Abstract
Preterm microbial colonization is affected by gestational age, antibiotic treatment, type of birth, but also by type of feeding. Breast milk has been acknowledged as the gold standard for human nutrition. In preterm infants breast milk has been associated with improved growth and cognitive development and a reduced risk of necrotizing enterocolitis and late onset sepsis. In the absence of their mother’s own milk (MOM), pasteurized donor human milk (DHM) could be the best available alternative due to its similarity to the former. However, little is known about the effect of DHM upon preterm microbiota and potential biological implications. Our objective was to determine the impact of DHM upon preterm gut microbiota admitted in a referral neonatal intensive care unit (NICU). A prospective observational cohort study in NICU of 69 neonates <32 weeks of gestation and with a birth weight ≤1,500 g was conducted. Neonates were classified in three groups according to feeding practices consisting in their MOM, DHM, or formula. Fecal samples were collected when full enteral feeding (defined as ≥150 cc/kg/day) was achieved. Gut microbiota composition was analyzed by 16S rRNA gene sequencing. Despite the higher variability, no differences in microbial diversity and richness were found, although feeding type significantly influenced the preterm microbiota composition and predictive functional profiles. Preterm infants fed MOM showed a significant greater presence of Bifidobacteriaceae and lower of Staphylococcaceae, Clostridiaceae, and Pasteurellaceae compared to preterm fed DHM. Formula fed microbial profile was different to those observed in preterm fed MOM. Remarkably, preterm infants fed DHM showed closer microbial profiles to preterm fed their MOM. Inferred metagenomic analyses showed higher presence of Bifidobacterium genus in mother’s milk group was related to enrichment in the Glycan biosynthesis and metabolism pathway that was not identified in the DHM or in the formula fed groups. In conclusion, DHM favors an intestinal microbiome more similar to MOM than formula despite the differences between MOM and DHM. This may have potential beneficial long-term effects on intestinal functionality, immune system, and metabolic activities.
Keywords: preterm infant, their mother’s own milk, donor human milk, formula milk, intestinal colonization, microbiota
Introduction
In preterm infants, neonatal microbial dynamics and alterations in early gut microbiota may precede and/or predispose to diseases such as NEC or LOS (The European Perinatal Health Report, 2010). In the newborn period differential microbial colonization clearly relates to weeks of gestation and mode of delivery (Lehtonen et al., 2017), but also to infant nutrition (Collado et al., 2015).
Human milk is the gold standard for infant nutrition in the first 12 months of life for term and preterm newborn infants (American Academy of Pediatrics, 2012). Beyond nutritional components, HM contains important bioactive compounds such oligosaccharides, cytokines, immunoglobulins, microbes, and proteins among others that directly influence the developing infant and shape the intestinal microbiota colonization. Those bioactive compounds are considered not only protective but also stimulate the development and maturation of the immature immune system (Agostoni, 2010; Ballard and Morrow, 2013). Moreover, breastfeeding practices have been associated with a risk reduction of NEC and LOS in preterm infants (Meinzen-Derr et al., 2009; Collado et al., 2012) and an improvement in growth and cognitive development and modulating metabolic and inflammatory conditions in later childhood and adulthood (Ballard and Morrow, 2013; Belfort et al., 2016). However, often mothers who deliver preterm are not able to successfully breastfeed (Wilson et al., 2017). In the absence of MOM, DHM has become the preferred alternative for preterm infants (ESPGHAN Committee on Nutrition et al., 2013). Despite the beneficial effects of DHM, little is known about its effect upon preterm gut microbiota colonization and its potential biological implications. Most HM bank guidelines recommended Holder pasteurization (62.5°C for 30 min) in order to inactivate viral and bacterial agents (Human Milk Banking Association of North America, 2000; Arslanoglu et al., 2010; National Institute for Health and Clinical Excellence, 2010; Peila et al., 2016). However, HM pasteurization causes the loss of several of the structural and functional properties of HM (Baro et al., 2001). Moreover, pasteurization also significantly reduces the cellular and bacterial constituents, enzymatic activities, and IgA, lactoferrin and lysozyme contents (Untalan et al., 2009; Christen et al., 2013; Espinosa-Martos et al., 2013; Sousa et al., 2014). Contrarily, other components with biological relevance such as oligosaccharides, nucleotides, and polyunsaturated and long chain fatty acids (LCPUFA) are preserved (Bertino et al., 2008; Coscia et al., 2015). As a consequence, pasteurization is still a matter of debate (Bertino et al., 2009; ESPGHAN Committee on Nutrition et al., 2013; Corpeleijn et al., 2016; Madore et al., 2017).
In this scenario, we hypothesized that DHM would promote a specific microbiota profile similar to the observed in the preterm infants who receive MOM. To pursue this objective we analyzed the impact of different nutritional approaches upon the gut microbiota composition of preterm infants born at ≤32 weeks of gestation.
Materials and Methods
Study Design
We conducted a prospective, observational unicentric cohort study including consecutively admitted preterm infants born at ≤32 weeks of gestation and birth weight ≤1,500 g in the Division of Neonatology of the University and Polytechnic Hospital La Fe (Valencia, Spain) during a 12-month period. The study protocol was approved by the hospital IRB (Comité de Ética e Investigación Médica) and parents approved and signed informed consent in all cases.
Patients’ Characteristics
Inclusion and exclusion criteria are shown in Table 1. Demographic, perinatal, clinical, and analytical data were recorded and matched according to the type of feeding (Table 2). Administration and duration of antibiotic therapy was also collected.
Table 1.
Inclusion and exclusion criteria for preterm infants receiving different types of nutrition and whose microbiota was studied.
| Inclusion criteria | Exclusion criteria |
|---|---|
| BW ≤ 1.500 g and/or GA ≤ 32 weeks | GA > 32 weeks |
| Enteral intake (≥ 150 mL/kg/day) | Parents refuse to participate/sign informed consent |
| The principal nutrient received (MOM, DHM, or formula) represents 80% of the intake | Mixed breastfeeding |
| DHM from just one donor to one premature or maximum of two different donors | Chromosomopathies |
| No additional treatments that could alter the microbiota (e.g., probiotics) or oxidative status (e.g., vitamins C, A, and E) | Major malformations or surgery of the digestive tract |
GA, gestational age; BW, birth weight; MOM, mother’s own milk; DHM, donor human milk.
Table 2.
Perinatal characteristics and confounders of preterm infants receiving different types of nutrition and whose microbiota was studied.
| MOM (n = 34) | DHM (n = 28) | p-Value | |
|---|---|---|---|
| GA weeks, mean (SD) | 28.85 (1.9) | 29.78 (2.42) | 0.09 |
| Antenatal steroids full course (%) | 97.1 | 92.8 | 0.44 |
| Type of delivery (%) | |||
| Vaginal | 58.8 | 39.3 | 0.126 |
| Cesarean section | 41.2 | 60.7 | |
| Birth weight (g), mean (SD) | 1,228 (301) | 1304.3 (262) | 0.3 |
| Race (%) | |||
| Caucasian | 85.3 | 67.8 | 0.1 |
| Non-Caucasian | 14.7 | 32.1 | |
| Apgar 1 min (median; 5–95% CI) | 7.3 (2.15) | 7.1 (1.81) | 0.68 |
| Apgar 5 min (median; 5–95% CI) | 9.02 (1.3) | 8.6 (1.4) | 0.24 |
| Age (days) at sample collection, mean (SD) | 9.7 (7.03) | 8.7 (6.2) | 0.52 |
| Chorioamnionitis (%) | 76.4 | 89.2 | 0.19 |
| Mechanical ventilation | 11.7 | 14.3 | 0.76 |
| Non-invasive ventilation | 75 | 85.3 | 0.3 |
| Persistent ductus arteriosus | 29.4 | 25 | 0.69 |
| Antibiotic therapy (%) | 38.2 | 39.2 | 0.93 |
Fresh DHM was collected and immediately frozen at -20°C until Holder pasteurization process (62.5°C for 30 min followed by fast cooling). After treatment, pasteurized DHM was frozen until the distribution to patients. The Division of Neonatology protocol involves strong support to breastfeeding and offering DHM as a supplement to preterm infants born below ≤32 weeks or ≤1,500 g birth weight. Type of feeding was a parents’ decision. The infants were fed at least with an 80% of either MOM or DHM and table intakes of 150 cc/kg/day.
The nutritional intake was prospectively monitored but never influenced by this observational study.
Fecal Samples, DNA Extraction, and 16S rDNA Sequencing
Fecal samples were directly collected from the diaper when full enteral feeding (defined as ≥150 cc/kg/day of MOM, DHM, or formula) was achieved. Samples were frozen and stored at -80°C for later analysis.
Total fecal DNA was isolated using the MasterPure Complete DNA & RNA Purification Kit (Epicentre, Madison, WI, United States) according to the manufacturer’s instructions with modifications that included a bead-beater step and enzyme incubation to increase DNA extraction as described elsewhere (Boix-Amoros et al., 2016). Total DNA concentration was measured using a Qubit® 2.0 Fluorometer (Life Technologies, Carlsbad, CA, United States) and normalized to 5 ng/μL for 16S rDNA gene (V3–V4 region) amplification using Nextera XT Index Kit. Amplicons were checked with a Bioanalyzer DNA 1000 chip and libraries were sequenced using a 2 × 300 bp paired-end run (MiSeq Reagent kit v3) on a MiSeq-Illumina platform (FISABIO sequencing service, Valencia, Spain). Controls during DNA extraction and PCR amplification were also included and sequenced.
Bioinformatics and Statistical Analysis
Data were obtained using an ad hoc pipeline written in RStatistics environment (R Core Team, 2012) and data processing was performed using a QIIME pipeline (version 1.9.0) (Caporaso et al., 2010). Chimeric sequences and sequences that could not be aligned were also removed from the data set. The clustered sequences were utilized to construct operational taxonomic units (OTUs) tables with 97% identity and representative sequences were taxonomically classified based on the Greengenes 16S rRNA gene database (version 13.8). Sequences that could not be classified to domain level, or classified as cyanobacteria and chloroplasts, were removed from the data set. Subsequently, alpha diversity indices (Chao1 and Shannon, Species richness estimates and diversity index, respectively), beta diversity based on UNIFRAC unweighted distance (phylogenetic) and Bray–Curtis distance (non-phylogenetic), and PERMANOVA with 999 permutations was used to test significance between groups. The DESeq2 method was used to determine differential abundances of specific bacteria between feeding groups. Calypso software1 version 7.36 was used with total sum normalization for the statistical analysis, and also, cumulative sum scaling (CSS) normalization for multivariate test. Furthermore, predictive inferred functional analysis was performed using PICRUSt (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States) approach as described (Langille et al., 2013). Linear discriminant analysis effect size (LEfSe) (Segata et al., 2011) was used to detect unique biomarkers in relative abundance of bacterial taxonomy and specific functions (KEGG pathways). A size effect cut-off of 3.0 on the logarithmic linear discriminant analysis (LDA) score was used. The 16S rRNA gene sequence data generated is available through GenBank Sequence Read Archive Database under project accession number PRJEB25948.
Results
Patients’ Characteristics
A total of 69 preterm infants ≤32 weeks of gestation pertaining to the MOM (n = 34), DHM (n = 28), and formula (n = 7) groups were recruited. No differences for prenatal demographic characteristics or confounders during the hospitalization between patients in the three feeding-type groups were found (Table 2). Individual information is available in Supplementary Table S1.
A small group of preterm neonates (n = 7) fed with formula milk was also included. The low number is explained because the protocol of our NICU recommends for all preterm ≤32 weeks of gestation and ≤1,500 g MOM and DHM as an alternative and therefore preterm fed with formula render exceptional. Characteristics of the formula group were as follows: 33 ± 2 weeks of gestation; 1,702 ± 321.6 g birth weight; 57.1% male; 14.3% born by vaginal delivery (85.7% by cesarean section); 57.1% received antenatal steroids.
Impact of DHM on Preterm Microbiota
Significant differences were found in preterm microbiota composition according to feeding type. We found lower relative abundance of Firmicutes (30.9 vs 45.5%, p-value = 0.029) and higher abundance of Actinobacteria (20.1 vs 10.2%, p-value = 0.040) in MOM compared to DHM group (Figure 1A). At family level, higher abundance of Bifidobacteriaceae (19.5 vs 9.0%, p-value = 0.027) and lower abundance of Clostridiaceae (3.7 vs 11.2%, p-value = 0.029) were observed in MOM compared to DHM (Figure 1B). At genus level, higher levels of Bifidobacterium (19.5 vs 8.98%, p-value = 0.027) and unclassified Enterobacteriaceae (29.77 vs 18.48%, p-value = 0.060) and lower levels of Citrobacter (2.60 vs 9.83%, p-value = 0.060), and unclassified Clostridiaceae (3.46 vs 9.43%, p-value = 0.062) were observed in fecal samples of MOM as compared to the DHM group.
FIGURE 1.
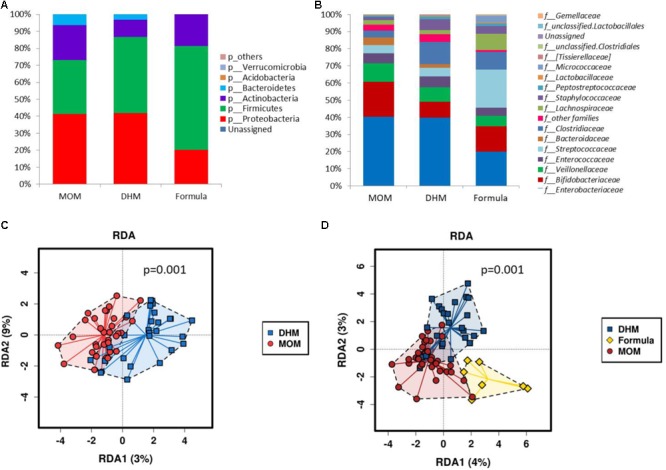
Microbial relative abundances (%) at phylum (A) and family level (B) of preterm gut microbiome according to diet (MOM, DHM, and formula). RDA plots of the preterm microbiota grouped by infant feeding type: MOM vs DHM (C) and MOM, DHM, and formula (D).
The effect of the diet on the preterm gut microbiota was explored by applying the multivariate method PERMANOVA with 999 permutations on the phylogenetic distances (p-value = 0.09 for unweighted UNIFRAC distance) and Bray–Curtis distance (non-phylogenetic; p-value = 0.0046). Furthermore, multivariate redundant discriminant analysis (RDA) based on the observed OTUs showed statistically significant differences in microbial composition between MOM and DHM groups (p = 0.001) (Figure 1C).
To explore the variation of the microbial community composition between MOM and DHM, we performed LEfSe tests to detect differences in relative abundance of bacterial taxa across fecal samples (Figure 2). At the family level, Bifidobacteriaceae family was significantly enriched in MOM compare to DHM samples (LDA = 4.90, p-value = 0.025) while Staphylococcaceae (LDA = 4.63, p-value ≤0.042). Pasteurellaceae family was enriched in DHM (LDA = 4.358, p-value = 0.050). Specific enriched features at genus and OTUs levels are shown in Figure 2.
FIGURE 2.
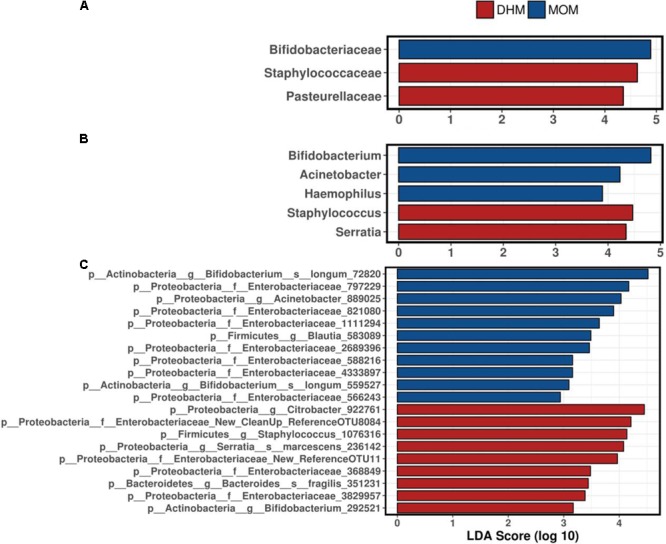
Linear discriminant analysis (LDA) combined with effect size measurements (LEfSe) revealed specific microbes at family (A), genus (B), and OTUs (C) between MOM and DHM feeding groups.
DESeq2 test was used to identify differential abundances of specific bacteria between feeding groups. Actinobacteria phylum was higher in MOM compared to DHM group (20.07 vs 10.25%, p-value = 0.0044, FDR = 0.013). The abundance of Staphylococcus (p-value <0.0001, FDR < 0.0001), Clostridium (p-value <0.0001, FDR = 0.0013), Serratia (p-value <0.0001, FDR = 0.0022), Coprococcus (p-value = 0.0021, FDR = 0.012), Aggregatibacter (p-value = 0.015, FDR = 0.059), and Lactobacillus (p-value = 0.056, FDR = 0.18) was significantly higher in DHM group than MOM group. However, Bacteroides (p-value <0.0001, FDR = 0.0044), Acinetobacter (p-value <0.0001, FDR = 0.002), and Haemophilus (p-value = 0.0014, FDR = 0.009) were significantly higher in the MOM than in DHM group.
Impact of Formula vs Human Milk Groups on Preterm Microbiome
Despite the low number of formula fed preterm infants, we analyzed the differences in microbiome between MOM, DHM, and FM groups. A multivariate RDA based on the observed OTUs showed statistically significant differences in microbial composition between groups (p = 0.001) (Figure 1D). Significantly higher relative abundance of Firmicutes (p-value = 0.027, FDR = 0.016) was observed in FM group compared to MOM and DHM (Figure 1A). At genus level, significant higher abundance of Blautia (p-value <0.001, adjusted p-value = 0.033, FDR = 0.033), Streptococcus (p-value = 0.0024, FDR = 0.054), Acidaminococcus (p-value = 0.0093, FDR = 0.099), Rothia (p-value = 0.0059, FDR = 0.088), and Dorea (p-value = 0.011, FDR = 0.099) were observed in the FM group compared to the MOM and DHM groups.
Preterm core microbiome was composed by a total of 15 shared genus independently of feeding-type diet (Figure 3A). Acinetobacter genus was exclusively present in MOM group; while Coprococcus genus was present in DHM and unclassified Peptostreptococcaceae genus in formula group.
FIGURE 3.
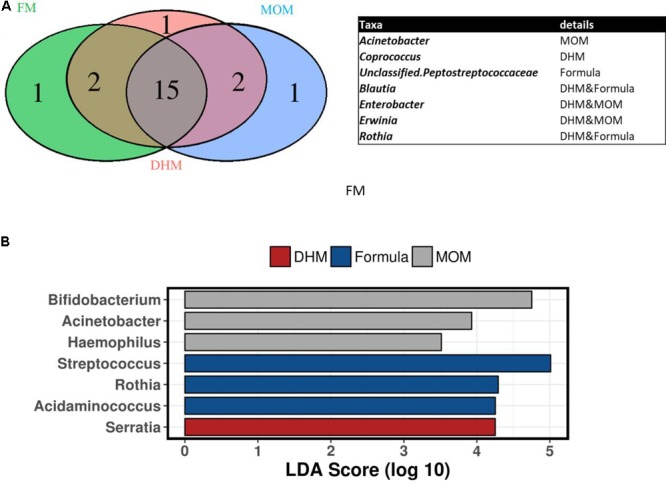
Venn diagram of shared genus between group (A) and linear discriminant analysis LEfSe test at genus level between MOM, DHM, and formula preterm microbiota (B).
Linear discriminant analysis effect size test showed that Rothia, Streptococcus, and Acidaminococcus genus were significantly enriched in formula group compared to MOM and DHM group, while Bifidobacterium, Acinetobacter, and Haemophilus genus were enriched significantly in MOM compared to DHM, representing a hallmark for breast fed preterm gut microbiota (Figure 3B).
We also applied discriminant analysis of principal components (DAPC) identifying specific feeding-type related clusters of preterm microbiota (Figure 4). These microbial shifts were attributed to subtle changes in the abundance of several bacterial OTUs. Clostridium, Bifidobacterium, unclassified Enterobacteriaceae and Veillonella related OTUs were the strongest indicator of the presence of distinct microbial clusters.
FIGURE 4.
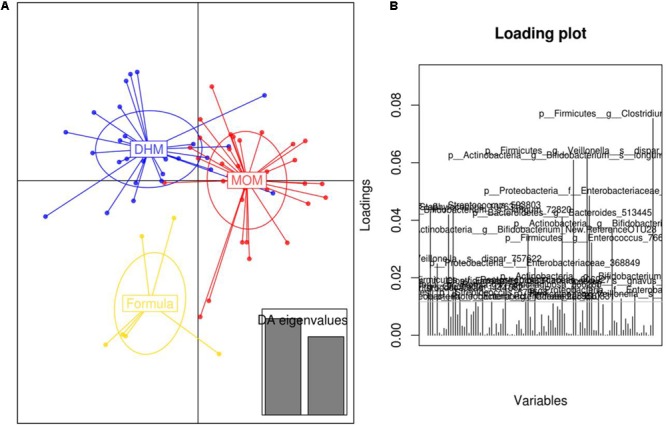
Discriminant analysis of principal components (DAPC) plot (A) at OTUs level revealed distinct clustering of the MOM (red), DHM (blue), and formula-fed group (yellow). Canonical loading plot (B) showing differentially abundant bacterial genera. The individual peaks show the magnitude of the influence of each variable on separation of the groups (0.05 threshold level).
Functional Assignment of the Preterm Microbiota
Inferred metagenomic PICRUSt prediction revealed significant differences in the main functional classes (Kyoto Encyclopedia of Genes and Genomes, KEGG categories at level 2), deriving from functional acquisitions associated with different diets (multivariate RDA test, p = 0.007) (Figure 5A). Moreover, no different metabolic profile was found (RDA test p-value >0.05) when MOM and DHM were compared, while formula functional profile was significantly different from those observed in MOM (RDA test p-value = 0.024) and DHM (RDA test p-value = 0.002).
FIGURE 5.
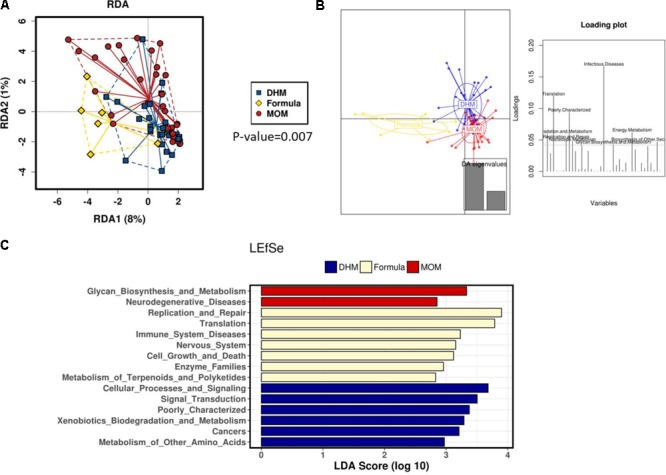
RDA plot (A). Discriminant analysis of principal components (DAPC) plot and loading plot (B) at KEGG pathways. The individual peaks in the loading plot show the magnitude of the influence of each variable on separation of the groups (0.05 threshold level). LEfSe test (C) revealed distinct clustering of the MOM (red), DHM (blue), and formula-fed group (yellow).
The DAPC to identify specific clusters of functional activity (KEGG level 2 and 3) of the gut microbiome in preterm groups according to type of diet (Figure 5B) suggesting a distinct KEGG activities DHM and MOM microbiome activities are similar than those observed in the FM group.
Linear discriminant analysis effect size analysis performed on PICRUSt output showed several KEGG (level 2, Figure 5C and level 3, Supplementary Figure S1) categories differentially present in the MOM, DHM, and formula groups. MOM functional profile is mostly represented by bacterial secretion system, lipopolysaccharide (LPS) biosynthesis, and biosynthesis. In particular, we observed a deprivation in functions involved in complex carbohydrate metabolism, deriving from HMO present in breast milk, such us Glycan biosynthesis and metabolism in the formula group (p-value = 0.019) compared to the other MOM and DHM groups. This glycan pathway was not different between MOM and DHM profiles (p-value >0.05). Interestingly, we observed a reduced LPS biosynthesis (p-value <0.0001) and LPS biosynthesis proteins (p-value <0.0001) in formula fed infants compared to MOM and DHM (Supplementary Figure S2). However, the most predominant function in DHM group was the two-component regulatory system followed by other functions related to amino acid metabolisms, fatty acids metabolisms (butanoate metabolism) and to sulfur metabolism and sulfur relay system and also, nitrogen metabolism in DHM group (Supplementary Figure S2). Methane metabolism pathway is also enriched in formula group as compared to MOM and DHM groups (Supplementary Figure S2). Furthermore, we also observed in formula group an enrichment of KEGG functions related to sugar metabolisms as galactose metabolism, and amino sugar and nucleotide sugar metabolisms compared to the observed ones in MOM and DHM (Supplementary Figure S2).
Discussion
In recent years, nutritional practices have shifted toward encouraging breastfeeding practices in preterm neonates (Keunen et al., 2015; O’Connor et al., 2016). In the absence of MOM, DHM has become the preferred nutritional alternative and a formula feeding remains the last option when the others are not available. The setting up of milk banks has rendered DHM the most widely prescribed alternative to MOM (Underwood, 2013).
In preterm infants, our results have demonstrated that the feeding type has an important impact on gut microbial composition in preterm infants ≤1,500 g. We found that MOM and DHM microbial profiles were different. MOM fed babies showed a significantly enriched and greater presence of Bifidobacteriaceae and lower of Staphylococcaceae, Clostridiaceae, and Pasteurellaceae compared to DHM fed babies. At genus level, higher levels of Bifidobacterium and unclassified Enterobacteriaceae and lower unclassified Clostridiaceae were observed in fecal samples from MOM group compared to DHM preterm group. It has been reported that preterm infant receiving MOM had a higher abundance of Clostridiales, Lactobacillales, and Bacillales compared to both the DHM and formula groups (Gregory et al., 2016). Both these groups had higher abundance of Enterobacteriales. After controlling for gender, postnatal age, weight, and birth gestational age, the diversity of gut microbiota increased over time and was constantly higher in infants fed MOM relative to infants with other feeding types. Finally, in the formula microbial profile was distinct than those observed in MOM and DHM, suggesting that DHM favors an intestinal microbiome more similar to MOM despite the differences between MOM and DHM.
In accordance to the microbiota shifts, we observed that KEGG profiles in DHM and MOM were similar than those microbial profile observed in formula. MOM functional profile is mostly represented by bacterial secretion system, LPS biosynthesis and biosynthesis protein which would be mainly related to the presence of Gram-negative bacteria. Interestingly, we observed a significant reduction on the LPS biosynthesis and proteins in formula fed infants compared to MOM and DHM (without difference between them). These data would suggest the potential link between LPS and immune response as reported previously (Cullen et al., 2015). Recent study has been shown that variation on the microbial LPS produced by specific microbiota groups as Enterobacteriaceae and Bacteroides spp., could either stimulate or actively inhibit inflammatory pathway and also, have a role on the autoimmune diseases risk (Vatanen et al., 2016). In our context, MOM and DHM modulate a preterm gut microbiota toward an enrichment in Bifidobacterium spp. and also, Bacteroides spp. that may promote the specific LPS signaling and its contribution to the immune system. Furthermore, we observed a significant reduction in functions derived from the presence of HMO and involved in complex carbohydrate metabolism (e.g., glycan biosynthesis) in the FM group as compared with the MOM and DHM groups. These differences could be explained by the abundance of HMO metabolizers as Bifidobacterium and Bacteroides spp. in preterm gut fed with MOM and DHM compared to formula fed preterm.
We also found enrichment on the functions related to the fatty acids metabolism and to sulfur and nitrogen metabolism in DHM and MOM groups. Several enteric bacteria and oral bacteria produce reduced sulfur and nitrogen and maybe some specific bacteria, as Deltaproteobacteria, Clostridium spp., Veillonella spp., Rothia spp., would be responsible for this functional contribution as they were enriched in DHM group although it was also observed in MOM group. In formula metagenome, we observed enrichment of KEGG functions related to sugar metabolisms as galactose metabolism, involved in conversion of galactose into glucose could arise from consumption of infant formula and/or dairy products, and amino sugar and nucleotide sugar metabolisms. In general, minor differences were observed in the functional profiles between MOM and DHM suggesting the potential effect of DHM in mimicking the microbiome functionality of own maternal milk feeding. These results would open new possibilities in future research where bigger studies should be carried out.
Two important factors influence the differences found in preterm gut microbiota depending on feeding type (MOM and DHM). The first one would be related to the timing of milk extraction in relation to gestational age and to the lactation stage. While MOM is the biological product of a prematurely interrupted gestation, DHM is composed mainly by donated mature milk from mothers who completed term gestations and were extracting milk for several weeks thereafter. Preterm milk has higher amount of proteins, fats, and energy (Underwood, 2013; Gidrewicz and Fenton, 2014). Hence, depending on the staging of lactation they can be also a great variability among donators of components such as essential fatty acids or amino acids indispensable not only for an adequate nutrition but also for promoting microbiota colonization. Although donor milk pooling tries to avoid these circumstances, a recent study has shown a shortage of docosahexaenoic acid or lysine in DHM. Targeted supplementation would be needed not only to optimize nutritional properties of DHM but also to improve bacterial colonization (Ballard and Morrow, 2013). The second factor would be related with the pasteurization procedure that inevitably alters essential thermolabile milk components. Hence, differences in the microbiota would be at least partially explained by the different composition in relation to nutritional parameters and bioactive compounds such as immune markers, microbiota, oligosaccharides, neurotrophic, and growth factors among others (Bertino et al., 2008; Untalan et al., 2009; Ballard and Morrow, 2013; Christen et al., 2013; Espinosa-Martos et al., 2013; Sousa et al., 2014; Coscia et al., 2015; Valentine et al., 2017). Altogether these findings could explain the similarities and differences in the microbiota profile between preterm infants fed DHM or MOM and influence health outcomes in preterm infants.
Finally, the number of formula fed infants included in this study was limited (n = 7) due mainly because milk bank provides with DHM to almost all preterm babies attended in our NICU (>90%) and therefore it is difficult to recruit preterm babies on formula feeding and randomization was not ethically acceptable. Despite these limitations, our results reveal a substantial impact of DHM feeding on the structure of the intestinal microbial community composition.
Conclusion
Feeding type modulates the preterm microbiome composition. DHM feeding had an impact on preterm microbiota that could have potential beneficial long-term effects on intestinal functionality, immune system, and metabolism. However, available pasteurization methods cause changes that may blunt many of the positives aspects derived from the use of MOM. Therefore, further studies are stringently need to understand the complex links between microbiome and DHM host, its impact on health programming, and to develop sensitive methods capable of providing promptly after birth preterm infants with HM as similar as possible to their MOM when the latter is not yet available.
Author Contributions
AP-L, MG, MC, MCC, and MV planned the experiments. AP-L, MG, MC, AN-R collected the samples and clinical data. AP-L, CA, and MCC performed the microbiological analysis and analyzed data. AP-L wrote first draft and all authors commented criticized, and reviewed the manuscript. All authors accepted the final version the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful for all the participant families providing biological samples for this study.
Abbreviations
- BPD
bronchopulmonary dysplasia
- DHM
donor human milk
- FM
special preterm formula
- HM
human milk
- LOS
late onset sepsis
- MOM
mother’s own milk
- NEC
necrotizing enterocolitis
- NICU
neonatal intensive care unit
- PVL
periventricular leukomalacia
- ROP
retinopathy of prematurity
Funding. CA and MCC would like to acknowledge the Spanish Government of Economy and Competitiveness (MINECO) (AGL2015-707487-P grant) and also, the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (ERC Starting Grant, project no. 639226). AP-L is a granted research fellow from the Health Research Institute La Fe (Valencia, Spain). MV and AN-R acknowledge research contract from RETICS funded by the PN 2018-2011 (Spain), ISCIII-Sub-Directorate General for Research Assessment and Promotion and the European Regional Development Fund (FEDER), reference RD12/0026. MC acknowledges a research grant JR16/00038 from the Health Research Institute Carlos III (Ministry of Economy, Industry and Competitiveness, Kingdom of Spain).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01376/full#supplementary-material
References
- Agostoni C. (2010). Enteral nutrient supply for preterm infants: cmomentary from the European society of paediatric gastroenterology, hepatology and nutrition committee on nutrition. J. Pediatr. Gastroenterol. Nutr. 50 85–91. 10.1097/MPG.0b013e3181adaee0 [DOI] [PubMed] [Google Scholar]
- American Academy of Pediatrics (2012). Breastfeeding and the use of human milk. Pediatrics 129 e827–e884. 10.1542/peds.2011-3552 [DOI] [PubMed] [Google Scholar]
- Arslanoglu S., Bertino E., Tonetto P., De Nisi G., Ambruzzi A. M., Biasini A., et al. (2010). Guidelines for the establishment and operation of a donor human milk bank. J. Matern. Fetal Neonat. Med. 23 1–20. 10.3109/14767058.2010.512414 [DOI] [PubMed] [Google Scholar]
- Ballard O., Morrow A. L. (2013). Human milk composition: nutrients and bioactive factors. Pediatr. Clin. North Am. 60 49–74. 10.1016/j.pcl.2012.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baro C., Giribaldi M., Arslanoglu S., Giuffrida M. G., Dellavalle G., Conti A., et al. (2001). Effect of two pasteurization methods on the protein content of human milk. Front. Biosci. 3 818–829. [DOI] [PubMed] [Google Scholar]
- Belfort M. B., Anderson P. J., Nowak V. A., Lee K. J., Molesworth C., Thompson D. K., et al. (2016). Breast milk feeding, brain development, and neurocognitive outcomes: A 7-year longitudinal study in infants born at less than 30 weeks’ gestation. J. Pediatr. 177 133–139. 10.1016/j.jpeds.2016.06.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertino E., Coppa G. V., Giuliani F., Coscia A., Gabrielli O., Sabatino G., et al. (2008). Effects of Holder pasteurization on human milk oligosaccharides. Int. J. Immunopathol. Pharmacol. 21 381–385. 10.1177/039463200802100216 [DOI] [PubMed] [Google Scholar]
- Bertino E., Giuliani F., Occhi L., Coscia A., Tonetto P., Marchino F., et al. (2009). Benefits of donor human milk for preterm infants: current evidence. Early Hum. Dev. 85 S9–S10. 10.1016/j.earlhumdev.2009.08.010 [DOI] [PubMed] [Google Scholar]
- Boix-Amoros A., Collado M. C., Mira A. (2016). Relationship between milk microbiota, bacterial load, macronutrients, and human cells during lactation. Front. Microbiol. 7:492 10.3389/fmicb.2016.00492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010). QIIME allows analysis of high-throughput cunity sequencing data. Nat. Methods 7 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen L., Lai C. T., Hartmann B., Hartmann P. E., Geddes D. T. (2013). The effect of UV-C pasteurization on bacteriostatic properties and immunological proteins of donor human milk. PLoS One 8:e85867. 10.1371/journal.pone.0085867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M. C., Cernada M., Baüerl C., Vento M., Pérez-Martínez G. (2012). Microbial ecology and host-microbiota interactions during early life stages. Gut Microbes 3 352–365. 10.4161/gmic.21215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M. C., Cernada M., Neu J., Pérez-Martínez G., Gormaz M., Vento M. (2015). Factors influencing gastrointestinal tract and microbiota immune interaction in preterm infants. Pediatr. Res. 77 726–731. 10.1038/pr.2015.54 [DOI] [PubMed] [Google Scholar]
- Corpeleijn W. E., de Waard M., Christmann V., van Goudoever J. B., Jansen-van, der Weide M. C.et al. (2016). Effect of donor milk on severe infections and mortality in very low-birth-weight infants: the early nutrition study randomized clinical trial. JAMA Pediatr. 1 654–661. 10.1001/jamapediatrics.2016.0183 [DOI] [PubMed] [Google Scholar]
- Coscia A., Peila C., Bertino E., Coppa G. V., Moro G. E., Gabrielli O., et al. (2015). Effect of Holder pasteurisation on human milk glycosaminoglycans. J. Pediatr. Gastroenterol. Nutr. 60 127–130. 10.1097/MPG.0000000000000570 [DOI] [PubMed] [Google Scholar]
- Cullen T. W., Schofield W. B., Barry N. A., Putnam E. E., Rundell E. A., Trent M. S., et al. (2015). Gut microbiota. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science 347 170–175. 10.1126/science.1260580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ESPGHAN Committee on Nutrition Arslanoglu S., Corpeleijn W., Moro G., Braegger C., Campoy C.et al. (2013). Donor human milk for preterm infants: current evidence and research directions. J. Pediatr. Gastroenterol. Nutr. 57 535–542. 10.1097/MPG.0b013e3182a3af0a [DOI] [PubMed] [Google Scholar]
- Espinosa-Martos I., Montilla A., de Segura A. G., Escuder D., Bustos G., Pallaìs C., et al. (2013). Bacteriological, biochemical, and immunological modifications in human colostrum after Holder pasteurisation. J. Pediatr. Gastroenterol. Nutr. 56 560–568. 10.1097/MPG.0b013e31828393ed [DOI] [PubMed] [Google Scholar]
- Gidrewicz D. A., Fenton T. R. (2014). A systematic review and meta-analysis of the nutrient content of preterm and term breast milk. BMC Pediatr. 14:216. 10.1186/1471-2431-14-216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory K. E., Samuel B. S., Houghteling P., Shan G., Ausubel F. M., Sadreyev R. I., et al. (2016). Influence of maternal breast milk ingestion on acquisition of the intestinal microbiome in preterm infants. Microbiome 4:68. 10.1186/s40168-016-0214-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Milk Banking Association of North America (2000). Guidelines for the Establishment and Operation of a Donor Human Milk Bank. Raleigh, NC: Human Milk Banking Association of North America. [Google Scholar]
- Keunen K., van Elburg R. M., van Bel F., Benders M. J. (2015). Impact of nutrition on brain development and its neuroprotective implications following preterm birth. Pediatr. Res. 77 148–155. 10.1038/pr.2014.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille M. G., Zaneveld J., Caporaso J. G., McDonald D., Knights D., Reyes J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31 814–821. 10.1038/nbt.2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtonen L., Gimeno A., Parra-Llorca A., Vento M. (2017). Early neonatal death: a challenge worldwide. Semin. Fetal Neonatal. Med. 22 153–160. 10.1016/j.siny.2017.02.006 [DOI] [PubMed] [Google Scholar]
- Madore L. S., Bora S., Erdei C., Jumani T., Dengos A. R., Sen S. (2017). Effects of donor breastmilk feeding on growth and early neurodevelopmental outcomes in preterm infants: an observational study. Clin. Ther. 39 1210–1220. 10.1016/j.clinthera.2017.05.341 [DOI] [PubMed] [Google Scholar]
- Meinzen-Derr J., Poindexter B., Wrage L., Morrow A. L., Stoll B., Donovan E. F. (2009). Role of human milk in extremely low birth weight infants’ risk of necrotizing enterocolitis or death. J. Perinatol. 29 57–62. 10.1038/jp.2008.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Institute for Health and Clinical Excellence (2010). Donor Breast Milk Banks: The Operation of Donor Breast Milk Bank Services. London: National Institute for Health and Clinical Excellence. [PubMed] [Google Scholar]
- O’Connor D. L., Gibbins S., Kiss A., Bando N., Brennan-Donnan J., Ng E., et al. (2016). Effect of supplemental donor human milk compared with preterm formula on neurodevelopment of very low-birth-weight infants at 18 months: a randomized clinical trial. JAMA 316 1897–1905. 10.1001/jama.2016.16144 [DOI] [PubMed] [Google Scholar]
- Peila C., Moro G. E., Bertino E., et al. (2016). The effect of holder pasteurization on nutrients and biologically-active components in donor human milk: a review. Nutrients 8:E477. 10.3390/nu8080477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2012). A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12: R60. 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa S. G., Santos M. D., Fidalgo L. G., Delgadillo I., Saraiva J. A. (2014). Effect of thermal pasteurisation and high-pressure processing on immunoglobulin content and lysozyme and lactoperoxidase activity in human colostrum. Food Chem. 151 79–85. 10.1016/j.foodchem.2013.11.024 [DOI] [PubMed] [Google Scholar]
- The European Perinatal Health Report (2010). >Available at: http://www.europeristat.com/reports/european-perinatal-health-report-2010.html [Google Scholar]
- Underwood M. A. (2013). Human milk for the premature infant. Pediatr. Clin. North Am. 60 189–207. 10.1016/j.pcl.2012.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untalan P. B., Keeney S. E., Palkowetz K. H., Rivera A., Goldman A. S. (2009). Heat susceptibility of interleukin-10 and other cytokines in donor human milk. Breastfeed Med. 4 137–144. 10.1089/bfm.2008.0145 [DOI] [PubMed] [Google Scholar]
- Valentine C. J., Morrow G., Reisinger A., Dingess K. A., Morrow A. L., Rogers L. K. (2017). Lactational stage of pasteurized human donor milk contributes to nutrient limitations for infants. Nutrients 9:E302. 10.3390/nu9030302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatanen T., Kostic A. D., d’Hennezel E., Siljander H., Franzosa E. A., Yassour M., et al. (2016). Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell 165 842–853. 10.1016/j.cell.2016.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson E., Edstedt Bonamy A. K., Bonet M., Toome L., Rodrigues C., Howell E. A., et al. (2017). Room for improvement in breast milk feeding after very preterm birth in Europe: results from the EPICE cohort. Matern. Child Nutr. 14:e12485. 10.1111/mcn.12485 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


