Abstract
Abstract
The apoptosome is a platform that activates apical procaspases in response to intrinsic cell death signals. Biochemical and structural studies in the past two decades have extended our understanding of apoptosome composition and structure, while illuminating the requirements for initiator procaspase activation. A number of studies have now provided high-resolution structures for apoptosomes from C. elegans (CED-4), D. melanogaster (Dark), and H. sapiens (Apaf-1), which define critical protein interfaces, including intra and interdomain interactions. This work also reveals interactions of apoptosomes with their respective initiator caspases, CED-3, Dronc and procaspase-9. Structures of the human apoptosome have defined the requirements for cytochrome c binding, which triggers the conversion of inactive Apaf-1 molecules to an extended, assembly competent state. While recent data have provided a detailed understanding of apoptosome formation and procaspase activation, they also highlight important evolutionary differences with functional implications for caspase activation.
Graphical Abstract
Comparison of the CARD/CARD disks and apoptosomes formed by CED-4, Dark and Apaf-1. Cartoons of the active states of the CARD-CARD disks, illustrating the two CED-4 CARD tetrameric ring layers (CED4a and CED4b; top row) and the binding of 8 Dronc CARDs and between 3-4 pc-9 CARDs, to the Dark and Apaf-1 CARD disk respectively (middle and lower rows). Ribbon diagrams of the active CED-4, Dark and Apaf-1 apoptosomes are shown (right column). 
Facts
The CED-4 apoptosome can interact with CED-3 in an 8:2 stoichiometry via CED-3 L2’ loop and CED-4 “hutch” interactions.
The octameric Dark apoptosome recruits Dronc in an 8:8 stoichiometry via CARD–CARD interactions.
The heptameric Apaf-1 apoptosome interacts directly with three or four procaspase-9 (pc-9) molecules via CARD–CARD interactions to form an acentric, disk-like spiral on the platform.
Procaspase-9 forms homodimers with itself and heterodimers with Apaf-1 in the apoptosome via its GCFNF406 motif.
Autocleavage of the pc-9 homodimer causes dissociation from the apoptosome.
Procaspase-9/Apaf-1 heterodimer formation precedes inter-subunit cleavage by caspase-3.
Procaspase activation mechanisms are quite divergent between worms, flies, and humans.
Open questions
What is the physiological consequence of recruiting three vs. four pc-9 molecules via the CARD disk?
Does formation of pc-9 homodimers and pc-9/Apaf-1 heterodimers on the apoptosome represent different active states that decide cell fate (i.e., non-apoptotic vs. death signaling), while exemplifying sequential stages in the intrinsic cell death pathway?
Is caspase-3 activated by recruitment to the apoptosome or by a flexibly-tethered pc-9 homodimer, or by both?
Introduction
Cloning of the core cell death genes in Caenorhabditis elegans 25 years ago [1] transformed the cell death research field. Along with the cell death initiator EGL-1 (the BH-3-only protein), the inhibitor CED-9 (the Bcl-2 homolog) and the executioner CED-3 (the initiator procaspase), the death machinery also required CED-4, an adapter protein for CED-3 activation. Subsequent studies identified orthologues of the core cell death machinery in more complex organisms and it became apparent that initiator procaspase activation uses CED-4-like platforms. Thus Apaf-1 [2] in mammals and Dark/Dapaf-1/Hac-1 [3–5] in Drosophila were found to function in an analogous way to CED-4 in C. elegans for the activation of the apical caspases, procaspase-9 (pc-9), and Dronc in their respective intrinsic cell death pathways.
In particular, the molecular and structural details by which Apaf-1 activates pc-9 have been a matter of intense research. The first indication that Apaf-1 forms a higher molecular weight complex with cytochrome c and dATP to form a pc-9 activating platform, came from biochemical studies nearly 20 years ago [6–8]. This complex was termed the Apaf-1 apoptosome [6]. Structural studies of the apoptosome during this time have been important in revealing critical features required for its function and in understanding the kinetics and recruitment of pc-9, which leads to activation of a proteolytic cascade during apoptosis. Despite these studies, the mechanism by which the heptameric apoptosome facilitates pc-9 activation has been a matter of considerable debate. Recent near atomic structures have now provided new insights into this mechanism, while also adding further controversy.
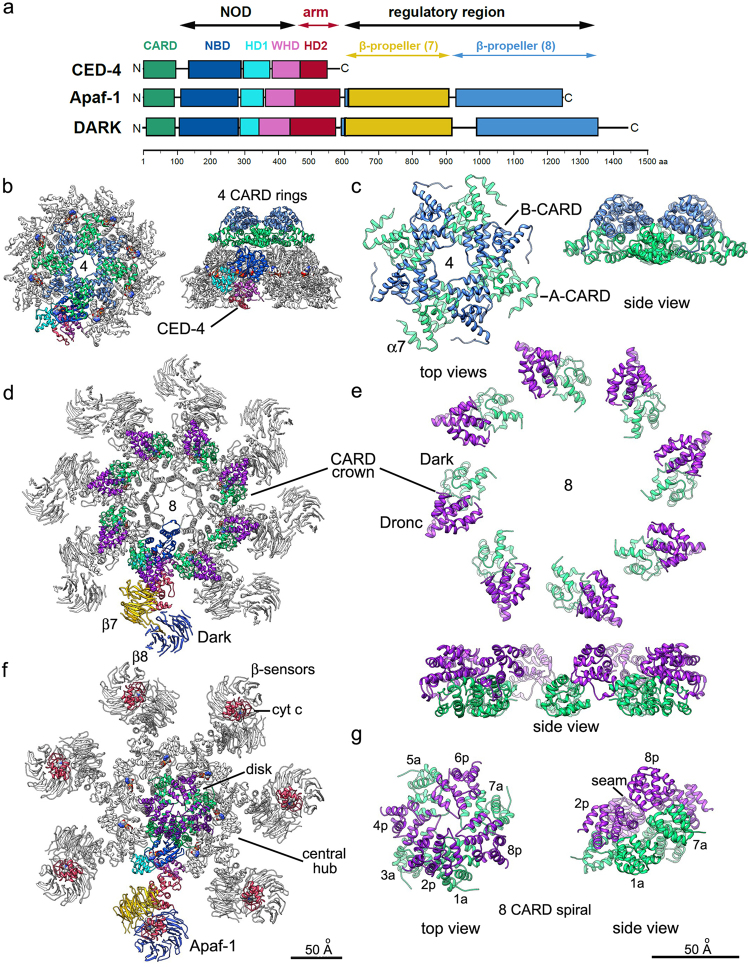
Protein domains in CED-4 are highly conserved in Apaf-1 and Dark, including an amino-terminal caspase recruitment domain (CARD), followed by: a nucleotide-dependent oligomerization domain (NOD) consisting of an α/β nucleotide-binding domain (NBD), helical domain HD1, and a winged helix domain (WHD), that forms a small three-stranded β-sheet. This module is followed by helical domain 2 (HD2) (Fig. 1a). The N-terminal CARD in these adapter molecules is responsible for binding to apical procaspases (CED-4, Dronc or pc-9) via CARD–CARD interactions. The NBD contains conserved Walker A and Walker B box motifs typical of the AAA+ family of ATPases [6, 9–13]. Unlike many AAA+ ATPases, CED-4, Apaf-1, and Dark do not have detectable ATPase activity, indicating that ATP hydrolysis is very inefficient [9, 11, 12, 14–18]. While ATP is constitutively bound to CED-4, the Apaf-1 apoptosome is kept in an autoinhibited state when bound to ADP [19, 20] and requires extensive conformational alterations for ATP/dATP binding and apoptosome activation. Assembly of the Dark apoptosome is also nucleotide-dependent (but only by dATP) [21], and can also be driven by addition of Dronc CARDs in a nucleotide-independent manner [16].
Fig. 1.
Evolutionarily conserved and divergent features of apoptosomes. a A schematic representation of C.elegans CED-4, Drosophila Dark and human Apaf-1. In Dark and Apaf-1, the last blade in the 8-blade β-propeller is formed by the HD2 linker (blue region). Protein domains are color-coded and are drawn approximately to scale. b The ground state CED-4 apoptosome is shown as a ribbon diagram with a color-coded subunit and two stacked, four CARD rings, in top and side views (PDB 3LQR). c Details of the interdigitated and stacked four CARD rings are shown with CARD-A in light green and CARD-B in blue. Linker helix 7 is marked (α7). d A top view of the Dark apoptosome is shown in a possible post-activation state (PDB 3J9K) with the Dark-Dronc CARD crown (Dronc CARD in purple). e The Dark-Dronc crown is shown at higher magnification in top and side views. f A top view of the active Apaf-1 apoptosome is shown with an 8 CARD disk (PDB 5JUY). g Top and side views are shown of the 8 CARD disk annotated with positions of Apaf-1 CARDs and pc-9 CARDs (in purple) that form a left-handed spiral for Apaf-1/pc-9 CARD pairs. Respective scale bars for b, d, f, and for c, e, g are shown
In contrast to CED-4, both Dark and Apaf-1 have two carboxy-terminal β-propeller regulatory domains that are joined to the central hub via the second helical domain. This region is comprised of a series of WD40 repeats that form 7-blade and 8-blade β-propellers and fold into a V-shaped region (compare Fig. 1b, d, f) [16, 18, 20, 22]. The number of WD40 repeats varies between the most commonly expressed Apaf-1 isoform (Apaf-1XL; 15 repeats) and a shorter variant (Apaf-1LN) in which the fifth repeat in the 7-blade β-propeller is missing [23]. This “defect” may affect cytochrome c binding and a full compliment of 15 WD40 repeats is required for pc-9 activation. The evolutionary role of the WD40 regulatory region is conflicting. In Apaf-1, the WD40 domains are required for binding cytochrome c, which is essential for apoptosome activation [2, 6, 11, 24]. CED-4 does not contain WD40 motifs, and activates CED-3 in a quite different manner [25–27]. Interestingly, the β-propeller regulatory regions in the Dark apoptosome do not bind cytochrome c [16, 18, 21, 22, 28], and cytochrome c is not required to activate the Drosophila apoptosome [29–31]. However, β-propellers in Dark appear to stabilize the single ring apoptosome and may play a role in assembly [22].
Recent high-resolution structures of the Apaf-1 apoptosome have been critical in developing models of conformational changes that occur during assembly and have revealed features that may account for interactions between cytochrome c and Apaf-1 [14, 22, 32]. Furthermore, they have provided additional information about the central hub and disk-like spiral formed by Apaf-1 and pc-9 CARDs, thereby giving a better understanding of how procaspases are activated [22, 32]. The high-resolution structures of CED-4, Dark, and Apaf-1 apoptosomes are discussed below, including relevant information on evolutionary differences and their functional implications.
CED-4: a tetrameric apoptosome with two chains in the asymmetric unit
The absence of β-propeller regulatory regions in CED-4 is correlated with an evolutionarily distinct regulation of apoptosome assembly in the worm. Instead, CED-4 apoptosome formation is downregulated by interactions with the anti-apoptotic CED-9 protein to form an inhibitory complex (PDB 2A5Y) [27]. At the structural level, binding of CED-9 to the CED-4 NBD blocks ring formation. While CED-4 does not have typical AAA+ ATPase activity, the NBD has a strong preference for ATP-Mg2+ over dATP. In addition, nucleotide exchange does not appear to be required for apoptosome assembly as ATP molecules are already bound to the constitutive CED-4 dimer [15, 33]. Thus, single CED-9 molecules interact with CED-4 dimers to prevent further assembly [27, 34, 35]. Interestingly, the AAA+ ATPase family member, MAC-1, can also interact with CED-4 to inhibit cell death, but the structural basis and functional importance of this interaction, including whether it may cooperate with CED-9 binding, is not known [36]. The NBD through HD2 regions of the CED-4 dimer do not undergo major conformational changes during apoptosome assembly when released from CED-9. This is distinct from autoinhibited Apaf-1 with bound ADP, which requires cytochrome c and ATP/dATP to induce an extended conformation for apoptosome assembly [11, 12, 15].
In a CED-4/CED-9 crystal structure, one CARD sits above the NBD of monomer-A and while the second CARD appears to be disordered, both CARDs are accessible to other proteins. During cell death stimulation, CED-4 interactions with CED-9 are counter-inhibited by EGL-1 binding (PDB 1TY4), which induces a conformational change that leads to CED-9 dissociation from CED-4 [27, 34]. Four CED-4 dimers then oligomerize [35] into a classic wheel-like apoptosome with eight CED-4 molecules that can activate CED-3 (PDB 3LQQ; Fig. 1b) [27, 35, 37]. Thus, CED-9 inhibits CED-4-mediated activation of CED-3 by sequestering CED-4 dimers to prevent further oligomerization.
Each CED-4 in the apoptosome contains a buried ATP and a Mg2+ ion and the platform domains (NBD-HD2) exhibit a nearly perfect eightfold symmetry. In addition, four pairs of CED-4 CARDs form two interdigitated, fourfold symmetric rings that are stacked on the platform (Fig. 1c) [26]. In this arrangement, the four CARDs of CED-4 monomer-B with extended CARD-NBD linkers, interact with each other to form an upper, closed tetrameric ring, while four CARDs from monomer-A with α-helical linkers (α7) are located in the lower ring and interact with the α/β-fold and HD1 [26]. As a consequence, the CED-4 apoptosome forms a funnel-shaped structure with CARDs forming two rings on the top of the funnel (narrow end), a middle layer comprised of the α/β-fold and HD1 and a bottom layer (wide neck) with the WHD and HD2 domains [26]. The interior of this funnel has been described as a “hutch” (Fig. 1b) and has been proposed as a binding site for CED-3 during activation [26].
CED-3 is predominantly a monomer in solution [26] and forms asymmetric homodimers at high concentration [38]. This asymmetry is similar to that observed with Dronc and pc-9 homodimers. A crystal structure of the CED-3 dimer at 2.65 Å resolution shows one active site in a productive conformation, while the active site on the adjacent monomer is disordered (PDB 4M9R) [38]. The active site interface between CED-3 dimers is formed by L2/L4 loop contacts with the L2’ loop on the adjacent CED-3 monomer [38]. Insights into the mechanism of CED-3 recruitment and activation were provided recently, by a co-crystal structure at 3.2 Å resolution of CED-4 with a small fragment of CED-3 in the apoptosome (PDB 4M9S) [38]. Importantly, the CED-4 octamer may accommodate only two CED-3 molecules to yield a final CED-4/CED-3 stoichiometry of 8:2 [26, 38]. This work also demonstrated a requirement for the L2’ CED-3 surface loop (residues 391–395) as an interaction site that is critical for activation of CED-3 by the CED-4 apoptosome [38]. Critical residues required for CED-3 /CED-4 interactions, include the CED-4 hydrophobic pocket formed by V382, L393, F463, and L464, while the point mutation A394W in CED-4 greatly reduced CED-3 activation [38].
The function of the CED-3 CARD domain is still unclear, as it does not form a stable complex with the isolated CED-4 CARD in solution but does associate with the CED-4 apoptosome [38]. It has been proposed that the CED-3 CARD may facilitate recruitment to the apoptosome, by stabilizing interactions with CED-4 and/or by promoting an optimal conformation of CED-3 for dimerization and activation [38]. Thus, additional CARD–CARD interacting surfaces may be required to recruit CED-3 to the CED-4 apoptosome, which may be provided by the geometric arrangement of CED-4 CARDs in two rings. In this scenario, the long linker (residues 91–234; Fig. 2a) between the CED-3 CARD and its catalytic domain may allow CED-3 to be tethered to the CARD ring(s) on the top surface of the CED-4 apoptosome, while catalytic domains are sequestered through L2’ interactions within the concave bottom surface of the hutch. This mechanism provides a rationale for the much longer linker between the CARD and first catalytic subunit (p17) in CED-3. Moreover, the finding that only two CED-3 molecules interact with CED-4 to form an active apoptosome presumably reflects an evolutionarily distinct mechanism for caspase recruitment and activation. As recently proposed for pc-9, the activation of CED-3 may rely to some extent on “induced" conformational changes [22, 38–40] and the CARD linker may inhibit the zymogen [32].
Fig. 2.
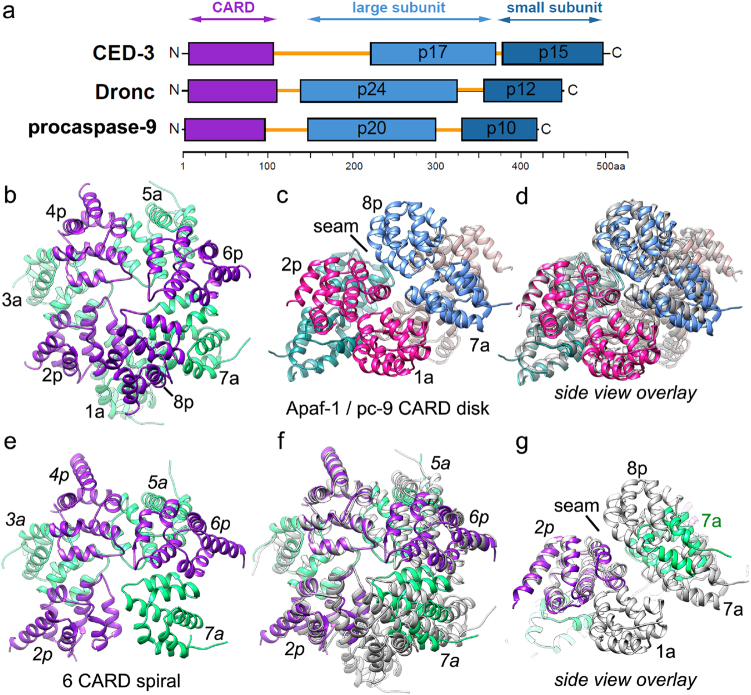
Apical procaspase diagrams and a comparison of CARD disks formed by Apaf-1 and pc-9 CARDs. a A schematic representation is shown for apical procaspases from C. elegans, D. melanogaster, and H. sapiens. Domains are color-coded with linker regions highlighted in gold. b, c Top and side views are shown of an Apaf-1/pc-9 8 CARD disk from an active human apoptosome (PDB 5JUY). CARD pairs 1a/2p, 3a/4p, 5a/6p, and 7a/8p are labeled. A seam or dislocation in the helical lattice between 1a/7a and 2p/8p is indicated in b. d An overlay is shown of two 8 CARD disk structures with the more recent model in gray (PDB 5WVE). e A top view is shown of a 6 CARD spiral from a crystal structure (PDB 5WVC) annotated with the relevant positions in the CARD disk from the apoptosome. f, g Overlays are shown between the 6 CARD spiral and an 8 CARD disk after aligning them on position 2p. The match between the two structures becomes somewhat worse as one moves around the left-handed spiral with the Apaf-1 CARD at position 7a in the 6 CARD spiral displaced vertically from its counterpart in the disk
The octameric Dark apoptosome
The Dark apoptosome is made up of eight molecules, similar to the CED-4 apoptosome, while the active human apoptosome contains seven Apaf-1 molecules. As a consequence, major structural differences in the Dark apoptosome may accommodate an additional subunit in the octagonal ring, when compared to the Apaf-1 apoptosome [16, 18, 21]. Moreover, cryo-EM structural studies demonstrated that the Dark apoptosome is comprised of two octameric complexes that assemble into a double ring mediated by interactions between N-terminal CARDs in the two opposing rings (PDB 3J9L, 5JUL) [16, 21, 22]. The N-terminal CARD of Dark recruits the Drosophila initiator caspase Dronc, leading to its activation and downstream activation of the executioner caspase, DrICE [28]. A Dark double ring is also evident following the addition of Dronc CARDs during assembly (PDB 3J9K) [16]. Remarkably, the Dronc CARD/Dark CARD heterodimer formation can promote Dark assembly in the absence of nucleotide exchange. When the Dronc CARD binds to Dark, the resulting CARD heterodimer forms a connection to the first β-propeller (β7. Fig. 1d) [16, 22]. This interaction creates a “bridge” between the NBD and the 7-bladed β-propeller in the V-shaped region that may stabilize an extended Dark conformation, thereby favoring ring assembly [22].
The resulting Dark single ring with an octagonal crown of Dark/Dronc CARD dimers (Fig. 1e) may then further dimerize through bridging interactions between Dronc CARDs to form a double ring. However, this double ring is unlikely to represent the active configuration as C-termini of the Dronc CARDs face inwards toward the ring center. In addition, the shorter CARD-p24 linker in Dronc (Fig. 2a) is not sufficiently long to be threaded back through the adjacent CARDs in the interface of the double ring to link up with catalytic domains, which would only be freely accessible to cleave the executioner caspase DrICE if they are positioned on the outside. Indeed, it is likely that the active Dark apoptosome with bound Dronc is a single octameric ring [18] and in this scenario, double rings may represent a post-activation complex that can accumulate at high protein concentration and/or be a result from cleavage of Dronc catalytic domains [16, 22]. Alternatively, single and double rings may represent transient active states that are dependent on the concentration of Dronc molecules [41]. Although somewhat speculative, a similar expanded Dronc CARD/Dark CARD crown may be present in both active and post-activation states (Fig. 1d, e). The proposed model of Dronc activation by the Dark apoptosome involves binding of four Dronc dimers that are flexibly tethered to Dark via CARD–CARD interactions [16, 22, 28]. This leads to proteolytic cleavage of Dronc and DrICE on the platform. Released Dronc dimers remain active and may then cleave and activate DrICE in solution.
A recent report of the near atomic structure of a Dark apoptosome in the ground state (PDB 5JUL) [22], also highlighted several observations and differences when compared to CED-4 and Apaf-1 apoptosomes. Cheng et al. (2017) [22] demonstrated an absence of the previously described canonical second α-helix in the Dark CARD domain, such that the CARD fold is comprised of five α-helices (not six) [22]. This does not prevent interactions between the Dark CARD and adjacent NBD and HD1 domains or block interactions with helix α8 of the NBD [22]. A second structural change is found in the HD1 fold, which contains only two α-helices, instead of four as evident in CED-4 and Apaf-1 apoptosomes. Two of the predicted helices form strands containing residues that may stabilize the domain fold and mediate interactions with the adjoining WHD [22].
Another distinct difference between Dark, CED-4 and the Apaf-1 apoptosomes is that Dark has a strict preference for dATP binding to the NBD, that is likely due to differences in HD1 side-chain interactions [22]. dATP binding appears to align the WHDs to facilitate interaction with adjacent monomers to form an octameric ring. The Dark apoptosome has low intrinsic dATPase activity [21] and residues critical for dATP binding include the NBD sensor I (Arg267) and the HD1 sensor (Arg322) [22]. A structural comparison indicates that residues that support Mg2+ binding in the CED-4 apoptosome, are lacking in Dark. In addition, the Walker B box in Dark lacks critical Glu/Asp residues required for catalysis of the gamma-phosphate and adjacent subunits do not provide the necessary structural features (Arg finger) required for accelerating catalysis [22].
Importantly, the improved resolution of the Drosophila apoptosome has provided additional information on the structural requirements for an active apoptosome. Interactions between the NBD of adjacent Dark molecules make up the central pore, which is framed by an inner ring of helix-loop-helix motifs stabilized by hydrophobic zipper interactions between adjacent α-helices. Each of these helix-loop-helix motifs contains a conserved initiator-specific motif (ISM) [18, 22, 42, 43]. An outer ring formed by WHD/HD1 inter-monomer interactions encircles the NBDs [22]. As a consequence of having an additional Dark subunit, the ISM ring is somewhat larger when compared to Apaf-1 and the angle formed by HD1-WHD is tighter. The HD2 arm has a more defined bend that shifts the β-propellers upward, which allows circumferential interactions between adjacent V-shaped domains. The recent structure also demonstrated a post-translational adenylylation of Dark at Lys251, which may provide a mechanism to stabilize the NBD ring [22], but this idea remains to be tested.
The new Dark apoptosome structure provides an improved model of β-propellers within the V-shaped region, that is based on homology with the Apaf-1 propellers [22]. The additional structural information on the β-propellers may explain the lack of interactions with cytochrome c [22]. In particular, the two β-propellers in Dark have an altered conformation which creates a larger cleft between them, when compared to human Apaf-1, along with a slight rotation to accommodate an additional Dark subunit [14, 44]. The 7-blade β-propeller also makes interactions with HD1 to stabilize the extended Dark conformation, which may facilitate apoptosome assembly. As a consequence of this altered positioning of β-propellers in the V-shaped region, stable interactions with a cytochrome c molecule are less likely [21]. This idea is supported by biological data, which shows that Dronc activation can occur in the absence of cytochrome c [21, 29, 30].
The heptameric Apaf-1 apoptosome
Early structural information was critical in revealing the domain organization of the apoptosome [10, 17, 19, 20, 41, 43, 45–47]. The human apoptosome contains seven Apaf-1 molecules symmetrically arranged in a wheel-shaped structure to form a central hub comprised of NODs with seven extended HD2 arms, each ending in a V-shaped region that is formed by the two β-propellers [19, 45]. The α/β-fold of the NBD and HD1 bind ADP/dATP between them, while the WD40 repeats form the 7- and 8-bladed β-propellers [19, 22, 43–45]. Contrary to earlier suggestions [43, 46] it was found that lateral interactions between the α/β-fold of adjacent Apaf-1 molecules are used to organize the central hub, while N-terminal CARDs sit above this ring, and are disordered in the absence of pc-9 (PDB 3J2T) [14, 26, 41, 43]. However, recent near atomic structures have demonstrated that Apaf-1 CARDs form a disk-like feature on the surface of the active apoptosome in the presence of pc-9 CARDs, and this arrangement differs from that found in CED-4 and Dark (Figs. 1 and 2) [16, 22, 41, 44, 48]. The stability of the central hub is maintained by a complex network of α-helical interactions, hydrogen bonds and salt bridges between adjacent NBD and HD1 modules from neighboring Apaf-1 molecules [14, 44]. The WHD/HD1 contacts on adjacent protomers appear to support apoptosome assembly with WHD domains bridging adjacent NBD and HD1 domains [14, 44]. As occurs in Dark, the ISM helix in Apaf-1 (α12) is paired with helix α13 through extensive hydrophobic interactions to form a helix-loop-helix motif in each subunit, which in turn, interact laterally with adjacent subunits to form a helical picket fence that lines the central pore in the ring [43, 44].
In healthy cells, Apaf-1 monomers exist in a closed, inactive conformation with ADP embedded in a crevice in the interface of the NBD-HD1 (PDB 1Z6T, 3SFZ) [6, 9, 12, 14, 20, 47]. This closed conformation presages the requirement for extensive conformational changes to the monomer to facilitate apoptosome assembly. In contrast to apoptosome formation in C. elegans and Drosophila, cytochrome c released from mitochondria during apoptosis signaling binds to monomeric Apaf-1, which triggers conformational changes leading to nucleotide exchange (ATP or dATP can replace ADP), and subsequent oligomerization of an extended Apaf-1 to form the apoptosome (Fig. 3) [2, 11, 19, 33, 43, 44]. In order to understand the conformational changes that occur, accurate models are needed of inactive and extended states of Apaf-1. Two crystal structures of inactive Apaf-1 monomers with bound ADP have been determined, with one lacking β-propellers [47] and the second lacking the N-terminal CARD [20] domains to facilitate crystallization. However, the NOD in these two crystal forms is virtually identical and this has allowed a consensus model to be constructed for the entire Apaf-1 monomer with bound ADP [14, 20, 44, 47]. Near atomic structures of the human apoptosome have now been achieved by several groups using single particle cryo-EM, to provide insights into the extended conformation of Apaf-1 in the absence and presence of pc-9 (PDB 3JBT, 5JUY, 5WVE; Fig. 3) [14, 44, 48]. As predicted [43], the conformation of Apaf-1 in the central hub, HD2 arms and regulatory region is essentially identical in both inactive and active apoptosomes [14, 44, 48]. These studies have provided important details of specific residues involved in interdomain van der Waals contacts within single Apaf-1 molecules, revealed residues critical for domain interactions that stabilize the active apoptosome, and showed interactions within the V-shaped sensor domain of the tandem 7- and 8-blade β- propellers, which forms the cytochrome c binding surface [14, 44, 48]
Fig. 3.
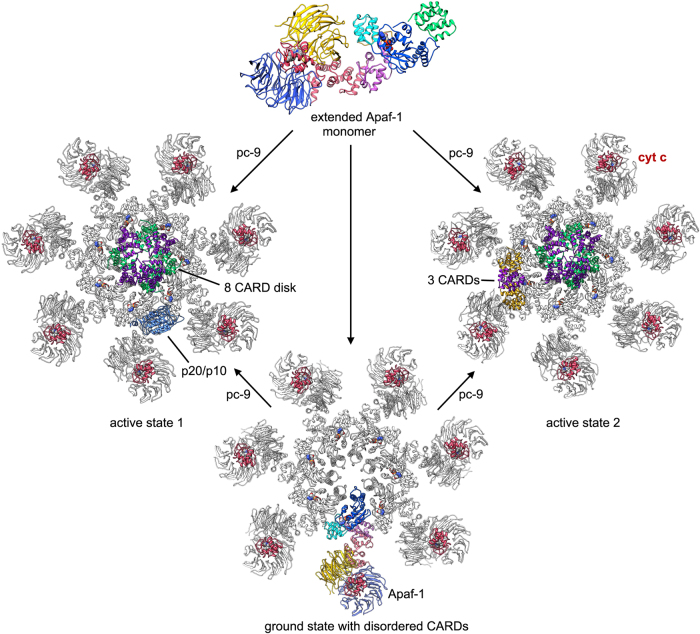
Assembly paths for ground state and active human apoptosomes. An extended Apaf-1 monomer (not shown to scale) may assemble with other monomers to form a heptameric apoptosome in the ground state with disordered Apaf-1 CARDs. However, in the presence of pc-9 two active platforms may be formed as shown (active state 1 and 2). In addition it may also be possible to form a third hybrid active platform with both p20/p10 and three CARD modules bound to the central hub (not shown, see Fig. 5e)
In the Apaf-1 apoptosome, the dATP-binding site is located at the NBD–HD1 interface, and is formed in part by the Walker A loop and the loop between HD1 and WHD. While Apaf-1 can bind ADP in the inactive state, there is a small preference to use dATP for apoptosome assembly in vitro (reviewed in ref. [41]), while ATP is much more abundant in vivo (~ 2 mM for ATP vs. 10 μM for dATP). [22, 45]. Some stabilization occurs through interactions between an arginine residue (Arg265) in the NBD, along with Ser325 and Tyr359 in the HD1 and the bound ATP/dATP molecule. A rotation of the NBD-HD1 pair about α-helix 20 in the WHD, positions the HD1-WHD loop at the bottom of the nucleotide pocket [14, 44], which allows the NBD-HD1 pair to form circumferential interactions within the central hub during assembly [43, 49]. This also breaks interactions with the WHD that normally stabilize a closed configuration [20].
Sensor β-propellers and cytochrome c binding
The “spokes” of the apoptosome wheel protrude outwards from the hub by the HD2 domains and each arm forms the base of the V-shaped region containing the two β-propeller sensor domains. In the longest Apaf-1 isoform, (Apaf-1XL) which is expressed in most tissues [23, 50], the fifteen WD40 repeats form tandem 7 and 8-blade β-propellers [9, 45, 51, 52] and a recent cryo-EM study indicated this has a novel closure mechanism [20]. This topology of the β-propellers is similar to that found in actin interacting protein (Aip1p), in which the linker from HD2 forms the d-strand of the last blade in the 8-blade β-propeller (indicated by the small blue region present at the beginning of the 7-blade propeller in Fig. 1a), before crossing over to form the 7-blade β-propeller [53]. This topology was verified by a crystal structure of murine Apaf-1 at 3.0 Å resolution [43] and appears to be conserved in the Dark β-propellers [22].
In the Apaf-1 apoptosome, the V-shaped β-propellers form the cytochrome c binding pocket, and interaction with cytochrome c appears to be stabilized by hydrogen bonds and salt bridges [18, 20, 22, 43, 45, 49, 54]. Both reduced and oxidized forms of cytochrome c can interact with the apoptosome to mediate its activation [55]. Interestingly, cytochrome c seems to interact preferentially with the 8-blade β-propeller, while a more limited contact surface is formed with the 7-blade β-propeller [14, 44]. Cytochrome c is resolved at ~ 6 Å resolution in the new maps, due to local motion and the molecule is rotated ~ 90°, relative to an earlier model based on molecular docking into a map where helices were not resolved [43, 49]. Directed mutagenesis revealed critical residues in cytochrome c required for stable association with Apaf-1 β-propellers, and some of these residues (G56, P76, I81) are important for apoptosome formation and subsequent caspase activation [14].
Truncated Apaf-1 molecules lacking WD-40 repeat regions can also assemble to form apoptosomes that are constitutively active but disassemble over time [56]. This has suggested that β-propellers may be somewhat inhibitory to assembly while also stabilizing the Apaf-1 apoptosome [28, 41, 56]. However, β-propellers in the Apaf-1 apoptosome are located at high radius and do not interact with adjacent subunits, nor do they interact with other domains in the monomer, with the exception of HD2. Hence, the destabilization mechanism remains mysterious. Although speculative, the lack of β-propellers in truncated apoptosomes could weaken HD2 interactions with the central hub leading to a time-dependent disassembly.
It is now clear that cytochrome c binding causes an upward rotation of the β-propeller region, which disrupts the connection between the 7-blade β-propeller and the NBD-HD1 pair in the inactive Apaf-1 monomer [14, 20], while the 7-blade β-propeller rotates toward the 8-blade β-propeller in a clamping motion [14, 32, 44, 49]. These events destabilize the autoinhibited Apaf-1 conformation and facilitate nucleotide exchange by promoting conformational changes that expose the ADP binding pocket to permit ATP/dATP binding [10, 14, 44, 47]. This stabilizes the Apaf-1 monomer in an extended conformation such that it can interact with neighboring protomers. Band shift experiments suggest that nucleotide exchange and associated conformational changes in Apaf-1 may occur in response to cytochrome c binding because no significant alterations in Apaf-1 conformation were detected with added dATP in the absence of cytochrome c [44]. When taken together, these data support a sequential model in which cytochrome c binding occurs before nucleotide exchange, to facilitate the transition from a closed to an extended Apaf-1 conformation. In addition, these findings indicate that WD40 repeats act as sensors that trigger initial apoptosome assembly by binding cytochrome c.
Possible mechanisms of procaspase-9 activation
Procaspase-9 is recruited by the apoptosome to form a holoenzyme that increases its catalytic activity [6, 7, 13]. In solution, pc-9 is a constitutive monomer when not bound to Apaf-1, whereas active caspases in general are dimers [57]. Remarkably, a dimeric interaction between two pc-9 monomers in a crystal lattice promotes the formation of an active site on one catalytic subunit, while the second subunit remains in an inactive configuration (PDB 1JXQ) [57] and a similar phenomenon occurs in CED-3 dimers at high concentration [38]. The N-terminal CARD in pc-9 interacts with Apaf-1 CARD(s) to recruit the apical procaspase to the apoptosome and this promotes activation of the catalytic domain, which is separated from the CARD by a long linker (Fig. 2a) [7, 58, 59]. In addition, the p20 and p10 subunits of the catalytic domain are also connected by a linker that is subject to self-cleavage [58]. Thus, in order to understand how pc-9 is activated we need to understand the role of CARDs and pc-9 linkers in the holo-apoptosome.
Different models that explain the mechanisms of pc-9 activation by the apoptosome have been proposed [9, 41, 46, 59–62]. The “proximity-induced dimerization model” predicts that recruitment of pc-9 molecules to the Apaf-1 apoptosome may facilitate homodimerization, which is thought to lead to pc-9 auto-activation [63]. However, until recently no structural or biochemical evidence had demonstrated that pc-9 is homodimeric when bound to the apoptosome (see below [50]). The “induced conformation model” proposed that the Apaf-1 apoptosome is a platform that binds pc-9 to facilitate its active conformation [39, 64]. Recent modeling suggested that the activation of pc-9 is mediated primarily via allosteric regulation by the apoptosome platform [65]. In all of these models, a primary function of the apoptosome is to assemble a multimeric complex between Apaf-1 and pc-9 CARDs to facilitate pc-9 activation by increasing the local concentration of the zymogen [44, 48]. In addition, data on an engineered pc-9 that is a constitutive dimer in solution suggests that the CARD and its linker may inhibit the catalytic domains [48]. Hence, formation of a CARD heterodimer on the apoptosome may release this inhibition, however, this idea remains to be tested.
While dimerization may play a key role in activation, how this occurs on the apoptosome has remained unexplained, and in fact recent data have suggested the possibility of a more complicated activation mechanism that involves both homodimerization of pc-9 molecules and heterodimer formation of a single pc-9 catalytic domain with the NBD of Apaf-1 [32, 44, 50]. Intriguingly, an inactive conformation of the Apaf-1 monomer does not preclude pc-9 binding via CARD–CARD interactions, as shown by band shift experiments [44]. Thus, pc-9/Apaf-1 heterodimers may be recruited to the assembling apoptosome as part of its activation (Fig. 3). It remains an open question whether assembly of Apaf-1 in the presence of pc-9 is guided by additional interactions between CARDs in the nascent disk.
An initial crystal structure revealed a stable 1:1 complex between the CARD domains of Apaf-1 and pc-9 with a complementary interface (known as Type I), which is indispensable for pc-9 activation [66]. However, this stable 1:1 complex was later shown to be insufficient to activate pc-9 [61]. Instead, Apaf-1 and pc-9 CARDs form higher order oligomeric complexes, stabilized primarily by two of three possible interfaces (known as TypeI, II and III), that are also found in other Death Domain complexes [61, 67]. Comparison of the interactions involved in the CARD-CARD disk in the CED-4, Dark and Apaf-1 apoptosomes, highlights some conservation of Type II and III interfaces, with Type I CARD interactions only found in the human apoptosome [42, 67]. A recent crystal structure at 2.1 Å resolution revealed a heterotrimeric complex comprised of two Apaf-1 CARDs with a pc-9 CARD sandwiched in between [61]. Type II interactions involving residues in pc-9 (Arg36/ Arg65) and Apaf-1 (Glu78) CARDs were shown to be critical for pc-9 activation [61], along with Type I interactions described previously [66]. Moreover, a single residue (Glu41 in the Apaf-1 CARD) can form a bifurcated interaction with a pc-9 CARD in a minimal Type III interface [61]. Additional interactions with Lys58 and Lys62 on the Apaf-1 CARD were also shown to be required for pc-9 activation and may help position Apaf-1 CARDs on the platform [48, 61]. Thus, formation of a heterotrimeric CARD complex provides a means to tether multiple pc-9 catalytic domains to the apoptosome platform [32, 43, 44, 48].
Two near atomic resolution structures of the active Apaf-1 apoptosome have revealed the overall architecture of the Apaf-1 and pc-9 CARD disk arrangement creates a tilted disk which sits in an above the central hub (Figs. 2 and 3) [44, 48]. The symmetry mismatch between the disk and platform results in an acentric positioning of the CARD disk when viewed from above (Fig. 3) [44, 48]. The disk can be described as a spiral of Apaf-1 and pc-9 CARD pairs with four Apaf-1 CARDs forming a bottom “layer” while three or four pc-9 CARDs are present on the top surface (Fig. 2b–d). In addition, the Apaf-1 CARDs move further from the surface of the central hub, as they spiral upwards in a left-handed manner and thus, have different linker conformations and different interactions with their cognate NBDs in the central hub. The Apaf-1 and pc-9 CARD pairs interact through Type II interfaces, and join laterally around the spiral primarily through Type I interactions. Remarkably, only four of the seven Apaf-1 CARDs interact with pc-9 CARDs in the disk [44, 48]. This is because the remaining three Apaf-1 CARD molecules do not readily fit into the subunit bonding pattern of the spiral and cannot continue the spiral because their CARD-NBD linkers are too short [44, 48]. Apaf-1 CARDs from adjacent subunits in the apoptosome are located at a seam (between positions 1a and 7a), where the spiral stops elongating vertically (Fig. 2c, d) [44]. Formation of the Apaf-1/pc-9 CARD disk is required for activation of the apoptosome and pc-9 catalytic domains are flexibly-tethered to the CARDs through a linker (Fig. 2a) [32, 44, 48]. In one structure, a disk with four Apaf-1 CARDs and three pc-9 CARDs was visualized, with weak density indicating that a fourth pc-9 CARD could bind to the top of the helical spiral at a lower occupancy [44]. In a second structure, the predominant configuration of the disk contained 8 CARDs [48] with the conformation of the CARDs in the spiral being very similar between the two independently solved structures (see overlay, Fig. 2d). Variations in pH and salt concentration may account for the different occupancy at the eighth position. Thus, apoptosome assembly promotes recruitment and local concentration of pc-9 CARDs, which allows formation of a novel helical oligomer with 7 or 8 CARDs.
A recent analysis (by MALS and SAXS) showed that CARD complex formation is concentration dependent and equal amounts of Apaf-1 and pc-9 CARDs associate as pairs through the stronger Type I interface, with a predominant 3:3 complex formed at high concentration in solution [67]. This CARD complex was crystallized and the structure determined at 3.0 Å resolution to reveal a left-handed spiral of three Apaf-1/pc-9 CARD pairs with a conformation similar to what is observed in the apoptosome but with some differences (PDB 5WVC; Fig. 2e) [67]. However, the environment in the crystal is quite different from that encountered on the assembling apoptosome, due in part to lattice contacts and the lack of a nucleating surface with restraints imposed by CARD-NBD linkers. This may account for observed differences between the helical CARD structures. This includes the addition of one or two extra CARDs to the disk-like spiral in the holo-apoptosome and having four Apaf-1 CARDs that form the base of the disk, rather than three as found in solution and in the crystal. After optimal alignment of the 8 CARD disk with the 6 CARD spiral from the crystal structure, there is a good congruence between CARDs at positions 2p, 3a, and 4p, with some divergence in the latter half of the 6 CARD spiral (Fig. 2f, g). In particular, the Apaf-1 CARD at position 7a is mislocated vertically to an intermediate position, in the absence of the Apaf-1 CARD at position 1a (Fig. 2g). Thus, interactions between the platform and CARD disk are critical to nucleate the larger 7 and 8 CARD spirals found on the apoptosome.
During disk assembly, nucleation of the spiral may occur in different ways, including formation of a CARD heterotrimer comprised of two Apaf-1 CARDs and one pc-9 CARD, with an Apaf-1 CARD located at the base of the spiral (at position 1a). Subsequent addition of two Apaf-1/pc-9 CARD pairs that interact through their Type I interfaces may then occur with a final capping of the spiral by a pc-9 CARD at position 8p in some cases. However, other permutations are possible given the flexible nature of the Apaf-1 CARD-NBD linker such that three Apaf-1/pc-9 CARD “Type I” pairs may be nucleated sequentially from the central hub, with the addition of an Apaf-1 CARD at the base of the spiral (at position 1a), during the early stages of assembly [67]. Importantly, assembly of the disk may be cooperative in order to maintain the proper spacing of the four Apaf-1 CARDs on the apoptosome, which form the base of the spiral [44].
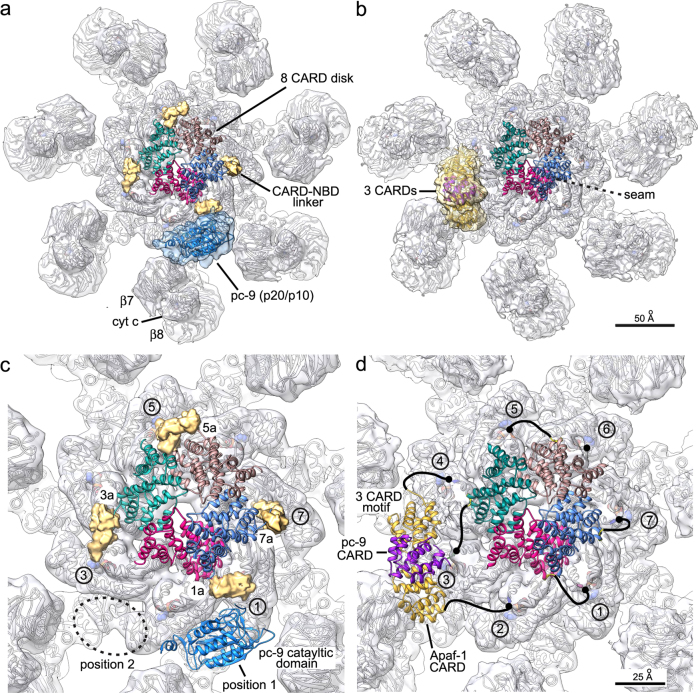
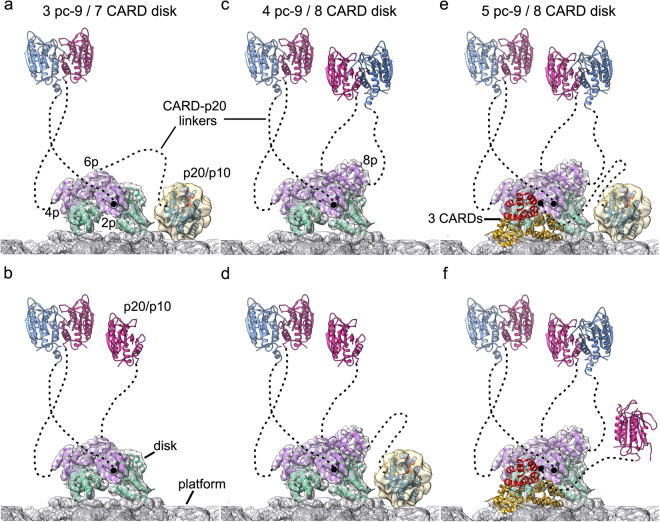
For the active apoptosome, one major difference is apparent on the central hub, when comparing the published structures (Figs. 3 and 4) [32, 44, 48]. In brief, density identified as a catalytic domain of pc-9 (p20/p10) is located on the central hub in a recent 3D map, and this feature was found to be present in about 50% of the complexes [44]. In addition, a similar density was visualized in a previous lower resolution map of the holo-apoptosome, which allowed a reasonable, though preliminary docking of the p20/p10 subunits [32]; hence, the observed feature is reproducible. However, this novel density has only been visualized at low resolution, which may reflect some flexibility in the attachment of the p20/p10 subunits to the central hub. In a near atomic resolution map determined by Yigong Shi’s group [48], an additional sickle-shaped density was resolved at ~ 7 Å resolution on the central hub, adjacent to the CARD disk. This density clearly contains an additional pc-9 CARD at the center, which interacts with two Apaf-1 CARDs located on either side, in a manner similar to that observed in two crystal structures [61, 67]. However, this heterotrimeric configuration could only be resolved in ~ 10% of the particles [48]. Thus, it seems that experimental conditions during sample preparation and grid freezing can influence how pc-9 interacts with potential-binding sites on the central hub, including the formation of a disk-like spiral with variable numbers of pc-9 CARDs and in the use of binding sites that are located adjacent to the acentric disk. When the two recent asymmetric structures of the holo-apoptosome are aligned based on their CARD disks, the novel density peaks located on the central hub are in quite distinct locations (Fig. 4). While the pc-9 catalytic domain (p20/p10) may be located at two positions (with the most likely one shown in Fig. 4c) [5], the three CARD density is rotated and has a different profile as viewed from the side (not shown), when compared to that for pc-9 catalytic domain (Fig. 4b, d). The pattern of Apaf-1 CARD-NBD linker densities allows a precise mapping of all relevant CARD-NBD linkers in the three CARD holo-apoptosome with six ordered Apaf-1 CARDs (Fig. 4d) [48]. Critically, the linker length (~ 25–30 Å) and the relative positions of the linkers may preclude the binding of a three CARD module to either position 1 or 2 on the central hub. Thus, it appears that there are multiple sites on the central hub that may be utilized in distinct ways to bind a pc-9 CARD or a catalytic domain.
Fig. 4.
Comparison of two structures of the active Apaf-1 apoptosome with a symmetry mismatch between the CARD disk and the platform. a A top view is presented of an active apoptosome with the platform in gray ribbons (PDB 5JUY) within the relevant density map. An acentric 8 CARD disk and density for the four CARD-NBD linkers (gold) are shown. A region of density (blue) identified as a p20/p10 catalytic domain of pc-9 is located on the central hub [5]. b A model for an active platform with an 8 CARD disk and a three CARD module on the central hub (PDB 5WVE) [7]. c A close-up view of a, with density for the p20/p10 catalytic domain omitted and a second possible position for this feature indicated (dashed oval; see text and ref. [5] for details). Linker densities in gold are positioned to connect Apaf-1 CARDs in positions 1a, 3a, 5a, and 7a with helix α8 in the NBDs of subunits 1, 3, 5, and 7. d A close-up view of b is shown with relevant CARD-NBD linkers as black lines to their respective Apaf-1 subunits. Apaf-1 CARDs in the three CARD module are shown in gold
Procaspase-9 has been estimated to bind to the apoptosome with ratios between 2–5 zymogens per 7 Apaf-1 molecules [32, 61, 68], so it seems that the exact number of pc-9 molecules bound to the apoptosome may vary depending upon conditions during assembly, which may reflect the dynamic nature of this proteolytic machine. By extension, it seems clear that a sixth or a seventh pc-9 CARD is not bound by their Apaf-1 counterparts, relative to the sites already documented on the apoptosome, which may reflect either a steric limitation or the necessity for a larger oligomer to stabilize the bound pc-9 CARDs. Importantly, some pc-9 catalytic domains are not seen in the current cryo-EM density maps, due to their flexible attachment through long CARD-p20 linkers, which makes it difficult to decipher and understand the precise mechanisms of activation, while highlighting a need for further single molecule studies.
The primary function of the apoptosome is to activate pc-9 so that it can execute apoptosis. While structural studies have provided insight into recruitment and requirements for pc-9 activation, a series of biochemical assays have now demonstrated that both proximity-induced dimerization and allosteric regulation of monomeric pc-9 are critical to regulate apoptosome function [50, 68] and may also govern the duration of apoptosome activity. Firstly, recruitment of pc-9 by the apoptosome increases its local concentration to allow homodimerization, which increases its affinity for the complex [50]. Interestingly, an uncleavable pc-9 has a higher propensity for homodimerization and shows an increased protease activity (caspase-3 cleavage) within the apoptosome, than does cleaved caspase-9 [50]. As a consequence, cleavage of the inter-subunit p20/p10 linker of the catalytic domain reduces pc-9 affinity for the apoptosome and once processed, caspase-9 is released and does not re-bind the apoptosome. Importantly, these studies have demonstrated that pc-9 homodimerization is the key event required for activation and that the apoptosome acts to facilitate this event [50]. This is consistent with biochemical studies of other CARD caspases, such as caspase-2 and Dronc, where initial activation requires dimerization, rather than cleavage of the zymogen [50, 69–71]. Stabilization of pc-9 binding to the apoptosome not only requires CARD–CARD interactions, but interactions of the pc-9 small subunit (p10) via its GCFNF404 motif to form homo- and heterodimers [50]. What is not clear at this point, is how pc-9 homodimer formation affects the stability of the CARD–CARD disk, as catalytic domain dimers seem to be quite flexible, since they can be released from the holo-apoptosome by site-directed thrombinolysis [32, 43] and are not resolved in cryo-EM maps refined without symmetry [32, 44, 48]. The GCFNF motif in pc-9 also interacts with an Apaf-1 NOD to form pc-9/Apaf-1 heterodimers that efficiently cleave caspase-3 [50]. This data suggests that a pc-9 catalytic domain, which interacts directly with the apoptosome, may be active. This putative active site for caspase-3 cleavage may correspond to the novel density observed on the central hub that has been interpreted as a p20/p10 molecule [22, 32] and further studies are needed on this point. When taken together, these findings indicate that pc-9 can have two different active conformations within the apoptosome, with one or possibly two homodimers tethered to the CARD disk and a pc-9/Apaf-1 heterodimer bound to the central hub [32, 43, 44, 50]. When holo-apoptosomes with a three CARD module bound to the central hub are factored in, then at least six permutations are possible for the disposition of three to five pc-9 catalytic domains (Fig. 5). Thus, the number of possible active pc-9 catalytic domains (as homodimers and heterodimers) will vary depending upon the number of pc-9 molecules recruited to the apoptosome, as they fill in possible binding sites mediated by Apaf-1 CARDs.
Fig. 5.
Models of procaspase-9 activation on the Apaf-1 apoptosome. At least six permutations are possible with pc-9 CARDs docked via 7 and 8 CARD disks, and a three CARD module on the central hub. a, b Three pc-9 molecules with a 7 CARD disk on the platform. c, d Four pc-9 molecules with an 8 CARD disk on the central hub. e, f Five pc-9 molecules with an 8 CARD disk on the platform
Finally, complete cleavage of pc-9 by caspase-3 removes the inter-subunit linker and restores full caspase-9 activity, indicating that caspase-3 also has a proteolytic feedback on pc-9 that may amplify the apoptotic signal [50]. A similar mechanism has been proposed for caspase-2 [69, 72, 73]. These biochemical studies have further presented a molecular timer model where the rate of caspase-9 dissociation from the apoptosome is determined by an initial cleavage of the p20-p10 linker which reduces its apoptosome binding affinity [68]. However, complete dissociation of cleaved caspase-9 from the holo-apoptosome would require release of its CARD from the disk or three CARD module. Thus, there may be a hierarchy for the release of active caspase-9 molecules that depends upon the local environment of their CARDs. The design of the CARD disk, with all pc-9 CARDs located on the top surface, may also facilitate the release of caspase-9.
Conclusions
Evolutionary differences in the structures of CED-4, Dark, and Apaf-1 apoptosomes are reflected in their oligomeric state and in the absence or presence of extended HD2 arms with β-propellers. Remarkably, there are also quite different mechanisms for regulating assembly in worm, fly, and human apoptosomes and different requirements for CARD–CARD interactions with their respective procaspases. Proteolytic activation appears to depend on the recruitment of a variable number of procaspase molecules to these platforms and in the Apaf-1 apoptosome, on the formation of a CARD-CARD disk and perhaps on a novel three CARD module that may assemble on the central hub. While near atomic structures and biochemical data have provided critical insights into pc-9 recruitment and activation by the apoptosome, there are clearly still some discrepancies with regards to Apaf-1/pc-9 stoichiometry. Transient states of the active apoptosome caused by rapid kinetics of its formation and caspase-9 dissociation may partly explain such differences. Nevertheless, further studies are necessary to fully delineate the mechanisms of apoptosome assembly especially in light of the possibility of co-assembly of Apaf-1/pc-9 heterodimers in cells where pc-9 may be a limiting factor [44, 64]. Furthermore, a better understanding is needed of how CARD-p20 linkers may avoid entanglement during assembly of an active apoptosome. In fact, the recruitment of only three pc-9 molecules to an apoptosome with a 7 CARD disk would minimize this problem.
So what are the biological and physiological implications of regulating the apoptosome and how does this modulate cell death execution? In particular, can apoptosome assembly and function be selectively enhanced to induce cell death, thereby aborting proliferation of aberrant cells? Targeting pc-9 specifically may prove difficult as other caspases may also be affected [74] and compounds that prevent apoptosome formation may also affect non-apoptotic roles of Apaf-1 [75–77]. Based on recent studies it may be feasible to regulate apoptosome function by targeting NOD interactions, procaspase-9 recruitment through CARD–CARD interactions and pc-9 activation through interfaces involved in homo- and hetero-dimerization, which in turn can mediate caspase-3 activation and apoptotic cell death. Knowledge of these interactions is critical to understanding cell death mechanisms, which are deficient in many cancers and/or can be exploited to prevent unwanted cell deaths in neurodegenerative diseases and chemotherapy-induced cytotoxicities mediated by active caspase-9.
Acknowledgements
This caspase work in our laboratory (LD, SK) is supported by the National Health and Medical Research Council (NHMRC) of Australia project grants 1021456 and 1043057, a Cancer Council Collaborative Research Fellowship to LD and a NHMRC Senior Principal Research Fellowship to SK (1103006). Apoptosome structure/function studies were supported by NIGMS GM63834 (CWA).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Footnotes
Edited by G. Melino
Contributor Information
Loretta Dorstyn, Email: Loretta.Dorstyn@unisa.edu.au.
Sharad Kumar, Email: Sharad.Kumar@unisa.edu.au.
References
- 1.Ellis HM, Horvitz HR. Genetic control of programmed cell death in the nematode C. elegans. Cell. 1986;44:817–29. doi: 10.1016/0092-8674(86)90004-8. [DOI] [PubMed] [Google Scholar]
- 2.Zou H, Henzel WJ, Liu X, Lutschg A, Wang X. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell. 1997;90:405–13. doi: 10.1016/S0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- 3.Kanuka H, Sawamoto K, Inohara N, Matsuno K, Okano H, Miura M. Control of the cell death pathway by Dapaf-1, a Drosophila Apaf-1/CED-4-related caspase activator. Mol Cell. 1999;4:757–69. doi: 10.1016/S1097-2765(00)80386-X. [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez A, Oliver H, Zou H, Chen P, Wang X, Abrams JM. Dark is a Drosophila homologue of Apaf-1/CED-4 and functions in an evolutionarily conserved death pathway. Nat Cell Biol. 1999;1:272–9. doi: 10.1038/12984. [DOI] [PubMed] [Google Scholar]
- 5.Zhou L, Song Z, Tittel J, Steller H. HAC-1, a Drosophila homolog of APAF-1 and CED-4 functions in developmental and radiation-induced apoptosis. Mol Cell. 1999;4:745–55. doi: 10.1016/S1097-2765(00)80385-8. [DOI] [PubMed] [Google Scholar]
- 6.Zou H, Li Y, Liu X, Wang X. An APAF-1 cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J Biol Chem. 1999;274:11549–56. doi: 10.1074/jbc.274.17.11549. [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez J, Lazebnik Y. Caspase-9 and APAF-1 form an active holoenzyme. Genes Dev. 1999;13:3179–84. doi: 10.1101/gad.13.24.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shiozaki EN, Chai J, Shi Y. Oligomerization and activation of caspase-9, induced by Apaf-1 CARD. Proc Natl Acad Sci USA. 2002;99:4197–202. doi: 10.1073/pnas.072544399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bao Q, Shi Y. Apoptosome: a platform for the activation of initiator caspases. Cell Death Differ. 2007;14:56–65. doi: 10.1038/sj.cdd.4402028. [DOI] [PubMed] [Google Scholar]
- 10.Bao Q, Riedl SJ, Shi Y. Structure of Apaf-1 in the auto-inhibited form: a critical role for ADP. Cell Cycle. 2005;4:1001–3. doi: 10.4161/cc.4.8.1849. [DOI] [PubMed] [Google Scholar]
- 11.Kim HE, Du F, Fang M, Wang X. Formation of apoptosome is initiated by cytochrome c-induced dATP hydrolysis and subsequent nucleotide exchange on Apaf-1. Proc Natl Acad Sci USA. 2005;102:17545–50. doi: 10.1073/pnas.0507900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reubold TF, Wohlgemuth S, Eschenburg S. A new model for the transition of APAF-1 from inactive monomer to caspase-activating apoptosome. J Biol Chem. 2009;284:32717–24. doi: 10.1074/jbc.M109.014027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saleh A, Srinivasula SM, Acharya S, Fishel R, Alnemri ES. Cytochrome c and dATP-mediated oligomerization of Apaf-1 is a prerequisite for procaspase-9 activation. J Biol Chem. 1999;274:17941–5. doi: 10.1074/jbc.274.25.17941. [DOI] [PubMed] [Google Scholar]
- 14.Zhou M, Li Y, Hu Q, Bai XC, Huang W, Yan C, et al. Atomic structure of the apoptosome: mechanism of cytochrome c- and dATP-mediated activation of Apaf-1. Genes Dev. 2015;29:2349–61. doi: 10.1101/gad.272278.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seiffert BM, Vier J, Hacker G. Subcellular localization, oligomerization, and ATP-binding of Caenorhabditis elegans CED-4. Biochem Biophys Res Commun. 2002;290:359–65. doi: 10.1006/bbrc.2001.6211. [DOI] [PubMed] [Google Scholar]
- 16.Pang Y, Bai XC, Yan C, Hao Q, Chen Z, Wang JW, et al. Structure of the apoptosome: mechanistic insights into activation of an initiator caspase from Drosophila. Genes Dev. 2015;29:277–87. doi: 10.1101/gad.255877.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi Y. Apoptosome assembly. Methods Enzymol. 2008;442:141–56. doi: 10.1016/S0076-6879(08)01407-9. [DOI] [PubMed] [Google Scholar]
- 18.Yuan S, Yu X, Topf M, Dorstyn L, Kumar S, Ludtke SJ, et al. Structure of the Drosophila apoptosome at 6.9 a resolution. Structure. 2011;19:128–40. doi: 10.1016/j.str.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Acehan D, Jiang X, Morgan DG, Heuser JE, Wang X, Akey CW. Three-dimensional structure of the apoptosome: implications for assembly, procaspase-9 binding, and activation. Mol Cell. 2002;9:423–32. doi: 10.1016/S1097-2765(02)00442-2. [DOI] [PubMed] [Google Scholar]
- 20.Reubold TF, Wohlgemuth S, Eschenburg S. Crystal structure of full-length Apaf-1: how the death signal is relayed in the mitochondrial pathway of apoptosis. Structure. 2011;19:1074–83. doi: 10.1016/j.str.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 21.Yu X, Wang L, Acehan D, Wang X, Akey CW. Three-dimensional structure of a double apoptosome formed by the Drosophila Apaf-1 related killer. J Mol Biol. 2006;355:577–89. doi: 10.1016/j.jmb.2005.10.040. [DOI] [PubMed] [Google Scholar]
- 22.Cheng TC, Akey IV, Yuan S, Yu Z, Ludtke SJ, Akey CW. A Near-atomic structure of the dark apoptosome provides insight into assembly and activation. Structure. 2017;25:40–52. doi: 10.1016/j.str.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benedict MA, Hu Y, Inohara N, Nunez G. Expression and functional analysis of Apaf-1 isoforms. Extra Wd-40 repeat is required for cytochrome c binding and regulated activation of procaspase-9. J Biol Chem. 2000;275:8461–8. doi: 10.1074/jbc.275.12.8461. [DOI] [PubMed] [Google Scholar]
- 24.Baliga B, Kumar S. Apaf-1/cytochrome c apoptosome: an essential initiator of caspase activation or just a sideshow? Cell Death Differ. 2003;10:16–18. doi: 10.1038/sj.cdd.4401166. [DOI] [PubMed] [Google Scholar]
- 25.Yuan J, Horvitz HR. The Caenorhabditis elegans cell death gene ced-4 encodes a novel protein and is expressed during the period of extensive programmed cell death. Development. 1992;116:309–20. doi: 10.1242/dev.116.2.309. [DOI] [PubMed] [Google Scholar]
- 26.Qi S, Pang Y, Hu Q, Liu Q, Li H, Zhou Y, et al. Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4. Cell. 2010;141:446–57. doi: 10.1016/j.cell.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 27.Yan N, Chai J, Lee ES, Gu L, Liu Q, He J, et al. Structure of the CED-4-CED-9 complex provides insights into programmed cell death in Caenorhabditis elegans. Nature. 2005;437:831–7. doi: 10.1038/nature04002. [DOI] [PubMed] [Google Scholar]
- 28.Wu CC, Bratton SB. DARK apoptosome secrets come to light. Structure. 2011;19:4–6. doi: 10.1016/j.str.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dorstyn L, Mills K, Lazebnik Y, Kumar S. The two cytochrome c species, DC3 and DC4, are not required for caspase activation and apoptosis in Drosophila cells. J Cell Biol. 2004;167:405–10. doi: 10.1083/jcb.200408054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dorstyn L, Read S, Cakouros D, Huh JR, Hay BA, Kumar S. The role of cytochrome c in caspase activation in Drosophila melanogaster cells. J Cell Biol. 2002;156:1089–98. doi: 10.1083/jcb.200111107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dorstyn L, Kumar S. A cytochrome c-free fly apoptosome. Cell Death Differ. 2006;13:1049–51. doi: 10.1038/sj.cdd.4401918. [DOI] [PubMed] [Google Scholar]
- 32.Yuan S, Yu X, Asara JM, Heuser JE, Ludtke SJ, Akey CW. The holo-apoptosome: activation of procaspase-9 and interactions with caspase-3. Structure. 2011;19:1084–96. doi: 10.1016/j.str.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES, et al. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–89. doi: 10.1016/S0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 34.Yan N, Gu L, Kokel D, Chai J, Li W, Han A, et al. Structural, biochemical, and functional analyses of CED-9 recognition by the proapoptotic proteins EGL-1 and CED-4. Mol Cell. 2004;15:999–1006. doi: 10.1016/j.molcel.2004.08.022. [DOI] [PubMed] [Google Scholar]
- 35.Yan N, Xu Y, Shi Y. 2:1 Stoichiometry of the CED-4-CED-9 complex and the tetrameric CED-4: insights into the regulation of CED-3 activation. Cell Cycle. 2006;5:31–4. doi: 10.4161/cc.5.1.2263. [DOI] [PubMed] [Google Scholar]
- 36.Wu D, Chen PJ, Chen S, Hu Y, Nunez G, Ellis RE. C. elegans MAC-1, an essential member of the AAA family of ATPases, can bind CED-4 and prevent cell death. Development. 1999;126:2021–31. doi: 10.1242/dev.126.9.2021. [DOI] [PubMed] [Google Scholar]
- 37.Yang X, Chang HY, Baltimore D. Essential role of CED-4 oligomerization in CED-3 activation and apoptosis. Science. 1998;281:1355–7. doi: 10.1126/science.281.5381.1355. [DOI] [PubMed] [Google Scholar]
- 38.Huang W, Jiang T, Choi W, Qi S, Pang Y, Hu Q, et al. Mechanistic insights into CED-4-mediated activation of CED-3. Genes Dev. 2013;27:2039–48. doi: 10.1101/gad.224428.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chao Y, Shiozaki EN, Srinivasula SM, Rigotti DJ, Fairman R, Shi Y. Engineering a dimeric caspase-9: a re-evaluation of the induced proximity model for caspase activation. PLoS Biol. 2005;3:e183. doi: 10.1371/journal.pbio.0030183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yin Q, Park HH, Chung JY, Lin SC, Lo YC, da Graca LS, et al. Caspase-9 holoenzyme is a specific and optimal procaspase-3 processing machine. Mol Cell. 2006;22:259–68. doi: 10.1016/j.molcel.2006.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan S, Akey CW. Apoptosome structure, assembly, and procaspase activation. Structure. 2013;21:501–15. doi: 10.1016/j.str.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Danot O, Marquenet E, Vidal-Ingigliardi D, Richet E. Wheel of life, wheel of death: a mechanistic insight into signaling by STAND proteins. Structure. 2009;17:172–82. doi: 10.1016/j.str.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 43.Yuan S, Yu X, Topf M, Ludtke SJ, Wang X, Akey CW. Structure of an apoptosome-procaspase-9 CARD complex. Structure. 2010;18:571–83. doi: 10.1016/j.str.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng TC, Hong C, Akey IV, Yuan S, Akey CW. A near atomic structure of the active human apoptosome. Elife. 2016;5:e17755. [DOI] [PMC free article] [PubMed]
- 45.Yu X, Acehan D, Menetret JF, Booth CR, Ludtke SJ, Riedl SJ, et al. A structure of the human apoptosome at 12.8 A resolution provides insights into this cell death platform. Structure. 2005;13:1725–35. doi: 10.1016/j.str.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 46.Riedl SJ, Salvesen GS. The apoptosome: signalling platform of cell death. Nat Rev Mol Cell Biol. 2007;8:405–13. doi: 10.1038/nrm2153. [DOI] [PubMed] [Google Scholar]
- 47.Riedl SJ, Li W, Chao Y, Schwarzenbacher R, Shi Y. Structure of the apoptotic protease-activating factor 1 bound to ADP. Nature. 2005;434:926–33. doi: 10.1038/nature03465. [DOI] [PubMed] [Google Scholar]
- 48.Li Y, Zhou M, Hu Q, Bai XC, Huang W, Scheres SH, et al. Mechanistic insights into caspase-9 activation by the structure of the apoptosome holoenzyme. Proc Natl Acad Sci USA. 2017;114:1542–7. doi: 10.1073/pnas.1620626114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuan S, Topf M, Reubold TF, Eschenburg S, Akey CW. Changes in Apaf-1 conformation that drive apoptosome assembly. Biochemistry. 2013;52:2319–27. doi: 10.1021/bi301721g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu CC, Lee S, Malladi S, Chen MD, Mastrandrea NJ, Zhang Z, et al. The Apaf-1 apoptosome induces formation of caspase-9 homo- and heterodimers with distinct activities. Nat Commun. 2016;7:13565. doi: 10.1038/ncomms13565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi Y. Mechanical aspects of apoptosome assembly. Curr Opin Cell Biol. 2006;18:677–84. doi: 10.1016/j.ceb.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 52.Cain K. Chemical-induced apoptosis: formation of the Apaf-1 apoptosome. Drug Metab Rev. 2003;35:337–63. doi: 10.1081/DMR-120026497. [DOI] [PubMed] [Google Scholar]
- 53.Voegtli WC, Madrona AY, Wilson DK. The structure of Aip1p, a WD repeat protein that regulates Cofilin-mediated actin depolymerization. J Biol Chem. 2003;278:34373–9. doi: 10.1074/jbc.M302773200. [DOI] [PubMed] [Google Scholar]
- 54.Adrain C, Slee EA, Harte MT, Martin SJ. Regulation of apoptotic protease activating factor-1 oligomerization and apoptosis by the WD-40 repeat region. J Biol Chem. 1999;274:20855–60. doi: 10.1074/jbc.274.30.20855. [DOI] [PubMed] [Google Scholar]
- 55.Mendez DL, Akey IV, Akey CW, Kranz RG. Oxidized or reduced cytochrome c and axial ligand variants all form the apoptosome in vitro. Biochemistry. 2017;56:2766–9. doi: 10.1021/acs.biochem.7b00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Azad T, Tashakor A, Rahmati F, Hemmati R, Hosseinkhani S. Oscillation of apoptosome formation through assembly of truncated Apaf-1. Eur J Pharmacol. 2015;760:64–71. doi: 10.1016/j.ejphar.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 57.Renatus M, Stennicke HR, Scott FL, Liddington RC, Salvesen GS. Dimer formation drives the activation of the cell death protease caspase 9. Proc Natl Acad Sci USA. 2001;98:14250–5. doi: 10.1073/pnas.231465798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bratton SB, Salvesen GS. Regulation of the Apaf-1-caspase-9 apoptosome. J Cell Sci. 2010;123:3209–14. doi: 10.1242/jcs.073643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pop C, Timmer J, Sperandio S, Salvesen GS. The apoptosome activates caspase-9 by dimerization. Mol Cell. 2006;22:269–75. doi: 10.1016/j.molcel.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 60.Cain K, Bratton SB, Cohen GM. The Apaf-1 apoptosome: a large caspase-activating complex. Biochimie. 2002;84:203–14. doi: 10.1016/S0300-9084(02)01376-7. [DOI] [PubMed] [Google Scholar]
- 61.Hu Q, Wu D, Chen W, Yan Z, Yan C, He T, et al. Molecular determinants of caspase-9 activation by the Apaf-1 apoptosome. Proc Natl Acad Sci USA. 2014;111:16254–61. doi: 10.1073/pnas.1418000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shi Y. Apoptosome: the cellular engine for the activation of caspase-9. Structure. 2002;10:285–8. doi: 10.1016/S0969-2126(02)00732-3. [DOI] [PubMed] [Google Scholar]
- 63.Boatright KM, Renatus M, Scott FL, Sperandio S, Shin H, Pedersen IM, et al. A unified model for apical caspase activation. Mol Cell. 2003;11:529–41. doi: 10.1016/S1097-2765(03)00051-0. [DOI] [PubMed] [Google Scholar]
- 64.Shi Y. Caspase activation: revisiting the induced proximity model. Cell. 2004;117:855–8. doi: 10.1016/j.cell.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 65.Wurstle ML, Rehm M. A systems biology analysis of apoptosome formation and apoptosis execution supports allosteric procaspase-9 activation. J Biol Chem. 2014;289:26277–89. doi: 10.1074/jbc.M114.590034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Qin H, Srinivasula SM, Wu G, Fernandes-Alnemri T, Alnemri ES, Shi Y. Structural basis of procaspase-9 recruitment by the apoptotic protease-activating factor 1. Nature. 1999;399:549–57. doi: 10.1038/21124. [DOI] [PubMed] [Google Scholar]
- 67.Su TW, Yang CY, Kao WP, Kuo BJ, Lin SM, Lin JY, et al. Structural insights into DD-fold assembly and caspase-9 activation by the apaf-1 apoptosome. Structure. 2017;25:407–20. doi: 10.1016/j.str.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 68.Malladi S, Challa-Malladi M, Fearnhead HO, Bratton SB. The Apaf-1*procaspase-9 apoptosome complex functions as a proteolytic-based molecular timer. EMBO J. 2009;28:1916–25. doi: 10.1038/emboj.2009.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Baliga BC, Read SH, Kumar S. The biochemical mechanism of caspase-2 activation. Cell Death Differ. 2004;11:1234–41. doi: 10.1038/sj.cdd.4401492. [DOI] [PubMed] [Google Scholar]
- 70.Dorstyn L, Kumar S. A biochemical analysis of the activation of the Drosophila caspase DRONC. Cell Death Differ. 2008;15:461–70. doi: 10.1038/sj.cdd.4402288. [DOI] [PubMed] [Google Scholar]
- 71.Read SH, Baliga BC, Ekert PG, Vaux DL, Kumar S. A novel Apaf-1-independent putative caspase-2 activation complex. J Cell Biol. 2002;159:739–45. doi: 10.1083/jcb.200209004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Butt AJ, Harvey NL, Parasivam G, Kumar S. Dimerization and autoprocessing of the Nedd2 (caspase-2) precursor requires both the prodomain and the carboxyl-terminal regions. J Biol Chem. 1998;273:6763–8. doi: 10.1074/jbc.273.12.6763. [DOI] [PubMed] [Google Scholar]
- 73.Schweizer A, Briand C, Grutter MG. Crystal structure of caspase-2, apical initiator of the intrinsic apoptotic pathway. J Biol Chem. 2003;278:42441–7. doi: 10.1074/jbc.M304895200. [DOI] [PubMed] [Google Scholar]
- 74.McStay GP, Salvesen GS, Green DR. Overlapping cleavage motif selectivity of caspases: implications for analysis of apoptotic pathways. Cell Death Differ. 2008;15:322–31. doi: 10.1038/sj.cdd.4402260. [DOI] [PubMed] [Google Scholar]
- 75.Ferraro E, Pesaresi MG, De Zio D, Cencioni MT, Gortat A, Cozzolino M, et al. Apaf1 plays a pro-survival role by regulating centrosome morphology and function. J Cell Sci. 2011;124:3450–63. doi: 10.1242/jcs.086298. [DOI] [PubMed] [Google Scholar]
- 76.Mouhamad S, Galluzzi L, Zermati Y, Castedo M, Kroemer G. Apaf-1 deficiency causes chromosomal instability. Cell Cycle. 2007;6:3103–7. doi: 10.4161/cc.6.24.5046. [DOI] [PubMed] [Google Scholar]
- 77.Zermati Y, Mouhamad S, Stergiou L, Besse B, Galluzzi L, Boehrer S, et al. Nonapoptotic role for Apaf-1 in the DNA damage checkpoint. Mol Cell. 2007;28:624–37. doi: 10.1016/j.molcel.2007.09.030. [DOI] [PubMed] [Google Scholar]