Abstract
Tumour metastasis is one of the most serious challenges of cancer as it is the major cause of mortality in patients with solid tumours, including osteosarcoma (OS). In this regard, anti-metastatic genes have potential for metastasis inhibition strategies. Recent evidence showed the importance of breast cancer metastasis suppressor 1 (BRMS1) in control of OS invasiveness, but the regulation of BRMS1 in OS remains largely unknown. Here, we used bioinformatics analyses to predict BRMS1-targeting microRNAs (miRNAs), and the functional binding of miRNAs to BRMS1 mRNA was evaluated using a dual luciferase reporter assay. Among all BRMS1-targeting miRNAs, only miR-151b, miR-7-5p and miR-3200-5p showed significant expression in OS specimens. Specifically, we found that only miR-3200-5p significantly inhibited protein translation of BRMS1 via pairing to the 3′-UTR of the BRMS1 mRNA. Moreover, we detected significantly lower BRMS1 and significantly higher miR-3200-5p in the OS specimens compared to the paired adjacent non-tumour bone tissues. Furthermore, BRMS1 and miR-3200-5p levels were inversely correlated to each other. Low BRMS1 was correlated with metastasis and poor patient survival. In vitro, overexpression of miR-3200-5p significantly decreased BRMS1 levels and promoted OS cell invasion and migration, while depletion of miR-3200-5p significantly increased BRMS1 levels and inhibited OS cell invasion and migration. Thus, our study revealed that miR-3200-5p may be a critical regulator of OS cell invasiveness.
Keywords: breast cancer metastasis suppressor 1 (BRMS1), miR-3200-5p, osteosarcoma (OS)
INTRODUCTION
Osteosarcoma (OS) is a notorious bone tumour in humans, largely due to its distal metastases (Ding et al., 2016; Ren et al., 2015; Wang et al., 2016). Despite years of continuous progress in cancer therapy, metastasis of OS remains the major driver of cancer mortality and is responsible for the majority of cancer-associated deaths (Catalano et al., 2013). Each step for cancer metastasis requires complex molecular regulation.
Breast cancer metastasis suppressor 1 (BRMS1) was originally identified in a breast carcinoma cell line (Seraj et al., 2000b). Then, BRMS1 was shown to suppress metastasis of multiple tumour types (Nagji et al., 2010; Samant et al., 2007; Seraj et al., 2000a; Smith et al., 2009) through regulation of the expression of metastasis-associated genes, including osteopontin and CXCR4 (Hurst et al., 2008; Huo et al., 2015; Meehan et al., 2004; Roesley et al., 2016). Nevertheless, the regulation of BRMS1 during tumourigenesis, especially in OS, remains largely unknown.
MicroRNAs (miRNAs), master regulators of gene expression, are small non-coding RNAs capable of post-transcriptional suppression of gene expression by sequence-specific interactions with the miRNA response elements (MREs) on the 3′ untranslated regions (3′-UTRs) of related mRNA targets (Han et al., 2015; Wang et al., 2015). Abnormal expression of miRNAs in different steps of malignancies has been discovered in a variety of cancers. However, previous studies on miR-3200-5p are very rare, and only a very recent report demonstrated that the serum levels of miR-3200 declined during treatment, and higher serum levels of miR-3200 appeared to be associated with lower residual breast cancer burden and relapse rates at time of diagnosis (Al-Khanbashi et al., 2016).
Here, we studied the regulation of BRMS1 by miRNAs in OS specimens and OS cells. We used bioinformatics analyses to predict BRMS1-targeting miRNAs, and the functional binding of miRNAs to BRMS1 mRNA was evaluated using a dual luciferase reporter assay. Among all BRMS1-targeting miRNAs, only miR-151b, miR-7-5p and miR-3200-5p showed significant expression in OS specimens. Specifically, we found that only miR-3200-5p significantly inhibited protein translation of BRMS1 via pairing to the 3′-UTR of the BRMS1 mRNA. Moreover, we detected significantly lower BRMS1 and significantly higher miR-3200-5p in the OS specimens. Furthermore, BRMS1 and miR-3200-5p levels were inversely correlated. In vitro, miR-3200-5p increased OS cell invasiveness by suppressing BRMS1.
MATERIALS AND METHODS
Experimental protocol approval and patient specimens
All experimental protocols were approved by the Research Bureau of Shanghai Tenth People’s Hospital. Surgically resected OS specimens and paired adjacent non-tumour bone tissues (NT) were obtained from 30 OS patients in Shanghai Tenth People’s Hospital from 2010 to 2012 (Table 1). All the involved patients provided informed consent and a signed agreement. The specimens were cryo-preserved.
Table 1.
Clinical-pathological characteristics
| Patients (n; %) | P value | |
|---|---|---|
| OS tissue/ Normal tumor-adjacent tissue (NT) | 30 (100%) /30 (100%) | |
| Age (>60/≥60 years old) | 12 (40%) /18 (60%) | 0.56 |
| Gender (male/female) | 22 (73%) /8 (27%) | |
| Tumor site (bone) | 30 (100%) | |
| Tumor grade (well or moderate/poor) | 0 (0%) /9 (30%) /21 (70%) | 0.011 |
| Tumor stage (I/II/III/IV) | 0 (0%) /0 (0%) /15 (50%) /15 (50%) | 0.008 |
| Lymph node metastasis (no/yes) | 0 (0%) /30 (100%) | 0.007 |
| Distal metastasis at diagnosis (no/yes) | 27 (90%) /3 (10%) | 0.009 |
Culturing and transfection of OS cells
U2OS and SaOS2, two widely used human OS cell lines, were purchased from the American Type Culture Collection (ATCC, USA) and were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen, USA) supplemented with 15% foetal bovine serum (FBS; Invitrogen) in a humidified chamber with 5% CO2 at 37°C. The U2OS cell line was established in 1964 from a moderately differentiated sarcoma (Ponten and Saksela, 1967). The SaOS2 cell line was established in the 1970s (Fogh et al., 1977). These two lines were chosen in the current study since SaOS2 cells express relatively low miR-3200-5p and relatively high BRMS1, while U2OS cells express relatively high miR-3200-5p and relatively low BRMS1. All plasmids used for cell transfection were purchased from Origene (China). Transfection was performed with Lipofectamine 3000 (Invitrogen).
Transwell cell invasion assay
Transwell cell invasion assays were performed using Matrigel-coated Transwell cell culture chambers. Briefly, transfected cells were harvested, after which 104 cells in 100 μl non-serum medium were placed in the upper chamber of an insert (pore size, 8 μm), which was pre-coated with Matrigel (Becton-Dickinson Biosciences, USA). The lower chamber was filled with 600 μl RPMI-1640 medium (Invitrogen) with 20% FBS (Invitrogen). After 24 h of incubation, cells were removed from the upper chamber of the filter using a cotton swab. Cells on the underside were washed with PBS and fixed with 5% glutaraldehyde, stained with 0.5% crystal violet, and counted in five randomly selected fields under a phase contrast microscope. Assays were performed in triplicate.
Scratch wound healing assay
Briefly, cells were seeded in 24-well plates at a density of 104 cells/well in complete media and cultured to confluence. The cell monolayer was serum starved overnight, after which the cell monolayer was scraped with a pipette tip to generate scratch wounds. Cells were incubated at 37°C for 24 h when the time lapse images were captured. Images were captured from five randomly selected fields in each sample, and the wound areas were calculated by NIH ImageJ software.
Cell growth/proliferation assay
The cell growth/proliferation rate was determined with an MTT Kit (MTT, Roche, USA).
Bioinformatics and 3′-UTR luciferase reporter assay
Target genes for miRNAs were predicted using TargetScan web tools, as previously described (Coronnello and Benos, 2013). The candidate miRNAs were determined using the context+ score method (Garcia et al., 2011), shown in Supplementary table. The BRMS1 3′-UTR reporter plasmid (pRL-BRMS1) and the BRMS1 3′-UTR reporter plasmid with a mutant at the corresponding miRNA binding site (pRL-BRMS1-mut) were purchased from Origene (China). Dual luciferase reporter assays (Promega, USA) were performed to examine the functional binding of the miRNAs to BRMS1 mRNA.
Quantitative RT-PCR (RT-qPCR)
Quantitative RT-PCR (RT-qPCR) was performed with a standard procedure. Briefly, total RNA, with efficient recovery of small RNAs, was obtained using a miRCURY RNA isolation kit (Exiqon, Denmark), and cDNA was synthesised by a Universal cDNA Synthesis Kit (Exiqon). Detection of the mature form of the miRNAs was performed using the miRCURY LNA Universal RT microRNA PCR Kit and LNA microRNA Primer Sets, according to the manufacturer’s instructions (Exiqon). Relative expression levels were determined using the comparative quantification characteristic of the Rotorgene software. The U6 small nuclear RNA was used as an internal control and the comparative Ct (ΔΔCt) method was used to determine the expression fold change. A melting curve analysis was performed for each of the primer sets utilised, and each showed a single peak indicating the specificity of each of the primers tested.
ELISA
Briefly, the protein was extracted using RIPA lysis buffer (1% NP40, 0.1% SDS, 100 μg/ml phenylmethylsulfonyl fluoride, 0.5% sodium deoxycholate, in PBS) on ice. The supernatants were collected after centrifugation at 12000× g at 4°C for 20 min. Protein concentration was determined using a BCA protein assay kit (Bio-Rad, China), and whole lysates were used to assess protein levels for human BRMS1 using a human BRMS1 ELISA kit (LSBio, USA), according to the kit instructions.
Animal manipulation
All the animal protocols were approved by the research committee at Tongji University School of Medicine and carried out in accordance with the guidelines. Twelve-week-old male immune-deficient nude mice (SLAC Laboratory Animal Co. Ltd., China) received tumour cell implantation. Tumour cells were grafted subcutaneously. The tumour was dissected out and weighted 4 weeks after transplantation.
Statistical analysis
Statistical analyses were performed with GraphPad Prism 6 (GraphPad Software, USA). Comparison among different groups was performed using a one-way analysis of variance (ANOVA) test followed by Tukey’s multiple comparison post hoc analysis. The correlation was evaluated using multivariate Cox regression analysis and calculated by Spearman’s rank correlation coefficients. Patients’ 5-year survival was recorded by Kaplan-Meier curve. All values represent the mean ± standard deviation (SD). A value of p < 0.05 was considered statistically significant.
RESULTS
MiR-3200-5p targets the 3′-UTR of BRMS1 mRNA to inhibit its translation in OS cells
First, we examined whether BRMS1 is regulated by miRNAs in OS cells. Thus, we performed bioinformatics analyses to identify the candidate miRNAs that bind to the 3′-UTR of BRMS1 mRNA. Among all BRMS1-targeting miRNAs (Supplementary Table), 3 miRNAs (miR-151b, miR-7-5p and miR-3200-5p) were expressed in the OS cell line U2OS. To determine whether the binding of these 3 miRNAs to BRMS1 mRNA functionally inhibits protein translation of BRMS1, we transfected U2OS cells with miR-151b, miR-7-5p and miR-3200-5p or antisense sequences (as-miR-151b, as-miR-7-5p and as-miR-3200-5p). The U2OS cells were also transfected with a null sequence as a control (null). Changes in the miRNA levels in cells were confirmed by RT-qPCR (Figs. 1A–1C). Then, the intact 3′-UTR of BRMS1 mRNA (BRMS1 3′-UTR), together with a 3′-UTR with a mutant at either miRNA binding site of BRMS1 mRNA (BRMS1 3′-UTR mut), was cloned into luciferase reporter plasmids. U2OS cells were co-transfected with one miRNA/as-miRNA/null plasmid and one BRMS1 3′-UTR or BRMS1 3′-UTR mut plasmid. The results suggest that among all 3 miRNAs, only miR-3200-5p specifically targets the 3′-UTR of BRMS1 mRNA to inhibit its translation in OS cells (Figs. 1D–1F), as shown by bioinformatics analyses of the binding site of miR-3200-5p on the 3′-UTR of BRMS1 mRNA (Fig. 1G).
Fig. 1. MiR-3200-5p targets 3′-UTR of BRMS1 mRNA to inhibit its translation in OS cells.
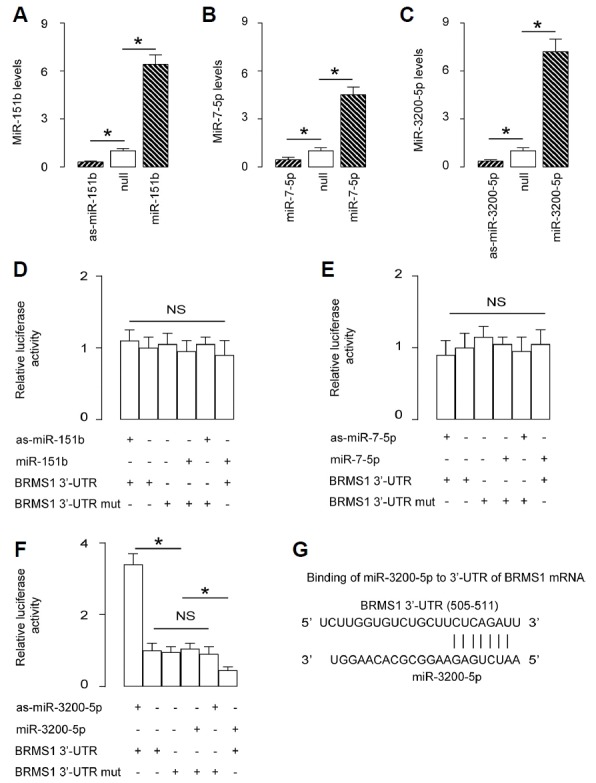
(A–C) U2OS cells were transfected with miR-151b, miR-7-5p and miR-3200-5p or antisense sequences (as-miR-151b, as-miR-7-5p and as-miR-3200-5p). The U2OS cells were also transfected with a null sequence as a control (null). RTqPCR showed changes in the miR-151b levels in miR-151b-modified cells (A), changes in the miR-7-5p levels in miR-7-5p-modified cells (B), and changes in the miR-3200-5p levels in miR-3200-5pmodified cells (C). (D–F) The intact 3′-UTR of BRMS1 mRNA (BRMS1 3′-UTR), together with a 3′-UTR with a mutant at either miRNA binding site of BRMS1 mRNA (BRMS1 3′-UTR mut), was then cloned into luciferase reporter plasmids. U2OS cells were co-transfected with one miRNA/as-miRNA/null plasmid and one BRMS1 3′-UTR or BRMS1 3′-UTR mut plasmid. (D) Assay for miR-151b. (E) Assay for miR-7-5p. (F) Assay for miR-3200-5p. (G) Bioinformatics analyses for binding sites of miR-3200-5p on the 3′-UTR of BRMS1 mRNA. *p < 0.05. NS: nonsignificant. N = 5.
Association of miR-3200-5p and BRMS1 levels in OS specimens
The levels of BRMS1 and miR-3200-5p in 30 pairs of resected OS tissues and adjacent non-tumour bone tissues (NT) were measured by ELISA and RT-qPCR, respectively (Table 1). We found that OS specimens contained significantly lower levels of BRMS1 (Fig. 2A) and significantly higher levels of miR-3200-5p (Fig. 2B). We then performed a correlation test using these 30 OS specimens and detected a strong inverse correlation between BRMS1 and miR-3200-5p (Fig. 1C, γ = −0.67, p < 0.0001), indicating a possible regulatory relationship between miR-3200-5p and BRMS1 in OS. Moreover, OS specimens with metastasis expressed significantly lower levels of BMSM1 than those without metastasis (Fig. 2D). The five-year-survival of these 30 cases was analysed based on the BRMS1 levels, in which a median cutoff point was selected to differentiate BRMS1-high (n = 15) from BRMS1-low cases (n = 15). The results showed that OS patients with lower BRMS1 levels had a significantly poorer overall survival (Fig. 2E).
Fig. 2. Association of miR-3200-5p and BRMS1 levels in OS specimens.
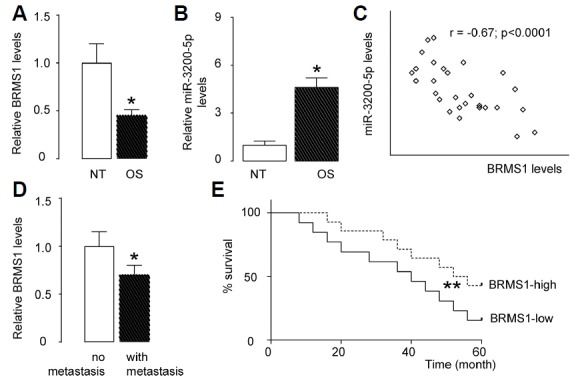
(A) The levels of BRMS1 in 30 pairs of OS tissues and adjacent non-tumour bone tissues (NT) were measured by ELISA. (B) The levels of miR-3200-5p in 30 pairs of OS tissues and NT were measured by RT-qPCR. (C) A correlation test was performed between BRMS1 and miR-3200-5p, using the 30 OS specimens. (D) The levels of BRMS1 in OS tissues from cases with or without metastasis, measured by ELISA. (E) The 30 patients were followed-up for 5 years. The median value was chosen as the cutoff point for BRMS1-high cases (n=15) from BRMS1-low cases (n=15). Kaplan-Meier curves were generated. *p < 0.05. N = 30.
Modification of miR-3200-5p levels in OS cells
Next, we examined the effects of miR-3200-5p on BRMS1 and cell invasion in OS cells. We chose U2OS and SaOS2 cell lines for this study, since SaOS2 cells express relatively low miR-3200-5p and relatively high BRMS1, while U2OS cells express relatively high miR-3200-5p and relatively low BRMS1 (Figs. 3A and 3B). We then transfected SaOS2 cells with miR-3200-5p and U2OS cells with antisense for miR-3200-5p (as-miR-3200-5p) and confirmed the alteration of miR-3200-5p levels in these cells by RT-qPCR (Figs. 3C and 3D).
Fig. 3. Modification of miR-3200-5p levels in OS cells.
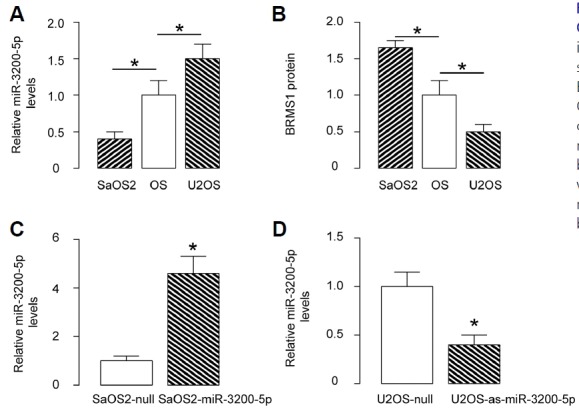
(A) MiR-3200-5p levels by RT-qPCR in U2OS and SaOS2 cells compared to OS specimens. (B) BRMS1 protein levels by ELISA in U2OS and SaOS2 cells compared to OS specimens. (C) Transfection of SaOS2 cells with miR-3200-5p or null control plasmids. MiR-3200-5p levels were determined by RT-qPCR. (D) Transfection of U2OS cells with as-miR-3200-5p or null control plasmids. MiR-3200-5p levels were determined by RT-qPCR. *p < 0.05. N = 5.
Overexpression of miR-3200-5p in OS cells promotes cell invasion via BRMS1 suppression
We found that overexpression of miR-3200-5p did not alter BRMS1 mRNA (Fig. 4A) but decreased BRMS1 protein in SaOS2 cells (Fig. 4B). The effects of miR-3200-5p modification on cell growth and invasiveness were then investigated. We found that miR-3200-5p overexpression in SaOS2 cells did not alter cell growth/proliferation in an MTT assay (Fig. 4C) but significantly increased cell invasion in a transwell cell invasion assay (Figs. 4D and 4E) and significantly increased cell migration in a scratch wound healing assay (Figs. 4F and 4G). To ascertain whether miR-3200-5p promotes OS cell invasion by suppressing BRMS1, we prepared plasmids for BRMS1 overexpression (BRMS1) and depletion (shBRMS1). SaOS2-miR-3200-5p cells were further transfected with BRMS1, which rescued expression of BRMS1 mRNA (Fig. 4A) and protein (Fig. 4B) in these cells. We found that overexpression of BRMS1 abrogated the effects of miR-3200-5p on cell invasion (Figs. 4D and 4E) and migration (Figs. 4F and 4G) in SaOS2 cells, without affecting cell growth (Fig. 4C). Hence, overexpression of miR-3200-5p in OS cells promotes cell invasion via BRMS1 suppression.
Fig. 4. Overexpression of miR-3200-5p in OS cells promotes cell invasion via BRMS1 suppression.
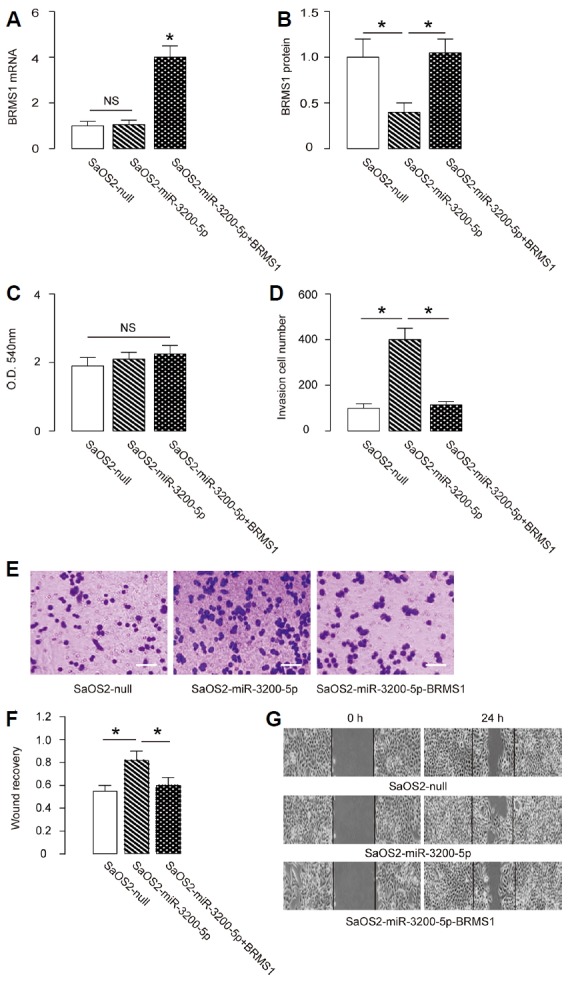
(A, B) The BRMS1 levels in miR-3200-5p-overexpressing (and BRMS1-overexpressing) SaOS2 cells by RT-qPCR (A) and by ELISA (B). (C) MTT assay. (D–E) SaOS2 cell invasion by miR-3200-5p overexpression (and BRMS1 overexpression) in a transwell cell invasion assay, shown by quantification (D) and by representative images (E). (F, G) SaOS2 cell migration by miR-3200-5p overexpression (and BRMS1 overexpression) in a scratch wound healing assay, shown by quantification (F) and by representative images (G). *p < 0.05. NS: non-significant. N = 5.
Depletion of miR-3200-5p in OS cells inhibits cell invasion via BRMS1 augmentation
Next, we found that depletion of miR-3200-5p did not alter BRMS1 mRNA (Fig. 5A) but increased BRMS1 protein in U2OS cells (Fig. 5B). The effects of miR-3200-5p modification on cell growth and invasiveness were then investigated. We found that miR-3200-5p depletion in U2OS cells did not alter cell growth in an MTT assay (Fig. 5C) but significantly decreased cell invasion in a transwell cell invasion assay (Figs. 5D and 5E) and cell migration in a scratch wound healing assay (Figs. 5F and 5G). To ascertain whether miR-3200-5p suppression inhibits OS cell invasion by increasing BRMS1, U2OS-as-miR-3200-5p cells were further transfected with shBRMS1, which abolished the induction of expression of BRMS1 mRNA (Fig. 5A) and protein (Fig. 5B) in these cells. We found that depletion of BRMS1 abrogated the effects of as-miR-3200-5p on cell invasion (Figs. 5D and 5E) and migration (Figs. 5F and 5G) in SaOS2 cells, without affecting cell growth (Fig. 5C). Hence, depletion of miR-3200-5p in OS cells inhibits cell invasion via BRMS1 augmentation.
Fig. 5. Depletion of miR-3200-5p in OS cells inhibits cell invasion via BRMS1 augmentation.
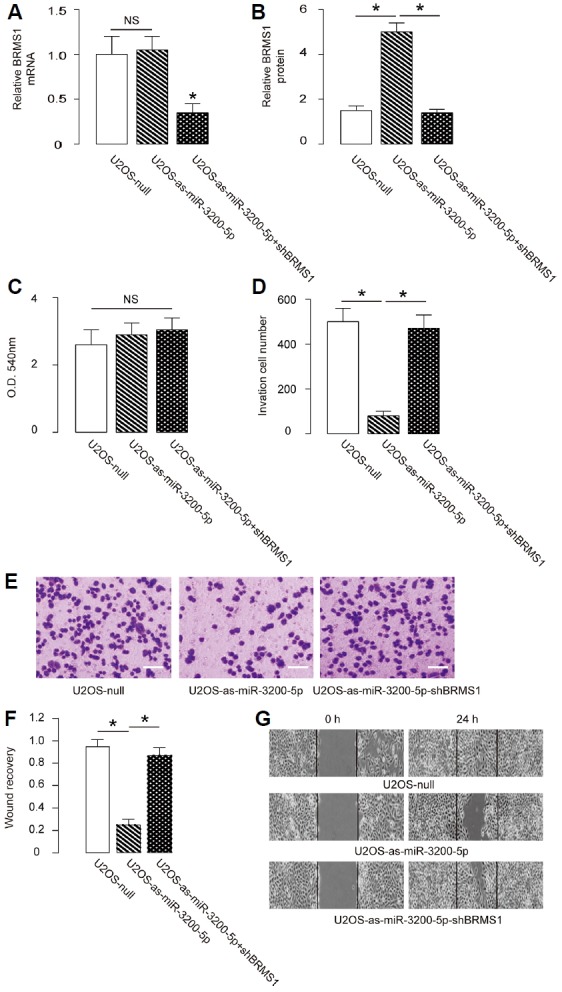
(A, B) The BRMS1 levels in miR-3200-5p-depleted (and BRMS1-depleted) U2OS cells by RT-qPCR (A) and by ELISA (B). (C) MTT assay. (D–E) U2OS cell invasion by miR-3200-5p depletion (and BRMS1 depletion) in a transwell cell invasion assay, shown by quantification (D), and by representative images (E). (F, G) U2OS cell migration by miR-3200-5p depletion (and BRMS1 depletion) in a scratch wound healing assay, shown by quantification (F) and by representative images (G). *p < 0.05. NS: nonsignificant. N = 5.
Modification of miR-3200-5p levels in OS cells affects tumour growth in vivo
The tumour growth of OS cells with different levels of miR-3200-5p was evaluated in vivo. The miR-3200-5p-modified U2OS cells with controls were subcutaneously implanted into nude mice at a density of 107 cells. Four weeks after tumour cell transplantation, the tumour was removed and weighed. These results suggest that overexpression of miR-3200-5p in OS cells significantly increased tumour growth in vivo, and depletion of miR-3200-5p in OS cells significantly decreased tumour growth in vivo (Figs. 6A and 6B). A schematic was thus made to summarise the current study, showing that miR-3200-5p may inhibit protein translation of BRMS1 via pairing to the 3′-UTR of the BRMS1 mRNA in OS cells to promote cell invasion (Fig. 6C).
Fig. 6. Modification of miR-3200-5p levels in OS cells affects tumour growth in vivo.
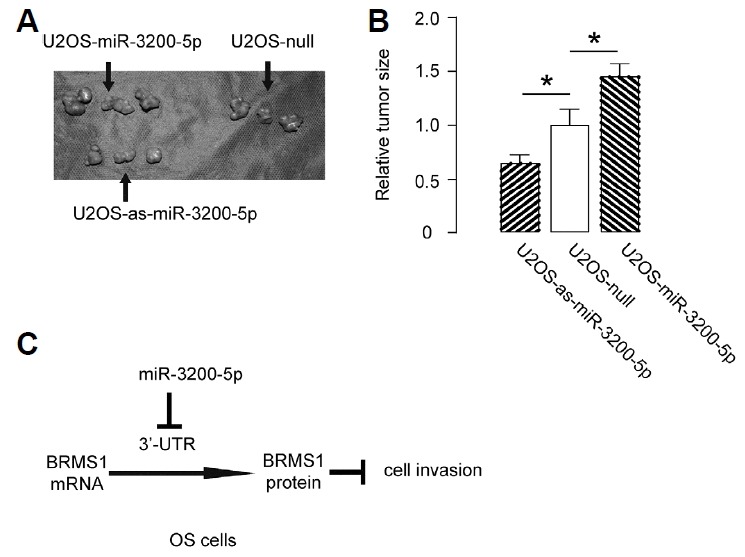
(A–B) The same number (107) of U2OS-null, U2OS-miR-3200-5p or U2OS-as-miR-3200-5p cells was subcutaneously transplanted into nude mice for 4 weeks, and then, the tumour was dissected out, and tumour weight was assessed, as shown by gross images (A) and by quantification (B). (C) Schematic: miR-3200-5p may inhibit protein translation of BRMS1 by pairing to the 3′-UTR of the BRMS1 mRNA in OS cells to promote cell invasion. *p < 0.05. N = 3.
DISCUSSION
The inhibitory role of BRMS1 in cancer invasion has been well documented in past studies. For example, Roesley et al. (2016) showed that BRMS1 is a substrate of Cyclin-Dependent Kinase 2 (CDK2), by which it is phosphorylated on serine 237 in breast cancer cells. Although the mutation of BRMS1 on serine 237 did not affect cell cycle progression and proliferation of cancer cells, it indeed changed the cell migration (Roesley et al., 2016). However, the regulation of BRMS1 by miRNAs was not reported in breast cancer but in nasopharyngeal carcinoma cells (Yan et al., 2016). In this study, Yan et al. (2016) showed that miR-346, a BRMS1-targeting miRNA, was upregulated in nasopharyngeal carcinoma tissues compared with adjacent non-tumourous nasopharyngeal tissues. Inhibition of miR-346 significantly attenuated the migration and invasion of nasopharyngeal carcinoma cells. Nevertheless, a role of BRMS1 in OS invasion and metastasis, as well as its regulation by miRNAs, has not been documented so far.
Here, we used bioinformatics analyses to screen all miRNAs that target BRMS1 in OS cells, and we focused on the expression levels of those that were detectable in OS cells. From the 3 candidates, only miR-3200-5p showed functional binding to the 3′-UTR of BRMS1 mRNA. To the best of our knowledge, this is the first study showing that BRMS1 protein levels could be regulated by a specific miRNA in OS. High level of miR-3200-5p in OS specimens was associated with low BRMS1 levels. We thus designed in vitro experiments to show a regulatory relationship between miR-3200-5p and BRMS1 in OS cells, which was consistent with the clinical findings showing an inverse correlation of these two factors in OS specimens.
In addition to regulation of BRMS1 by miRNAs, BRMS1 protein levels may be modulated at the level of degradation, such as through protein ubiquitination. Moreover, miR-3200-5p may have targets other than BRMS1, and these targets should be analysed in the future to provide an overview of the effects of miR-3200-5p in the OS cell invasion. Furthermore, future studies may also address the regulation of miR-3200-5p in OS and confirm this model in vivo.
Compared with overexpression of BRMS1, using as-miR-3200-5p to increase BRMS1 levels has an advantage, since overexpression of BRMS1 in OS cells may result in a further increase in the levels of miR-3200-5p as a feedback mechanism, to decrease the efficacy of the treatment. To summarise, the current study sheds light on miR-3200-5p as a crucial factor that enhances OS cell invasiveness and provides evidence for using miR-3200-5p as a promising therapeutic target for OS treatment.
Supplementary data
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Al-Khanbashi M., Caramuta S., Alajmi A.M., Al-Haddabi I., Al-Riyami M., Lui W.O., Al-Moundhri M.S. Tissue and serum miRNA profile in locally advanced breast cancer (LABC). in response to neo-adjuvant chemotherapy (NAC). Treatment. PLoS One. 2016;11:e0152032. doi: 10.1371/journal.pone.0152032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalano V., Turdo A., Di Franco S., Dieli F., Todaro M., Stassi G. Tumor and its microenvironment: a synergistic interplay. Semi Cancer Biol. 2013;23:522–532. doi: 10.1016/j.semcancer.2013.08.007. [DOI] [PubMed] [Google Scholar]
- Coronnello C., Benos P.V. ComiR: Combinatorial microRNA target prediction tool. Nucleic Acids Res. 2013;41:W159–164. doi: 10.1093/nar/gkt379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Congwei L., Bei Q., Tao Y., Ruiguo W., Heze Y., Bo D., Zhihong L. mTOR: An attractive therapeutic target for osteosarcoma? Oncotarget. 2016;7:50805–50813. doi: 10.18632/oncotarget.9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogh J., Wright W.C., Loveless J.D. Absence of HeLa cell contamination in 169 cell lines derived from human tumors. J Natl Cancer Inst. 1977;58:209–214. doi: 10.1093/jnci/58.2.209. [DOI] [PubMed] [Google Scholar]
- Garcia D.M., Baek D., Shin C., Bell G.W., Grimson A., Bartel D.P. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol. 2011;18:1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K., Chen X., Bian N., Ma B., Yang T., Cai C., Fan Q., Zhou Y., Zhao T.B. MicroRNA profiling identifies MiR-195 suppresses osteosarcoma cell metastasis by targeting CCND1. Oncotarget. 2015;6:8875–8889. doi: 10.18632/oncotarget.3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo X., Li S., Shi T., Suo A., Ruan Z., Guo H., Yao Y. Cullin3 promotes breast cancer cells metastasis and epithelial-mesenchymal transition by targeting BRMS1 for degradation. Oncotarget. 2015;6:41959–41975. doi: 10.18632/oncotarget.5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst D.R., Xie Y., Vaidya K.S., Mehta A., Moore B.P., Accavitti-Loper M.A., Samant R.S., Saxena R., Silveira A.C., Welch D.R. Alterations of BRMS1-ARID4A interaction modify gene expression but still suppress metastasis in human breast cancer cells. J Biol Chem. 2008;283:7438–7444. doi: 10.1074/jbc.M709446200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan W.J., Samant R.S., Hopper J.E., Carrozza M.J., Shevde L.A., Workman J.L., Eckert K.A., Verderame M.F., Welch D.R. Breast cancer metastasis suppressor 1 (BRMS1). forms complexes with retinoblastoma-binding protein 1 (RBP1). and the mSin3 histone deacetylase complex and represses transcription. J Biol Chem. 2004;279:1562–1569. doi: 10.1074/jbc.M307969200. [DOI] [PubMed] [Google Scholar]
- Nagji A.S., Liu Y., Stelow E.B., Stukenborg G.J., Jones D.R. BRMS1 transcriptional repression correlates with CpG island methylation and advanced pathological stage in non-small cell lung cancer. J Pathol. 2010;221:229–237. doi: 10.1002/path.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponten J., Saksela E. Two established in vitro cell lines from human mesenchymal tumours. Int J Cancer. 1967;2:434–447. doi: 10.1002/ijc.2910020505. [DOI] [PubMed] [Google Scholar]
- Ren L., Mendoza A., Zhu J., Briggs J.W., Halsey C., Hong E.S., Burkett S.S., Morrow J., Lizardo M.M., Osborne T., et al. Characterization of the metastatic phenotype of a panel of established osteosarcoma cells. Oncotarget. 2015;6:29469–29481. doi: 10.18632/oncotarget.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesley S.N., Suryadinata R., Morrish E., Tan A.R., Issa S.M., Oakhill J.S., Bernard O., Welch D.R., Sarcevic B. Cyclin-dependent kinase-mediated phosphorylation of breast cancer metastasis suppressor 1 (BRMS1). affects cell migration. Cell Cycle. 2016;15:137–151. doi: 10.1080/15384101.2015.1121328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samant R.S., Clark D.W., Fillmore R.A., Cicek M., Metge B.J., Chandramouli K.H., Chambers A.F., Casey G., Welch D.R., Shevde L.A. Breast cancer metastasis suppressor 1 (BRMS1). inhibits osteopontin transcription by abrogating NF-kappaB activation. Mol Cancer. 2007;6:6. doi: 10.1186/1476-4598-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seraj M.J., Harding M.A., Gildea J.J., Welch D.R., Theodorescu D. The relationship of BRMS1 and RhoGDI2 gene expression to metastatic potential in lineage related human bladder cancer cell lines. Clin Exp Metastasis. 2000a;18:519–525. doi: 10.1023/a:1011819621859. [DOI] [PubMed] [Google Scholar]
- Seraj M.J., Samant R.S., Verderame M.F., Welch D.R. Functional evidence for a novel human breast carcinoma metastasis suppressor, BRMS1, encoded at chromosome 11q13. Cancer Res. 2000b;60:2764–2769. [PubMed] [Google Scholar]
- Smith P.W., Liu Y., Siefert S.A., Moskaluk C.A., Petroni G.R., Jones D.R. Breast cancer metastasis suppressor 1 (BRMS1). suppresses metastasis and correlates with improved patient survival in non-small cell lung cancer. Cancer Lett. 2009;276:196–203. doi: 10.1016/j.canlet.2008.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Zhou X., Wei M. MicroRNA-144 suppresses osteosarcoma growth and metastasis by targeting ROCK1 and ROCK2. Oncotarget. 2015;6:10297–10308. doi: 10.18632/oncotarget.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wang C., Zhou Z., Sun M., Zhou C., Chen J., Yin F., Wang H., Lin B., Zuo D., et al. CD151-mediated adhesion is crucial to osteosarcoma pulmonary metastasis. Oncotarget. 2016;7:60623–60638. doi: 10.18632/oncotarget.11380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H.L., Li L., Li S.J., Zhang H.S., Xu W. miR-346 promotes migration and invasion of nasopharyngeal carcinoma cells via targeting BRMS1. J Biochem Mol Toxicol. 2016;30:602–607. doi: 10.1002/jbt.21827. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


