing Group.
Recently, Wunsch et al. demonstrated a nanoscale lateral displacement (DLD) array to sort, separate and enrich EVs.108 The critical design and nature of particle trajectories are shown in Figure 8c. At the micrometer scale, particles with diameter smaller than the critical diameter (Dc) will be displaced laterally across the array in a bumping mode, while particles with diameter larger than Dc will follow the laminar flow in a zigzag mode. The authors used silicon processes to produce nansocale DLD arrays of uniform gap size ranging from 25 to 235 nm to enable vesicle separation (Figure 8d). In operation, larger vesicles are displaced by the nano-DLD array to the right side of the channel and are collected at a side channel, while smaller vesicles will follow a zigzag mode and flow out of the array into a large channel (Figure 8e). The authors further applied the DLD array to separate fluorescent labelled human-urine-derived EVs into two fractions: fully bumped, and zigzag and partially bumped (Figure 8f). Most EVs exhibited a partial bumping mode in the exosome displacement distribution, in agreement with their size distribution (60–70 nm).108 The device, however, required high pressure due to the dense array structure, and had a limited throughout.
4.2.2. Contact-Free Sorting
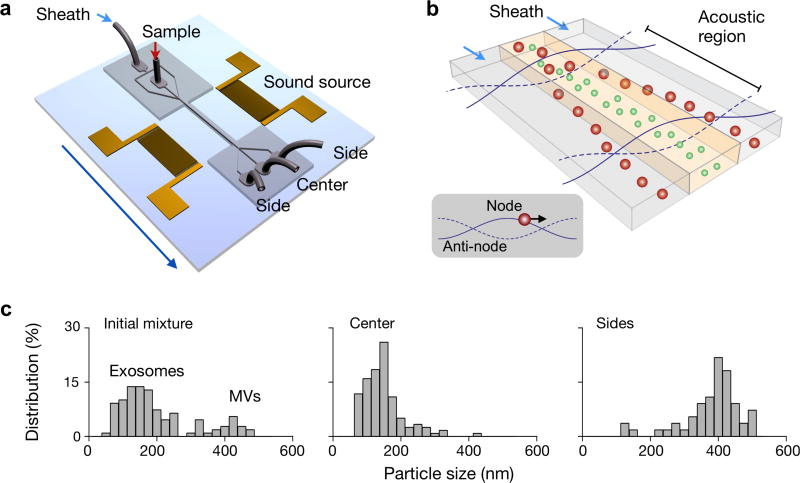
Lee et al. recently used acoustic waves to fractionate EVs in a contact-free manner. The separation uses ultrasound standing waves to exert differential acoustic force on vesicles according to their size and density.109 The device consists of a pair of interdigitated transducer (IDT) electrodes to generate a standing surface acoustic wave across the flow channel (Figure 9a). The operation principle is shown in Figure 9b. Particles in an acoustic region experience radiation pressure and migrate toward the pressure nodes. Larger vesicles move faster as the acoustic force is proportional to the vesicle volume. Moreover, the filtering cut-off size could be controlled electronically in situ, to enable on-demand versatile size selection. By optimizing the design of the ultrasound transducers and underlying electronics, the authors were able to achieve a high separation yield to isolate nanoscale vesicles from other types of EVs (Figure 9c). With the capacity for rapid and contact-free EV isolation, the developed system could become a versatile preparatory tool for EV analyses.
Figure 9. Contact-free sorting of EVs.
(a) Schematic of the acoustic wave sorter. The device consists of a pair of interdigitated transducers to generate standing ultrasound wave to exert differential forces on vesicles of different sizes. (b) During operation, vesicles in an acoustic region experience radiation pressure that is proportional to the vesicle size and move towards the pressure node. Larger vesicles move faster than smaller vesicle, thereby forming differential separation trajectories. (c) By in situ tuning of cut-off size, vesicles could be separated with versatile size selection and a good separation yield. Reprinted with permission from Ref 109. Copyright 2015 American Chemical Society.
4.2.3. Immunoaffinity Enrichment
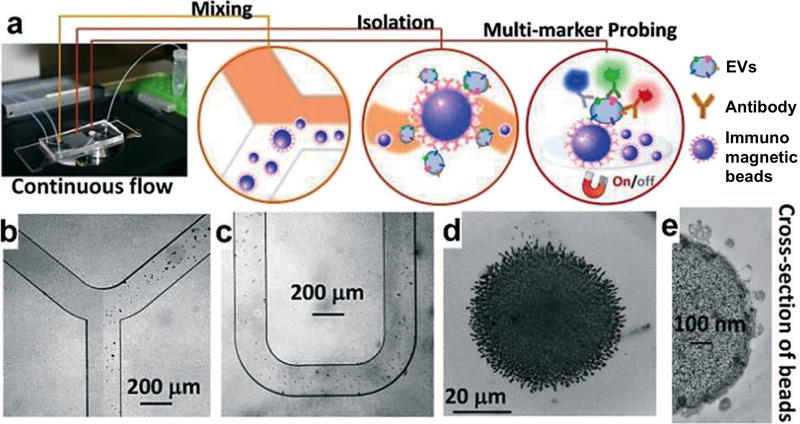
Due to its simplicity, immunoaffinity capture is an attractive approach for point-of-care applications.22, 110–113 In this approach, EVs are captured with specific antibodies against common EV markers including the tetraspanins (e.g., CD9, CD63, CD81) as well as tumor-associated markers (e.g., EGFR, EpCAM). Various microfluidic devices have been implemented for selective isolation of EVs. Zhao et al. recently developed a microfluidic ExoSearch chip that enables continuous mixing and isolation of EVs using immunomagnetic beads.112 As shown in Figure 10, the chip consists of a Y-shaped injector and a serpentine fluidic mixer for incubation and binding of EVs with immunomagnetic beads. Magnetic beads with bound EVs can then be retained as tight aggregates by magnetic forces for downstream optical detection. The approach can be readily applied in point-of-care settings for small scale enrichment, as they provide a fast and easy approach for chip-integration and detection. However, these methods are primarily marker-dependent and thus tend to underestimate counts. The strong affinity of antibody-antigen binding also makes it challenging to dissociate captured EVs for subsequent functional analyses.
Figure 10. Immunoaffinity enrichment.
(a) Schematics of a microfluidic chip that enables continuous mixing and isolation of EVs using immunomagnetic beads. EVs are enriched by immunomagnetic selection and retained as tight aggregates by magnetic force. The retained clusters could be subsequently probed with secondary markers for optical detection. Microscopy images of the device: (b) Y-shaped injector, (c) serpentine fluidic mixer for immunomagnetic binding, (d) magnetic aggregates and (e) bound EVs on immunomagnetic beads. Reprinted with permission from Ref 112. Copyright 2016 The Royal Society of Chemistry.
5. EV PROTEIN ANALYSIS
5.1. Proteins Enriched in EVs
EV proteins derive mainly from cellular plasma membrane, cytosol, but not from other intracellular organelles (e.g., Golgi apparatus, endoplasmic reticulum, and nucleus).7, 15, 47 This protein constitution of EV is indicative of vesicle biogenesis and cargo sorting.98 The International Society for Extracellular Vesicles therefore recommends careful characterization of EV proteins, especially transmembrane proteins and cytosolic proteins.26
5.1.1. Membrane Proteins
In mammalian vesicles, both transmembrane and lipid-bound extracellular proteins (e.g., lactadherin) are found associated with microvesicles and exosomes.26 Within the group of transmembrane proteins, exosomes are enriched with tetraspanins (e.g., CD9, CD63, CD81), a superfamily of proteins with four transmembrane domains.51, 52 Tetraspanins are involved in membrane trafficking and biosynthetic maturation,114, 115 and thereby are highly expressed in exosomes; this property has led to the use of tetraspanins for exosome quantification and characterization. It should be noted, however, that tetraspanins are not uniquely expressed in exosomes alone.26 Microvesicles, on the other hand, are enriched with integrins, selectins and CD40 ligands. Reflecting their derivation from the plasma membrane of cells, EVs are enriched with specific transmembrane protein receptors (e.g., epidermal growth factor receptors/EGFRs6, 116) and adhesion proteins (e.g., epithelial cell adhesion molecule/EpCAM21, 117). As many of these transmembrane proteins are involved in normal physiology and disease pathogenesis, they are used as important pathophysiological EV biomarkers.
5.1.2. Intravesicular Proteins
EV-associated intravesicular proteins have diverse functions. They include cytosolic proteins that have membrane- or receptor binding capacity, such as TSG101, ALIX, annexins and Rabs, which are involved in vesicle trafficking. EVs are also enriched with cytoskeletal proteins (e.g., actins, myosins, tubulins), molecular chaperones (e.g., heat-shock proteins/HSPs), metabolic enzymes (e.g., enolases, glyceraldehyde 3-phosphate dehydrogenase/GAPDH) and ribosomal proteins.26, 47 Interestingly, recently studies have identified that EV protein cargoes can be effectively transported to and received by recipient cells to elicit potent cellular responses in vitro and in vivo.61, 118 This introduces new opportunities of utilizing EVs as therapeutics and drug carriers.32
5.2. Conventional Protein Analyses
EV protein quantification and characterization is important not only for shedding light on EV biogensis and cargo sorting,4, 37 but also for identifying physiological and pathological markers.119, 120 Conventional protein analyses, including western blotting and enzyme-linked immunosorbent assay (ELISA), however, typically require a large sample volume, extensive processing and/or bulky, specialized instrumentation.25 These procedures are thus less suitable for clinical uses, especially for studies that involve a large patient cohort or quantification of rare molecular markers.
5.2.1. Western Blotting and ELISA
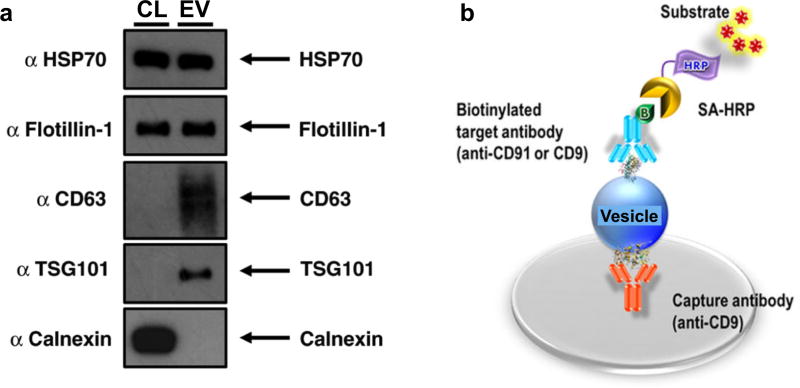
Western blotting, also known as immunoblotting, is a common protein analytical technique employed in many molecular biology disciplines. In EV protein evaluation, western blotting is arguably the most commonly used technique for demonstrating the presence of target proteins reportedly associated with EVs. In this process, purified vesicle preparations (discussed in Section 2, typically prepared through current gold standard of gradient ultracentrifugation) may be treated with buffered lysis solutions which contain denaturants and protease inhibitors. The protein lysates are then separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), before being transferred over to a membrane for immunoblotting of specific protein targets (Figure 11a).121 While the approach has a significant preparatory and processing time (>10 hrs), western blotting can provide useful information on the size of the different proteins.
Figure 11. Conventional EV protein analysis.
(a) Western blotting. EV protein lysate is separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), before being transferred over to a membrane for immunoblotting of specific EV protein targets (e.g., HSP70, Flotillin-1, CD61). (b) Enzyme-linked immunosorbent assay (ELISA). In the specific “sandwich” configuration, vesicles or lysates could be applied to a solid support that has been pre-treated with an immobilized capturing antibody. Captured vesicle targets are then exposed to an detecting target antibody. Reprinted with permission from Ref 121. Copyright 2015 Lobb et al. Reprinted with permission from Ref 122. Copyright 2014 Nature Publishing Group.
As in western blotting, enzyme-linked immunosorbent assay (ELISA) is another established technique for protein quantification and could be executed in multiple different assay formats. In the specific “sandwich” configuration, purified vesicle preparations or vesicle lysates could be applied directly to a solid support that has been pre-treated with an immobilized capturing antibody; captured vesicular targets are then exposed to another detecting antibody (Figure 11b).122 This requirement for a pair of non-interacting antibodies improves detection specificity but makes it difficult for developing new assays and executing simultaneous, multiplexed measurements. Both western blotting and ELISA have similar limits of detection. ELISA, however, can be significantly faster than western blotting, and be scaled up for higher throughput measurements.
5.2.2. Mass Spectrometry
Unlike western blotting and ELISA which quantify targeted proteins in a relatively small scale, mass spectrometry enables high throughput peptide profiling.123, 124 Purified EV preparations undergo enzymatic digestion and peptide separation before being ionized and analyzed by mass spectrometer. Along this complex processing, multiple steps critically affect EV proteomic profiling. Aside from effective EV purification (discussed in Section 3), peptide fractionation prior to mass spectrometry analysis is considered an important prerequisite for identifying vesicular proteins with high confidence. This is commonly achieved through three main approaches: 1) SDS-PAGE,125, 126 2) two-dimensional liquid chromatography,127 and 3) isoelectric-focusing based fractionation.128
It is worth noting that since mass spectrometric analysis identifies digested peptide fragments, proper protein identification, quantitation and validation is necessary. Two technical methodologies have been developed for quantitation: label-based and label-free.47 In label-based quantitation, tags (isobaric or isotopic) are used in comparative analyses.129 In label-free quantitation, spectral counting of chromatogram intensity could be applied.130 Identified protein candidates can then be validated using other conventional protein technologies such as western blotting. In terms of detection sensitivity, mass spectrometry methods are typically not as sensitive as antibody-based technologies.
While mass spectrometry requires significant preparatory and processing time (days), it can provide high-throughput, quantitative and comparative EV proteomic analyses. To date, several thousands of vesicular proteins have been catalogued for systematic, protein-protein interaction analyses.131, 132 Detailed discussions of mass spectrometry-based proteomic analyses of mammalian47, 123, 124 and bacterial133, 134 EVs have been highlighted in several reviews. Such network and interaction studies could shed light on functional activities of EV cargos and their important roles in mediating long-distance intercellular communication.
5.3. New Protein Analyses
To address the technical challenges associated with protein quantification in EVs, new generations of biosensors are under development. In comparison to conventional protein detection methods, these biosensors leverage on distinct sensing mechanisms and are designed specifically to detect a wide range of EVs of varying sizes and molecular contents (Table 3). Many of these technologies also require significantly smaller sample volumes and minimal sample processing, and are thus well-suited for point-of-care biomedical applications.
Table 3.
Comparison of new technologies for EV protein detection.
| Platform | Sensing mechanism |
Detection Limit |
Advantages | Limitations |
|---|---|---|---|---|
| Bead-based flow cytometry | EVs captured on beads and fluorescently labeled | - |
|
|
| Small particle flow cytometry | Scattering and fluorescence | - |
|
|
| ExoScreen | Photosensitizer beads | - |
|
|
| MicroNMR | NMR | ~ 104 EVs |
|
|
| nPLEX | SPR | ~ 103 EVs |
|
|
| iMEX | Electrochemical | ~ 104 EVs |
|
|
5.3.1. Small Particle Flow Cytometry
Flow cytometry is a powerful technique for characterizing single large particles, e.g., cells or larger micrometer-size entity, based on light scattering and fluorescence activation; however, conventional flow cytometry has limited sensitivity and resolution to detect small particles that have a diameter < 500 nm.135 In addition, it also suffers from a high optical background due to the presence of small particles (~ 200 nm) in sheath fluids. When conventional flow cytometry is applied for EV quantification, a significant number of small EVs may be missed, or if they are detected they are underestimated, as multiple small vesicles need to be simultaneously illuminated to trigger a count and are thus counted as a single event. This latter phenomenon is described as the “Swarm Theory”.136
To adapt conventional flow cytometry for EV profiling, micrometer-sized latex beads have been used to bind to multiple vesicles. The bound EVs are then stained with fluorescent antibodies and characterized for their protein markers.137, 138 However, this approach lacks single vesicle profiling capabilities and does not differentiate between different vesicular subsets, which may result in the loss of distinctive signatures.
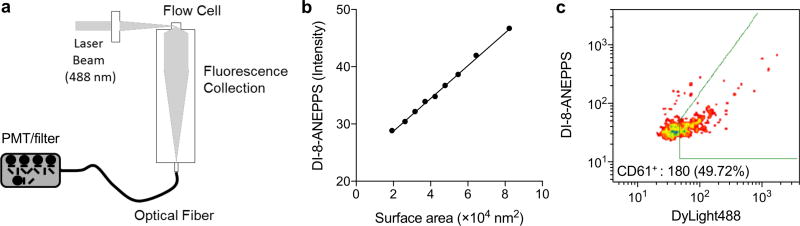
To address this challenge, highly sensitive flow cytometry instruments are under development, that can discriminate particles as small as 100 nm in diameter.139–142 For example, Stoner et al. recently developed a highly sensitive flow cytometer and EV measurement approaches to enumerate, size, as well as molecularly characterize individual EVs.140 The developed system, termed vesicle flow cytometry, systematically incorporates and optimizes laser excitation, laser beam shaping optics, flow cell, forward angle obscuration bar, orthogonal collection optics, and optical relay fibers, and fluorescence detector (Figure 12a). Employing the system, the author evaluated different fluorescent probes and labeling protocols for their effectiveness in staining the vesicle membrane and surface markers, and compared the fluorescence intensity distributions of vesicles to their diameter distributions. Interestingly, the authors found that the voltage sensing dye di-8ANEPPS could produce vesicle fluorescence in proportion to vesicle surface area, allowing for the measurements of vesicle size and concentrations (Figure 12b). In a poof-of-concept study, the authors used vesicle flow cytometer to detect EVs in platelet-rich plasma samples. EVs were isolated, and co-labeled with di-8ANEPPS for size measurement and fluorescent-antibodies against platelet-surface antigen. The vesicle flow cytometer detected EVs based on di-8ANEPPS signal, and resolved platelet-specific EV sub-populations (Figure 12c).
Figure 12. Small particle flow cytometry.
(a) To discriminate vesicles as small as 100 nm in diameter, a highly sensitive flow cytometry instrument, termed vesicle flow cytometry, was developed. (b) Fluorescent intensity from liposomes, labeled with di-8-ANEPPS, were calibrated for the vesicle diameter. The surface area of liposomes were estimated from NTA analysis. The linear regression provided coefficients for calibration of vesicle size. (c) EVs were isolated from platelet-rich plasma samples, and labeled with di-8-ANEPPS (vesicle size measurements) and fluorescent (DyLight488) antibodies against CD61 (platelet-specific). Vesicle flow cytometer detected EVs in plasma, and resolved EV sub-populations expressing cell surface markers. Reprinted with permission from Ref 140. Copyright 2015 International Society for Advancement of Cytometry.
5.3.2. Micro-Nuclear Magnetic Resonance
Magnetic sensing based on specific magnetic nanoparticles (MNPs) have recently received considerable attention.143, 144 Such sensing experiences little interference from native biological samples as most biological entities are naturally devoid of ferromagnetic background. Even optically turbid samples will thus appear as transparent to magnetic fields; however, when they are targeted with specific MNPs, they attain a high contrast against the native biological background. In the case of magnetic detection based on nuclear magnetic resonance (NMR), when MNPs are placed in NMR magnetic fields, they create local magnetic fields which change the transverse relaxation rate of surrounding water molecules to amplify the analytical signal.145, 146 NMR thus represents an attractive sensing mechanism; it reduces sample processing and improves detection sensitivity, and has been developed for multiple point-of-care applications, e.g., detection of circulating tumor cells and bacteria directly from blood samples.
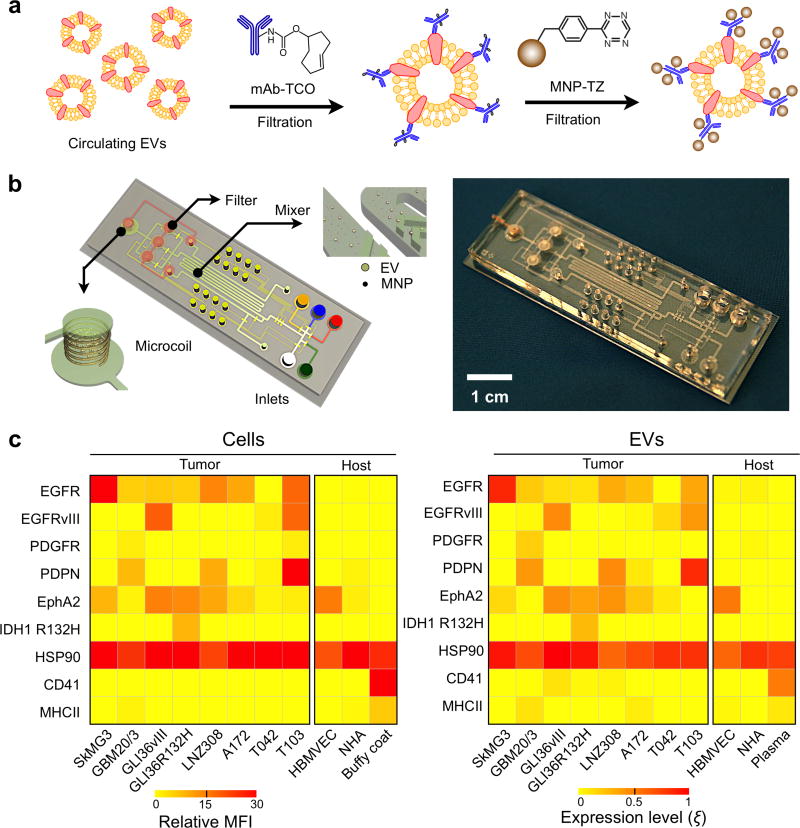
Adapting NMR to EV detection, however, has presented considerable engineering challenges because these vesicles are significantly smaller than tumor cells by one to two orders of magnitude. To bridge this size gap, Shao et al. developed a new analytical technology specifically dedicated for EV detection and protein profiling.20 In this approach, a two-step bio-orthogonal click chemistry was used to label EVs with MNPs (Figure 13a). This small molecule (< 200Da) labeling strategy did not appreciably increase the size of the antibody or the MNP, thereby improving the efficiency in retaining the targeted vesicles from unbound antibodies and MNPs. Targeted EVs were then directly measured on-chip using a microfluidic micro-nuclear magnetic resonance (µNMR) to determine the abundance of EV biomarkers (Figure 13b).
Figure 13. Micro-nuclear magnetic resonance.
(a) Assay schematics to maximize magnetic nanoparticle (NMP) binding onto EVs. A two-step bio-orthogonal click chemistry was used to label EVs with MNPs. (b) The microfluidic system for on-chip detection of circulating EVs is designed to detect MNP-targeted vesicles, concentrate MNP-tagged vesicles (while removing unbound MNPs) and provide in-line NMR detection. (c) GBM markers (EGFR, EGFRvIII, PDGFR, PDPN, EphA2 and IDH1 R132H), a positive EV control marker (HSP90), as well as host cell markers (CD41, MHCII) were profiled in both parental cells (left) and their corresponding EVs (right). Using a four-GBM marker combination (EGFR, EGFRvIII, PDPN and IDH1 R132H), GBM derived EVs could be distinguished from host cell–derived EVs. MFI, mean fluorescence intensity; HBMVEC, human brain microvascular endothelial cell; NHA, normal human astrocyte; buffy coat and plasma were isolated from whole blood donated by healthy volunteers. Reprinted with permission from Ref 20. Copyright 2012 Nature Publishing Group.
As compared to conventional protein technologies, the developed µNMR system demonstrated a significantly better detection sensitivity, ~103 fold more sensitive than western blotting and ELISA.20 Using this integrated technology, Shao et al. further profiled EVs from glioblastoma multiforme (GBM) cell lines grown in culture (Figure 13c). Comparative protein analyses confirmed that EVs indeed reflect the protein profiles of their parental cells for the tested markers, and a four-GBM marker combination (EGFR, EGFRvIII, PDPN, IDH1 R132H) was identified to distinguish cancer EVs from host cell-derived EVs.
5.3.3. Nano-plasmonic Exosome (nPLEX) Sensor
In view of the small dimensions of EVs, surface plasmon resonance (SPR) represents a novel sensing scheme for rapid, label-free profiling of EVs. SPR refers to a collective oscillation of conduction electrons at the metal-dielectric interface when illuminated by incident light.147 Unlike other optical detection methods based on time-sensitive fluorescent and chemiluminescent probes, SPR sensing detects a change in the local refractive index associated with biomolecular binding near a metal-dielectric interface, and can be applied in a label-free and real-time manner.
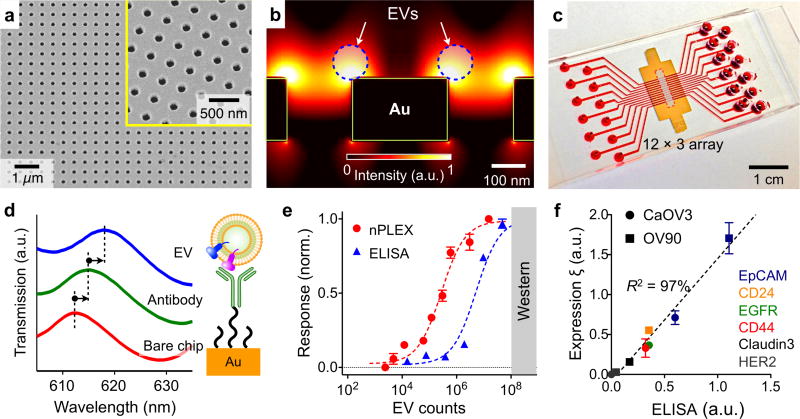
Im et al. recently developed a new SPR platform, termed nano-plasmonic exosome (nPLEX) sensor, for EV protein analysis.21 The sensing is based on transmission SPR through periodic nanohole arrays (Figure 14a). This transmission-type SPR offers significant advantages over the conventional reflection configuration for EV profiling: 1) the probing depth (<200 nm) can be easily tuned to match EV size to enhance the detection sensitivity, and 2) the collinear transmission optical setup simplifies device miniaturization.148 The geometry of the nanoholes was further optimized through three-dimensional simulation studies to match the sensing range with the mean diameter of exosomes (~ 100 nm) (Figure 14b). The authors further integrated the nanohole sensing arrays with multichannel microfluidics for independent and parallel analyses (Figure 14c).
Figure 14. Surface plasmon resonance.
(a) The nPLEX sensing is based on transmission SPR through periodic nanohole arrays. The hole diameter is 200 nm with a periodicity of 450 nm. The structure was patterned in a gold film (200-nm thick) deposited on a glass substrate. (b) Finite-difference time-domain simulation shows the enhanced electromagnetic fields tightly confined near a periodic nanohole surface. The field distribution overlaps with the size of EVs captured onto the sensing surface, maximizing the detection sensitivity. (c) The sensing array can be integrated with multichannel microfluidics for independent and parallel analyses. (d) Assay schematic of changes in transmission spectra showing EV detection. The gold surface is pre-functionalized by a layer of polyethylene glycol (PEG), and antibody conjugation and specific EV binding were monitored by transmission spectral shifts as measured by sensor. (e) In comparison to gold standard methods, the nPLEX assay demonstrated excellent sensitivity, being 104 and 102 more sensitive than Western blotting and chemiluminescence ELISA, respectively. (f) Correlation between nPLEX and ELISA measurements. The marker protein expression level was determined by normalizing the marker signal with that of anti-CD63, which accounted for variation in exosomal counts across samples. a.u., arbitrary unit. Reprinted with permission from Ref 21. Copyright 2014 Nature Publishing Group.
To confer molecular specificity, the sensing surface was functionalized with different capturing antibodies.21 Upon specific binding of EVs, the optical transmission spectral peaks would red shift due to a change in the local refractive index (Figure 14d). The magnitude of spectral shift correlated with molecular mass density covering the sensor surface and thus enabled quantitative analysis of EV proteins. Using a series of titration studies, this label-free nPLEX assay demonstrated a superior limit of detection of ~ 3000 vesicles (670 aM), which is 104 and 102 more sensitive than western blotting and chemiluminescence ELISA, respectively (Figure 14e). Importantly, in comparison with gold standard ELISA measurements, the nPLEX showed excellent accuracy across different protein markers (Figure 14f). The entire assay could be accomplished in < 30 minutes with minimally processed samples, making the system attractive for rapid clinical uses.21 This system is now under development for commercial applications.
5.3.4. Integrated Magnetic-electrochemical Exosome (iMEX) Sensor
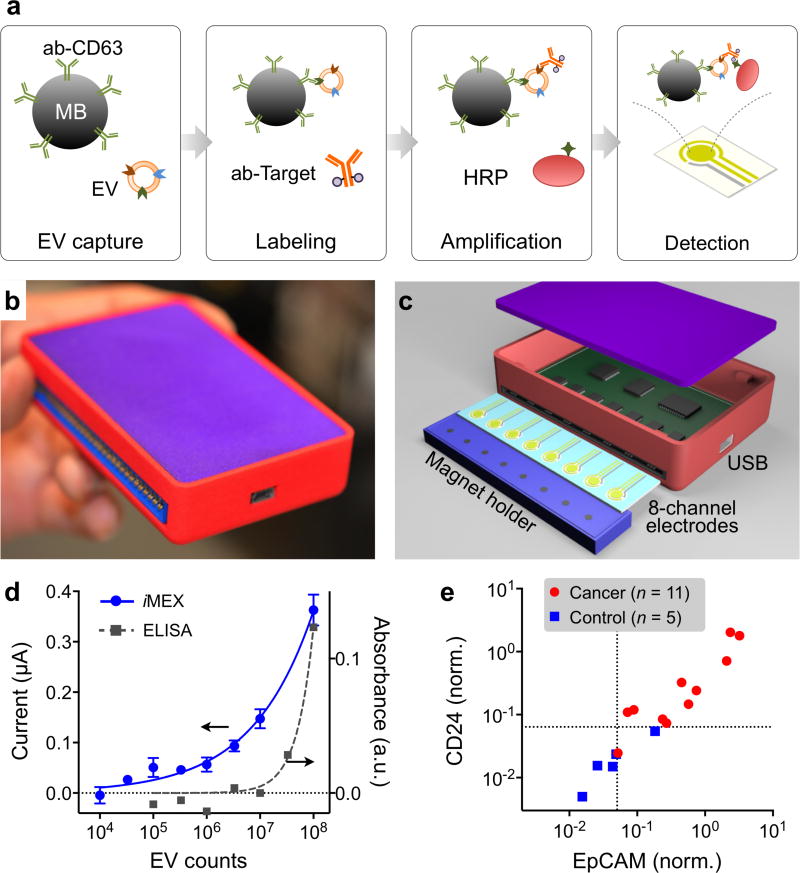
Bringing EV analysis to point-of-care settings, Jeong et al. recently developed a new portable sensor, integrated magnetic-electrochemical exosome (iMEX), for fast, streamlined EV analyses.149 A unique feature of iMEX is the integration of exosome isolation and detection into a single platform: magnetic beads are used for EV capture and labeling, and bead-bound EVs are detected through electrochemical sensing (Figure 15a). This approach offers many practical advantages: i) cell-specific exosomes can be isolated directly from complex media without the need for extensive filtration or centrifugation; ii) the assay can achieve a high detection sensitivity, by combining the merits of both magnetic enrichment and enzymatic amplification; and iii) through the electrical detection scheme, sensors can be easily miniaturized and expanded for parallel measurements.
Figure 15. Electrochemical detection.
(a) Assay schematics of the iMEX platform. EVs are captured on magnetic beads directly in plasma and labeled with HRP enzyme for electrochemical detection. The magnetic beads are coated with antibodies against CD63, an enriched surface marker in exosomes. (b) A photograph of the developed iMEX platform. (c) Sensor schematic. The sensor can simultaneously measure signals from eight electrodes. Small cylindrical magnets are located below the electrodes to concentrate immunomagnetically captured EVs. (d) Varying number of EVs were spiked into human plasma and assayed by iMEX and ELISA. The detection limits were 3 × 104 (iMEX) and 3 × 107 (ELISA). (e) Plasma samples from ovarian cancer patients (n = 11) and healthy controls (n = 5) were analyzed with the iMEX assay. EpCAM and CD24 levels were much higher in cancer patients. The EpCAM and CD24 expression levels (ξEpCAM vs ξCD24) were highly correlated (R2 = 0.870). Reprinted with permission from Ref 149. Copyright 2016 American Chemical Society.
The first iMEX system had eight independent detection channels, and was packaged as a handheld unit (Figure 15b). A card-edge connector was used for quick attachment of the electrodes, and a magnet holder, containing 8 cylindrical magnets, was placed underneath the electrodes to concentrate magnetic beads to the sensor surface (Figure 15c). The iMEX effectively provided simultaneous readouts from all electrodes through rapid polling of each channel. The iMEX showed high detection sensitivity with a limit of detection (LOD) of ~104 exosomes, and importantly offered a wide dynamic range spanning over four orders of magnitude (Figure 15d).149 EV protein profiles, measured by ELISA and iMEX, were highly correlated, which confirmed iMEX’s analytical capacity.149
In a proof-of-concept clinical application, the iMEX system was used to detect EVs in blood derived from ovarian cancer patients. Clinical plasma samples were aliquoted without any purification, and each aliquot (10 µL per marker) was incubated with magnetic beads for EV capture, and consecutively labeled for target markers. The parallel nature of iMEX detection enabled simultaneous measurements of four putative cancer markers (CD63, EpCAM, CD24, CA125).149 The iMEX assay revealed that the expression levels of EpCAM and CD24 in EVs were higher (P < 0.05) in ovarian cancer patients than healthy controls (Figure 15e). These experiments demonstrated the iMEX’s clinical potential for on-spot EV detection. The iMEX did not require any specialized systems, and was implemented as one portable system, and the entire assay was completed within 1 hour while consuming only 10 µL of non-purified clinical samples.
5.3.5. ExoScreen
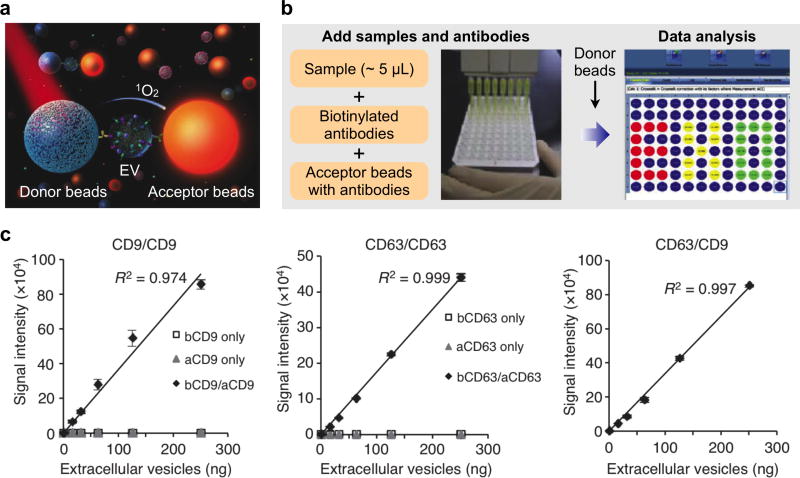
Aside from the planar sensors described above, Yoshioka et al. recently developed an amplified, solution-based luminescent proximity homogenous assay for rapid, sensitive analysis of EVs as a liquid biopsy.150 The authors used photosensitizer beads as direct reporters and did not require any purification step before protein analysis in serum. In a scheme akin to ELISA, this assay requires two types of immunobeads: 1) donor beads, which are excited at 680 nm to release singlet oxygen, and 2) acceptor beads, which can be excited by the released singlet oxygen to emit at 615 nm, but only when they are situated within 200 nm away from the donor beads (Figure 16a). Simultaneous binding of donor beads and acceptor beads on a single EV (i.e., only for vesicles with diameter < 200 nm) enables signal generation.
Figure 16. ExoScreen technology.
(a) Assay principle of ExoScreen. This proximity assay requires two types of immunobeads: 1) donor beads, which are excited at 680 nm to release singlet oxygen, and 2) acceptor beads, which can be only excited by the released singlet oxygen when they are situated within 200 nm away from the donor beads. (b) Assay workflow. Biological samples are first treated with biotinylated antibodies and acceptor beads conjugated with a second antibody. Streptavidin-coated donor beads were then added to complete the proximity assay for data acquisition. (c) Correlation between ExoScreen measurements for CD9 positive EVs, CD63 positive EVs or CD63/CD9 double-positive EVs and EV protein concentration in a dilution series. The addition of biotinylated antibodies and acceptor beads conjugated antibodies is denoted ‘bCD9/aCD9’ or ‘bCD63/aCD63’. Right panel shows the addition of biotinylated CD63 antibodies and acceptor beads conjugated CD9 antibodies. Reprinted with permission from Ref 150. Copyright 2014 Nature Publishing Group.
This assay was thus termed “ExoScreen” as it targeted smaller EVs (e.g., exosomes) and could be implemented for biomarker screening in a variety of diseases.150 The assay has a mix-and-read format: native biological samples are first treated with biotinylated antibodies and acceptor beads conjugated with a second antibody. Streptavidin-coated donor beads were then added to complete the proximity assay for data acquisition (Figure 16b). Starting with as little as 5µL of serum sample, the assay could be established in a multi-well plate format and accomplished within 2 hours.
As different antibodies could be conjugated with the donor and acceptor beads, respectively, the assay was used for screening of double-positive EVs. ExoScreen was able to quantify different concentrations of double-positive EVs, while negative controls (i.e., assayed with only the biotinylated antibody or acceptor bead-conjugated antibody) showed minimal fluorescent signals (Figure 16c). As a proof-of-concept clinical study, the assay was applied for detecting colorectal cancer EV biomarkers and identified that CD147/CD9 double-positive EVs could be used to differentiate between healthy donors and colorectal cancer patients.150
5.3.6. Comparison
Unlike nucleic acid detection wherein target numbers can be amplified, EV protein assays are limited to directly cope with low concentration of relevant EV biomarkers (e.g., tumor-derived EVs in blood). Conventional assays thus often require extensive purification steps and large sample volumes to achieve sufficient analytical power. The new biosensors summarized in Table 3 leverage on EV-compatible sensing mechanisms to overcome these challenged. A common key advantage of these methods is the sparing use of EVs per marker, which would facilitate multiplexed molecular validation in volume-limited clinical samples. Differences can be summarized as below. Bead-based flow cytometry or nPLEX are well-suited for high throughput screening, and have superior sensitivity to other methods. These methods, however, are costly and require specialized instrumentation. The iMEX and microNMR are complementary systems, allowing for point-of-use detection at lower device cost and biofluid throughput. Both nPLEX and iMEX have been validated through numerous clinical studies, and they are now being commercialized. Small particle flow cytometry140, 142 is still in the development phase, and has intrinsically low throughput; individual EVs should pass the detection zone slowly to ensure good signal-to-noise ratio through long integration time.
6. EV NUCLEIC ACID ANALYSIS
6.1. Nucleic Acids in EVs
In addition to protein cargoes, EVs contain different forms of RNA and DNA (Table 4). RNAs represent the major nucleic acid cargo of EVs. As compared to the cellular RNA fraction,151 RNAs transported by EVs are generally shorter in size (typically < 200 nucleotides, but can extend out to 5 kb).152, 153 They are predominately noncoding RNAs and include microRNA (miRNA), transfer RNA (tRNA), as well as long noncoding RNA (lnRNA) and mostly fragmented mRNAs.154–156 In the pool of longer transcripts (> 200 nucleotides), coding messenger RNA (mRNA) has been identified,152, 154 typically up to a size of 1 kb.58 Several studies have shown that RNAs can be transferred to and remain functional in recipient cells.9, 10, 157 mRNA can be translated into proteins, and miRNAs transferred may regulate the translation of target mRNA in recipient cells.58 The amount and nature of RNA in EVs can vary according to the cell types of origin, even though some RNAs are systematically enriched.158–160 Because of their retained functionality in recipient cells, this RNA sorting raises interesting hypotheses that dedicated mechanisms may exist for RNA partition into EVs, and the potential that these machineries can be exploited to deliver therapeutic RNA cargoes. This is an area of active research and has been overviewed in other biology reviews.32, 161–163
Table 4.
Different types of RNA found in EVs.
| RNA | Functions | Coding | Typical Size |
|---|---|---|---|
| Messenger RNA (mRNA) | Protein translation | Yes | 400 – 12,000 nt average ~ 2,100 nt |
| MicroRNA (miRNA) | Post-transcriptional gene silencing | No | 17 – 24 nt |
| Y RNA | Component of Ro60 ribonucleoprotein particle; initiation factor for DNA replication | No | ~ 100 nt |
| Signal Recognition Particle RNA (SRP RNA) | Component of SRP ribonucleoprotein complex that directs protein trafficking | No | ~ 280 nt |
| Transfer RNA (tRNA) | Adapter for matching amino acid to mRNA | No | 76 – 90 nt |
| Ribosomal RNA (rRNA) | RNA component of ribosome | No | 18S (1.9 kb) 28S (5.0 kb) |
| Small nuclear RNA (snRNA) | RNA processing such as mRNA splicing | No | ~ 150 nt |
| Small nucleolar RNA (snoRNA) | Guiding chemical modifications of other RNAs | No | 20 – 24 nt |
| Long noncoding RNA (lncRNA) | Many putative functions, including in-transcription and post-transcription regulations | No | > 100 nt |
nt: nucleotides, kb: kilobases
6.1.1. mRNA
mRNAs are a large family of coding RNA molecules that specify protein sequence information. Recent studies have found that EVs contain a substantial proportion of their parent cells’ mRNA pool, many of which are cell type-specific mRNA.58, 164 These mRNA molecules, often in fragmented form, reside within EVs and are protected from RNase degradation, making them robust circulating biomarkers. Furthermore, the fraction of polyadenylated mRNA molecules in EVs suggest that some of them (<2 kb) are capable of encoding polypeptides in support of protein synthesis (i.e., functionality in protein translation). This has been confirmed in multiple studies through different translation assays in recipient cells.9, 10, 60 These studies highlight the multifaceted role of EVs as specific cellular messengers in influencing the recipient cells and facilitating intercellular communication.32, 61, 165
6.1.2. miRNA
miRNAs are a class of small, noncoding RNAs (typically 17–24 nucleotides) which mediate post-transcriptional gene silencing usually by targeting the 3’ untranslated region of mRNAs. By suppressing protein translation, EV miRNAs are powerful regulators for a wide range of biological processes.157, 165 Unlike circulating mRNAs in EVs, miRNAs can also exist in multiple stable forms when circulating in bodily fluids. In addition to being packaged into EVs, circulating miRNAs can also be loaded onto high-density lipoprotein,166, 167 or bound to AGO2 protein outside the vesicles.168, 169 Current evidence is accumulating that while the majority of circulating miRNAs are bound to RNA-binding proteins, a small proportion of miRNAs can also be found in EVs. Nevertheless, it remains unclear on the distribution of miRNAs within EVs.170–172 As in the case of mRNA, miRNA profiles in EVs reflect their cell of origin but differs somewhat from their parental cells. Some miRNAs have been found preferentially sorted into EVs and remaining functional in recipient cells to regulate protein translation.159, 160, 173, 174 Recent studies also found that fetal bovine serum, commonly used in mammalian cell culture, could contribute to miRNA artifacts in in vitro EV preparations.175, 176
6.1.3. Other RNA Types
In addition to mRNA and miRNA, many noncoding RNA types have been identified in EVs through next generation sequencing.152, 177 These RNAs include transfer RNA (tRNA), ribosomal RNA (rRNA), small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), as well as long noncoding RNA (lncRNA).58, 152, 178. These RNA types are summarized in Table 4.
6.1.4. DNA
Recent studies have shown that certain EVs may contain DNA fragments.11, 57, 179–181 These DNA are double-stranded fragments which range from 100 base pairs (bp) to 2.5 kbp.57 The larger-sized population (> 2.5 kbp) was found to be predominately external DNA associated with EVs and smaller-sized population (100 bp – 2.5 kbp) as internal DNA confined within EVs. These fragments represent the whole genomic DNA and could be used to identify mutations present in parental tumor cells.57, 180 While there is compelling evidence for the presence of DNAs in EVs, their functional roles have yet to be determined.
6.2. Conventional Extraction and Detection Tools
EV nucleic acids have been extensively researched as a potential circulating biomarker as well as an intercellular regulator of recipient cells. Conventional nucleic acid extraction and analysis tools have been successfully used to lay important foundation in our understanding of EV nucleic acids. Because the intrinsic amount of nucleic acids in EVs is low, it is important to develop efficient extraction procedures and sensitive detection strategies, particularly to interrogate rare molecular targets from small sample volume.182, 183
6.2.1. Precipitation and Spin Columns
Different methods have been used for extracting exosomal nucleic acids. These commonly include phenol-chloroform extraction and spin column techniques.10, 11, 184 As in cellular RNA extraction, the phenol-based method relies on phase separation upon centrifugation; nucleic acid partitions into the aqueous phase and is recovered through precipitation with ethanol. This approach is laborious and lengthy, but could potentially provide RNA of superior purity. Spin columns, on the other hand, is a solid phase extraction method to enable rapid RNA purification (Figure 17).184 This method relies on the strong binding of nucleic acids onto silica in the presence of chaotropic agents, and could be implemented after phenol-chloroform extraction to to facilitate processing. Both of these extraction approaches have been developed and marketed under different commercial names, with varying degrees of reported success.185
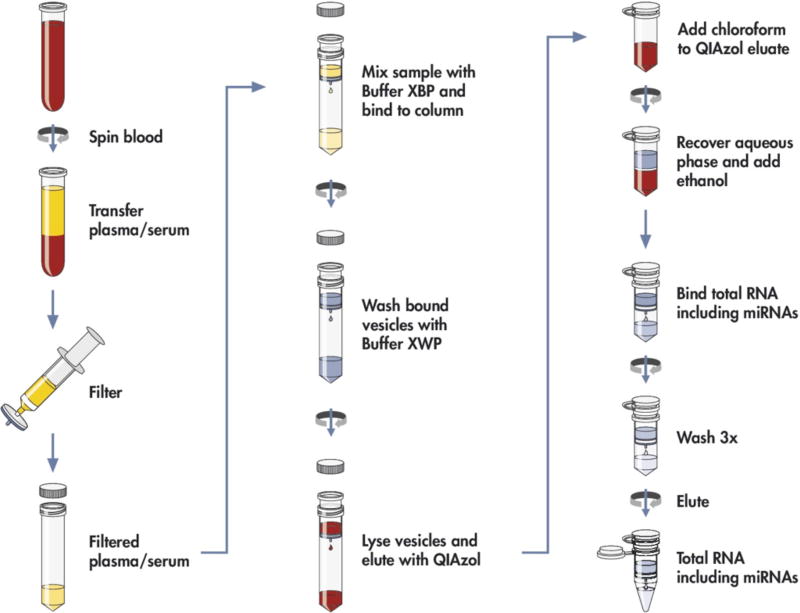
Figure 17. Workflow of RNA extraction using spin column.
EV RNA is isolated from whole blood by separating the plasma or serum, pre-filtering the sample to exclude cell-contamination, and loading on the membrane affinity column followed by a brief wash. The bound vesicles are lysed and eluted with QIAzol; the RNA extracted by addition of chloroform, precipitated by ethanol and further purified using an RNeasy column. Reprinted with permission from Ref 184. Copyright 2015 Enderle et al.
6.2.2. Amplification and Sequencing
Extracted exosomal nucleic acids are subjected to different modes of analyses. In addition to verification of nucleic acid quality, yield, and size, amplification and sequencing approaches are typically employed to detect and quantify in a sequence-dependent manner.25 For example, a target sequence can be selectively amplified through polymerase chain reaction (PCR) and detected with either end-point electrophoresis or real-time fluorescence measurements (RT-PCR) (Figures 18a, 18b).10, 11 These approaches, albeit of limited throughput, help to quantitate known target sequences in exosomes. For high-throughput discovery and quantitation of unknown exosomal RNA transcripts, recent advances in next generation sequencing (NGS) have made significant contributions to our understanding of exosomal RNA contents and their distribution (Figure 18c).152 Importantly, sequencing-based RNA profiling analysis generates millions of reads with good read depth and coverage to facilitate discovery and characterization of the whole transcriptome (including known and unknown RNAs).152, 177 Such comprehensive analysis may provide further insights on the mechanisms of exosome-mediated molecular effects and contribute to biomarker discovery.
Figure 18. Amplification and sequencing of EV nucleic acids.
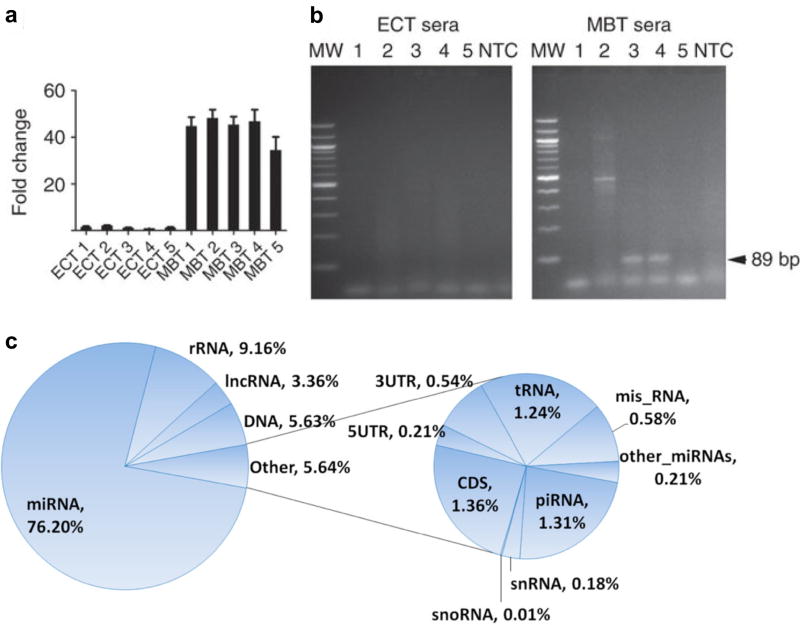
(a) Amplification and detection of c-Myc, a tumor oncogene, on epidermoid carcinoma (ECT) and medulloblastoma (MBT) xenografted tumors with qPCR. Values were normalized to GAPDH, a housekeeping control. (b) EV RNA were isolated from corresponding serum samples. c-Myc PCR product was amplified using human specific primers. Amplified DNA was resolved by electrophoresis in a 2% agarose gel and visualized with ethidium bromide staining. c-Myc is shown as an 89 bp fragment (arrow). MW, molecular weight; NTC, no template control. (c) Pie chart of RNA species and their distributions in the plasma-derived exosomes. Misc RNAs are the RNA sequences that mapped to the human genome but not in any of the categories listed. The DNA category represents the novel transcripts that have no annotation in the human RNA database. Reprinted with permission from Ref 11. Copyright 2011 Nature Publishing Group. Reprinted with permission from Ref 152. Copyright 2013 BioMed Central Ltd.
6.3. New Extraction and Detection Technologies
With a growing interest in utilizing EV nucleic acids as minimally-invasive diagnostic markers, new biosensor technologies have been developed to enable more efficient and rapid extraction and analysis. Many of these new platforms provide sensitive quantitation of target nucleic acid markers and are capable of distinguishing disease markers against a complex biological background, down to even single nucleotide point mutations. These thus open many new clinical opportunities for next generation personalized molecular medicine.
6.3.1. Droplet PCR
While conventional PCR is powerful technology to detect large gene/transcript changes (e.g., EGFRvIII deletion mutation10), its application for detecting single nucleotide mutations can be challenging due to its limited sensitivity. This issue is particularly relevant for EVs, as the fraction of mutated transcripts is low amongst a large background of wild-type transcripts.94 To improve the detection sensitivity of EV RNAs as tumor biomarkers, Chen et al. recently adapted a droplet digital PCR (ddPCR) technology to detect rare mutations in EVs (Figure 19).94
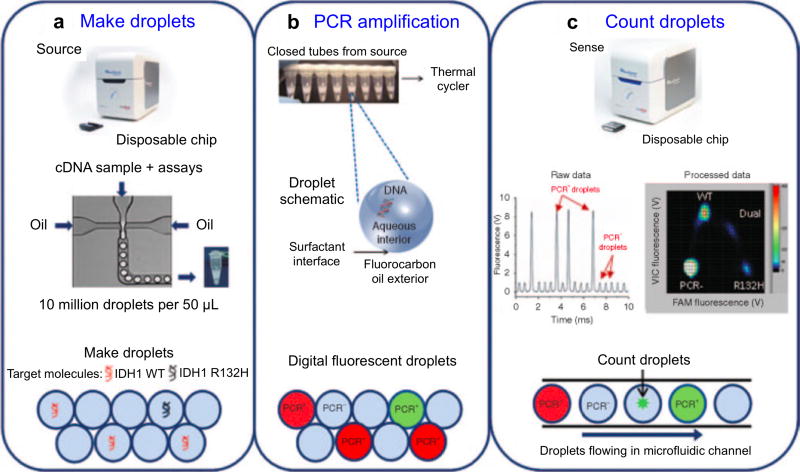
Figure 19. Application of droplet digital PCR for EV analyses.
The workflow consists of three steps: (a) Making PCR droplets. Aqueous PCR reaction mixture is injected through a microfluidic droplet generator along with surfactant-containing fluorocarbon oil to produce five picoliter droplets. (b) PCR amplification. The droplets are loaded into a standard thermal cycler for endpoint PCR amplification, with single-target-molecule-containing droplets resulting in specific probe hydrolysis (PCR+) and bright fluorescence and the majority of droplets, containing no target molecule, resulting in only background probe fluorescence (PCR−). (c) Each droplet's fluorescence is detected and processed into a two-dimensional scatter plot display. Reprinted with permission from Ref 94. Copyright 2013 American Society of Gene & Cell Therapy.
In this approach, EVs were isolated from frozen biobanked serum and cerebrospinal fluid (CSF) samples from GBM patients as well as controls. Upon conventional RNA extraction and reverse transcription, every sample was partitioned into millions of 5 picoliter-volume aqueous droplets in an oil emulsion. This setup ensures that a single droplet contains no more than one copy of a nucleic acid target. The authors then subjected the droplet mixture to regular PCR amplification and detected single droplet fluorescence intensity to measure the concentration of mutant transcripts in clinical samples.94 Using the developed ddPCR technology, the study identified mutant mRNA transcripts for isocitrate dehydrogenase 1 (IDH1) in EVs from GBM patients, but not from healthy controls. IDH1 mutations have been reported as a novel prognostic marker and molecular stratifier for GBM.186 Interestingly, the mutant mRNAs was most readily detected in CSF-derived EVs,187 from patients bearing IDH1-mutant GBM tumors, which presents an important biofluid for brain disorders.
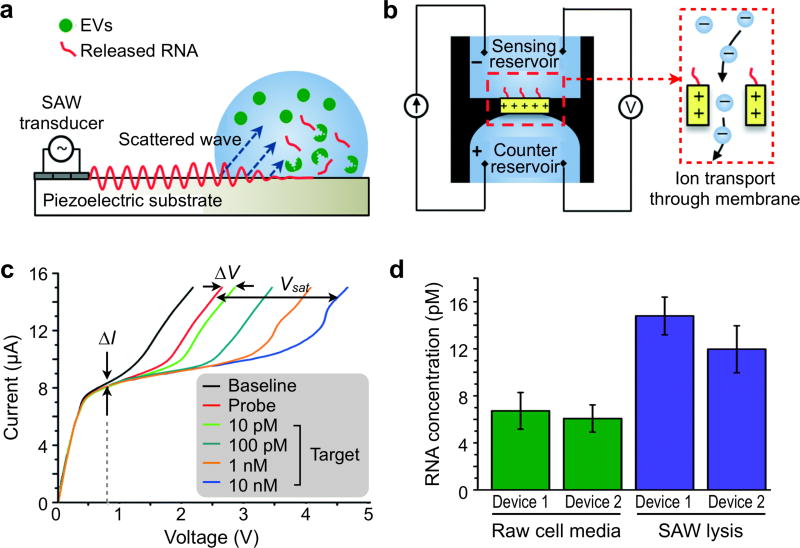
6.3.2. Microfluidics for On-Chip Extraction and Detection
For point-of-care EV nucleic acid analysis, Shao et al. recently developed a comprehensive microfluidic platform that integrates three functional modules: targeted enrichment of EVs, on-chip RNA isolation and real-time RNA analysis (Figure 20a).22 This platform, termed immunomagnetic exosome RNA (iMER) analysis, utilizes antibody-functionalized magnetic beads to separate cancer-specific EVs from host-derived vesicles (Figure 20b). The immunomagnetically enriched vesicles are then lysed on chip. As the EV lysate passes through a glass-bead filter, EV RNA is selectively adsorbed and eluted from the filter for reverse transcription and qPCR analysis. To streamline the assay processing, all key components were integrated onto a chip cartridge. The fluidic flow was controlled through torque-activated valves and the cartridge was loaded into a custom-designed PCR system equipped with a thermal cycler and a fluorescence detector (Figure 20c).
Figure 20. Microfluidics for on-chip EV RNA extraction and detection.
(a) Assay schematics of the immuno-magnetic exosome RNA (iMER) analysis. Cancer exosomes in serum are first captured onto magnetic microbeads containing affinity ligands (for example, anti-CD63 and anti-EGFR). The immuno-enriched exosomal population is then lysed and the lysate flows through a glass bead filter, where RNA efficiently adsorbs onto packed glass beads. Finally, the collected RNA is eluted and reverse-transcribed for real-time amplification and quantitation. (b) Scanning electron micrographs of magnetic microbeads after immunoaffinity capture. Microbeads (left, 3 mm) functionalized with antibodies against EGFRvIII, a cancer-specific deletion mutant, captured innumerable tumor vesicles from GLI36vIII conditioned medium. (c) Photograph of the microfluidic iMER prototype. (d) Exosomal MGMT and APNG mRNA levels correlate with in vitro TMZ sensitivity (ED50). Cell lines were treated with varying doses of TMZ to determine their respective drug sensitivities (top panel). iMER analysis revealed that the levels of MGMT, APNG or both were elevated in resistant cell lines, whereas they were both low in sensitive ones (bottom panel). (e) Higher average levels of exosomal MGMT/APNG were observed in resistant cell lines than in sensitive ones. However, there was overlap in exosomal mRNA levels between resistant and sensitive cell lines, demonstrating that a single marker was unable to distinguish drug resistance. Dotted line indicates the mean. Reprinted with permission from Ref 22. Copyright 2015 Nature Publishing Group.
With the system developed, the authors investigated two mRNA targets of nuclear proteins, MGMT (O6-methylguanine DNA methyltransferase) and APNG (alkylpurine-DNA-N-glycosylase), both of which encode DNA repair functionalities that can abrogate the effects of alkylating chemotherapies.22 While these nuclear proteins are key determinants of GBM treatment efficacy, they are rarely found in EVs due to their nucleus localization. The authors, however, found that the mRNAs encoding these nuclear proteins could be detected within EVs, presumably because the proteins are translated in the cytoplasm before being translocated to the nucleus. Employing this platform, the study identified that key EV mRNA markers could differentiate between cancer cells that were resistant and sensitive to treatment with temozolomide.22 Resistant cancer cells were associated with high levels of MGMT mRNA and/or APNG mRNA in their released EVs (Figure 20d), indicating downregulated DNA repair enzymes. Importantly, the platform further enabled rapid analysis of a small volume of clinical sample (~100 uL blood in < 3 hours). Longitudinal monitoring of clinical blood samples showed that EV mRNA profiles change over treatment course and could be serially correlated with treatment response (Figure 20e).
6.3.3. Ion-Exchange Nanodetector
To reduce sample loss and processing time for EV miRNA detection, Taller et al. developed two serial microfluidic platforms, a lysis device and a detection device, to reduce total analysis time to ~ 1.5 hour, which included ~ 30 minutes for EV lysis and ~ 1 hour for detection.188 In this study, EV lysis was achieved via surface acoustic waves (SAWs) which were generated on the surface of a piezoelectric crystal with alternating current applied through an interdigitated transducer (Figure 21a). The electromechanical coupling of SAWs produces a strong electric field; this application of both the electric (dielectrophoretic force) and acoustic radiation forces aide in EV lysis. Importantly, SAW lysis represents an excellent alternative to traditional chemical lysis approaches, which can interfere with RNA detection downstream by changing pH and ionic strength of the overall solution.188
Figure 21. Ion-exchange nanodetector.
(a) Schematic of surface acoustic wave (SAW) device and SAW-induced lysing of exosomes to release RNA for detection. SAWs generated at the transducer refract into the liquid bulk, inducing fluid motion, and electromechanical coupling also generates a complimentary electric wave at the surface of the substrate. (b) Schematic of ion-exchange nano-membrane sensor consisting of two reservoirs separated by the membrane. RNA in the sensing reservoir hybridize to complimentary oligos immobilized on the surface of the membrane. The inset shows the ion transport through the device to generate current. (c) Representative current voltage characteristic (CVC) for nano-membrane sensor. The black, red, and blue curves indicate a CVC taken with the bare membrane, a CVC taken with the probe attached to the membrane, and a CVC taken with the probes on the membrane surface fully saturated with target RNA, respectively. (d) Target RNA concentration as detected by the nano-membrane sensor and determined using the universal calibration curve before and after SAW lysis for two different nano-membrane devices. Reprinted with permission from Ref 188. Copyright 2015 The Royal Society of Chemistry.
RNA detection was accomplished through an ion-exchange nano-membrane sensor (Figure 21b). The sensor was comprised of an anion-exchange membrane bridging across two reservoirs; when an electric current is applied across the membrane, anions are driven through the membrane, producing a corresponding current-voltage characteristic (CVC) measured across the membrane.188 By functionalizing the membrane surface with capturing oligonucleotide probes, target RNA binding onto the membrane surface can be sensitively and quantitatively measured through changes in the CVC measurements (Figure 21c).
Using this microfluidic system, the author demonstrated that the SAW device could achieve a lysis efficiency of 38 ± 10% and further developments could be implemented to achieve a higher lysis rate (Figure 21d). In addition, using a universal calibration curve developed for miRNA quantification, detection of the target miRNA (miR-550) was accomplished both in cell culture media (before lysis) and after SAW lysis, indicating the presence of the miRNA as both free floating and EV-encapsulated.
6.3.4. LSPR-based Assay
As previously discussed, SPR is a sensitive technique for label-free detection of biomolecular binding events at a metal-dielectric interface. Specifically, localized SPR (LSPR) occurs due to the confinement of a surface plasmon in nanoparticles with size comparable or smaller than the incident light wavelength.189–191 In this aspect, LSPR differs from SPR as the induced plasmons oscillate locally to the nanostructure rather than along the metal-dielectric interface. As a result, the electromagnetic field decays much more rapidly in LSPR. This shorter field decay length (< 100 nm) reduces sensor interference from bulk refractive index changes and provides increased sensitivity to small biomolecular binding at the surface.
Leveraging on this advantage of LSPR for detection of small biomolecules, Joshi et al. recently developed a sensitive miRNA sensor using chemically synthesized gold nanoprisms attached onto a solid substrate (Figure 22a).192 The sensor was fabricated using two steps: 1) ~ 40 nm edge-length gold nanoprisms were chemically synthesized and covalently attached to a glass substrate; 2) the immobilized nanoprisms were functionalized with capturing DNA probes and polyethylene glycol spacers.192 Upon hybridization of the miRNA target (miR-10b) with the capturing DNA probes, the LSPR resonant peak shifted (Figure 22b). Due to its atomically flat surface for homogenous packing as well as strong electromagnetic field enhancement at its sharp nanoprism tips, the sensor achieved high detection sensitivity, with a limit of detection of 91 aM.
Figure 22. Localized surface plasmonic resonance for detecting EV RNA.
(a) Scanning electron microscopy (SEM) image of gold nanoprisms for LSPR-based sensors fabrication. (b) Assay schematic of the LSPR assay. The immobilized nanoprisms are functionalized with capturing DNA probes and polyethylene glycol spacers. Upon hybridization of the miRNA target (miR-10b) with the capturing DNA probes, the LSPR resonant peak shifts. (c) miR-10b detection in clinical samples from pancreatic ductal adenocarcinoma (PDAC; left) or chronic pancreatitis (CP; right) patients. Determination of the miRNA levels was performed in plasma, exosomes and supernatants, respectively. Reprinted with permission from Ref 192. Copyright 2015 American Chemical Society.
The authors further employed the developed biosensor for miR-10b detection in clinical samples from pancreatic ductal adenocarcinoma (PDAC) and chronic pancreatitis (CP) patients (Figure 22c). Determination of the miRNA levels was performed in plasma, exosomes and supernatants, respectively. Interestingly, miR-10b level was found to be significantly higher in PDAC patients than in CP patients.192 Importantly, a very high level of the miRNA was associated with pancreatic cancer exosomes.
7. CLINICAL APPLICATIONS OF EV ANALYSES
With the growing evidence that EVs carry diverse representative biomolecules and effect important functions in intercellular communication, their clinical applications have grown tremendously.16, 31, 42, 119, 193, 194 Disease-derived EVs are being studied to yield improved diagnostic detection (Table 5). Furthermore, they can also be engineered as effective vehicles for therapeutics.32, 195 These potential applications have attracted significant interests from both clinical as well as biotechnology industries. Several EV-based companies are formed (e.g., Exosome Diagnostics), to develop innovative, personalized disease detection, monitoring and treatment.
Table 5.
Application of EVs as circulating biomarkers for various diseases.
| Disease type | Organs | References |
|---|---|---|
| Cancer | Glioblastoma multiforme | 6, 10, 22, 116, 196 |
| Head and neck cancer | 197, 198 | |
| Breast cancer | 199–202 | |
| Lung cancer | 203–206 | |
| Pancreatic cancer | 207–211 | |
| Endometrial cancer | 212 | |
| Gastric cancer | 213 | |
| Liver cancer | 214–216 | |
| Bladder cancer | 216–218 | |
| Colorectal cancer | 219, 220 | |
| Prostate cancer | 221–224 | |
| Ovarian cancer | 21, 225–228 | |
|
| ||
| Neuro-degenerative disease | Alzheimer’s disease | 229–233 |
| Parkinson disease | 234–236 | |
|
| ||
| Acute organ injury | Cardiovascular disease | 237, 238 |
| Liver injury | 215, 216 | |
| Kidney injury | 95, 239–242 | |
| Preeclampsia | 243–245 | |
7.1. Cancer Diagnostics
Tumors are complex structures that contain both malignant cells and surrounding stroma cells, such as endothelial cells, fibroblasts and immune cells.246–248 Recent studies have shown that EVs play an important role in facilitating intercellular communication in the tumor microenvironment, thereby modulating disease initiation, progression and treatment response.14, 249, 250 Leveraging the rich diversity of passive and active cargo that EVs carry, researchers have analyzed EVs to uncover specific molecular signatures for a wide variety of cancers to advance disease detection, treatment monitoring and resistance surveillance.14, 27, 251
7.1.1. Glioblastoma Multiforme
Glioblastoma multiforme (GBM) is the most common primary malignancy of the central nervous system. GBM poses significant challenges for repeat biopsies, especially due to the extensive complexities and morbidity associated with invasive brain biopsies. There thus remains a critical unmet need for minimally invasive biomarkers to characterize the primary disease and monitor treatment response.252, 253
Prior research has shown that GBM shed large quantities of cancer-specific EVs into the circulation.6, 10, 116, 196 Early work by Skog et al. discovered that these vesicles could pass out throurgh the blood-brain-barrier.10 GBM EVs were found to contain mRNA, miRNA (Figure 23a) as well as angiogenic proteins (Figure 23b). The authors further treated brain microvascular endothelial cells with GBM-derived EVs, and found that the EVs could not only stimulate tubule formation in endothelial cells, an indication of their angiogenic ability, but also deliver functional RNA to the recipient cells. Importantly, tumor specific mRNA mutation (EGFRvIII variant) and miRNA characteristics of gliomas could be detected in serum samples derived from GBM patients, indicating the potential of EVs to provide molecular information about a patient’s cancer through a blood test.10
Figure 23. Glioblastoma multiforme detection.
(a) Levels of mature miRNAs in EVs and glioblastoma cells from patient (GBM1) were analyzed using quantitative miRNA RT–PCR. (b) Total protein from primary glioblastoma cells and EVs from them was analyzed on a human angiogenesis antibody array. (c) Longitudinal EV MGMT and APNG mRNA analyses were performed in seven GBM patients. Clinical assessments (NR, S, R) were based on radiological findings, clinical examination and lab values. (d) Sequential EV mRNA changes between two time points were analyzed in GBM patients (n = 7) undergoing temozolomide treatment. All changes were normalized to their initial values and plotted according to clinical evaluation at the end of the assessment period (the later time point). All changes were independent of initial tissue MGMT methylation status. Reprinted with permission from Ref 10. Copyright 2008 Nature Publishing Group. Reprinted with permission from Ref 22. Copyright 2015 Nature Publishing Group.
More recently, Shao et al. developed a microfluidic chip to analyze exosomal mRNA levels for monitoring drug resistance in GBM.22 While temozolomide (TMZ) is the main chemotherapy for GBM, tumor response varies during treatment and depends on two key nuclear enzymes MGMT and APNG, whose levels correlate inversely to treatment efficacy. In this study, the authors developed a microfluidic platform to enrich for GBM-specific EVs and analyze the mRNA levels of MGMT and APNG.22 They showed that the exosomal mRNA levels for these targets correlated with cellular levels. In a proof-of-concept clinical study, the mRNA levels were found to change considerably during treatment (Figure 23c).22 When sequential exosomal mRNA changes between two adjacent time points were analyzed in patients undergoing TMZ treatment (Figure 23d), mRNA changes could be used to independently correlate to treatment response (e.g., radiological findings, clinical examination and laboratory values).
7.1.2. Ovarian Cancer
Ovarian cancer is the most lethal gynecological malignancies and possesses a high capacity for metastasis. Because of the significant role EVs have in carcinogenicity and cancer progression, research efforts have been directed to discover and apply EVs for disease diagnostics and prognostics.225, 226 Im et al. has applied the nPLEX platform for quantitative profiling of ovarian cancer EVs.21 The detection is based on transmission surface plasmon resonance, as described in Chapter 4. The authors started with antibody profiling of ovarian cancer and other host cell (non-cancer) markers, chosen based on prior studies. Through clustering analysis and antibody selection, the following marker assays were implemented in cells and EVs (Figure 24a): epithelial cell adhesion molecule (EpCAM), CD24, cancer antigen 125 (CA-125), cancer antigen 19-9 (CA19-9), human epidermal growth factor receptor 2 (HER2), mucin 18 (MUC18), epidermal growth factor receptor (EGFR), claudin 3 (CLDN3). For non-cancer cell identification, CD45 (leukocyte), CD41 (platelet) and D2-40 (mesothelial cells) were selected. Comparative protein expression analysis indicated an excellent correlation (Pearson coefficient >0.95), supporting the use of EVs as a circulating cellular surrogate.21
Figure 24. Ovarian cancer detection.
(a) Putative ovarian cancer markers (EpCAM, CD24, CA19-9, CLDN3, CA-125, MUC18, EGFR, HER2), immune host cell markers (CD41, CD45) and a mesothelial marker (D2-40) were profiled on EVs (left) and their parental ovarian cell lines (right). MFI, mean fluorescence intensity. a.u., arbitrary unit. (b) Exosomal protein levels of EpCAM and CD24 in ascites samples from patients were measured by nPLEX. Ovarian cancer patient samples (n = 20) were associated with elevated EpCAM and CD24 levels, whereas non-cancer patients (n = 10) showed negligible signals. (c) Longitudinal monitoring of treatment responses. Ascites samples were collected from ovarian cancer patients before and after chemotherapy (n = 8) and profiled with nPLEX. The bars represent the changes in CD24 and EpCAM levels per exosome after treatment. Reprinted with permission from Ref 21. Copyright 2014 Nature Publishing Group.
Importantly, the authors also identified that the marker combination of EpCAM and CD24 could be used as a minimal signature set to distinguish EVs from cancer versus benign cells. This marker signature was further tested on EVs derived from patient ascites fluid (i.e., excess fluid accumulation in the peritoneal space). While EV concentrations were highly heterogenous, the levels of EpCAM and CD24 were significantly higher in ovarian cancer patient samples than in controls (i.e., ascites from nonmalignant conditions such as liver cirrhosis; Figure 24b). By pairing the protein level profiles, the EV diagnostics achieved the accuracy of 97% for detection of ovarian cancer. Finally, using the same marker combination, the authors explored EV profiling to monitor clinical response or progression during treatment. Using longitudinal clinical samples derived from the same patients, serial changes in exosomal EpCAM and CD24 level were measured (Figure 24c) and levels of EV EpCAM, CD24 or both were found to decrease among responding patients.
7.1.3. Pancreatic Cancer
Pancreatic cancer is highly metastatic with a dismal prognosis, mainly due to delayed detection. A significant patient population (~80%) presents late with metastasis at diagnosis.254 The 5-year survival is <5%, making mortality and incidence rates almost identical. Progress in treatment options has thus motivated extensive research in identifying novel circulating biomarkers for early detection.207–209, 211, 255 Currently, CA19-9 is the only serum biomarker in routine clinical use for managing pancreatic cancer; however, this marker has a limited specificity as its expression is also elevated in related diseases, such as chronic pancreatitis and obstructive jaundice.256, 257
Yang et al. recently used an advanced nanoplasmonic sensing (NPS) platform to analyze circulating tumor-derived EVs (tEVs) and identified a set of EV biomarkers for detection of pancreatic ductal adenocarcinoma (PDAC).211 Compared to the precedent nPLEX system, the new platform incorporated a larger number of sensing arrays (> 100 sensing spots) and automatic operation for routine clinical operation. The authors first tested the platform with tEVs from PDAC cell lines as well as 11 patient-derived tumor xenograft models in PDAC. The NPS analysis showed good correlation of protein expression between tEVs and their parental cells (Spearman correlation coefficient r = 0.86) for selected cancer markers prescreened by whole cell analyses.
In the following clinical studies, the authors analyzed tEV markers in plasma samples of 32 patients as a training cohort consisting of 22 cases of PDAC and 10 healthy controls. In this training cohort, they identified a marker combination (EGFR, EPCAM, MUC1, GPC1 and WNT2) that showed a 100% detection accuracy while no single markers achieved sufficiently high sensitivity and accuracy. The marker combination, called PDACEV signature, was then applied to a prospective cohort of 43 patients of PDAC (n = 22), pancreatitis (n = 8), benign cyst (n = 5) and age-matched controls (n = 8; Figure 25a). In the prospective cohort, the accuracy for the PDACEV signature was 84%, outperforming all other single markers as well as serum CA19-9 (Figure 25b). The PDACEV signature showed a moderate correlation with tumor size (r = 0.58; P = 0.018; Figure 25c) and lower values for PDAC patients treated with neoadjuvant regimen than the untreated PDAC group. The EV analysis and PDACEV signature could be applied to different types of pancreatic diseases, potentially for early detection of intraductal papillary mucinous neoplasms (IPMNs) that have a high risk for progressing to invasive cancer (Figure 25d).
Figure 25. Pancreatic cancer detection.
(a) Heatmap analysis of EV markers for pancreatic cancer detection. The PDACEV signature is defined as a combined marker panel of EGFR, EPCAM, MUC1, GPC1, and WNT2. (b, c) Correlation of the PDACEV signature values with a serum biomarker CA 19-9 (b) and the tumor diameter (c) for patients with PDAC. tx, treatment. The dashed red line in (b) indicates the threshold values for positivity (CA 19-9, 37 U/ml; PDACEV signature, 0.87). (d) EV analyses for patients with different types of pancreatic diseases. The PDACEV signature values were measured for patient cohorts (n = 103) including (i) PDAC without treatment (n = 22), (ii) PDAC treated with neoadjuvant regimen (n = 24), (iii) IPMN (n = 13), (iv) other GI cancers mimicking the symptoms of pancreaticoduodenal cancers (n = 11), (v) pancreatic NET (n = 12), (vi) pancreatitis (n = 8), (vii) benign cystic tumors (n = 5), and (viii) age-matched controls (n = 18). Reprinted with permission from Ref 211. Copyright 2017 American Association for the Advancement of Science AAAS.
7.1.4. Prostate Cancer
Prostate cancer is the most common solid malignant disease worldwide. Prostate-specific antigen (PSA) is the most widely used blood-borne biomarker for screening prostate cancer, but faces much controversy and uncertainty. Unlike other cancers where early detection can save lives, experts believe that prostate cancer may be an interesting exception.258, 259 New evidence strongly suggests that most prostate cancers detected, especially the low grade tumors, will remain indolent for the patient’s lifetime. Overdiagnosis and overtreatment of indolent prostate cancer, contributed primarily by a lack of specificity of PSA for high grade tumors, is a serious health issue in most developed countries.
To address this challenge, Exosome Diagnostics has recently developed a novel urine exosome expression assay (the ExoDx Prostate IntelliScore/EPI urine exosome assay). This test is the first commercial EV diagnostic product and utilizes a three-gene signature, in combination with a proprietary algorithm. Recently, McKiernan and colleagues validated this gene expression assay to differentiate high-grade prostate cancer and avoid unnecessary biopsies.221 The EV-derived gene expression signature was first derived from normalized PCA 3 and ERG (V-ets erythroblastosis virus E26 oncogene homologs) RNA. To determine the performance of this urine EV gene expression assay, in addition and in comparison to standard of care (SOC, i.e., PSA level, age, race, and family history), the researchers first performed reverse-transcriptase polymerase chain reaction in a training set of patients. In this training cohort, the urine EV expression assay was performed in 255 men with known biopsy outcomes, and EPI demonstrated superior performance for predicting high grade disease.221 The derived prognostic score was further validated in an independent cohort of 512 patients (Figure 26a).260 As a stand-alone diagnostic test, the EPI test does not take into account other standard of care parameters in the score thus making it a complement to the current PSA test.
Figure 26. Prostate cancer and lung cancer detection.
(a) A urine-based EV gene expression assay, ExoDx™ Prostate IntelliScore (EPI), was used to evaluate high-grade prostate cancer at initial biopsy (n = 512). Area under receiver operating characteristic curve (AUC) analysis indicated the EPI test in combination with standard of care (SOC) significantly outperforms SOC along for predicating high-grade disease. (b) A protein marker panel was developed to diagnose lung cancers. AUC generated for the number of markers is given with the 95% confidence interval for the entire lung cancer cohort. Reprinted with permission from Ref 260. Copyright 2017 Exosome Diagnostics, Inc. Reprinted with permission from Ref 203. Copyright 2016 International Association for the Study of Lung Cancer.
7.1.5. Lung Cancer
Lung cancer is the main cause of cancer-related death worldwide.261 Several factors are associated with the poor patient outcome; a primary reason is delayed diagnosis, as survival rates decrease dramatically from early to late stages.204 The current standard diagnostic procedures typically involve sophisticated imaging methods (e.g., computed axial tomography, positron emission tomography) as well as invasive transbronchial needle aspiration or transthoracic biopsy.262, 263
Sandfeld-Paulsen et al. recently explored the potential of EV protein profiling in diagnosing lung cancer of all stages and various histological subtypes in patients.253 The authors reported that by using a vesicle array that contained 49 antibodies for EV capture and detection, they could identify multimarker panels and optimized the multmarker model by area under the curve (AUC) and random forest analysis (Figure 26b). In their study, the markers CD151, CD171, and tetraspanin 8 were the strongest separators of patients with lung cancer of all histological subtypes versus healthy controls.
7.1.6. Breast Cancer
Breast cancer is the most common type of cancer in women, accounting to 500,000 deaths yearly worldwide.264 Identifying a biomarker for early detection of breast cancer could enable significant clinical benefits, including better treatment outcome as well as less-aggressive treatment. CA 15-3, the most widely used serum marker in patients with metastatic breast cancer, however, has a low sensitivity (<70%).265
Hannafon and colleagues recently developed a molecular profile of exosome miRNAs secreted from breast cancer cells.266 The team collected exosomes from the conditioned media of human breast cancer cell lines, mouse plasma of patient-derived orthotopic xenograph models (PDX), as well as human plasma samples. Cellular and exosome miRNAs from cancer cell lines were profiled by next-generation small RNA sequencing, while plasma exosome miRNA expression was analyzed by qPCR.
By comparing the miRNA profiles across cancer cell lines, the authors discovered that several miRNAs were highly enriched in breast cancer exosomes (Figures 27a, b). In particular, miR-1246 was also detected at a significantly higher level in exosomes isolated from PDX mouse plasma. The authors further validated the finding by extending the miRNA profile to human plasma (Figures 27c, d). miR-1246 and miR-21 were detected at a higher level in plasma exosomes of breast cancer patients (n = 16), as compared to that of healthy control subjects. Receiver operating characteristic curve analysis further indicated that the combination of plasma exosome miR-1246 and miR-21 formed a better indicator of breast cancer than individual marker measurements.266
Figure 27. Breast cancer detection.
(a, b) qPCR analysis of miRNA in exosomes derived from mammary epithelial cells (MCF10A) and breast cancer cell lines (MCF7, ZR-75-1, T47D, MDA-MB-231, BT-20, BT-474, SK-BR-3). Fold change in expression is shown for the exosome miRNA relative to their cellular miRNA levels and normalized against spike-in miRNA control. (c, d) qPCR analysis of exosome miRNA expression in normal plasma and plasma from breast cancer patients (n =16). Exosomes were isolated from the plasma samples using commercial reagents and total RNA was extracted for qPCR analysis. Reprinted with permission from Ref 266. Copyright 2016 Hannafon et al.
7.2. Neurodegenerative Diseases
A similar model of disease progression is thought to occur for a majority of neurodegenerative diseases (e.g., Alzheimer’s disease, Parkinson’s disease, frontotemporal dementia), wherein a misfolded protein self-associates to form an ordered aggregate with the ability to propagate in cells.231, 267–270 In Alzheimer’s disease (AD), the Abeta peptide that forms amyloids is perhaps the best known of these protein aggregates.271 In Parkinson’s disease, a separate type of aggregate forms intracellularly composed mainly of alpha-synuclein and termed Lewy body.272 Recent studies have revealed that many misfolded proteins involved in neurodegenerative diseases are transported in EVs.229, 273 These vesicles thus present new hope for detection and monitoring of neurodegenerative diseases.
AD is a late-onset neurological disorder causing progressive loss of memory and cognitive abilities as a result of abnormal neurodegeneration. While the exact etiology of AD remains a topic of debate, it is clear that the accumulation of amyloid Abeta peptides in plaques and the neurofibrillary tangles of tau are important for the disease progression.232 These amyloid peptides are derived from proteolytic processing of the amyloid precursor protein (APP). This processing can occur in multiple locations in the cells, notably in pathways regulating endosomal vesicle recycling, which also coincide with EV formation.233, 274 Rajendran et al. determined that Abeta peptides are transported into multivesicular bodies.230 A small fraction of the Abeta peptides were found to be sorted into the intraluminal vesicles, and exported extracellularly in EVs (Figure 28a). The authors further showed that exosomal proteins could accumulate in the plaques of AD patient brains, suggesting a role for EVs in the spread of AD pathogenesis.
Figure 28. Detection of neurodegenerative diseases.
(a) EVs obtained from Alzheimer’s disease (AD) model cell line (N2A) were negatively stained with 1% uranyl acetate and immunolabeled with antibodies for the exosomal marker Alix. Exosomes also were immunolabeled for Aβ40 or Aβ42 and cholera toxin B subunit (CTx-B), which binds to the ganglioside GM1. (b) Longitudinal analysis of the development of altered levels of phosphorylated IRS-1 in AD. PC-AD, preclinical values 1 to 10 yr before diagnosis for patients with AD; control, values for cognitively normal healthy subjects matched by age and gender with each patient with AD at the time of diagnosis. Reprinted with permission from Ref 230. Copyright 2006 The National Academy of Sciences of the USA. Reprinted with permission from Ref 275. Copyright 2015 FASEB.
More recently, Kapogiannis et al. found that insulin resistance pathway markers could also be used in AD detection.275 Insulin resistance causes diminished glucose uptake in similar regions of the brain in AD and type 2 diabetes. By enriching for neural EVs from plasma samples through immunoaffinity capture, the researchers investigated the exosomal expression of phosphorylated serine-type 1 insulin receptor substrate (IRS-1) in control and preclinical AD patients (PC-AD) whose samples were obtained one to ten years before AD diagnosis, as well as AD patients (Figure 28b). Interestingly, the marker could be found in EVs at levels that were similar in the preclinical and manifest stages of AD, both of which were elevated in comparison to the control samples.
7.3. Acute Organ Injury
Due to their role in tissue homeostasis, EVs have emerged as an attractive circulating biomarker for noninvasive assessment of organ response to acute and chronic injuries. To date, EVs have been investigated to examine injuries to the lung199, 276 heart,277, 278 and kidney239, 240, as well as indications of preeclampsia, a pregnancy complication characterized by high blood pressure and signs of liver and kidney damages.243–245 For example, in an acute lung injury models, Moon et al. demonstrated that lung epithelial cell-derived EVs contain caspase-3, a pro-apoptotic factor, which activates macrophages. In acute myocardial injury, serum levels of cardiac-associated EV miRNAs (miR-1 and miR-133a) rapidly increased in the early hours after the onset of injury.199
Acute kidney injury (AKI) has a high prevalence in intensive care units, and incurs a high mortality and morbidity. Zhou et al. recently aimed to use urinary EV markers for detection and monitoring of AKI.241 The authors employed two-dimensional gel electrophoresis and mass spectrometry (LC-MS/MS) to identify a panel of candidate protein biomarkers isolated from urinary EVs. Fetuin-A, the most promising candidate biomarker, was validated through immunogold labeling and transmission electron microscopy to be localized inside urinary EVs (Figure 29a). Temporal release of urinary exosomal Fetuin-A was further measured in an AKI animal model (Figure 29b) and human subjects (Figure 29c) through Western blotting analysis. Notably, exosomal Fetuin-A increased over 52 fold at day 2 (a day before conventional serum creatinine increased) and remained elevated at day 5 after cisplatin-induced AKI, indicating the effectiveness of the urinary exosomal marker as a predictive marker of structural renal injury.241
Figure 29. Detection of acute kidney injury.
(a) Fetuin-A, a novel kidney injury marker, could be found inside rat urinary vesicles after cisplatin-induced acute kidney injury (AKI). Inset shows a magnified image of a urinary vesicle labeled with gold-conjugated anti-Fetuin-A. (b) Temporal secretion of urinary exosomal Fetuin-A in different AKI animal models. Western blotting analysis of urinary vesicles obtained from three rat models: one rat pre-, day 0, day 1, and day 2 after cisplatin injection; one rat from pre- and 8 h after bilateral ischemia and reperfusion (I/R); one rat from pre- and 24–30 h after volume depletion (VD). (c) Level of Fetuin-A in urinary vesicles of intensive care unit (ICU) patients with and without AKI, as compared to healthy volunteers. Reprinted with permission from Ref 241. Copyright 2006 International Society of Nephrology.
7.4. Therapeutic Potential
EVs are endogenous carriers of bioactive materials (e.g., proteins, mRNA, miRNA). Recent studies have shown that EVs participate in the transfer of materials and exchange of functional information, thereby mediating pathophysiological processes and activating long range intercellular communication.32, 279. While current EV classification is based on vesicle biogenesis, little is known about the correlation between the vesicles’ bioactivity and their classification, or if there are subpopulations of vesicles which harbor distinct functionalities. Better understanding, through molecular characterization proffered by technological improvements, will likely improve their diagnostic potential as circulating biomarkers as well as bioactive functionalities for therapeutic delivery.
To date, EVs have been used directly or engineered as therapeutic agents with multiple applications, such as in regenerative medicine280–282, cancer therapy,283 and immune modulation (Figure 30a).29, 284, 285 We will highlight some key examples, and direct readers to a more comprehensive review.32 Recent studies have found that EVs derived from mesenchymal stem cells possess beneficial paracrine effects in models of myocardial infraction, kidney injury and skeletal muscle repair.281, 286–288 This paracrine effect depends on the transfer of growth factors, proteins, bioactive lipids, and genetic materials to recipient cells via EVs, and provides potential in obviating the safety concerns related to direct stem cell transplantation.
Figure 30. Therapeutic applications of EVs.
(a) EVs could be used as therapeutic agents in antigen presentation, immune modulation, and tissue repair, through the transfer of EV proteins and nucleic acids. (b) As EVs naturally contain regulatory RNAs, they can be utilized for delivering oligonucleotide drugs of choice. EVs can be engineered to have targeting ligands present on their surface. Drug loading can be carried out either endogenously or exogenously, before being purified and applied for treatment. Reprinted with permission from Ref 32. Copyright 2013 Nature Publishing Group.
EVs can also be engineered for therapeutic delivery (Figure 30b).289 For example, two different approaches have been exploited in using EVs for gene therapy. In the first study, adeno-associated virus (AAV) vectors encapsulated in vesicles were shown to be substantially more efficient and less immunogenic than free vectors for the delivery of genetic cargo to recipient cells.290 In the second study, vesicles harboring suicide gene mRNA and protein were derived from transfected parental cells.291 The resultant vesicles were injected repeatedly into Schwannoma tumors in an orthotropic mouse model, which combined with systemic prodrug treatment led to tumor regression.
8. CONCLUSIONS AND OUTLOOK
Collectively, EVs contain diverse cellular constituents that not only mirror parent cells but also influence tumor microenvironment and disease progression.292, 293 The high heterogeneity and small size of EVs, however, pose technical challenges that preclude seamless interrogation of their molecular information and interactions. As reviewed here, many new analytical platforms are being developed and optimized for EVs, to render their analyses more convenient and sensitive than conventional methods (see Table 2). These platforms could provide insights into emerging EV biology and position EVs as 1) a potential non-invasive biomarker to guide effective strategies for personalized medicine and 2) a novel treatment to deliver endogenous modulation (e.g., immune activation) or exogenous therapeutics (e.g., drugs and proteins).
We conclude by proposing key aspects for further improvements in EV assays. First, a standard sample for assay calibration should be established. As with any other analytical tests, EV assays are highly susceptible to how samples are handled; EV counts, types, and molecular contents tend to vary per collection methods, storage media, and the assay itself.25, 96, 183 Having EV and assay standards will reduce inconsistencies stemming from protocol differences, promote assay reproducibility, and enable unbiased cross-platform comparison. As a part of this effort, The National Institutes of Health is developing a reference database for extracellular RNA as biomarkers (see https://exrna.org/). Second, to set robust baselines for disease status, it is important to analyze matched control samples, because human physiological factors (e.g., gender, age) could affect EV production and composition.24, 119 Statistical analyses should also consider these factors as biological variables, along with EV molecular information. Finally, we note the need to develop a technology for single EV profiling to unravel EVs’ biogenesis, molecular compositions, and diversity. Most of new analytical platforms, although superior to conventional methods, still requires large number of EVs, effectively measuring bulk properties from an ensemble of vesicles. Analyzing individual EVs could reveal unique molecular profiles of cell-specific EVs, which will further promote innovative clinical use of these vesicles (e.g., diagnostic, drug carriers), and allow us to construct a comprehensive EV atlas per different physical/biological parameters (e.g., vesicle size, origin, cell state).
Acknowledgments
The authors were supported in part by NIH grants R21-CA205322 (H.L.), R01-HL113156 (H.L.), R01-CA204019 (R.W.), R01-EB010011 (R.W.), R01-EB00462605A1 (R.W.), P01-CA069246 (X.O.B., R.W., H.L.), CA179563 (X.O.B), K99-CA201248-02 (H.I.); Liz Tilberis Award - Ovarian Cancer Research Fund (C.M.C.); a pilot grant from the Andrew L. Warshaw, M.D. Institute for Pancreatic Cancer Research at MGH (H.I.); the Lustgarten Foundation (R.W.); MGH Scholar Fund (H.L.); and Singapore government research grants R-722-000-005-133 (H.S.), NMRC/TCR/016-NNI/2016 (H.S.), and NUS Early Career Research Award (H.S.).
Biographies
Huilin Shao is Assistant Professor of Biomedical Engineering, and Principal Investigator at the Biomedical Institute for Global Health Research and Technology (BIGHEART), National University of Singapore. She received her BA from Cornell University, with a double major in Biological Sciences and in Physics, and completed her dual PhD (Biophysics) at Harvard University and PhD (Medical Engineering) from Harvard-MIT Health Sciences and Technology (HST). Her research focuses on developing integrated nanotechnology-based platforms for molecular analyses of novel biomarkers. She is a recipient of the NUS Early Career Research Award and the L’Oreal-UNESCO For Women in Science National Fellowship.
Hyungsoon Im is Assistant Professor in Radiology at Harvard Medical School (HMS) and at the Center for Systems Biology, Massachusetts General Hospital (MGH). He received his Ph.D. in Electrical Engineering from the University of Minnesota, Twin Cities and completed postdoctoral training at MGH. His research focuses on developing next generation diagnostics to understand the make-up of human cancers and changes associated with therapy. He has exploited uniquely designed nanomaterials and sensors to quantitatively analyze biomarkers and overcome clinical unmet needs. He is a recipient of NIH Pathway to Independence Award and International Society for Extracellular Vesicles (ISEV) Scholarship.
Cesar M. Castro, is an HMS Assistant Professor of Medicine and Director of the Cancer Program at the MGH Center for Systems Biology. He received his M.D. from the University of California - San Francisco (UCSF) School of Medicine where he also completed his residency in Internal Medicine. Dr. Castro completed fellowship in Medical Oncology at the Dana Farber / Partners Cancer Care program. His research interests comprise the development and translation of nanosensing and molecular imaging tools to propel cancer related proof-of-concept and early phase translational studies. His work has included creation and validation of novel technologies for EV characterization notably nPLEX and iMEX.
Xandra O. Breakefield is a Professor of Neurology at HMS and Director of the Molecular Neurogenetics Unit at MGH. She received her Ph.D. from Georgetown University in 1971 and did postdoctoral training with Dr. Marshall Nirenberg at the National Institutes of Health. She was on the faculty of Yale Medical School from 1974–1984 and has been on the faculty at HMS since 1984. She has published over 500 publications in peer reviewed journals. She served as President of the American Society of Gene and Cell Therapy. Her work has been honored by several awards, including the Mika Salpeter LIfetime Achievement Award (Society for Neuroscience), William Silen Lifetime Achievement in Mentoring Award (HMS), Special Achievement Award (International Society of Extracellular Vesicles), and Prize for Excellence in Dystonia Research (Bachmann-Strauss and Michael J Fox Foundation). She is a member of the American Academy of Arts and Sciences.
Ralph Weissleder is the Thrall Professor of Radiology and Professor of Systems Biology at Harvard Medical School (HMS), Director of the Center for Systems Biology at Massachusetts General Hospital (MGH), senior faculty in the HMS Department of Systems Biology, and Attending Clinician (Interventional Radiology) at MGH. He graduated from Heidelberg University in 1985, obtained his postdoctoral and residency training at MGH (1986-91) and has been on faculty at HMS since 1991. He has published over >750 publications in peer-reviewed journals, authored several textbooks and been named one of the “The World’s Most Influential Scientific Minds” by Thomson Reuters in 2014. His work has been honored with numerous awards including the J. Taylor International Prize in Medicine, the Millenium Pharmaceuticals Innovator Award, the AUR Memorial Award, the ARRS President's Award, The Society for Molecular Imaging Lifetime Achievement Award, the Academy of Molecular Imaging 2006 Distinguished Basic Scientist Award among others. In 2009 he was elected member of the US National Institute of Medicine.
Hakho Lee is Hostetter MGH Research Scholar, HMS Associate Professor of Radiology, Director of the Biomedical Engineering Program at the MGH Center for Systems Biology. He received his Ph.D. in Physics from Harvard University, and completed his post-doctoral training at MGH. Dr. Lee has expertise in nanomaterials, biophysics, microfluidics, and electronics. His research focuses on developing novel biomedical sensors for clinical applications, for example, the world’s smallest portable NMR device, integrated circuit (IC) chips for cancer cell detection, and a point-of-use device for allergen detection. He also pioneered new analytical technologies for EV characterization, including nPLEX, iMEX, iMER, iKEA, and SEA.
References
- 1.Théry C, Zitvogel L, Amigorena S. Exosomes: Composition, Biogenesis and Function. Nat. Rev. Immunol. 2002;2:569–79. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 2.Théry C, Ostrowski M, Segura E. Membrane Vesicles as Conveyors of Immune Responses. Nat. Rev. Immunol. 2009;9:581–93. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 3.Raposo G, Stoorvogel W. Extracellular Vesicles: Exosomes, Microvesicles, and Friends. J. Cell Biol. 2013;200:373–83. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colombo M, Raposo G, Théry C. Biogenesis, Secretion, and Intercellular Interactions of Exosomes and Other Extracellular Vesicles. Annu. Rev. Cell Dev. Biol. 2014;30:255–89. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 5.Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle Formation During Reticulocyte Maturation. Association of Plasma Membrane Activities With Released Vesicles (Exosomes) J. Biol. Chem. 1987;262:9412–20. [PubMed] [Google Scholar]
- 6.Graner MW, Alzate O, Dechkovskaia AM, Keene JD, Sampson JH, Mitchell DA, Bigner DD. Proteomic and Immunologic Analyses of Brain Tumor Exosomes. FASEB J. 2009;23:1541–57. doi: 10.1096/fj.08-122184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Exosomes: Proteomic Insights and Diagnostic Potential. Expert Rev. Proteomics. 2009;6:267–83. doi: 10.1586/epr.09.17. [DOI] [PubMed] [Google Scholar]
- 8.Mathivanan S, Fahner CJ, Reid GE, Simpson RJ. Exocarta 2012: Database of Exosomal Proteins, RNA and Lipids. Nucleic Acids Res. 2012;40:D1241–4. doi: 10.1093/nar/gkr828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-Mediated Transfer of mRNAs and MicroRNAs is a Novel Mechanism of Genetic Exchange Between Cells. Nat. Cell Biol. 2007;9:654–9. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 10.Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma Microvesicles Transport RNA and Proteins That Promote Tumour Growth and Provide Diagnostic Biomarkers. Nat. Cell Biol. 2008;10:1470–6. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balaj L, Lessard R, Dai L, Cho YJ, Pomeroy SL, Breakefield XO, Skog J. Tumour Microvesicles Contain Retrotransposon Elements and Amplified Oncogene Sequences. Nat Commun. 2011;2:180. doi: 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalra H, et al. Vesiclepedia: A Compendium for Extracellular Vesicles With Continuous Community Annotation. PLoS Biol. 2012;10:e1001450. doi: 10.1371/journal.pbio.1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keerthikumar S, et al. Exocarta: A Web-Based Compendium of Exosomal Cargo. J. Mol. Biol. 2016;428:688–92. doi: 10.1016/j.jmb.2015.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pucci F, et al. Scs Macrophages Suppress Melanoma By Restricting Tumor-Derived Vesicle-B Cell Interactions. Science. 2016;352:242–6. doi: 10.1126/science.aaf1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raimondo F, Morosi L, Chinello C, Magni F, Pitto M. Advances in Membranous Vesicle and Exosome Proteomics Improving Biological Understanding and Biomarker Discovery. Proteomics. 2011;11:709–20. doi: 10.1002/pmic.201000422. [DOI] [PubMed] [Google Scholar]
- 16.van der Pol E, Böing AN, Harrison P, Sturk A, Nieuwland R. Classification, Functions, and Clinical Relevance of Extracellular Vesicles. Pharmacol. Rev. 2012;64:676–705. doi: 10.1124/pr.112.005983. [DOI] [PubMed] [Google Scholar]
- 17.Gerlinger M, et al. Intratumor Heterogeneity and Branched Evolution Revealed By Multiregion Sequencing. N. Engl. J. Med. 2012;366:883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Logozzi M, et al. High Levels of Exosomes Expressing Cd63 and Caveolin-1 in Plasma of Melanoma Patients. PLoS One. 2009;4:e5219. doi: 10.1371/journal.pone.0005219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hosseini M, Khatamianfar S, Hassanian SM, Nedaeinia R, Shafiee M, Maftouh M, Ghayour-Mobarhan M, Sales SS, Avan A. Exosome-Encapsulated MicroRNAs as Potential Circulating Biomarkers in Colon Cancer. Curr. Pharm. Des. 2016;23:1705–1709. doi: 10.2174/1381612822666161201144634. [DOI] [PubMed] [Google Scholar]
- 20.Shao H, Chung J, Balaj L, Charest A, Bigner DD, Carter BS, Hochberg FH, Breakefield XO, Weissleder R, Lee H. Protein Typing of Circulating Microvesicles Allows Real-Time Monitoring of Glioblastoma Therapy. Nat. Med. 2012;18:1835–40. doi: 10.1038/nm.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Im H, Shao H, Park YI, Peterson VM, Castro CM, Weissleder R, Lee H. Label-Free Detection and Molecular Profiling of Exosomes With a Nano-Plasmonic Sensor. Nat. Biotechnol. 2014;32:490–5. doi: 10.1038/nbt.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shao H, Chung J, Lee K, Balaj L, Min C, Carter BS, Hochberg FH, Breakefield XO, Lee H, Weissleder R. Chip-Based Analysis of Exosomal mRNA Mediating Drug Resistance in Glioblastoma. Nat Commun. 2015;6:6999. doi: 10.1038/ncomms7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.György B, et al. Membrane Vesicles, Current State-of-the-art: Emerging Role of Extracellular Vesicles. Cell. Mol. Life Sci. 2011;68:2667–88. doi: 10.1007/s00018-011-0689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yáñez-Mó M, et al. Biological Properties of Extracellular Vesicles and Their Physiological Functions. J. Extracell. Vesicles. 2015;4:27066. doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Witwer KW, et al. Standardization of Sample Collection, Isolation and Analysis Methods in Extracellular Vesicle Research. J. Extracell. Vesicles. 2013;2 doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lötvall J, et al. Minimal Experimental Requirements for Definition of Extracellular Vesicles and Their Functions: A Position Statement From the International Society for Extracellular Vesicles. J. Extracell. Vesicles. 2014;3:26913. doi: 10.3402/jev.v3.26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Meel R, Krawczyk-Durka M, van Solinge WW, Schiffelers RM. Toward Routine Detection of Extracellular Vesicles in Clinical Samples. Int. J. Lab Hematol. 2014;36:244–53. doi: 10.1111/ijlh.12247. [DOI] [PubMed] [Google Scholar]
- 28.Shao H, Chung J, Issadore D. Diagnostic Technologies for Circulating Tumour Cells and Exosomes. Biosci. Rep. 2015;36:e00292. doi: 10.1042/BSR20150180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robbins PD, Morelli AE. Regulation of Immune Responses By Extracellular Vesicles. Nat. Rev. Immunol. 2014;14:195–208. doi: 10.1038/nri3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maas SL, Breakefield XO, Weaver AM. Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell. Biol. 2017;27:172–188. doi: 10.1016/j.tcb.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: Current Knowledge of Their Composition, Biological Functions, and Diagnostic and Therapeutic Potentials. Biochim. Biophys. Acta. 2012;1820:940–8. doi: 10.1016/j.bbagen.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 32.El Andaloussi S, Mäger I, Breakefield XO, Wood MJ. Extracellular Vesicles: Biology and Emerging Therapeutic Opportunities. Nat. Rev. Drug Discov. 2013;12:347–57. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 33.Akers JC, Gonda D, Kim R, Carter BS, Chen CC. Biogenesis of Extracellular Vesicles (Ev): Exosomes, Microvesicles, Retrovirus-Like Vesicles, and Apoptotic Bodies. J. Neurooncol. 2013;113:1–11. doi: 10.1007/s11060-013-1084-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feingold KR, Grunfeld C. Introduction to Lipids and Lipoproteins. Endotext 2000 [Google Scholar]
- 35.Barteneva NS, Fasler-Kan E, Bernimoulin M, Stern JN, Ponomarev ED, Duckett L, Vorobjev IA. Circulating Microparticles: Square the Circle. BMC Cell Biol. 2013;14:23. doi: 10.1186/1471-2121-14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nguyen HP, Simpson RJ, Salamonsen LA, Greening DW. Extracellular Vesicles in the Intrauterine Environment: Challenges and Potential Functions. Biol. Reprod. 2016;95:109. doi: 10.1095/biolreprod.116.143503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kowal J, Tkach M, Théry C. Biogenesis and Secretion of Exosomes. Curr. Opin. Cell Biol. 2014;29:116–25. doi: 10.1016/j.ceb.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 38. [accessed Nov 5, 2017];Secreted Extracellular Vesicles. abcam Home Page http://docs.abcam.com/pdf/general/secreted_extracellular_vesicles_web.pdf.
- 39.Piper RC, Katzmann DJ. Biogenesis and Function of Multivesicular Bodies. Annu. Rev. Cell Dev. Biol. 2007;23:519–47. doi: 10.1146/annurev.cellbio.23.090506.123319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hurley JH, Hanson PI. Membrane Budding and Scission By the Escrt Machinery: It’s All in the Neck. Nat. Rev. Mol. Cell Biol. 2010;11:556–66. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luzio JP, Gray SR, Bright NA. Endosome-Lysosome Fusion. Biochem. Soc. Trans. 2010;38:1413–6. doi: 10.1042/BST0381413. [DOI] [PubMed] [Google Scholar]
- 42.Théry C. Exosomes: Secreted Vesicles and Intercellular Communications. F1000 Biol. Rep. 2011;3:15. doi: 10.3410/B3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zwaal RF, Schroit AJ. Pathophysiologic Implications of Membrane Phospholipid Asymmetry in Blood Cells. Blood. 1997;89:1121–32. [PubMed] [Google Scholar]
- 44.Hugel B, Martinez MC, Kunzelmann C, Freyssinet JM. Membrane Microparticles: Two Sides of the Coin. Physiology (Bethesda) 2005;20:22–7. doi: 10.1152/physiol.00029.2004. [DOI] [PubMed] [Google Scholar]
- 45.Muralidharan-Chari V, Clancy J, Plou C, Romao M, Chavrier P, Raposo G, D’Souza-Schorey C. Arf6-Regulated Shedding of Tumor Cell-Derived Plasma Membrane Microvesicles. Curr. Biol. 2009;19:1875–85. doi: 10.1016/j.cub.2009.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nabhan JF, Hu R, Oh RS, Cohen SN, Lu Q. Formation and Release of Arrestin Domain-Containing Protein 1-Mediated Microvesicles (Armms) At Plasma Membrane By Recruitment of Tsg101 Protein. Proc. Natl. Acad. Sci. U S A. 2012;109:4146–51. doi: 10.1073/pnas.1200448109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choi DS, Kim DK, Kim YK, Gho YS. Proteomics of Extracellular Vesicles: Exosomes and Ectosomes. Mass Spectrom. Rev. 2015;34:474–90. doi: 10.1002/mas.21420. [DOI] [PubMed] [Google Scholar]
- 48.Henne WM, Buchkovich NJ, Emr SD. The Escrt Pathway. Dev. Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 49.Wollert T, Hurley JH. Molecular Mechanism of Multivesicular Body Biogenesis By Escrt Complexes. Nature. 2010;464:864–9. doi: 10.1038/nature08849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Colombo M, Moita C, van Niel G, Kowal J, Vigneron J, Benaroch P, Manel N, Moita LF, Théry C, Raposo G. Analysis of Escrt Functions in Exosome Biogenesis, Composition and Secretion Highlights the Heterogeneity of Extracellular Vesicles. J. Cell Sci. 2013;126:5553–65. doi: 10.1242/jcs.128868. [DOI] [PubMed] [Google Scholar]
- 51.van Niel G, Charrin S, Simoes S, Romao M, Rochin L, Saftig P, Marks MS, Rubinstein E, Raposo G. The Tetraspanin Cd63 Regulates Escrt-Independent and - Dependent Endosomal Sorting During Melanogenesis. Dev. Cell. 2011;21:708–21. doi: 10.1016/j.devcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Verweij FJ, et al. Lmp1 Association With Cd63 in Endosomes and Secretion Via Exosomes Limits Constitutive Nf-Κb Activation. EMBO J. 2011;30:2115–29. doi: 10.1038/emboj.2011.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vanlandingham PA, Ceresa BP. Rab7 Regulates Late Endocytic Trafficking Downstream of Multivesicular Body Biogenesis and Cargo Sequestration. J. Biol. Chem. 2009;284:12110–24. doi: 10.1074/jbc.M809277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ostrowski M, et al. Rab27a and Rab27b Control Different Steps of the Exosome Secretion Pathway. Nat. Cell Biol. 2010;12:19–30. doi: 10.1038/ncb2000. sup pp 1–13. [DOI] [PubMed] [Google Scholar]
- 55.Zeigerer A, et al. Rab5 is Necessary for the Biogenesis of the Endolysosomal System in Vivo. Nature. 2012;485:465–70. doi: 10.1038/nature11133. [DOI] [PubMed] [Google Scholar]
- 56.Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brügger B, Simons M. Ceramide Triggers Budding of Exosome Vesicles Into Multivesicular Endosomes. Science. 2008;319:1244–7. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 57.Thakur BK, et al. Double-Stranded DNA in Exosomes: A Novel Biomarker in Cancer Detection. Cell Res. 2014;24:766–9. doi: 10.1038/cr.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wei Z, et al. Coding and Noncoding Landscape of Extracellular RNA Released By Human Glioma Stem Cells. Nat Commun. 2017;8:1145. doi: 10.1038/s41467-017-01196-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nolte-’t Hoen EN, Buermans HP, Waasdorp M, Stoorvogel W, Wauben MH, t Hoen PA. Deep Sequencing of RNA From Immune Cell-Derived Vesicles Uncovers the Selective Incorporation of Small Non-Coding RNA Biotypes With Potential Regulatory Functions. Nucleic Acids Res. 2012;40:9272–85. doi: 10.1093/nar/gks658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lai CP, Kim EY, Badr CE, Weissleder R, Mempel TR, Tannous BA, Breakefield XO. Visualization and Tracking of Tumour Extracellular Vesicle Delivery and RNA Translation Using Multiplexed Reporters. Nat Commun. 2015;6:7029. doi: 10.1038/ncomms8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lo Cicero A, Stahl PD, Raposo G. Extracellular Vesicles Shuffling Intercellular Messages: For Good or for Bad. Curr. Opin. Cell Biol. 2015;35:69–77. doi: 10.1016/j.ceb.2015.04.013. [DOI] [PubMed] [Google Scholar]
- 62.György B, Hung ME, Breakefield XO, Leonard JN. Therapeutic Applications of Extracellular Vesicles: Clinical Promise and Open Questions. Annu. Rev. Pharmacol. Toxicol. 2015;55:439–64. doi: 10.1146/annurev-pharmtox-010814-124630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gould SJ, Raposo G. As We Wait: Coping With an Imperfect Nomenclature for Extracellular Vesicles. J. Extracell. Vesicles. 2013;2:20389. doi: 10.3402/jev.v2i0.20389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van der Pol E, Hoekstra AG, Sturk A, Otto C, van Leeuwen TG, Nieuwland R. Optical and Non-Optical Methods for Detection and Characterization of Microparticles and Exosomes. J. Thromb. Haemost. 2010;8:2596–607. doi: 10.1111/j.1538-7836.2010.04074.x. [DOI] [PubMed] [Google Scholar]
- 65.Sokolova V, Ludwig AK, Hornung S, Rotan O, Horn PA, Epple M, Giebel B. Characterisation of Exosomes Derived From Human Cells By Nanoparticle Tracking Analysis and Scanning Electron Microscopy. Colloids Surf. B Biointerfaces. 2011;87:146–50. doi: 10.1016/j.colsurfb.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 66.Théry C, Amigorena S, Raposo G, Clayton A. Isolation and Characterization of Exosomes From Cell Culture Supernatants and Biological Fluids. Curr. Protoc. Cell Biol. 2006;(Unit 3.22) doi: 10.1002/0471143030.cb0322s30. Chapter 3. [DOI] [PubMed] [Google Scholar]
- 67.Hakulinen J, Sankkila L, Sugiyama N, Lehti K, Keski-Oja J. Secretion of Active Membrane Type 1 Matrix Metalloproteinase (MMP-14) Into Extracellular Space in Microvesicular Exosomes. J. Cell. Biochem. 2008;105:1211–8. doi: 10.1002/jcb.21923. [DOI] [PubMed] [Google Scholar]
- 68.Street JM, Barran PE, Mackay CL, Weidt S, Balmforth C, Walsh TS, Chalmers RT, Webb DJ, Dear JW. Identification and Proteomic Profiling of Exosomes in Human Cerebrospinal Fluid. J. Transl. Med. 2012;10:5. doi: 10.1186/1479-5876-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tatischeff I, Larquet E, Falcón-Pérez JM, Turpin PY, Kruglik SG. Fast Characterisation of Cell-Derived Extracellular Vesicles By Nanoparticles Tracking Analysis, Cryo-Electron Microscopy, and Raman Tweezers Microspectroscopy. J. Extracell. Vesicles. 2012;1:19179. doi: 10.3402/jev.v1i0.19179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yuana Y, Koning RI, Kuil ME, Rensen PC, Koster AJ, Bertina RM, Osanto S. Cryo-Electron Microscopy of Extracellular Vesicles in Fresh Plasma. J. Extracell. Vesicles. 2013;2:21494. doi: 10.3402/jev.v2i0.21494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sharma S, Rasool HI, Palanisamy V, Mathisen C, Schmidt M, Wong DT, Gimzewski JK. Structural-Mechanical Characterization of Nanoparticle Exosomes in Human Saliva, Using Correlative Afm, Fesem, and Force Spectroscopy. ACS Nano. 2010;4:1921–6. doi: 10.1021/nn901824n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yuana Y, Oosterkamp TH, Bahatyrova S, Ashcroft B, Garcia Rodriguez P, Bertina RM, Osanto S. Atomic Force Microscopy: A Novel Approach to the Detection of Nanosized Blood Microparticles. J. Thromb. Haemost. 2010;8:315–23. doi: 10.1111/j.1538-7836.2009.03654.x. [DOI] [PubMed] [Google Scholar]
- 73.Lawrie AS, Albanyan A, Cardigan RA, Mackie IJ, Harrison P. Microparticle Sizing By Dynamic Light Scattering in Fresh-Frozen Plasma. Vox Sang. 2009;96:206–12. doi: 10.1111/j.1423-0410.2008.01151.x. [DOI] [PubMed] [Google Scholar]
- 74.Xu Y, Nakane N, Maurer-Spurej E. Novel Test for Microparticles in Platelet-Rich Plasma and Platelet Concentrates Using Dynamic Light Scattering. Transfusion. 2011;51:363–70. doi: 10.1111/j.1537-2995.2010.02819.x. [DOI] [PubMed] [Google Scholar]
- 75.Sitar S, Kejžar A, Pahovnik D, Kogej K, Tušek-Žnidarič M, Lenassi M, Žagar E. Size Characterization and Quantification of Exosomes By Asymmetrical-Flow Field-Flow Fractionation. Anal. Chem. 2015;87:9225–33. doi: 10.1021/acs.analchem.5b01636. 15. [DOI] [PubMed] [Google Scholar]
- 76.Dragovic RA, et al. Sizing and Phenotyping of Cellular Vesicles Using Nanoparticle Tracking Analysis. Nanomedicine. 2011;7:780–8. doi: 10.1016/j.nano.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gardiner C, Ferreira YJ, Dragovic RA, Redman CW, Sargent IL. Extracellular Vesicle Sizing and Enumeration By Nanoparticle Tracking Analysis. J. Extracell. Vesicles. 2013;2:19671. doi: 10.3402/jev.v2i0.19671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Coumans FA, van der Pol E, Böing AN, Hajji N, Sturk G, van Leeuwen TG, Nieuwland R. Reproducible Extracellular Vesicle Size and Concentration Determination With Tunable Resistive Pulse Sensing. J. Extracell. Vesicles. 2014;3:25922. doi: 10.3402/jev.v3.25922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vogel R, et al. A Standardized Method to Determine the Concentration of Extracellular Vesicles Using Tunable Resistive Pulse Sensing. J. Extracell. Vesicles. 2016;5:31242. doi: 10.3402/jev.v5.31242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Akers JC, Ramakrishnan V, Nolan JP, Duggan E, Fu CC, Hochberg FH, Chen CC, Carter BS. Comparative Analysis of Technologies for Quantifying Extracellular Vesicles (EVs) in Clinical Cerebrospinal Fluids (CSF) PLoS One. 2016;11:e0149866. doi: 10.1371/journal.pone.0149866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Maas SL, Broekman ML, de Vrij J. Tunable Resistive Pulse Sensing for the Characterization of Extracellular Vesicles. Methods Mol. Biol. 2017;1545:21–33. doi: 10.1007/978-1-4939-6728-5_2. [DOI] [PubMed] [Google Scholar]
- 82.Lee K, Fraser K, Ghaddar B, Yang K, Kim E, Balaj L, Chiocca EA, Breakefield XO, Lee H, Weissleder R. Multiplexed Profiling of Single Extracellular Vesicles. ACS Nano. doi: 10.1021/acsnano.7b07060. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schubert W, Bonnekoh B, Pommer AJ, Philipsen L, Böckelmann R, Malykh Y, Gollnick H, Friedenberger M, Bode M, Dress AW. Analyzing Proteome Topology and Function By Automated Multidimensional Fluorescence Microscopy. Nat. Biotechnol. 2006;24:1270–8. doi: 10.1038/nbt1250. [DOI] [PubMed] [Google Scholar]
- 84.Lin JR, Fallahi-Sichani M, Sorger PK. Highly Multiplexed Imaging of Single Cells Using a High-Throughput Cyclic Immunofluorescence Method. Nat Commun. 2015;6:8390. doi: 10.1038/ncomms9390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Revenfeld AL, Bak R, Nielsen MH, Stensballe A, Varming K, Jørgensen M. Diagnostic and Prognostic Potential of Extracellular Vesicles in Peripheral Blood. Clin. Ther. 2014;36:830–46. doi: 10.1016/j.clinthera.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 86.Arraud N, Linares R, Tan S, Gounou C, Pasquet JM, Mornet S, Brisson AR. Extracellular Vesicles From Blood Plasma: Determination of Their Morphology, Size, Phenotype and Concentration. J. Thromb. Haemost. 2014;12:614–27. doi: 10.1111/jth.12554. [DOI] [PubMed] [Google Scholar]
- 87.Andre F, et al. Malignant Effusions and Immunogenic Tumour-Derived Exosomes. Lancet. 2002;360:295–305. doi: 10.1016/S0140-6736(02)09552-1. [DOI] [PubMed] [Google Scholar]
- 88.Bard MP, Hegmans JP, Hemmes A, Luider TM, Willemsen R, Severijnen LA, van Meerbeeck JP, Burgers SA, Hoogsteden HC, Lambrecht BN. Proteomic Analysis of Exosomes Isolated From Human Malignant Pleural Effusions. Am. J. Respir. Cell Mol. Biol. 2004;31:114–21. doi: 10.1165/rcmb.2003-0238OC. [DOI] [PubMed] [Google Scholar]
- 89.Ginestra A, Miceli D, Dolo V, Romano FM, Vittorelli ML. Membrane Vesicles in Ovarian Cancer Fluids: A New Potential Marker. Anticancer Res. 1999;19:3439–45. [PubMed] [Google Scholar]
- 90.Graves LE, Ariztia EV, Navari JR, Matzel HJ, Stack MS, Fishman DA. Proinvasive Properties of Ovarian Cancer Ascites-Derived Membrane Vesicles. Cancer Res. 2004;64:7045–9. doi: 10.1158/0008-5472.CAN-04-1800. [DOI] [PubMed] [Google Scholar]
- 91.Zonneveld MI, Brisson AR, van Herwijnen MJ, Tan S, van de Lest CH, Redegeld FA, Garssen J, Wauben MH, Nolte-’t Hoen EN. Recovery of Extracellular Vesicles From Human Breast Milk is Influenced By Sample Collection and Vesicle Isolation Procedures. J. Extracell. Vesicles. 2014;3:24215. doi: 10.3402/jev.v3.24215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pieters BC, et al. Commercial Cow Milk Contains Physically Stable Extracellular Vesicles Expressing Immunoregulatory Tgf-Β. PLoS One. 2015;10:e0121123. doi: 10.1371/journal.pone.0121123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yang J, Wei F, Schafer C, Wong DT. Detection of Tumor Cell-Specific mRNA and Protein in Exosome-Like Microvesicles From Blood and Saliva. PLoS One. 2014;9:e110641. doi: 10.1371/journal.pone.0110641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen WW, et al. Beaming and Droplet Digital Pcr Analysis of Mutant Idh1 mRNA in Glioma Patient Serum and Cerebrospinal Fluid Extracellular Vesicles. Mol. Ther. Nucleic Acids. 2013;2:e109. doi: 10.1038/mtna.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Salih M, Zietse R, Hoorn EJ. Urinary Extracellular Vesicles and the Kidney: Biomarkers and Beyond. Am. J. Physiol. Renal Physiol. 2014;306:F1251–9. doi: 10.1152/ajprenal.00128.2014. [DOI] [PubMed] [Google Scholar]
- 96.Gardiner C, Di Vizio D, Sahoo S, Théry C, Witwer KW, Wauben M, Hill AF. Techniques Used for the Isolation and Characterization of Extracellular Vesicles: Results of a Worldwide Survey. J. Extracell. Vesicles. 2016;5:32945. doi: 10.3402/jev.v5.32945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vidal M, Mangeat P, Hoekstra D. Aggregation Reroutes Molecules From a Recycling to a Vesicle-Mediated Secretion Pathway During Reticulocyte Maturation. J. Cell Sci. 1997;110:1867–77. doi: 10.1242/jcs.110.16.1867. [DOI] [PubMed] [Google Scholar]
- 98.Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, Dingli F, Loew D, Tkach M, Théry C. Proteomic Comparison Defines Novel Markers to Characterize Heterogeneous Populations of Extracellular Vesicle Subtypes. Proc. Natl. Acad. Sci. U S A. 2016;113:E968–77. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Alvarez ML, Khosroheidari M, Kanchi Ravi R, DiStefano JK. Comparison of Protein, MicroRNA, and mRNA Yields Using Different Methods of Urinary Exosome Isolation for the Discovery of Kidney Disease Biomarkers. Kidney Int. 2012;82:1024–32. doi: 10.1038/ki.2012.256. [DOI] [PubMed] [Google Scholar]
- 100.Rekker K, Saare M, Roost AM, Kubo AL, Zarovni N, Chiesi A, Salumets A, Peters M. Comparison of Serum Exosome Isolation Methods for MicroRNA Profiling. Clin. Biochem. 2014;47:135–8. doi: 10.1016/j.clinbiochem.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 101.Böing AN, van der Pol E, Grootemaat AE, Coumans FA, Sturk A, Nieuwland R. Single-Step Isolation of Extracellular Vesicles By Size-Exclusion Chromatography. J. Extracell. Vesicles. 2014;3:23430. doi: 10.3402/jev.v3.23430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nordin JZ, et al. Ultrafiltration With Size-Exclusion Liquid Chromatography for High Yield Isolation of Extracellular Vesicles Preserving Intact Biophysical and Functional Properties. Nanomedicine. 2015;11:879–83. doi: 10.1016/j.nano.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 103.Lozano-Ramos I, Bancu I, Oliveira-Tercero A, Armengol MP, Menezes-Neto A, Del Portillo HA, Lauzurica-Valdemoros R, Borràs FE. Size-Exclusion Chromatography-Based Enrichment of Extracellular Vesicles From Urine Samples. J. Extracell. Vesicles. 2015;4:27369. doi: 10.3402/jev.v4.27369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gámez-Valero A, Monguió-Tortajada M, Carreras-Planella L, Franquesa M, Beyer K, Borràs FE. Size-Exclusion Chromatography-Based Isolation Minimally Alters Extracellular Vesicles’ Characteristics Compared to Precipitating Agents. Sci. Rep. 2016;6:33641. doi: 10.1038/srep33641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kreimer S, Ivanov AR. Rapid Isolation of Extracellular Vesicles From Blood Plasma With Size-Exclusion Chromatography Followed By Mass Spectrometry-Based Proteomic Profiling. Methods Mol. Biol. 2017;1660:295–302. doi: 10.1007/978-1-4939-7253-1_24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kang D, Oh S, Ahn SM, Lee BH, Moon MH. Proteomic Analysis of Exosomes From Human Neural Stem Cells By Flow Field-Flow Fractionation and Nanoflow Liquid Chromatography-Tandem Mass Spectrometry. J. Proteome Res. 2008;7:3475–80. doi: 10.1021/pr800225z. [DOI] [PubMed] [Google Scholar]
- 107.Rho J, Chung J, Im H, Liong M, Shao H, Castro CM, Weissleder R, Lee H. Magnetic Nanosensor for Detection and Profiling of Erythrocyte-Derived Microvesicles. ACS Nano. 2013;7:11227–33. doi: 10.1021/nn405016y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wunsch BH, Smith JT, Gifford SM, Wang C, Brink M, Bruce RL, Austin RH, Stolovitzky G, Astier Y. Nanoscale Lateral Displacement Arrays for the Separation of Exosomes and Colloids Down to 20 Nm. Nat. Nanotechnol. 2016;11:936–940. doi: 10.1038/nnano.2016.134. [DOI] [PubMed] [Google Scholar]
- 109.Lee K, Shao H, Weissleder R, Lee H. Acoustic Purification of Extracellular Microvesicles. ACS Nano. 2015;9:2321–7. doi: 10.1021/nn506538f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chen C, Skog J, Hsu CH, Lessard RT, Balaj L, Wurdinger T, Carter BS, Breakefield XO, Toner M, Irimia D. Microfluidic Isolation and Transcriptome Analysis of Serum Microvesicles. Lab Chip. 2010;10:505–11. doi: 10.1039/b916199f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yoo CE, Kim G, Kim M, Park D, Kang HJ, Lee M, Huh N. A Direct Extraction Method for MicroRNAs From Exosomes Captured By Immunoaffinity Beads. Anal. Biochem. 2012;431:96–8. doi: 10.1016/j.ab.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 112.Zhao Z, Yang Y, Zeng Y, He M. A Microfluidic Exosearch Chip for Multiplexed Exosome Detection Towards Blood-Based Ovarian Cancer Diagnosis. Lab Chip. 2016;16:489–96. doi: 10.1039/c5lc01117e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Reátegui E, et al. Engineered Nanointerfaces for Microfluidic Isolation and Molecular Profiling of Tumor-Specific Extracellular Vesicles. Nat Commun. 2017 doi: 10.1038/s41467-017-02261-1. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Perez-Hernandez D, Gutiérrez-Vázquez C, Jorge I, López-Martin S, Ursa A, Sánchez-Madrid F, Vázquez J, Yáñez-Mó M. The Intracellular Interactome of Tetraspanin-Enriched Microdomains Reveals Their Function as Sorting Machineries Toward Exosomes. J. Biol. Chem. 2013;288:11649–61. doi: 10.1074/jbc.M112.445304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Andreu Z, Yáñez-Mó M. Tetraspanins in Extracellular Vesicle Formation and Function. Front. Immunol. 2014;5:442. doi: 10.3389/fimmu.2014.00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Al-Nedawi K, Meehan B, Kerbel RS, Allison AC, Rak J. Endothelial Expression of Autocrine Vegf Upon the Uptake of Tumor-Derived Microvesicles Containing Oncogenic Egfr. Proc. Natl. Acad. Sci. U S A. 2009;106:3794–9. doi: 10.1073/pnas.0804543106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tauro BJ, Greening DW, Mathias RA, Mathivanan S, Ji H, Simpson RJ. Two Distinct Populations of Exosomes Are Released From Lim1863 Colon Carcinoma Cell-Derived Organoids. Mol. Cell. Proteomics. 2013;12:587–98. doi: 10.1074/mcp.M112.021303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mittelbrunn M, Sánchez-Madrid F. Intercellular Communication: Diverse Structures for Exchange of Genetic Information. Nat. Rev. Mol. Cell Biol. 2012;13:328–35. doi: 10.1038/nrm3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yuana Y, Sturk A, Nieuwland R. Extracellular Vesicles in Physiological and Pathological Conditions. Blood Rev. 2013;27:31–9. doi: 10.1016/j.blre.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 120.Katsuda T, Kosaka N, Ochiya T. The Roles of Extracellular Vesicles in Cancer Biology: Toward the Development of Novel Cancer Biomarkers. Proteomics. 2014;14:412–25. doi: 10.1002/pmic.201300389. [DOI] [PubMed] [Google Scholar]
- 121.Lobb RJ, Becker M, Wen SW, Wong CS, Wiegmans AP, Leimgruber A, Moller A. Optimized Exosome Isolation Protocol for Cell Culture Supernatant and Human Plasma. J. Extracell. Vesicles. 2015;4:27031. doi: 10.3402/jev.v4.27031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ueda K, Ishikawa N, Tatsuguchi A, Saichi N, Fujii R, Nakagawa H. Antibody-Coupled Monolithic Silica Microtips for Highthroughput Molecular Profiling of Circulating Exosomes. Sci. Rep. 2014;4:6232. doi: 10.1038/srep06232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kreimer S, Belov AM, Ghiran I, Murthy SK, Frank DA, Ivanov AR. Mass-Spectrometry-based Molecular Characterization of Extracellular Vesicles: Lipidomics and Proteomics. J. Proteome Res. 2015;14:2367–84. doi: 10.1021/pr501279t. [DOI] [PubMed] [Google Scholar]
- 124.Pocsfalvi G, Stanly C, Vilasi A, Fiume I, Capasso G, Turiák L, Buzas EI, Vékey K. Mass Spectrometry of Extracellular Vesicles. Mass Spectrom. Rev. 2016;35:3–21. doi: 10.1002/mas.21457. [DOI] [PubMed] [Google Scholar]
- 125.Utleg AG, Yi EC, Xie T, Shannon P, White JT, Goodlett DR, Hood L, Lin B. Proteomic Analysis of Human Prostasomes. Prostate. 2003;56:150–61. doi: 10.1002/pros.10255. [DOI] [PubMed] [Google Scholar]
- 126.Pisitkun T, Shen RF, Knepper MA. Identification and Proteomic Profiling of Exosomes in Human Urine. Proc. Natl. Acad. Sci. U S A. 2004;101:13368–73. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gonzalez-Begne M, Lu B, Han X, Hagen FK, Hand AR, Melvin JE, Yates JR. Proteomic Analysis of Human Parotid Gland Exosomes By Multidimensional Protein Identification Technology (Mudpit) J. Proteome Res. 2009;8:1304–14. doi: 10.1021/pr800658c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Choi DS, Choi DY, Hong BS, Jang SC, Kim DK, Lee J, Kim YK, Kim KP, Gho YS. Quantitative Proteomics of Extracellular Vesicles Derived From Human Primary and Metastatic Colorectal Cancer Cells. J. Extracell. Vesicles. 2012;1:18704. doi: 10.3402/jev.v1i0.18704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bachi A, Bonaldi T. Quantitative Proteomics as a New Piece of the Systems Biology Puzzle. J. Proteomics. 2008;71:357–67. doi: 10.1016/j.jprot.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 130.Lu P, Vogel C, Wang R, Yao X, Marcotte EM. Absolute Protein Expression Profiling Estimates the Relative Contributions of Transcriptional and Translational Regulation. Nat. Biotechnol. 2007;25:117–24. doi: 10.1038/nbt1270. [DOI] [PubMed] [Google Scholar]
- 131.Choi DS, et al. The Protein Interaction Network of Extracellular Vesicles Derived From Human Colorectal Cancer Cells. J. Proteome Res. 2012;11:1144–51. doi: 10.1021/pr200842h. [DOI] [PubMed] [Google Scholar]
- 132.Choi DS, Kim DK, Kim YK, Gho YS. Proteomics, Transcriptomics and Lipidomics of Exosomes and Ectosomes. Proteomics. 2013;13:1554–71. doi: 10.1002/pmic.201200329. [DOI] [PubMed] [Google Scholar]
- 133.Lee EY, et al. Gram-Positive Bacteria Produce Membrane Vesicles: Proteomics-Based Characterization of Staphylococcus Aureus-Derived Membrane Vesicles. Proteomics. 2009;9:5425–36. doi: 10.1002/pmic.200900338. [DOI] [PubMed] [Google Scholar]
- 134.Kim JH, Lee J, Park J, Gho YS. Gram-Negative and Gram-Positive Bacterial Extracellular Vesicles. Semin. Cell Dev. Biol. 2015;40:97–104. doi: 10.1016/j.semcdb.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 135.Orozco AF, Lewis DE. Flow Cytometric Analysis of Circulating Microparticles in Plasma. Cytometry A. 2010;77:502–14. doi: 10.1002/cyto.a.20886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.van der Pol E, van Gemert MJ, Sturk A, Nieuwland R, van Leeuwen TG. Single Vs. Swarm Detection of Microparticles and Exosomes By Flow Cytometry. J. Thromb. Haemost. 2012;10:919–30. doi: 10.1111/j.1538-7836.2012.04683.x. [DOI] [PubMed] [Google Scholar]
- 137.Pospichalova V, et al. Simplified Protocol for Flow Cytometry Analysis of Fluorescently Labeled Exosomes and Microvesicles Using Dedicated Flow Cytometer. J. Extracell. Vesicles. 2015;4:25530. doi: 10.3402/jev.v4.25530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Arraud N, Gounou C, Turpin D, Brisson AR. Fluorescence Triggering: A General Strategy for Enumerating and Phenotyping Extracellular Vesicles By Flow Cytometry. Cytometry A. 2016;89:184–95. doi: 10.1002/cyto.a.22669. [DOI] [PubMed] [Google Scholar]
- 139.Nolte-’t Hoen EN, et al. Quantitative and Qualitative Flow Cytometric Analysis of Nanosized Cell-Derived Membrane Vesicles. Nanomedicine. 2012;8:712–20. doi: 10.1016/j.nano.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Stoner SA, Duggan E, Condello D, Guerrero A, Turk JR, Narayanan PK, Nolan JP. High Sensitivity Flow Cytometry of Membrane Vesicles. Cytometry A. 2016;89:196–206. doi: 10.1002/cyto.a.22787. [DOI] [PubMed] [Google Scholar]
- 141.Kibria G, et al. A Rapid, Automated Surface Protein Profiling of Single Circulating Exosomes in Human Blood. Sci. Rep. 2016;6:36502. doi: 10.1038/srep36502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Nolan JP, Jones JC. Detection of Platelet Vesicles By Flow Cytometry. Platelets. 2017;28:256–262. doi: 10.1080/09537104.2017.1280602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Shao H, Yoon TJ, Liong M, Weissleder R, Lee H. Magnetic Nanoparticles for Biomedical Nmr-Based Diagnostics. Beilstein J. Nanotechnol. 2010;1:142–54. doi: 10.3762/bjnano.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Shao H, Min C, Issadore D, Liong M, Yoon TJ, Weissleder R, Lee H. Magnetic Nanoparticles and MicroNMR for Diagnostic Applications. Theranostics. 2012;2:55–65. doi: 10.7150/thno.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lee H, Sun E, Ham D, Weissleder R. Chip-Nmr Biosensor for Detection and Molecular Analysis of Cells. Nat. Med. 2008;14:869–74. doi: 10.1038/nm.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lee H, Yoon TJ, Figueiredo JL, Swirski FK, Weissleder R. Rapid Detection and Profiling of Cancer Cells in Fine-Needle Aspirates. Proc. Natl. Acad. Sci. U S A. 2009;106:12459–64. doi: 10.1073/pnas.0902365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Brolo AG. Plasmonics for Future Biosensors. Nat. Photonics. 2012;6:709–713. [Google Scholar]
- 148.Im H, Shao H, Weissleder R, Castro CM, Lee H. Nano-Plasmonic Exosome Diagnostics. Expert Rev. Mol. Diagn. 2015;15:725–33. doi: 10.1586/14737159.2015.1041378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Jeong S, Park J, Pathania D, Castro CM, Weissleder R, Lee H. Integrated Magneto-Electrochemical Sensor for Exosome Analysis. ACS Nano. 2016;10:1802–9. doi: 10.1021/acsnano.5b07584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yoshioka Y, et al. Ultra-Sensitive Liquid Biopsy of Circulating Extracellular Vesicles Using Exoscreen. Nat Commun. 2014;5:3591. doi: 10.1038/ncomms4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ravasi T, et al. Experimental Validation of the Regulated Expression of Large Numbers of Non-Coding RNAs From the Mouse Genome. Genome Res. 2006;16:11–9. doi: 10.1101/gr.4200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Huang X, et al. Characterization of Human Plasma-Derived Exosomal RNAs By Deep Sequencing. BMC Genomics. 2013;14:319. doi: 10.1186/1471-2164-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Eirin A, Riester SM, Zhu XY, Tang H, Evans JM, O’Brien D, van Wijnen AJ, Lerman LO. MicroRNA and mRNA Cargo of Extracellular Vesicles From Porcine Adipose Tissue-Derived Mesenchymal Stem Cells. Gene. 2014;551:55–64. doi: 10.1016/j.gene.2014.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bellingham SA, Coleman BM, Hill AF. Small RNA Deep Sequencing Reveals a Distinct miRNA Signature Released in Exosomes From Prion-Infected Neuronal Cells. Nucleic Acids Res. 2012;40:10937–49. doi: 10.1093/nar/gks832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Di Vizio D, et al. Large Oncosomes in Human Prostate Cancer Tissues and in the Circulation of Mice With Metastatic Disease. Am. J. Pathol. 2012;181:1573–84. doi: 10.1016/j.ajpath.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Morello M, et al. Large Oncosomes Mediate Intercellular Transfer of Functional MicroRNA. Cell Cycle. 2013;12:3526–36. doi: 10.4161/cc.26539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Mittelbrunn M, Gutiérrez-Vázquez C, Villarroya-Beltri C, González S, Sánchez-Cabo F, González MÁ, Bernad A, Sánchez-Madrid F. Unidirectional Transfer of MicroRNA-Loaded Exosomes From T Cells to Antigen-Presenting Cells. Nat Commun. 2011;2:282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bolukbasi MF, Mizrak A, Ozdener GB, Madlener S, Ströbel T, Erkan EP, Fan JB, Breakefield XO, Saydam O. Mir-1289 and “zipcode”-Like Sequence Enrich mRNAs in Microvesicles. Mol. Ther. Nucleic Acids. 2012;1:e10. doi: 10.1038/mtna.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, Martin-Cofreces N, Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M, Sánchez-Madrid F. Sumoylated Hnrnpa2b1 Controls the Sorting of Mirnas Into Exosomes Through Binding to Specific Motifs. Nat Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Koppers-Lalic D, et al. Nontemplated Nucleotide Additions Distinguish the Small RNA Composition in Cells From Exosomes. Cell Rep. 2014;8:1649–58. doi: 10.1016/j.celrep.2014.08.027. [DOI] [PubMed] [Google Scholar]
- 161.Lee Y, El Andaloussi S, Wood MJ. Exosomes and Microvesicles: Extracellular Vesicles for Genetic Information Transfer and Gene Therapy. Hum. Mol. Genet. 2012;21:R125–34. doi: 10.1093/hmg/dds317. [DOI] [PubMed] [Google Scholar]
- 162.Zhang J, Li S, Li L, Li M, Guo C, Yao J, Mi S. Exosome and Exosomal MicroRNA: Trafficking, Sorting, and Function. Genomics Proteomics Bioinformatics. 2015;13:17–24. doi: 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Tkach M, Théry C. Communication By Extracellular Vesicles: Where We Are and Where We Need to Go. Cell. 2016;164:1226–32. doi: 10.1016/j.cell.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 164.Batagov AO, Kurochkin IV. Exosomes Secreted By Human Cells Transport Largely mRNA Fragments That Are Enriched in the 3’-Untranslated Regions. Biol Direct. 2013;8:12. doi: 10.1186/1745-6150-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Redzic JS, Balaj L, van der Vos KE, Breakefield XO. Extracellular RNA Mediates and Marks Cancer Progression. Semin. Cancer Biol. 2014;28:14–23. doi: 10.1016/j.semcancer.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs Are Transported in Plasma and Delivered to Recipient Cells By High-Density Lipoproteins. Nat. Cell Biol. 2011;13:423–33. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wagner J, Riwanto M, Besler C, Knau A, Fichtlscherer S, Röxe T, Zeiher AM, Landmesser U, Dimmeler S. Characterization of Levels and Cellular Transfer of Circulating Lipoprotein-Bound MicroRNAs. Arterioscler. Thromb. Vasc. Biol. 2013;33:1392–400. doi: 10.1161/ATVBAHA.112.300741. [DOI] [PubMed] [Google Scholar]
- 168.Arroyo JD, et al. Argonaute2 Complexes Carry a Population of Circulating MicroRNAs Independent of Vesicles in Human Plasma. Proc. Natl. Acad. Sci. U S A. 2011;108:5003–8. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of Extracellular Circulating MicroRNA. Nucleic Acids Res. 2011;39:7223–33. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Min PK, Chan SY. The Biology of Circulating MicroRNAs in Cardiovascular Disease. Eur. J. Clin. Invest. 2015;45:860–74. doi: 10.1111/eci.12475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Turchinovich A, Weiz L, Burwinkel B. Isolation of Circulating MicroRNA Associated With RNA-Binding Protein. Methods Mol Biol. 2013;1024:97–107. doi: 10.1007/978-1-62703-453-1_8. [DOI] [PubMed] [Google Scholar]
- 172.Chevillet JR, et al. Quantitative and Stoichiometric Analysis of the MicroRNA Content of Exosomes. Proc. Natl. Acad. Sci. U S A. 2014;111:14888–93. doi: 10.1073/pnas.1408301111. 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Santangelo L, Giurato G, Cicchini C, Montaldo C, Mancone C, Tarallo R, Battistelli C, Alonzi T, Weisz A, Tripodi M. The RNA-Binding Protein Syncrip is a Component of the Hepatocyte Exosomal Machinery Controlling MicroRNA Sorting. Cell Rep. 2016;17:799–808. doi: 10.1016/j.celrep.2016.09.031. [DOI] [PubMed] [Google Scholar]
- 174.Teng Y, et al. MVP-Mediated Exosomal Sorting of miR-193a Promotes Colon Cancer Progression. Nat Commun. 2017;8:14448. doi: 10.1038/ncomms14448. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Tosar JP, Cayota A, Eitan E, Halushka MK, Witwer KW. Ribonucleic Artefacts: Are Some Extracellular RNA Discoveries Driven By Cell Culture Medium Components. J. Extracell. Vesicles. 2017;6:1272832. doi: 10.1080/20013078.2016.1272832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wei Z, Batagov AO, Carter DR, Krichevsky AM. Fetal Bovine Serum RNA Interferes With the Cell Culture Derived Extracellular RNA. Sci. Rep. 2016;6:31175. doi: 10.1038/srep31175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Conley A, et al. High-Throughput Sequencing of Two Populations of Extracellular Vesicles Provides an mRNA Signature That Can be Detected in the Circulation of Breast Cancer Patients. RNA Biol. 2017;14:305–316. doi: 10.1080/15476286.2016.1259061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Crescitelli R, Lässer C, Szabó TG, Kittel A, Eldh M, Dianzani I, Buzás EI, Lötvall J. Distinct RNA Profiles in Subpopulations of Extracellular Vesicles: Apoptotic Bodies, Microvesicles and Exosomes. J. Extracell. Vesicles. 2013;2:20677. doi: 10.3402/jev.v2i0.20677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Guescini M, Genedani S, Stocchi V, Agnati LF. Astrocytes and Glioblastoma Cells Release Exosomes Carrying mtDNA. J. Neural. Transm. (Vienna) 2010;117:1–4. doi: 10.1007/s00702-009-0288-8. [DOI] [PubMed] [Google Scholar]
- 180.Kahlert C, et al. Identification of Double-Stranded Genomic DNA Spanning All Chromosomes With Mutated KRAS and P53 DNA in the Serum Exosomes of Patients With Pancreatic Cancer. J. Biol. Chem. 2014;289:3869–75. doi: 10.1074/jbc.C113.532267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Takahashi A, et al. Exosomes Maintain Cellular Homeostasis By Excreting Harmful DNA From Cells. Nat Commun. 2017;8:15287. doi: 10.1038/ncomms15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Laurent LC, et al. Meeting Report: Discussions and Preliminary Findings on Extracellular RNA Measurement Methods From Laboratories in the NIH Extracellular RNA Communication Consortium. J. Extracell. Vesicles. 2015;4:26533. doi: 10.3402/jev.v4.26533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Mateescu B, et al. Obstacles and Opportunities in the Functional Analysis of Extracellular Vesicle RNA - an ISEV Position Paper. J. Extracell. Vesicles. 2017;6:1286095. doi: 10.1080/20013078.2017.1286095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Enderle D, et al. Characterization of RNA From Exosomes and Other Extracellular Vesicles Isolated By a Novel Spin Column-Based Method. PLoS One. 2015;10:e0136133. doi: 10.1371/journal.pone.0136133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Van Deun J, Mestdagh P, Sormunen R, Cocquyt V, Vermaelen K, Vandesompele J, Bracke M, De Wever O, Hendrix A. The Impact of Disparate Isolation Methods for Extracellular Vesicles on Downstream RNA Profiling. J. Extracell. Vesicles. 2014;3:24858. doi: 10.3402/jev.v3.24858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A Summary. Acta Neuropathol. 2016;131:803–20. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 187.Saugstad JA, et al. Analysis of Extracellular RNA in Cerebrospinal Fluid. J. Extracell. Vesicles. 2017;6:1317577. doi: 10.1080/20013078.2017.1317577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Taller D, Richards K, Slouka Z, Senapati S, Hill R, Go DB, Chang HC. On-Chip Surface Acoustic Wave Lysis and Ion-Exchange Nanomembrane Detection of Exosomal RNA for Pancreatic Cancer Study and Diagnosis. Lab Chip. 2015;15:1656–66. doi: 10.1039/c5lc00036j. [DOI] [PubMed] [Google Scholar]
- 189.SepúAlveda B, Angelomé PC, Lechuga LM, Liz-Marzán LM. LSPR-Based Nanobiosensors. Nano Today. 2009;4:244–251. [Google Scholar]
- 190.Petryayeva E, Krull UJ. Localized Surface Plasmon Resonance: Nanostructures, Bioassays and Biosensing--a Review. Anal. Chim. Acta. 2011;706:8–24. doi: 10.1016/j.aca.2011.08.020. [DOI] [PubMed] [Google Scholar]
- 191.Szunerits S, Boukherroub R. Sensing Using Localised Surface Plasmon Resonance Sensors. Chem. Commun. 2012;48:8999–9010. doi: 10.1039/c2cc33266c. [DOI] [PubMed] [Google Scholar]
- 192.Joshi GK, Deitz-McElyea S, Liyanage T, Lawrence K, Mali S, Sardar R, Korc M. Label-Free Nanoplasmonic-Based Short Noncoding RNA Sensing At Attomolar Concentrations Allows for Quantitative and Highly Specific Assay of MicroRNA-10b in Biological Fluids and Circulating Exosomes. ACS Nano. 2015;9:11075–89. doi: 10.1021/acsnano.5b04527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Théry C. Cancer: Diagnosis By Extracellular Vesicles. Nature. 2015;523:161–2. doi: 10.1038/nature14626. [DOI] [PubMed] [Google Scholar]
- 194.Zhang Y, Wang XF. A Niche Role for Cancer Exosomes in Metastasis. Nat. Cell Biol. 2015;17:709–11. doi: 10.1038/ncb3181. [DOI] [PubMed] [Google Scholar]
- 195.Lener T, et al. Applying Extracellular Vesicles Based Therapeutics in Clinical Trials - an Isev Position Paper. J. Extracell. Vesicles. 2015;4:30087. doi: 10.3402/jev.v4.30087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Touat M, Duran-Pena A, Alentorn A, Lacroix L, Massard C, Idbaih A. Emerging Circulating Biomarkers in Glioblastoma: Promises and Challenges. Expert Rev. Mol. Diagn. 2015;15:1311–23. doi: 10.1586/14737159.2015.1087315. [DOI] [PubMed] [Google Scholar]
- 197.Bergmann C, Strauss L, Wieckowski E, Czystowska M, Albers A, Wang Y, Zeidler R, Lang S, Whiteside TL. Tumor-Derived Microvesicles in Sera of Patients With Head and Neck Cancer and Their Role in Tumor Progression. Head Neck. 2009;31:371–80. doi: 10.1002/hed.20968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Principe S, Hui AB, Bruce J, Sinha A, Liu FF, Kislinger T. Tumor-Derived Exosomes and Microvesicles in Head and Neck Cancer: Implications for Tumor Biology and Biomarker Discovery. Proteomics. 2013;13:1608–23. doi: 10.1002/pmic.201200533. [DOI] [PubMed] [Google Scholar]
- 199.Moon PG, et al. Identification of Developmental Endothelial Locus-1 on Circulating Extracellular Vesicles as a Novel Biomarker for Early Breast Cancer Detection. Clin. Cancer Res. 2016;22:1757–66. doi: 10.1158/1078-0432.CCR-15-0654. [DOI] [PubMed] [Google Scholar]
- 200.Lowry MC, Gallagher WM, O’Driscoll L. The Role of Exosomes in Breast Cancer. Clin. Chem. 2015;61:1457–65. doi: 10.1373/clinchem.2015.240028. [DOI] [PubMed] [Google Scholar]
- 201.Ma X, et al. Essential Role for Trpc5-Containing Extracellular Vesicles in Breast Cancer With Chemotherapeutic Resistance. Proc. Natl. Acad. Sci. U S A. 2014;111:6389–94. doi: 10.1073/pnas.1400272111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Akagi T, Kato K, Kobayashi M, Kosaka N, Ochiya T, Ichiki T. On-Chip Immunoelectrophoresis of Extracellular Vesicles Released From Human Breast Cancer Cells. PLoS One. 2015;10:e0123603. doi: 10.1371/journal.pone.0123603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Sandfeld-Paulsen B, Jakobsen KR, Bæk R, Folkersen BH, Rasmussen TR, Meldgaard P, Varming K, Jørgensen MM, Sorensen BS. Exosomal Proteins as Diagnostic Biomarkers in Lung Cancer. J. Thorac. Oncol. 2016;11:1701–10. doi: 10.1016/j.jtho.2016.05.034. [DOI] [PubMed] [Google Scholar]
- 204.Vansteenkiste J, et al. 2nd ESMO Consensus Conference on Lung Cancer: Early-Stage Non-Small-cell Lung Cancer Consensus on Diagnosis, Treatment and Follow-Up. Ann. Oncol. 2014;25:1462–74. doi: 10.1093/annonc/mdu089. [DOI] [PubMed] [Google Scholar]
- 205.Frydrychowicz M, Kolecka-Bednarczyk A, Madejczyk M, Yasar S, Dworacki G. Exosomes - Structure, Biogenesis and Biological Role in Non-Small-cell Lung Cancer. Scand. J. Immunol. 2015;81:2–10. doi: 10.1111/sji.12247. [DOI] [PubMed] [Google Scholar]
- 206.Fujita Y, Kosaka N, Araya J, Kuwano K, Ochiya T. Extracellular Vesicles in Lung Microenvironment and Pathogenesis. Trends Mol. Med. 2015;21:533–42. doi: 10.1016/j.molmed.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 207.Allenson K, et al. High Prevalence of Mutant KRAS in Circulating Exosome-Derived DNA From Early-Stage Pancreatic Cancer Patients. Ann. Oncol. 2017;28:741–747. doi: 10.1093/annonc/mdx004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Melo SA, et al. Glypican-1 Identifies Cancer Exosomes and Detects Early Pancreatic Cancer. Nature. 2015;523:177–82. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Nuzhat Z, Kinhal V, Sharma S, Rice GE, Joshi V, Salomon C. Tumour-Derived Exosomes as a Signature of Pancreatic Cancer - Liquid Biopsies as Indicators of Tumour Progression. Oncotarget. 2017;8:17279–17291. doi: 10.18632/oncotarget.13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Costa-Silva B, et al. Pancreatic Cancer Exosomes Initiate Pre-Metastatic Niche Formation in the Liver. Nat. Cell Biol. 2015;17:816–26. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Yang KS, et al. Multiparametric Plasma EV Profiling Facilitates Diagnosis of Pancreatic Malignancy. Sci. Transl. Med. 2017;9:eaal3226. doi: 10.1126/scitranslmed.aal3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Ng YH, Rome S, Jalabert A, Forterre A, Singh H, Hincks CL, Salamonsen LA. Endometrial Exosomes/microvesicles in the Uterine Microenvironment: A New Paradigm for Embryo-Endometrial Cross Talk At Implantation. PLoS One. 2013;8:e58502. doi: 10.1371/journal.pone.0058502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Li Q, Shao Y, Zhang X, Zheng T, Miao M, Qin L, Wang B, Ye G, Xiao B, Guo J. Plasma Long Noncoding RNA Protected By Exosomes as a Potential Stable Biomarker for Gastric Cancer. Tumour Biol. 2015;36:2007–12. doi: 10.1007/s13277-014-2807-y. [DOI] [PubMed] [Google Scholar]
- 214.Mohankumar S, Patel T. Extracellular Vesicle Long Noncoding RNA as Potential Biomarkers of Liver Cancer. Brief Funct. Genomics. 2016;15:249–56. doi: 10.1093/bfgp/elv058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Szabo G, Momen-Heravi F. Extracellular Vesicles in Liver Disease and Potential as Biomarkers and Therapeutic Targets. Nat. Rev. Gastroenterol. Hepatol. 2017;14:455–466. doi: 10.1038/nrgastro.2017.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Maji S, Matsuda A, Yan IK, Parasramka M, Patel T. Extracellular Vesicles in Liver Diseases. Am. J. Physiol. Gastrointest. Liver Physiol. 2017;312:G194–G200. doi: 10.1152/ajpgi.00216.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Welton JL, Khanna S, Giles PJ, Brennan P, Brewis IA, Staffurth J, Mason MD, Clayton A. Proteomics Analysis of Bladder Cancer Exosomes. Mol. Cell. Proteomics. 2010;9:1324–38. doi: 10.1074/mcp.M000063-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Andreu Z, Otta Oshiro R, Redruello A, López-Martin S, Gutiérrez-Vázquez C, Morato E, Marina AI, Olivier Gómez C, Yáñez-Mó M. Extracellular Vesicles as a Source for Non-Invasive Biomarkers in Bladder Cancer Progression. Eur. J. Pharm. Sci. 2017;98:70–79. doi: 10.1016/j.ejps.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 219.Dong L, et al. Circulating Long RNAs in Serum Extracellular Vesicles: Their Characterization and Potential Application as Biomarkers for Diagnosis of Colorectal Cancer. Cancer Epidemiol. Biomarkers Prev. 2016;25:1158–66. doi: 10.1158/1055-9965.EPI-16-0006. [DOI] [PubMed] [Google Scholar]
- 220.Matsumura T, et al. Exosomal MicroRNA in Serum is a Novel Biomarker of Recurrence in Human Colorectal Cancer. Br. J. Cancer. 2015;113:275–81. doi: 10.1038/bjc.2015.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.McKiernan J, et al. A Novel Urine Exosome Gene Expression Assay to Predict High-Grade Prostate Cancer At Initial Biopsy. JAMA Oncol. 2016;2:882–9. doi: 10.1001/jamaoncol.2016.0097. [DOI] [PubMed] [Google Scholar]
- 222.Russo LM, et al. Urinary Exosomes as a Stable Source of mRNA for Prostate Cancer Analysis. J. Clin. Oncol. 2012;30:174. [Google Scholar]
- 223.Principe S, et al. In-Depth Proteomic Analyses of Exosomes Isolated From Expressed Prostatic Secretions in Urine. Proteomics. 2013;13:1667–1671. doi: 10.1002/pmic.201200561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Moltzahn F, Olshen AB, Baehner L, Peek A, Fong L, Stöppler H, Simko J, Hilton JF, Carroll P, Blelloch R. Microfluidic-Based Multiplex qRT-PCR Identifies Diagnostic and Prognostic MicroRNA Signatures in the Sera of Prostate Cancer Patients. Cancer Res. 2011;71:550–60. doi: 10.1158/0008-5472.CAN-10-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Dorayappan KD, Wallbillich JJ, Cohn DE, Selvendiran K. The Biological Significance and Clinical Applications of Exosomes in Ovarian Cancer. Gynecol. Oncol. 2016;142:199–205. doi: 10.1016/j.ygyno.2016.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Tang MK, Wong AS. Exosomes: Emerging Biomarkers and Targets for Ovarian Cancer. Cancer Lett. 2015;367:26–33. doi: 10.1016/j.canlet.2015.07.014. [DOI] [PubMed] [Google Scholar]
- 227.Nawaz M, et al. Extracellular Vesicles in Ovarian Cancer: Applications to Tumor Biology, Immunotherapy and Biomarker Discovery. Expert Rev. Proteomics. 2016;13:395–409. doi: 10.1586/14789450.2016.1165613. [DOI] [PubMed] [Google Scholar]
- 228.Beach A, Zhang HG, Ratajczak MZ, Kakar SS. Exosomes: An Overview of Biogenesis, Composition and Role in Ovarian Cancer. J. Ovarian Res. 2014;7:14. doi: 10.1186/1757-2215-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Coleman BM, Hill AF. Extracellular Vesicles--their Role in the Packaging and Spread of Misfolded Proteins Associated With Neurodegenerative Diseases. Semin. Cell Dev. Biol. 2015;40:89–96. doi: 10.1016/j.semcdb.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 230.Rajendran L, Honsho M, Zahn TR, Keller P, Geiger KD, Verkade P, Simons K. Alzheimer’s Disease Beta-Amyloid Peptides Are Released in Association With Exosomes. Proc. Natl. Acad. Sci. U S A. 2006;103:11172–7. doi: 10.1073/pnas.0603838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Ross CA, Poirier MA. Protein Aggregation and Neurodegenerative Disease. Nat. Med. 2004;10:S10–7. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 232.Spires-Jones TL, Hyman BT. The Intersection of Amyloid Beta and Tau At Synapses in Alzheimer’s Disease. Neuron. 2014;82:756–71. doi: 10.1016/j.neuron.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Zhang H, Ma Q, Zhang YW, Xu H. Proteolytic Processing of Alzheimer’s Β-Amyloid Precursor Protein. J. Neurochem. 2012;120 Suppl 1:9–21. doi: 10.1111/j.1471-4159.2011.07519.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Vella LJ, Hill AF, Cheng L. Focus on Extracellular Vesicles: Exosomes and Their Role in Protein Trafficking and Biomarker Potential in Alzheimer’s and Parkinson’s Disease. Int. J. Mol. Sci. 2016;17:173. doi: 10.3390/ijms17020173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Shi M, et al. Plasma Exosomal Α-Synuclein is Likely CNS-Derived and Increased in Parkinson’s Disease. Acta Neuropathol. 2014;128:639–650. doi: 10.1007/s00401-014-1314-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Chistiakov DA, Chistiakov AA. Α-Synuclein-carrying Extracellular Vesicles in Parkinson’s Disease: Deadly Transmitters. Acta Neurol. Belg. 2017;117:43–51. doi: 10.1007/s13760-016-0679-1. [DOI] [PubMed] [Google Scholar]
- 237.Bank IE, et al. The Diagnostic and Prognostic Potential of Plasma Extracellular Vesicles for Cardiovascular Disease. Expert Rev. Mol. Diagn. 2015;15:1577–88. doi: 10.1586/14737159.2015.1109450. [DOI] [PubMed] [Google Scholar]
- 238.Gaceb A, Martinez MC, Andriantsitohaina R. Extracellular Vesicles: New Players in Cardiovascular Diseases. Int. J. Biochem. Cell Biol. 2014;50:24–8. doi: 10.1016/j.biocel.2014.01.018. [DOI] [PubMed] [Google Scholar]
- 239.Gámez-Valero A, Lozano-Ramos SI, Bancu I, Lauzurica-Valdemoros R, Borràs FE. Urinary Extracellular Vesicles as Source of Biomarkers in Kidney Diseases. Front. Immunol. 2015;6:6. doi: 10.3389/fimmu.2015.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.van Balkom BW, Pisitkun T, Verhaar MC, Knepper MA. Exosomes and the Kidney: Prospects for Diagnosis and Therapy of Renal Diseases. Kidney Int. 2011;80:1138–45. doi: 10.1038/ki.2011.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Zhou H, et al. Exosomal Fetuin-a Identified By Proteomics: A Novel Urinary Biomarker for Detecting Acute Kidney Injury. Kidney Int. 2006;70:1847–57. doi: 10.1038/sj.ki.5001874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Zhang W, Zhou X, Zhang H, Yao Q, Liu Y, Dong Z. Extracellular Vesicles in Diagnosis and Therapy of Kidney Diseases. Am. J. Physiol. Renal Physiol. 2016;311:F844–F851. doi: 10.1152/ajprenal.00429.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Baig S, et al. Proteomic Analysis of Human Placental Syncytiotrophoblast Microvesicles in Preeclampsia. Clin. Proteomics. 2014;11:40. doi: 10.1186/1559-0275-11-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Escudero CA, Herlitz K, Troncoso F, Acurio J, Aguayo C, Roberts JM, Truong G, Duncombe G, Rice G, Salomon C. Role of Extracellular Vesicles and MicroRNAs on Dysfunctional Angiogenesis During Preeclamptic Pregnancies. Front. Physiol. 2016;7:98. doi: 10.3389/fphys.2016.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Pillay P, Maharaj N, Moodley J, Mackraj I. Placental Exosomes and Pre-Eclampsia: Maternal Circulating Levels in Normal Pregnancies and, Early and Late Onset Pre-Eclamptic Pregnancies. Placenta. 2016;46:18–25. doi: 10.1016/j.placenta.2016.08.078. [DOI] [PubMed] [Google Scholar]
- 246.Mueller MM, Fusenig NE. Friends or Foes - Bipolar Effects of the Tumour Stroma in Cancer. Nat. Rev. Cancer. 2004;4:839–49. doi: 10.1038/nrc1477. [DOI] [PubMed] [Google Scholar]
- 247.Pietras K, Ostman A. Hallmarks of Cancer: Interactions With the Tumor Stroma. Exp. Cell Res. 2010;316:1324–31. doi: 10.1016/j.yexcr.2010.02.045. [DOI] [PubMed] [Google Scholar]
- 248.Turley SJ, Cremasco V, Astarita JL. Immunological Hallmarks of Stromal Cells in the Tumour Microenvironment. Nat. Rev. Immunol. 2015;15:669–82. doi: 10.1038/nri3902. [DOI] [PubMed] [Google Scholar]
- 249.D’Souza-Schorey C, Clancy JW. Tumor-Derived Microvesicles: Shedding Light on Novel Microenvironment Modulators and Prospective Cancer Biomarkers. Genes Dev. 2012;26:1287–99. doi: 10.1101/gad.192351.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Webber J, Yeung V, Clayton A. Extracellular Vesicles as Modulators of the Cancer Microenvironment. Semin. Cell Dev. Biol. 2015;40:27–34. doi: 10.1016/j.semcdb.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 251.Fais S, et al. Evidence-Based Clinical Use of Nanoscale Extracellular Vesicles in Nanomedicine. ACS Nano. 2016;10:3886–99. doi: 10.1021/acsnano.5b08015. [DOI] [PubMed] [Google Scholar]
- 252.Hochberg FH, Atai NA, Gonda D, Hughes MS, Mawejje B, Balaj L, Carter RS. Glioma Diagnostics and Biomarkers: An Ongoing Challenge in the Field of Medicine and Science. Expert Rev. Mol. Diagn. 2014;14:439–52. doi: 10.1586/14737159.2014.905202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Santiago-Dieppa DR, Steinberg J, Gonda D, Cheung VJ, Carter BS, Chen CC. Extracellular Vesicles as a Platform for ‘Liquid Biopsy’ in Glioblastoma Patients. Expert Rev Mol Diagn. 2014;14:819–25. doi: 10.1586/14737159.2014.943193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Ilic M, Ilic I. Epidemiology of Pancreatic Cancer. World J. Gastroenterol. 2016;22:9694–9705. doi: 10.3748/wjg.v22.i44.9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Liang K, et al. Nanoplasmonic Quantification of Tumour-Derived Extracellular Vesicles in Plasma Microsamples for Diagnosis and Treatment Monitoring. Nat. Biomed. Eng. 2017;1:0021. doi: 10.1038/s41551-016-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Poruk KE, Gay DZ, Brown K, Mulvihill JD, Boucher KM, Scaife CL, Firpo MA, Mulvihill SJ. The Clinical Utility of Ca 19-9 in Pancreatic Adenocarcinoma: Diagnostic and Prognostic Updates. Curr. Mol. Med. 2013;13:340–51. doi: 10.2174/1566524011313030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Chari ST, et al. Early Detection of Sporadic Pancreatic Cancer: Summative Review. Pancreas. 2015;44:693–712. doi: 10.1097/MPA.0000000000000368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Loeb S, Bjurlin MA, Nicholson J, Tammela TL, Penson DF, Carter HB, Carroll P, Etzioni R. Overdiagnosis and Overtreatment of Prostate Cancer. Eur. Urol. 2014;65:1046–55. doi: 10.1016/j.eururo.2013.12.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Lee DJ, Mallin K, Graves AJ, Chang SS, Penson DF, Resnick MJ, Barocas DA. Recent Changes in Prostate Cancer Screening Practices and Prostate Cancer Epidemiology. J. Urol. 2017;198:1230–1240. doi: 10.1016/j.juro.2017.05.074. [DOI] [PubMed] [Google Scholar]
- 260.Donovan M, Torkler P, Bentis C, Noerholm M, Skog J, McKiernan J. Radical prostatectomy outcomes from a validated urine exosome gene expression assay which predicts high-grade (GS7) prostate cancer suggests utility for men enrolled in active surveillance. Presented at the American Urological Association Annual Meeting; Boston, MA. May 12–16, 2017; Paper 17-7028. [Google Scholar]
- 261.Siegel R, Ma J, Zou Z, Jemal A. Cancer Statistics, 2014. CA Cancer J. Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 262.MacMahon H, Austin JH, Gamsu G, Herold CJ, Jett JR, Naidich DP, Patz EF, Swensen SJ, Fleischner S. Guidelines for Management of Small Pulmonary Nodules Detected on Ct Scans: A Statement From the Fleischner Society. Radiology. 2005;237:395–400. doi: 10.1148/radiol.2372041887. [DOI] [PubMed] [Google Scholar]
- 263.Wiener RS, Schwartz LM, Woloshin S, Welch HG. Population-Based Risk for Complications After Transthoracic Needle Lung Biopsy of a Pulmonary Nodule: An Analysis of Discharge Records. Ann. Intern. Med. 2011;155:137–44. doi: 10.1059/0003-4819-155-3-201108020-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Grayson M. Breast Cancer. Nature. 2012;485:S49. doi: 10.1038/485S49a. [DOI] [PubMed] [Google Scholar]
- 265.Duffy MJ, Evoy D, McDermott EW. Ca 15-3: Uses and Limitation as a Biomarker for Breast Cancer. Clin. Chim. Acta. 2010;411:1869–74. doi: 10.1016/j.cca.2010.08.039. [DOI] [PubMed] [Google Scholar]
- 266.Hannafon BN, Trigoso YD, Calloway CL, Zhao YD, Lum DH, Welm AL, Zhao ZJ, Blick KE, Dooley WC, Ding WQ. Plasma Exosome MicroRNAs Are Indicative of Breast Cancer. Breast Cancer Res. 2016;18:90. doi: 10.1186/s13058-016-0753-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Taylor JP, Hardy J, Fischbeck KH. Toxic Proteins in Neurodegenerative Disease. Science. 2002;296:1991–5. doi: 10.1126/science.1067122. [DOI] [PubMed] [Google Scholar]
- 268.Ross CA, Poirier MA. Opinion: What is the Role of Protein Aggregation in Neurodegeneration. Nat. Rev. Mol, Cell Biol. 2005;6:891–8. doi: 10.1038/nrm1742. [DOI] [PubMed] [Google Scholar]
- 269.Irvine GB, El-Agnaf OM, Shankar GM, Walsh DM. Protein Aggregation in the Brain: The Molecular Basis for Alzheimer’s and Parkinson’s Diseases. Mol. Med. 2008;14:451–64. doi: 10.2119/2007-00100.Irvine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Goedert M. Neurodegeneration. Alzheimer’s and Parkinson’s Diseases: The Prion Concept in Relation to Assembled aβ, Tau, and Α-Synuclein. Science. 2015;349:1255555. doi: 10.1126/science.1255555. [DOI] [PubMed] [Google Scholar]
- 271.Haass C, Selkoe DJ. Soluble Protein Oligomers in Neurodegeneration: Lessons From the Alzheimer’s Amyloid Beta-Peptide. Nat. Rev. Mol. Cell Biol. 2007;8:101–12. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 272.Wakabayashi K, Tanji K, Odagiri S, Miki Y, Mori F, Takahashi H. The Lewy Body in Parkinson’s Disease and Related Neurodegenerative Disorders. Mol. Neurobiol. 2013;47:495–508. doi: 10.1007/s12035-012-8280-y. [DOI] [PubMed] [Google Scholar]
- 273.Quek C, Hill AF. The Role of Extracellular Vesicles in Neurodegenerative Diseases. Biochem. Biophys. Res. Commun. 2017;483:1178–1186. doi: 10.1016/j.bbrc.2016.09.090. [DOI] [PubMed] [Google Scholar]
- 274.Haass C, Kaether C, Thinakaran G, Sisodia S. Trafficking and Proteolytic Processing of App. Cold Spring Harb. Perspect. Med. 2012;2:a006270. doi: 10.1101/cshperspect.a006270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Kapogiannis D, Boxer A, Schwartz JB, Abner EL, Biragyn A, Masharani U, Frassetto L, Petersen RC, Miller BL, Goetzl EJ. Dysfunctionally Phosphorylated Type 1 Insulin Receptor Substrate in Neural-Derived Blood Exosomes of Preclinical Alzheimer’s Disease. FASEB J. 2015;29:589–96. doi: 10.1096/fj.14-262048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Povero D, Eguchi A, Li H, Johnson CD, Papouchado BG, Wree A, Messer K, Feldstein AE. Circulating Extracellular Vesicles With Specific Proteome and Liver MicroRNAs Are Potential Biomarkers for Liver Injury in Experimental Fatty Liver Disease. PLoS One. 2014;9:e113651. doi: 10.1371/journal.pone.0113651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Cheow ES, Cheng WC, Lee CN, de Kleijn D, Sorokin V, Sze SK. Plasma-Derived Extracellular Vesicles Contain Predictive Biomarkers and Potential Therapeutic Targets for Myocardial Ischemic (MI) Injury. Mol. Cell. Proteomics. 2016;15:2628–40. doi: 10.1074/mcp.M115.055731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Boulanger CM, Loyer X, Rautou PE, Amabile N. Extracellular Vesicles in Coronary Artery Disease. Nat. Rev. Cardiol. 2017;14:259–272. doi: 10.1038/nrcardio.2017.7. [DOI] [PubMed] [Google Scholar]
- 279.Gho YS, Lee C. Emergent Properties of Extracellular Vesicles: A Holistic Approach to Decode the Complexity of Intercellular Communication Networks. Mol. Biosyst. 2017;13:1291–1296. doi: 10.1039/c7mb00146k. [DOI] [PubMed] [Google Scholar]
- 280.De Jong OG, Van Balkom BW, Schiffelers RM, Bouten CV, Verhaar MC. Extracellular Vesicles: Potential Roles in Regenerative Medicine. Front. Immunol. 2014;5:608. doi: 10.3389/fimmu.2014.00608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Rani S, Ryan AE, Griffin MD, Ritter T. Mesenchymal Stem Cell-Derived Extracellular Vesicles: Toward Cell-Free Therapeutic Applications. Mol. Ther. 2015;23:812–23. doi: 10.1038/mt.2015.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.György B, et al. Rescue of Hearing By Gene Delivery to Inner-Ear Hair Cells Using Exosome-Associated Aav. Mol. Ther. 2017;25:379–391. doi: 10.1016/j.ymthe.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Kamerkar S, LeBleu VS, Sugimoto H, Yang S, Ruivo CF, Melo SA, Lee JJ, Kalluri R. Exosomes Facilitate Therapeutic Targeting of Oncogenic KRAS in Pancreatic Cancer. Nature. 2017;546:498–503. doi: 10.1038/nature22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Robbins PD, Dorronsoro A, Booker CN. Regulation of Chronic Inflammatory and Immune Processes By Extracellular Vesicles. J. Clin. Invest. 2016;126:1173–80. doi: 10.1172/JCI81131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Whiteside TL. Exosomes and Tumor-Mediated Immune Suppression. J. Clin. Invest. 2016;126:1216–23. doi: 10.1172/JCI81136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Katsuda T, Kosaka N, Takeshita F, Ochiya T. The Therapeutic Potential of Mesenchymal Stem Cell-Derived Extracellular Vesicles. Proteomics. 2013;13:1637–53. doi: 10.1002/pmic.201200373. [DOI] [PubMed] [Google Scholar]
- 287.Yeo RW, Lai RC, Zhang B, Tan SS, Yin Y, Teh BJ, Lim SK. Mesenchymal Stem Cell: An Efficient Mass Producer of Exosomes for Drug Delivery. Adv. Drug Deliv. Rev. 2013;65:336–41. doi: 10.1016/j.addr.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 288.Akyurekli C, Le Y, Richardson RB, Fergusson D, Tay J, Allan DS. A Systematic Review of Preclinical Studies on the Therapeutic Potential of Mesenchymal Stromal Cell- Derived Microvesicles. Stem Cell Rev. 2015;11:150–60. doi: 10.1007/s12015-014-9545-9. [DOI] [PubMed] [Google Scholar]
- 289.Vader P, Mol EA, Pasterkamp G, Schiffelers RM. Extracellular Vesicles for Drug Delivery. Adv. Drug Deliv. Rev. 2016;106:148–156. doi: 10.1016/j.addr.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 290.Maguire CA, et al. Microvesicle-Associated AAV Vector as a Novel Gene Delivery System. Mol. Ther. 2012;20:960–71. doi: 10.1038/mt.2011.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Mizrak A, Bolukbasi MF, Ozdener GB, Brenner GJ, Madlener S, Erkan EP, Ströbel T, Breakefield XO, Saydam O. Genetically Engineered Microvesicles Carrying Suicide mRNA/protein Inhibit Schwannoma Tumor Growth. Mol. Ther. 2013;21:101–8. doi: 10.1038/mt.2012.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Peinado H, et al. Pre-Metastatic Niches: Organ-Specific Homes for Metastases. Nat. Rev. Cancer. 2017;17:302–317. doi: 10.1038/nrc.2017.6. [DOI] [PubMed] [Google Scholar]
- 293.Becker A, Thakur BK, Weiss JM, Kim HS, Peinado H, Lyden D. Extracellular Vesicles in Cancer: Cell-to-cell Mediators of Metastasis. Cancer Cell. 2016;30:836–848. doi: 10.1016/j.ccell.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]