Abstract
Purpose
To assess expression of the histone demethylases KDM4A and KDM4B in granulosa collected from women undergoing oocyte retrieval and to determine if expression was related to pregnancy outcome.
Methods
Cumulus and mural granulosa cells were obtained from women undergoing oocyte retrieval. KDM4A and KDM4B mRNA expression was determined by qRT-PCR. KDM4A and KDM4B proteins were immunohistochemically localized in ovarian tissue sections obtained from archival specimens.
Results
KDM4A and KDM4B protein was localized to oocytes, granulosa cells, and theca and luteal cells in ovaries from reproductive-aged women. KDM4A and KDM4B mRNA expression was overall higher in cumulus compared to mural granulosa. When comparing granulosa demethylase gene expression, KDM4A and KDM4B mRNA expression was higher in both cumulus and mural granulosa from not pregnant patients compared to patients in the pregnant-live birth group.
Conclusions
Histone demethylases KDM4A and KDM4B mRNA are differentially expressed in cumulus and mural granulosa. Expression of both KDM4A and KDM4B mRNA was lower in cumulus granulosa and mural granulosa from pregnant compared to not pregnant patients. These findings suggest that altered expression of histone demethylases may impact epigenetic changes in granulosa cells associated with pregnancy.
Electronic supplementary material
The online version of this article (10.1007/s10815-018-1151-3) contains supplementary material, which is available to authorized users.
Keywords: Ovary, Granulosa, KDM4, Histone demethylase, Pregnancy, IVF
Introduction
The dynamics of ovarian follicular development and ovulation are tightly regulated events occurring in response to changing patterns of gene expression [1–3]. The importance of some genes within this complex network is understood and is evidenced by altered follicle development and/or loss of ovulation and luteal function when disrupted [4]. However, the trend towards delayed fertility in Westernized societies has resulted in a significant growth in the number of patients requiring assisted reproductive technologies. A more complete understanding of the processes governing follicle maturation and ovulation is essential for improving outcomes of in vitro fertility treatments.
One mechanism controlling gene expression occurs at the level of histones that function in packaging DNA. The nucleosome, considered the building block of chromatin, is composed of eight histones, two each H2A, H2B, H3, and H4. Histones regulate the availability of DNA to the transcriptional machinery through post-translational modifications of histone tails which can include phosphorylation, acetylation, ubiquitination, and methylation [5, 6]. Histone lysine methylation is a dynamic process that can both regulate transcriptional activation and repression and has been shown to play important roles in normal development, and transformation and progression of cancer [7]. The methylation state of lysine residues is controlled by enzymes, methyltransferases, and demethylases that function in the addition and removal of methylation marks. Two classes of methyltransferase, a family of SET-domain-containing enzymes and DotL1/Dot1p, mediate the transfer of a methyl group from S-adenosyl-L-methionine to histone lysine residues [8, 9]. Demethylation is carried out by two families of demethylases, the amine oxidases (lysine-specific demethylase, LSD/KDM1A, and LSD2/KDM1B) which remove mono- and di-methyl histone lysine marks and Jumonji C domain-containing iron-dependent dioxygenases which remove mono-, di-, and tri-methyl lysine residues (JmjC-KDMs). The methylation state of different lysine residues of histone 3 and 4 is associated with transcriptional activation (H3K4, H3K36, H3K79) or repression (H3K9, H3K27, H3K56, H4K20) [7]. Although the role of histone modifications in controlling gene expression has recently been an area of intense investigation, thus far there has been little attention given to this process as it may relate to follicle development and ovulation.
The JmjC-containing family of histone demethylases consists of approximately 24 genes that can be categorized into seven functionally divergent subfamilies. The highly conserved KDM4 subfamily (KDM4A-E) demethylates H3K9 and H3K36 di- and tri-methyl marks [10]. Diverse roles for KDM4 demethylases have been described, ranging from directing differentiation during embryogenesis, to promoting tumor progression, to maintaining an undifferentiated pattern of gene expression in stem cells [11]. A recent study reported a meta-analysis of next-generation sequencing profiles using the RNA-Seq Atlas and GENT databases. That study indicated robust expression of KDM4 subfamily members in the ovary [11]. Although discrete roles for KDM4 members in ovarian biology have yet to be described, many functions already ascribed to KDM4 have a correlation to processes occurring in the ovary.
Both KDM4A and KDM4B have roles in nuclear hormone receptor function. KDM4B is induced in an estrogen-dependent manner in breast cancer cells and can bind to the estrogen receptor and facilitate expression of estrogen-dependent genes [12–14]. Estrogen receptors are essential for normal ovarian follicular development, ovulation, and luteal function [15–20]. In prostate cancer, KDM4B stabilizes the androgen receptor (AR) and interacts to induce AR-mediated transcription [21, 22]. KDM4A and KDM4B both promote endometrial cancer progression by regulating AR and subsequent androgen-dependent expression of c-myc and p27kip1 [23]. Androgen receptor in the ovary functions in normal follicle development [24–26]. Inactivation of the AR in women is associated with premature ovarian failure [25], and AR polymorphisms have been associated with polycystic ovary syndrome [27–29] substantiating the importance of AR-mediated processes in normal ovarian biology. Progesterone receptor (PR) is another steroid receptor essential for normal follicle development, ovulation, and luteal function [30–33], and a role for Jumanji family histone demethylase KDM5A/B in PR-mediated gene expression has been demonstrated [34–36]. Most recently, a role for KDM4A as a maternal effect gene has been described in a murine knockout model [37].
Hypoxia and HIF-1α stimulate KDM4B which in turn functions to support HIF-1α-dependent gene expression to mediate the hypoxic response [38]. KDM4B expression in ovarian cancer correlated with expression of the hypoxic marker CA-IX and facilitated the expression of metastatic gene expression in hypoxic growth conditions [39]. The ovary receives a robust vascular supply yet the degree of hypoxia and the significance of hypoxia within the avascular granulosal compartment of the follicle continues to be debated [40]. Recent studies have demonstrated a role for HIF-1α within the ovary during follicle development, ovulation, and establishment of the corpus luteum [41–45]. KDM4A protein is induced independently of HIF stabilization [46] but can also regulate the expression of HIF-1α mRNA [47]. The potential interaction between hormone signaling, hypoxia, and KDM4 expression therefore provides rationale to investigate their contributions to fertility in human patients.
The current study examines the presence of KDM4A and KDM4B in cumulus and mural granulosa cells collected from women undergoing oocyte retrieval and seeks to determine if KDM4A/B expression is related to pregnancy outcome.
Materials and methods
Patients
Patients undergoing IVF by a single provider were recruited and informed consent was obtained. Ovarian follicle development was stimulated in patients using standard microdose flare or antagonist protocols and monitored by serial ultrasound and estradiol levels. Briefly, participants underwent controlled ovarian stimulation with recombinant FSH and human menopausal gonadotropins. When at least two follicles reached ≥ 18 mm in size, human chorionic gonadotropin was administered and 34–36 h later transvaginal ultrasound-guided oocyte retrieval was performed. This study was approved by the Institutional Review Board (#13637) at the University of Kansas Medical Center. All patients were consented for collection of granulosa cells prior to oocyte retrieval.
Granulosa cell preparation
Granulosa cells were collected and prepared as previously described with minor modification [48]. Mural granulosa from all follicles of an individual patient were pooled after removal of cumulus-oocyte complexes. Mural granulosa were enriched by sequential centrifugation over 75% percoll followed by 35% percoll and were washed in Hanks Balanced Salt Solution (HBSS) prior to RNA isolation. Cumulus-oocyte complexes were transferred to a clean dish where cumulus granulosa were cut from oocytes. Cumulus cells were pooled from a single patient and washed three times in HBSS prior to RNA isolation. Contamination by red blood cells was reduced by incubation of the cell fractions in ACK lysis buffer. This preparation method results in enriched cell populations. Any contamination with non-granulosa such as white blood cells or cross contamination of mural with cumulus or cumulus with mural cells was not specifically measured.
RNA isolation and PCR
Total RNA was isolated using Tri-reagent (ThermoFisher) according to manufacturer’s instructions, and integrity was confirmed by analysis of 18S and 28S RNA. cDNA was synthesized using total RNA (3 μg) from each sample and SuperScript II reverse transcriptase (Invitrogen). The resulting cDNA was diluted 10-fold in sterile water, and aliquots were subjected to qRT-PCR to quantify mRNA levels. Primers were designed using Roche Primer design tool and tested using NCBI Primer-BLAST/Primer3 [39, 49]. Primer sequences were as follows: 18S rRNA forward gcccgaagcgtttactttga and reverse tccattattcctagctgcggtatc, hKDM4A forward gccgctagaagtttcagtgag and reverse gcgtcccttggacttcttatt, hKDM4B forward ggactgacggcaacctctac and reverse cgtcctcaaactccacctg, hPGR forward ctctcaggctggcatggt and reverse ccactggctgtgggagag, and hStAR forward ccacccctagcacgtggat and reverse tcctggtcactgtagagagtctcttc. Primers were assessed by melt-curve analysis and generated only a single peak. Real-time PCR was carried out using SYBR GREEN PCR Master Mix (Applied Biosystems, Foster City, CA) and specific primers (250 nM each). Amplification and fluorescence detection were carried out using the ABI Prism 7500 Real Time PCR System (Applied Biosystems) and the following cycling conditions: initial hold step (95 °C for 10 min), 40 cycles of two-step PCR (92 °C for 15 s, then 60 °C for 1 min), and a dissociation step (95 °C for 15 s, 60 °C for 15 s, and a sequential increase to 95 °C). The comparative cycle threshold method was used for relative quantification of the amount of mRNA for each sample normalized to 18S RNA (∆CT). Data are presented as fold change (2ΔΔCt) in expression relative to the pregnant-live birth cohort.
Immunohistochemical staining
Ovaries from eight individual women were obtained through the University of Kansas Cancer Center Biospecimen Repository. Ovaries examined in the present study were removed for reasons unrelated to ovarian pathology such as cyst on the contralateral ovary, fallopian tube rupture due to ectopic pregnancy, oophorectomy and hysterectomy following pregnancy, and prophylactic oophorectomy due to history of breast cancer. The ovaries were histopathologically assessed as having no diagnostic abnormalities. Ovaries were from women of reproductive age (age 29–35 years) and were otherwise obtained de-identified. Immunostaining was carried out using standard peroxidase/DAB methods and hematoxylin counterstain [39]. Rabbit monoclonal KDM4A (#5328, Cell Signaling) and KDM4B antibody (#8638, Cell Signaling) were each used at dilutions of 1:50 and 1:100, respectively. Rabbit monoclonal IgG (#3900S, Cell Signaling) at the same concentration was used as a control for staining. Immunostaining was carried out on a single representative section from each ovary. Representative images were captured using an Olympus BX40 upright microscope equipped with an Olympus DP72 camera and CellSens software version 1.4 (Olympus, with permission from Dr. Mary Zelinski, ONPRC).
Statistical analysis
Patient demographics and cycle characteristics in pregnant and not pregnant groups were analyzed using a Mann-Whitney test to determine if population medians differed. qRT-PCR was carried out on each sample and run in duplicate or triplicate for each primer pair. The mean ∆CT for each primer pair (specific gene–18S) was determined for all patient samples in cumulus and mural samples individually. The median fold change between pregnant-live birth and not pregnant groups (2ΔΔCt) was determined and compared by Mann-Whitney test (GraphPad Prism7); p < 0.05 was considered different. Correlation analysis was determined using Spearman’s rank correlation coefficient (GraphPad Prism7). Spearman’s correlation (rs) = 1.0–0.8 very strong, 0.79–0.6 strong, 0.59–0.4 moderate, 0.39–0.2 weak, 0.19–0 very weak correlation.
Results
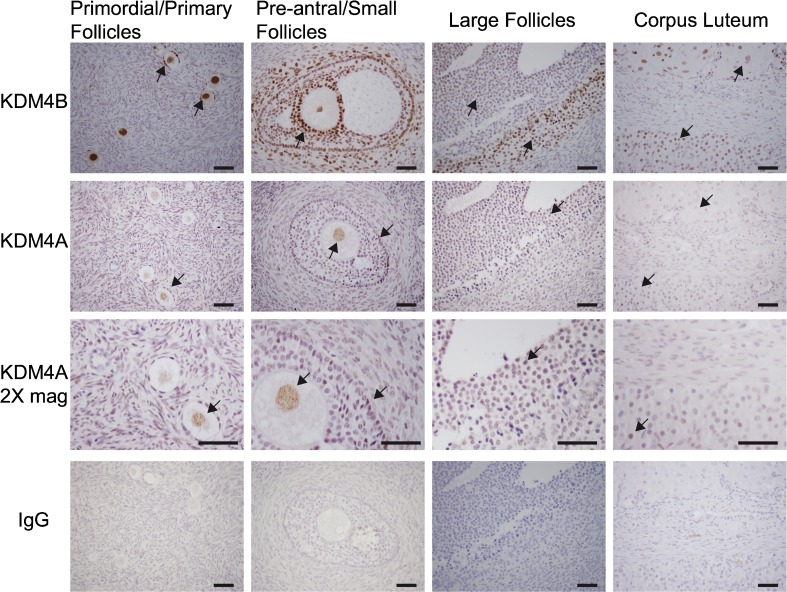
Immunostaining for KDM4B in ovarian follicles was initially noted during the analysis of human ovarian surface epithelial cells in a related study [39]. This observation prompted a more thorough investigation. Cellular localization of KDM4A and KDM4B was examined by immunohistochemical staining of normal ovaries collected from reproductive-aged women. Intense immunostaining for KDM4B was observed in theca, granulosa cells, corpora lutea, and oocytes. KDM4B staining in granulosa was apparent in follicles of all developmental stages from squamous granulosa of primordial follicles to both mural and cumulus granulosa of large antral follicles (Fig. 1). KDM4A immunostaining appeared diffuse across the ovary with specific localization to oocytes, granulosa cells, and luteal cells (Fig. 1). In all cases, immunostaining appeared to be confined to the nucleus.
Fig. 1.
Immunolocalization of KDM4A and KDM4B in human ovaries. KDM4B was immunolocalized to granulosa cells and theca in large and small follicles, corpora lutea, and granulosa and oocyte in primordial and primary follicles. In all cells, localization of KDM4B was confined to the nucleus. KDM4A immunostaining was localized to granulosa and luteal cells and was weakly and diffusely noted throughout the ovary. Due to the weaker level of staining, ×2 magnified images are presented for KDM4A. Control immunostaining using IgG-stained sections are presented. The figure includes results from four different ovaries and is representative of eight ovaries analyzed. Black arrowheads denote staining for KDM4B and KDM4A. Bar indicates 50 μm
Patient demographics and cycle characteristics of the patients included in the current analysis are described in Table 1. A total of 84 patients undergoing oocyte retrieval were categorized based on pregnancy outcome. Groups included patients undergoing embryo transfer that became pregnant and resulted in a live birth (pregnant-live birth) and patients that did not become pregnant following embryo transfer and who did not demonstrate a chemical pregnancy (not pregnant). Women in the pregnant-live birth group were younger, had higher anti-mullerian hormone (AMH) levels, were administered less FSH, achieved higher peak estradiol levels, had more oocytes at retrieval, and had greater numbers of two pronuclei (2PN) embryos compared to women in the not pregnant group (Table 1).
Table 1.
Patient demographics
| Pregnant-live birtha | Not pregnantb | |
|---|---|---|
| Number | 31 | 53 |
| Age, mean (range) | 32.2 (27–46) * | 34.6 (24–44) |
| BMI, mean ± sem | 25.59 ± 0.96 | 26.85 ± 0.69 |
| AMH, ng/ml mean ± sem | 4.27 ± 0.64 ** | 2.83 ± 0.50 |
| Estradiol, peak pg/ml mean ± sem | 3107 ± 291 * | 2285 ± 161 |
| FSH, IU administered mean ± sem | 3468 ± 351.6 * | 4640 ± 255.7 |
| Number oocytes retrieved, mean ± sem | 17.83 ± 1.63 ** | 12.34 ± 0.79 |
| Number 2PN embryos, mean ± sem | 12.17 ± 1.11 ** | 6.53 ± 0.58 |
| Infertility etiology | ||
| Male factor | 13 | 20 |
| Combined male/female factor | 3 | 11 |
| Female factor | 18 | 33 |
| Endometriosis | 2 | 8 |
| PCOS | 7 | 4 |
| DOR | 3 | 12 |
| Tubal | 5 | 22 |
| Tubal ligation | 2 | 1 |
| Uterine | 0 | 2 |
| Other | 2 | 10 |
| Unexplained | 3 | 5 |
PCOS polycystic ovarian syndrome, DOR diminished ovarian reserve, BMI body mass index, AMH anti-mullerian hormone, 2PN two pronuclei
aPregnant live birth includes patients undergoing embryo transfer and confirmed pregnancy and live birth. Includes 25 singleton and 6 twin births
bNot pregnant includes patients undergoing embryo transfer and not pregnant, no biochemical pregnancy, and no spontaneous abortion
*p < 0.02
**p < 0.006 pregnant live birth vs. not pregnant
Expression of mRNA-coding histone demethylase KDM4A and KDM4B in cumulus and mural granulosa was examined for all patients. In general, expression of both histone demethylases was higher in cumulus granulosa cells compared to mural granulosa. Expression of KDM4A mRNA was approximately 5-fold higher and expression of KDM4B mRNA was approximately 2-fold higher in cumulus granulosa compared to mural granulosa cells.
Analysis of histone demethylase mRNA revealed differential expression in granulosa from women pregnant-live birth and women not pregnant. KDM4A mRNA expression level was higher in both cumulus granulosa and mural granulosa from women in the not pregnant group compared to the pregnant-live birth group. In cumulus cells, KDM4A mRNA was 12.82-fold higher in the not pregnant group compared to the pregnant-live birth group (p = 0.0022; Fig. 2). In mural granulosa cells, KDM4A mRNA levels were 5.34-fold higher in the not pregnant group compared to the pregnant-live birth group (p = 0.0332; Fig. 2).
Fig. 2.
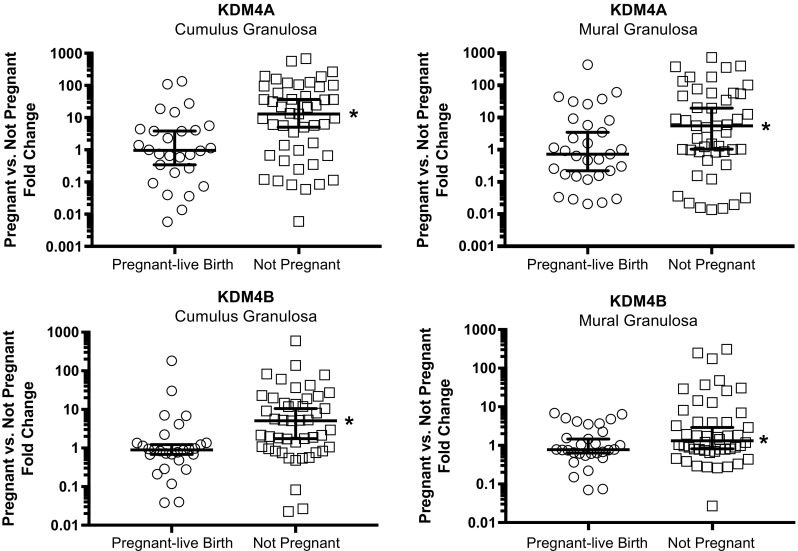
Expression of KDM4A and KDM4B mRNA in cumulus and mural granulosa. Granulosa cells were collected at the time of oocyte retrieval; RNA was isolated and expression of KDM4A and KDM4B was assessed by qRT-PCR. Normalized data (∆CT) is expressed as fold change from the mean of the pregnant cohort. The median line ± 95% confidence intervals are shown. Pregnant-live birth (n = 31) versus not pregnant (n = 53) were compared by Mann-Whitney. *p < 0.04 pregnant-live birth versus not pregnant
Similar to KDM4A, KDM4B mRNA expression level was higher in both cumulus and mural granulosa from women in the not pregnant group compared to the pregnant-live birth group (Fig. 2). In cumulus cells, KMD4B mRNA levels were 5.07-fold higher and in mural granulosa 1.32-fold higher in the not pregnant group compared to the pregnant-live birth group (p = 0.0012 cumulus and p = 0.04 mural; Fig. 2).
Spearman’s correlation was run to determine the general relationship between cumulus and mural granulosal expression of the KDM4 histone demethylases within each experimental group. Expression levels of KDM4A and KDM4B mRNA were very strongly correlated in cumulus cells and in mural cells from patients in the not pregnant group. In comparison, the correlation of KDM4A and KDM4B mRNA expression levels was weaker in cumulus and mural cells from patients in the pregnant-live birth group (Fig. 3).
Fig. 3.
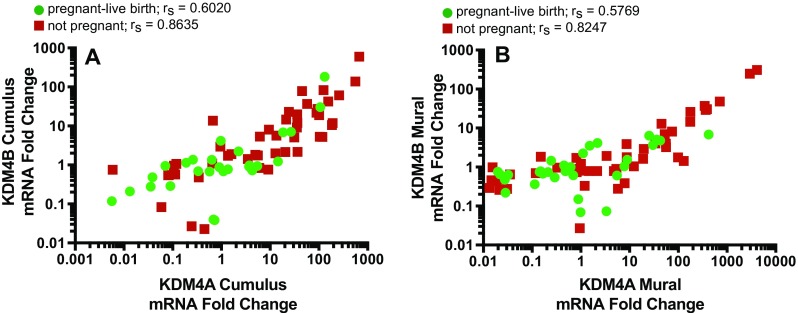
Correlation of KDM4A and KDM4B mRNA expression in cumulus and mural granulosa cells. The correlation of the fold change values for expression of KDM4A and KDM4B mRNA in the cumulus granulosa (a) and mural granulosa (b) was determined for patients in the not pregnant (red) and pregnant-live birth (green) groups. Spearman’s rs values are indicated for each correlation. Spearman’s correlation (rs) = 1.0–0.8 very strong, 0.79–0.6 strong, 0.59–0.4 moderate
As described, several differences were noted in the pregnant-live birth group and not pregnant group including age, AMH level, and peak estradiol levels (Table 1). Correlation of these parameters and KDM4A or KDM4B mRNA expression was analyzed. No correlation was observed between KDM4A or KDM4B mRNA expression within each cell type and pregnancy status and age, BMI, AMH, peak estradiol, FSH administered, number of oocytes retrieved, or number of 2PN embryos (Fig. 4 and Supplementary Figures 1–6).
Fig. 4.
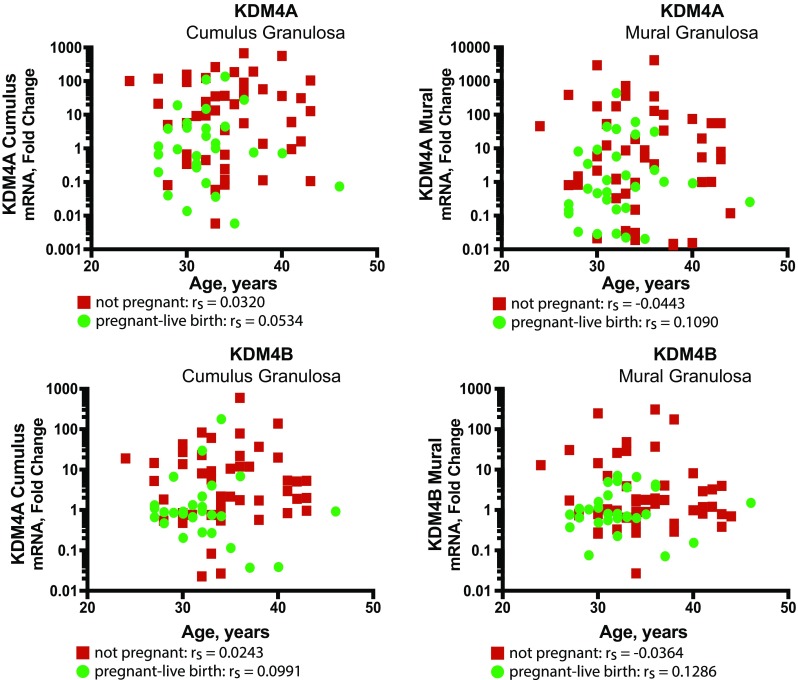
Correlation of KDM4 mRNA expression in cumulus and mural granulosa cells with patient age. The fold change values for expression of KDM4A and KDM4B mRNA in the cumulus and mural granulosa in pregnant-live birth (green) and not pregnant (red) groups did not correlate with patient age. Spearman’s correlation values are indicated (rs) = 1.0–0.8 very strong, 0.79–0.6 strong, 0.59–0.4 moderate, 0.39–0.2 weak, 0.19–0 very weak correlation
Discussion
The present studies are the first to demonstrate expression of mRNA for the histone demethylases KDM4A and KDM4B in granulosa cells. Compared to patients who become pregnant following ART, expression of both histone demethylases was higher in the patients who did not become pregnant, indicating that these epigenetic regulators may play a role in directing proper granulosal function around the time of ovulation and luteal formation.
Ongoing studies are exploring demethylase activity and specific genes affected by KDM4A and KDM4B in cumulus and mural granulosa to correlate mRNA expression with biological function. However, the current findings provide some evidence to suggest differential expression of histone demethylases may impact epigenetic changes and gene expression in granulosa associated with pregnancy.
As an initial approach to examine the potential significance of histone demethylase expression in granulosa cell function, patients were categorized based on pregnancy outcome and expression of KDM4A and KDM4B mRNA in each group was compared. The general demographics of the patients segregating into the pregnant-live birth and not pregnant groups was similar to many previous reports. As anticipated, women that became pregnant following embryo transfer were younger and exhibited higher levels of AMH compared to women that did not become pregnant [50–52]. Age and AMH data extended to anticipated outcomes of less FSH administered, higher peak estradiol, more oocytes retrieved, and greater numbers of 2PN embryos in the pregnant-live birth group [53–55]. It is interesting to note there was no segregation of BMI in the two groups. Several studies have indicated increased BMI inversely correlates with pregnancy outcome [56, 57]. Mean BMI for patients in the pregnant-live birth group was 25.59 (range 19.15–44.8) and was not different from the not pregnant group, 26.85 (range 17.14–40.59); however, both groups were within the overweight category as defined by the World Health Organization (WHO. Obesity and overweight. Fact sheet; Updated June 2016 http://www.who.int/mediacentre/factsheets/fs311/en/). The diagnosed cause of infertility for the current study group is indicated in Table 1. Due to the relatively small sample size of this study, the significance of KDM4A or KDM4B expression as an indicator of infertility within a specific etiology cannot be assessed. However, it will be of future interest to determine, for example, if ovulatory dysfunction is associated with altered expression of KDM4A and or KDM4B.
The ovulatory surge of LH initiates significant change in the expression patterns of many genes within the ovary leading to ovulation and establishment of the corpus luteum. The importance of many of these genes has been further demonstrated by knock-out and knock-down studies in rodents [18, 33, 58–65] and through the identification of mutations associated with infertility in women [66–72]. Recent studies have now begun to tie the expression of several of these critical genes to methylation state providing greater insight to the mechanisms controlling the final maturation of the follicle and ovulation. Increased expression of CYP11A1 in rat granulosa following an ovulatory dose of hCG was associated with increased H3K4me3 and decreased H3K9me3 and H3K23me3 [73]. Similarly, H3K9me3 marks in the StAR promoter were decreased after hCG [74]. These data indicate rapid changes in histone methylation following the ovulatory surge occurring with rapid changes in gene expression. Additional studies highlight altered patterns in methylation of critical genes found in pathological conditions associated with altered fertility. Cumulus granulosa from women with endometriosis have lower CYP19 gene expression compared to cumulus from women without endometriosis. Hypermethylation of H3K9 in the CYP19 promoter was associated with the lower expression [75]. CYP19 expression is also reduced in women with PCOS [76–78]. Studies have demonstrated differences of methylation state in PCOS ovarian tissue compared to controls where some genes were hypermethylated (such as IGFBP2, CYP19A1, AMHR2) and others hypomethylated (INSR, AMH) in PCOS ovaries [79]. Ovarian function and fertility can be impacted by exposure to chemicals in the environment and these effects have also been linked to altered DNA methylation of key genes. Exposure to methoxychlor (MXC), a synthetic insecticide, during critical phases of ovary development during the late prenatal and early postnatal period results in altered ovarian function and reduced fertility in adult mice [80, 81]. Recent studies indicate altered DNA methylation as a mechanism leading to MCX effects. Following MCX dosing up to postnatal day 7, hypermethylation of many genes involved in normal folliculogenesis was found. In addition, altered methylation was observed at postnatal day 60, long after the final dose of MXC [82]. Therefore, patterns of DNA and histone methylation in genes critical for follicular development, ovulation, and luteal formation change as part of normal biology. Methylation patterns different from that observed in normal ovarian tissues have been associated with conditions such as PCOS and altered patterns of methylation in the ovary occur following exposure to environmental reproductive toxins. The current data suggest that the histone demethylase KDM4A and KDM4B may play a role in mediating some of these events. Future studies focused on demonstrating KDM4A and KDM4B demethylase activity on specific target genes in ovarian cell types will substantiate this finding.
Although specific roles for KDM4A and KDM4B in granulosa cell function are yet to be determined, an importance may be inferred from the expression data observed in this study. The correlation in expression level of the histone demethylase KDM4A and KDM4B mRNA was very strong in both cumulus and mural granulosa cells in the not pregnant group. It is interesting to note the correlation in expression was weaker in cells from patients in the pregnant-live birth group. This observation indicates a differential expression of these histone demethylases occurring in cells associated with successful pregnancy compared to cells associated with no pregnancy. Additional analysis was carried out to determine if other characteristics differentiating the pregnant and not pregnant group correlated to KDM4 gene expression. For example, age was significantly lower in the pregnant-live birth group, and therefore, it is possible that KMD4 mRNA expression may correlate with age. However, no strong correlation was observed in KDM4 gene expression and age (Fig. 4) or other parameters assessed including BMI, AMH, peak estradiol, FSH administered, number of oocyte retrieved, or number of 2PN embryos (Supplementary Figures 1–6). Together, these data suggest changes in the expression of KDM4A and KDM4B are not related to factors such as age but may be related to specific cellular functions associated with successful pregnancy.
Currently, it is unclear if the elevated expression in non-pregnant patients represents aberrant induction of KDM4A and KDM4B relative to the pregnant cohort or if there is a failure to repress KDM4A and KDM4B expression at the time of ovulation. It may be that elevated expression of KDM4 in granulosa of the not pregnant group is representative of premature luteinization; KDM4A and KDM4B immunoreactivity was observed in luteal cells (Fig. 3). However, expression of StAR or PGR mRNA, two marker genes of luteinization, did not correlate with KDM4 expression in the current data set (see Supplementary Figures 7 and 8). Further, it is possible a subgroup may exist within either the non-pregnant or pregnant patient groups providing weight to the data due to unidentified pathological conditions. In addition, expression determined here may be related to infertility in general and may not reflect the expression patterns in cells from fertile women. Continued analysis of larger patient cohorts will provide insight toward these possibilities.
Immunostaining confirmed the cellular localization of KDM4A and KDM4B in specific cells within the normal human ovary. This observation is in general agreement with two other reports identifying KDM4 expression in the ovary. First, a meta-analysis of next-generation sequencing conducted on normal human tissues using RNA-Seq Atlas and GENT databases indicated KDM4A, KDM4B, and KDM4C are broadly expressed across several tissue types with high expression in the human ovary [11]. That study further described that based on reads per kilobase per million (RPKM), the expression level of KDM4A and KDM4C was higher than KDM4B in the ovary [11]. In another recent study, generation of KDM4A null mice lead to the identification of a role for KDM4A in preimplantation embryo development and in the maintenance of a uterine environment advantageous to implantation and embryo development [37]. That study further explored general expression levels in the mouse ovary where b-galactosidase staining from KMD4A, B, or C-promotor driven LacZ was illustrated from the authors data (KDM4A) and data from the International Mouse Phenotyping Consortium (KDM4B and KDM4C). In that case, both KDM4A and B expression were noted in the female reproductive tract, with greater overall KDM4A expression described and little to no expression of KDM4C observed [37]. Additionally, KDM4A was detected in the granulosa and oocytes of early stage follicles [37]. In the present study, KDM4A immunostaining was overall weak and appeared diffuse throughout the ovary. However, specific localization was observed in the oocytes, granulosa of growing follicles, and luteal cells. Expression of KDM4B was more robust and was specifically observed in oocytes, granulosa, and theca cells. Differences in the intensity of immunostaining between KDM4A and KDM4B may be a reflection of different levels of protein expression in the human ovary or may reflect variation in the affinity of the antibodies used. Thus, the present data, although not quantitative, demonstrate KDM4A and KDM4B proteins are localized to specific cells within the human follicle and corpus luteum. Considering the newly described role of KDM4A as a maternal effect gene that contributes to the oocyte-embryo transition in the mouse [37], it is also of significant interest that the supporting granulosa cells in humans express both KDM4A and KDM4B.
The present study demonstrates the histone demethylase KMD4A and KDM4B are localized within the ovary to oocytes, granulosa cells, theca, and luteal cells. Differential expression of KDM4A/B mRNA was found with pregnancy in cumulus and mural granulosa cells. These data indicate KDM4A and KDM4B may have a role in control of gene expression important for function and differentiation of granulosa cells supportive of successful pregnancy.
Electronic supplementary material
(JPEG 359 kb)
(JPEG 340 kb)
(JPEG 375 kb)
(JPEG 390 kb)
(JPEG 391 kb)
(JPEG 387 kb)
(JPEG 383 kb)
(JPEG 368 kb)
(DOCX 157 kb)
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s10815-018-1151-3) contains supplementary material, which is available to authorized users.
References
- 1.Field SL, Dasgupta T, Cummings M, Orsi NM. Cytokines in ovarian folliculogenesis, oocyte maturation and luteinisation. Mol Reprod Dev. 2014;81:284–314. doi: 10.1002/mrd.22285. [DOI] [PubMed] [Google Scholar]
- 2.Monniaux D, Clement F, Dalbies-Tran R, Estienne A, Fabre S, Mansanet C, Monget P. The ovarian reserve of primordial follicles and the dynamic reserve of antral growing follicles: what is the link? Biol Reprod. 2014;90:85. doi: 10.1095/biolreprod.113.117077. [DOI] [PubMed] [Google Scholar]
- 3.Richards JS, Pangas SA. The ovary: basic biology and clinical implications. J Clin Invest. 2010;120:963–972. doi: 10.1172/JCI41350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roy A, Matzuk MM. Deconstructing mammalian reproduction: using knockouts to define fertility pathways. Reproduction. 2006;131:207–219. doi: 10.1530/rep.1.00530. [DOI] [PubMed] [Google Scholar]
- 5.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 6.Tessarz P, Kouzarides T. Histone core modifications regulating nucleosome structure and dynamics. Nat Rev Mol Cell Biol. 2014;15:703–708. doi: 10.1038/nrm3890. [DOI] [PubMed] [Google Scholar]
- 7.Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell. 2012;48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dillon SC, Zhang X, Trievel RC, Cheng X. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 2005;6:227. doi: 10.1186/gb-2005-6-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McLean CM, Karemaker ID, van Leeuwen F. The emerging roles of DOT1L in leukemia and normal development. Leukemia. 2014;28:2131–2138. doi: 10.1038/leu.2014.169. [DOI] [PubMed] [Google Scholar]
- 10.Hillringhaus L, Yue WW, Rose NR, Ng SS, Gileadi C, Loenarz C, Bello SH, Bray JE, Schofield CJ, Oppermann U. Structural and evolutionary basis for the dual substrate selectivity of human KDM4 histone demethylase family. J Biol Chem. 2011;286:41616–41625. doi: 10.1074/jbc.M111.283689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Labbe RM, Holowatyj A, Yang ZQ. Histone lysine demethylase (KDM) subfamily 4: structures, functions and therapeutic potential. Am J Transl Res. 2013;6:1–15. [PMC free article] [PubMed] [Google Scholar]
- 12.Berry WL, Janknecht R. KDM4/JMJD2 histone demethylases: epigenetic regulators in cancer cells. Cancer Res. 2013;73:2936–2942. doi: 10.1158/0008-5472.CAN-12-4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawazu M, Saso K, Tong KI, McQuire T, Goto K, Son DO, Wakeham A, Miyagishi M, Mak TW, Okada H. Histone demethylase JMJD2B functions as a co-factor of estrogen receptor in breast cancer proliferation and mammary gland development. PLoS One. 2011;6:e17830. doi: 10.1371/journal.pone.0017830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi L, Sun L, Li Q, Liang J, Yu W, Yi X, Yang X, Li Y, Han X, Zhang Y, Xuan C, Yao Z, Shang Y. Histone demethylase JMJD2B coordinates H3K4/H3K9 methylation and promotes hormonally responsive breast carcinogenesis. Proc Natl Acad Sci U S A. 2011;108:7541–7546. doi: 10.1073/pnas.1017374108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Couse JF, Yates MM, Deroo BJ, Korach KS. Estrogen receptor-beta is critical to granulosa cell differentiation and the ovulatory response to gonadotropins. Endocrinology. 2005;146:3247–3262. doi: 10.1210/en.2005-0213. [DOI] [PubMed] [Google Scholar]
- 16.Drummond AE, Fuller PJ. Ovarian actions of estrogen receptor-beta: an update. Semin Reprod Med. 2012;30:32–38. doi: 10.1055/s-0031-1299595. [DOI] [PubMed] [Google Scholar]
- 17.Emmen JM, Couse JF, Elmore SA, Yates MM, Kissling GE, Korach KS. In vitro growth and ovulation of follicles from ovaries of estrogen receptor (ER){alpha} and ER{beta} null mice indicate a role for ER{beta} in follicular maturation. Endocrinology. 2005;146:2817–2826. doi: 10.1210/en.2004-1108. [DOI] [PubMed] [Google Scholar]
- 18.Rumi MK, Singh P, Roby KF, Zhao X, Iqbal K, Ratri A, Lei T, Cui W, Borosha S, Dhakal P, Kubota K, Chakraborty D, Vivian JL, Wolfe MW, Soares MJ. Defining the role of estrogen receptor beta in the regulation of female fertility. Endocrinology. 2017;158:2330–2343. doi: 10.1210/en.2016-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stouffer RL. Progesterone as a mediator of gonadotrophin action in the corpus luteum: beyond steroidogenesis. Hum Reprod Update. 2003;9:99–117. doi: 10.1093/humupd/dmg016. [DOI] [PubMed] [Google Scholar]
- 20.Su EJ, Xin H, Monsivais D. The emerging role of estrogen receptor-beta in human reproduction. Semin Reprod Med. 2012;30:62–70. doi: 10.1055/s-0031-1299598. [DOI] [PubMed] [Google Scholar]
- 21.Chu CH, Wang LY, Hsu KC, Chen CC, Cheng HH, Wang SM, Wu CM, Chen TJ, Li LT, Liu R, Hung CL, Yang JM, Kung HJ, Wang WC. KDM4B as a target for prostate cancer: structural analysis and selective inhibition by a novel inhibitor. J Med Chem. 2014;57:5975–5985. doi: 10.1021/jm500249n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duan L, Rai G, Roggero C, Zhang QJ, Wei Q, Ma SH, Zhou Y, Santoyo J, Martinez ED, Xiao G, Raj GV, Jadhav A, Simeonov A, Maloney DJ, Rizo J, Hsieh JT, Liu ZP. KDM4/JMJD2 histone demethylase inhibitors block prostate tumor growth by suppressing the expression of AR and BMYB-regulated genes. Chem Biol. 2015;22:1185–1196. doi: 10.1016/j.chembiol.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qiu MT, Fan Q, Zhu Z, Kwan SY, Chen L, Chen JH, Ying ZL, Zhou Y, Gu W, Wang LH, Cheng WW, Zeng J, Wan XP, Mok SC, Wong KK, Bao W. KDM4B and KDM4A promote endometrial cancer progression by regulating androgen receptor, c-myc, and p27kip1. Oncotarget. 2015;6:31702–31720. doi: 10.18632/oncotarget.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Gendt K, Verhoeven G. Tissue- and cell-specific functions of the androgen receptor revealed through conditional knockout models in mice. Mol Cell Endocrinol. 2012;352:13–25. doi: 10.1016/j.mce.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 25.Kimura S, Matsumoto T, Matsuyama R, Shiina H, Sato T, Takeyama K, Kato S. Androgen receptor function in folliculogenesis and its clinical implication in premature ovarian failure. Trends Endocrinol Metab. 2007;18:183–189. doi: 10.1016/j.tem.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 26.Prizant H, Gleicher N, Sen A. Androgen actions in the ovary: balance is key. J Endocrinol. 2014;222:R141–R151. doi: 10.1530/JOE-14-0296. [DOI] [PubMed] [Google Scholar]
- 27.Lin LH, Baracat MC, Maciel GA, Soares JM, Jr, Baracat EC. Androgen receptor gene polymorphism and polycystic ovary syndrome. Int J Gynaecol Obstet. 2013;120:115–118. doi: 10.1016/j.ijgo.2012.08.016. [DOI] [PubMed] [Google Scholar]
- 28.Wang R, Goodarzi MO, Xiong T, Wang D, Azziz R, Zhang H. Negative association between androgen receptor gene CAG repeat polymorphism and polycystic ovary syndrome? A systematic review and meta-analysis. Mol Hum Reprod. 2012;18:498–509. doi: 10.1093/molehr/gas024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang T, Liang W, Fang M, Yu J, Ni Y, Li Z. Association of the CAG repeat polymorphisms in androgen receptor gene with polycystic ovary syndrome: a systemic review and meta-analysis. Gene. 2013;524:161–167. doi: 10.1016/j.gene.2013.04.040. [DOI] [PubMed] [Google Scholar]
- 30.Akison LK, Robker RL. The critical roles of progesterone receptor (PGR) in ovulation, oocyte developmental competence and oviductal transport in mammalian reproduction. Reprod Domest Anim. 2012;47(Suppl 4):288–296. doi: 10.1111/j.1439-0531.2012.02088.x. [DOI] [PubMed] [Google Scholar]
- 31.Kim J, Bagchi IC, Bagchi MK. Control of ovulation in mice by progesterone receptor-regulated gene networks. Mol Hum Reprod. 2009;15:821–828. doi: 10.1093/molehr/gap082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kubota K, Cui W, Dhakal P, Wolfe MW, Rumi MA, Vivian JL, Roby KF, Soares MJ. Rethinking progesterone regulation of female reproductive cyclicity. Proc Natl Acad Sci U S A. 2016;113:4212–4217. doi: 10.1073/pnas.1601825113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peluso JJ. Multiplicity of progesterone's actions and receptors in the mammalian ovary. Biol Reprod. 2006;75:2–8. doi: 10.1095/biolreprod.105.049924. [DOI] [PubMed] [Google Scholar]
- 34.Stratmann A, Haendler B. The histone demethylase JARID1A regulates progesterone receptor expression. FEBS J. 2011;278:1458–1469. doi: 10.1111/j.1742-4658.2011.08058.x. [DOI] [PubMed] [Google Scholar]
- 35.Vicent GP, Nacht AS, Font-Mateu J, Castellano G, Gaveglia L, Ballare C, Beato M. Four enzymes cooperate to displace histone H1 during the first minute of hormonal gene activation. Genes Dev. 2011;25:845–862. doi: 10.1101/gad.621811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vicent GP, Nacht AS, Zaurin R, Font-Mateu J, Soronellas D, Le Dily F, Reyes D, Beato M. Unliganded progesterone receptor-mediated targeting of an RNA-containing repressive complex silences a subset of hormone-inducible genes. Genes Dev. 2013;27:1179–1197. doi: 10.1101/gad.215293.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sankar A, Kooistra SM, Gonzalez JM, Ohlsson C, Poutanen M, Helin K. Maternal expression of the histone demethylase Kdm4a is crucial for pre-implantation development. Development. 2017;144:3264–3277. doi: 10.1242/dev.155473. [DOI] [PubMed] [Google Scholar]
- 38.Salminen A, Kaarniranta K, Kauppinen A. Hypoxia-inducible histone lysine demethylases: impact on the aging process and age-related diseases. Aging Dis. 2016;7:180–200. doi: 10.14336/AD.2015.0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson C, Qiu L, Hong Y, Karnik T, Tadros G, Mau B, Ma T, Mu Y, New J, Louie RJ, Gunewardena S, Godwin AK, Tawfik OW, Chien J, Roby KF, Krieg AJ. The histone demethylase KDM4B regulates peritoneal seeding of ovarian cancer. Oncogene. 2017;36:2565–2576. doi: 10.1038/onc.2016.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson JG, Brown HM, Kind KL, Russell DL. The ovarian antral follicle: living on the edge of hypoxia or not? Biol Reprod. 2015;92:153. doi: 10.1095/biolreprod.115.128660. [DOI] [PubMed] [Google Scholar]
- 41.Fadhillah YS, Nishimura R, Okuda K. Hypoxia promotes progesterone synthesis during luteinization in bovine granulosa cells. J Reprod Dev. 2014;60:194–201. doi: 10.1262/jrd.2014-014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kowalewski MP, Gram A, Boos A. The role of hypoxia and HIF1alpha in the regulation of STAR-mediated steroidogenesis in granulosa cells. Mol Cell Endocrinol. 2015;401:35–44. doi: 10.1016/j.mce.2014.11.023. [DOI] [PubMed] [Google Scholar]
- 43.Rico C, Dodelet-Devillers A, Paquet M, Tsoi M, Lapointe E, Carmeliet P, Boerboom D. HIF1 activity in granulosa cells is required for FSH-regulated Vegfa expression and follicle survival in mice. Biol Reprod. 2014;90:135. doi: 10.1095/biolreprod.113.115634. [DOI] [PubMed] [Google Scholar]
- 44.Tam KK, Russell DL, Peet DJ, Bracken CP, Rodgers RJ, Thompson JG, Kind KL. Hormonally regulated follicle differentiation and luteinization in the mouse is associated with hypoxia inducible factor activity. Mol Cell Endocrinol. 2010;327:47–55. doi: 10.1016/j.mce.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 45.Yalu R, Oyesiji AE, Eisenberg I, Imbar T, Meidan R. HIF1A-dependent increase in endothelin 2 levels in granulosa cells: role of hypoxia, LH/cAMP, and reactive oxygen species. Reproduction. 2015;149:11–20. doi: 10.1530/REP-14-0409. [DOI] [PubMed] [Google Scholar]
- 46.Black JC, Atabakhsh E, Kim J, Biette KM, Van Rechem C, Ladd B, Burrowes PD, Donado C, Mattoo H, Kleinstiver BP, Song B, Andriani G, Joung JK, Iliopoulos O, Montagna C, Pillai S, Getz G, Whetstine JR. Hypoxia drives transient site-specific copy gain and drug-resistant gene expression. Genes Dev. 2015;29:1018–1031. doi: 10.1101/gad.259796.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dobrynin G, McAllister TE, Leszczynska KB, Ramachandran S, Krieg AJ, Kawamura A, Hammond EM. KDM4A regulates HIF-1 levels through H3K9me3. Sci Rep. 2017;7:11094. doi: 10.1038/s41598-017-11658-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roby KF, Weed J, Lyles R, Terranova PF. Immunological evidence for a human ovarian tumor necrosis factor-a. J Clin Endocrinol Metab. 1990;71:1096–1102. doi: 10.1210/jcem-71-5-1096. [DOI] [PubMed] [Google Scholar]
- 49.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinf. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bolat SE, Ozdemirci S, Kasapoglu T, Duran B, Goktas L, Karahanoglu E. The effect of serum and follicular fluid anti-Mullerian hormone level on the number of oocytes retrieved and rate of fertilization and clinical pregnancy. North Clin Istanb. 2016;3:90–96. doi: 10.14744/nci.2016.02418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keane K, Cruzat VF, Wagle S, Chaudhary N, Newsholme P, Yovich J. Specific ranges of anti-Mullerian hormone and antral follicle count correlate to provide a prognostic indicator for IVF outcome. Reprod Biol. 2017;17:51–59. doi: 10.1016/j.repbio.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 52.Zebitay AG, Cetin O, Verit FF, Keskin S, Sakar MN, Karahuseyinoglu S, Ilhan G, Sahmay S. The role of ovarian reserve markers in prediction of clinical pregnancy. J Obstet Gynaecol. 2017;37:492–497. doi: 10.1080/01443615.2016.1269730. [DOI] [PubMed] [Google Scholar]
- 53.Barton SE, Missmer SA, Ashby RK, Ginsburg ES. Multivariate analysis of the association between oocyte donor characteristics, including basal follicle stimulating hormone (FSH) and age, and IVF cycle outcomes. Fertil Steril. 2010;94:1292–1295. doi: 10.1016/j.fertnstert.2009.07.1672. [DOI] [PubMed] [Google Scholar]
- 54.Ben-Haroush A, Farhi J, Zahalka Y, Sapir O, Meizner I, Fisch B. Correlations between antral follicle count and ultrasonographic ovarian parameters and clinical variables and outcomes in IVF cycles. Gynecol Endocrinol. 2012;28:432–435. doi: 10.3109/09513590.2011.634935. [DOI] [PubMed] [Google Scholar]
- 55.Hughes EG, Robertson DM, Handelsman DJ, Hayward S, Healy DL, de Kretser DM. Inhibin and estradiol responses to ovarian hyperstimulation: effects of age and predictive value for in vitro fertilization outcome. J Clin Endocrinol Metab. 1990;70:358–364. doi: 10.1210/jcem-70-2-358. [DOI] [PubMed] [Google Scholar]
- 56.Provost MP, Acharya KS, Acharya CR, Yeh JS, Steward RG, Eaton JL, Goldfarb JM, Muasher SJ. Pregnancy outcomes decline with increasing body mass index: analysis of 239,127 fresh autologous in vitro fertilization cycles from the 2008–2010 Society for Assisted Reproductive Technology registry. Fertil Steril. 2016;105:663–669. doi: 10.1016/j.fertnstert.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 57.Rittenberg V, Seshadri S, Sunkara SK, Sobaleva S, Oteng-Ntim E, El-Toukhy T. Effect of body mass index on IVF treatment outcome: an updated systematic review and meta-analysis. Reprod BioMed Online. 2011;23:421–439. doi: 10.1016/j.rbmo.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 58.Bennett J, Baumgarten SC, Stocco C. GATA4 and GATA6 silencing in ovarian granulosa cells affects levels of mRNAs involved in steroidogenesis, extracellular structure organization, IGF-I activity, and apoptosis. Endocrinology. 2013;154:4845–4858. doi: 10.1210/en.2013-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bennett J, Wu YG, Gossen J, Zhou P, Stocco C. Loss of GATA-6 and GATA-4 in granulosa cells blocks folliculogenesis, ovulation, and follicle stimulating hormone receptor expression leading to female infertility. Endocrinology. 2012;153:2474–2485. doi: 10.1210/en.2011-1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim J, Sato M, Li Q, Lydon JP, Demayo FJ, Bagchi IC, Bagchi MK. Peroxisome proliferator-activated receptor gamma is a target of progesterone regulation in the preovulatory follicles and controls ovulation in mice. Mol Cell Biol. 2008;28:1770–1782. doi: 10.1128/MCB.01556-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nagashima T, Kim J, Li Q, Lydon JP, DeMayo FJ, Lyons KM, Matzuk MM. Connective tissue growth factor is required for normal follicle development and ovulation. Mol Endocrinol. 2011;25:1740–1759. doi: 10.1210/me.2011-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pelusi C, Ikeda Y, Zubair M, Parker KL. Impaired follicle development and infertility in female mice lacking steroidogenic factor 1 in ovarian granulosa cells. Biol Reprod. 2008;79:1074–1083. doi: 10.1095/biolreprod.108.069435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robker RL, Russell DL, Espey LL, Lydon JP, O’Malley BW, Richards JS. Progesterone-regulated genes in the ovulation process: ADAMTS-1 and cathepsin L proteases. Proc Natl Acad Sci U S A. 2000;97:4689–4694. doi: 10.1073/pnas.080073497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rumi MA, Dhakal P, Kubota K, Chakraborty D, Lei T, Larson MA, Wolfe MW, Roby KF, Vivian JL, Soares MJ. Generation of Esr1-knockout rats using zinc finger nuclease-mediated genome editing. Endocrinology. 2014;155:1991–1999. doi: 10.1210/en.2013-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yazawa T, Kawabe S, Kanno M, Mizutani T, Imamichi Y, Ju Y, Matsumura T, Yamazaki Y, Usami Y, Kuribayashi M, Shimada M, Kitano T, Umezawa A, Miyamoto K. Androgen/androgen receptor pathway regulates expression of the genes for cyclooxygenase-2 and amphiregulin in periovulatory granulosa cells. Mol Cell Endocrinol. 2013;369:42–51. doi: 10.1016/j.mce.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 66.Arnhold IJ, Latronico AC, Batista MC, Izzo CR, Mendonca BB. Clinical features of women with resistance to luteinizing hormone. Clin Endocrinol. 1999;51:701–707. doi: 10.1046/j.1365-2265.1999.00863.x. [DOI] [PubMed] [Google Scholar]
- 67.Artini PG, Ruggiero M, Papini F, Valentino V, Uccelli A, Cela V, Genazzani AR. Chromosomal abnormalities in women with premature ovarian failure. Gynecol Endocrinol. 2010;26:717–724. doi: 10.3109/09513590.2010.500427. [DOI] [PubMed] [Google Scholar]
- 68.Bentov Y, Kenigsberg S, Casper RF. A novel luteinizing hormone/chorionic gonadotropin receptor mutation associated with amenorrhea, low oocyte yield, and recurrent pregnancy loss. Fertil Steril. 2012;97:1165–1168. doi: 10.1016/j.fertnstert.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 69.Desai SS, Achrekar SK, Paranjape SR, Desai SK, Mangoli VS, Mahale SD. Association of allelic combinations of FSHR gene polymorphisms with ovarian response. Reprod BioMed Online. 2013;27:400–406. doi: 10.1016/j.rbmo.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 70.Khoury K, Barbar E, Ainmelk Y, Ouellet A, Lehoux JG. Gonadal function, first cases of pregnancy, and child delivery in a woman with lipoid congenital adrenal hyperplasia. J Clin Endocrinol Metab. 2009;94:1333–1337. doi: 10.1210/jc.2008-1694. [DOI] [PubMed] [Google Scholar]
- 71.Matsuzaki S, Yanase T, Murakami T, Uehara S, Nawata H, Yajima A. Induction of endometrial cycles and ovulation in a woman with combined 17alpha-hydroxylase/17,20-lyase deficiency due to compound heterozygous mutations on the p45017alpha gene. Fertil Steril. 2000;73:1183–1186. doi: 10.1016/S0015-0282(00)00500-8. [DOI] [PubMed] [Google Scholar]
- 72.Mitri F, Bentov Y, Behan LA, Esfandiari N, Casper RF. A novel compound heterozygous mutation of the luteinizing hormone receptor -implications for fertility. J Assist Reprod Genet. 2014;31:787–794. doi: 10.1007/s10815-014-0249-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Okada M, Lee L, Maekawa R, Sato S, Kajimura T, Shinagawa M, Tamura I, Taketani T, Asada H, Tamura H, Sugino N. Epigenetic changes of the Cyp11a1 promoter region in granulosa cells undergoing luteinization during ovulation in female rats. Endocrinology. 2016;157:3344–3354. doi: 10.1210/en.2016-1264. [DOI] [PubMed] [Google Scholar]
- 74.Lee L, Asada H, Kizuka F, Tamura I, Maekawa R, Taketani T, Sato S, Yamagata Y, Tamura H, Sugino N. Changes in histone modification and DNA methylation of the StAR and Cyp19a1 promoter regions in granulosa cells undergoing luteinization during ovulation in rats. Endocrinology. 2013;154:458–470. doi: 10.1210/en.2012-1610. [DOI] [PubMed] [Google Scholar]
- 75.Pruksananonda K, Wasinarom A, Sereepapong W, Sirayapiwat P, Rattanatanyong P, Mutirangura A. Epigenetic modification of long interspersed elements-1 in cumulus cells of mature and immature oocytes from patients with polycystic ovary syndrome. Clin Exp Reprod Med. 2016;43:82–89. doi: 10.5653/cerm.2016.43.2.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jakimiuk AJ, Weitsman SR, Brzechffa PR, Magoffin DA. Aromatase mRNA expression in individual follicles from polycystic ovaries. Mol Hum Reprod. 1998;4:1–8. doi: 10.1093/molehr/4.1.1. [DOI] [PubMed] [Google Scholar]
- 77.Soderlund D, Canto P, Carranza-Lira S, Mendez JP. No evidence of mutations in the P450 aromatase gene in patients with polycystic ovary syndrome. Hum Reprod. 2005;20:965–969. doi: 10.1093/humrep/deh690. [DOI] [PubMed] [Google Scholar]
- 78.Yang F, Ruan YC, Yang YJ, Wang K, Liang SS, Han YB, Teng XM, Yang JZ. Follicular hyperandrogenism downregulates aromatase in luteinized granulosa cells in polycystic ovary syndrome women. Reproduction. 2015;150:289–296. doi: 10.1530/REP-15-0044. [DOI] [PubMed] [Google Scholar]
- 79.Yu YY, Sun CX, Liu YK, Li Y, Wang L, Zhang W. Genome-wide screen of ovary-specific DNA methylation in polycystic ovary syndrome. Fertil Steril. 2015;104:145–53.e6. doi: 10.1016/j.fertnstert.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 80.Patel S, Zhou C, Rattan S, Flaws JA. Effects of endocrine-disrupting chemicals on the ovary. Biol Reprod. 2015;93:20. doi: 10.1095/biolreprod.115.130336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Paulose T, Tannenbaum LV, Borgeest C, Flaws JA. Methoxychlor-induced ovarian follicle toxicity in mice: dose and exposure duration-dependent effects. Birth Defects Res B Dev Reprod Toxicol. 2012;95:219–224. doi: 10.1002/bdrb.21007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zama AM, Uzumcu M. Targeted genome-wide methylation and gene expression analyses reveal signaling pathways involved in ovarian dysfunction after developmental EDC exposure in rats. Biol Reprod. 2013;88:52. doi: 10.1095/biolreprod.112.104802. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(JPEG 359 kb)
(JPEG 340 kb)
(JPEG 375 kb)
(JPEG 390 kb)
(JPEG 391 kb)
(JPEG 387 kb)
(JPEG 383 kb)
(JPEG 368 kb)
(DOCX 157 kb)