Abstract
The microRNA-34a is a well-studied tumor suppressor microRNA (miRNA) and a direct downstream target of TP53 with roles in several pathways associated with oncogenesis, such as proliferation, cellular growth, and differentiation. Due to its broad tumor suppressive activity, it is not surprising that miR34a expression is altered in a wide variety of solid tumors and hematological malignancies. However, the mechanisms by which miR34a is regulated in these cancers is largely unknown. In this study, we find that a long noncoding RNA transcribed antisense to the miR34a host gene, is critical for miR34a expression and mediation of its cellular functions in multiple types of human cancer. We name this long noncoding RNA lncTAM34a, and characterize its ability to facilitate miR34a expression under different types of cellular stress in both TP53-deficient and wild-type settings.
Introduction
In recent years advances in functional genomics have revolutionized our understanding of the human genome. Evidence now points to the fact that approximately 75% of the genome is transcribed but only ~1.2% of this is responsible for encoding proteins1,2. Of these recently identified elements, long noncoding (lnc) RNAs are defined as transcripts exceeding 200 base pairs (bp) in length with a lack of a functional open reading frame. Some lncRNAs are dually classified as antisense (as) RNAs that are expressed from the same locus as a sense transcript in the opposite orientation. Current estimates using high-throughput transcriptome sequencing, indicate that up to 20–40% of the approximately 20,000 protein-coding genes exhibit antisense transcription3–5.
Systematic large-scale studies have shown aberrant expression of asRNAs to be associated with tumorigenesis6 and, although characterization of several of these has identified asRNA-mediated regulation of multiple well-known tumorigenic factors7,8, the vast majority of potential tumor-associated asRNAs have not yet been characterized. The known mechanisms by which asRNAs accomplish their regulatory functions are diverse, and include recruitment of chromatin modifying factors8,9, acting as microRNA (miRNA) sponges10, and causing transcriptional interference11.
Responses to cellular stress, e.g., DNA damage, sustained oncogene expression, and nutrient deprivation, are all tightly controlled cellular pathways that are almost universally dysregulated in cancer. Cellular signaling, in response to these types of stresses, often converges on the transcription factor TP53 that regulates transcription of coding and noncoding downstream targets. One important noncoding target of TP53 is the tumor suppressor miRNA known as miR34a12. Upon TP53 activation miR34a expression is increased allowing it to down-regulate target genes involved in cellular pathways such as growth factor signaling, apoptosis, differentiation, and cellular senescence13,14. Thus, miR34a is a crucial factor in mediating activated TP53 response and, the fact that it is often deleted or down-regulated in human cancers indicates its tumor suppressive effect and makes it a valuable prognostic marker15–19. Reduced miR34a transcription is mediated via epigenetic regulation in many solid tumors, including colorectal-, pancreatic-, and ovarian cancer20, as well as numerous types of hematological malignancies21. In addition, miR34a has been shown to be transcriptionally regulated via TP53 homologs, TP63 and TP73, other transcription factors, e.g., STAT3 and MYC, and, in addition, posttranscriptionally through miRNA sponging by the NEAT1 lncRNA22–26. Despite these findings, the mechanisms underlying miR34a regulation in the context of oncogenesis have not yet been fully elucidated.
Studies across multiple cancer types have reported a decrease in oncogenic phenotypes when miR34a expression is induced in a TP53-null background, although endogenous mechanisms for achieving this have not yet been discovered18,27–30. In addition, previous reports from large-scale studies interrogating global TP53-mediated regulation of lncRNAs have identified a lncRNA (known as RP3-510D11.2 and LINC01759) originating in the antisense orientation from the miR34a locus that is induced upon numerous forms of cellular stress31–35. Despite this, none of these studies have functionally characterized this transcript, which we name Long-Non-Coding Transcriptional Activator of MiR34a (lncTAM34a). In this study we functionally characterize the lncTAM34a transcript, and find that it positively regulates miR34a expression resulting in a decrease of several tumorigenic phenotypes. Furthermore, we find that lncTAM34a-mediated up-regulation of miR34a is sufficient to induce endogenous cellular mechanisms counteracting several types of stress stimuli in a TP53-deficient background. Finally, similar to the functional roles of antisense transcription at protein-coding genes, we identify a rare example of an antisense RNA capable of regulating a cancer-associated miRNA.
Results
lncTAM34a is a broadly expressed noncoding transcript whose levels correlate with miR34a expression
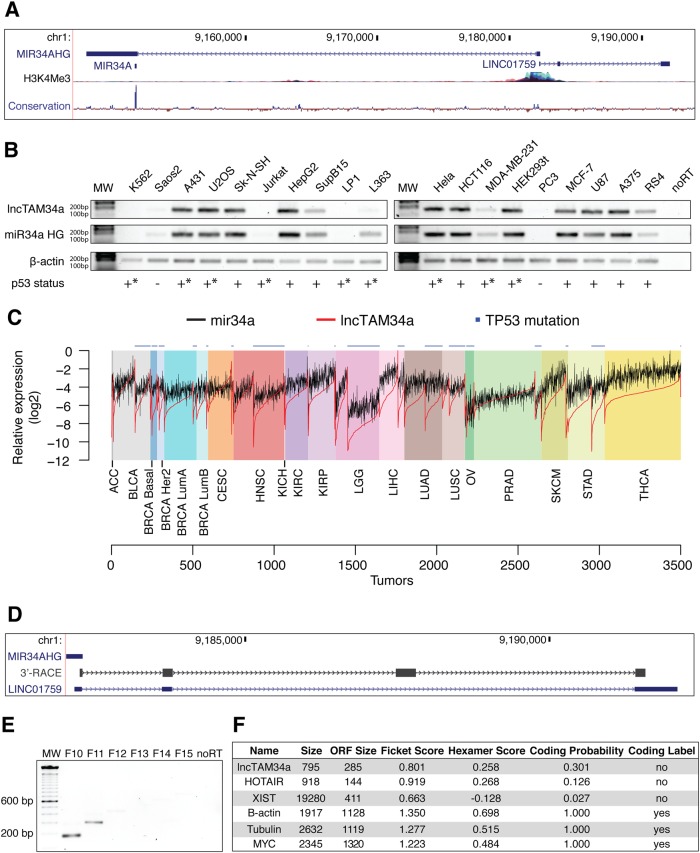
lncTAM34a is transcribed in a “head-to-head” orientation with approximately 100 bp overlap with the miR34a host gene (HG) (Fig. 1a). Due to the fact that sense/antisense pairs can be both concordantly and discordantly expressed, we sought to evaluate this relationship in the case of miR34a HG and its asRNA. Using a diverse panel of cancer cell lines, we detected co-expression of both the miR34a HG and lncTAM34a (Fig. 1b). We used cell lines with a known TP53 status in the panel due to previous reports that miR34a and lncTAM34a are known downstream targets of TP53. These results indicate that miR34a HG and lncTAM34a are co-expressed and that their expression levels are related to TP53 status, with TP53−/− cells tending to have decreased or undetectable expression of both transcripts.
Fig. 1. Characterization of the lncTAM34a transcript.
a Architecture of the miR34a locus (hg38, RefSeq) including miR34a HG, mature miR34a, and lncTAM34a (LINC01759). H3K4me3 ChIP-seq data, indicating the active promoter region, and conservation are also shown. b Semiquantitative PCR data from the screening of a panel of cancer cell lines. Wild-type TP53 is indicated with +, − indicates null, and +* represents either a nonnull TP53 mutation or wild-type TP53 with mechanisms present that inhibit its function (e.g., SV40 large T antigen in HEK293T cells). c TCGA correlation analysis. Expression was log2 normalized to the maximum expression value. Nonsynonymous TP53 mutations are indicated on the top of the plot (cancer type abbreviation definitions and corresponding statistics are in Fig. 1-Supplement 1). d 3′-RACE sequencing results and the annotated lncTAM34a (LINC01759) are shown. e Semiquantitative PCR results from the primer walk assay (i.e., common reverse primer (exon 2) and forward primers (F10–F15) staggered upstream of lncTAM34a’s annotated start site) performed using HEK293T cells (Fig. 1-Supplement 2a details primer placement). f Coding-potential analysis assessed via the coding-potential assessment tool including lncTAM34a, two known noncoding RNAs (HOTAIR and XIST), and three protein-coding RNAs (β-actin, Tubulin, and MYC)
We next sought to analyze primary cancer samples to examine whether a correlation between lncTAM34a and miR34a expression levels could be identified. We utilized RNA sequencing data from The Cancer Genome Atlas (TCGA) after stratifying patients by cancer type, TP53 status, and, in the case of breast cancer, cancer subtypes. The results indicate that lncTAM34a and miR34a expression are strongly correlated in the vast majority of cancer types examined, both in the presence and absence of wild-type TP53 (Fig. 1c, Supplementary Figure 1A). The results also further confirm that the expression levels of both miR34a and lncTAM34a are significantly reduced in patients with nonsynonymous TP53 mutations (Supplementary Figure 1B).
Next, we aimed to gain a thorough understanding of lncTAM34a’s molecular characteristics and cellular localization. To experimentally determine the 3′ termination site for the lncTAM34a transcript we performed 3′ rapid amplification of cDNA ends (RACE) using the U2OS osteosarcoma cell line that exhibited high endogenous levels of lncTAM34a in the cell panel screening. Sequencing the cloned cDNA indicated that the transcripts 3′ transcription termination site is 525 bp upstream of the lncTAM34a transcript’s annotated termination site (Fig. 1d). Next, we characterized the lncTAM34a 5′ transcription start site by carrying out a primer walk assay, i.e., a common reverse primer was placed in exon 2 and forward primers were gradually staggered upstream of lncTAM34a’s annotated start site (Supplementary Figure 2A). Our results indicated that the 5′ start site for lncTAM34a is in fact approximately 90 (F11 primer)–220 bp (F12 primer) upstream of the annotated start site (Fig. 1e). Polyadenylation status was evaluated via cDNA synthesis with either random nanomers or oligo(DT) primers followed by semiquantitative PCR, which showed that the lncTAM34a is polyadenylated although the unspliced form seems to only be present in a polyadenylation negative state (Supplementary Figure 2B). Furthermore, we investigated the propensity of lncTAM34a to be alternatively spliced in U2OS cells, using PCR cloning followed by sequencing and found that the transcript is posttranscriptionally spliced to form multiple isoforms (Supplementary Figure 2C). In order to evaluate the subcellular localization of lncTAM34a, we made use of RNA sequencing data from five cancer cell lines included in the ENCODE36 project that had been fractionated into cytosolic and nuclear fractions. The analysis revealed that the lncTAM34a transcript primarily localizes to the nucleus with only a minor fraction in the cytosol (Supplementary Figure 2D).
Lastly, we utilized several approaches to evaluate the coding potential of the lncTAM34a transcript. The Coding-Potential Assessment Tool is a bioinformatics-based tool that uses a logistic regression model to evaluate coding-potential by examining open reading frame (ORF) length, ORF coverage, Fickett score, and hexamer score37. Results indicated that lncTAM34a has a similar low coding capacity to known noncoding transcripts such as HOTAIR and XIST (Fig. 1f). We further confirmed these results using the Coding-Potential Calculator that uses a support vector machine-based classifier and accesses an alternate set of discriminatory features (Supplementary Figure 2E)38. Finally, we downloaded mass spectrometry spectra for 11 cancer cell lines39, 7 of which were also present in the cell line panel above (Fig. 1b), and searched it against a database of human protein sequences which also contained the 6 frame translation of lncTAM34a. However, we did not manage to detect any peptides matching the sequence in any of the 11 cell lines. Taken together our results indicate that lncTAM34a is not a coding transcript and that it is not translated to any significant degree.
TP53-mediated regulation of lncTAM34a expression
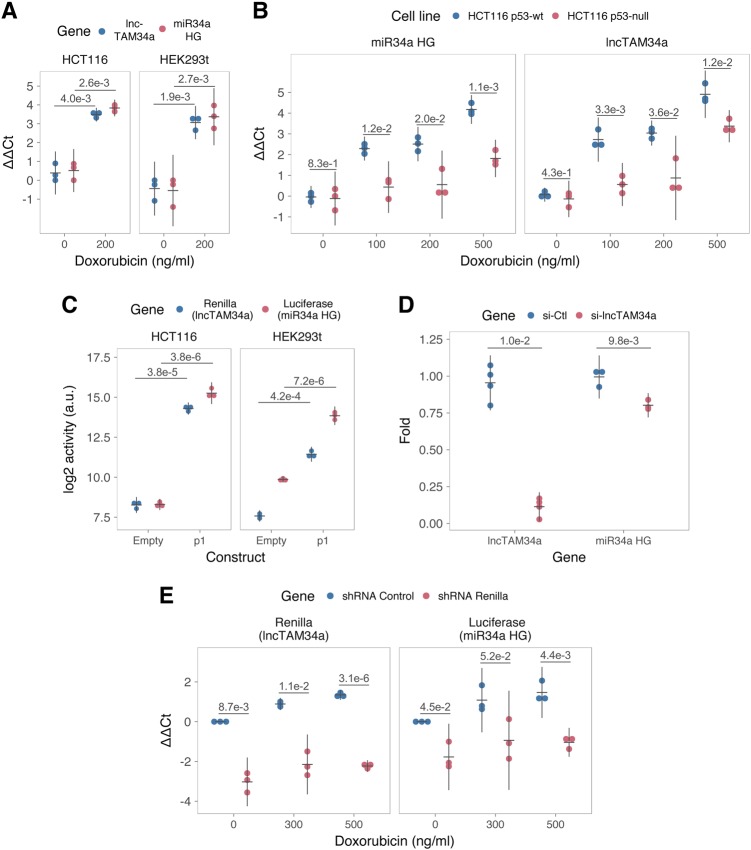
miR34a is a known downstream target of TP53 and has been previously shown to exhibit increased expression within multiple contexts of cellular stress. Several global analyses of TP53-regulated lncRNAs have also shown lncTAM34a to be induced upon TP53 activation31–35. To confirm these results in our biological systems, we treated HEK293T, embryonic kidney cells, and HCT116, colorectal cancer cells, with the DNA damaging agent doxorubicin to activate TP53. QPCR-mediated measurements of both miR34a HG and lncTAM34a indicated that their expression levels were increased in response to doxorubicin treatment in both cell lines (Fig. 2a). To assess whether TP53 was responsible for the increase in lncTAM34a expression upon DNA damage, we treated TP53+/+ and TP53−/− HCT116 cells with increasing concentrations of doxorubicin and monitored the expression of both miR34a HG and lncTAM34a. We observed a dose-dependent increase in both miR34a HG and lncTAM34a expression levels with increasing amounts of doxorubicin, revealing that these two transcripts are co-regulated, although, this effect was largely abrogated in TP53−/− cells (Fig. 2b). These results indicate that TP53 activation increases lncTAM34a expression upon DNA damage. Nevertheless, TP53−/− cells also showed a dose-dependent increase in both miR34a HG and lncTAM34a, suggesting that additional factors, other than TP53 are capable of initiating an increase in expression of both of these transcripts upon DNA damage.
Fig. 2. TP53-mediated regulation of the miR34a locus.
a Evaluating the effects of 24 h of treatment with 200 ng/ml doxorubicin on lncTAM34a and miR34a HG in HCT116 and HEK293T cells.* b Monitoring miR34a HG and lncTAM34a expression levels during 24 h of doxorubicin treatment in TP53+/+ and TP53−/− HCT116 cells.* c Quantification of luciferase and renilla levels after transfection of HCT116 and HEK293T cells with the p1 construct (Fig. 2-Supplement 2 contains a schematic representation of the p1 construct).* d lncTAM34a (n = 4) and miR34a HG (n = 3) levels after 48 h siRNA-mediated knockdown of lncTAM34a in U2OS cells.* e HCT116 cells were co-transfected with the p1 construct and shRNA renilla or shRNA control and subsequently treated with increasing doses of doxorubicin. Twenty-four hour posttreatment, cells were harvested and renilla and luciferase levels were measured using QPCR.* *Individual points represent results from independent experiments, error bars show the 95% CI, black horizontal lines represent the mean, and p values are shown over long horizontal lines indicating the comparison tested. All experiments in Fig. 2 were performed in biological triplicate unless otherwise stated
The head-to-head orientation of miR34a HG and lncTAM34a, suggests that transcription is initiated from a single promoter in a bidirectional manner (Fig. 1a). To investigate whether miR34a HG and lncTAM34a are transcribed from the same promoter as divergent transcripts, we cloned the previously reported miR34a HG promoter, a ~300 bp region including the TP53 binding site and the majority of the first exon of both transcripts, into a luciferase/renilla dual reporter vector (Supplementary Figure 3A, B)12. We, hereafter, refer to this construct as p1. Upon transfection of p1 into HCT116 and HEK293T cell lines we observed increases in both luciferase and renilla indicating that miR34a HG and lncTAM34a expression can be regulated by a single promoter contained within the p1 construct (Fig. 2c).
lncTAM34a facilitates miR34a induction in response to DNA damage
We hypothesized that lncTAM34a may regulate miR34a HG levels and investigated this via short interfering (si) RNA-mediated knockdown of lncTAM34a and examining the effects on miR34a HG. The results show that a decrease in lncTAM34a levels causes a concordant decrease in miR34a HG (Fig. 2d) indicating that lncTAM34a positively regulates miR34a HG levels.
Knockdown of endogenous lncTAM34a is complicated by its various isoforms (Supplementary Figure 2C) and targeting individual isoforms is not possible due to the structure of the locus. We hypothesized that the overlapping regions of the sense and antisense transcripts may mediate the observed regulation and, for these reasons, we utilized the p1 construct to further evaluate the regulatory role of lncTAM34a on miR34a HG. Accordingly, we first cotransfected the p1 construct, containing the overlapping region of the two transcripts, and two different short hairpin (sh) RNAs targeting renilla into HEK293T cells and subsequently measured luciferase and renilla expression. The results indicated that shRNA-mediated knockdown of the p1-renilla transcript (corresponding to lncTAM34a) caused p1-luciferase (corresponding to miR34a HG) levels to concomitantly decrease (Supplementary Figure 3C). These results further confirm that lncTAM34a positively regulates levels of miR34a HG and, moreover, that the transcriptional product of lncTAM34a within the p1 construct contributes to inducing a miR34a response.
To further support these conclusions and better understand the role of lncTAM34a during TP53 activation, TP53+/+ HCT116 cells were cotransfected with p1 and shRNA renilla (2.1) and subsequently treated with increasing doses of doxorubicin. Again, the results showed a concomitant reduction in luciferase levels upon knockdown of p1-renilla, i.e., the lncTAM34a corresponding segment of the p1 transcript (Fig. 2e). Furthermore, the results showed that in the absence of p1-renilla the expected induction of p1-luciferase in response to TP53 activation by DNA damage is abrogated. Collectively these results indicate that lncTAM34a positively regulates miR34a expression and furthermore, suggests that it plays an important role in TP53-mediated miR34a response to DNA damage.
lncTAM34a can regulate miR34a HG independently of TP53
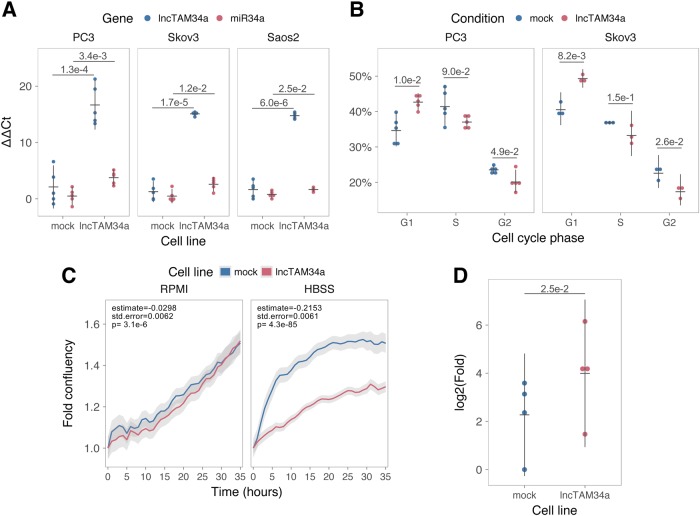
Despite the fact that TP53 regulates miR34a HG and lncTAM34a expression, our results showed that other factors are also able to regulate this locus (Fig. 2b). Utilizing a lentiviral system, we stably overexpressed the most abundant isoform (Supplementary Figure 7) of lncTAM34a in three TP53-null cell lines, PC3 (prostate cancer), Saos2 (osteogenic sarcoma), and Skov3 (ovarian adenocarcinoma). We first analyzed the levels of lncTAM34a in these stable cell lines, compared to HEK293T cells, which have high endogenous levels of lncTAM34a. On average, the overexpression was approximately 30-fold higher in the overexpression cell lines than in HEK293T cells, roughly corresponding to physiologically relevant levels in cells encountering a stress stimulus, such as DNA damage (Supplementary Figure 4A). Analysis of miR34a levels in the lncTAM34a overexpressing cell lines showed that this overexpression resulted in a concomitant increase in the expression of miR34a in all three cell lines (Fig. 3a). These results indicate that, in the absence of TP53, miR34a expression may be rescued by activating lncTAM34a expression.
Fig. 3. lncTAM34a positively regulates miR34a and its associated phenotypes.
a QPCR-mediated quantification of miR34a expression in cell lines stably overexpressing lncTAM34a.* b Cell-cycle analysis comparing stably overexpressing lncTAM34a cell lines to the respective mock control.* c Analysis of cellular growth over time in lncTAM34a overexpressing PC3 cells. Fold confluency (% confluency/time 0 % confluency) is indicated on the y-axis. Colored lines indicate the polynomial regression model where mean fold confluency from three independent experiments was modeled as a function of time and cell line. The grey shadows illustrate the 95% confidence interval for the regression. Estimates, standard error (std.error), and p values for the cell line covariate for each model are indicated in the upper left hand corner. d Differential phosphorylated polymerase II binding in lncTAM34a overexpressing PC3 cells.* *Individual points represent results from independent experiments, error bars show the 95% CI, black horizontal lines represent the mean, and p values are shown over long horizontal lines indicating the comparison tested
miR34a has been previously shown to regulate cell-cycle progression, with miR34a induction causing G1 arrest12,40. Cell-cycle analysis via determination of DNA content showed a significant increase in G1 phase cells and a concomitant decrease in G2 phase cells in the PC3 and Skov3 lncTAM34a overexpressing cell lines, indicating G1 arrest (Fig. 3b, Supplementary Figure 4B). The effects of miR34a on the cell cycle are mediated by its ability to target cell-cycle regulators such as cyclin D1 (CCND1)41. Quantification of both CCND1 RNA expression (Supplementary Figure 4B) and protein levels (Supplementary Figure 4C) in the PC3 lncTAM34a overexpressing cell line showed a significant decrease of CCND1 levels compared to the mock control. Collectively, these results indicate that lncTAM34a-mediated induction of miR34a is sufficient to result in the corresponding miR34a-directed effects on cell cycle.
miR34a is also a well-known inhibitor of cellular growth via its ability to negatively regulate growth factor signaling. Furthermore, starvation has been shown to induce miR34a expression causing inactivation of numerous prosurvival growth factors13. We further interrogated the effects of lncTAM34a overexpression by monitoring the growth of the PC3 stable cell lines in both normal and starvation conditions via confluency measurements over a 35-h period. Under normal growth conditions there is a small but significant reduction (P = 3.1e-6; polynomial regression, Fig. 3c, Supplementary Figure 5A) in confluency in the lncTAM34a overexpressing cell lines compared to mock control. However, these effects on cell growth are clearly increased in starvation conditions (P = 4.3e-85; polynomial regression; Fig. 3c, Supplementary Figure 5A). Specifically measuring proliferation rate, as opposed to confluency, largely confirms the observation that increases in lncTAM34a expression mediate decreases in cell cycle, although the degree to which cellular stress effects these changes differs when measuring confluency or proliferation (Supplementary Figure 5B). This suggests that additional cellular phenotypes, such as morphology or cell adhesion capability, may also be affected by increased lncTAM34a expression in contexts of cellular stress. In summary, we find that overexpression of lncTAM34a is sufficient to increase miR34a expression and gives rise to known phenotypes observed upon induction of miR34a.
lncTAM34a transcriptionally activates miR34a HG
Antisense RNAs have been reported to mediate their effects both via transcriptional and posttranscriptional mechanisms. Due to the fact that miR34a expression is undetected in wild-type PC3 cells (Fig. 1b) but, upon overexpression of lncTAM34a, increases to detectable levels, we hypothesized that lncTAM34a is capable of regulating miR34a expression via a transcriptional mechanism. To substantiate this hypothesis, we performed chromatin immunoprecipitation (ChIP) for phosphorylated polymerase II (polII) at the miR34a HG promoter in both lncTAM34a overexpressing and mock control cell lines. Indeed, we found a clear increase in phosphorylated polII binding at the miR34a promoter upon lncTAM34a overexpression indicating the ability of lncTAM34a to transcriptionally regulate miR34a levels (Fig. 3d).
Low-lncTAM34a expression levels are associated with decreased survival
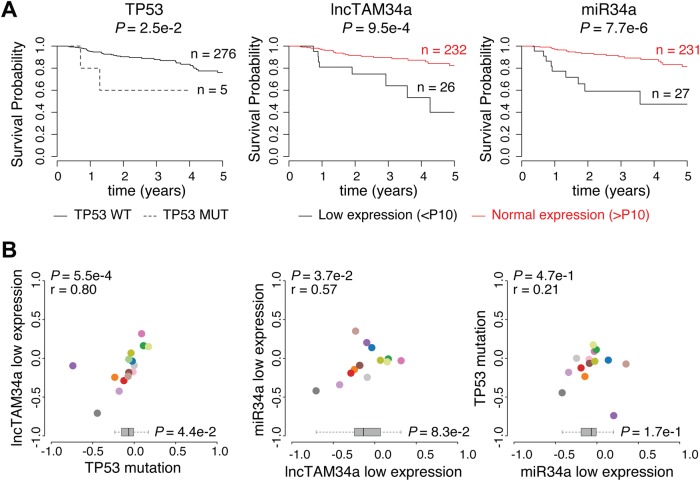
As TP53 mutations and low expression of miR34a have been associated with worse prognosis in cancer, we compared survival rates of samples with low expression of lncTAM34a (bottom 10th percentile) to control samples in 17 cancer types from TCGA (Supplementary Figure 6)17–19. To correct for the effect of TP53 mutations we focused on non-TP53 mutated samples, and noted a worse survival for the low-expression group in several cancers. This effect was most pronounced in papillary kidney cancer (unadjusted P = 0.00095; Fig. 4a). By systematically comparing 5-year survival probabilities between the low-expression group and the control group for each cancer we found a median reduction of 5-year survival probability of 9.6% (P = 0.083; Wilcoxon signed rank test; Fig. 4b). Furthermore, we found that lncTAM34a expression showed similar patterns in terms of direction and strength of association with 5-year survival probability as miR34a expression (r = 0.57, P = 0.037) and TP53 mutations (r = 0.80, P = 0.00054) across the different cancer types (Fig. 4b). Although these results do not implicate any causal relationship, they do indicate a striking similarity between the association of worse prognosis and TP53 mutations, low miR34a, and low-lncTAM34a expression.
Fig. 4. Survival analysis in TCGA cancers.
a Kaplan-Meier survival curves comparing the effects of TP53-mutated samples (left), low-lncTAM34a expression (middle) and low-miR34a expression (right) to control samples in papillary kidney cancer (results for other cancers in Fig. 4-Supplement 1). Middle and right panel include only TP53 wild-type patients where RNAseq data exists. b Correlation analysis between the effects on the 5-year survival probability of TP53-mutated samples, low-lncTAM34a expression, and low-miR34a expression as indicated. For each variable the 5-year survival probability was compared to the control group (negative values indicate lower survival and positive values indicate higher survival). Spearman correlation coefficients are given on top left of each plot. Each dot indicates one cancer type (see Fig.1c for legend). Boxplots on the bottom summarize the effects for the parameter on the x-axis, with indication of p values, as calculated using paired Wilcoxon signed rank test. Low expression was defined as TP53 nonmutated samples having expression values in the bottom 10th percentile
Discussion
Multiple studies have previously shown asRNAs to be crucial for the appropriate regulation of cancer-associated protein-coding genes and that their dysregulation can lead to perturbance of tumor suppressive and oncogenic pathways, as well as, cancer-related phenotypes6,7,42,43. Here we show that asRNAs are also capable of regulating cancer-associated miRNAs resulting in similar consequences as protein-coding gene dysregulation (Fig. 5). Interestingly, we show that, both in the presence and absence of TP53, lncTAM34a provides an additional regulatory level to control miR34a expression in both homeostasis and upon encountering various forms of cellular stress. Furthermore, we find that lncTAM34a-mediated increase in miR34a expression is sufficient to drive the appropriate cellular responses to these stress stimuli (Figs. 2d and 3c). Previous studies have exploited various molecular biology methods to up-regulate miR34a expression in cells lacking wild-type TP5318,27–30. In this study, we demonstrate a novel, endogenous mechanism of miR34a regulation that has similar phenotypic outcomes as has been previously shown for miR34a induction in a TP53-deficient background.
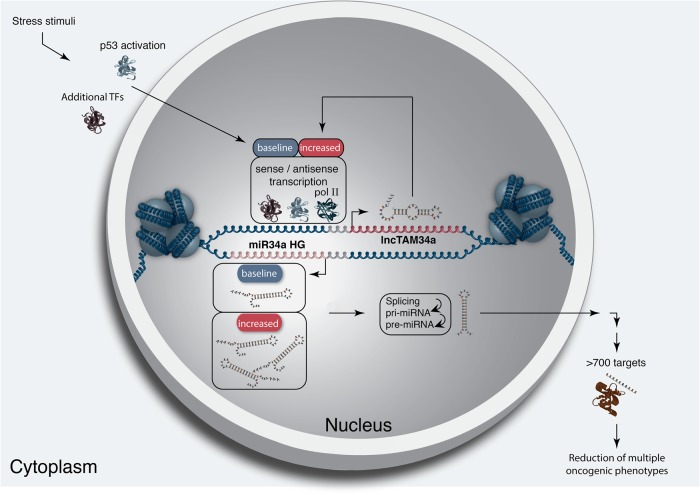
Fig. 5. A graphical summary of the proposed lncTAM34a function.
Stress stimuli, originating in the cytoplasm or nucleus, activate TP53 as well as additional factors. These factors then bind to the miR34a promoter and drive baseline transcription levels of the sense and antisense strands. lncTAM34a serves to further increase miR34a HG transcription levels resulting in enrichment of polymerase II at the miR34a promoter and a positive feed-forward loop. miR34a HG then, in turn, is spliced and processed in multiple steps before the mature miR34a binds to the RISC complex allowing it to repress its targets and exert its tumor suppressive effects
In agreement with previous studies, we demonstrate that upon encountering various types of cellular stress, TP53 in concert with additional factors initiates transcription at the miR34a locus, thus increasing the levels of lncTAM34a and miR34a31–35. We found that overexpression of lncTAM34a leads to recruitment of polII to the miR34a promoter and hypothesize that lncTAM34a may provide positive feedback for miR34a expression whereby it serves as a scaffold for the recruitment of additional factors that facilitate polII-mediated transcription. In this manner, miR34a expression is induced, driving a shift toward senescence, a reduction in growth factor signaling, and in some cases, apoptosis. On the other hand, in cells without functional TP53, other factors, which typically act independently or in concert with TP53, may initiate transcription of the miR34a locus. Due to the fact that lncTAM34a can alter miR34a expression in these cells, we speculate that it is interacting with one of these additional factors, possibly recruiting it to the miR34a locus in order to drive miR34a transcription, similar to mechanisms described for other lncRNAs44–46. The head-to-head orientation of the miR34a HG and lncTAM34a causes sequence complementarity between the RNA and the promoter DNA, making targeting by direct binding an attractive mechanism. Previous reports have also illustrated the ability of asRNAs to form hybrid DNA:RNA R-loops and, thus, facilitate an open chromatin structure and the transcription of the sense gene47. The fact that the p1 construct only contains a small portion (~300 bp) of the lncTAM34a transcript indicates that this portion is sufficient to give rise to at least a partial miR34a inducing response and therefore, that lncTAM34a may be able to facilitate miR34a expression independent of additional factors (Fig. 2d, Supplementary Figure 3C). Nevertheless, further work will need to be performed to explore the exact mechanism whereby lncTAM34a regulates miR34a gene expression.
An antisense transcript arising from the miR34a locus, Lnc34a, has been previously reported to negatively regulate the expression of miR34a48. Although the Lnc34a and lncTAM34a transcripts share some sequence similarity, we believe them to be separate RNAs that are, potentially, different isoforms of the same gene. We utilized CAGE and RNAseq data from the ENCODE project to evaluate the presence of lncTAM34a and Lnc34a in 28 and 36 commonly used cancer cell lines, respectively. Although the results show the presence of lncTAM34a in these cell lines, we find no evidence for Lnc34a transcription (Supplementary Figures 7 and 8). These results are in line with the findings of Wang et al. indicating that Lnc34a is highly expressed in colon cancer stem cell spheres compared to all other cell types used in their study and may not be broadly expressed in other tissues or tumor types. The fact that lncTAM34a and Lnc34a would appear to have opposing roles in their regulation of miR34a, further underlines the complexity of the regulation at this locus.
Clinical trials utilizing miR34a replacement therapy have previously been conducted but, disappointingly, were terminated after adverse side effects of an immunological nature were observed in several of the patients14. Although it is not presently clear if these side effects were caused by miR34a or the liposomal carrier used to deliver the miRNA, the multitude of evidence indicating miR34a’s crucial role in oncogenesis still makes its therapeutic induction an interesting strategy and needs further investigation. Our results indicate an association between survival probability and low-lncTAM34a expression making it an attractive candidate for controlled preclinical studies. Due to the lncTAM34a-mediated positive feedback on miR34a expression, initiation of this feedback mechanism may provide a sustained miR34a induction in a relatively more robust manner than miR34a replacement alone. In summary, our results have identified lncTAM34a as a vital component in the regulation of miR34a and its particular importance in typical examples of cellular stress encountered in cancer. On a broader level, the conclusions drawn in this study provide an example of asRNA-mediated regulation of a clinically relevant cancer-associated miRNA and contribute to fundamental knowledge concerning miR34a regulation.
Materials and methods
Cell culture
All cell lines were cultured at 5% CO2 and 37 °C with HEK293T, Saos2, and Skov3 cells cultured in DMEM high glucose (GE Healthcare Life Sciences, Hyclone, Amersham. UK, Cat# SH30081), HCT116 and U2OS cells in McCoy’s 5a (ThermoFisher Scientific, Pittsburgh, MA, USA, Cat# SH30200), and PC3 cells in RPMI (GE Healthcare Life Sciences, Hyclone, Cat# SH3009602) and 2 mM l-glutamine (GE Healthcare Life Sciences, Hyclone, Cat# SH3003402). All growth mediums were supplemented with 10% heat-inactivated FBS (ThermoFisher Scientific, Gibco, Cat# 12657029) and 50 μg/ml of streptomycin (ThermoFisher Scientific, Gibco, Cat# 15140122) and 50 μg/ml of penicillin (ThermoFisher Scientific, Gibco, Cat# 15140122). All cell lines were purchased from ATCC, tested negative for mycoplasma, and their identity was verified via STR profiling.
Bioinformatics, data availability, and statistical testing
The USCS genome browser49 was utilized for the bioinformatic evaluation of antisense transcription utilizing the RefSeq50 gene annotation track.
All raw experimental data, code used for analysis, and supplementary methods are available for review at51 and are provided as an R package. All analysis took place using the R statistical programming language52 using external packages that are documented in the package associated with this article53–64. The package facilitates replication of the operating system and package versions used for the original analysis, reproduction of each individual figure and figure supplement included in the article, and easy review of the code used for all steps of the analysis, from raw-data to figure.
The significance threshold (alpha) in this study was set to 0.05. Statistical testing was performed using an unpaired two sample Student’s two-sided t test unless otherwise specified. Data were either approximated to be normally distributed or transformed to be so in cases where a parametric test was utilized. In addition, variance was not assumed to be equal between groups and, therefore, the Welch (or Satterthwaite) approximation to the degrees of freedom was used.
Coding potential
Protein-coding capacity was evaluated using the Coding-Potential Assessment Tool37 and Coding-Potential Calculator38 with default settings. Transcript sequences for use with Coding-Potential Assessment Tool were downloaded from the UCSC genome browser using the Ensembl accessions: HOTAIR (ENST00000455246), XIST (ENST00000429829), β-actin (ENST00000331789), Tubulin (ENST00000427480), and MYC (ENST00000377970). Transcript sequences for use with Coding-Potential Calculator were downloaded from the UCSC genome browser using the following IDs: HOTAIR (uc031qho.1) and β-actin (uc003soq.4).
Peptide identification in MS/MS spectra
Orbitrap raw MS/MS files for 11 human cell lines were downloaded from the PRIDE repository (PXD002395;39) converted to mzML format using msConvert from the ProteoWizard tool suite65. Spectra were then searched using MSGF + (v10072)66 and Percolator (v2.08)67. All searches were done against the human protein subset of Ensembl 75 in the Galaxy platform68 supplemented with the 6 frame translation of both the annotated (LOC102724571; hg38) and PCR cloned sequence of lncTAM34a (Supplementary data;51). MSGF + settings included precursor mass tolerance of 10 ppm, fully tryptic peptides, maximum peptide length of 50 amino acids and a maximum charge of 6. Fixed modification was carbamidomethylation on cysteine residues; a variable modification was used for oxidation on methionine residues. Peptide Spectral Matches found at 1% FDR (false discovery rate) were used to infer peptide identities. The output from all searches are available in Ref. 51.
si- and sh-RNAs
shRNA-expressing constructs were cloned into the U6M2 construct using the BglII and KpnI restriction sites as previously described69. shRNA constructs were transfected using Lipofectamine 2000 or 3000 (ThermoFisher Scientific, Cat# 12566014 and L3000015). The sequences targeting renilla is as follows: shRenilla 1.1 (AAT ACA CCG CGC TAC TGG C) and shRenilla 2.1 (TAA CGG GAT TTC ACG AGG C). si-lncTAM34a (GGG AGA AGA CGA UUC UUU, Eurofins) and si-Control (Qiagen, Cat# 1027310) were transfected using Lipofektamine 3000 at 10 nM.
Bidirectional promoter cloning
The overlapping region (p1) corresponds with the sequence previously published as the TP53 binding site in12 which we synthesized, cloned into the pLucRluc construct70, and sequenced to verify its identity.
Promoter activity
Cells were cotransfected with the p1 renilla/firefly bidirectional promoter construct70 and GFP by using Lipofectamine 2000 (Life Technologies, Cat# 12566014). The expression of GFP and luminescence was measured 24 h posttransfection by using the Dual-Glo Luciferase Assay System (Promega, Cat# E2920) and detected by the GloMax-Multi + Detection System (Promega, Cat# SA3030). The expression of luminescence was normalized to GFP.
Generation of U6-expressed lncTAM34a lentiviral constructs
The U6 promoter was amplified from the U6M2 cloning plasmid69, and ligated into the Not1 restriction site of the pHIV7-IMPDH2 vector71. lncTAM34a was PCR amplified and subsequently cloned into the Nhe1 and Pac1 restriction sites in the pHIV7-IMPDH2-U6 plasmid.
Lentiviral particle production, infection, and selection
Lentivirus production was performed as previously described in Ref. 71. Briefly, HEK293T cells were transfected with viral and expression constructs using Lipofectamine 2000 (ThermoFisher Scientific, Cat# 12566014), after which viral supernatants were harvested 48 and 72 h posttransfection. Viral particles were concentrated using PEG-IT solution (Systems Biosciences, Palo Alto, CA, USA, Cat# LV825A-1) according to the manufacturer’s recommendations. HEK293T cells were used for virus titration and GFP expression was evaluated 72 h postinfection via flow cytometry (LSRII, BD Biosciences, San Jose, CA, USA) after which TU/ml was calculated.
Stable lines were generated by infecting cells with a multiplicity of infection of 1 and subsequently initiating 1–2 μM mycophenolic acid-based (Merck, Kenilworth, NJ, USA, Cat# M5255) selection 48–72 h postinfection. Cells were expanded as the selection process was monitored via flow cytometry analysis (LSRII, BD Biosciences) of GFP and selection was terminated once >90% of the cells were GFP positive. Quantification of lncTAM34a overexpression and miR34a was performed in biological quintuplet for all cell lines.
Western blotting
Samples were lysed in 50 mM Tris-HCl (Sigma Aldrich, St. Louis, MO, USA, Cat# T2663), pH 7.4, 1% NP-40 (Sigma Aldrich, Cat# I8896), 150 mM NaCl (Sigma Aldrich, Cat# S5886), 1 mM EDTA (Promega, Madison, WI, USA, Cat# V4231), 1% glycerol (Sigma Aldrich, Cat# G5516), 100 μM vanadate (Sigma Aldrich, Cat# S6508), protease inhibitor cocktail (Roche Diagnostics, Basel, Switzerland, Cat# 004693159001), and PhosSTOP (Roche Diagnostics, Cat# 04906837001). Lysates were subjected to SDS-PAGE and transferred to PVDF membranes. The proteins were detected by western blot analysis by using an enhanced chemiluminescence system (Western Lightning–ECL, PerkinElmer, Waltham, MA, USA, Cat# NEL103001EA). Antibodies used were specific for CCND1 1:1000 (Cell Signaling, Danvers, MA, USA, Cat# 2926), and GAPDH 1:5000 (Abcam, Cambridge, UK, Cat# ab9485). All western blot quantifications were performed using ImageJ72.
RNA extraction and cDNA synthesis
For downstream SYBR green applications, RNA was extracted using the RNeasy mini kit (Qiagen, Venlo, Netherlands, Cat# 74106) and subsequently treated with DNase (Ambion Turbo DNA-free, ThermoFisher Scientific, Cat# AM1907). 500 ng RNA was used for cDNA synthesis using MuMLV (ThermoFisher Scientific, Cat# 28025013) and a 1:1 mix of oligo(dT) and random nanomers.
For analysis of miRNA expression with Taqman, samples were isolated with TRIzol reagent (ThermoFisher Scientific, Cat# 15596018) and further processed with the miRNeasy kit (Qiagen, Cat# 74106). cDNA synthesis was performed using the TaqMan MicroRNA Reverse Transcription Kit (ThermoFisher Scientific, Cat# 4366597) using the corresponding oligos according to the manufacturer’s recommendations.
qPCR and PCR
PCR was performed using the KAPA2G Fast HotStart ReadyMix PCR Kit (Kapa Biosystems, Wilmington, MA, USA, Cat# KK5601) with corresponding primers. Quantitative polymerase chain reaction (qPCR) was carried out using KAPA 2G SYBRGreen (Kapa Biosystems, Cat# KK4602) using the Applied Biosystems 7900HT machine with the cycling conditions: 95 °C for 3 min, 95 °C for 3 s, and 60 °C for 30 s.
QPCR for miRNA expression analysis was performed according to the primer probe set manufacturers recommendations (ThermoFisher Scientific) and using the TaqMan Universal PCR Master Mix (ThermoFisher Scientific, Cat# 4304437) with the same cycling scheme as above. Primer and probe sets for TaqMan were also purchased from ThermoFisher Scientific (Life Technologies at time of purchase, TaqMan® MicroRNA Assay, hsa-miR-34a, human, Cat# 4440887, Assay ID: 000426 and Control miRNA Assay, RNU48, human, Cat# 4440887, Assay ID: 001006).
The ΔΔCt method was used to quantify gene expression. All qPCR-based experiments were performed in at least technical duplicate. Primers for all PCR-based experiments are listed in Supplementary Document 2 and arranged by figure.
Cell-cycle distribution
Cells were washed in PBS and fixed in 4% paraformaldehyde at room temperature overnight. Paraformaldehyde was removed, and cells were resuspended in 95% EtOH. The samples were then rehydrated in distilled water, stained with DAPI and analyzed by flow cytometry on a LSRII (BD Biosciences) machine. Resulting cell-cycle phases were quantified using the ModFit software (Verity Software House, Topsham, ME, USA). Experiments were performed in biological quadruplet (PC3) or triplicate (Skov3). The log2 fraction of cell-cycle phase was calculated for each replicate and a two sample t test was utilized for statistical testing.
3′ Rapid amplification of cDNA ends
3′-RACE was performed as described as previously in Ref. 8. Briefly, U2OS cell RNA was polyA-tailed using yeast polyA polymerase (ThermoFisher Scientific, Cat# 74225Z25KU) after which cDNA was synthesized using oligo(dT) primers. Nested-PCR was performed first using a forward primer in lncTAM34a exon 1 and a tailed oligo(dT) primer followed by a second PCR using an alternate lncTAM34a exon 1 primer and a reverse primer binding to the tail of the previously used oligo(dT) primer. PCR products were gel purified and cloned the Strata Clone Kit (Agilent Technologies, Santa Clara, CA, USA, Cat# 240205), and sequenced.
Chromatin immunoprecipitation
The ChIP was performed as previously described in Ref. 8 with the following modifications. Cells were crosslinked in 1% formaldehyde (Merck, Cat# 1040039025), quenched with 0.125 M glycine (Sigma Aldrich, Cat# G7126), and lysed in cell lysis buffer comprised of: 5 mM PIPES (Sigma Aldrich, Cat# 80635), 85 mM KCL (Merck, Cat# 4936), 0.5% NP40 (Sigma Aldrich, Cat# I8896), protease inhibitor (Roche Diagnostics, Cat# 004693159001). Samples were then sonicated in 50 mM TRIS-HCL pH 8.0 (Sigma Aldrich, MO, USA, Cat# T2663) 10 mM EDTA (Promega, WI, USA, Cat# V4231), 1% SDS (ThermoFisher Scientific, Cat# AM9822), and protease inhibitor (Roche Diagnostics, Cat# 004693159001) using a Bioruptor Sonicator (Diagenode, Denville, NJ, USA). Samples were incubated over night at 4 °C with the polII antibody (Abcam, Cat# ab5095) and subsequently pulled down with Salmon Sperm DNA/Protein A Agarose (Millipore, Cat# 16-157) beads. DNA was eluted in an elution buffer of 1% SDS (ThermoFisher Scientific, Cat# AM9822) 100 mM NaHCO3 (Sigma Aldrich, Cat# 71631), followed by reverse crosslinking, RNaseA (ThermoFisher Scientific, Cat# 1692412) and protease K (New England Biolabs, Ipswich, MA, USA, Cat# P8107S) treatment. The DNA was eluted using Qiagen PCR purification kit (Cat# 28106) and quantified via qPCR. qPCR was performed in technical duplicate using the standard curve method and reported absolute values. The fraction of input was subsequently calculated using the mean of the technical replicates followed by calculating the fold over the control condition. Statistical testing was performed using four biological replicates with the null hypothesis that the true log2 fold change values were equal to zero.
Confluency and proliferation analysis
Cells were incubated in the Spark Multimode Microplate (Tecan, Männedorf, Switzerland) reader for 48 h at 37 °C with 5% CO2 in a humidity chamber in either normal medium or HBSS (ThermoFisher Scientific, Cat# 14025092). Confluency was measured every hour using bright-field microscopy and the percentage of confluency was reported via the plate reader’s inbuilt algorithm. Fold confluency was then calculated as % confluency/% confluency time 0 for each condition and cell line and the mean of the three technical replicates was subsequently calculated for each of the three biological replicates. A polynomial regression model was then constructed modeling the fold confluency as the dependent variable and time and cell line as independent variables. Reported p values are derived from the t test, testing the null hypothesis that the coefficient estimate of the cell line covariate is equal to 0.
Analysis of the proliferation rate of PC3 stable cell lines was performed using the CellTrace Violet assay (ThermoFisher Scientific, Cat# C34557). CellTrace Violet is a fluorescent dye that binds covalently to all free amines on the surface and inside of cells. As cells divide the dye is diluted and hence fluorescence intensity is decreased. PC3 stable cell lines, either miR34a asRNA overexpressing or mock, were harvested and stained in 1 ml PBS with 5 µM CellTrace Violet for 20 min and subsequently seeded in 12 well plates at 2 × 104 cells per well. Time 0 measurements were taken once cells had attached and treatments with RPMI (Gibco, life technology) 10% FBS, RPMI 0.1% FBS, or HBSS were simultaneously initiated in the remaining cells. RPMI mediums were all additionally supplemented with 2 mM l-glutamine and 50 µg/ml Penicillin–Streptomycin. Cells were incubated for the indicated times before harvesting and CellTrace Violet was quantified via flow cytometry (Sony SH800S Cell Sorter). Time 0 was performed in biological triplicate and technical duplicate whereas all other time points were performed in biological triplicates with one technical replicate. Analysis was performed by first, subsampling each replicate and condition so that each had a total of 10,000 cells. The mean of each technical replicate for time point 0 was calculated and, subsequently, the time 0 fluorescence intensity was subtracted from each sample. The mean difference in fluorescence intensity was then calculated for each biological replicate and condition and used to build a polynomial regression model per condition (i.e., RPMI, 0.1% FBS, and HBSS) where difference in fluorescence intensity was modeled as a function of time and cell line. Reported P values are derived from the t test, testing the null hypothesis that the coefficient estimate of the cell line covariate is equal to 0.
Pharmacological compounds
Doxorubicin was purchased from Teva (Petah Tikva, Israel, cat. nr. 021361).
Cellular localization analysis
Quantified RNAseq data from 11 cell lines from the GRCh38 assembly was downloaded from the ENCODE project database and quantifications for lncTAM34a (ENSG00000234546), GAPDH (ENSG00000111640), and MALAT1 (ENSG00000251562) were extracted. Cell lines for which data was downloaded include: A549, GM12878, HeLa-S3, HepG2, HT1080, K562 MCF-7, NCI-H460, SK-MEL-5, SK-N-DZ, and SK-N-SH. Initial exploratory analysis revealed that several cell lines should be removed from the analysis due to (a) a larger proportion of GAPDH in the nucleus than cytoplasm, (b) variation of lncTAM34a expression is too large to draw conclusions, or (c) they have no or low (<6 TPM) lncTAM34a expression. Furthermore, only polyadenylated libraries were used in the final analysis, due to the fact that the cellular compartment enrichment was improved in these samples. All analyzed genes are reported to be polyadenylated. In addition, only samples with 2 biological replicates were retained. For each cell type, gene, and biological replicate the fraction of transcripts per million (TPM) in each cellular compartment was calculated as the fraction of TPM in the specific compartment by the total TPM. The mean and standard deviation for the fraction was subsequently calculated for each cell type and cellular compartment and this information was represented in the final figure.
CAGE analysis
All available CAGE data from the ENCODE project36 for 36 cell lines was downloaded from the UCSC genome browser49 for genome version hg19. Of these, 28 cell lines had CAGE transcription start sites (TSS) mapping to the plus strand of chromosome 1 and in regions corresponding to 200 base pairs upstream of the Lnc34a start site (9241796 − 200) and 200 base pairs upstream of the GENCODE annotated lncTAM34a start site (9242263 + 200). These cell lines included: HFDPC, H1-hESC, HMEpC, HAoEC, HPIEpC, HSaVEC, GM12878, hMSC-BM, HUVEC, AG04450, hMSC-UC, IMR90, NHDF, SK-N-SH_RA, BJ, HOB, HPC-PL, HAoAF, NHEK, HVMF, HWP, MCF-7, HepG2, hMSC-AT, NHEM.f_M2, SkMC, NHEM_M2, and HCH. In total 74 samples were included. Seventeen samples were polyA−, 47 samples were polyA+, and 10 samples were total RNA. In addition, 34 samples were whole cell, 15 enriched for the cytosolic fraction, 15 enriched for the nucleolus, and 15 enriched for the nucleus. All CAGE transcription start sites were plotted and the RPKM of the individual reads was used to color each read to indicate their relative abundance. In cases where CAGE TSS spanned identical regions, the RPMKs of the regions were summed and represented as one CAGE TSS in the figure. In addition, a density plot shows the distribution of the CAGE reads in the specified interval.
Splice junction analysis
All available whole cell (i.e., nonfractionated) spliced read data originating from the Cold Spring Harbor Lab in the ENCODE project36 for 38 cell lines was downloaded from the UCSC genome browser49. Of these cell lines, 36 had spliced reads mapping to the plus strand of chromosome 1 and in the region between the Lnc34a start (9241796) and transcription termination (9257102) site (note that lncTAM34a resides totally within this region). Splice junctions from the following cell lines were included in the final figure: A549, Ag04450, Bj, CD20, CD34 mobilized, Gm12878, H1-hesc, Haoaf, Haoec, Hch, Helas3, Hepg2, Hfdpc, Hmec, Hmepc, Hmscat, Hmscbm, Hmscuc, Hob, Hpcpl, Hpiepc, Hsavec, Hsmm, Huvec, Hvmf, Hwp, Imr90, Mcf7, Monocd14, Nhdf, Nhek, Nhemfm2, Nhemm2, Nhlf, Skmc, and Sknsh. All splice junctions were included in the figure and colored according to the number of reads corresponding to each. In cases where identical reads were detected multiple times, the read count was summed and represented as one read in the figure.
TCGA data analysis
RNAseq data and copy number data were downloaded from TCGA and processed as described previously34. Briefly, RNAseq data were aligned to the human hg19 assembly and quantified using GENCODE (v19) annotated HTSeq-counts and FPKM normalizations. Expression data from miR34a and lncTAM34a (identified as RP3-510D11.2) were used for further analysis. Copy number amplitudes for GENCODE genes were determined from segmented copy number data. Samples that were diploid for lncTAM34a were identified as those samples that had copy number amplitudes between −0.1 and 0.1.
Somatic mutation data were downloaded from the Genomics Data Commons data portal (GDC) as mutation annotation format (maf) files, called using Mutect2 on 30/10/2017 (v7)73.
Survival analysis was performed on TCGA vital state and follow-up data, downloaded from GDC on 27/10/2017 using the R survival package64.
Electronic supplementary material
Acknowledgements
The authors would like to kindly thank Martin Enge for his critical review of the manuscript and fruitful discussions. Dan Grandér †, who played a significant role in the conceptualization and supervision of this project, sadly passed away before the initial submission of the manuscript. May he rest in peace. This work has been supported by the Swedish Research Council [521-2012-2037], Swedish Cancer Society [150768], Cancer Research Foundations of Radiumhemmet [144063], and the Swedish Childhood Cancer Foundation [PR2015-0009].
Competing interests
The authors declare no competing interests.
Footnotes
Edited by A. Oberst
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41419-018-0777-5).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 2.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ozsolak F, et al. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell. 2010;143:1018–1029. doi: 10.1016/j.cell.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J, et al. Over 20% of human transcripts might form sense-antisense pairs. Nucleic Acids Res. 2004;32:4812–4820. doi: 10.1093/nar/gkh818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katayama S, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 6.Balbin OA, et al. The landscape of antisense gene expression in human cancers. Genome Res. 2015;25:1068–1079. doi: 10.1101/gr.180596.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yap KL, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnsson P, et al. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat. Struct. Mol. Biol. 2013;20:440–446. doi: 10.1038/nsmb.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Memczak S, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 11.Conley AB, Jordan IK. Epigenetic regulation of human cis-natural antisense transcripts. Nucleic Acids Res. 2012;40:1438–1445. doi: 10.1093/nar/gkr1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raver-Shapira N, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol. Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 13.Lal A, et al. Capture of microRNA-bound mRNAs identifies the tumor suppressor miR-34a as a regulator of growth factor signaling. PLoS Genet. 2011;7:e1002363. doi: 10.1371/journal.pgen.1002363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Slabakova E, Culig Z, Remsik J, Soucek K. Alternative mechanisms of miR-34a regulation in cancer. Cell Death Dis. 2017;8:e3100. doi: 10.1038/cddis.2017.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng J, et al. The impact of miR-34a on protein output in hepatocellular carcinoma HepG2 cells. Proteomics. 2010;10:1557–1572. doi: 10.1002/pmic.200900646. [DOI] [PubMed] [Google Scholar]
- 16.Cole KA, et al. A functional screen identifies miR-34a as a candidate neuroblastoma tumor suppressor gene. Mol. Cancer Res. 2008;6:735–742. doi: 10.1158/1541-7786.MCR-07-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallardo E, et al. miR-34a as a prognostic marker of relapse in surgically resected non-small-cell lung cancer. Carcinogenesis. 2009;30:1903–1909. doi: 10.1093/carcin/bgp219. [DOI] [PubMed] [Google Scholar]
- 18.Liu C, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat. Med. 2011;17:211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zenz T, et al. miR-34a as part of the resistance network in chronic lymphocytic leukemia. Blood. 2009;113:3801–3808. doi: 10.1182/blood-2008-08-172254. [DOI] [PubMed] [Google Scholar]
- 20.Vogt M, et al. Frequent concomitant inactivation of miR-34a and miR-34b/c by CpG methylation in colorectal, pancreatic, mammary, ovarian, urothelial, and renal cell carcinomas and soft tissue sarcomas. Virchows Arch.: Int. J. Pathol. 2011;458:313–322. doi: 10.1007/s00428-010-1030-5. [DOI] [PubMed] [Google Scholar]
- 21.Chim CS, et al. Epigenetic inactivation of the miR-34a in hematological malignancies. Carcinogenesis. 2010;31:745–750. doi: 10.1093/carcin/bgq033. [DOI] [PubMed] [Google Scholar]
- 22.Agostini M, et al. Neuronal differentiation by TAp73 is mediated by microRNA-34a regulation of synaptic protein targets. Proc. Natl. Acad. Sci. USA. 2011;108:21093–21098. doi: 10.1073/pnas.1112061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding N, Wu H, Tao T, Peng E. NEAT1 regulates cell proliferation and apoptosis of ovarian cancer by miR-34a-5p/BCL2. Onco Targets Ther. 2017;10:4905–4915. doi: 10.2147/OTT.S142446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rokavec M, et al. Corrigendum. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J. Clin. Invest. 2015;125:1362. doi: 10.1172/JCI81340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su X, et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature. 2010;467:986–990. doi: 10.1038/nature09459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang TC, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahn YH, et al. ZEB1 drives prometastatic actin cytoskeletal remodeling by downregulating miR-34a expression. J. Clin. Invest. 2012;122:3170–3183. doi: 10.1172/JCI63608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stahlhut C, Slack FJ. Combinatorial action of microRNAs let-7 and miR-34 effectively synergizes with Erlotinib to suppress non-small cell lung cancer cell proliferation. Cell Cycle. 2015;14:2171–2180. doi: 10.1080/15384101.2014.1003008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang P, et al. TGF-beta-miR-34a-CCL22 signaling-induced Treg cell recruitment promotes venous metastases of HBV-positive hepatocellular carcinoma. Cancer Cell. 2012;22:291–303. doi: 10.1016/j.ccr.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, et al. Tumor suppressor miR-34a targets PD-L1 and functions as a potential immunotherapeutic target in acute myeloid leukemia. Cell Signal. 2015;27:443–452. doi: 10.1016/j.cellsig.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 31.Leveille N, et al. Genome-wide profiling of p53-regulated enhancer RNAs uncovers a subset of enhancers controlled by a lncRNA. Nat. Commun. 2015;6:6520. doi: 10.1038/ncomms7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rashi-Elkeles S, et al. Parallel profiling of the transcriptome, cistrome, and epigenome in the cellular response to ionizing radiation. Sci. Signal. 2014;7:rs3. doi: 10.1126/scisignal.2005032. [DOI] [PubMed] [Google Scholar]
- 33.Hunten S, et al. p53-Regulated Networks of Protein, mRNA, miRNA, and lncRNA Expression Revealed by Integrated Pulsed Stable Isotope Labeling With Amino Acids in Cell Culture (pSILAC) and Next Generation Sequencing (NGS) Analyses. Mol. Cell Proteom. 2015;14:2609–2629. doi: 10.1074/mcp.M115.050237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ashouri A, et al. Pan-cancer transcriptomic analysis associates long noncoding RNAs with key mutational driver events. Nat. Commun. 2016;7:13197. doi: 10.1038/ncomms13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim KH, Kim HJ, Lee TR. Epidermal long noncoding RNAs are regulated by ultraviolet irradiation. Gene. 2017;637:196–202. doi: 10.1016/j.gene.2017.09.043. [DOI] [PubMed] [Google Scholar]
- 36.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L, et al. CPAT: coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013;41:e74. doi: 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kong L, et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35:W345–W349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geiger T, Wehner A, Schaab C, Cox J, Mann M. Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol. Cell Proteom. 2012;11:M111 014050. doi: 10.1074/mcp.M111.014050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tarasov V, et al. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 41.Sun F, et al. Downregulation of CCND1 and CDK6 by miR-34a induces cell-cycle arrest. FEBS Lett. 2008;582:1564–1568. doi: 10.1016/j.febslet.2008.03.057. [DOI] [PubMed] [Google Scholar]
- 42.Yu W, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Serviss JT, Johnsson P, Grander D. An emerging role for long noncoding RNAs in cancer metastasis. Front. Genet. 2014;5:234. doi: 10.3389/fgene.2014.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hung T, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ng SY, Johnson R, Stanton LW. Human long noncoding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012;31:522–533. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ng SY, Bogu GK, Soh BS, Stanton LW. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol. Cell. 2013;51:349–359. doi: 10.1016/j.molcel.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 47.Boque-Sastre R, et al. Head-to-head antisense transcription and R-loop formation promotes transcriptional activation. Proc. Natl. Acad. Sci. USA. 2015;112:5785–5790. doi: 10.1073/pnas.1421197112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, L. et al. A long noncoding RNA targets microRNA miR-34a to regulate colon cancer stem cell asymmetric division. Elife5:e14620, 10.7554/eLife.14620 (2016). [DOI] [PMC free article] [PubMed]
- 49.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O’Leary NA, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.miR34AasRNAproject https://github.com/GranderLab/miR34a_asRNA_project (2017).
- 52.Team, R. C. R: A Language and Environment for Statistical Computing https://www.R-project.org/ (2017).
- 53.liftr: Containerize R Markdown Documents v. R package version 0.7 (2017).
- 54.tidyverse: Easily Install and Load the ‘Tidyverse’ v. R package version 1.2.1 (2017).
- 55.rlang: Functions for Base Types and Core R and “Tidyverse” Features v. R package version 0.1.4 (2017).
- 56.magrittr: A Forward-Pipe Operator for R v. R package version 1.5 (2014).
- 57.knitr: A General-Purpose Package for Dynamic Report Generation in R v. R package version 1.17 (2017).
- 58.rmarkdown: Dynamic Documents for R v. R package version 1.8 (2017).
- 59.ggthemes: Extra Themes, Scales and Geoms for “ggplot2” v. R package version 3.4.0 (2017).
- 60.gggenes: Draw Gene Arrow Maps in “ggplot2” v. R package version 0.2.0.9003.
- 61.extrafont: Tools for using fonts v. R package version 0.17 (2014).
- 62.scales: Scale Functions for Visualization v. R package version 0.5.0 (2017).
- 63.gtable: Arrange “Grobs” in Tables v. R package version 0.2.0 (2016).
- 64.A Package for Survival Analysis in S v. version 2.38 (2015).
- 65.Holman JD, Tabb DL, Mallick P. Employing ProteoWizard to convert raw mass spectrometry data. Curr. Protoc. Bioinformatics. 2014;46:13. doi: 10.1002/0471250953.bi1324s46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim S, Pevzner PA. MS-GF+makes progress towards a universal database search tool for proteomics. Nat. Commun. 2014;5:5277. doi: 10.1038/ncomms6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Granholm V, et al. Fast and accurate database searches with MS-GF+Percolator. J. Proteome Res. 2014;13:890–897. doi: 10.1021/pr400937n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boekel J, et al. Multi-omic data analysis using Galaxy. Nat. Biotechnol. 2015;33:137–139. doi: 10.1038/nbt.3134. [DOI] [PubMed] [Google Scholar]
- 69.Amarzguioui M, Rossi JJ, Kim D. Approaches for chemically synthesized siRNA and vector-mediated RNAi. FEBS Lett. 2005;579:5974–5981. doi: 10.1016/j.febslet.2005.08.070. [DOI] [PubMed] [Google Scholar]
- 70.Polson A, Durrett E, Reisman D. A bidirectional promoter reporter vector for the analysis of the p53/WDR79 dual regulatory element. Plasmid. 2011;66:169–179. doi: 10.1016/j.plasmid.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 71.Turner AM, Ackley AM, Matrone MA, Morris KV. Characterization of an HIV-targeted transcriptional gene-silencing RNA in primary cells. Human. Gene Ther. 2012;23:473–483. doi: 10.1089/hum.2011.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grossman RL, et al. Toward a shared vision for cancer genomic data. N. Engl. J. Med. 2016;375:1109–1112. doi: 10.1056/NEJMp1607591. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.