Abstract
The rapid increase in carbapenem resistance among gram-negative bacteria has renewed focus on the importance of polymyxin antibiotics (colistin or polymyxin E). However, the recent emergence of plasmid-mediated colistin resistance determinants (mcr-1, -2, -3, -4, -5, -6, and -7), especially mcr-1, in carbapenem-resistant Enterobacteriaceae is a serious threat to global health. Here, we characterized a novel mobile colistin resistance gene, mcr-8, located on a transferrable 95,983-bp IncFII-type plasmid in Klebsiella pneumoniae. The deduced amino-acid sequence of MCR-8 showed 31.08%, 30.26%, 39.96%, 37.85%, 33.51%, 30.43%, and 37.46% identity to MCR-1, MCR-2, MCR-3, MCR-4, MCR-5, MCR-6, and MCR-7, respectively. Functional cloning indicated that the acquisition of the single mcr-8 gene significantly increased resistance to colistin in both Escherichia coli and K. pneumoniae. Notably, the coexistence of mcr-8 and the carbapenemase-encoding gene blaNDM was confirmed in K. pneumoniae isolates of livestock origin. Moreover, BLASTn analysis of mcr-8 revealed that this gene was present in a colistin- and carbapenem-resistant K. pneumoniae strain isolated from the sputum of a patient with pneumonia syndrome in the respiratory intensive care unit of a Chinese hospital in 2016. These findings indicated that mcr-8 has existed for some time and has disseminated among K. pneumoniae of both animal and human origin, further increasing the public health burden of antimicrobial resistance.
Introduction
Klebsiella pneumoniae, belonging to the family Enterobacteriaceae, is a bacterial pathogen commonly associated with nosocomial infections, including pneumonia, bloodstream infection, urinary tract infection, and hepatic abscess, especially among immunocompromised patients1. Carbapenem was used as the drug of choice for the treatment of infections caused by multidrug-resistant K. pneumoniae. However, the increased prevalence of carbapenem-resistant K. pneumoniae (CRKP) greatly compromised the efficacy of carbapenem antibiotics2. Colistin has been considered a last resort antibiotic, used either alone or in combination with other drugs, for the treatment of serious infections caused by CRKP3. In recent years, multiple studies have indicated that the prevalence of colistin resistance has increased rapidly among Enterobacteriaceae4. In 2015, Liu et al.5 identified the first mobile colistin resistance gene, mcr-1, in Enterobacteriaceae, mainly Escherichia coli and K. pneumoniae. In a short time, mcr-1 and its slightly altered gene variants (mcr-1.1 to mcr-1.12) were ubiquitously identified in various Enterobacteriaceae of different origins in over 40 countries across five different continents6–10. Several other MCR homologs (MCR-2, MCR-3, MCR-4, and MCR-5) were subsequently identified in Enterobacteriaceae. Very recently, two MCR homologs (MCR-6 and MCR-7) were annotated and deposited into GenBank, and the mcr-7.1 gene was detected in K. pneumoniae of chicken origin in China11. To date, mcr-2 has only been detected in E. coli and Salmonella isolates from European countries12,13, whereas mcr-3 has been widely identified in Enterobacteriaceae (mainly E. coli) and Aeromonas spp. from Asia, Europe, and North America14. mcr-4 and mcr-5 were first characterized in Salmonella and E. coli from European countries15,16 and further detected in Enterobacter cloacae and E. coli isolates from Asia (Singapore and Japan)17,18. Thus far, only mcr-1 and mcr-3 have been reported in K. pneumoniae, and the prevalence of these two genes is relatively low5,6. In this study, we characterized a novel mobile colistin resistance gene, mcr-8, in a K. pneumoniae isolate and then identified the coexistence of mcr-8 and blaNDM in K. pneumoniae isolates from both animals and humans.
Results
A novel transferrable colistin resistance determinant in Klebsiella pneumoniae
During the period from 2015 to 2017, our routine surveillance indicated an increased number of colistin-resistant K. pneumoniae isolates in which the recently characterized plasmid-mediated colistin resistance genes mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 were absent. One K. pneumoniae isolate, designated KP91, was obtained from a swine fecal sample. The isolate exhibited a multidrug resistance profile, including resistance to colistin (MIC = 16 μg/ml) (Table 1). Whole-genome sequencing indicated that KP91 belonged to the ST42 group and contained multiple antimicrobial resistance genes, including the β-lactam resistance genes blaSHV-1 and blaCTX-M-14, aminoglycoside resistance genes strA, strB, armA, and aph(4)-Ia, macrolide resistance genes mph(E) and msr(E), quinolone resistance genes oqxA and qnrB4, sulfonamide resistance genes sul1, sul2, and sul3, tetracycline resistance genes tet(A), tet(B), and tet(34), and trimethoprim resistance gene dfrA12 (Table 1). KP91 also carried multiple virulence genes, including kfuC, mrkABCDF, irp2, iucA and iucB, which have been associated with urinary tract infections, septicemia, and pneumonia19–21.
Table 1.
Minimum inhibitory concentrations of tested antimicrobial agents for the studied bacterial isolates
| Species | MIC (µg/ml) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CST | PB | TET | MEM | TAZ | AMC | SXT | AN | GEN | STR | FFC | CIP | |
| K.pneumoniae KP91 | 16 | 8 | >256 | 0.06 | 16 | 32/16 | 16 | >256 | 512 | 256 | 256 | 256 |
| E. coli J53 + pKP91 | 4 | 4 | 1 | 0.015 | 0.5 | 4/2 | 0.5 | 1 | <0.125 | 4 | 4 | 0.015 |
| E. coli J53 | 0.25 | 0.25 | 1 | 0.03 | 0.5 | 4/2 | 0.5 | 1 | <0.125 | 4 | 4 | 0.015 |
| DH5ɑ + pUC19-mcr-8 | 1 | 0.5 | – | – | – | – | – | – | – | – | – | – |
| DH5ɑ + pUC19 | 0.25 | 0.25 | – | – | – | – | – | – | – | – | – | – |
| K. pneumoniae_ | 8 | 8 | – | – | – | – | – | – | – | – | – | – |
| ATCC13883+pUC19-mcr-8 | ||||||||||||
| K. pneumoniae_ | 1 | 2 | – | – | – | – | – | – | – | – | – | – |
| ATCC13883+pUC19 | ||||||||||||
Antimicrobial susceptibilities, reported as MIC (µg/ml), were interpreted as resistant (bold) or susceptible (plain text) in accordance with established breakpoints. CST colistin, PB polymyxin B, TET tetracycline, MEM meropenem, TAZ ceftazidime, AMC amoxicillin-clavulanate, SXT trimethoprim-sulfamethoxazole, AN amikacin, GEN gentamycin, STR streptomycin, FFC florfenicol, CIP ciprofloxacin. “–” indicates that the MIC was not measured
Conjugation assays showed that the colistin resistance determinant in K. pneumoniae KP91 can be transferred into the recipient E. coli strain J53. The transconjugant J53-pKP91 had a pulsed-field gel electrophoresis (PFGE) pattern identical to that of E. coli J53 but distinct from that of KP91. S1-PFGE indicated that J53-pKP91 differed from E. coli J53 by the presence of a single ~90-kb plasmid, designated pKP91 (Fig. 1). These results confirmed the presence of an uncharacterized colistin resistance determinant located on a conjugative plasmid in K. pneumoniae KP91.
Fig. 1. Location of mcr-8 in Klebsiella pneumoniae KP91 and its transconjugants.
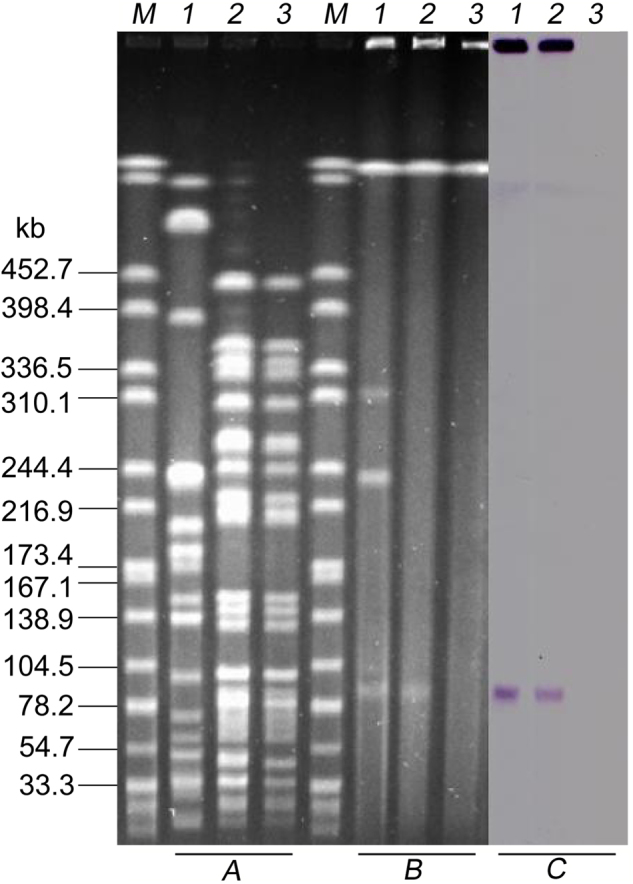
a XbaI-digested PFGE of K. pneumoniae KP91, transconjugants, and the recipient E. coli strain J53. b S1-PFGE and c the corresponding Southern hybridization using the mcr-8-specific probe. Lane M, marker H9812; lane 1, K. pneumoniae KP91; lane 2, transconjugant E. coli J53-pKP91; lane 3, recipient E. coli J53
Identification and functional confirmation of mcr-8 in K. pneumoniae and E. coli
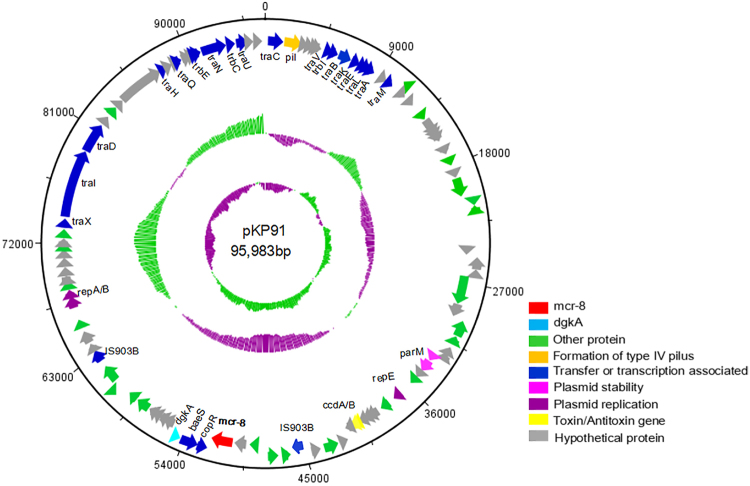
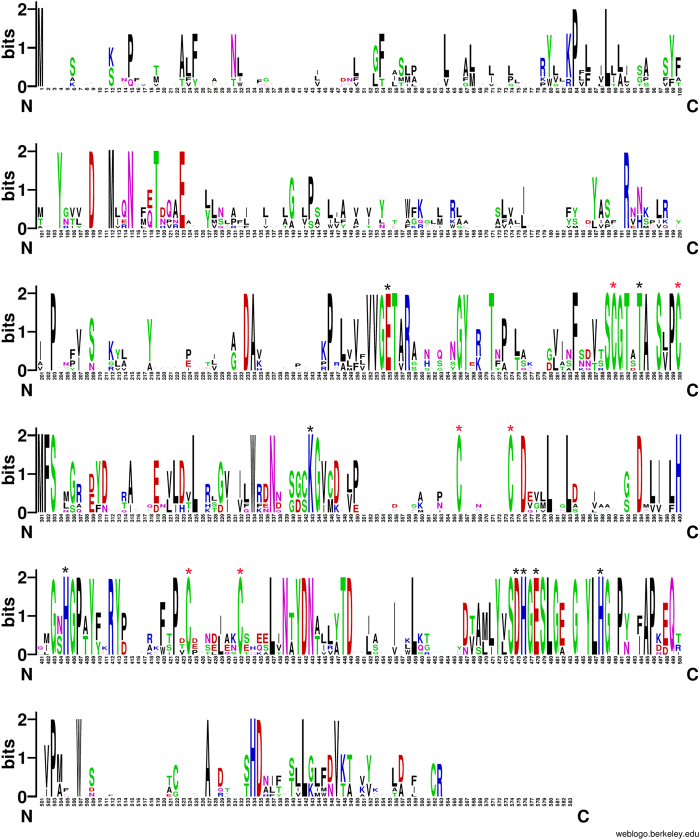
To characterize the colistin resistance determinants, we used Single Molecule Real-Time (SMRT) sequencing to generate the complete sequence of the plasmid isolated from transconjugant J53-pKP91. SMRT sequencing revealed that plasmid pKP91 was 95,983 bp in size, with a GC content of 50.16%, which differed from the GC content of the genome (56.33%) (Fig. 2). pKP91 was identified as a typical IncFII-type plasmid and contained genes involved in plasmid stability, formation of a type-IV pilus, and conjugative transfer (Fig. 2). BLASTn analysis identified a 1698-bp open-reading frame (ORF) encoding a putative phosphoethanolamine transferase, which showed 50.23% nucleotide sequence identity to the corresponding region of mcr-3. The amino-acid sequence of this ORF showed 31.08%, 30.26%, 39.96%, 37.85%, 33.51%, 30.43%, and 37.46% amino-acid sequence identity to MCR-1, MCR-2, MCR-3, MCR-4, MCR-5, MCR-6 and MCR-7, respectively (Fig. S1). Sequence alignment indicated the previously identified active sites (E246, T285, K333, H395, D465, H466, E468, H478) and six cysteine residues that form three disulfide bonds between residues Cys281/Cys291, Cys356/Cys364, and Cys414/Cys422 in the catalytic domain of MCR-1 are highly conserved among these protein ORFs (Fig. 3, Fig. S1)22–25. These results suggested that the corresponding gene might encode a functional phosphoethanolamine transferase that mediates resistance to colistin in both K. pneumoniae and its E. coli transconjugant. In addition, the expression of this single ORF in E. coli DH5ɑ and K. pneumoniae ATCC13883 resulted in four- and eightfold increases, respectively, in the minimum inhibitory concentration of colistin (Table 1), which further confirmed the function of the gene. This gene was designated mcr-8 based on the paper regarding the naming or assignment of allele numbers for mobile colistin resistance (mcr) genes (Partridge, S. et al Proposal for assignment of allele numbers for mobile colistin resistance (mcr) genes. J. Antimicrob. Chemother) (under revision).
Fig. 2. The genetic contents of plasmid pKP91.
Circles display (outside to inside) (i) size in bp and (ii) the positions of predicted coding sequences transcribed in a clockwise orientation
Fig. 3. WebLogo of amino-acid sequences of MCR-1–8.
A logo represents each column of the alignment in a stack of letters, with the height of each letter proportional to the observed frequency of the corresponding amino acid, and the overall height of each stack proportional to the sequence conservation, measured in bits, at that position. Red asterisks indicate the six conserved cysteine residues that form three disulfide bonds. Black asterisks indicate the conserved active sites among MCR-1–8. The logo was built using WebLogo software (http://weblogo.berkeley.edu/logo.cgi)
The presence of mcr-8 in K. pneumoniae of human origin
BLASTp analysis of MCR-8 against the NCBI database revealed a protein sequence with 99% amino-acid identity and 99% query coverage to MCR-8 in a clinical CRKP strain WCHKP1845, originally isolated from the sputum of a patient with pneumonia in the respiratory intensive care unit at West China Hospital, Chengdu, China, in May 2016. In the corresponding paper, the authors claimed that colistin resistance was conferred by an unknown mechanism in strain WCHKP184526. These analyses suggest that mcr-8 may be responsible for the colistin resistance of this strain, and the clinical isolate WCHKP1845 with ST1 was not related to the pig-origin KP91 with ST42.
Structure prediction and genetic environment analysis of MCR-8
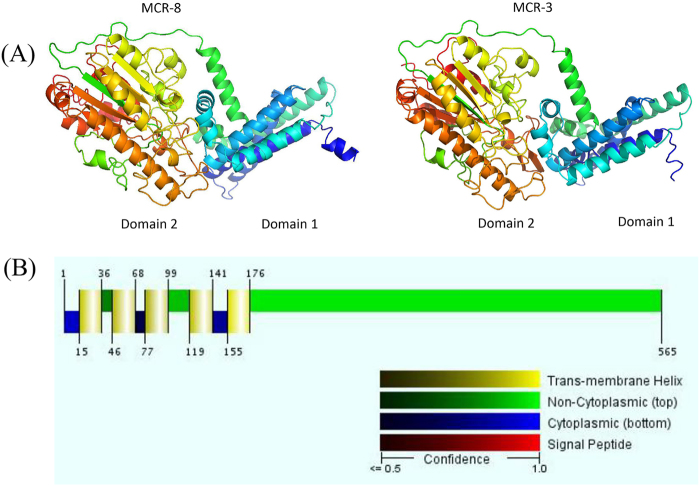
Similar to previously characterized MCR proteins, MCR-8 was predicted using RaptorX (Xu Group, Chicago, IL, USA) to contain two domains (Fig. 4a). Domain 1 (residues 1–234) contained five transmembrane α-helices (Fig. 4b), whereas domain 2 (residues 235–565) contained the putative catalytic center. i-Tasser homology modeling revealed that the best-fit structure in the Protein Data Bank for MCR-8 was 5FGN, which was the first characterized full-length lipid A phosphoethanolamine transferase from Neisseria meningitidis27.
Fig. 4. Predicted crystal structure and transmembrane domain of MCR-8.
a Structure prediction for MCR-8 and reference protein MCR-3. Domain 1 was predicted to be a transmembrane domain, while domain 2 was predicted to be a phosphoethanolamine transferase. b The five transmembrane α-helices predicted by the Philius transmembrane prediction server (type confidence, 0.99; topology confidence, 0.88)
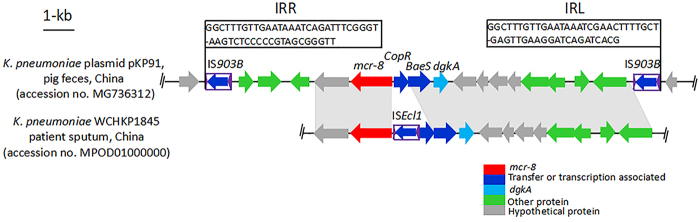
Genetic environment analysis revealed that two complete insertion sequences of IS903B, originating from E. coli, were located up- and downstream of mcr-8 in pKP91. In addition, the 50-bp IRR and 50-bp IRL (Fig. 5) were identical to those surrounding IS903B described in the ISfinder database (https://www-is.biotoul.fr/index.php). IS903B was absent from the 13.47-kb NCBI contig from clinical WCHKP1845, whereas a complete insertion sequence, ISEcl1, originating from Enterobacter cloacae, was present downstream of mcr-8 in WCHKP1845 (Fig. 5). These findings revealed that several IS elements (IS903B, ISEcl1) may play a pivotal role in the dissemination of mcr-8 among Enterobacteriaceae.
Fig. 5. Comparison of the genetic environments of mcr-8.
K. pneumoniae WCHKP1845 was isolated from the sputum of patients in China (accession no. MPOD01000000). The positions and orientations of the genes are indicated by arrows, with the direction of transcription shown by the arrowhead. Gray shading indicates > 90% nucleotide sequence identity
Coexistence of mcr-8 and carbapenemase genes in K. pneumoniae isolates
In total, we examined 53 colistin- and carbapenem-resistant K. pneumoniae isolates derived from pigs (n = 28) and chickens (n = 25). Of these, four (pig = 3, chicken = 1) isolates were positive for mcr-8, whereas five (pig = 3, chicken = 2) were positive for mcr-1. In addition, we detected the presence of blaNDM but not the blaVIM, blaIMP, blaOXA-48, or blaKPC genes in 47 of the carbapenem-resistant K. pneumoniae isolates. mcr-8 and blaNDM were shown to coexist in three isolates. These findings indicated that the novel mcr-8 gene has disseminated among K. pneumoniae and that the prevalence and coexistence of mcr-8 and blaNDM in K. pneumoniae may further threaten public health through either the food chain or environmental routes.
Phylogenetic typing analysis
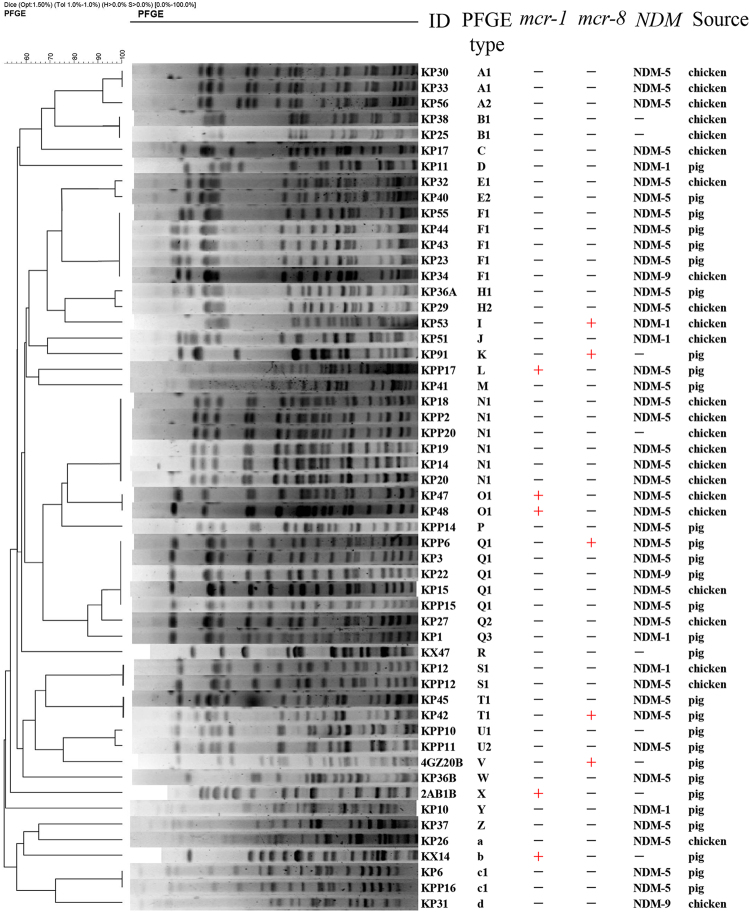
To determine whether the mcr-1- and mcr-8-positive K. pneumoniae isolates were genetically related, five mcr-1- and five mcr-8-positive isolates, including KP91, along with 44 mcr-negative carbapenem-resistant K. pneumoniae isolates, were selected for PFGE analysis with XbaI digestion. Using a cutoff of 80% pattern similarity, the 54K. pneumoniae isolates were grouped into 13 clusters (PFGE patterns represented by multiple strains) and 17 unique PFGE patterns (PFGE patterns represented by a single strain) (Fig. 6). Overall, the K. pneumoniae isolates from different regions were genetically diverse, suggesting that the mcr-8-positive K. pneumoniae isolates were also genetically diverse and that mcr-8 could disseminate among different K. pneumoniae isolates, mainly by horizontal transmission.
Fig. 6. PFGE analysis of mcr-1- and mcr-8-positive Klebsiella pneumoniae isolates as well as other K. pneumoniae isolates.
XbaI was used for digestion of the genomic DNA. All K. pneumoniae isolates were collected from pig fecal matter and chicken cloacae samples from Shandong, China
Discussion
The rapid emergence of mobile colistin resistance genes represents a paradigm shift in colistin-resistant mechanisms, which were long dominated by chromosomal mutations and vertical transmission. To date, mcr-1 and mcr-3 have frequently been detected in Enterobacteriaceae from different regions28, whereas mcr-2, mcr-4, and mcr-5 have been rarely reported. Although all of these mcr genes have been identified in various Enterobacteriaceae and non-Enterobacteriaceae species, E. coli remains the predominant host. A small number of reports have indicated the presence of mcr-1 and mcr-3 in K. pneumoniae at relatively low detection rates6,18,29. In this study, we identified a novel mobile colistin gene, mcr-8, carried on an IncFII-type conjugative plasmid in K. pneumoniae. The presence of mcr-8 (7.54%, 4/53) among the colistin-resistant CRKP isolates indicates that this novel mobile colistin resistance gene may already be widely disseminated among K. pneumoniae isolates of livestock origin.
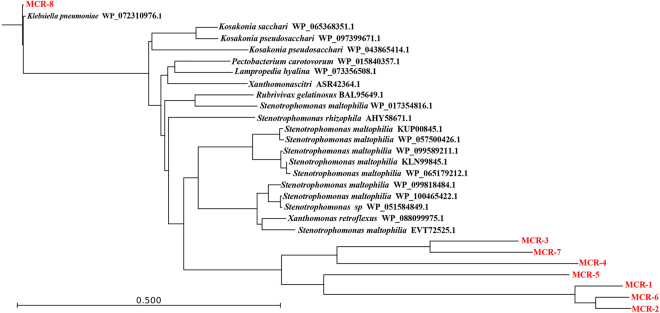
MCR-8 homologs are present in various other species. BLASTp analysis also indicated that a number of proteins in the NCBI database showed 60–70% amino-acid identity to MCR-8. These putative phosphoethanolamine-lipid A transferase proteins include sequences from Kosakonia sacchari (WP_065368351.1), Kosakonia pseudosacchari (WP_097399671.1), Klebsiella aerogenes (WP_043865414.1), Pectobacterium carotovorum (WP_015840357.1), Lampropedia hyalina (WP_073356508.1) Acidovorax avenae subsp. avenae (WP_013595040.1), Rubrivivax gelatinosus (WP_088099975.1), two Xanthomonas isolates, and 11 Stenotrophomonas sp. isolates (Fig. 7, Table S2). The majority of these bacteria are pathogens of plants, including sweet potato, sugarcane root, pumpkin, carrot, rice, potato, rapeseed, and shrub willow, but some of the strains were isolated from manure, soil, and cerebrospinal fluid. In total, the bacteria came from 15 countries across Asia, Europe, South America, and Oceania. These findings indicate that MCR-8-like phosphoethanolamine transferases are widely distributed among various bacteria from different environments and pose a potential risk of transfer to human pathogens.
Fig. 7.
Phylogenetic tree of the deduced amino-acid sequences of 20 putative phosphoethanolamine transferases from different bacterial species and MCR-1–7 with MCR-8 using CLC Genomics Workbench 9 (CLC Bio-Qiagen, Aarhus, Denmark)
In conclusion, we identified a novel gene, mcr-8, conferring resistance to colistin, which is considered the last resort drug for the treatment of carbapenem-resistant Enterobacteriaceae, especially CRKP. The coexistence of mcr-8 and blaNDM was noted in K. pneumoniae from both food-producing animals and human clinical samples and poses a serious public health concern. Therefore, further surveillance may help us understand the prevalence and dissemination of this novel antibiotic resistance gene.
Materials and methods
Bacterial isolation and susceptibility testing
All K. pneumoniae isolates were recovered from pig fecal matter and chicken cloacae samples collected from Shandong Province, China, according to a previously described protocol30. Isolates were identified as described previously27. The MICs of the K. pneumoniae isolates to colistin, tetracycline, meropenem, ceftazidime, amoxicillin-clavulanate, trimethoprim-sulfamethoxazole, amikacin, gentamicin, streptomycin, florfenicol, and ciprofloxacin were determined using the broth microdilution method according to the guidelines of the Clinical and Laboratory Standards Institute (document VET01-A4), and the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (http://www.eucast.org).
Conjugation assay
Conjugation assays were performed according to a previously described method31. In brief, the azide-resistant E. coli strain J53 was used as the recipient, and donor and recipient strains were mixed at a ratio of 1:3 on a microporous membrane for 12 h. The mixtures were collected and then plated on Luria-Bertani agar containing azide (100 μg/ml) and colistin (2 μg/ml).
Genome sequencing and assembly
Genomic DNA was extracted from K. pneumoniae isolate KP91 using a Wizard Genomic DNA Purification Kit (Promega, Beijing, China) and used as template for 150-bp paired-end whole-genome sequencing using the Illumina HiSeq 2500 System (Annoroad, Beijing, China). The draft genome was assembled using the CLC Genomics Workbench 9.0 (CLC Bio-Qiagen, Aarhus, Denmark). The conjugative plasmid was extracted from E. coli transconjugant strain J53-pKP91 using a Plasmid Midi Kit (OMEGA, Norcross, GA, USA) and then sequenced using both the Illumina HiSeq platform and the PacBio RSII System (Sinobiocore, Beijing, China). Plasmid assembly was performed using the Hierarchical Genome Assembly Process (HGAP) and Quiver as part of the SMRT analysis program (version 2.3) using the HGAP3 protocol and then corrected using Pilon.
Functional cloning of mcr-8
A 2631-bp DNA fragment including the mcr-8 coding sequence and its flanking regions was amplified from KP91 using the primers CMCR8-F (5′-CCCAAGCTTTTGATTGTCCCTGTCGCCAT-3′) and CMCR8-R (5′-CACCGATAAGAGGAACCAGTGAATTCCGG-3′) and then ligated into cloning vector pUC19 to yield pUC19-mcr-8. The recombinant vector was then transferred into E. coli DH5α and K. pneumoniae ATCC13883 via electroporation.
Sequence analysis and structure prediction of MCR-8
Plasmid replicon typing and multilocus sequence typing were carried out using tools available from the Center for Genomic Epidemiology website (http://whileereas genomicepidemiology.org/), whereas coding sequence prediction was completed using Prodigal (http://compbio.ornl.gov/prodigal/). Genomic sequences were annotated using the RFam (http://rfam.xfam.org), nr (http://www.ncbi.nlm.nih.gov/RefSeq/), KEGG (http://www.genome.jp/kegg/), and SwissProt databases32. Reference sequences of antibiotic resistance genes were obtained from the ARG-ANNOT database33. Prediction of the MCR-8 structure was carried out using i-Tasser homology modeling (https://zhanglab.ccmb.med.umich.edu/I-TASSER/), and the structure was depicted in cartoon form using PyMOL software 1.6.0 (PyMOL, DeLano Scientific LLC, San Carlos, America). Transmembrane α-helices were predicted using the Philius transmembrane prediction server (http://yeastrc.org/philius/showResults.do).
Detection of genes conferring resistance to colistin and carbapenem
The presence of the five colistin resistance genes, mcr-1 to -5, in the K. pneumoniae isolates was determined by PCR and Sanger sequencing. We also detected the presence of the carbapenem resistance genes blaNDM, blaVIM, blaIMP, blaOXA-48, and blaKPC in the K. pneumoniae isolates by PCR analysis. The primers and conditions used for the PCR assays are listed in Table S1.
PFGE, S1 nuclease-PFGE, and Southern blotting
PFGE analysis of K. pneumoniae isolates was performed using XbaI as the restriction endonuclease, as previously described34,35. Salmonella enterica serovar Braenderup H9812 digested with XbaI was used as the reference marker. The PFGE results were analyzed using InfoQuest software version 4.5 (Bio-Rad Laboratories, Hercules, CA, USA). S1 nuclease-PFGE was performed to determine the plasmid profiles. The genomic location of mcr-8 was indicated by Southern hybridization using a digoxigenin-labeled mcr-8 probe according to the manufacturer’s instructions for the DIG-High Prime DNA Labeling and Detection Starter Kit II (Roche Diagnostics, Basel, Switzerland).
Accession number
The complete nucleotide sequence of plasmid pKP91 has been deposited in GenBank under accession no. MG736312.
Electronic supplementary material
Acknowledgements
The study was supported by grants from the National Key Research and Development Program of China (2018YFD0500300) and the National Natural Science Foundation of China (31722057 and 31530076). We are grateful to professor Ming-Gui Wang (Institute of Antibiotics, Huashan Hospital, Fudan University, Shanghai, China) for kindly providing the reference strain E. coli J53AzR.
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Zhangqi Shen, Email: szq@cau.edu.cn.
Yang Wang, Email: wangyang@cau.edu.cn.
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41426-018-0124-z).
References
- 1.Wang Z, Qin RR, Huang L, Sun LY. Risk factors for carbapenem-resistant Klebsiella pneumoniae infection and mortality of Klebsiella pneumoniae infection. Chin. Med. J. 2018;131:56–62. doi: 10.4103/0366-6999.221267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Su, C. et al. Treatment outcome of non-carbapenemase-producing carbapenem-resistant Klebsiella pneumoniae infections: a multicenter study in Taiwan. Eur. J. Clin. Microbiol. Infect. Dis. 10.1007/s10096-017-3156-8 (2017). [DOI] [PubMed]
- 3.Falagas ME, Karageorgopoulos DE, Nordmann P. Therapeutic options for infections with Enterobacteriaceae producing carbapenem-hydrolyzing enzymes. Future Microbiol. 2011;6:653–666. doi: 10.2217/fmb.11.49. [DOI] [PubMed] [Google Scholar]
- 4.Poirel L, Jayol A, Nordmann P. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 2017;30:557–596. doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, et al. Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1-positive Enterobacteriaceae in patients and healthy adults from China: an epidemiological and clinical study. Lancet Infect. Dis. 2017;17:390–399. doi: 10.1016/S1473-3099(16)30527-8. [DOI] [PubMed] [Google Scholar]
- 7.Yi L, et al. mcr-1-harboring salmonella enterica serovar typhimurium sequence type 34 in pigs, china. Emerg. Infect. Dis. 2017;23:291–295. doi: 10.3201/eid2302.161543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo, J. et al. Emergence of mcr-1 in Raoultella ornithinolytica and Escherichia coli from retail vegetables, China. Antimicrob. Agents Chemother. 61, pii:e01139–17 (2017). [DOI] [PMC free article] [PubMed]
- 9.Mendes A, et al. mcr-1 in carbapenemase-producing Klebsiella pneumoniae with hospitalized patients, Portugal, 2016-2017. Emerg. Infect. Dis. 2018;24:762–766. doi: 10.3201/eid2404.171787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Principe L, et al. Multicenter prospective study on the prevalence of colistin resistance in Escherichia coli: relevance of mcr-1-positive clinical isolates in Lombardy, Northern Italy. Infect. Drug. Resist. 2018;11:377–385. doi: 10.2147/IDR.S160489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang, Y. et al. Novel plasmid-mediated colistin resistance gene mcr-7.1 in Klebsiella pneumoniae. J. Antimicrob. Chemother. 10.1093/jac/dky111 (2018). [DOI] [PubMed]
- 12.Xavier, B. B. et al. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, 2016. Euro. Surveill. 21 (27):pii=30280 (2016). [DOI] [PubMed]
- 13.Garcia-Graells C, et al. Detection of plasmid-mediated colistin resistance, mcr-1 and mcr-2 genes, in Salmonella spp. isolated from food at retail in Belgium from 2012 to 2015. Foodborne Pathog. Dis. 2018;15:114–117. doi: 10.1089/fpd.2017.2329. [DOI] [PubMed] [Google Scholar]
- 14.Yin W, et al. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. MBio. 2017;8:e00543. doi: 10.1128/mBio.00543-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carattoli, A. et al. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro. Surveill. 22, pii: 30589 (2017). [DOI] [PMC free article] [PubMed]
- 16.Borowiak M, et al. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J. Antimicrob. Chemother. 2017;72:3317–3324. doi: 10.1093/jac/dkx327. [DOI] [PubMed] [Google Scholar]
- 17.Teo J, et al. mcr-3 and mcr-4 variants in carbapenemase-producing clinical Enterobacteriaceae do not confer phenotypic polymyxin resistance. J. Clin. Microbiol. 2018;56:e01562–17. doi: 10.1128/JCM.01562-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukuda A, et al. High prevalence of mcr-1, mcr-3 and mcr-5 in Escherichia coli derived from diseased pigs in Japan. Int. J. Antimicrob. Agents. 2018;51:163–164. doi: 10.1016/j.ijantimicag.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 19.Bailey D, et al. Structural and functional characterization of aerobactin synthetase iucA from a hypervirulent pathotype of Klebsiella pneumoniae. Biochemistry. 2016;55:3559–3570. doi: 10.1021/acs.biochem.6b00409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Cassia A, et al. Presence of fimH, mrkD, andirp2 virulence genes in KPC-2-producing Klebsiella pneumoniae isolates in Recife-PE, Brazil. Curr. Microbiol. 2014;69:824–831. doi: 10.1007/s00284-014-0662-0. [DOI] [PubMed] [Google Scholar]
- 21.Huang Y, et al. mrkF is a component of type 3 fimbriae in Klebsiella pneumoniae. Res. Microbiol. 2009;160:71–79. doi: 10.1016/j.resmic.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 22.Hinchliffe P, et al. Insights into the mechanistic basis of plasmid-mediated colistin resistance from crystal structures of the catalytic domain of MCR-1. Sci. Rep. 2017;7:39392. doi: 10.1038/srep39392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stojanoski, V. et al. Structure of the catalytic domain of the colistin resistance enzyme MCR-1. BMC. Biology. 14, 81(2016). [DOI] [PMC free article] [PubMed]
- 24.Ma G, Zhu Y, Yu Z, Ahmad A, Zhang H. High resolution crystal structure of the catalytic domain of MCR-1. Sci. Rep. 2016;6:39540. doi: 10.1038/srep39540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu M, et al. Crystal structure of Escherichia coli originated MCR-1, a phosphoethanolamine transferase for colistin resistance. Sci. Rep. 2016;6:38793. doi: 10.1038/srep38793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao, Z., Feng, Y., Lin, J. & Zong, Z. Draft genome sequence of a colistin resistant Klebsiella pneumoniae clinical strain carrying the blaNDM-1 carbapenemase gene. Genome. Announc. 10.1128/genomeA.01654-16 (2017). [DOI] [PMC free article] [PubMed]
- 27.Anandan A, et al. Structure of a lipid A phosphoethanolamine transferase suggests how conformational changes govern substrate binding. Proc. Natl. Acad. Sci. USA. 2017;114:2218–2223. doi: 10.1073/pnas.1612927114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quan J, et al. Prevalence of mcr-1 in Escherichia coli and Klebsiella pneumoniae recovered from bloodstream infections in China: a multicentre longitudinal study. Lancet Infect. Dis. 2017;17:400–410. doi: 10.1016/S1473-3099(16)30528-X. [DOI] [PubMed] [Google Scholar]
- 29.Singh S, et al. Emergence of chromosome-borne colistin resistance gene mcr-1 in clinical isolates of Klebsiella pneumoniae from India. Antimicrob. Agents Chemother. 2018;62:e01885–17. doi: 10.1128/AAC.00195-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quan J, et al. Prevalence of mcr-1 in Escherichia coli and Klebsiella pneumoniae recovered from bloodstream infections in China a multicentre longitudinal study. Lancet Infect. Dis. 2017;4:400–410. doi: 10.1016/S1473-3099(16)30528-X. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, et al. Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat. Microbiol. 2017;2:16260. doi: 10.1038/nmicrobiol.2016.260. [DOI] [PubMed] [Google Scholar]
- 32.Boutet E, et al. UniProtKB/Swiss-Prot, the manually annotated section of the UniProt knowledgebase: how to use the entry view. Methods Mol. Biol. 2016;1374:23–54. doi: 10.1007/978-1-4939-3167-5_2. [DOI] [PubMed] [Google Scholar]
- 33.Gupta SK, et al. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 2014;58:212–220. doi: 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lei L, et al. mcr-1 in Enterobacteriaceae from companion animals, Beijing, China, 2012-2016. Emerg. Infect. Dis. 2017;23:710–711. doi: 10.3201/eid2304.161732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koroglu M, et al. Investigation of clonal relationships of K. pneumoniae isolates from neonatal intensive care units by PFGE and rep-PCR. J. Infect. Dev. Ctries. 2015;9:829–836. doi: 10.3855/jidc.6326. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.