Abstract
Active cysteinyl protease Caspase-6 is associated with early Alzheimer and Huntington diseases. Higher entorhinal cortex and hippocampal Caspase-6 levels correlate with lower cognitive performance in aged humans. Caspase-6 induces axonal degeneration in human primary neuron cultures and causes inflammation and neurodegeneration in mouse hippocampus, and age-dependent memory impairment. To assess whether Caspase-6 causes damage to another neuronal system, a transgenic knock-in mouse overexpressing a self-activated form of Caspase-6 five-fold in the striatum, the area affected in Huntington disease, and 2.5-fold in the hippocampus and cortex, was generated. Detection of Tubulin cleaved by Caspase-6 confirmed Caspase-6 activity. The Caspase-6 expressing mice and control littermates were subjected to behavioral tests to assess Huntington disease-relevant psychiatric, motor, and cognitive deficits. Depression was excluded with the forced swim and sucrose consumption tests. Motor deficits were absent in the nesting, clasping, rotarod, vertical pole, gait, and open field analyzes. However, Caspase-6 mice developed age-dependent episodic and spatial memory deficits identified by novel object recognition, Barnes maze and Morris water maze assays. Neuron numbers were maintained in the striatum, hippocampus, and cortex. Microglia and astrocytes were increased in the hippocampal stratum lacunosum molecular and in the cortex, but not in the striatum. Synaptic mRNA profiling identified two differentially expressed genes in transgenic hippocampus, but none in striatum. Caspase-6 impaired synaptic transmission and induced neurodegeneration in hippocampal CA1 neurons, but not in striatal medium spiny neurons. These data revealed that active Caspase-6 in the striatal medium spiny neurons failed to induce inflammation, neurodegeneration or behavioral abnormalities, whereas active Caspase-6 in the cortex and hippocampus impaired episodic and spatial memories, and induced inflammation, neuronal dysfunction, and neurodegeneration. The results indicate age and neuronal subtype-dependent Caspase-6 toxicity and highlight the importance of targeting the correct neuronal subtype to identify underlying molecular mechanisms of neurodegenerative diseases.
Introduction
Caspase-6 (Casp6) activity is associated with axonal degeneration in human primary neuron cultures [1], in mouse dorsal root ganglia, sympathetic cervical, and commissural neurons [2–4], and in ischemic [5] and ethanol-mediated [6] brain damage. Casp6 activity is also associated with age-dependent human cognitive impairment and Alzheimer disease (AD) pathologies [7–9]. Expression of a self-activated form of Casp6 in the hippocampal CA1 region results in inflammation, neurodegeneration, and age-dependent episodic and spatial memory impairment in ACL mice [10]. Despite being grouped with the effector caspases, Caspase-3 and Caspase-7, active Casp6 does not induce cell death in various peripheral cell lines [11, 12]. Furthermore, Casp6 has a protective role in the amyotrophic lateral sclerosis mutant G93A super oxide dismutase transgenic mouse model [13]. Therefore, Casp6 is not always detrimental to brain neurons. To determine if active Casp6 is sufficient to induce neurodegeneration in all neurons, here, we assessed the effect of active Casp6 expression in the medium spiny neurons (MSN) principally affected by Huntington disease (HD).
HD is an autosomal dominant neurodegenerative disorder caused by CAG repeat expansion in the human huntingtin (HTT) gene that expresses mutant huntingtin protein (mHtt) containing N-terminal extended polyglutamine repeats [14]. HD pathology is characterized by intracellular inclusions of cleaved mHtt (∆mHtt) aggregates, neurodegeneration, and neuronal loss of the MSN, and inflammation, mainly in the striatopallidal pathway [15–17]. At advanced stages, atrophy, loss of neurons and mHtt inclusions occur in the deep layers of the cortex [15, 18] and hippocampus [19, 20]. HD causes motor impairments such as chorionic movements with gait disturbance, bradykinesia and rigidity [21], and psychiatric and cognitive impairments characterized by depression, apathy, irritability, impulsivity and social disinhibition, and decreased attention and learning abilities [21], detected up to ten years before motor deficits [14, 21]. Casp6 is thought to also be involved in HD pathogenesis [22–24]. Increased Casp6 mRNA levels [22] and Casp6 activation [23] are reported in the cortex and striatum of early-grade human HD. Casp6 activation in human striatum is proportional to the number of mHtt CAG repeats and correlates with the age of disease onset in HD patients [24]. Casp6 cleaves Htt at amino acid D586 in vitro, ex-vivo, and in vivo [24, 25]. This fragment is generated in murine HD brains [26, 27] and co-localizes with active Casp6 in the nucleus of cultured striatal cells under apoptotic conditions [28]. In BACHD or YAC128 mutant HTT transgenic mice, disease severity correlates with cortical and striatal active Casp6 levels [24]. Mice expressing the 586 aa ∆mHtt fragment with 82 polyglutamine repeats, but not with normal 23 polyglutamine repeats, display several HD-like features including astrogliosis, motor and cognitive deficits, early mortality, and age-dependent brain atrophy [29, 30]. Prevention of Casp6-induced mHtt cleavage by expression of the dominant-negative catalytically inactive Casp6-C163A, mutagenesis of mHtt to D586A, or treatment with a peptide inhibitor based on the Htt Casp6 cleavage site, decreases the sensitivity of cells to several excitotoxic insults, and prevents body weight gain, brain atrophy, depression, and cognitive and motor disorders in BACHD and YAC128 HD mice models [23, 25, 26, 31, 32]. In addition, YAC128 and BACHD mice lacking Casp6 are resistant to mHtt-induced body weight gain and depression-like phenotype [33, 34]. Together, these results indicate that Casp6 plays an important role in HD pathogenesis.
Therefore, a new transgenic knock-in mouse (ACK) was generated to express a self-activating form of human Casp6 under the control of the adenosine A2A receptor (Adora2a) promoter in the striatum, and to a lesser extent in the cortex and hippocampus. The ACK mice do not develop motor or psychiatric disturbances, but show cortical and hippocampal neuroinflammation and age-dependent memory impairments. Furthermore, Caspase-6 impaired synaptic transmission and induced neurodegeneration in hippocampal CA1 neurons, but not in striatal MSN. Consequently, these results demonstrate a differential brain region-specific vulnerability of neurons to Casp6 activity.
Results
Expression of a self-activated form of human Casp6 in the MSN of the indirect striatopallidal pathway
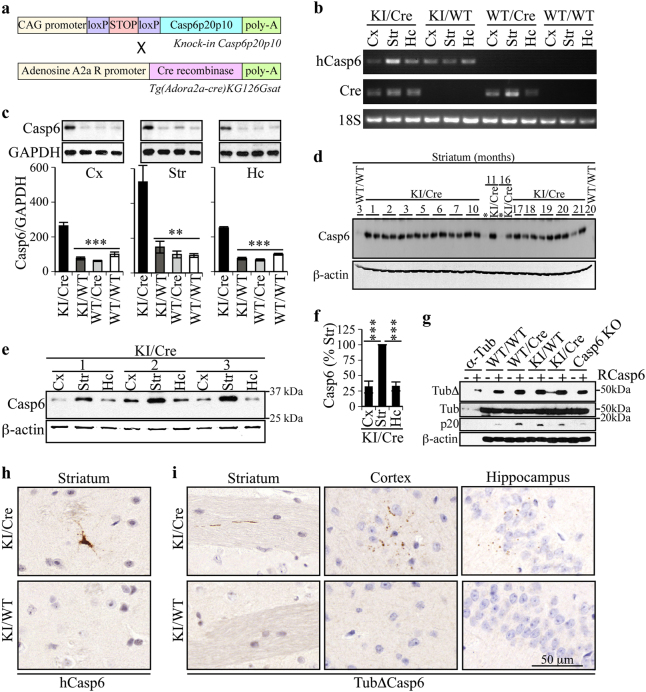
To determine if active Casp6 induces neurodegeneration in striatal neurons, a transgenic mouse line (ACK) expressing a self-activating form of human Casp6 in the MSN of the indirect striatopallidal pathway was generated by crossing the knocked-in (KI) CASP6 transgenic mouse with a mouse expressing Cre recombinase under the Adora2a promoter (Fig. 1a). These ACK KI/Cre mice were compared to wild type (WT/WT), KI CASP6 transgene in the absence of Cre (KI/WT), or Cre only (WT/Cre) control littermates. Viability and fertility were normal in the four genotypes. Similar body weight and rectal temperature in the four genotypes excluded potential effects of the transgene on basal metabolism (Supplementary Fig. 1). Human Casp6 mRNA levels were higher in the KI/Cre striatum and present in the cortex and hippocampus (Fig. 1b). Casp6 mRNA was not detected in WT/WT or WT/Cre brains, but was observed in KI/WT brains, indicating leakiness of the transgenic mRNA expression. Nevertheless, Casp6 protein was not detected above endogenous mouse Casp6 by western blotting (Fig. 1c). Casp6 protein levels in the KI/Cre striatum, cortex, and hippocampus were 5.2, 2.5, and 2.6-fold higher than in the WT/WT tissues, respectively. ACK hippocampal Casp6 levels were only 35% of that in the hippocampal ACL Casp6 transgenic [10] (Supplementary Fig. 2a, b). Transgenic Casp6 protein expression levels were maintained with aging (Fig. 1d). Casp6 protein levels were ~three-fold higher in the striatum compared to the cortex and hippocampus (Fig. 1e, f). Tubulin cleaved by Casp6 (Tub∆Casp6), migrating with that generated by the addition of recombinant active Casp6 in brain protein extracts, was detected in KI/Cre, but not in control or Casp6 null (Casp6 KO) striatal protein extracts (Fig. 1g). The presence of Tub∆Casp6 in the KI/Cre striatum confirmed Casp6 activation. Consistent with our previous observations [10], active p20 Casp6 levels remained undetectable. Immunopositive anti-active Casp6 was observed only in striatal neurons due to the lower levels expressed in cortex and hippocampus (Fig. 1h). Immunopositive anti-Tub∆Casp6 staining (Fig. 1i) confirmed the presence of active Casp6 in ACK KI/Cre striatum, cortex, and hippocampus, but not in control brains (Supplementary Fig. 2c, d). Tub∆Casp6 immunostaining as punctate structures in neuritic cytoplasm indicated degenerating neurons. Together, these results confirm that the ACK KI/Cre mouse expresses Casp6 predominantly in the striatum, and to a lesser extent in the cortex and the hippocampus.
Fig. 1.
Casp6 ACK KI/Cre mice express active Casp6. a Schematic diagram showing Casp6 cDNA and Adora2a-Cre constructs in transgenic mice crossed to generate the Casp6-expressing ACK KI/Cre, and control littermates KI/WT, WT/Cre and WT/WT mice. b Ethidium bromide-stained agarose gel of human Casp6, Cre recombinase, and ribosomal 18S RT-PCR amplicons from striatum (Str), cortex (Cx) and hippocampus (Hc) of KI/Cre, KI/WT and WT/WT mice. c Western blot of 15 µg total protein extracts from KI/Cre, KI/WT and WT/WT Cx, Str, and Hc with anti-Casp6 and GAPDH antibodies. The histograms represent densitometric analyzes of ratios of Casp6 over GAPDH levels and relative to WT/WT (n = 3/genotype, 15–22 months of age). One-way ANOVA followed by Dunnett’s post-hoc test against KI/Cre was performed. **p < 0.01, ***p < 0.001 d Western blot analysis of Casp6 in striatal protein extracts from KI/Cre mice aged from 1 to 21 months with anti-Casp6 and anti-β-actin antibodies. WT/WT (3 and 20 month old) and KI/WT (11 and 16 month old, marked with asterisks) striatal protein extracts were used as controls for endogenous Casp6 expression. e, f Western blot and densitometric analyzes of Casp6 (15 µg/lane) in cortical, striatal and hippocampal protein extracts from three independent KI/Cre mice. Results are represented as ratios of Casp6 over β-actin levels and are relative to Str. One-way ANOVA followed by Dunnett’s post-hoc test against striatum were performed. ***p < 0.001. g Western blot analysis of Tub∆Casp6 in 17 µg striatal protein extracts from 6–7 month old mice digested or not with recombinant human Casp6. h, i Representative micrograph of 6 KI/Cre and 5 KI/WT striatum (8 months of age), cortex and hippocampus (16–21 month old) tissue sections immunostained with anti-human Casp6 (h) or anti-Tub∆Casp6 (i)
Casp6 expressing mice do not display HD-like depressive behavior or locomotor impairments
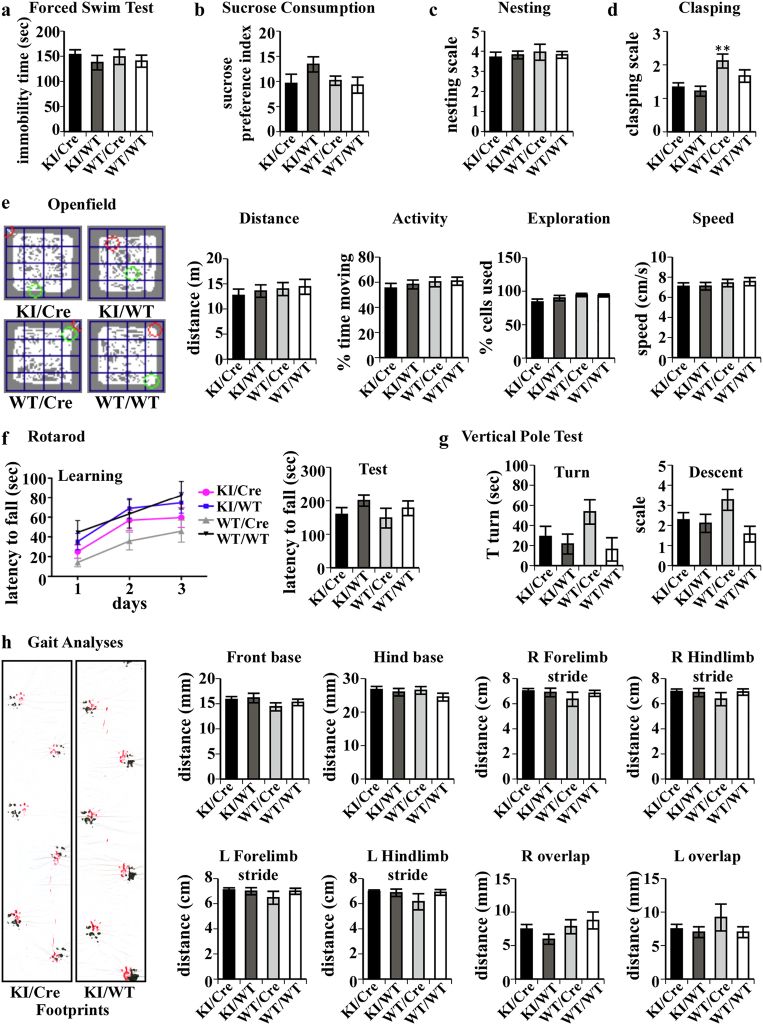
To evaluate if the ACK KI/Cre mouse develops HD-related features, psychiatric, and motor functions of aged mice were compared to those of control littermates. 18 to 21-months-old KI/Cre and control littermates spent equivalent time in an immobile state in the forced swim test indicating the absence of depressive behavior (Fig. 2a; p = 0.769). Evaluation of anhedonia, a major component of depression, by sucrose consumption, was normal in KI/Cre (Fig. 2b; p = 0.3020), indicating the absence of depressive behavior in the Casp6-expressing KI/Cre mice.
Fig. 2.
Assessment of HD-like depressive and locomotor behaviors of ACK mice. a Time in an immobile state during the forced swim test. Data represent the mean and SEM from 16 KI/Cre, 9 KI/WT, 8 WT/Cre and 9 WT/WT aged from 18–21 months. b Sucrose preference index of 10 KI/Cre, 9 KI/WT, 4 WT/Cre and 13 WT/WT aged from 16–20 months. c Average nesting score ±SEM of 25 KI/Cre, 6 KI/WT, 10 WT/Cre and 25 WT/WT aged from 16–22 months. d Average clasping score ±SEM measured by tail suspension in 33 KI/Cre, 21 KI/WT, 22 WT/Cre and 18 WT/WT aged from 15 to 19 months. Statistical evaluation was done with ANOVA followed by a Dunnett’s post-hoc analysis against KI/Cre. **: p < 0.01. e Openfield performances of mice assessed by evaluating the path, the total distance, % time in activity, % of cells used and average speed of 31 KI/Cre, 25 KI/WT, 17 WT/Cre, and 27 WT/WT aged from 12 to 18 months. The green circle is the start position and the red is the circle end position of the mouse. f Motor coordination and balance were evaluated using a rotarod test. The latencies during training and test were evaluated on 17 KI/Cre, 13 KI/WT, 11 WT/Cre and 10 WT/WT aged from 15 to 18 months. Locomotor learning was assessed by two-way ANOVA followed by a Bonferroni post-hoc test. g The time to turn downward and to completely descend the pole during the vertical pole test was evaluated on 17 KI/Cre, 13 KI/WT, 11 WT/Cre, and 10 WT/WT aged from 15 to 18 months. h Gait analysis on 16 KI/Cre, 10 KI/WT, 9 WT/Cre and 9 WT/WT aged from 18 to 21 months was performed with the footprint test. Red represents hind paws; black represents front paws
Gross motor dysfunctions were assessed by the nesting and clasping tests. Evaluation of the nest quality revealed that the aged mice made nests achieving normal scores [35] of approximately 3.8, independent of the genotype (Fig. 2c). Abnormal involuntary movements evaluated with the clasping test were significantly increased in WT/Cre mice compared to control littermates (Fig. 2d), confirming previous observations of Cre toxicity in the brain [36]. Nevertheless, no significant difference was observed in the clasping score of KI/Cre mice compared to control littermates, suggesting that human Casp6 expression reduces Cre-induced abnormal involuntary movements. Locomotor activity of aged KI/Cre mice (Fig. 2e), 5–7 month old, and 12–15 month old mice (Supplementary Fig. 3), analyzed by the openfield test distance traveled, percentage of time moving, percentage of cells used and average speed, was normal. Fine locomotor problems including motor coordination and balance were evaluated with a rotarod protocol (Fig. 2f) commonly used to characterize HD mouse models [31] and the vertical pole test (Fig. 2g). The time spent on the rotarod significantly increased over time for all four genotypes, indicating the ability to learn the task (Fig. 2f; days, F(2,94) = 46.82, p < 0.0001). The groups did not differ in overall level of performance or change across days (genotype, F(3,47) = 2.07; genotype x day, F(6,94) = 0.64, both p > 0.05). During the testing session, the KI/Cre mice spent the same amount of time on the accelerating rotarod as the control littermates (p = 0.3794). In the vertical pole test, the performances of the four genotypes were equivalent (Fig. 2g). The latency before mice turned downward (p = 0.1341) and completely descended the pole (p = 0.1341) did not differ significantly between groups. Lastly, gait abnormalities tested by the footprint test did not reveal walking pattern or gait differences in the KI/Cre mice when compared to control mice (Fig. 2h). Taken together, the results indicate that the presence of active human Casp6 in mouse striatum does not modify their overall activity, exploratory behavior, and motor function.
ACK Casp6-expressing KI/Cre mice display age-dependent cognitive deficits
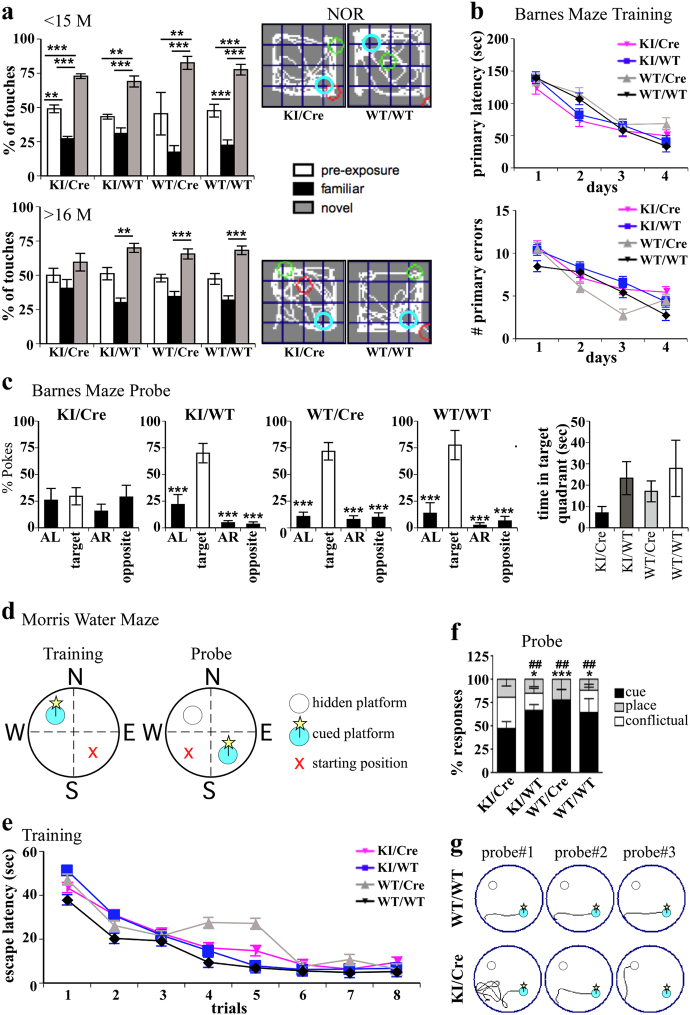
The KI/Cre mice were assessed for cognitive function since the human Casp6 was expressed and activated in the brain striatum, cortex and hippocampus, three regions involved in cognition. Adult (12–15 months of age) and old (16–19 months of age) mice were submitted to the novel object recognition (NOR) test to assess episodic memory function. The adult KI/Cre and control littermates significantly spent more time touching the novel object than the familiar one, indicating normal episodic memory (Fig. 3a). However, the old KI/Cre mice spent an equal amount of time touching the familiar and the novel objects, revealing memory recall impairment, whereas control littermates performed normally.
Fig. 3.
Assessment of ACK Casp6 mice cognitive function. a NOR analyzes of adult (<15 M, upper panel) or old (>16 M, lower panel) mice showing the percentage of touches of two identical objects during the familiarization phase (pre-exposure) or of a familiar and novel object during the test phase, two hours later (left panels) and a representative path of KI/Cre versus WT/WT mice (right panels). The light blue circle represents the position of the novel object during the test phase, green circle is the start position and the red circle is the end position of the mouse. Data represent the mean ± SEM of 13 KI/Cre, 10 KI/WT, 4 WT/Cre, and 15 WT/WT adult mice aged from range 12 to 15 months or 16 KI/Cre, 14 KI/WT, 11 WT/Cre, and 10 WT/WT old mice aged from 16 to 19 months. Statistical evaluations were done with one way ANOVA followed by a Tukey post-hoc analysis. **p < 0.01; ***p < 0.001. b Barnes maze training performance of 9 KI/Cre, 10 KI/WT, 9 WT/Cre, and 8 WT/WT aged from 14 to 18 months. Data show the mean ± SEM for the latency to reach the target hole (primary latency) and number of errors to the first encounter of the escape hole (primary errors) during training. Data were analyzed by repeated-measures ANOVAs, with genotype and days as factors. c Barnes maze probe test shows mean ± SEM of pokes in the five holes of the target quadrant, its adjacent left (AL), adjacent right (AR), or opposite quadrants during the probe test performed 24 h after the last training trial. Statistical evaluations were done with one way ANOVA followed by a Dunnett’s post-hoc analysis against the target quadrant. ***p < 0.001. (Right panel) Total time spent in the target quadrant during the probe test. d–f Performance of 12 KI/Cre, 11 KI/WT, 6 WT/Cre and 14 WT/WT mice aged from 15 to 20 months in the search strategy preference test of the Morris Water Maze. d Schematic representation of the position of the cued platform (blue circle with a yellow star), hidden platform (clear circle) and starting position (red cross) during the training and the probe tests. e Latency to reach the platform during training. Data represent the mean ± SEM of each genotype for each trial. f The histogram represents mean % ± SEM of cue (black bar), place (gray bar) and conflictual (white bar) responders. Statistical evaluations were done with two-way ANOVA followed by a Bonferroni post-hoc analysis. *p < 0.05; ***p < 0.001 for cue versus spatial. ##p < 0.01 for cue versus conflictual. g Representative swim paths during the probe test
The Barnes maze further characterized spatial memory function in the old KI/Cre mice (Fig. 3b, c). During the acquisition phase (Fig. 3b), the primary errors (days, F(3,93) = 8.42, p < 0.0001; genotypes x days, F(9,93) = 0.40, p = 0.9340) and the primary latency (primary latency: days, F(3,93) = 47.90, p < 0.0001; genotypes x days, F(9,93) = 1.12, p = 0.3553) to find the escape hole decreased significantly over time and to the same extent for the four genotypes. Short-term memory was assessed by a single probe, 24 h after the last trial (Fig. 3c). Contrary to the control mice, the number of pokes made by KI/Cre mice was not significantly higher in the target area compared to other areas. Although the KI/Cre mice spent a bit less time in the target quadrant than the control littermates, this measure did not reach statistical significance. These results indicate impaired spatial memory in the KI/Cre mice.
Mice performances in the search strategy preference test of the Morris water maze (Fig. 3d) were also evaluated to analyze the dynamic interplay between cortico-striatal and hippocampal memories [37]. All genotypes learned to find the platform during the training sessions (Fig. 3e) since the escape latency decreased significantly over time (days, F(7,273) = 49.82, p < 0.0001; genotypes x days, F(21,273) = 1.13, p = 0.3142). During the probe test (Fig. 3f, g), the KI/Cre used the cued (cortico-striato-related) or spatial (hippocampal-related) strategy equally, contrary to the control littermates who preferred the cued strategy. More conflictual responses were observed in the KI/Cre compared to the control mice, confirming that the dynamic interplay between the hippocampal-cortico-striatal systems was impaired in the Casp6 expressing mice.
Casp6 overexpression selectively increases the expression of synaptic plasticity-related genes in the hippocampus, but not the striatum, of ACK mice
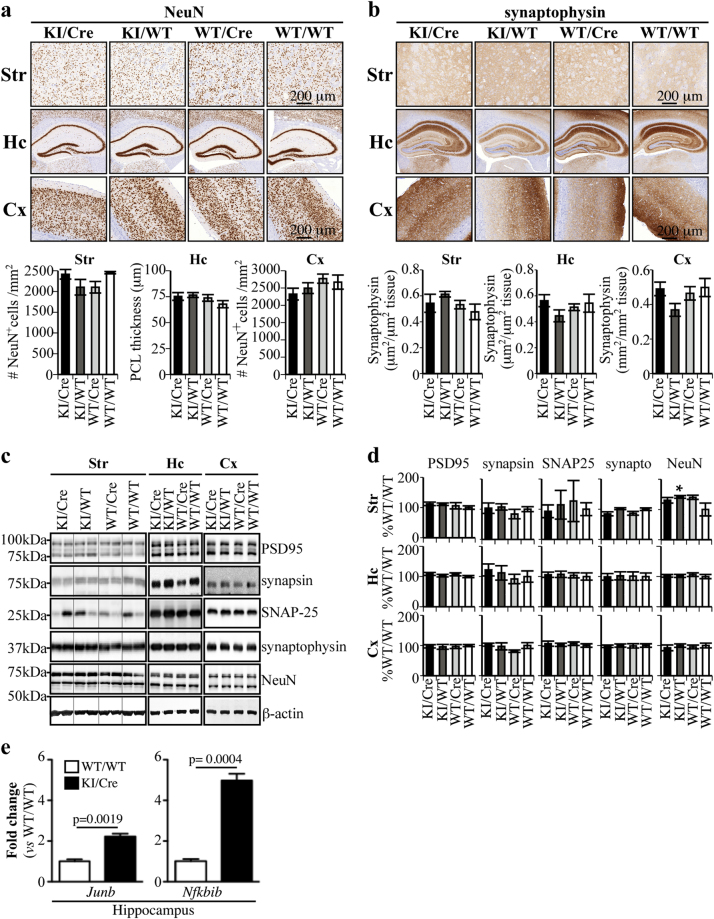
To investigate the mechanisms implicated in age-dependent Casp6-induced cognitive impairments, we evaluated the levels of the neuronal marker NeuN and synaptophysin staining in the striatum, hippocampus, and cortex of aged mice (Fig. 4a, b). Quantitative analyzes of the immunostaining revealed that the number of neurons and synaptophysin staining were equivalent across the four genotypes in the striatum, the hippocampus, and the cortex. By western blot analysis, NeuN, synaptophysin, PSD-95, synapsin, and SNAP-25 levels of KI/Cre mice were unchanged in the striatum, hippocampus, and cortex (Fig. 4c, d) compared to control littermates. mRNA profiling of 84 synaptic plasticity-related genes identified JunB and Nfkbib to be differentially expressed 2.2 and 5-fold, respectively, in the aged KI/Cre hippocampus (Fig. 4e). No differentially expressed genes were detected in the striatum. These results indicate greater susceptibility of hippocampal neurons to Casp6 than striatal neurons.
Fig. 4.
Differential synaptic profile mRNA expression in Casp6-expressing hippocampus, but not in striatum tissues. a, b Representative micrograph of 20–22-month-old KI/Cre, KI/WT, WT/Cre, or WT/WT hippocampus tissue sections immunostained with anti-NeuN (a) or anti-synaptophysin (b). Histograms represent the quantification of NeuN (a) and synaptophysin (b) immunostaining in the brain of 5 KI/Cre, 5 KI/WT, 5 WT/Cre, and 5 WT/WT mice. c, d Western blot and densitometric analyzes of synaptic and neuronal markers in the brains of aged (16–20 months) KI/Cre, KI/WT, WT/Cre, and WT/WT hippocampal protein extracts with antibodies against PSD-95, synapsin, SNAP-25, synaptophysin and NeuN. Quantitative results are represented as ratios of protein levels over β-actin and are arbitrarily expressed as percentage ± SEM of WT/WT. Statistical evaluations were done with ANOVA followed by a Dunnett’s post-hoc analysis against KI/Cre values. **p < 0.01, ***p < 0.001. e Differential synaptic plasticity profile mRNA expression of Junb and Nfkbib in KI/Cre versus WT/WT hippocampal tissues. No change was observed in striatum. Student’s t-test shows a significant difference between WT/WT and KI/Cre (n = 3 each)
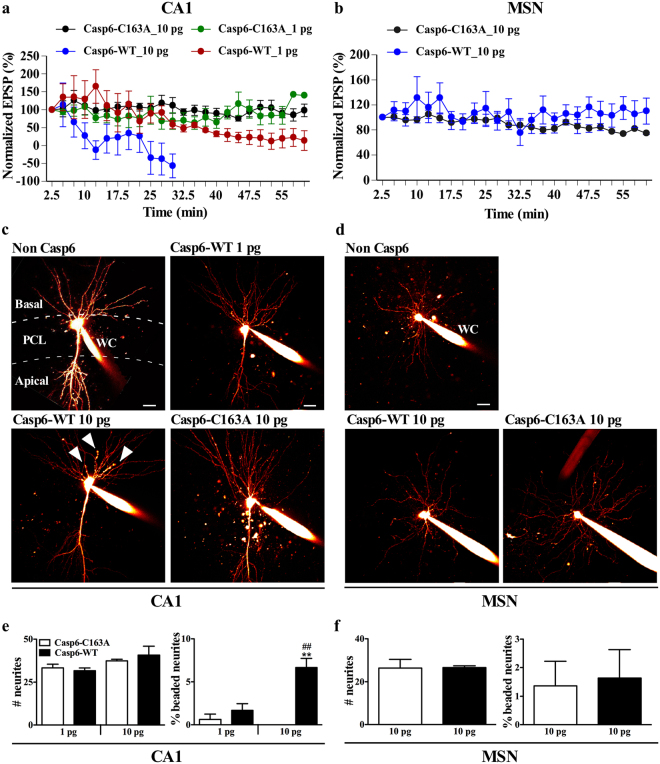
Casp6 impairs excitatory post-synaptic potential (EPSP) and induces neuritic degeneration in hippocampal CA1 pyramidal neurons but not in striatal MSN
To compare the effects of Casp6 on hippocampal CA1 pyramidal neuron and striatal MSN synaptic function, synaptic transmission in the neurons patched with recombinant active Casp6-WT or catalytically inactive Casp6-C163A was studied. The activity of Casp6 in the internal solution was stable for 2 h, and decreased to 80% between 2–5 h (Supplementary Fig. 4). Decreased EPSP amplitude in hippocampal CA1 pyramidal neurons occurred rapidly after patching 10 pg Casp6-WT, but not 10 pg Casp6-C163A (Fig. 5a). Decreased EPSP amplitude occurred 40 min after patching pyramidal neurons with 1 pg Casp6-WT. EPSP recordings in MSN patched with 10 pg of Casp6-WT or Casp6-C163A were stable for one hour (Fig. 5b). Moreover, CA1 pyramidal neurons exhibited dose–response sensitive neuronal degeneration evidenced by Alexa 594 positive axonal beading (Fig. 5c), whereas no obvious morphological changes were observed in MSNs with 10 pg of Casp6-WT (Fig. 5d). Quantitation showed a 6 fold increase in beaded neurites in the 10 pg Casp6 patched CA1 neurons (Fig. 5e) but no change in MSN (Fig. 5f). The total number of neurites per patched neuron were similar in Casp6-WT and Casp6-C163A patched CA1 neurons and MSN (Fig. 5e, f). These results reinforce the notion that CA1 pyramidal neurons are more vulnerable to active Casp6 than the MSN.
Fig. 5.
Effect of recombinant Casp6 on synaptic transmission and neurodegeneration in hippocampal and striatal neurons. a, b EPSP synaptic transmission of CA1 pyramidal neurons (a) and MSN (b) patched with 1 or 10 pg recombinant Casp6-WT or Casp6-C163A. Averaged EPSPs were expressed as the percentage of the first EPSP amplitude for each condition (CA1 pyramidal neurons: n = 5/condition; MSN: n = 5 for 10 pg Casp6-C163A and n = 6 for 10 pg Casp6-WT). c, d Representative flattened two-photon-imaging stack of hippocampal c and striatal d neurons patched by whole-cell recording electrode (WC) with Alexa 594. Neuritic degeneration in the basal side of pyramidal neurons with 10 pg Casp6-WT in hippocampal CA1 pyramidal cell layer (PCL) was indicated by white arrows. Scar bar = 50 µm. e, f Total number of neurites and percentage of beaded neurites of patched CA1 pyramidal neurons (e) and MSN (f). Statistical evaluations were done with one-way ANOVA followed by Bonferroni post-hoc test. **p < 0.01 vs. 10 pg Casp6-C163A; ## p < 0.01 vs. 1 pg Casp6-WT
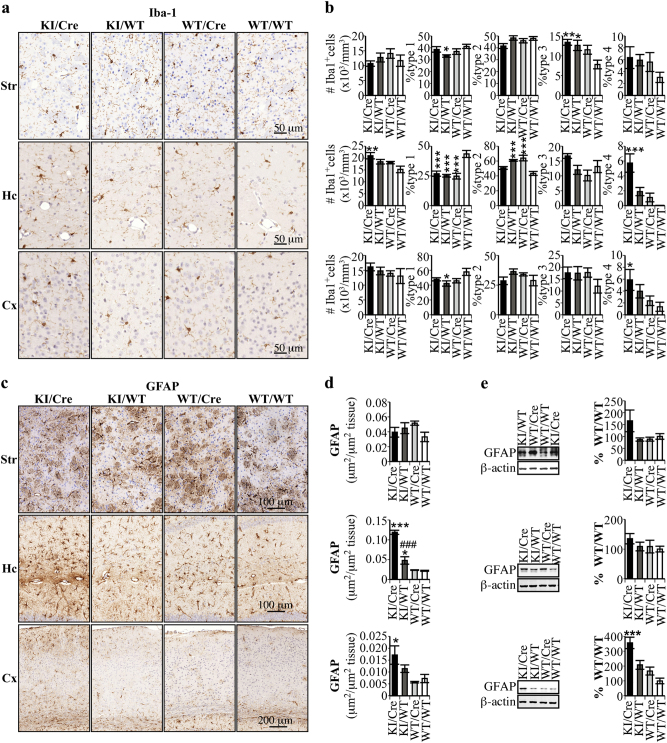
ACK KI/Cre mice display neuroinflammation in the hippocampus and cortex, but not in striatum
Analyzes of immunohistological striatal sections indicated that the total number of Iba-1 positive microglia was similar for all four genotypes (Fig. 6a, b). Analysis of type one (ramified), type two (shorter abundant ramifications), type three (amoeboid) and type four (enlarged) microglia [38] indicated that only the percentage of type three microglia was significantly increased in the KI/Cre and KI/WT striatum compared to the WT/WT. In the hippocampus stratum lacunosum molecular, the total number of Iba-1 positive microglia and the % of type four microglia were specifically increased in the KI/Cre compared to the WT/WT. No difference in the number but an increase % type four microglia was observed in the cortex. The area of GFAP astrocytic immunostaining was not increased in the KI/Cre striatum, but was increased in the hippocampal SLM and cortex, compared to the WT/WT (Fig. 6c, d). Western blotting confirmed the increase in GFAP in the cortex of KI/Cre mice (Fig. 6e). Together, these results indicate that active human Casp6 induces neuroinflammation specifically in the hippocampus and cortex, but not in the striatum, of aged ACK mice.
Fig. 6.
Evaluation of neuroinflammation in the striatum, hippocampus and cortex of old ACK mice. Immunohistochemical analysis of Iba-1 (a, b) and GFAP (c, d) staining of 19–21 months old KI/Cre, KI/WT, WT/Cre, or WT/WT striatum tissue sections (n = 5/genotype). a Representative micrograph of Iba-1 immunostaining. b Number of Iba-1 immunopositive cells and stereological quantification of type 1, type 2, type 3 and type 4 Iba-1 positive cells in the striatum, hippocampus and cortex sections of mice. Results are expressed as number of Iba-1 positive cells per volume of tissue or as the average percentage distribution ± SEM per genotype. c Representative micrograph of GFAP immunostaining. d Quantification of the area immunostained by GFAP. Results are expressed as means ± SEM. e Western blot analysis of striatum, hippocampus and cortex protein extracts of 5 KI/Cre, 3 KI/WT, 3 WT/Cre, and 3 WT/WT aged from 20 to 22 months with antibodies against GFAP and β-actin. The histogram represents densitometric measurements of the protein bands. Results are represented as ratios of GFAP over β-actin levels and are arbitrarily expressed as percentage ± SEM of WT/WT. Statistical evaluation were done with ANOVA followed by a Dunnett’s post-hoc analysis against KI/Cre values. *p < 0.05, **p < 0.01, ***p < 0.001
Casp6 immunopositive AD-related pathologies are present in the hippocampus of HD and AD brains, but not in the caudate nucleus
To determine the extent of Casp6 activation in HD brains, active Casp6, and Tau∆Casp6 immunostaining were compared in HD and AD brains (Supplementary Fig. 5). Strong anti-active Casp6 and anti-Tau∆Casp6 immunostaining were detected in pre-tangles and mature neurofibrillary tangles, and in neuropil threads of three Alzheimer and three stage 2 HD hippocampi, although less widespread in HD as expected (Supplementary Fig. 5a–c). By contrast, no immunoreactivity was detected in the striatal caudate nucleus, the main brain area pathologically affected in HD (Supplementary Fig. 5d). These results suggest that active Casp6 accumulation and damage is predominant in AD pathologies, but not in HD pathologies.
Discussion
Our data expose the differential CNS neuronal vulnerability to Casp6 expression and activation, show the inability of Casp6 to induce pathogenesis in striatum, and confirm the age-dependent memory impairment caused by Casp6.
Our results expose differential striatal and hippocampal neuronal vulnerability to Casp6. Indeed, 5.2-fold over-expression of active Casp6 in the transgenic striatum, a level equivalent to that found in Huntington disease striatum [24], did not alter striatal behavioral functions, synaptic protein gene expression profiles, nor induce inflammation and neurodegeneration. Furthermore, a 30-fold higher dose of active Casp6 did not induce neurodegeneration or synaptic transmission deficits in MSN. By contrast, 2.5-fold overexpression of Casp6 in the hippocampus was associated with episodic and spatial memory impairment, differential expression of two genes involved in synaptic plasticity, and increased inflammation. Additionally, CA1 pyramidal neurons exhibited dose-dependent Casp6-mediated synaptic transmission impairment and neurodegeneration. This differential neuronal vulnerability is surprising since Casp6 activation is associated with axonal degeneration in primary human cortical neurons [1], NGF-deprived mouse sensory neurons [2, 4], and ischemia and stroke-mediated cell death [5, 39]. The reason for this differential vulnerability is not clear at this time. The cleavage of specific protein substrates with more relevant functions in specific neuronal subtypes [40, 41] could explain increased vulnerability in hippocampal neurons. Protective factors could prevent Casp6-mediated neurodegeneration in MSN, although these are not inhibitors since Tub∆Casp6 is observed in striatal tissues. Alternatively, the vulnerability of hippocampal neurons to Casp6 may be related to inflammation, consistent with recent reports showing brain region-dependent diversity in the gene expression profile of isolated mouse brain microglia [42] and higher astrocyte density in the cortex and hippocampus compared to striatum [43].
Our results are consistent with the differential neuronal vulnerability of familial neurodegenerative diseases. Familial mutations in AD, HD, and Parkinson disease cause selective degeneration of neuronal subtypes, despite mutant proteins being expressed in most neurons. Prion protein mutations highlight the exquisite difference in neuronal vulnerability to a specific mutation [44]. In these, the differential vulnerability of neurons may be related to the activation of specific neurodegenerative pathways in the susceptible neurons. However, here, we placed the degeneration-inducing active Casp6 directly in the neurons and still, we observe selective neuronal vulnerability. Our results highlight the importance of identifying neuronal subtype specific mechanism of pathogenesis in each neurodegenerative disease.
Despite expression and activation of Casp6 in the MSN of the indirect striatopallidal pathway, no depressive or motor impairments, nor striatal neurodegeneration or inflammation, normally associated with HD, were observed. The inability of Casp6 to induce HD pathogenesis was unexpected given the strong evidence for the implication of Casp6 in HD. Casp6 expression and activation has been reported in human and mouse HD brains [23, 24, 28]. Truncation of mHtt protein cleaved by Casp6 at amino acid 586 (mHtt∆Casp6) is observed in HD mouse models and is toxic in cultured cells and in mice [25, 26]. Expression of the mHtt∆Casp6 N-terminal fragment induces Htt immunopositive nuclear and cytoplasmic inclusion bodies in cerebellum and striatal neurons, astrogliosis, brain atrophy, progressive movement disorder, and cognitive impairment accompanied by further proteolytic processing of mHtt∆Casp6, weight gain, and decreased lifespan in mice [29, 30, 34]. Failure of Casp6 to induce HD pathogenesis is thus likely due to the absence of mHtt. Indeed, WT Htt is cleaved by Casp6 but is not toxic, whereas mHtt∆Casp6 is detrimental [29, 30]. Our data agree with other evidence that disputes the role of Casp6 in HD pathogenesis. Neuronal population-specific expression analyzes reveal no difference in Casp6 mRNA levels between HD and normal brains [45], and the levels of Casp6 mRNA in the striatum, the cortex and mouse embryonic fibroblasts cells derived from YAC128 mice are similar to those of WT mice [33, 46, 47]. Furthermore, neurodegeneration, locomotor deficits and production of mHtt586 fragments are unaffected by Casp6 ablation in HD mice [33, 34, 48], suggesting that mHtt cleavage at D586 residue is not specific to Casp6 in vivo and that Casp6 activity is not required for the pathogenic aspects of mHtt. Moreover, other caspases generate mHtt cleaved at D586 [23, 33, 34], and shorter Casp2-generated N-terminal Htt fragments are significantly more toxic than Casp6-generated fragments in mice [49]. These and our results indicate that in the absence of mHtt, Casp6 is not detrimental to MSN and striatal tissues.
Lastly, as previously observed [10], spatial and episodic memory impairments caused by the expression of Casp6 in the ACK mouse occur only in aging, consistent with age-dependent human neurodegenerative diseases. However, in these cases, there is cell type-specific expression of mutant proteins that must initiate precise degenerative pathways, explaining the selective vulnerability. In our model, the active Casp6 would have been expected to cause age-independent neurodegeneration because it is expressed three weeks after birth [50]. Therefore, young neurons are likely protected by higher levels of neurotropic factors, healthier and more active microglia, less cumulated damaged molecules due to active proteasome and lysosomal systems, and healthier mitochondria to provide the required energy for neurotransmission [51]. This ACK mouse model provides a novel tool to study age-dependent neurodegeneration.
In conclusion, we demonstrate age and neuronal subtype-dependent Casp6 toxicity indicating that it is essential to carefully consider the right neuronal subtype when investigating underlying molecular mechanisms of human neurodegenerative diseases and establishing therapeutic targets. Specifically, “Omics” studies should strive to specifically study vulnerable neurons. Mutant gene expression in the right neuronal subtype should be achieved from its endogenous promoter in animal models [52]. Even if the neuropathology is not quite reproduced in the model, mechanisms of disease may be better reproduced and could provide more appropriate therapeutic targets for the development of efficient medications against human disease.
Materials and methods
Mice models
All animal procedures followed the Canadian Council on Animal Care guidelines and were approved by the McGill Animal care committee. Mice were bred and aged in the Goodman Cancer Research Centre Mouse Transgenic Facility at McGill University and transferred to the Lady Davis Institute for behavioral testing. Mice were housed in a temperature-controlled room at 22 °C and were kept on a 12/12 h light/dark cycle with ad libitum access to food and water. Casp6 Knock-Out (KO) mice were obtained from Jackson Laboratories (Bar Harbor, ME, USA). ACL mice expressing human active Casp6 in the CA1 region of the hippocampus were generated previously [10].
To express human active Casp6 in the MSN of the indirect striatopallidal pathway, knock-in mice conditionally expressing a self-activating form of human Casp6 [10] were crossed with the BAC-transgenic adenosine A2a receptor-Cre mouse line [Tg(Adora2a-cre)KG126Gsat, stock number # 034744-UCD, Mutant Mouse Resource and Research Center (MMRRC; Davis, California, USA)]. Resulting ACK mice were viable and fertile. Mice were monitored daily following the McGill University Animal Resources Center’s protocol. Because Casp6 expressed specifically in the CA1 of the mouse hippocampus causes age-related neurodegeneration in transgenic mice [10], phenotyping was conducted with aged mice of over 15 months of age and any aberrant phenotype re-assessed in 12–15 month-old mice.
Total RNA extraction and reverse-transcriptase polymerase chain reaction
Total RNA was isolated with Trizol (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. Five hundred ng of total RNA was treated with DNAse RQ1 (Promega, Madison, WI, USA) prior to cDNA synthesis with avian myeloblastosis reverse transcriptase (Roche, Mannheim, Germany). The following primers were used to amplify a 229 bp human Casp6 amplicon: 5′-CGATGTGCCAGTCATTCCTT-3′ and 5′-CTCTAAGGAGGAGCCATAT-3′, a 386 bp Cre recombinase (Cre) amplicon: 5′-TCTCAGGTACTGACGGTGGACCAGCTTGCATGATCTCC-3′ and 5′-GTGAAACAGCATTGCTGTCACTT-3′, and a 151 bp 18S amplicon: 5′-GTAACCCGTTGAACCCCAT-3′ and 5′-CCATCCAATCGGTAGTAGCG-3′. Amplifications were conducted for 27 (18S), 30 (human Casp6) and 35 (Cre) cycles of 95 °C for 30 s, 61.1 °C (human Casp6)/70 °C (Cre)/58 °C (18S) for 30 s, and 68 °C for 60 s, with a final 68 °C extension phase for 5 min.
Mouse synaptic plasticity RT2-profiler PCR array
The expression of 84 key genes related to synaptic plasticity were assessed by RT-qPCR with the Mouse Synaptic Plasticity RT2 Profiler PCR Array (PAMM-126Z, Qiagen), according to the manufacturer’s instructions. Data was normalized to the geometric mean of the housekeeping genes and analyzed by the comparative Ct-method (2−ΔCT). Genes with fold change >1.5 and p-value < 0.05 were considered as biologically and significantly altered.
Behavioral assessment
Mice were handled daily during 5 min for one week before testing. Because the human Casp6 transgene was inserted in the X chromosome Hprt locus, only males were used to avoid a lionization effects. The experimenter was blinded to the genotype. Mice performances were recorded using the HVS 2100 automated video tracking system (HSV Image, Hampton, UK) or recorded manually.
Psychiatric analyzes
Forced swim test
Mice were placed 6 min in a cylinder (25 cm tall, 19 cm wide) filled with room temperature water to a depth of 15 cm. Immobility was timed during the last 4 min of the session.
Sucrose consumption test
Individually housed animals were provided ad libitum access to two bottles of water for four days before replacing the water in one of the bottles with 2% sucrose/H2O. Liquid intake was estimated by weighing the bottles daily for an additional four days. The sucrose preference index was calculated as the ratio of 2% sucrose solution consumed over total liquid consumed normalized to kilogram body weight [31].
Sensory and motor analyzes
Nesting score
Mice were separated into individual cages containing one Nestlet (#NES3600, Ancare, Bellmore, NY, USA). The next day, the quality of the nests was scored using the defined 5-points nest-rating scale [35].
Clasping score
Mice were suspended by the tail at a height of 30 cm, for two 20 s intervals, and limb movements were evaluated with a rating scale [53].
Openfield
Mice were placed in an empty box (Stoelting Co, # 60111, Wood Dale, IL, USA) and were allowed to explore for 5 min, three days in a row. The HVS 2100 analysis software divided the field in 16 virtual zones (4 rows × 4 columns) and analyzed the total distance traveled (m), the percentage of time moving, and the number of entries in each zone. The average speed was estimated by dividing the total distance traveled per time moving.
Rotarod
Mice were given three daily trials of 2 min at a fixed speed (15 rpm) rotarod (IITC Life Science, Woodland Hills, CA, USA) during three consecutive days (nine trials total) with an inter-trial interval of 2 h. Mice falling from the rod were returned on the rod until a maximum of 10 falls/trial. The time to first fall and the total number of falls per trial were recorded. On the fourth day, mice were tested on a rod accelerating from 5 to 40 rpm over 300 s. Three trials were performed, with an inter-trial interval of 2 h. The mean latency to fall from the rod was recorded.
Vertical pole test
Mice were placed head upwards at the top of a vertical rough-surfaced pole (diameter: 20 mm; height: 95 cm) and the mouse ability to turn downwards and descend the pole measured. Mice were subjected to tree trials of 180 s maximum with an inter-trial interval of 2 h. The performance was scored as described (http://neurobehaviour.lunenfeld.ca/DEFAULT.ASP?page=Motor%20Functions).
Footprints
The hind and fore paws were coated with non-toxic paint of different color and mice were allowed to walk along a narrow corridor lined with white paper. Light was used at one end of the corridor to encourage the mouse to walk to the other end. Eight parameters were measured: the left and right foot overlap (distance between right/left hindlimb and forelimb), the right and left forelimb strides, the right and the left hindlimb strides, the front base (distance between the forelimbs) and the hind base (distance between the hindlimbs). Each parameter was evaluated on a set of four consecutive footprints.
Cognitive analyzes
Novel object recognition test
Twenty-four hours after the openfield assessment, mice were allowed to explore two identical objects for 5 min (pre-exposure), returned to their home cage and room for 2 h, and presented with the familiar and a novel object (#62020, Stoelting Co, Wood Dale, IL, USA). The position of the novel object was counterbalanced between the animals to avoid any bias related to a preference in the location of the new object and the use of potential founding spatial cues. The number of times touching each object was recorded.
Search strategy preference test of the Morris water maze
Morris water maze test was performed as previously described [37] with minor modifications, using a pool (150 cm diameter × 60 cm height, UgoBasile, Varese, Italy) filled with 21–22 °C water made opaque by adding white nontoxic paint (Tempera white #46 gouache, DemCo, Quebec, QC, CA), and platforms of 10 cm diameter. Mice were submitted to four trials per day for two consecutive days; 60 s each, followed by a series of three test trials, 24 h later. During the acquisition task, the platform was marked by a visible static colored cylinder suspended above the platform, and mice were released at a constant starting position. During the test trials, 24 h later, the cued platform was located in the opposite quadrant and a hidden platform was placed in the training quadrant. The mouse responses were analyzed for place response: the mouse swam directly to the original platform; cue-guided response: the mouse swam directly to the cued platform; conflictual response: the mouse first swam in a quadrant but escaped the water by using the platform located in the opposite quadrant.
Barnes maze
Barnes maze testing was performed as described [54]. Mice were given four trials per day, with an intertrial interval of 15 min, for four consecutive days. Twenty-four hours after the last training trial, the mice were submitted to a single probe test. Mouse movements were continuously tracked and the latency to reach the target hole (primary latency), the number of errors to the first encounter of the escape hole (primary errors), the total error and the latency to enter the escape hole, and the number of nose pokes in each hole were recorded and analyzed.
Tissue preparation
Mice were sacrificed and tissues were processed as previously described [55]. Frozen tissues were homogenized in five volume of radioimmunoprecipitation assay buffer [(50 mM Tris–HCl, pH 7.4, 1% NP-40, 150 mM NaCl, 0.25% Na-deoxycholate, 1 mM EDTA, 1 mM Na3VO4, 1 mM NaF, 1 mM PMSF, 10 µL/mL of Proteases Inhibitors Cocktail (P8340, Sigma–Aldrich, St. Louis, MO, USA)] with a mechanical homogenizer (TH, Omni International, Marietta, GA), and centrifuged for 20 min at 15,000 rpm at 4 °C. The supernatants were stored at −80 °C until processing.
In vitro digestion of brain extracts by recombinant active Casp6
Proteins were precipitated with acetone (1:4 ratio, −20 °C overnight) [56]. The pellets were resuspended in Stennicke’s buffer (SB) (20 mM piperazine-N, N-bis (2-ethanesulfonic acid (BioShop Canada Inc., Burlington, Ontario, CA) pH 7.2, 30 mM NaCl, 1 mM EDTA, 0.1% CHAPS, 10% sucrose) [57]. The reaction mix consisted of 400 ng recombinant active Casp6, prepared from the pET23b cDNA construct [58], 200 μg brain protein extract, SB and freshly added 10 μM DTT and was incubated 4 h at 37 °C [40].
Western blots analyzes
Samples were prepared in Laemli buffer, as described [55]. Five to twenty microgram of proteins were separated on SDS-PAGE gels and blotted onto PVDF membranes. Membranes were probed with primary antibodies diluted in SuperBlock® blocking buffer (Thermoscientific, Rockford, USA): 1/10000 GN20622 anti-Tubulin∆Casp6 [40], 1/1000 2125 anti-α-Tubulin (Cell Signaling, Danvers, MA, USA), 1/5000 SAB4200544 anti-synaptophysin (Sigma-Aldrich, Oakville, ON, CA), 1/5000 AB1543 anti-synapsin (Cell Signaling Tech. Danvers, MA, USA), 1/1000 SMI-81R anti-SNAP-25 (Biolegend, San Diego, CA, USA), 1/1000 MAB377 anti-NeuN (Millipore, Etobicook, ON, CA), 1/5000 73448 anti-PSD-95 (UC Davis/NIH NeuroMab Facility), 1/10000 MAB374 anti-GAPDH (Millipore), 1/3000 G3893 anti-GFAP (Sigma-Aldrich), 1/5000 10630 anti-p20 Casp6 [7], 1/1000 9762 anti-Casp6 (Cell Signaling Tech.), 1/5000 A5441 anti-β-actin (Sigma-Aldrich). Immunoreactivity was detected with horseradish peroxidase-linked secondary antibodies (Jackson Immunoresearch Laboratories, West Grove, PA, USA for anti-mouse; and Dako Cytomation Pascal, Burlington, Ontario, CA for anti-rabbit) diluted in 5% nonfat dry milk in PBS-T. Membranes were revealed by enhanced chemiluminescence (ECL Prime, GE Healthcare Biosciences, Piscataway, NJ) in a Fujifilm LAS4000 imaging system (Fujifilm USA, Valhalla, NY) or by scanning the X-ray film on the ChemiGenius apparatus (Syngene, Frederik, MD, USA) using a lower light setting in transmission mode. The β-actin immunoreactivity was detected with anti-mouse secondary antibody conjugated with alkaline phosphatase (Jackson ImmunoResearch Laboratories). Densitometric analyzes were performed with Image Gauge analysis software 3.0. (Fujifilm USA).
Immunohistochemistry
Mice were anesthetized with isoflurane, transcardially perfused with saline and 4% paraformaldehyde. The dissected brains were postfixed for 24 h in 10% neutral-buffered formalin (Thermo Fisher Scientific, Ottawa, ON, CA), embedded in paraffin blocks, and 4 μm sections cut at the IRIC histology core facility (Université de Montréal, Montreal, CA). Postmortem human brain tissues were obtained from the Douglas-Bell Canada Brain Bank (Douglas Mental Health University Institute, Montreal, Quebec, Canada). Eight micron sections of formalin-fixed, paraffin-embedded brains from three HD patients (Vonsattel grade II; case #325: female, 60 years; case # 1723: male, 66 years; case #1020: female, 75 years) and three aged-matched non-neurodegenerative controls (case #770: female, 77 years; case # 1670: female, 51 years; case # 1722: male, 59 years) and one AD patient (case #725: female, 75 years) were used. Paraformaldehyde-fixed, paraffin-embedded 4 μm thick tissue sections of two Alzheimer cases from the Religious Orders Study (case #10101327, male 89 years and case #20965598: female, 90 years) were kindly provided by Dr D.A. Bennett (Rush Medical School, Chicago, Il). Immunohistological staining were performed with an automated Dako immunostainer (Burlington, ON, CA) as described [9], with the following primary antibodies: 1/8000 Z0334 anti-GFAP (Dako), 1/2000 019-19741 anti-Iba-1 (Wako, Richmond, VA, USA), 1/5000 LSB477 anti-human Casp6 (Lifespan Bioscience, Seattle, WA, USA), 1/2000 MAB377 anti-NeuN (Millipore), 1/8000 SAB4200544 anti-synaptophysin (Sigma-Aldrich), 1/5000 GN20622 anti-Tubulin cleaved by Casp6 (Tub∆Casp6) [40] and 1/25000 anti-Tau cleaved by Casp6 (Tau∆Casp6) [7]. Immunostaining was revealed with anti-rabbit-HRP or -mouse-HRP antibodies and diaminobenzidine (Dako, Burlington, ON, CA). The tissue sections were mounted in Permount mounting medium (Fisher Scientific, Ottawa, ON, CA) and digitally scanned with the MIRAX SCAN scanner (Zeiss, Don Mills, ON, CA). The area of GFAP and synaptophysin staining was quantified with Image J software (NIH, Bethesda, MD, USA) and expressed as μm2 staining/μm2 of tissue. NeuN staining was analyzed by quantifying the number of NeuN positive nuclei with the Image J software. Two parameters were used to analyze Iba-1 staining: quantification of total positive cell/mm3 tissue and morphological characterization of Iba-1+ cells according to scoring schemes described [38]. An observer blinded to genotypes did quantifications.
Preparation of recombinant Casp6 for electrophysiological experiments
Casp6 activity assays were performed with 5–500 nM recombinant active Casp6 (Casp6-WT) and 100 mM Ac-VEID-AFC in Stennick buffer as described [11]. Recombinant Casp6 were active site titrated using z-VAD-fmk. The ratio between reaction velocity with inhibitor over reaction velocity without inhibitor was plotted against the inhibitor concentration, and the concentration for y = 0 was determined as the active site concentration of the Casp6.
Titrated recombinant Casp6-WT was pre-activated in Stennick buffer for 15 min at 37 °C, then diluted at 1:1000 in internal solution for cell patching. The catalytically inactive Casp6-C163A, purified and prepared as Casp6-WT, was used as a negative control.
Electrophysiology
Hippocampal whole-cell recordings were performed on 300 µm transverse hippocampal slices from 10 to 14-day-old C57BL/6 mice prepared with artificial cerebrospinal fluid (ACSF) (125 mM NaCl, 2.5 mM KCl, 1.25 mM NaH2PO4, 26 mM NaHCO3, 1 mM MgCl2, 2 mM CaCl2, 45 mM glucose, ~338 mOsm) as described [59]. The patch pipettes were filled with internal solution (125 mM KCl, 2.5 mM K gluconate, 10 mM HEPES, 4 mM MgATP, 0.3 mM NaGTP, and 10 mM Na-phosphocreatine, pH 7.3–7.4 adjusted by KOH, ~310 mOsm adjusted with sucrose) with 1–10 pg/10 µL recombinant Casp6-WT or Casp6-C163A and 0.05 mM Alexa 594. Cell-attached whole-cell current-clamp recordings were performed using standard techniques [59]. CA1 pyramidal neurons with large pyramidal soma and thick apical dendrite were identified using Dodt contrast and confirmed by morphology as visualized by two-photon microscopy of Alexa 594 fluorescence, their resting membrane potential and action potential properties. The stimulation electrode was placed in the CA1 stratum oriens and neurons were given five biphasic pulses of 25–50 V every 30 s to evoke EPSPs.
Striatal whole-cell recording was performed on 300 µm horizontal brain slices from 10–14 day old mice. Striatal MSNs were identified post hoc by their morphology, resting membrane potential, and action potential properties, and input-output curve. To obtain the EPSPs in striatal MSN, the dorsal MSN near the sensory-motor cortex was patched and the cortico-striatal afferents were stimulated by placing the stimulation electrode in layer 5 of the sensory-motor cortex. The stimulation pattern was given as described for CA1 neurons.
Each neuron used in the data was required to have stable membrane potential (±4 mV), input resistance (±15%) and temperature (31–33 °C) throughout the recordings. Analysis of EPSPs was performed using custom software running with Igor Pro (WaveMetrics Inc., Lake Oswego, OR, USA). Peak amplitudes of EPSPs were measured, and averaged every 2.5 min (5 traces). All averaged EPSPs were normalized to the first 2.5 min EPSP in each neuronal recording.
Two-photon imaging
Two-photon excitation was achieved using a Chameleon XR (Coherent, Santa Clara, CA, USA) at 820 nm for Alexa 594. Two-photon microscopes were custom-built as described [60]. Imaging data were acquired using Matlab (The MathWorks, Natick, MA, USA) via PCI-6110 boards (National Instruments). After recording, images of neurons were taken from two-photon laser scanning microscope image stacks. The whole visible neuron was scanned at 2 ms/line and 512 × 512 pixels and three frames were averaged for each optical section. Flattened two-photon-imaging stack was used for morphological identification and quantification.
Statistical analyzes
Data are expressed as means ± SEM. Statistical analyzes were performed with one-way analysis of variance (one-way ANOVA) followed by the Dunnett’s post hoc test. For repeated measures, the two-way ANOVA with genotype status as one factor and time for the other, followed by the Bonferroni post hoc test was used. Statistical calculations were performed using GraphPad Prism 5.0 software (GraphPad, La Jolla, CA, USA). Results were considered significantly different at p < 0.05.
Electronic supplementary material
Acknowledgements
We would like to thank the animal quarters staff, especially Véronique Michaud, for maintaining the mice, Dr Joseph Flores for mice perfusions, the IRIC Université de Montréal pathology core for embedding and cutting brain tissue sections, Dr Agne Tubeleviciute-Aydin for the preparation of recombinant Casp6, and Dr Yves De Koninck, Dr Sonia Goulet and Dr François Y Doré (Université Laval, QC) for lending us the rotarod apparatus. The Douglas – Bell Canada Brain Bank is supported by the Quebec Suicide Research Network of the Fonds de Recherche du Québec - Santé (FRQS) and by the Douglas Institute Foundation. We are very grateful to Dr Steffen Albrecht (McGill University, Montreal, QC) for help with the interpretation of the human immunostaining in HD brains, and to Dr David A. Bennett (Rush Medical School, Chicago, Il) for the AD tissues. We thank Christina You Chien Chou, Jennifer Brock, and Elvis Cela for help and guidance with electrophysiology and 2-photon imaging. Anastasia Noël received a postdoctoral scholarship from the Alzheimer Society of Canada. Libin Zhou is a recipient of an Alzheimer Society of Canada Dr and Mrs Spatz doctoral award. This work was supported by funds from the Canadian Institutes of Health Research (CIHR) MOP-243413-BCA-CGAG-45097, and the JGH Foundation to ALB. P.J.S. was funded by CIHR OG 126137, CIHR NIA 288936, NSERC DG 2017-04730, NSERC DAS 2017-507818.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by J.M. Hardwick
Electronic supplementary material
The online version of this article (10.1038/s41418-017-0043-x) contains supplementary material, which is available to authorized users.
References
- 1.Sivananthan SN, Lee AW, Goodyer CG, LeBlanc AC. Familial amyloid precursor protein mutants cause caspase-6-dependent but amyloid beta-peptide-independent neuronal degeneration in primary human neuron cultures. Cell Death Dis. 2010;1:e100. doi: 10.1038/cddis.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nikolaev A, McLaughlin T, O’Leary DD, Tessier-Lavigne M. APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature. 2009;457:981–9. doi: 10.1038/nature07767. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.Uribe V, Wong BK, Graham RK, Cusack CL, Skotte NH, Pouladi MA, et al. Rescue from excitotoxicity and axonal degeneration accompanied by age-dependent behavioral and neuroanatomical alterations in caspase-6-deficient mice. Hum Mol Genet. 2012;21:1954–67. doi: 10.1093/hmg/dds005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cusack CL, Swahari V, Hampton Henley W, Michael Ramsey J, Deshmukh M. Distinct pathways mediate axon degeneration during apoptosis and axon-specific pruning. Nat Comm. 2013;4:1876. doi: 10.1038/ncomms2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akpan N, Serrano-Saiz E, Zacharia BE, Otten ML, Ducruet AF, Snipas SJ, et al. Intranasal delivery of caspase-9 inhibitor reduces caspase-6-dependent axon/neuron loss and improves neurological function after stroke. J Neurosci. 2011;31:8894–904. doi: 10.1523/JNEUROSCI.0698-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sokolowski JD, Gamage KK, Heffron DS, LeBlanc AC, Deppmann CD, Mandell JW. Caspase-mediated cleavage of actin and tubulin is a common feature and sensitive marker of axonal degeneration in neural development and injury. Acta Neuropathol Commun. 2014;2:16. doi: 10.1186/2051-5960-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo H, Albrecht S, Bourdeau M, Petzke T, Bergeron C, LeBlanc AC. Active caspase-6 and caspase-6-cleaved tau in neuropil threads, neuritic plaquwill talk like that in hindies, and neurofibrillary tangles of Alzheimer’s disease. Am J Pathol. 2004;165:523–31. doi: 10.1016/S0002-9440(10)63317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Albrecht S, Bourdeau M, Bennett D, Mufson EJ, Bhattacharjee M, LeBlanc AC. Activation of caspase-6 in aging and mild cognitive impairment. Am J Pathol. 2007;170:1200–9. doi: 10.2353/ajpath.2007.060974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramcharitar J, Afonso VM, Albrecht S, Bennett DA, LeBlanc AC. Caspase-6 activity predicts lower episodic memory ability in aged individuals. Neurobiol Aging. 2013;34:1815–24. doi: 10.1016/j.neurobiolaging.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.LeBlanc AC, Ramcharitar J, Afonso V, Hamel E, Bennett DA, Pakavathkumar P, et al. Caspase-6 activity in the CA1 region of the hippocampus induces age-dependent memory impairment. Cell Death Differ. 2014;21:696–706. doi: 10.1038/cdd.2013.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klaiman G, Champagne N, LeBlanc AC. Self-activation of caspase-6 in vitro and in vivo: caspase-6 activation does not induce cell death in HEK293T cells. Biochim Biophys Acta. 2009;1793:592–601. doi: 10.1016/j.bbamcr.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 12.Gray DC, Mahrus S, Wells JA. Activation of specific apoptotic caspases with an engineered small-molecule-activated protease. Cell. 2010;142:637–46. doi: 10.1016/j.cell.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hogg MC, Mitchem MR, Konig HG, Prehn JH. Caspase 6 has a protective role in SOD1(G93A) transgenic mice. Biochim Biophys Acta. 2016;1862:1063–73. doi: 10.1016/j.bbadis.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 14.Saudou F, Humbert S. The biology of Huntingtin. Neuron. 2016;89:910–26. doi: 10.1016/j.neuron.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 15.Sieradzan KA, Mechan AO, Jones L, Wanker EE, Nukina N, Mann DM. Huntington’s disease intranuclear inclusions contain truncated, ubiquitinated huntingtin protein. Exp Neurol. 1999;156:92–99. doi: 10.1006/exnr.1998.7005. [DOI] [PubMed] [Google Scholar]
- 16.Reiner A, Albin RL, Anderson KD, D’Amato CJ, Penney JB, Young AB. Differential loss of striatal projection neurons in Huntington disease. Proc Natl Acad Sci USA. 1988;85:5733–7. doi: 10.1073/pnas.85.15.5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vonsattel JP, Myers RH, Stevens TJ, Ferrante RJ, Bird ED, Richardson EP., Jr. Neuropathological classification of Huntington’s disease. J Neuropathol Exp Neurol. 1985;44:559–77. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- 18.Hedreen JC, Peyser CE, Folstein SE, Ross CA. Neuronal loss in layers V and VI of cerebral cortex in Huntington’s disease. Neurosci Lett. 1991;133:257–61. doi: 10.1016/0304-3940(91)90583-F. [DOI] [PubMed] [Google Scholar]
- 19.Spargo E, Everall IP, Lantos PL. Neuronal loss in the hippocampus in Huntington’s disease: a comparison with HIV infection. J Neurol Neurosurg Psychiatry. 1993;56:487–91. doi: 10.1136/jnnp.56.5.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maat-Schieman ML, Dorsman JC, Smoor MA, Siesling S, Van Duinen SG, Verschuuren JJ, et al. Distribution of inclusions in neuronal nuclei and dystrophic neurites in Huntington disease brain. J Neuropathol Exp Neurol. 1999;58:129–37. doi: 10.1097/00005072-199902000-00003. [DOI] [PubMed] [Google Scholar]
- 21.Nance MA. Huntington disease: clinical, genetic, and social aspects. J Geriatr Psychiatry Neurol. 1998;11:61–70. doi: 10.1177/089198879801100204. [DOI] [PubMed] [Google Scholar]
- 22.Hodges A, Strand AD, Aragaki AK, Kuhn A, Sengstag T, Hughes G, et al. Regional and cellular gene expression changes in human Huntington’s disease brain. Hum Mol Genet. 2006;15:965–77. doi: 10.1093/hmg/ddl013. [DOI] [PubMed] [Google Scholar]
- 23.Hermel E, Gafni J, Propp SS, Leavitt BR, Wellington CL, Young JE, et al. Specific caspase interactions and amplification are involved in selective neuronal vulnerability in Huntington’s disease. Cell Death Differ. 2004;11:424–38. doi: 10.1038/sj.cdd.4401358. [DOI] [PubMed] [Google Scholar]
- 24.Graham RK, Deng Y, Carroll J, Vaid K, Cowan C, Pouladi MA, et al. Cleavage at the 586 amino acid caspase-6 site in mutant huntingtin influences caspase-6 activation in vivo. J Neurosci. 2010;30:15019–29. doi: 10.1523/JNEUROSCI.2071-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wellington CL, Singaraja R, Ellerby L, Savill J, Roy S, Leavitt B, et al. Inhibiting caspase cleavage of huntingtin reduces toxicity and aggregate formation in neuronal and nonneuronal cells. J Biol Chem. 2000;275:19831–8. doi: 10.1074/jbc.M001475200. [DOI] [PubMed] [Google Scholar]
- 26.Graham RK, Deng Y, Slow EJ, Haigh B, Bissada N, Lu G, et al. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006;125:1179–91. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 27.Landles C, Sathasivam K, Weiss A, Woodman B, Moffitt H, Finkbeiner S, et al. Proteolysis of mutant huntingtin produces an exon 1 fragment that accumulates as an aggregated protein in neuronal nuclei in Huntington disease. J Biol Chem. 2010;285:8808–23. doi: 10.1074/jbc.M109.075028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warby SC, Doty CN, Graham RK, Carroll JB, Yang YZ, Singaraja RR, et al. Activated caspase-6 and caspase-6-cleaved fragments of huntingtin specifically colocalize in the nucleus. Hum Mol Genet. 2008;17:2390–404. doi: 10.1093/hmg/ddn139. [DOI] [PubMed] [Google Scholar]
- 29.Tebbenkamp AT, Green C, Xu G, Denovan-Wright EM, Rising AC, Fromholt SE, et al. Transgenic mice expressing caspase-6-derived N-terminal fragments of mutant huntingtin develop neurologic abnormalities with predominant cytoplasmic inclusion pathology composed largely of a smaller proteolytic derivative. Hum Mol Genet. 2011;20:2770–82. doi: 10.1093/hmg/ddr176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waldron-Roby E, Ratovitski T, Wang X, Jiang M, Watkin E, Arbez N, et al. Transgenic mouse model expressing the caspase 6 fragment of mutant huntingtin. J Neurosci. 2012;32:183–93. doi: 10.1523/JNEUROSCI.1305-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pouladi MA, Graham RK, Karasinska JM, Xie Y, Santos RD, Petersen A, et al. Prevention of depressive behaviour in the YAC128 mouse model of Huntington disease by mutation at residue 586 of huntingtin. Brain. 2009;132:919–32. doi: 10.1093/brain/awp006. [DOI] [PubMed] [Google Scholar]
- 32.Aharony I, Ehrnhoefer DE, Shruster A, Qiu X, Franciosi S, Hayden MR, et al. A Huntingtin-based peptide inhibitor of caspase-6 provides protection from mutant Huntingtin-induced motor and behavioral deficits. Hum Mol Genet. 2015;24:2604–14. doi: 10.1093/hmg/ddv023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gafni J, Papanikolaou T, Degiacomo F, Holcomb J, Chen S, Menalled L, et al. Caspase-6 activity in a BACHD mouse modulates steady-state levels of mutant huntingtin protein but is not necessary for production of a 586 amino acid proteolytic fragment. J Neurosci. 2012;32:7454–65. doi: 10.1523/JNEUROSCI.6379-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong BK, Ehrnhoefer DE, Graham RK, Martin DD, Ladha S, Uribe V, et al. Partial rescue of some features of Huntington Disease in the genetic absence of caspase-6 in YAC128 mice. Neurobiol Dis. 2015;76:24–36. doi: 10.1016/j.nbd.2014.12.030. [DOI] [PubMed] [Google Scholar]
- 35.Deacon RM. Assessing nest building in mice. Nat Protoc. 2006;1:1117–9. doi: 10.1038/nprot.2006.170. [DOI] [PubMed] [Google Scholar]
- 36.Forni PE, Scuoppo C, Imayoshi I, Taulli R, Dastru W, Sala V, et al. High levels of Cre expression in neuronal progenitors cause defects in brain development leading to microencephaly and hydrocephaly. J Neurosci. 2006;26:9593–602. doi: 10.1523/JNEUROSCI.2815-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dagnas M, Guillou JL, Prevot T, Mons N. HDAC inhibition facilitates the switch between memory systems in young but not aged mice. J Neurosci. 2013;33:1954–63. doi: 10.1523/JNEUROSCI.3453-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahmed Z, Sheng H, Xu YF, Lin WL, Innes AE, Gass J, et al. Accelerated lipofuscinosis and ubiquitination in granulin knockout mice suggest a role for progranulin in successful aging. Am J Pathol. 2010;177:311–24. doi: 10.2353/ajpath.2010.090915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shabanzadeh AP, D’Onofrio PM, Monnier PP, Koeberle PD. Targeting caspase-6 and caspase-8 to promote neuronal survival following ischemic stroke. Cell Death Dis. 2015;6:e1967. doi: 10.1038/cddis.2015.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klaiman G, Petzke TL, Hammond J, LeBlanc AC. Targets of caspase-6 activity in human neurons and Alzheimer disease. Mol Cell Proteom. 2008;7:1541–55. doi: 10.1074/mcp.M800007-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cho JH, Lee PY, Son WC, Chi SW, Park BC, Kim JH, et al. Identification of the novel substrates for caspase-6 in apoptosis using proteomic approaches. BMB Rep. 2013;46:588–93. doi: 10.5483/BMBRep.2013.46.12.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grabert K, Michoel T, Karavolos MH, Clohisey S, Baillie JK, Stevens MP, et al. Microglial brain region-dependent diversity and selective regional sensitivities to aging. Nat Neurosci. 2016;19:504–16. doi: 10.1038/nn.4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pickrell AM, Fukui H, Wang X, Pinto M, Moraes CT. The striatum is highly susceptible to mitochondrial oxidative phosphorylation dysfunctions. J Neurosci. 2011;31:9895–904. doi: 10.1523/JNEUROSCI.6223-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jackson WS. Selective vulnerability to neurodegenerative disease: the curious case of prion protein. Dis Model Mech. 2014;7:21–29. doi: 10.1242/dmm.012146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuhn A, Thu D, Waldvogel HJ, Faull RL, Luthi-Carter R. Population-specific expression analysis (PSEA) reveals molecular changes in diseased brain. Nat Methods. 2011;8:945–7. doi: 10.1038/nmeth.1710. [DOI] [PubMed] [Google Scholar]
- 46.Ehrnhoefer DE, Skotte NH, Ladha S, Nguyen YT, Qiu X, Deng Y, et al. p53 increases caspase-6 expression and activation in muscle tissue expressing mutant huntingtin. Hum Mol Genet. 2014;23:717–29. doi: 10.1093/hmg/ddt458. [DOI] [PubMed] [Google Scholar]
- 47.Ehrnhoefer DE, Caron NS, Deng Y, Qiu X, Tsang M, Hayden MR. Laquinimod decreases Bax expression and reduces caspase-6 activation in neurons. Exp Neurol. 2016;283:121–8. doi: 10.1016/j.expneurol.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 48.Landles C, Weiss A, Franklin S, Howland D, Bates G. Caspase-6 does not contribute to the proteolysis of mutant huntingtin in the HdhQ150 knock-in mouse model of Huntington’s disease. PLoS Curr. 2012;4:e4fd085bfc9973. doi: 10.1371/4fd085bfc9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O’Brien R, DeGiacomo F, Holcomb J, Bonner A, Ring KL, Zhang N, et al. Integration-independent transgenic Huntington. J Biol Chem. 2015;290:19287–306. doi: 10.1074/jbc.M114.623561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsien JZ, Chen DF, Gerber D, Tom C, Mercer EH, Anderson DJ, et al. Subregion- and cell type-restricted gene knockout in mouse brain. Cell. 1996;87:1317–26. doi: 10.1016/S0092-8674(00)81826-7. [DOI] [PubMed] [Google Scholar]
- 51.Mattson MP, Magnus T. Ageing and neuronal vulnerability. Nat Rev Neurosci. 2006;7:278–94. doi: 10.1038/nrn1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saito T, Matsuba Y, Mihira N, Takano J, Nilsson P, Itohara S, et al. Single App knock-in mouse models of Alzheimer’s disease. Nat Neurosci. 2014;17:661–3. doi: 10.1038/nn.3697. [DOI] [PubMed] [Google Scholar]
- 53.Samadi P, Boutet A, Rymar VV, Rawal K, Maheux J, Kvann JC, et al. Relationship between BDNF expression in major striatal afferents, striatum morphology and motor behavior in the R6/2 mouse model of Huntington’s disease. Genes Brain Behav. 2013;12:108–24. doi: 10.1111/j.1601-183X.2012.00858.x. [DOI] [PubMed] [Google Scholar]
- 54.Whittington RA, Virag L, Gratuze M, Petry FR, Noel A, Poitras I, et al. Dexmedetomidine increases tau phosphorylation under normothermic conditions in vivo and in vitro. Neurobiol Aging. 2015;36:2414–28. doi: 10.1016/j.neurobiolaging.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petry FR, Pelletier J, Bretteville A, Morin F, Calon F, Hebert SS, et al. Specificity of anti-tau antibodies when analyzing mice models of Alzheimer’s disease: problems and solutions. PLoS One. 2014;9:e94251. doi: 10.1371/journal.pone.0094251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Botelho D, Wall MJ, Vieira DB, Fitzsimmons S, Liu F, Doucette A. Top-down and bottom-up proteomics of SDS-containing solutions following mass-based separation. J Proteome Res. 2010;9:2863–70. doi: 10.1021/pr900949p. [DOI] [PubMed] [Google Scholar]
- 57.Stennicke HR, Salvesen GS. Biochemical characteristics of caspases-3, -6, -7, and -8. J Biol Chem. 1997;272:25719–23. doi: 10.1074/jbc.272.41.25719. [DOI] [PubMed] [Google Scholar]
- 58.Denault J, Salvesen G. Expression, purification, and characterization of caspases. Curr Protoc Prot Sci 2003;3:21.13.1–21.13.15. [DOI] [PubMed]
- 59.Lalanne T, Abrahamsson T, Sjostrom PJ. Using multiple whole-cell recordings to study spike-timing-dependent plasticity in acute neocortical slices. Cold Spring Harb Protoc. 2016;2016:pdbprot091306. doi: 10.1101/pdb.prot091306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Buchanan KA, Blackman AV, Moreau AW, Elgar D, Costa RP, Lalanne T, et al. Target-specific expression of presynaptic NMDA receptors in neocortical microcircuits. Neuron. 2012;75:451–66. doi: 10.1016/j.neuron.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.