Abstract
The incidence of Plasmodium vivax infection has declined markedly in Malaysia over the past decade despite evidence of high-grade chloroquine resistance. Here we investigate the genetic changes in a P. vivax population approaching elimination in 51 isolates from Sabah, Malaysia and compare these with data from 104 isolates from Thailand and 104 isolates from Indonesia. Sabah displays extensive population structure, mirroring that previously seen with the emergence of artemisinin-resistant P. falciparum founder populations in Cambodia. Fifty-four percent of the Sabah isolates have identical genomes, consistent with a rapid clonal expansion. Across Sabah, there is a high prevalence of loci known to be associated with antimalarial drug resistance. Measures of differentiation between the three countries reveal several gene regions under putative selection in Sabah. Our findings highlight important factors pertinent to parasite resurgence and molecular cues that can be used to monitor low-endemic populations at the end stages of P. vivax elimination.
Plasmodium vivax incidence in Malaysia has declined markedly over the last decade, despite evidence of chloroquine resistance. Here, Auburn et al. compare population structure of P. vivax in Malaysia to regions with intermediate and high transmission and identify genetic regions under putative selection.
Introduction
Outside of sub-Saharan Africa, Plasmodium vivax has become the predominant cause of malaria1. Reports of antimalarial drug resistance and life-threatening complications in children and pregnant women underline the importance of containing this species2–6. However, whilst the incidence of P. falciparum has declined in most endemic countries, the proportion of P. vivax infections in co-endemic regions has demonstrated a consistent rise, highlighting the remarkable adaptive potential and relative resilience of this species7. The re-emergence of P. vivax in many regions following successful elimination programmes serves as an important reminder of the parasite’s propensity to resurge in areas where it was once almost eliminated8. In order for malaria programmes to interrupt transmission successfully, a relentless focus is required on surveillance to identify foci of infection and instigate appropriate control responses to eliminate these. A better understanding of the transmission dynamics and adaptive processes of the parasite is essential. Whilst several genomic studies have explored the molecular dynamics of P. vivax populations with stable transmission9–13, aside from a small selection of microsatellite-based studies14–16, little is known about the molecular epidemiology during the critical, end stages of malaria elimination.
Malaysia has experienced a sharp decline in P. vivax prevalence over the past decade17,18, providing a unique endemic setting from which to assess the changing transmission dynamics and selective pressures as the parasite population enters the vulnerable malaria pre-elimination phase. A recent clinical trial demonstrated high-grade chloroquine resistance (CQR) with parasite recurrence reported within 28 days of starting treatment in 60% of patients19. Despite declining treatment efficacy, the number of reported P. vivax cases has fallen from approximately 3000 in 2005 to under 100 in 20151. Most P. vivax malaria is reported from the island of Borneo, where control efforts are challenged by constraints in case detection and accessibility to effective diagnosis and treatment. A recent microsatellite-based study of isolates collected in Sabah between 2010 and 2012 demonstrated local fragmentation and evidence of unstable transmission14. Since then, indigenous cases of P. vivax have continued to decline, but the threat of resurgence remains high, driven by an increase in imported cases1, and high levels of CQR19.
To explore the critical transition to P. vivax elimination, genome-wide population genetic analysis was undertaken on clinical isolates from Sabah, Malaysia and compared with isolates from western Thailand and Indonesia. In Thailand, P. vivax transmission is falling and 5 to 15% of patients treated with CQ have recurrent parasitaemia by day 28 (low-grade CQR)20. In Papua, Indonesia P. vivax transmission remains high and stable, with more than 50% of patients treated with CQ having recurrent parasitaemia by day 28 (high-grade CQR)21. The parasite population structure in Sabah was examined for evidence of unstable transmission and bottlenecking, and comparative analyses were undertaken against Thailand and Indonesia to identify genomic regions under putative selection. Further investigation of the temporal dynamics and epidemiological risk factors of specific parasite strains were investigated in a broader selection of samples using new and published microsatellite data14 on isolates collected between 2010 and 2015.
Our results reveal that the P. vivax population in Sabah is highly structured with high relatedness amongst infections, declining diversity and increased inbreeding, consistent with declining but also increasingly unstable transmission. A large clonal expansion is observed, potentially reflecting the emergence of an adaptive new strain. A high prevalence of known resistance determinants is observed in Sabah, and several new signals of selection have been revealed, requiring further investigation with clinical phenotypes. These findings highlight the substantial adaptive potential in a pre-elimination P. vivax population, and the need for a stronger framework for molecular surveillance to track the early emergence of adaptive new strains.
Results
Genomic data summary
A total of 259 independent samples had high-quality genomic data, with <5% missing genotype calls at 527,107 high-quality typeable single-nucleotide polymorphisms (SNPs). Corresponding genotype failure rates were below 5%. The high-quality samples included 51 isolates from Sabah collected between May 2011 and December 2014; 3 (5.9%) of which came from the West Coast, with the remaining 48 (94.1%) from Kudat Division. Additional high-quality samples were available from Thailand (n = 104) and Indonesia (n = 104).
Low complexity of infection in Sabah
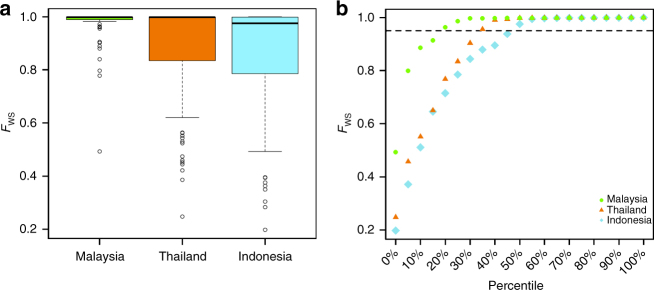
Within-sample parasite diversity was characterised using FWS analysis, and revealed a significantly higher proportion of monoclonal infections in Sabah (84.3%, 43/51) relative to Thailand (65.4%, 68/104) and Papua Indonesia (51.9%, 54/104) (Pearson’s χ2 test, p = 0.0234; p = 1.8 × 10−4), indicative of less frequent superinfection and/or co-transmission (Fig. 1). Individual FWS scores are presented in Supplementary Data 1.
Fig. 1.
FWS plots illustrating trends in within-sample infection complexity in Sabah Malaysia, Thailand and Indonesia. Boxplots (a) and scatter plots (b) of the FWS measure, which provides a gauge of the diversity within individual infections scaled from 0 (high diversity) to 1 (no diversity). The boxplot boxes present the median and interquartile range, with the lower whiskers presenting max(min(x), quartile 1–1.5 × interquartile range). The dotted line in (b) illustrates the FWS = 0.95, above which infections are essentially monoclonal. A higher proportion of monoclonal infections are observed in Sabah, Malaysia, relative to Thailand and Indonesia. The plots were generated using genomic data on 259 high-quality samples
Marked population structure in Sabah
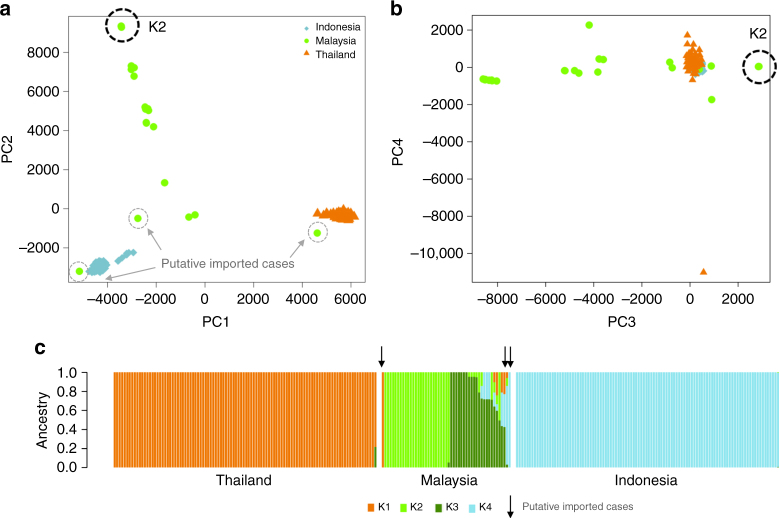
At the population-level, principal coordinate analysis (PCoA) illustrated that, other than three putative imported cases, the isolates from Sabah were genetically distinct from Thailand and Indonesia, and exhibited striking population structure (Fig. 2a, b). Extensive population structure was observed in Sabah, in marked contrast to Thailand and Indonesia, which displayed tight clusters with no evidence of structure. Whilst the first principal component (PC1) separated Thailand and Indonesia, located over 4500 km apart, PC2 was driven by the variation within Sabah, where the maximal geographic distance between sites was <200 km. ADMIXTURE analysis confirmed the separation between countries and distinct structure within Sabah, identifying four sub-populations in the country-wide sample set (Supplementary Fig. 2). In accordance with the PCoA plots, Thailand and Indonesia formed two distinct sub-populations (K1 and K4), while Sabah was split into two major sub-populations, K2 (n = 26, 51%) and K3 (n = 17, 33%), as well as a subset of admixed samples with <70% ancestry to any given K (n = 5, 10%), and 3 (6%) cases defined as putatively imported on the basis of ancestry ranging from 80 to 100% to K1 or K4 (Fig. 2c). Whilst the K2 sub-population was clearly diverged, there was also extensive divergence among the K3 and mixed infections. Despite the K2 and K3 isolates being derived from the same Division in Sabah (Kudat), the genetic divergence between these sub-populations (FST = 0.479) was greater than that observed between K1 and K4 (FST = 0.295).
Fig. 2.
Plasmodium vivax population structure in Sabah relative to Thailand and Indonesia. a, b PCoA plots illustrating the genetic differentiation within and between populations. Principal components 1–4 reflect 17.6%, 11.7%, 3% and 1.3% of the variance, respectively. The genetically identical Malaysian K2 isolates are circled in black and the three putatively imported infections are circled in grey; the unlabelled green dots reflect the K3 and mixed ancestry infections which constitute the baseline Sabah population. Aside from the K2 strain, there is clear evidence of divergence among the baseline Sabah isolates. c An ADMIXTURE bar plot illustrating the population structure within and among populations at K = 4, highlighting the clear distinction between Thailand, Malaysia and Indonesia, and the marked sub-structure within Malaysia. All plots were generated using genomic data derived from 259 high-quality samples
Genetically identical K2 infections
Neighbour-joining analysis confirmed the extensive structure within Sabah, illustrating clusters of highly related infections, in marked contrast to more panmictic structure in Thailand and Indonesia (Fig. 3). Strikingly, all 26 K2 infections were genetically near-identical (Fig. 3). The median difference between pairs of K2 isolates across the 527,107 SNPs was five nucleotides (i.e. differing at 0.0009% SNPs on average). After excluding the imported isolates and using a single representative of the K2 strain, the median SNP-based nucleotide difference across Sabah was 11,333 (2.15% SNPs), demonstrating a moderately diverse underlying reservoir of infection despite the high prevalence of identical isolates. Higher levels of pairwise nucleotide divergence were observed in Thailand (15,433 nucleotide differences, 2.93% SNPs) and Indonesia (13,423 nucleotide differences, 2.55% SNPs).
Fig. 3.
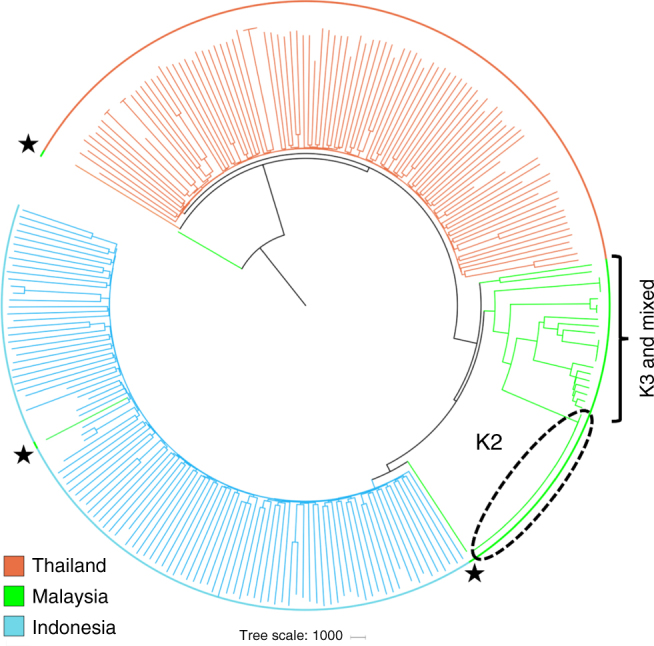
Neighbour-joining tree illustrating the relatedness between the P. vivax isolates from Sabah relative to Thailand and Indonesia The plot was created from genomic data on 259 high-quality infections and illustrates a rooted tree highlighting the genetically identical K2 cluster in Sabah, Malaysia. The K3 and mixed infections, which form the baseline Sabah population, are also annotated. Three putatively imported cases presenting in Sabah are annotated with black stars. One of the putatively imported infections (PY0045-C) aligned with the Papua Indonesian isolates, suggestive of importation from this region, whilst the other two cases aligned between Papua Indonesia and Sabah (PY0004-C), and close to Thailand (PY0120-C), presumably reflecting importation from regions not represented by the available sample set
Large region of sequence homology in Sabah
To explore the chromosome-level structure in the Sabah isolates, patterns of identity by descent (IBD) were explored across the genome of Sabah (both with all isolates and with a single K2 representative), Thailand and Indonesia (Supplementary Data 2). Sabah exhibited higher fractions of pairwise IBD across the genome (median = 0.446, range 0.061–0.768) than Thailand (median = 0.012, range 0.004–0.221) and Indonesia (median = 0.022, range 0.001–0.273) (Wilcoxon's rank-sum test, both p < 2.2 × 10−16) reflecting higher relatedness. Levels of pairwise IBD were further lower in the baseline Sabah population, where the K2 strain was represented once (median = 0.267, range 0.133–0.642). Illustrations of the IBD along each chromosome are presented in Supplementary Fig. 3. The top 5% IBD positions in Malaysia (all or baseline samples) were distributed across 10 regions on chromosomes 3, 4, 5, 8, 10 and 12. The largest stretch of IBD was observed on chromosome 12 (1942–2599 kb), with high fractions of IBD between both K2 and K3 isolates. The region encompasses 153 genes including multidrug resistance 2 (MDR2, PVP01_1259100), and gamete egress and sporozoite traversal (GEST, PVP01_1258000). Notable genes in other IBD regions include multidrug resistance 1 (MDR1) on chromosome 10, and dihydrofolate synthase/folylpolyglutamase synthase (DHFS-FPGS, PVP01_1229800) on chromosome 12. Details on the top IBD regions in Thailand and Indonesia, where a higher threshold of 1% was applied, can be found in Supplementary Fig. 3.
High prevalence of drug resistance mutations in Sabah
To assess whether the structure within Sabah reflected drug selection, the prevalence of mutations previously associated with clinical or ex vivo antimalarial resistance2 was investigated (Table 1). Both the K2 and K3 sub-populations exhibited 100% prevalence of the MDR1 F1076L and Y976F variants and the dihydropteroate synthase (DHPS) A553G and A383G variants. The prevalence of reference alleles differed between K2 and K3 at DHPS G626A, A619T, E618D and E132G (0% in K2 vs. 76–82% in K3, Pearson’s χ2 test, p = 4.4 × 10−4–4.6 × 10−4) (Supplementary Data 3); the clinical relevance of these variants is unknown. Although differences were observed in the prevalence of dihydrofolate reductase (DHFR-TS) quadruple mutants (0% in K2 vs. 67% in K3, Pearson’s χ2 test, p = 1.0 × 10−5), overall there was a 100% prevalence of one or more mutations in both sub-populations. None of the Sabah isolates had increased copy number (CN) of MDR1.
Table 1.
Prevalence of variants that have previously been associated with P. vivax drug resistance
| Gene | Chr. | Positions | Mutation | Drug | Sabah frequency | Sabah K2 frequency | Sabah K3 frequency | Thailand frequency | Indonesia frequency |
|---|---|---|---|---|---|---|---|---|---|
| MDR1 | 10 | 479,908 | F1076L | CQ | 98% (50/51) | 100% (26/26) | 100% (17/17) | 50% (54/104) | 100% (104/104) |
| (PVP01_1010900) | 10 | 480,207 | Y976F | CQ, AQ + SP | 94% (48/51) | 100% (26/26) | 100% (17/17) | 13% (14/104) | 100% (104/104) |
| 10 | Copy number variant | 2+ copies | MQ | 0% (0/49) | 0% (0/26) | 0% (0/17) | 19% (20/104) | 0% (0/84) | |
| DHFR-TS | 5 | 1,077,530; 1,077,532 | F57L/I | Antifolate, AQ + SP | 96% (46/48) | 100% (26/26) | 100% (15/15) | 91% (88/97) | 81% (71/88) |
| (PVP01_0526600) | 5 | 1,077,533; 1,077,534; 1,077,535 | S58R | Antifolate, AQ + SP | 34% (17/50) | 0% (0/26) | 69% (11/16) | 100% (104/104) | 99% (97/98) |
| 5 | 1,077,543 | T61M | Antifolate, AQ + SP | 25% (13/51) | 0% (0/26) | 67% (10/15) | 91% (90/99) | 81% (71/88) | |
| 5 | 1,077,711 | S117N/T | Antifolate, AQ + SP | 25% (12/48) | 100% (26/26) | 100% (17/17) | 100% (102/102) | 99% (87/88) | |
| Double mutant | Antifolate, AQ + SP | 73% (35/48) | 100% (26/26) | 33% (5/15) | 1% (1/89) | 17% (15/87) | |||
| Triple mutant | Antifolate, AQ + SP | 2% (1/48) | 0% (0/26) | 0% (0/15) | 0% (0/89) | 0% (0/87) | |||
| Quadruple mutant | Antifolate, AQ + SP | 25% (12/48) | 0% (0/26) | 67% (10/15) | 99% (88/89) | 82% (71/87) | |||
| DHPS | 14 | 1,270,401 | A553G | Antifolate | 92% (46/50) | 100% (26/26) | 100% (17/17) | 98% (98/100) | 18% (16/91) |
| (PVP01_1429500) | 14 | 1,270,911 | A383G | Antifolate | 96% (48/50) | 100% (26/26) | 100% (17/17) | 100% (104/104) | 97% (99/102) |
Mutation prevalence was calculated with homozygous calls only. Genotype failures were <5% at all markers in all populations
CQ chloroquine, AQ amodiaquine, SP sulfadoxine-pyrimethamine, MQ mefloquine
The prevalence of non-synonymous variants in orthologues of genes implicated in drug resistance in P. falciparum was also investigated (Supplementary Data 3). Plasmepsin IV (PVP01_1340900) is an orthologue of P. falciparum plasmepsin II (PF3D7_1408000), in which CN amplification has been associated with piperaquine resistance22,23. The frequency of a plasmepsin IV I165V variant differed significantly between Sabah (90%) and Thailand (40%, Pearson’s χ2 test, p = 0.0052), but not Indonesia (89%, Pearson’s χ2 test, p = 1.000). There was no significant difference between the three countries in the prevalence of variants in pvcrt-o (PVP01_0109300) or kelch-13 (PVP01_1211100), whose P. falciparum orthologues (PF3D7_0709000 and PF3D7_1343700) are associated with CQ and artemisinin resistance respectively24,25. Details of the non-synonymous mutations of the P. vivax orthologues of P. falciparum multidrug resistance-associated proteins 1 and 2, and multidrug resistance protein 2 (PVP01_020300, PVP01_1447300 and PVP01_1259100 respectively) are presented in Supplementary Data 3.
Highly differentiated variants in drug resistance candidates
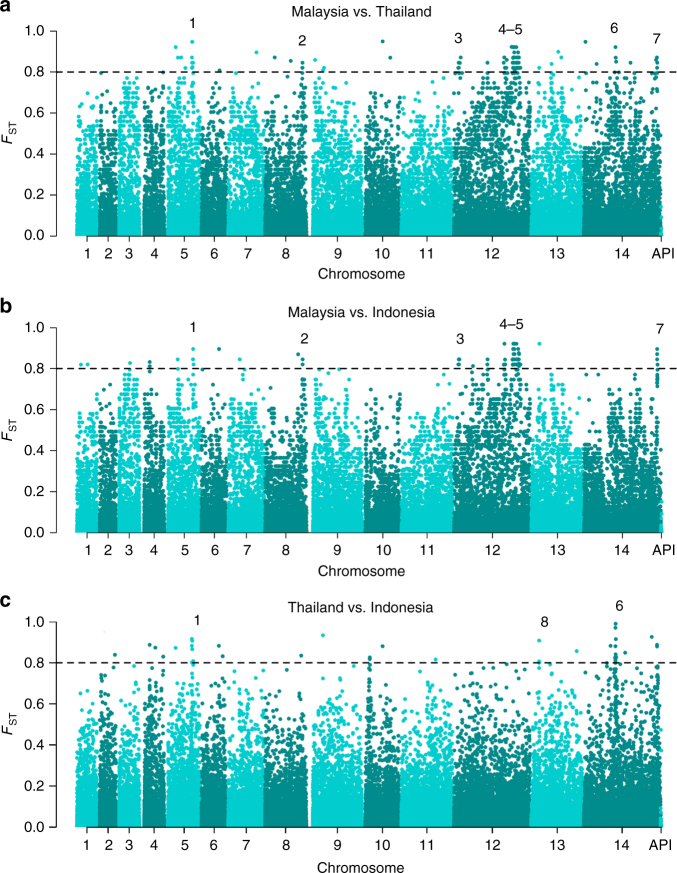
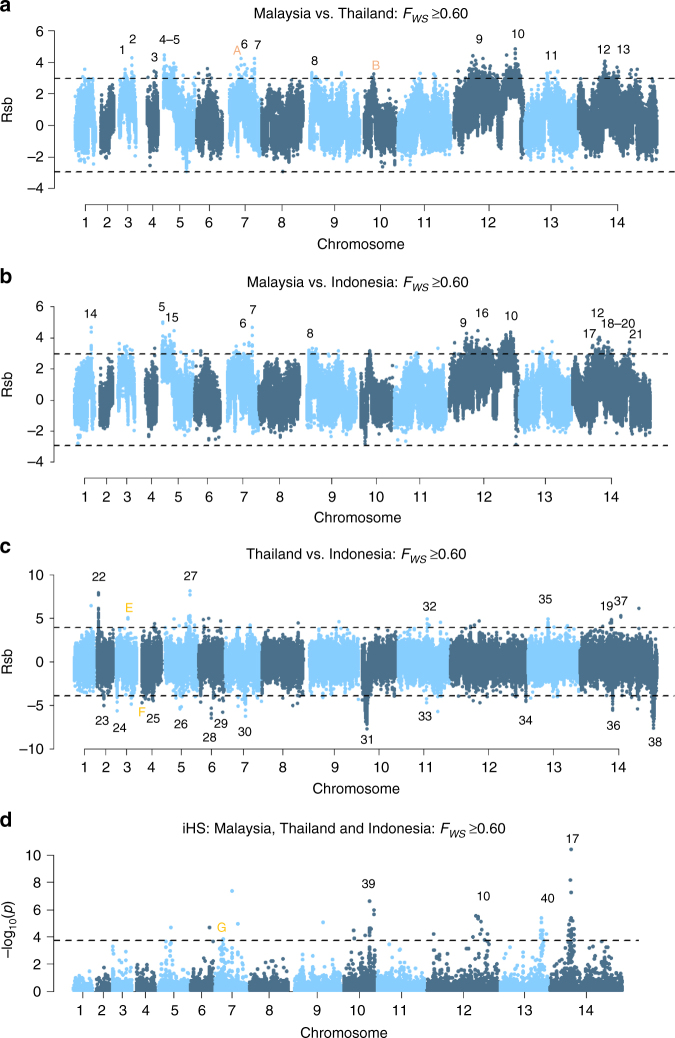
To identify potential new modulators of drug resistance, measures of genetic differentiation (FST) and Rsb-based cross-population extended haplotype homozygosity were undertaken to detect gene regions with evidence of selection. The extensive homology among the K2 isolates obscured signal detection in Sabah; hence, analysis was undertaken with a single representative of this strain. A high-level summary of the multi-SNP FST and Rsb signals is presented in Supplementary Data 4, with detailed results provided in Supplementary Data 5 and 6, respectively. Seven Sabah-specific regions exhibited multiple highly differentiated (FST > 0.8) SNPs (Fig. 4a, b). In Rsb-based comparisons between Sabah and each of Thailand and Indonesia, the top 0.5% of SNPs all comprised regions with extended homozygosity in Sabah (Fig. 5a, b), with 13 and 15 multi-SNP regions identified, respectively.
Fig. 4.
Genetic differentiation between Malaysia, Thailand and Indonesia. a–c Manhattan plots illustrating the FST between Malaysia (Sabah), Thailand and Indonesia. The Sabah population included a single K2 representative and excluded the three imported cases. Horizontal dashed lines illustrate FST = 0.8. A total of 97 and 76 positions exhibited FST scores ≥0.8 between Sabah and Thailand and Indonesia, respectively; 27 and 19 conferring non-synonymous changes. Seven multi-SNP regions identified in comparisons against Sabah are labelled: these regions comprise ≥3 SNPs with FST≥0.8 within 80 kb of one another. Region 1 encompasses DHFR-TS. Region 2 contains two genes on chromosome 8 with unknown function (PVP01_0834900 and PVP01_0835000). Region 3 is located on chromosome 12 and includes a nucleoside transporter (NT1) with a highly differentiated L188M mutation between Sabah and Thailand, but not Sabah and Indonesia. Regions 4 and 5 are located adjacently on chromosome 12, corresponding with the highly homologous Sabah sequence. The regions contain biologically plausible drug resistance candidates including MDR2 and haeme detoxification protein (HDP). Another notable gene is GEST, implicated in transmission between the parasite and both its Anopheline and human hosts37. Region 6 encompasses DHPS. Region 7 is located on chromosome 14, encompassing 7 genes but with no clear driver of selection. Region 8 contains several SNPs on chromosome 13 exhibiting differentiation between Thailand and Indonesia only. The region encompasses the cysteine repeat modular protein 3 (CRMP3). In addition to GEST and CRMP3, two other genes with roles in host cell invasion or egress displayed highly differentiated variants; CRMP1 and glideosome-associated protein 40 (GAP40). Signatures of selection between southeast Asian and South American P. vivax populations have previously been observed in CRMP126, but this is the first report of selection in GEST and GAP40
Fig. 5.
Genome-wide scans to identify regions with extended haplotype homozygosity using low-complexity samples. a–c Manhattan plots of the Rsb index for the given populations, and d the iHS p value for the pooled populations. Analyses were conducted on low-complexity samples (FWS≥0.60). The dashed black lines demark the top 0.5% SNPs with the most significant p values. Regions with ≥3 SNPs within 80 kb of one another and overall SNP density <10 kb per SNP amongst the top 0.5% SNPs are numbered in black. Several previously described signals in known or putative drug resistance candidates were identified including DHPS (region 19), DHFR-TS (region 27) and MRP1 (region 22)10–13. A previously described signal on chromosome 10 (region 31) is downstream of MDR111. The signal in region 10 reflects the highly homologous region on chromosome 12. Putative genetic drivers in the chromosome 12 region are MDR213, GEST and HDP, but the latter has no directly supporting SNPs. Other drug resistance candidates include nucleoside transporter 2 and folate transporter 1 in regions 4 and 5, drug/metabolite transporter 1 (DMT1) in region 12, CG2-related protein in region 21, a previously described voltage-dependent anion-selective channel (region 35)11, and plasmepsin IV in region 40. The putative drivers in regions 3, 7, 9 and 39 include several merozoite surface proteins: MSP5, MSP1, MSP7-like gene cluster and MSP3.2, respectively12, 26. Another surface protein presenting a putative driver is the duffy-binding protein in region 29. In other gene classes, candidate drivers include zinc finger protein (region 6), calcium/calmodulin-dependent protein kinase (region 8), filament assembling protein (region 14), Plasmodium exported proteins (region 15), an AP2 domain transcription factor (region 17)13, DNA-binding protein (region 23), 3’ exonuclease (region 25) and JmjC domain-containing protein (region 36). The putative genetic drivers in regions 13, 18, 24, 28, 34 and 38 have unknown function, and those in regions 1, 2, 11, 16, 20, 26, 30, 32, 33 and 37 remain unclear. Orange letters indicate regions with 1–2 SNPs amongst the top 0.5% SNPs that were represented by ≥3 SNPs in the monoclonal samples (FWS≥0.95) and are detailed in Supplementary Fig. 4
Evidence of differential selection was detected with the FST metric at multiple SNPs in the DHFR-TS and DHPS regions. Both the FST and Rsb measures found evidence of differential selection in the chromosome 12 region with high homology in Sabah. Two non-synonymous MDR2 variants (V43L and A603T) were among the top 0.5% of SNPs identified with Rsb analysis in comparisons against Sabah. The chromosome 5 region with high homology in Sabah also demonstrated evidence of differential selection supported by the FST and Rsb measures. In other genomic regions, putative drug resistance candidates included a putative drug/metabolite transporter (DMT1, PVP01_1424900) in a region of chromosome 14 supported by multiple SNPs amongst the top 0.5% of Rsb scores exhibited a V217I variant displaying high differentiation between Sabah and Thailand (FST = 0.71), and Thailand and Indonesia (FST = 0.88), but not between Sabah and Indonesia (FST = 0.02). Also on chromosome 14, a CG2-related protein (PVP01_1450700) displayed multiple SNPs amongst the top 0.5% of Rsb scores.
The putative drug resistance-associated signals in the comparisons of Thailand vs. Indonesia (Figs. 4c and 5c) have been described previously11. These include SNPs in DHFR-TS and DHPS demonstrating high differentiation by FST analysis, and regions with extended haplotype homozygosity in Thailand encompassing DHFR-TS and MRP1. A region with extended haplotype homozygosity in Indonesia was also observed downstream of MDR1.
Several genes with functions unrelated to antimalarial drugs displayed highly differentiated variants, including GEST, which exhibited two non-synonymous mutations, E68D and K144R, displaying high differentiation between Sabah and both Thailand and Indonesia (all FST >0.90), and a Q230L mutation among the top 0.5% Rsb scores in comparisons of Malaysia to Thailand. In other regions, the merozoite surface proteins 1 (MSP1) and 5 (MSP5) were notable, exhibiting 15 and 3 variants, respectively, among the top 0.5% Rsb scores in comparisons of Malaysia to Thailand and Indonesia.
Signals of extended haplotype homozygosity using the iHS
Further haplotype-based scans were undertaken using the integrated haplotype score (iHS) measure to identify regions with differential levels of linkage disequilibrium (LD) surrounding the ancestral (P. cynomolgi) relative to the derived allele. Derived alleles could be confidently called at only 10,167 loci and, after filtering out non-informative loci, the homology in Sabah proved too extensive to permit iHS analysis in this population alone. The analysis was therefore conducted on the pooled Sabah, Thai and Indonesian samples. Four regions were supported by multiple SNPs amongst the top 0.5% of scores (Fig. 5d and Supplementary Data 4). The first region overlaps with a previously described signal on chromosome 10, potentially reflecting selection on MSP3.226. The second region comprised SNPs in the chromosome 12 region with high homology in Sabah. The third region presents a signal on chromosome 13, possibly driven by selection on plasmepsin IV. The fourth region overlaps with a previously described signal on chromosome 14 postulated to reflect selection on an AP2 domain transcription factor (PVP01_1418100)13. Further details are presented in Supplementary Data 7.
Assessment of the iHS and of the Rsb tests using strictly monoclonal samples demonstrated no major differences in the key trends, confirming the accuracy of the results on the low-complexity samples (Supplementary Fig. 4).
CN amplifications in Sabah
Investigation of CN variation was undertaken on 46 Sabah, 104 Thai and 84 Indonesian isolates: excluding 22 isolates with uneven coverage. Three regions demonstrated evidence of CN amplification in Sabah; a 28S ribosomal RNA gene on chromosome 5 (PVP01_0504500), duffy-binding protein 1 on chromosome 6 (DBP1, PVP01_0623800)11,27,28 and an exported Plasmodium protein with unknown function on chromosome 14 (PVP01_1470400)11 (Table 2 and Supplementary Data 8). The PVP01_0504500 and PVP01_1470400 amplifications were highly prevalent in Sabah (82–96% in K2 and K3), and also demonstrated moderate to high frequency in Thailand (92 and 49%) and Indonesia (95 and 46%).
Table 2.
Summary of copy number variants observed in Sabah
| Gene | Description | Chr. | Sabah frequency | Sabah K2 frequency | Sabah K3 frequency | Thailand frequency | Indonesia frequency |
|---|---|---|---|---|---|---|---|
| PVP01_0504500 | 28S ribosomal RNA | 5 | 89% (41/46) | 96% (25/26) | 82% (14/17) | 92% (96/104) | 95% (80/84) |
| PVP01_0623800 | Duffy-binding protein 1 | 6 | 4% (2/46) | 0% (0/26) | 6% (1/17) | 30% (31/104) | 6% (5/84) |
| PVP01_1470400 | Plasmodium exported protein | 14 | 89% (41/46) | 96% (25/26) | 82% (14/17) | 49% (51/104) | 46% (39/84) |
Twenty-two samples with an excess of CNVs (18+) were excluded from the analysis
Increasingly unstable transmission in Sabah over time
To explore the epidemiology of P. vivax in Sabah further, genetic data were obtained on a larger collection of samples using short tandem repeat (STR) genotyping. A total of 212 independent PCR-confirmed P. vivax mono-species or mixed-species infections were successfully genotyped (<50% marker fails), with 187 isolates having complete data. Demographic details of the patients are summarised in Supplementary Table 1.
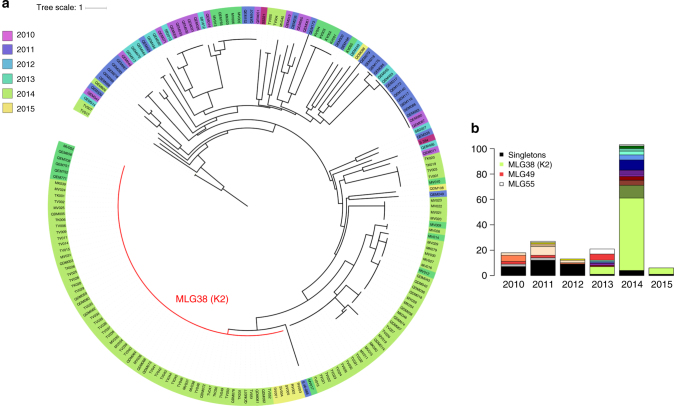
Among the 51 genomic samples, 47 could be genotyped successfully, with 46 having complete data. Consistent with the genomic data, all 25 K2 infections exhibited the same multi-locus genotype (MLG38). A further 44 samples carried MLG38, comprising 69 K2 isolates in total (Fig. 6a). Assessment of the temporal dynamics of the K2 strain revealed epidemic-like transmission. K2 was first observed in 2013 (29%), rose sharply in prevalence in 2014 (55%) and persisted into 2015 (83%) up to the end of the collection period (Fig. 6b).
Fig. 6.
Temporal trends in the relatedness of Sabah P. vivax isolates using the microsatellite data. a A neighbour-joining tree illustrating trends in the relatedness of P. vivax infections between 2010 and 2015. Data are presented on 187 P. vivax isolates with complete MLGs across the nine microsatellite loci. b A bar plot illustrating temporal trends in the frequency of different MLGs between 2010 and 2015. Colour coding reflects distinct MLGs with the exception of singly observed (singleton) MLGs, which were grouped together for simple visual representation (back bars). MLG38 (genomic K2 strain) was the most frequent MLG observed in the dataset. MLGs 49 and 55 persisted for at least 4 years
In addition to K2, several smaller clusters of highly related infections were observed in Sabah, indicative of broadly unstable transmission in the region (Fig. 6a). Collectively, the results of several population genetic measures indicated a state of declining transmission intensity concurrent with increasing instability. First, a gradual decline was observed in the percentage of polyclonal infections, dropping from 50% in 2010 to 8% in 2014 (p = 1.16 × 10−6), suggestive of reduced rates of superinfection (Supplementary Table 2), and this was associated with a strong, positive correlation between the annual number of P. vivax cases and the proportion of polyclonal infections (Pearson’s correlation, r = 0.98, p = 0.004). Second, there was a trend of declining diversity, as measured by allelic richness (Rs), over the study years (Supplementary Table 2). Third, investigations of LD revealed high allelic associations (ISA range 0.167–0.536), reflecting high rates of inbreeding owing to a composite of bottlenecking and clonal expansions (Supplementary Table 3). On restricting analysis to unique MLGs, LD remained significant in all years, but the ISA scores dropped between 1.5-fold and 10-fold in each year, indicative of epidemic-like transmission. The trends in Rs and LD remained the same when analysis was restricted to low-complexity infections (Supplementary Tables 2 and 3).
High proportion of outbreak (K2) infections among students
To characterise the epidemiology surrounding the expansion of the K2 strain (MLG38), we explored the demographics of individuals carrying K2 vs. low-frequency strains (Supplementary Table 4). No significant difference was observed in the proportion of males (62 vs. 73%, Pearson's chi-squared test, p > 0.05) or median parasite density (4180 vs. 4186 parasites µl−1, Wilcoxon's rank-sum test, p > 0.05). However, a significant difference was observed in the median age of patients carrying K2 (15 years) vs. low-frequency MLGs (24 years, p = 0.0137). Occupational details were available for 182 (97%) of the patients with complete MLGs. Two categories were investigated; agricultural/foresty-related occupations and students. A higher proportion of students were observed among the patients carrying K2 (37%) vs. low-frequency MLGs (16%, Pearson's chi-squared test, p = 0.012). No significant differences were observed in the proportion of patients with agricultural/forestry-related occupations (Pearson's chi-squared test, p > 0.05).
Discussion
Multiple genomic studies have been conducted in the Asia-Pacific region10,11,13, but none have examined the molecular epidemiology of P. vivax during the critical transition phase to elimination. Our investigation of the genetic and genomic epidemiology of P. vivax in Sabah, Malaysia, provides important insights into the molecular changes taking place in a parasite population in the late stages of malaria elimination. Our analysis reveals a highly structured population with high relatedness, declining diversity and increased inbreeding demonstrative of marked interruption to local transmission. A continual flux was observed in the genetic make-up of the parasite population, including several small and one large, rapidly emerging clonal expansion. Whilst some of these changes may have resulted from demographic or neutral processes, others may reflect more concerning adaptive changes as the parasite population is bottlenecked under heavy pressure from antimalarial drugs and other selective forces.
Antimalarial drug pressure in P. falciparum has been shown to have an important role in shaping the genetic make-up in Cambodian isolates29. We postulate that selection may have had a similar impact on P. vivax in Sabah. Genomic studies have shown that antimalarial drugs confer some of the strongest selective pressures on the parasite genome10–13. Intense drug selection is a highly likely consequence of the final stages of elimination, providing an environment in which only the most drug-resistant infections survive30. Furthermore, the population structure in low transmission settings may foster the emergence of multigenic resistance phenotypes. In highly structured populations where inbreeding is common, the resistance alleles are likely to be common and unlikely to be separated during recombination. The Sabah P. vivax population exhibited a highly structured pattern, with divergence not just of the K2 strain, but also among the K3 and mixed infections. The extreme divergence in Sabah relative to the other Asia-Pacific populations mirrors that observed in P. falciparum in Cambodia as artemisinin-resistant founder populations emerged29, suggesting that founder populations may already be emerging in response to selective pressures. However, additional phenotypic data are needed to confirm this hypothesis as the extensive divergence in Sabah may also reflect extensive bottlenecking alone.
Until recently, CQ was recommended for treating P. vivax blood-stage infections in Malaysia. However, the vivax population in Sabah has also been exposed to artemisinin, mefloquine, lumefantrine and sulfadoxine plus pyrimethamine, which have been used to treat co-endemic Plasmodium species. In 2016, the efficacy of CQ against P. vivax in Sabah was shown to have fallen below 40%, and antimalarial guidelines were changed to artemether-lumefantrine for all species of malaria19. Prior to this date there had been no formal clinical evaluation of CQR in P. vivax in Malaysia, although a retrospective hospital-based study from peninsular Malaysia between 1994 and 2010 suggested low levels of CQR31.
We were able to compare the parasite genomes collected in Sabah with those collected from western Thailand and Papua, Indonesia. Previous clinical surveys between 2007 and 2010 have shown low levels of CQR (<15% recurrence by day 28) in P. vivax populations on the Thai-Myanmar border32,33. In Papua, Indonesia high-grade CQR (>60% recurrence by day 28) was documented in 2005 following which antimalarial policy was changed to DHA-piperaquine21. Over the past 12 years nearly all patients with uncomplicated malaria, in both the public and private sector, are treated with DHA-piperaquine, with very little use of CQ34. The prevalence of CQ resistant P. vivax isolates may therefore have declined in recent years; however, no clinical data are available to confirm this.
We observed a high prevalence (95–100%) of polymorphisms previously associated with drug resistance in P. vivax, including double, triple and quadruple mutations in DHFR-TS and DHPS in Sabah. Signals of differential selection reflecting different haplotypic backgrounds were also observed in the DHFR-TS and DHPS regions in comparisons between Sabah and Thailand and Indonesia. A high prevalence of MDR1 Y976F and F1076L variants was also observed in Sabah. However, MDR1 appears to be, at best, a modulator of CQR, with definitive markers of resistance to this drug remaining elusive2.
On investigation of polymorphisms previously associated with drug resistance in P. vivax in K2 vs. K3, we observed differences in the prevalence of several DHPS and DHFR-TS alleles, potentially reflecting sulfadoxine-pyrimethamine pressure. The extensive homology in the K2 sub-population did not permit genome-wide investigations of differential selection and haplotype homozygosity to identify any novel selective determinants. Nonetheless, although the K2 sub-population was notable for its extreme genetic identity, the extensive divergence across Sabah was not restricted to this strain. Therefore, rather than limiting our investigation of drug-related selective pressure to the K2 strain, we undertook genome-wide tests of selection in the baseline Sabah population using a single K2 representative.
Several new genes identified using genome-wide tests of selection in the current study present plausible molecular determinants of CQR. The MDR2 gene is a biologically plausible resistance determinant owing to its function as a cellular transporter, and was located on a specific haplotype background in the majority of Sabah isolates in a region with evidence of extended haplotype homozygosity based on iHS analysis, and differential selection based on FST and Rsb measures. The MDR2 gene also demonstrated differential selection in Sabah relative to Thailand and Indonesia at non-synonymous variants supported by both the FST and Rsb metrics.
The P. vivax CG2-related protein, on chromosome 14, presents another putative determinant of CQ efficacy. In P. falciparum, CG2 protein is involved in the uptake and digestion of haemoglobin in erythrocytic stage parasites35. Other biologically plausible genes identifed as potential modulators of resistance to CQ or other antimalarials on the basis of intragenic supporting SNPs with evidence of genetic differentiation or extended haplotype homozygosity included plasmepsin IV and drug/metabolite transporter 1. The plasmepsin IV gene on chromosome 13, which encodes a hemoglobinase, and exhibited an I165V variant amongst the top 1% of iHS scores, is intriguing owing to the role of CN variation in P. falciparum plasmepsin II in piperaquine resistance22,23.
The candidates identified add several new genes for consideration in future molecular studies of P. vivax drug resistance, although these require further investigation and validation with clinical or ex vivo phenotypes. Whilst we were conservative in our selection of regions under positive selection, and identified signatures in previously reported regions, the population structure in Sabah presents a challenge in this low-endemic setting. Furthermore, comparison with the Indonesian isolates, where high-grade CQR has been reported previously, may have been confounded by the high rate of local transmission36, which may have rapidly reduced haplotype homozygosity surrounding any drug resistance alleles in this population.
Selective forces other than antimalarial drug pressure may have also shaped the genetic structure in Sabah. There was evidence of differential selection in multiple genes involved in gamete egress and sporozoite traversal, including GEST. The P. berghei orthologue of GEST has important roles in mediating transmission between the parasite and both its Anopheline and human hosts37, and hence certain variants of this gene may influence transmission. In addition, there was evidence of extended haplotypes in Sabah in regions encompassing MSP1 and MSP5, potentially reflecting immune-related pressure.
In addition to selective forces, the genetic dynamics in Sabah are likely to have been influenced by demographic factors. Using STR-based genotyping data, we were able to explore the contribution of several host demographic factors to the expansion of the K2 strain. The data revealed that students relative to other occupational groups assessed were more likely to carry K2 alleles than low-frequency strains. This finding might reflect factors related to the immune status of this particular age group, who had a median age of 15 years. Alternatively, it might reflect a high-frequency of K2 transmission events taking place in or around school, college or home, highlighting occupation as a potentially important epidemiological link in reactive case detection strategies. However, further investigation of transmission networks is required with more detailed epidemiological data.
Based on clustering patterns in the genomic data, we were able to determine putative imported infections, identifying three cases in Sabah. The K2 strain did not display any evidence of importation. Rather, the large stretches of homologous sequence between K2 and other Sabah infections in regions such as chromosome 12 suggests recent common ancestry. However, we cannot discard the possibility that the K2 lineage derives from a local recombination event between an isolate from Sabah and an imported infection, bringing together genetic features that were highly adaptive to the local environment.
The moderate diversity in the baseline Sabah population supports the local adaptive potential. Aside from imported infections, it will be important to determine the contribution of a hidden reservoir of asymptomatic or subpatent infections in maintaining local diversity, and whether the genetic structure of these infections is comparable to the clinical cases. A genetic study of actively and passively detected P. vivax cases from Timika, Papua Indonesia, revealed similar genetic diversity between the two reservoirs, although a higher frequency of polyclonal infections in the actively detected cases38. However, there are no data available on the genetic make-up of passively vs. actively detected P. vivax in low-endemic settings.
Rebound or resurgence of infection remains a great concern for malaria elimination programs. The continual flux in the genetic make-up of the Sabah vivax population, rapid expansion of new strains and high prevalence of known resistance-associated determinants highlight the strong adaptive potential in a population on the verge of elimination. These findings have important implications for other vivax endemic countries striving to achieve malaria elimination, underlining critical areas for further research. A stronger foundation for molecular surveillance is needed, to track the early emergence of adaptive parasites that will guide appropriate and timely public health interventions.
Methods
Ethics
All samples were collected with written informed consent from patients or their legal guardians. Ethical approval was provided by the Human Research Ethics Committee of Northern Territory Department of Health and Families (HREC-2010-1431, HREC-2012-1815 and HREC-2010-1396), the Medical Research Ethics Committee, Ministry of Health, Malaysia (NMMR-10-754-6684, NMRR-12-511-12579), the Mahidol University Faculty of Medical Technology (MUTM 2011-043-03), the Eijkman Institute Research Ethics Committee (EIREC-47) and the Oxford Tropical Research Ethics Committee (OXTREC-45-10).
Study site
The focal study site was in Sabah, Malaysia (Supplementary Fig. 1a). Details on the epidemiology of malaria in Sabah have been published previously17. Four species of malaria are endemic in the region: P. falciparum, P. vivax, P. malariae and P. knowlesi. Whilst the incidence of P. vivax and P. falciparum has declined substantially over the past decade, P. knowlesi cases have risen markedly, to become the most common cause of malaria (Supplementary Fig. 1b). There is no significant seasonal variation in malaria transmission, with the region experiencing moderately constant temperature and high humidity throughout the year. Until 2016, the national policy for treating uncomplicated P. vivax infection was CQ plus primaquine (14-day regimen), but following documentation of high-grade CQR, first-line treatment was changed to artemether-lumefantrine plus primaquine.
Sample details
Details of the genomic samples are presented in Supplementary Data 1. The Sabah samples were sourced from clinical and epidemiological studies conducted at the Queen Elizabeth Hospital in Kota Kinabalu, Kudat District Hospital or Kota Marudu District Hospital between September 2010 and June 201519,39,40. Two to five millilitres of venous blood was collected from symptomatic patients with Plasmodium spp.-positive thick blood smears. A 200 µl aliquot of blood was set aside for PCR-based assays. The remaining aliquot was subject to leukocyte depletion by cellulose filtration in preparation for whole-genome sequencing41. DNA extraction was performed using commercial kits (Qiagen). Plasmodium species was confirmed by PCR, using previously described assays42,43. For comparative analysis, previously published genomic data were derived from P. vivax isolates from Thailand and Indonesia11.
Whole-genome sequencing and data analysis
Leukocyte-depleted samples with ≥50 ng DNA comprising <90% human DNA were subject to whole-genome sequencing, read alignment and variant calling within the framework of a community study in the Malaria Genomic Epidemiology Network (MalariaGEN)44. Briefly, sequencing was undertaken on the Illumina GAII or Hi-Seq 2000 platform at the Wellcome Trust Sanger Institute. Paired-end libraries were prepared according to the manufacturer’s protocol. Cluster generation and sequencing were undertaken following the manufacturer’s protocols for generating standard paired-end 75–150 bp reads. Reads aligning to the human reference genome were removed before any analyses were undertaken. The remaining reads were aligned against the P. vivax P01 reference45 using bwa-mem version 0.7.15 with -M parameter46. Improvements of the bam files were undertaken using Picard version 2.6.0 tools CleanSam, FixMateInformation and MarkDuplicates, and GATK version 3.6 base quality score recalibration. Standard alignment metrics were generated for each sample using the flagstat utility from samtools version 1.2 and GATK’s CallableLoci version 3.547,48. SNP discovery and genotype calling were undertaken using methods defined previously for P. vivax field isolates11, with data derived from the MalariaGEN P. vivax Genome Variation Project release 3.0. A set of 532,751 high-quality bi-allelic SNPs with VQSLOD score >3 were derived from an initial set of 4,084,419 discovered variants. High-quality samples were defined as those with ≥95% calls at the high-quality SNPs. A set of 527,107 high-quality typeable SNPs was then derived by excluding SNPs with >5% missing calls in the high-quality samples. Missing calls were defined as positions with <5 reads. For haplotype-based analyses, either the major allele (highest read depth) or reference allele (at positions with equal allele depths) were used to reconstruct haplotypes at heterozygote positions. Where missing data were not permitted, haplotypes were reconstructed using information from calls with <5 reads.
For assessment of parasite relatedness, neighbour-joining (NJ) and PCoA were conducted using a pairwise distance matrix calculated using the R statistical package. The NJ plot was created using the iTOL software49. Assessment of population structure was performed using ADMIXTURE version 1.3.0, with the most likely number of sub-populations (K) determined by the cross-validation error50. ADMIXTURE was also used to determine the genetic differentiation (FST) (combined across all SNPs) between the derived sub-populations.
Within-host infection complexity was assessed using the within-sample F statistic (FWS)51,52, which estimates the fixation of alleles within each infection relative to the diversity observed in the total population on a scale from 0 to 1. Previous studies have demonstrated that an FWS ≥0.95 is highly indicative of clonal infection51,52. The significance of differences in the proportion of infections with FWS ≥0.95 between populations was assessed using Pearson’s χ2 test.
Measures of pairwise IBD were undertaken using hmmIBD with the default parameters for recombination rate (based on P. falciparum estimates), and genotyping error rate, and using allele frequencies estimated by the program53. The pairwise IBD outputs were then determined at 1 kb intervals across the accessible regions of each chromosome and average fractions of IBD derived for each position in each population. Differences in the median IBD between populations were determined using the Wilcoxon's rank-sum test.
Pairwise measures of the genetic differentiation (FST) at individual SNPs were calculated using Weir and Cockerham’s formula using the R software54, and results illustrated as Manhattan plots implemented with the qqman package. Analyses of cross-population haplotype diversity and genetic differentiation were restricted to high-quality typable SNPs with minor allele frequency >1% across Sabah, Thailand and Indonesia.
Population-based measures of haplotype diversity were used to scan the genome for regions under putative positive directional selection55–57. The Rsb measure of cross-population extended haplotype homozygosity and the iHS were measured using the R-based rehh package58. For iHS analysis, ancestral alleles were derived by mapping P. cynomolgi reads59 against the P. vivax P01 reference. Only positions where P. cynomolgi calls were homozygote and matched either the reference or alternative P. vivax allele were included in analysis.
Large CN variations (CNVs) were detected using pysamstats (http://github.com/alimanfoo/pysamstats), which we have previously used and validated in a study of P. vivax MDR1 CNV60. For each sample, coverage in non-overlapping 300 bp bins was calculated and normalised by dividing by the median coverage across all bins with the same integer percentage GC content. A hidden Markov model implemented with the python package sklearn.hmm.GaussianHMM was used to call individual CNVs, and all variants >3 kb were recorded. Further validation of CN amplifications was undertaken using face-away mapping reads27,28.
Aside from the FWS and FST tests, all genomic analyses were undertaken using the major allele call at heterozygous positions. IBD analysis was undertaken on monoclonal samples (FWS ≥0.95). The Rsb and iHS tests were undertaken on low-complexity samples (defined as FWS≥0.6), and additionally assessed with monoclonal samples (FWS≥0.95). All other genomic analyses were conducted without FWS filters.
STR genotyping and data analysis
Genotyping was undertaken at nine STR markers (Pv3.27, msp1F3, MS1, MS5, MS8, MS10, MS12, MS16 and MS20) using methods described previously61. The labelled PCR products were sized by denaturing capillary electrophoresis on an ABI-3100 Genetic Analyser with GeneScan LIZ-600 (Applied Biosystems) internal size standards. Genotype calling was undertaken using VivaxGEN version 1.062. A threshold of 100 relative fluorescence units was applied for peak detection, and minor alleles were called if they had ≥33% height of the predominant allele.
The multiplicity of infection (MOI) for a given sample was defined as the maximum number of alleles observed at any of the nine markers. Population-level diversity was assessed using allelic richness (Rs)63. In the isolates with complete genotyping data, MLGs were reconstructed from the predominant alleles for assessment of the genetic relatedness between sample pairs using (1-ps) as a measure of genetic distance64. An unrooted neighbour-joining tree was generated with the R-based ape package65. The MLGs were also used to assess multi-locus LD. LD was measured using the standardised index of association (IAS), with significance estimates determined using 10,000 random permutations66. Apart from measures of polyclonality and MOI, only the predominant allele at each locus in each isolate was included in th analysis67. Rs and LD analyses were conducted on all infections and additionally in low-complexity samples (maximum 1 multi-allelic locus) to assess the potential impact of MLG reconstruction errors.
Differences in proportions were examined using Pearson’s χ2 test. Pearson’s product–moment correlation and Spearman’s rank correlation was used to test correlations in parametric and non-parametric data, respectively. All tests were performed using R, and assuming a significance threshold of 0.05.
Data availability
The Illumina sequence reads generated on the new P. vivax samples from Sabah, Malaysia has been deposited in the European Nucleotide Archive (ENA) under the accession codes listed in Supplementary Table 5A vcf file containing the genotyping data used in the study is available on the MalariaGEN website (https://www.malariagen.net/resource/24). The authors declare that all other data supporting the findings of this study are available within the article and its Supplementary Information files, or are available from the authors upon request.
Electronic supplementary material
Description of Additional Supplementary Files
Acknowledgements
We thank the patients who contributed their samples to the study, and the health workers and field teams who assisted with the sample collections. We also thank the staff of the Wellcome Sanger Institute Sample Logistics, Sequencing, and Informatics facilities for their contribution. We thank the Director General of Health, Malaysia, for permission to publish this study. Financial support for the study was provided by the Wellcome Trust (Senior Fellowship in Clinical Science awarded to R.N.P., 200909), the National Health and Medical Research Council, Australia (‘Improving Health Outcomes in the Tropical North: A multidisciplinary collaboration ‘HOT North’ Career Development Fellowship awarded to S.A. (1131932)) and the Bill and Melinda Gates Foundation (OPP1164105). The patient sampling and metadata collection was funded by the Asia-Pacific Malaria Elimination Network (108-07), the Malaysian Ministry of Health (BP00500420) and the Australian National Health and Medical Research Council (1037304 and 1045156; Fellowships to N.M.A. (1042072 and 1135820), B.E.B. (1088738) and M.J.G. (1074795)). M.J.G. was also supported by a ‘Hot North’ Early Career Fellowship (1131932). The whole-genome sequencing component of the study was supported by grants from the Medical Research Council and UK Department for International Development (M006212) and the Wellcome Trust (206194, 204911) awarded to D.P.K. T.G.C. is supported by the Medical Research Council UK (MR/K000551/1, MR/M01360X/1, MR/N010469/1, MC_PC_15103). S.C. is funded by the Medical Research Council UK (MR/M01360X/1, MC_PC_15103).
Author contributions
S.A., E.D.B., O.M., R.D.P., R.A., F.N., S.C., T.G.C., N.M.A., D.P.K. and R.N.P. contributed to study design and management. M.J.G., B.E.B., T.W., I.H., J.M., R.N. and K.S. carried out field and laboratory work to obtain P. vivax samples, clinical metadata and microsatellite genotyping data. O.M., R.D.P., R.A. and D.P.K. managed the genomic data production. S.A., E.D.B., R.D.P., R.A. and H.T. performed data analyses. S.A. and R.N.P. wrote the first draft of the manuscript, and all authors contributed to and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary Information accompanies this paper at 10.1038/s41467-018-04965-4.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sarah Auburn, Email: Sarah.Auburn@menzies.edu.au.
Ric N. Price, Email: Ric.Price@menzies.edu.au
References
- 1.WHO. World Malaria Report 2016. Geneva: World Health Organization; 2016. [Google Scholar]
- 2.Price RN, Auburn S, Marfurt J, Cheng Q. Phenotypic and genotypic characterisation of drug-resistant Plasmodium vivax. Trends Parasitol. 2012;28:522–529. doi: 10.1016/j.pt.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tjitra E, et al. Multidrug-resistant Plasmodium vivax associated with severe and fatal malaria: a prospective study in Papua, Indonesia. PLoS Med. 2008;5:e128. doi: 10.1371/journal.pmed.0050128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poespoprodjo JR, et al. Vivax malaria: a major cause of morbidity in early infancy. Clin. Infect. Dis. 2009;48:1704–1712. doi: 10.1086/599041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poespoprodjo JR, et al. Adverse pregnancy outcomes in an area where multidrug-resistant Plasmodium vivax and Plasmodium falciparum infections are endemic. Clin. Infect. Dis. 2008;46:1374–1381. doi: 10.1086/586743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Price RN, et al. Global extent of chloroquine-resistant Plasmodium vivax: a systematic review and meta-analysis. Lancet Infect. Dis. 2014;14:982–991. doi: 10.1016/S1473-3099(14)70855-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cotter C, et al. The changing epidemiology of malaria elimination: new strategies for new challenges. Lancet. 2013;382:900–911. doi: 10.1016/S0140-6736(13)60310-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sattabongkot J, Tsuboi T, Zollner GE, Sirichaisinthop J, Cui L. Plasmodium vivax transmission: chances for control? Trends Parasitol. 2004;20:192–198. doi: 10.1016/j.pt.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 9.de Oliveira TC, et al. Genome-wide diversity and differentiation in New World populations of the human malaria parasite Plasmodium vivax. PLoS Negl. Trop. Dis. 2017;11:e0005824. doi: 10.1371/journal.pntd.0005824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hupalo DN, et al. Population genomics studies identify signatures of global dispersal and drug resistance in Plasmodium vivax. Nat. Genet. 2016;48:953–958. doi: 10.1038/ng.3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pearson RD, et al. Genomic analysis of local variation and recent evolution in Plasmodium vivax. Nat. Genet. 2016;48:959–964. doi: 10.1038/ng.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Winter DJ, et al. Whole genome sequencing of field isolates reveals extensive genetic diversity in Plasmodium vivax from Colombia. PLoS Negl. Trop. Dis. 2015;9:e0004252. doi: 10.1371/journal.pntd.0004252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parobek CM, et al. Selective sweep suggests transcriptional regulation may underlie Plasmodium vivax resilience to malaria control measures in Cambodia. Proc. Natl. Acad. Sci. USA. 2016;113:E8096–E8105. doi: 10.1073/pnas.1608828113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abdullah NR, et al. Plasmodium vivax population structure and transmission dynamics in Sabah Malaysia. PLoS One. 2013;8:e82553. doi: 10.1371/journal.pone.0082553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wangchuk S, et al. Where chloroquine still works: the genetic make-up and susceptibility of Plasmodium vivax to chloroquine plus primaquine in Bhutan. Malar. J. 2016;15:277. doi: 10.1186/s12936-016-1320-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waltmann A, et al. Increasingly inbred and fragmented populations of Plasmodium vivax associated with the eastward decline in malaria transmission across the Southwest Pacific. PLoS Negl. Trop. Dis. 2018;12:e0006146. doi: 10.1371/journal.pntd.0006146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.William T, et al. Changing epidemiology of malaria in Sabah, Malaysia: increasing incidence of Plasmodium knowlesi. Malar. J. 2014;13:390. doi: 10.1186/1475-2875-13-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajahram GS, et al. Falling Plasmodium knowlesi malaria death rate among adults despite rising incidence, Sabah, Malaysia, 2010–2014. Emerg. Infect. Dis. 2016;22:41–48. doi: 10.3201/eid2201.151305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grigg MJ, et al. Efficacy of artesunate-mefloquine for chloroquine-resistant Plasmodium vivax malaria in Malaysia: an open-label, randomized, controlled trial. Clin. Infect. Dis. 2016;62:1403–1411. doi: 10.1093/cid/ciw121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phyo AP, et al. Antimalarial activity of artefenomel (OZ439), a novel synthetic antimalarial endoperoxide, in patients with Plasmodium falciparum and Plasmodium vivax malaria: an open-label phase 2 trial. Lancet Infect. Dis. 2016;16:61–69. doi: 10.1016/S1473-3099(15)00320-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ratcliff A, et al. Therapeutic response of multidrug-resistant Plasmodium falciparum and P. vivax to chloroquine and sulfadoxine-pyrimethamine in southern Papua, Indonesia. Trans. R. Soc. Trop. Med. Hyg. 2007;101:351–359. doi: 10.1016/j.trstmh.2006.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amato R, et al. Genetic markers associated with dihydroartemisinin-piperaquine failure in Plasmodium falciparum malaria in Cambodia: a genotype–phenotype association study. Lancet Infect. Dis. 2017;17:164–173. doi: 10.1016/S1473-3099(16)30409-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Witkowski B, et al. A surrogate marker of piperaquine-resistant Plasmodium falciparum malaria: a phenotype–genotype association study. Lancet Infect. Dis. 2017;17:174–183. doi: 10.1016/S1473-3099(16)30415-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ariey F, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Djimde A, et al. A molecular marker for chloroquine-resistant falciparum malaria. N. Engl. J. Med. 2001;344:257–263. doi: 10.1056/NEJM200101253440403. [DOI] [PubMed] [Google Scholar]
- 26.Diez Benavente E, et al. Genomic variation in Plasmodium vivax malaria reveals regions under selective pressure. PLoS ONE. 2017;12:e0177134. doi: 10.1371/journal.pone.0177134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hostetler JB, et al. Independent origin and global distribution of distinct Plasmodium vivax duffy binding protein gene duplications. PLoS Negl. Trop. Dis. 2016;10:e0005091. doi: 10.1371/journal.pntd.0005091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Menard D, et al. Whole genome sequencing of field isolates reveals a common duplication of the Duffy binding protein gene in Malagasy Plasmodium vivax strains. PLoS Negl. Trop. Dis. 2013;7:e2489. doi: 10.1371/journal.pntd.0002489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miotto O, et al. Multiple populations of artemisinin-resistant Plasmodium falciparum in Cambodia. Nat. Genet. 2013;45:648–655. doi: 10.1038/ng.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maude RJ, et al. The last man standing is the most resistant: eliminating artemisinin-resistant malaria in Cambodia. Malar. J. 2009;8:31. doi: 10.1186/1475-2875-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jamaiah I, et al. Malaria: a 10-year (1994–2003) retrospective study at University Malaya Medical Center (UMMC), Kuala Lumpur, Malaysia. Southeast. Asian J. Trop. Med. Public Health. 2005;36(Suppl. 4):60–63. [PubMed] [Google Scholar]
- 32.Phyo AP, et al. Dihydroartemisinin-piperaquine versus chloroquine in the treatment of Plasmodium vivax malaria in Thailand: a randomized controlled trial. Clin. Infect. Dis. 2011;53:977–984. doi: 10.1093/cid/cir631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muhamad P, Ruengweerayut R, Chacharoenkul W, Rungsihirunrat K, Na-Bangchang K. Monitoring of clinical efficacy and in vitro sensitivity of Plasmodium vivax to chloroquine in area along Thai Myanmar border during 2009–2010. Malar. J. 2011;10:44. doi: 10.1186/1475-2875-10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Devine A, et al. Treatment-seeking behavior after the implementation of a unified policy of dihydroartemisinin-piperaquine for the treatment of uncomplicated malaria in Papua, Indonesia. Am. J. Trop. Med. Hyg. 2017;98:543–550. doi: 10.4269/ajtmh.17-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cooper RA, et al. PfCG2, a Plasmodium falciparum protein peripherally associated with the parasitophorous vacuolar membrane, is expressed in the period of maximum hemoglobin uptake and digestion by trophozoites. Mol. Biochem. Parasitol. 2005;144:167–176. doi: 10.1016/j.molbiopara.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 36.Pava Z, et al. Genetic micro-epidemiology of malaria in Papua Indonesia: extensive P. vivax diversity and a distinct sub-population of asymptomatic P. falciparum infections. PLoS ONE. 2017;12:e0177445. doi: 10.1371/journal.pone.0177445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Talman AM, et al. PbGEST mediates malaria transmission to both mosquito and vertebrate host. Mol. Microbiol. 2011;82:462–474. doi: 10.1111/j.1365-2958.2011.07823.x. [DOI] [PubMed] [Google Scholar]
- 38.Pava Z, et al. Passively versus actively detected malaria: similar genetic diversity but different complexity of infection. Am. J. Trop. Med. Hyg. 2017;97:1788–1796. doi: 10.4269/ajtmh.17-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barber BE, et al. Parasite biomass-related inflammation, endothelial activation, microvascular dysfunction and disease severity in vivax malaria. PLoS Pathog. 2015;11:e1004558. doi: 10.1371/journal.ppat.1004558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grigg MJ, et al. Individual-level factors associated with the risk of acquiring human Plasmodium knowlesi malaria in Malaysia: a case–control study. Lancet Planet Health. 2017;1:e97–e104. doi: 10.1016/S2542-5196(17)30031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Auburn S, et al. Effective preparation of Plasmodium vivax field isolates for high-throughput whole genome sequencing. PLoS ONE. 2013;8:e53160. doi: 10.1371/journal.pone.0053160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Padley D, Moody AH, Chiodini PL, Saldanha J. Use of a rapid, single-round, multiplex PCR to detect malarial parasites and identify the species present. Ann. Trop. Med. Parasitol. 2003;97:131–137. doi: 10.1179/000349803125002977. [DOI] [PubMed] [Google Scholar]
- 43.Imwong M, et al. Spurious amplification of a Plasmodium vivax small-subunit RNA gene by use of primers currently used to detect P. knowlesi. J. Clin. Microbiol. 2009;47:4173–4175. doi: 10.1128/JCM.00811-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malaria Genomic Epidemiology N. A global network for investigating the genomic epidemiology of malaria. Nature. 2008;456:732–737. doi: 10.1038/nature07632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Auburn S, et al. A new Plasmodium vivax reference sequence with improved assembly of the subtelomeres reveals an abundance of pir genes. Wellcome Open Res. 2016;1:4. doi: 10.12688/wellcomeopenres.9876.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Auburn S, et al. Characterization of within-host Plasmodium falciparum diversity using next-generation sequence data. PLoS ONE. 2012;7:e32891. doi: 10.1371/journal.pone.0032891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Manske M, et al. Analysis of Plasmodium falciparum diversity in natural infections by deep sequencing. Nature. 2012;487:375–379. doi: 10.1038/nature11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schaffner, S. F., Taylor, A. R., Wong, W., Wirth, D. F. & Neafsey, D. E. hmmIBD: software to infer pairwise identity by descent between haploid genotypes. Malar. J. 17, 196 (2018). [DOI] [PMC free article] [PubMed]
- 54.Holsinger KE, Weir BS. Genetics in geographically structured populations: defining, estimating and interpreting F(ST) Nat. Rev. Genet. 2009;10:639–650. doi: 10.1038/nrg2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sabeti PC, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 56.Tang K, Thornton KR, Stoneking M. A new approach for using genome scans to detect recent positive selection in the human genome. PLoS Biol. 2007;5:e171. doi: 10.1371/journal.pbio.0050171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gautier M, Vitalis R. rehh: an R package to detect footprints of selection in genome-wide SNP data from haplotype structure. Bioinformatics. 2012;28:1176–1177. doi: 10.1093/bioinformatics/bts115. [DOI] [PubMed] [Google Scholar]
- 59.Pasini EM, et al. An improved Plasmodium cynomolgi genome assembly reveals an unexpected methyltransferase gene expansion. Wellcome Open Res. 2017;2:42. doi: 10.12688/wellcomeopenres.11864.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Auburn S, et al. Genomic analysis reveals a common breakpoint in amplifications of the Plasmodium vivax multidrug resistance 1 locus in Thailand. J. Infect. Dis. 2016;15:1235–1242. doi: 10.1093/infdis/jiw323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hamedi Y, et al. Molecular epidemiology of P. vivax in Iran: high diversity and complex sub-structure using neutral markers, but no evidence of Y976F mutation at pvmdr1. PLoS ONE. 2016;11:e0166124. doi: 10.1371/journal.pone.0166124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trimarsanto H, et al. VivaxGEN: An open access platform for comparative analysis of short tandem repeat genotyping data in Plasmodium vivax populations. PLoS Negl. Trop. Dis. 2017;11:e0005465. doi: 10.1371/journal.pntd.0005465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hurlbert SH. The nonconcept of species diversity: a critique and alternative parameters. Ecology. 1971;52:577–586. doi: 10.2307/1934145. [DOI] [PubMed] [Google Scholar]
- 64.Bowcock AM, et al. High resolution of human evolutionary trees with polymorphic microsatellites. Nature. 1994;368:455–457. doi: 10.1038/368455a0. [DOI] [PubMed] [Google Scholar]
- 65.Paradis E, Claude J, Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 66.Haubold B, Hudson RR. LIAN 3.0: detecting linkage disequilibrium in multilocus data. Linkage Analysis. Bioinformatics. 2000;16:847–848. doi: 10.1093/bioinformatics/16.9.847. [DOI] [PubMed] [Google Scholar]
- 67.Anderson TJ, Su XZ, Bockarie M, Lagog M, Day KP. Twelve microsatellite markers for characterization of Plasmodium falciparum from finger-prick blood samples. Parasitology. 1999;119(Part 2):113–125. doi: 10.1017/S0031182099004552. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The Illumina sequence reads generated on the new P. vivax samples from Sabah, Malaysia has been deposited in the European Nucleotide Archive (ENA) under the accession codes listed in Supplementary Table 5A vcf file containing the genotyping data used in the study is available on the MalariaGEN website (https://www.malariagen.net/resource/24). The authors declare that all other data supporting the findings of this study are available within the article and its Supplementary Information files, or are available from the authors upon request.