Abstract
Objectives
In vitro fertilization (IVF) has been linked to an increased risk for imprinting disorders in offspring. The data so far have predominantly been retrospective, comparing the rate of IVF conceptions in affected patients with controls. We describe a series of fetuses with omphalocele that were tested for Beckwith-Wiedemann syndrome (BWS) and subsequently ascertained as to whether pregnancies were conceived by assisted reproductive technologies (ART).
Methods
Fetuses were tested for BWS by Southern blot, PCR based methods, and methylation analysis to identify the imprinting status at primarily the IC2 locus, KCNQ1OT1, as well as IC1, H19/IGF-2. Some fetuses were also tested for uniparental disomy of chromosome 11p.
Results
We tested 301 fetuses with omphalocele for BWS. Forty samples were positive. Sixteen were from IVF pregnancies, for an overall rate of 40%. Such as high proportion of IVF pregnancies in a series of BWS-positive fetuses has not been described previously. Possible factors such as twinning and ascertainment bias are discussed.
Conclusion
We found about a 20-fold overrepresentation of IVF cases in fetuses with BWS/omphalocele when compared with the rate of ART pregnancies in the USA (p < .0001). Our series provides support for an association of IVF and BWS. Patients should be counseled about these risks and made aware of the availability of prenatal diagnosis for detection.
Keywords: Omphalocele, Beckwith-Wiedemann syndrome, Methylation, Imprinting, IVF
Introduction
In the USA, approximately 1–3% of births involve the use of assisted reproductive technology (ART). In 2002, Cox et al. [1] reported two Angelman syndrome (AS) patients from their clinic who happened to be born of pregnancies conceived by artificial reproductive techniques (ART). Both were positive for abnormal SNRPN methylation [2] and did not have chromosome 15q11 deletions or maternal uniparental disomy (UPD) of chromosome 15q (presumed imprinting error). Both were born following intracytoplasmic sperm injection (ICSI) due to oligospermia and poor motility.
Following this report, it has been postulated that imprinting disorders may be more prevalent in children born after ART which is biologically plausible as in the mouse model embryo culture media has been shown to affect gene imprinting [2–4]. To date, there have been numerous observations suggesting an association between ART and Beckwith-Wiedemann syndrome (BWS) as well as AS. Orstavik et al. [3] also observed a case of Angelman syndrome associated with ART, specifically ICSI, with laboratory identical to what Cox et al. had reported. ICSI was done following failed traditional in vitro fertilization (IVF) though neither parent had obvious causes for infertility.
To further evaluate a possible association of ART with an imprinting disorder, DeBaun et al. [4] in 2003 studied another imprinting syndrome, BWS, by inquiring their BWS patient registries located in two centers. In one center, they found 7 cases, five conceived by ICSI. Of these, 6 were tested by molecular means, and 4 had alterations of IC1, 1 of both IC1 and IC2, and one tested normal [5]. For four of the cases from one registry, the denominator was unknown. From the other center, 3 of 65 (4.6%) cases in their registry were conceived by ART. The population frequency of ART at that time was 0.76%, so the increased risk was approximately 6-fold.
Maher et al. [6] studied their BWS clinic patients from 1995 to 2000 (n = 149). They found that 6 of the 149 were conceived by ART: 3 by standard IVF and 3 by ICSI. During the study period, there were 43,037 births in the area, about 1% conceived by ART. From this, they would have expected only 1.7 cases of ART in the 149 BWS patients. The difference was significant by Poisson distribution, p = .018, with an estimated increased risk of BWS following IRT of 1.5- to 8.8-fold.
Similarly, Giquel et al. [7] examined their BWS patient cohort of 149 and found 6 who were conceived by ART, with 1.3% being the population incidence at the time. By their calculations, they would have expected 1.54 BWS patients with a history of ART, and the difference from the expectation was significant at .018. The odds ratio from the study predicts a 3.2-fold increase in BWS following ART.
xPopulation surveys followed these anecdotal and small BWS patient series. Halliday et al. [8] studied all births in Australia between 1983 and 2003. There were 1,316,500 births in this period, with 37 babies having BWS. This yielded a population prevalence of BWS of 1/35,580 births. Of these BWS cases, 4 were conceived by IVF. They matched 148 control infants and found 1 positive for IVF. The comparison of 1/148 with 4/37 showed an odds ratio for an increased risk of BWS following IVF of 17.8-fold, with a very wide confidence interval of 1.8–437.9. All of the 37 patients were evaluated clinically and were proven by molecular methods, KVDMR (IC2) and H19 (IC1) [5].
Chang et al. [9] reported on 19 BWS/ART patients. Of 12 further evaluated, 5 were from ICSI pregnancies, and 5 were from standard IVF. The other 2 were products of in utero insemination. There did not seem to be an effect of culture media or embryo transfer method upon the incidence of BWS following ART. The causes of infertility in the involved couples were varied.
Sutcliffe et al. [10] contacted families having children with four imprinting syndromes: BWS, AS, Prader-Willi syndrome, and transient neonatal diabetes mellitus. They found that 2.9%, 95% confidence interval (CI) of 1.4–6.4%, were conceived by ART, as compared with a population ART incidence of 0.8%.
Lidegaard et al. [11] reviewed 6052 ART births in Denmark. They did not find a case of an imprinting syndrome following ART.
Doonbos et al. [12] evaluated their Dutch imprinting syndrome (AS, BWS) clinic patients. They found 220, all molecularly proven. Those born via ART represented 6.4% of the group. This compares with a population rate of 2.1%. Of 71 BWS patients, 4 were conceived by ART. The rate of births following infertility was 6.8% in the BWS group and 3.4% in the general population. However, correcting for the infertility, there did not appear to be an increased risk of BWS following ART.
Bowdin et al. [13] evaluated IVF births in England by sending out a survey with about a 60% return rate. Of 1524 questionnaires, the group was able to evaluate 47. They found one BWS case in this small group.
Kallen et al. [14] studied 31,850 births following ART from 1986 to 2006. They found 4 cases of PWS, with an expected population prevalence of 1/15,000 to1/22,000 births [15, 16]. In their sample, they would have expected 1 case. Three of the PWS births were conceived by ICSI and 1 from standard IVF. Two cases of Russell-Silver syndrome were observed, with an expected incidence of only 1/30,000 to 1/100,000 [17]. One case was born after ICSI and one IVF. Finally, one BWS case (ICSI) was found. Therefore, seven imprinting syndrome cases were found in the sample of about 32,000 ART births (1/4570). However, no molecular data were available to verify if all cases resulted from primary imprinting errors.
In Italy, Mussa et al. [18] analyzed a total of 379,872 live births delivered between 2005 and 2014 for incidence of BWS. They reported a significantly greater proportion of ART conceived births (7 of 7884 or 1:1126) than in those conceived naturally (31 of 371,957, 1:12,254). The significance of this difference is p < .001, with a relative risk of 10.7 (95% CI, 4.7 to 24.2).
The prenatal literature on this topic is scarce. Wilson-Haug and colleagues [19] studied a group of 30 fetuses with omphalocele. Six had BWS clinically or by DNA testing. Three were conceived by IVF and the difference between the unaffected and the BWS group for IVF reached a p value < .05.
The available case reports, registry studies, population studies, and questionnaire studies suggest a link between ART and increased incidence of BWS and AS and other hypomethylation syndromes (see below). The predicted increased incidence varies from none to 18-fold. Given the disparity in the above results and the wide confidence intervals, it is difficult to define what the increased risk of BWS following ART really is.
To further investigate this question, we tested by molecular means fetuses with prenatally diagnosed omphalocele. In a majority of BWS cases, omphalocele is observed, approximately 80% [20]. When omphalocele is observed prenatally, and trisomies have been excluded by chromosome or array testing, BWS is an important consideration meriting prenatal diagnosis if desired by the parents [5, 19]. A significant percentage of fetuses with omphalocele have BWS [19].
We retrospectively studied prenatal samples sent to our laboratory between 2002 and 2015 for BWS testing. We identified 301 pregnancies affected by omphalocele and found 40 (13.3%) positive cases. The genetic mechanism causing BWS was abnormal imprinting of KCNQ1OT1 (IC2) in 36 cases, H19 (IC1) in 2 cases, or both loci due to paternal uniparental isodisomy 11 (patUPD11) in 2 cases. Among the 40 fetuses that tested positive for BWS, 16 (40.0%) were conceived by IVF. This compares with a population incidence of about 1–2% for IVF pregnancies [21]. This study provides supporting evidence to the existing anecdotal and registry data suggesting an increased risk of BWS following IVF.
Methods and materials
Beckwith syndrome genetics are complex [5]. The majority of cases result from either overexpression of IGF2 (usually paternal expressed only) or reduced expression of CDKN1C (usually maternally expressed). About 15–20% results from mosaic paternal uniparental disomy (UPD) 11p, which causes abnormalities of both imprinted regions. Other cases result from CDKN1C mutations (perhaps 5–10%, more commonly in familial cases) and 1–2% from chromosomal rearrangements. Between 2002 and 2011, our laboratory tested only for abnormal imprinting of LIT1/KCNQ1OT1 (IC2) [22–24], which detects primary imprinting errors at that locus. Overall, detection rate of BWS is approximately 70–75% with assays for the LIT1/KCNQ1OT1 locus. An abnormal result is usually caused by a primary imprinting error at this locus, but with a possibility of associated paternal UPD 11 (patUPD11). As described below, since 2011, we have added H19/IC1 testing which allows for both, detection of isolated gain of methylation of H19 (IC1) on the maternal chromosome (approximately 5% of BWS) and of patUPD11 when combined with KCNQ1OT1/IC2 analysis.
The Shodair Genetics Laboratory is a clinical laboratory certified by the Centers for Medicare and Medicaid Services (CMS) through Clinical Laboratory Improvement Amendments (CLIA; certificate number: 27D0652530). Between 2002 and 2015, we tested 301 prenatal samples obtained by CVS or amniocentesis from pregnancies affected by fetuses with omphalocele and normal chromosomes or chromosomal microarrays sent to our laboratory for BWS testing. The work reported here is the result of routine provision of medical attention at the standard of care, with data collected solely for patient care purposes. No additional data were collected, or medical intervention undertaken for the purposes of this study. As such, a Research Involving Human Subjects Committee was not convened.
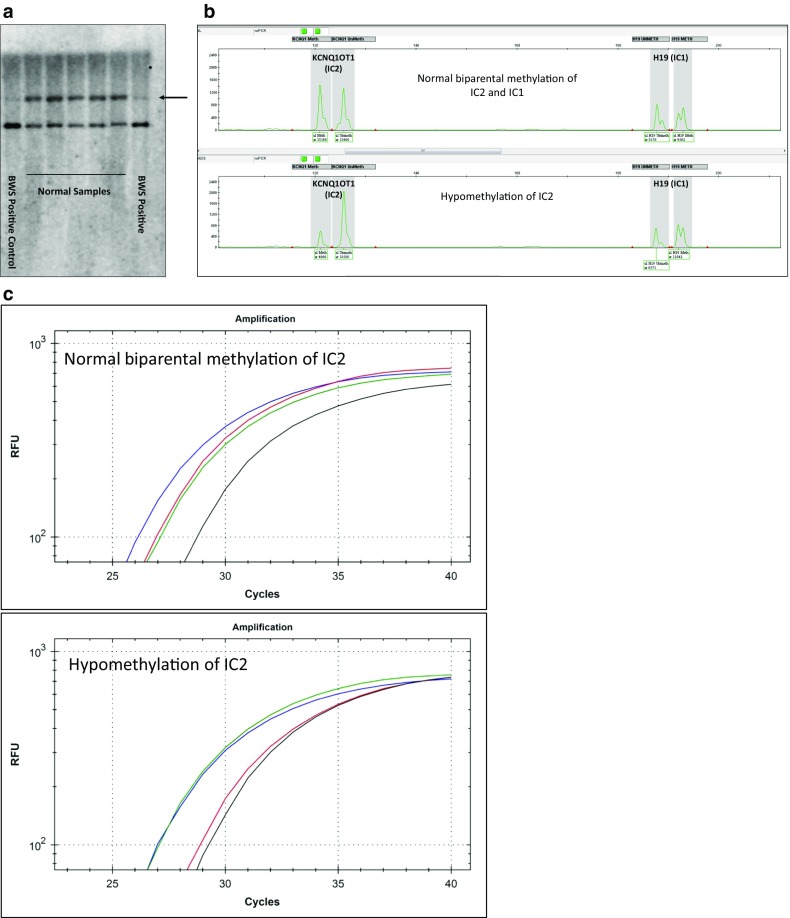
Since initiation of clinical testing for BWS, methods in our laboratory have evolved. Initially, Southern blot analysis was performed. For Beckwith-Wiedemann syndrome, isolated fetal DNA was digested with BamHI/NotI and probed with a LIT1/KCNQ1OT1 sequence [22–24] (IC2, Fig. 1a). Later, we added a bisulfite-treated methylation PCR assay adapted from Coffee et al. [25] (Fig. 1b). For details, see Johnson et al. [26]
Fig. 1.
Methods and results of fetal DNA analysis for Beckwith-Wiedemann syndrome. a Southern blot results on leukocyte DNA digested with BamHI and NotI hybridized with a LIT1/KCNQ1OT1 probe. Note the absence or reduced intensity of the maternal fragment in the BWS positive (arrow). b Methylation-sensitive (MS) polymerase chain reaction for LIT1/KCNQOT1 (IC2) and H19 (IC1). DNA was treated with bisulfite and then amplified. Normal results show roughly equal size/area of the methylated (Meth) and unmethylated (UnMeth) alleles (upper lane). Patients with mosaic hypomethylation of the (maternal) LIT1/KCNQOT1 (IC2) locus show skewed results (lower lane), consistent with mosaic Beckwith-Wiedemann syndrome (BWS). c Restriction digest PCR (RDPCR) of the LIT1/KCNQOT1 (IC2) locus shows that DNA digested with methylation-sensitive or methylation-dependent enzymes amplifies similarly (upper panel) and that in an affected patient, the sample digested with a methylation-dependent enzyme amplifies like a control sample (undigested), whereas the sample digested with a methylation-sensitive enzyme digests normally (the DNA is not methylated) and amplifies late like a double digest control (bottom panel). Blue, DNA not digested (Mock digest); green, DNA digested with a methylation-dependent restriction enzyme; red, DNA digested with a methylation-sensitive enzyme; black, DNA double digested with both enzymes
Subsequently, we replaced Southern blot with a restriction digest PCR (RDPCR). As this is not described elsewhere, we will describe the technique here. In this 4-reaction procedure, sample DNA is either digested with EaeI, which is methylation sensitive (not cutting methylated CpG in the T/C GGCC A/G recognition sequence), with LpnPI, which preferentially recognizes methylated CpG at the center of a 32-base pair sequence, with both enzymes, or with neither (Mock digest). PCR for the KCNQ1OT1 locus is then performed. Expected results include roughly equal maternal and paternal allele quantitative PCR results (analyzed using either a Light Scanner 32 [Idaho Technologies, Salt Lake City, UT] or a CFX 96 real-time PCR machine [Bio-Rad, Hercules, CA]) for the two digests, indicating a methylation index [27] near 0.5. For the undigested sample, the PCR reaches maximum product one cycle before the single digests (which amplify only one intact copy), and a product amplifying much later is seen in the double digest samples (Fig. 1c).
As noted above, in 2011, we added H19 (IC1) methylation PCR [28] and a similar RDPCR test. We use similar PCR methods for the H19 assay. The use two different assays for the above tests is to improve estimates of the MI. Samples tested prior to 2011 were initially only tested for the KCNQ1OT1 (IC2) locus (n = 182). Among those, 18 of the 23 samples that tested positive for the KCNQ1OT1 (IC2) locus were re-analyzed on the H19 (IC1) locus for this study to evaluate for possible paternal UPD11. For 4 of these samples, the H19 (IC1) locus could not be analyzed due to insufficient DNA.
Results
Between 2002 and 2015, our laboratory performed methylation testing for Beckwith-Wiedemann syndrome (BWS) in 185 amniocentesis and 102 CVS samples from pregnancies affected by fetal omphalocele. During the same time period, 9 samples from isolated fetal DNA, and 5 from fetal losses, were analyzed (Table 1).
Table 1.
Prenatal BWS test results of 301 fetuses with omphalocele
| Sample type | Total number | BWS positivea | BWS negativeb |
|---|---|---|---|
| Amniocytes (cultured) | 184 | 32 | 152 |
| Amniocytes (direct) | 1 | 0 | 1 |
| CVS (cultured) | 101 | 5 | 96 |
| CVS (direct) | 1 | 0 | 1 |
| POC/fetal tissue (cultured) | 5 | 1 | 4 |
| Extracted DNA (CVS or amniocytes) | 9 | 2 | 7 |
| Total | 301 (100%) | 40 (13.3%) | 261 (86.7%) |
BWS Beckwith-Wiedemann syndrome
aAmong the 40 positive samples, 23 were initially only tested for the IC2 locus and IC1 methylation was determined retrospectively for this study; of these, 4 did not have sufficient DNA to analyze the IC1 locus
bOf the 261 negative samples, 103 were assessed for both IC2 and IC1 loci, whereas 158 were tested for the IC2 locus only
Of the 301 samples, 40 tested positive for BWS (Table 1). We found 36 positives by Southern blot and/or methylation PCR assays for LIT1/KCNQ1OT1. Two samples revealed isolated gain of methylation of H19 (IC1), whereas another 2 samples showed abnormal methylation of both LIT1/KCNQ1OT1 (IC2) and H19 (IC1) loci consistent with paternal uniparental disomy 11 (patUPD11). Methylation of H19 (IC1) was normal in 32 of the 36 IC2-positive BWS samples and could not be verified in 4 samples due to insufficient DNA (Table 2). Among the 40 BWS-positive samples, 32 were cultured amniocytes and 5 were cultured CVS. One of the POC as well as 2 isolated fetal DNA samples tested positive as well (Table 1).
Table 2.
Genetic mechanisms among BWS-positive samples
| Causes of BWS by genetic mechanism | Total number (percentage) | ART pregnancies number (percentage) | Non-ART pregnancies number (percentage) |
|---|---|---|---|
| Loss of methylation of IC2 on maternal chromosome | 36a | 15 | 21 |
| Gain of methylation of IC1 on maternal chromosome | 2 | 0 | 2 |
| Paternal UPD11 | 2 | 1 | 1 |
| Total | 40 (100%) | 16 (40.0%) | 24 (60.0%) |
BWS Beckwith-Wiedemann syndrome
aIn 4 of 36 samples, only IC2 was analyzed
Maternal age, available for 231 amniocentesis/CVS samples, was < 35 years in 137 samples and ≥ 35 years for 94 samples (Table 3). The average maternal age for the CVS and amniocentesis samples was 34 and 32.5, respectively. Of the 231 samples for which maternal age was available, 19/137 samples with maternal age < 35 years (13.9%) and 14/94 (14.9%) with maternal age 35 years and older tested positive indicating no significant difference in the positive rate between these age groups. Note that only for 33 of the 40 BWS-positive samples, maternal age was available (Table 3).
Table 3.
Maternal age distribution among BWS-positive samples
| Samples | Number of samples by maternal agea | |
|---|---|---|
| < 35 | ≥ 35 | |
| CVS | 50 | 36 |
| Amniocentesis | 87 | 58 |
| CVS and amniocentesis | 137 | 94 |
| Positive (percentage) | 19 (13.9%) | 14 (14.9%) |
BWS Beckwith-Wiedemann syndrome
aNumber of samples for which maternal age data was available
Twenty-four of the 301 analyzed samples were from pregnancies conceived by ART (Table 4), and 13 were from twin pregnancies (Table 5). Among the 40 BWS-positive samples, 16 (40.0%) were from IVF pregnancies (Table 4). Of these, 4 were cultured CVS samples subsequently confirmed by amniocentesis in 3 cases, and the remaining 12 samples were cultured amniocytes. Hypomethylation of the LIT1/KCNQ1OT1 (IC2) locus was present in 15 of the 16 positive samples from IVF pregnancies, whereas one was found positive due to patUPD showing a paternal methylation pattern for both IC2 (hypomethylation) and IC1 (hypermethylation). None of the samples from pregnancies conceived with ART showed isolated hypermethylation of H19 (IC1) as genetic mechanism for BWS (Table 2). Among the BWS 261 negative samples, 8 (3.1%) were from pregnancies conceived by ART (Table 4).
Table 4.
Pregnancies conceived by ART among BWS-positive and BWS-negative prenatal cases
| Sample type | ART pregnancies | |
|---|---|---|
| BWS-positive samples (n = 40) | BWS-negative samples (n = 261) | |
| Amniocytes | 12 | 2 |
| CVS | 4 | 6 |
| POC/fetal tissue | 0 | 0 |
| Total | 16 (40.0%) | 8 (3.1%) |
ART assisted reproductive technology, BWS Beckwith-Wiedemann syndrome
Table 5.
Twin pregnancies by BWS diagnosis and ART status
| BWS result and ART status | Twin pregnancies (percentage)a |
|---|---|
| BWS positive (n = 40) | 7 (17.5%) |
| • Conceived with ART (n = 16) | 5 (31.3%) |
| • Conceived without ART (n = 24) | 2 (8.3%) |
| BWS negative (n = 261) | 6 (2.3%) |
| Total (n = 301) | 13 (4.3%) |
ART assisted reproductive technology, BWS Beckwith-Wiedemann syndrome
aThe twinning rate in the USA in 2015 was 33.5 per 1000 deliveries corresponding to 3.35% (Martin et al. 2018)
Seven of the 40 BWS-positive samples were from twin pregnancies (17.5%) of which 5 were conceived by ART. Thus, twinning was present in 5 of the 16 BWS-positive samples from IVF pregnancies (31.3%), compared to 2 of the 24 BWS-positive samples conceived without ART (8.3%). In comparison, 6 of the 261 BWS-negative samples were from twin pregnancies (2.3%) (Table 5).
Discussion
Assisted reproductive technologies (ART) including in vitro fertilization (IVF) are generally considered safe infertility treatments but have been associated with birth defects and adverse pregnancy outcomes in some studies. Specifically, an increased risk following ART of fetal imprinting disorders including BWS and Angelman syndrome has been suggested. Postulated biological explanations range from direct effects of ART caused by embryo culture techniques, culture media, or the use of intracytoplasmic sperm injection (ICSI), to heritable traits associated with subfertility itself that are bypassed through ART as further discussed below.
Omphalocele is a major manifestation of BWS readily identifiable on prenatal ultrasound. Here, we reviewed prenatal cases with fetal omphalocele referred to our laboratory for molecular testing of BWS between 2002 and 2015. The percentage of fetuses with omphalocele found to have BWS was 13.3% based on the diagnostic methods used during that period. Considering that H19 (IC1) methylation testing was not established in our laboratory until 2011 and given that analysis of CDKN1C was not included, our testing up until 2011 would have missed cases caused by gain of methylation at H19 (IC1) estimated to make up 5% of alterations in BWS, and those cases caused by maternal pathogenic variants (5% of BWS cases in the absence of a family history). In contrast, loss of methylation at IC2 (maternal) and patUPD11 accounting for approximately 50% and 20% of BWS, respectively, would have been detected. Also, microdeletions and duplications that alter the methylation status of the IC2 (since 2011 also IC1) would have been detected. Overall, our diagnostic rate is comparable with previously reported prenatal series [19].
What is striking about our results is the high percentage of BWS-positive fetuses who were conceived by IVF (Table 4). We reviewed charts and called referring centers to verify that 16 of 40 BWS-positive fetuses (40.0%) were conceived by IVF. In contrast, only 8 of 261 BWS-negative fetuses (3.1%) were conceived using ART. For comparison, the population incidence for IVF conception in the USA is 1–2%. Thus, while the rate of ART-related pregnancies observed among BWS-negative fetuses with omphalocele was similar to the rate of ART-related pregnancies in the general population, fetuses conceived by IVF were overrepresented among BWS-positive fetuses with omphalocele. Using a 1–2% estimate for ART conception in the USA, the increase in risk with ART could approximate 20-fold, the highest estimate so far and the first from a prenatal series. This would create an overall risk of BWS with omphalocele following IVF of about 1/850 (BWS/omphalocele prevalence assumed at 1/17,000, about 80% of 1/13,700 [5]). The chi-square difference of proportions calculator indicates the difference in the observed (.366) vs expected number (.02) for this sample size reaches significance at p < .0001 [29].
The BWS-positive fetuses conceived with (16) and without (24) ART each included one case caused by patUPD11 and provide no observable evidence for a relationship between conception method and genetic mechanism, although our numbers are too small to allow for statistical comparison (Table 2). Of note, an association of UPD 11p and omphalocele/BWS has previously been suggested [30].
A contributing factor to the disproportionally high number of BWS-positive IVF pregnancies could be an overrepresentation of twin gestations in that group (Table 5): In our entire cohort of 301 fetuses with omphalocele, 13 (4.3%) were from twin gestations compared to a twinning rate of 3.35% in the USA in 2015. [31] Five of the 15 BWS-positive cases conceived by ART were twin gestations (31.3%) compared to 2 of 26 (8.3%) non-ART pregnancies that tested positive for BWS. However, the difference of 23.6% did not quite reach statistical significance (p = .055, 95% CI − 0.7 to 48.6 [32]) indicating that the high twinning rate alone is unlikely to be the only factor driving the overrepresentation of ART cases among the BWS positives. In contrast, 6 twin pregnancies were among the 261 BWS-negative fetuses with omphalocele (2.3%), similar to the twinning rate in the USA.
Many BWS twins have been described (summarized in Elalaoui SC et al. [33]). The majority of BWS twins are female MZ discordant, and most of them present hypomethylation of IC2. This has led to the hypothesis that the methylation defect and the twinning process are correlated. [34] In a cohort of 250 BWS patients described by Weksberg et al., the frequency of monozygotic (MZ) twinning was 8% as compared with 1% dizygotic (DZ) twinning. [35] Bliek et al. observed an excess of monozygotic twins among BWS patients with IC2 hypomethylation in their cohort of 400 BWS patients: 3% were twin pairs of which about two-thirds were MZ twins. [36] The high twining rate of a third among our BWS-positive cases conceived by IVF suggest additional factors at play that might in part derive from the ART process. If methylation failure precedes and possibly triggers the twinning process as previously suggested, [34, 36]ART might create conditions that allow for a higher rate of aberrant methylation.
Manipalviratn et al. [37] and Hiura et al. [38] discussed possible mechanisms within ART procedures which may lead to an increased incidence of imprinting syndromes. These include the hormones given to the prospective mother, oocyte collection and maturation in vitro, possible use of immature sperm, the injection procedure (ICSI), and embryo culture. Perhaps relevant are the observations that the early embryo undergoes a wave of demethylation and then the parental patterns are reset [39]. A maternal hypomethylation syndrome has been observed [40] with abnormal methylation at multiple known imprinting sites with varying manifestations of the associated syndromes, first described in infants with transient neonatal diabetes syndrome. Finally, the methylation status of early embryos and placentas revealed multiple variable findings at different loci and different developmental stages following ART in a recent study [41]. Lastly, the underlying problem which causes couples to seek out ART, infertility, may be a primary driver for the increased risk of associated imprinting syndromes[42].
Our study has several weaknesses. The referrals were likely non-random and the overall numbers are small. It is possible that prenatal centers were more likely to request BWS testing because of IVF conception. We think that the main reason for referral was simply the fetal omphalocele but cannot be certain. In our postnatal series of affected patients, a significant number have resulted from IVF pregnancies.
In summary, it is apparent there are no specific mechanisms known to disturb imprinting in ART embryos and no consistent estimates of the increased risk of associated syndromes (primarily BWS and AS). However, from our data and review of the literature, we recommend that patients be counseled about the increased risks for these imprinting syndromes and that prenatal testing be considered at the parents’ request.
Footnotes
The work reported here is the result of routine provision of medical attention at the standard of care, with data collected solely for patient care purposes. No additional data were collected or medical intervention undertaken for the purposes of this study. As such, a Research Involving Human Subjects Committee was not convened.
References
- 1.Cox GF, Burger J, Lip V, et al. Intracytoplasmic sperm injection may increase the risk of imprinting defects. Am J Hum Genet. 2002;71:162–164. doi: 10.1086/341096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glenn CC, Porter KA, Jong MT, et al. Functional imprinting and epigenetic modification of the human SNRPN gene. Hum Mol Genet. 1993;2(12):2001–2005. doi: 10.1093/hmg/2.12.2001. [DOI] [PubMed] [Google Scholar]
- 3.Orstavik KH, Eiklid K, van der Hagen CB, Spetalen S, Kierulf K, Skjeldal O, et al. Another case of imprinting defect in a girl with Angelman syndrome who was conceived by intracytoplasmic semen injection. Am J Hum Genet. 2003;72(1):218–221. doi: 10.1086/346030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeBaun MR, Niemitz EL, Feinberg AP. Association of in vitro fertilization with Beckwith-Wiedemann syndrome and epigenetic alteration of LIT1 and H19. Am J Hum Genet. 2003;72:156–160. doi: 10.1086/346031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shuman S, Beckwith JB, Smith AC et al. Beckwith-Wiedemann syndrome, in Gene Reviews, Pagon RA, Adam MP, Ardinger HH, et al., editors. Seattle: University of Washington; 1993–2015.
- 6.Maher ER, Brueton LA, Bowdin SC, Luharia A, Cooper W, Cole TR, et al. Beckwith-Wiedemann syndrome and assisted reproductive technology (ART). J Med Genet. 2003;40:62–4. [DOI] [PMC free article] [PubMed]
- 7.Gicquel C, Gaston V, Mandelbaum J, Siffroi JP, Flahault A, le Bouc Y. In vitro fertilization may increase the risk of Beckwith-Weidemann syndrome related to the abnormal imprinting of the KCNQ1OT gene. Am J Hum Genet. 2003;72:1338–1341. doi: 10.1086/374824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halliday J, Oke K, Breheny S, Algar E, Amor DJ. Beckwith-Wiedemann syndrome and IVF: a case control study. Am J Hum Genet. 2005;75:526–528. doi: 10.1086/423902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang AS, Moley KH, Wangler M, Feinberg AP, DeBaun MR. Association between Beckwith-Wiedemann syndrome and assisted reproductive technology: a case series of 19 patients. Fertil Steril. 2005;83:349–354. doi: 10.1016/j.fertnstert.2004.07.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sutcliffe AG, Peters CJ, Bowdin S, Temple K, Reardon W, Wilson L, et al. Assisted reproductive therapies and imprinting disorders—a preliminary British survey. Hum Reprod. 2006;21:1009–11. [DOI] [PubMed]
- 11.Lidegaard O, Pinborg A, Andersen AN. Imprinting diseases and IVF: Danish National IVF cohort study. Hum Reprod. 2005;20:950–954. doi: 10.1093/humrep/deh714. [DOI] [PubMed] [Google Scholar]
- 12.Doornbos ME, Maas SM, McDonnell J, Vermeiden JP, Hennekam RC. Infertility, assisted reproduction technologies and imprinting disturbances: a Dutch study. Hum Reprod. 2007;22(9):2476–2480. doi: 10.1093/humrep/dem172. [DOI] [PubMed] [Google Scholar]
- 13.Bowdin S, Allen C, Kirby G, Brueton L, Afnan M, Barratt C, et al. A survey of assisted reproductive technology births and imprinting disorders. Hum Reprod. 2007;22(12):3237–40. [DOI] [PubMed]
- 14.Kallen, Finnström O, Lindam A, Nilsson E, Nygren KG, Otterblad Olausson P. Trends in delivery and neonatal outcome after in vitro fertilization in Sweden: data for 25 years. Hum Reprod. 2010;4:1026–1034. doi: 10.1093/humrep/deq003. [DOI] [PubMed] [Google Scholar]
- 15.Whittington JE, Butler JV, Holland AJ. Short report: changing rates of genetic subtypes of Prader–Willi syndrome in the UK. Eur J Hum Genet. 2007;15:127–130. doi: 10.1038/sj.ejhg.5201716. [DOI] [PubMed] [Google Scholar]
- 16.Bittel DC, Butler MG. Prader-Willi syndrome: clinical genetics, cytogenetics and molecular biology. Expert Rev Mol Med. 2005;7(14):1–20. doi: 10.1017/S1462399405009531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.https://ghr.nlm.nih.gov/condition/russell-silver-syndrome
- 18.Mussa A, Molinatto C, Cerrato F, Palumbo O, Carella M, Baldassarre G, et al. Assisted reproductive techniques and risk of Beckwith-Wiedemann syndrome. Pediatrics. 2017;140(1):e20164311. [DOI] [PubMed]
- 19.Wilkins-Haug L, Porter A, Hawley P, Benson CB. Isolated fetal omphalocele, Beckwith-Wiedemann syndrome, and assisted reproductive technologies. Birth Defects Res A Clin Mol Teratol. 2009;85(1):58–62. doi: 10.1002/bdra.20547. [DOI] [PubMed] [Google Scholar]
- 20.Elliott M, Bayly R, Cole T, Temple IK, Maher ER, Elliott M. Clinical features and natural history of Beckwith-Wiedemann syndrome: presentation of 74 new cases. Clin Genet. 1994;46(2):168–174. doi: 10.1111/j.1399-0004.1994.tb04219.x. [DOI] [PubMed] [Google Scholar]
- 21.www.cdc.gov/art accessed 4/21/2018.
- 22.Lee MP, DeBaun MR, Mitsuya K, et al. Loss of imprinting of a paternally expressed transcript, with antisense orientation to KVLQT1, occurs frequently in Beckwith-Wiedemann syndrome and is independent of insulin-like growth factor II imprinting. Proc Natl Acad Sci U S A. 1999;96(9):5203–5208. doi: 10.1073/pnas.96.9.5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitsuya K, Meguro M, Lee MP, Katoh M, Schulz TC, Kugoh H, Yoshida MA, Niikawa N, Feinberg AP, Oshimura M LIT1, an imprinted antisense RNA in the human KvLQT1 locus identified by screening for differentially expressed transcripts using monochromosomal hybrids. Hum Mol Genet 1999;8(7):1209–17.16, 1217. [DOI] [PubMed]
- 24.Smilinich NJ, Day C, Fitzpatrick GV, et al. A maternally methylated CpG island in KvLQT1 is associated with an antisense paternal transcript and loss of imprinting in Beckwith-Wiedemann syndrome. Proc Natl Acad Sci U S A. 1999;96:8064–8069. doi: 10.1073/pnas.96.14.8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coffee B, Muralidharan K, Highsmith WE, Jr, et al. Molecular diagnosis of Beckwith-Wiedemann syndrome using quantitative methylation-sensitive polymerase chain reaction. Genet Med. 2006;8(10):628–634. doi: 10.1097/01.gim.0000237770.42442.cc. [DOI] [PubMed] [Google Scholar]
- 26.Johnson JP, Waterson J, Schwanke C, Schoof J. Genome-wide androgenetic mosaicism. Clin Genet. 2014;85:282–285. doi: 10.1111/cge.12146. [DOI] [PubMed] [Google Scholar]
- 27.Bliek J, Maas SM, Ruijter JM, Hennekam RC, Alders M, Westerveld A, et al. Increased tumour risk for BWS patients correlates with H19 and KCNQ1OT1 methylation: occurrence of KCNQ1OT1 hypomethylation in in familial cases of BWS. Hum Mol Genet. 2001;10:467–76. [DOI] [PubMed]
- 28.Zeschnigk M, Albrecht B, Buiting K, Kanber D, Eggermann T, Binder G, Gromoll J, Prott EC, Seland S, Horsthemke B. IGF2/H19 hypomethylation in Silver-Russell syndrome and isolated hemihypoplasia. Eur J Hum Genet. 2008;16(3):328–334. doi: 10.1038/sj.ejhg.5201974. [DOI] [PubMed] [Google Scholar]
- 29.www.medcalc.org/calc/comparison_of_proportions.php. Accessed 9/15/2017.
- 30.Grati FR, Turolla L, D’Ajello P, Ruggeri M, Miozzo M, Bracalente G, et al. Chromosome 11 segmental paternal isodisomy in amniocytes from two fetuses with omphalocele: new highlights on phenotype-genotype correlations in Beckwith-Wiedemann syndrome. J Med Genet. 2007;44:257–63. [DOI] [PMC free article] [PubMed]
- 31.Martin JA, Hamilton BE, Osterman MJK, Driscoll AK, Drake P. Births: final data for 2016. National Vital Statistics Reports; vol 67 no 1. Hyattsville, MD: National Center for Health Statistics. 2018. [PubMed]
- 32.http://www.sample-size.net/confidence-interval-proportion/. Accessed 9/15/2017.
- 33.Elalaoui SC, Garin I, Sefiani A, Perez de Nanclares G. Maternal hypomethylation of KvDMR in a monozygotic male twin pair discordant for Beckwith-Wiedemann syndrome. Molecular Syndromology. 2014;5(1):41–6. [DOI] [PMC free article] [PubMed]
- 34.Bestor TH. Imprinting errors and developmental asymmetry. Philos Trans R Soc Lond Ser B Biol Sci. 2003;358:1411–1415. doi: 10.1098/rstb.2003.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weksberg R, Shuman C, Caluseriu O, Smith AC, Fei YL, Nishikawa J, et al. Discordant KCNQ1OT1 imprinting in sets of monozygotic twins discordant for Beckwith-Wiedemann syndrome. Hum Mol Genet. 2002;11:1317–25. [DOI] [PubMed]
- 36.Bliek J, Alders M, Maas SM, Oostra RJ, Mackay DM, van der Lip K, et al. Lessons from BWS twins: complex maternal and paternal hypomethylation and a common source of haematopoietic stem cells. Eur J Hum Genet. 2009;17(12):1625–34. [DOI] [PMC free article] [PubMed]
- 37.Manipalviratn S, De Cherney A, Segars J. Imprinting disorders and assisted reproductive technology. Reprod Med Biol. 2014;13:193–202. doi: 10.1007/s12522-014-0183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hiura H, Okae H, Chiba H, Miyauchi N, Sato F, Sato A, et al. Imprinting and methylation errors in ART. Reprod Med Biol. 2014;13:193–202. [DOI] [PMC free article] [PubMed]
- 39.Kafri T, Ariel M, Brandeis M, Shemer R, Urven L, McCarrey J, et al. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes Dev. 1992;6:705–14. [DOI] [PubMed]
- 40.Mackay DJ, Boonen SE, Clayton-Smith J, Goodship J, Hahnemann JM, Kant SG, et al. A maternal hypomethylation syndrome presenting as transient neonatal diabetes mellitus. Hum Genet. 2006 Sep;120(2):262–9. [DOI] [PubMed]
- 41.Sakian S, Louie K, Wong EC, Havelock J, Kashyap S, Rowe T, et al. Altered gene expression of H19 and IGF2 in placentas from ART pregnancies. Placenta. 2015;36:1100–5. [DOI] [PubMed]
- 42.Marchesi DE, Qiao J, Feng HL. Embryo manipulation and imprinting. Semin Reprod Med. 2012;30:323–34. [DOI] [PubMed]