Abstract
Epithelial-mesenchymal transition (EMT) is a conserved cellular plasticity program that is reactivated in carcinoma cells and drives metastasis. While EMT is well studied its regulatory mechanisms remain unclear. Therefore, to identify novel regulators of EMT, a data mining approach was taken using published microarray data and a group of deubiquitinases (DUBs) were found to be upregulated in cells that have undergone EMT. Here, it is demonstrated that one DUB, ubiquitin specific peptidase 11 (USP11), enhances TGFβ-induced EMT and self-renewal in immortalized human mammary epithelial cells. Furthermore, modulating USP11 expression in human breast cancer cells altered the migratory capacity in vitro and metastasis in vivo. Moreover, elevated USP11 expression in human breast cancer patient clinical specimens correlated with decreased survival. Mechanistically, modulating USP11 expression altered the stability of TGFβ receptor type 2 (TGFBR2) and TGFβ downstream signaling in human breast cancer cells. Together, these data suggest that deubiquitination of TGFBR2 by USP11 effectively spares TGFBR2 from proteasomal degradation to promote EMT and metastasis.
Keywords: EMT, metastasis, TGFβ, USP11, proteasome
Introduction
Metastasis is the major cause of cancer-related deaths; despite intense investigation, it continues to be a poorly understood aspect of cancer progression (1). While deaths due to metastasis are likely to be abrogated by increased efforts in early detection of cancer, there is a need for more effective metastasis-specific therapies in the clinic (2).
Metastasis is fueled by disseminating tumor cells following a complex cascade of events starting with translocation of primary tumor cells to a distant site and culminating with the development into a lethal outgrowth in distant organs. Two well-studied drivers of metastasis are the epithelial-mesenchymal transition (EMT) and microenvironment-derived transforming growth factor-β (TGFβ). The EMT is a cellular developmental program that becomes reactivated during cancer progression. Previous work has demonstrated that the EMT endows neoplastic epithelial cells with mesenchymal and stem-like characteristics needed for subsequent steps in the metastatic cascade (3).
TGFβ is a cytokine that is known to promote cancer metastasis through multiple mechanisms, including enhancing invasive properties, stemness, and permissive effects on the tumor microenvironment (4,5). Interestingly, TGFβ is a potent inducer of EMT; thus, metastasis relies, in part, on TGFβ’s ability to induce an EMT program (6). The link between TGFβ and EMT is mechanistically clear: upon TGFβ type I and type II receptor (TGFBR1 and TGFBR2) heterocomplex activation, SMAD2/3 are phosphorylated and shuttled to the nucleus with SMAD4 to regulate expression of EMT transcription factors (7). However, what remain unclear are the ubiquitin-proteasome regulatory mechanisms of TGFβ signaling in the context of EMT and metastasis. Importantly, a deeper understanding of this mode of regulation may reveal novel enzymes that can be subject to rational and specific drug design.
There is increasing support for the importance of the ubiquitin-proteasome system in regulating EMT and cancer progression. Low proteasome activity is associated with the cancer stem cell state and EMT, and treatment of epithelial cells with proteasome inhibitors can induce EMT (8,9). In addition, a number of deubiquitinases (DUBs) have been implicated in cancer progression and metastasis, such as DUB3, USP4, USP13, USP15, USP28, and USP51 (10–15). Based on this work and the context-specific nature of EMT, we hypothesize that there exists a set of proteins within the ubiquitin-proteasome pathway that regulate EMT specifically in normal and neoplastic human mammary epithelial cells.
In this study, we show that specific DUBs are upregulated during EMT, and that Ubiquitin specific peptidase 11 (USP11) regulates TGFβ-induced EMT in immortalized human mammary epithelial cells. Further, we demonstrate that USP11 regulates human breast cancer cell behavior, including migration, self-renewal, and experimental metastasis. Finally, we show that USP11 exerts its effects on human breast cancer cell behavior by stabilizing the TGFβ type II receptor (TGFBR2) and enhancing TGFβ signaling. While this relationship has been previously shown in lung fibroblast cells (16), we demonstrate the relevance of this pathway in human breast cancer in vitro and in vivo models implicating USP11 as a useful therapeutic target in the clinic.
Materials & Methods
Animal studies
Animal studies were conducted using procedures approved by the IACUC at the University of California, San Diego (protocol # S09264). Studies were conducted in accordance to the ARRIVE guidelines. NOD-scid IL2Rgammanull (NSG) mice were obtained from Jackson Laboratories and UCSD Animal Care Program. For the tail vein metastasis assay, SUM159 or MDA-MB-231 cells were resuspended in PBS and injected into the tail vein of 6–8 week old female NSG mice. A total of 1×106–1.5×106 cells were injected in a volume of 100 µl. Mice were sacrificed after 6–8 weeks and lung metastases were quantified. Tumors located within vascular spaces in the lung were excluded from analysis.
Metastasis quantification
Lungs were perfused with PBS and then removed from the thoracic cavity. The lung lobes were fixed in Bouin’s solution for 6 hours, and tissue was processed for sectioning and H&E staining. H&E step sections were analyzed by a pathologist (M. V. Estrada) at the Tissue Technology Shared Resource (Moores Cancer Center, UCSD). Tumor burden was assessed by whole section tumor cellularity.
Cell culture
All cell cultures were maintained at 37°C with 5% CO2. Human mammary epithelial cell lines (HMLE) were cultured in MEGM (Lonza). shRNA and overexpression constructs were retrovirally transduced into HMLE cells. HEK293T, T47D, and MDA-MB-231 cells were cultured in DMEM with 10% FBS. SUM159 cells were cultured in Ham’s F12 supplemented with 10 mM HEPES, 5% FBS, 5 µg/ml insulin, and 1 µg/ml hydrocortisone. T47D cells were obtained from Li Ma (University of Texas, MD Anderson Cancer Center). All cell lines were authenticated by short tandem repeat (STR) analysis at ATCC. Since it was not present in any STR database as a basis of comparison, the HMLE cell line was authenticated on the basis of morphology and epithelial marker expression to the original cell line, as other studies have reported (17). In addition, all cell lines used in the manuscript tested negative for mycoplasma using the service provided by the Human Embryonic Stem Cell Core Facility at UCSD. Cells used for experiments were between 2 and 7 passages from thawing.
Vectors
The following retroviral vectors were gifts from Wade Harper: Flag-HA-GFP (Addgene # 22612), Flag-HA-USP13 (Addgene # 22568), Flag-HA-UCHL1 (Addgene # 22563) (18). Full length human USP11 was amplified from a cDNA library and cloned into a retroviral pDEST-Flag-HA vector. Catalytically inactive USP11 was generated by site-directed mutagenesis. shRNA hairpin sequences targeting firefly luciferase or USP11 were cloned into pINDUCER10 (miR-RUP) (19). Stable expression of DUBs and shRNAs was achieved by retroviral infection for 5–7 hours and selection with 2 µg/ml puromycin 24–48 hours later. Retroviruses were produced with HEK293T cells as previously described (8). CAGA12-firefly luciferase reporter was a gift from Peter ten Dijke (Leiden University Medical Center, Netherlands) (14). pGL4.74-renilla luciferase construct was obtained from Maryan Rizk (Guatelli Lab, University of California, San Diego). See Supplementary Data for shRNA hairpin sequences.
RNA extraction and RT-qPCR
Total RNA was extracted using TRIzol (Thermo-Fisher Scientific) and was reverse-transcribed with High-Capacity cDNA Reverse Transcription Kit (Thermo-Fisher Scientific). The resulting cDNAs were used for RT-qPCR using SsoAdvance SYBR Green Supermix (Bio-Rad) in triplicate. RT-qPCR and data collection were performed on CFX96 Touch Real-Time PCR Detection System (Bio-Rad). All the values were normalized to an internal control GAPDH. Relative expression for each target gene was compared to that of cells expressing Ctrl or shCtrl. See Supplementary Data for primer oligonucleotide sequences.
Microarray and Kaplan-Meier analysis
Microarray data from GEO accession GSE24202 (20) were analyzed with IPA (Qiagen). Briefly, 56 genes associated with the ubiquitin-proteasome pathway were chosen from the list of differentially expressed genes as determined by IPA. Venn diagram analysis was performed using the list of 56 genes. Kaplan-Meier survival curves for overall survival were generated from The Cancer Genome Atlas Breast Invasive Carcinoma gene expression dataset (n = 1215). Patients were stratified based on the expression of USP11, USP13, or UCHL1. High and low expression were defined as the top and bottom ~25% of the patient distribution. Expression, overall survival, and patient tumor subtype data were downloaded from the UCSC Cancer Genomics Browser (https://genome-cancer.ucsc.edu/). Prism (GraphPad) was used to test the statistical significance between the overall survival curves using the Log-rank (Mantel-Cox) Test. Kaplan-Meier survival curves for overall survival and distant metastasis free survival were generated with the web tool KMplotter (21).
Flow cytometry analysis
Suspensions of HMLE cell lines were stained with anti-CD44 antibody (Biolegend) and analyzed by flow cytometry on a BD Accuri C6 (BD Biosciences). Suspensions of MDA-MB-231 and SUM159 cell lines were fixed and permeabilized with the fixation/permeabilization concentrate kit (eBioscience), and stained with anti-TGFBR2 antibody (R&D). Data were analyzed using FlowJo software. Cell sorting was done using a FACS Aria 2 (BD Biosciences) at the UCSD Human Embryonic Stem Cell Core Facility. For apoptosis analysis, suspensions of HMLE cells from dissociated mammospheres were resuspended in Annexin V Binding Buffer (Biolegend) and stained with Annexin V-APC (Biolegend) and 7-AAD (Biolegend) and analyzed on a BD Accuri C6.
Mammosphere assay
HMLE mammosphere culture was performed by plating 1,000–5,000 cells in Mammocult medium (Stemcell Technologies) supplemented with 4 µg/ml heparin, 0.5 µg/ml hydrocortisone, and 1% methylcellulose. For serial passage into secondary mammosphere formation, mammospheres were dissociated into single cells by trypsinization and then plated in mammosphere culture conditions. Spheres were counted 10–12 days later. T47D spheres were formed by plating 1,000–4,000 cells in DMEM/F12 media supplemented with hEGF (20 ng/ml, Sigma), bFGF (20 ng/ml, Thermo-Fisher Scientific), heparin (4 µg/ml, Sigma), 1% methylcellulose (R&D), and B27 Supplement (Thermo-Fisher Scientific). All sphere assays were performed in 24-well ultra low-attachment plates (Corning).
Western blot analysis
Cells were lysed on ice in RIPA buffer supplemented with Protease/Phosphatase Inhibitor Cocktail (Cell Signaling Technology). 30 µg of total protein from each sample was resolved on Novex 4–20% Tris-Glycine Mini Protein Gels and transferred onto nitrocellulose membranes. Blots were probed with the appropriate antibodies: anti-USP11 (Bethyl), anti-USP13 (Bethyl), anti-phospho-SMAD2 (Cell Signaling Technology), anti-total-SMAD2 (Cell Signaling Technology), anti-HA (eBioscience), anti-Fibronectin (eBioscience), anti-ZEB1 (Cell Signaling), anti-SNAI1 (Cell Signaling), or anti-β-Actin (Sigma-Aldrich). Signals were detected using fluorescent secondary antibodies compatible with Odyssey infrared imaging system (Li-Cor Biosciences). For Figure 7C and Supplementary Figure 6, all western blots images are taken from the same membrane with the intervening irrelevant samples removed for clarity.
Cell migration assays
Scratch wound assays were performed in 6-well plates. Cells were grown to ~90% confluency and wound was created with a pipette tip. Media was replenished with media containing TGFβ (2.5 ng/ml). Images were taken in three separate locations along the scratch wound at 0 and 24 hours after wound was made. Extent of wound closure was analyzed with TScratch software (22). Transwell assays were performed in 24-well polycarbonate inserts (Falcon, 8 µm pore size). Cells were serum starved overnight, then plated in transwell inserts with serum-free media and complete media in the bottom chamber. Cells in the upper part of the transwells were removed with a cotton swab; migrated cells were fixed in 4% paraformaldehyde and stained with crystal violet 0.5%. Three random fields were photographed and the number of cells was counted with ImageJ and averaged. Every experiment was repeated independently at least three times.
Cell proliferation analysis
Cell proliferation was measured using the TetraZ Cell Counting Kit (BioLegend) following manufacturer’s instructions.
Dual luciferase reporter assay
SUM159 cell lines were transfected with the TGFβ signaling reporter CAGA12-firefly luciferase and pGL4.74-renilla luciferase (Promega) constructs using Fugene HD (Promega). After 24–48 hours, cells were treated with TGFβ (0.25 ng/ml) for 6 hours. Luciferase activity was then measured using the Dual-Glo Luciferase Assay Kit (Promega).
Statistical analysis
Statistical analyses were performed with an unpaired t test, one-way ANOVA, two-way ANOVA, or Log-rank (Mantel-Cox) Test (for Kaplan-Meier survival curves) using GraphPad Software. The resulting statistics are indicated in each figure as follows: ns = not significant (P > 0.05), * = (P ≤ 0.05), ** = (P ≤ 0.01), *** = (P ≤ 0.001).
Results
DUB expression is associated with EMT and decreased survival in human breast cancer patients
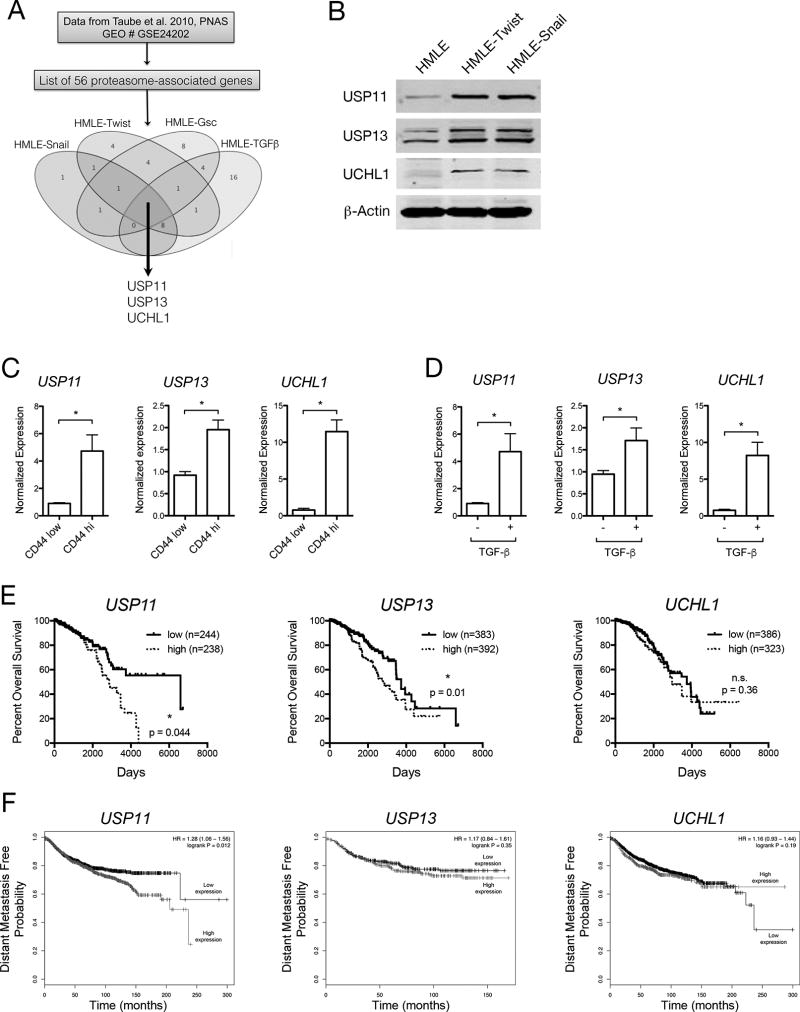
To identify novel regulators of EMT within the ubiquitin-proteasome pathway, we examined previously published microarray data. Taube et al. compared the transcriptomes of immortalized human mammary epithelial (HMLE) cells that have undergone EMT via stable overexpression of EMT-inducing factors (EMT-TFs) Twist, Snail, Gsc, or TGFβ (20). We filtered differentially expressed genes to a curated list of genes within the ubiquitin-proteasome pathway, including the 26S proteasome subunits, ubiquitin ligases, and deubiquitinases. Venn diagram analysis revealed three deubiquitinases – USP11, USP13, and UCHL1 – that were upregulated across all HMLE lines (Fig. 1A). We confirmed the upregulation of DUBs at the protein level (Fig. 1B) in HMLE-Twist and HMLE-Snail compared to the parental line. To show that DUB expression was also differentially expressed without forced overexpression of EMT-TFs, parental HMLE cells were sorted based on their level of cell surface CD44, a previously established EMT marker (23). In the ~1% of cells that were CD44high within the parental HMLE cell line, DUB expression was markedly increased compared to the rest of the population (CD44low) (Fig. 1C). USP11, USP13, and UCHL1 also appear to be a part of the TGFβ-induced EMT pathway, as their expression is upregulated upon TGFβ treatment and EMT induction (Fig. 1D). These results suggest that DUB expression is associated with the EMT, whether cells are induced with TGFβ or stable overexpression of EMT-TFs.
Figure 1. DUB expression is associated with EMT and decreased survival in human breast cancer patients.
A, Venn diagram analysis of microarray data from GEO accession GSE24202. Ubiquitin-proteasome system-associated genes were selected from the list of differentially expressed genes. B, Western blot analysis of DUB protein levels in HMLE cells with or without retroviral overexpression of EMT transcription factors. C, RT-qPCR analysis of DUB mRNA levels in HMLE cells sorted based on CD44 cell surface expression level. D, RT-qPCR analysis of DUB mRNA levels in HMLE cells treated or not with TGFβ for 7 days. E, Kaplan-meier analysis of overall survival in relation to the level of DUB gene expression in TCGA breast invasive carcinoma cases. F, Kaplan-meier analysis (KMplotter) of distant metastasis free survival in relation to DUB expression in patient tumor samples.
To establish a potential role for DUB expression in human disease progression, we examined their expression in cohorts of breast cancer patients. Using the TCGA Breast Cancer patient dataset, we found that patients with high expression of USP11 and USP13, but not UCHL1, had decreased overall survival (OS) compared to patients with low expression (Fig. 1E). Using the web tool Kmplot, we found that only patients with high expression of USP11, not USP13 or UCHL1, exhibited a decreased distant metastasis-free survival (DMFS) (Fig. 1F). These results are in line with previous work (24) and suggest that DUB expression may contribute to both disease progression and metastasis in human breast cancer patients.
To understand USP11’s role in disease progression in a more specific manner, we used the TCGA and Kmplot databases to correlate USP11 expression with clinically relevant patient subsets and tumor stages. In the TCGA Breast Cancer patient dataset, we found that high USP11 expression correlated with decreased OS in ER+ patients and patients with a Luminal A tumor subtype, but not other tumor subtypes (Supp. Fig. 1A). Using the Kmplot breast cancer database, we found that only patients a with Luminal B tumor subtype showed a correlation with decreased OS and high USP11 expression (Supp. Fig 1B). When using DMFS as the clinical outcome, Kmplot analysis showed that high USP11 expression correlated with decreased survival in patients with ER+, Luminal A, and HER2+ tumor subtypes (Supp. Fig. 1B). These survival analyses suggest that patients with tumors of epithelial nature (Luminal A/B, ER+) are more vulnerable to decreased survival and increased metastasis that is associated with high USP11 expression. Lastly, we found that USP11 expression was not significantly enriched in any tumor stage, suggesting that high USP11 expression is likely a risk factor present at tumor initiation (Supp. Fig. 1C).
USP11 and its catalytic activity are necessary for a complete acquisition of EMT characteristics in response to TGFβ
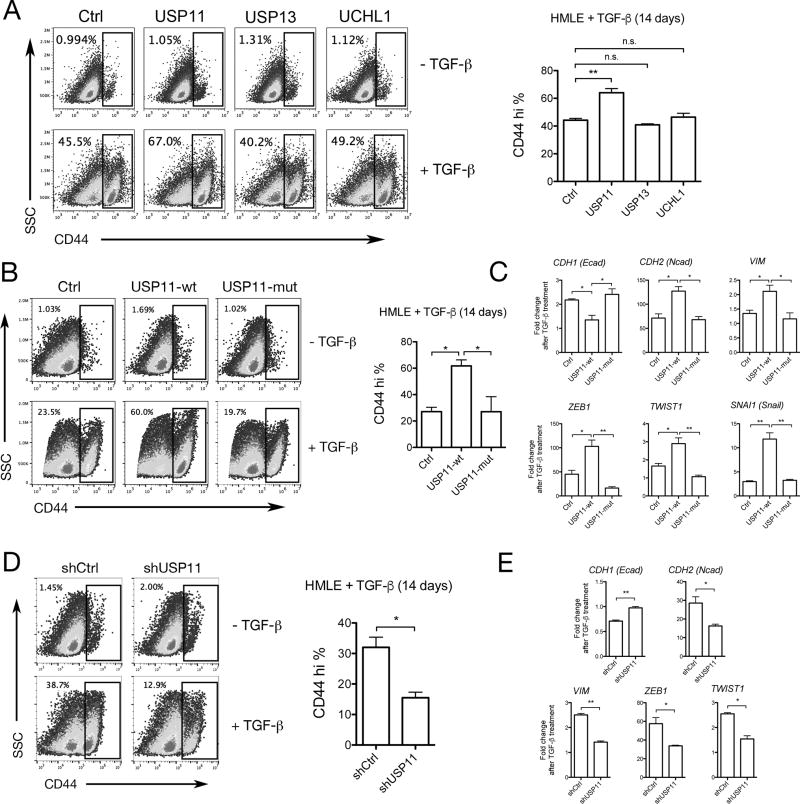
To test whether DUBs play a role in regulating EMT, USP11, USP13, and UCHL1 were retrovirally overexpressed in HMLE cells (Supp. Fig. 2A). We found that overexpression of these DUBs alone did not cause the cells to undergo EMT. However, when EMT was induced via treatment with TGFβ for 14 days, we observed that cells overexpressing USP11, but not USP13 or UCHL1, exhibited a higher percentage of CD44high cells (Fig. 2A). Based on this result, we decided to focus on the role USP11 plays in regulating TGFβ-induced EMT.
Figure 2. USP11 and its catalytic activity are necessary for a complete TGFβ-induced EMT.
A, B, D, Representative flow cytometry analysis of CD44 expression in HMLE cells treated or not with TGFβ for 14 days. (A) Retroviral overexpression of DUBs compared to GFP expressing control (Ctrl), (B) retroviral overexpression of wild type USP11 (USP11-wt) compared to C318S catalytic mutant USP11 (USP11-mut) and Ctrl, (D) retroviral expression of shRNA targeting USP11 (shUSP11) compared to a non-targeting control shRNA (shCtrl). Three independent experiments are represented in the bar graphs. C, E, RT-qPCR analysis of mRNA levels of EMT markers in HMLE cells treated or not with TGFβ for 14 days. (C) Retroviral overexpression of wild type USP11 (USP11-wt) compared to C318S catalytic mutant USP11 (USP11-mut) and Ctrl, (E) retroviral expression of shRNA targeting USP11 compared to a non-targeting control shRNA (shCtrl). Three independent experiments are represented in the bar graphs.
To determine if the effect of USP11 on EMT induction in HMLE cells was due to its deubiquitinating activity, we overexpressed either wild-type USP11 (USP11-wt) or a C318S catalytic mutant of USP11 (USP11-mut) in HMLE cells (Supp. Fig 2B). Unlike USP11-wt, expression of the C318S catalytic mutant failed to enhance EMT, based on CD44 cell surface expression (Fig. 2B) and mRNA expression of EMT markers CDH1 (E-cadherin), CDH2 (N-cadherin), VIM, ZEB1, TWIST1, and SNAI1 (Fig. 2C). These results suggest that USP11’s deubiquitinating activity is necessary to enhance TGFβ-induced EMT.
We next stably expressed USP11 shRNA in HMLE cells (HMLE-shUSP11) to determine if USP11 is necessary for EMT induction (Supp. Fig 2C). After 14 days of TGFβ treatment, HMLE-shUSP11 cells were unable to undergo EMT to the same extent as cells expressing a non-targeting control shRNA (HMLE-shCtrl). HMLE-shUSP11 cells displayed a significantly lower percentage of CD44high events (Fig. 2D) and failed to upregulate EMT markers to the same extent at HMLE-shCtrl cells (Fig. 2E). Together, these data suggest that while USP11 alone does not induce EMT, it is nonetheless required for TGFβ-induced EMT and is dependent on its catalytic activity to do so.
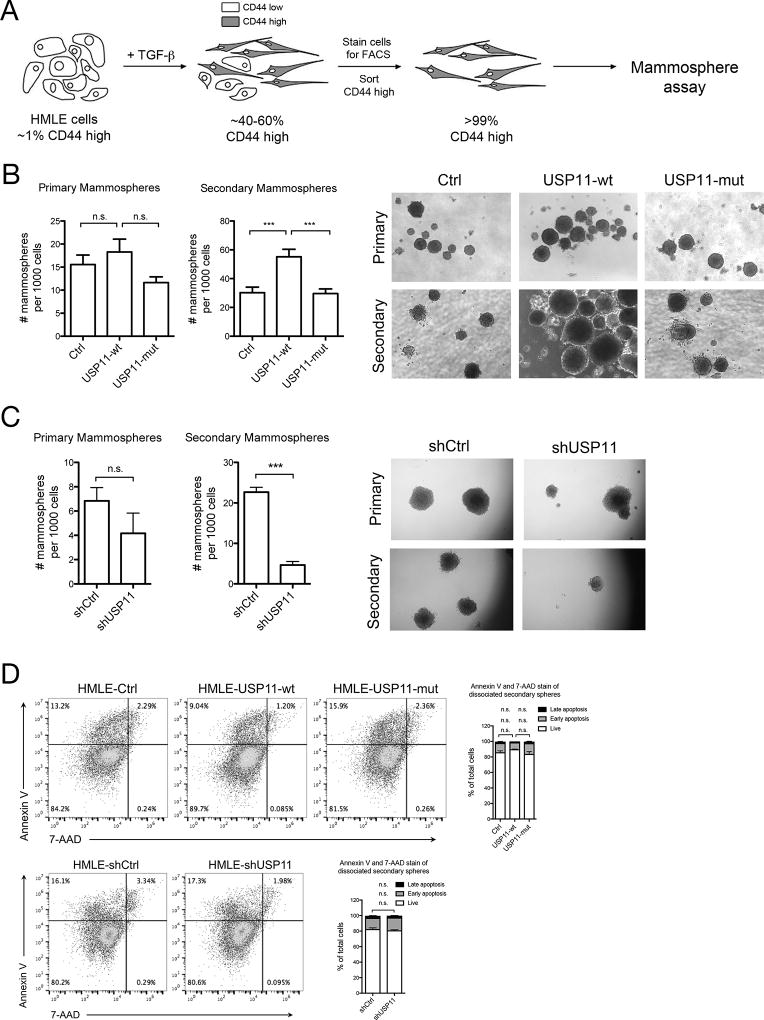
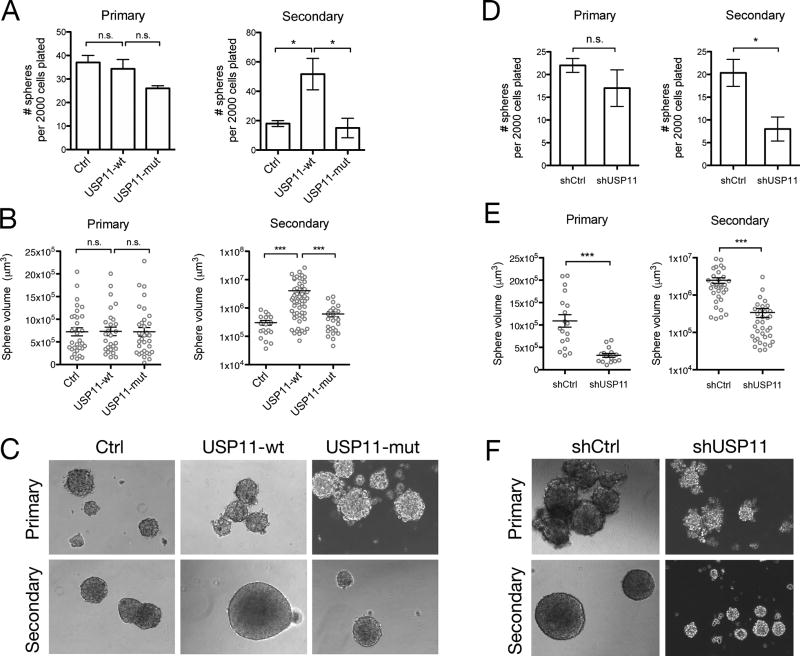
Cells that have undergone EMT have been shown to possess stem-like characteristics, namely growth in suspension and self-renewal in the mammosphere formation assay (23). We induced EMT with TGFβ treatment for 14 days in the USP11 overexpression and USP11 shRNA HMLE cell lines and then plated them in the mammosphere formation assay (Supp. Fig. 3A). We found that overexpression of USP11-wt, but not USP11-mut, increased the number of spheres formed compared to control cells (Supp. Fig. 3B), while shRNA knockdown of USP11 reduced the number of spheres formed compared to shCtrl cells (Supp. Fig. 3C). This result was expected since the percentage of CD44high cells correlates with the extent of sphere formation, and that CD44low cells do not possess mammosphere-forming capacity (23). To determine the role of USP11 in self-renewal, we first sorted cells based on CD44 expression, and then plated them in the sphere assay (Fig. 3A). Neither USP11 overexpression nor USP11 shRNA knockdown affected the number of primary spheres formed when only CD44high cells were plated suggesting that USP11 does not alter the number of stem-like cells in the CD44hi population (Fig. 3B, 3C). However, upon serial passage, USP11-wt, but not USP11-mut cells formed more secondary spheres when compared to control cells (Fig. 3B). Conversely, serial passage of shUSP11 spheres resulted in fewer secondary spheres compared to shCtrl (Fig. 3C). Notably, the differences observed in sphere formation are not due to apoptosis (Fig. 3D). These results suggest that USP11 and its deubiquitinating activity are necessary for the self-renewal capacity of human mammary epithelial cells. This is likely due to USP11’s effect on the stem-like characteristics of the daughter progenitor cells formed in primary spheres that are then passaged on to the secondary sphere formation experiment. It appears that an induction of EMT also induces heritable self-renewal characteristics that continues in subsequent passages of mammospheres, and the extent of this phenomenon is affected by USP11 expression, despite the lack of EMT stimulus (TGFβ).
Figure 3. USP11 regulates self-renewal in normal human epithelial cells.
A, HMLE cells were treated with TGFβ for 14 days, and then stained for cell surface CD44. CD44high cells were sorted by FACS and plated in the mammosphere assay. B, C, Primary mammosphere formation was quantified after 10–12 days. Primary spheres were dissociated and replated in the secondary mammosphere assay. Secondary sphere formation was quantified after 10–12 days. Three independent experiments are represented in the bar graphs. Representative images of spheres are shown. D, Secondary mammospheres were dissociated and single cells were stained for apoptosis markers (Annexin V and 7-AAD) and analyzed by flow cytometry. Live cells (Annexin V−/7-AAD−, lower left quadrant), early apoptotic cells (Annexin V+/7-AAD−, upper left quadrant), and late apoptotic cells (Annexin V+/7-AAD+, upper right quadrant) were quantified as a percentage of total cells. Two independent experiments are represented in the bar graphs.
USP11 and its catalytic activity influence human breast cancer cell migration and self-renewal in vitro
Since we found that USP11 is upregulated in a panel of human breast cancer cell lines compared to normal mammary epithelial cells (HMLE) (Supp. Fig. 4), we next sought to determine the effects of modulating USP11 expression on the behavior of human breast cancer cells. We chose two human breast cancer cell lines, MDA-MB-231 and SUM159, which exhibited the highest and lowest extent of USP11 expression, respectively (Supp. Fig. 4). We performed shRNA knockdown experiments in MDA-MB-231 cells and retroviral overexpression experiments in SUM159 cells and (Supp. Fig. 2D, 2E).
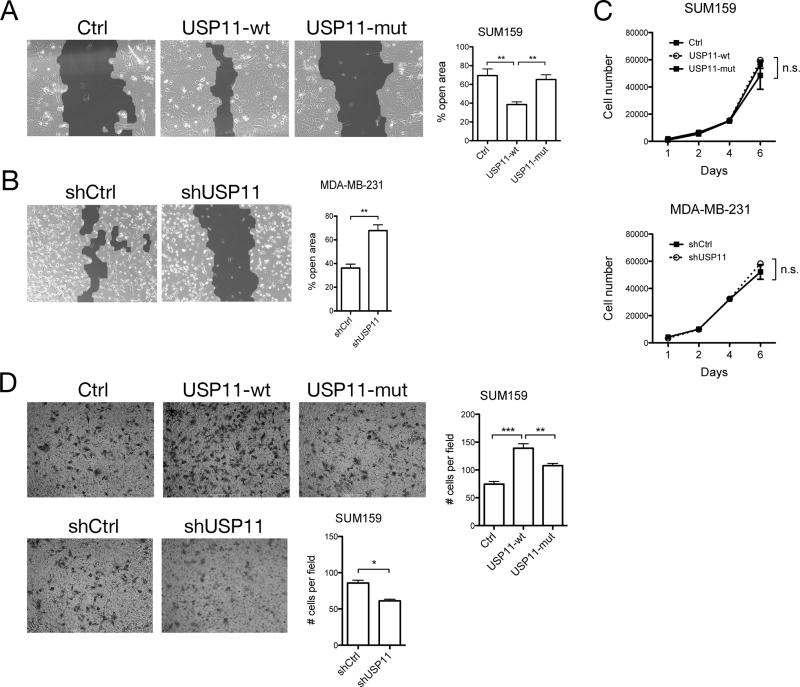
To determine the involvement of USP11 on the in vitro behavior of human breast cancer cells, we performed scratch wound and transwell cell migration assays. In a scratch wound closure assay, overexpression of USP11-wt, but not USP11-mut, in SUM159 cells resulted in significantly faster wound closure when compared to control cells (Fig. 4A). USP11 depletion caused MDA-MB-231 cells to close the wound significantly slower when compared to control cells, as evidenced by a significantly larger open area 24 hours after the scratch wound was made (Fig. 4B). The differences seen in wound closure did not appear to be due to proliferative differences, since proliferation was not significantly different across cell lines (Fig. 4C). In a transwell migration assay, overexpression of USP11-wt, but not USP11-mut, caused more SUM159 cells to migrate through the transwell membrane (Fig. 4D). Conversely, shRNA knockdown of USP11 in SUM159 cells caused fewer cells to migrate through the transwell membrane (Fig. 4D). These results suggest that deubiquitination by USP11 regulates collective cell migration in response to a wound (scratch wound assay) and single cell migration in response to a chemoattractant (transwell assay).
Figure 4. USP11 regulates human breast cancer cell migration, but not proliferation.
A, B, Scratch wound away was performed with (A) SUM159 cells overexpressing GFP (Ctrl), USP11-wt, or USP11-mut and (B) MDA-MB-231 cells expressing shCtrl or shUSP11, in media containing TGFβ. Percent open area was quantified by measuring the surface area of the scratch wound at 0 and 24 hours after scratch wound was made. Shown are representative images of the scratch wound 24 hours after scratch was made. Light gray area indicates area covered by cells, while dark gray area indicates the scratch wound. Three independent experiments are represented in the bar graph. C, Cell proliferation assay with SUM159 and MDA-MB-231 cells. D, Transwell migration assay in SUM159 cells. For each experiment, five random microscope fields were photographed and the number of cells per field was quantified using ImageJ. Two independent experiments are represented in the bar graphs.
In light of our observation that USP11 regulates self-renewal in normal mammary epithelial cells (Fig. 3), we next sought to determine if USP11 also regulates self-renewal in human breast cancer cells. Neither USP11 overexpression nor shRNA knockdown affected primary sphere formation in T47D human breast cancer cells (Fig. 5A, 5D). However, upon serial passage, overexpression of USP11-wt, but not USP11-mut, increased the number of secondary spheres compared to control cells (Fig. 5A). Conversely, USP11 depletion in T47D cells decreased the number of secondary spheres formed compared to shCtrl cells (Fig. 5D). Notably, sphere volume was significantly larger in USP11-wt cells compared to control and USP11-mut cells at the secondary passage, but not the primary passage (Fig. 5B, 5C). We also observed smaller spheres at both primary and secondary sphere passages in T47D cells lacking USP11 compared to shCtrl cells (Fig. 5E, 5F). Together, these results show that while USP11 may not affect primary sphere formation, it does regulate secondary sphere formation at subsequent passages suggesting a role in self-renewal. Strikingly, USP11 also affects sphere size, suggesting a role in the proliferative capacity of progenitor cells.
Figure 5. USP11 regulates human breast cancer self-renewal.
A, B, C, Primary and secondary sphere formation of T47D cells overexpressing GFP (Ctrl), USP11-wt, or USP11-mut. Sphere number (A) and sphere volume (B) were quantified. Representative sphere images are shown (C). D, E, F, Primary and secondary sphere formation of T47D cells expressing shCtrl or shUSP11. Sphere number (D) and sphere volume (E) were quantified. Representative sphere images are shown (F).
The extent of metastasis of human breast cancer xenografts is dependent on USP11 expression
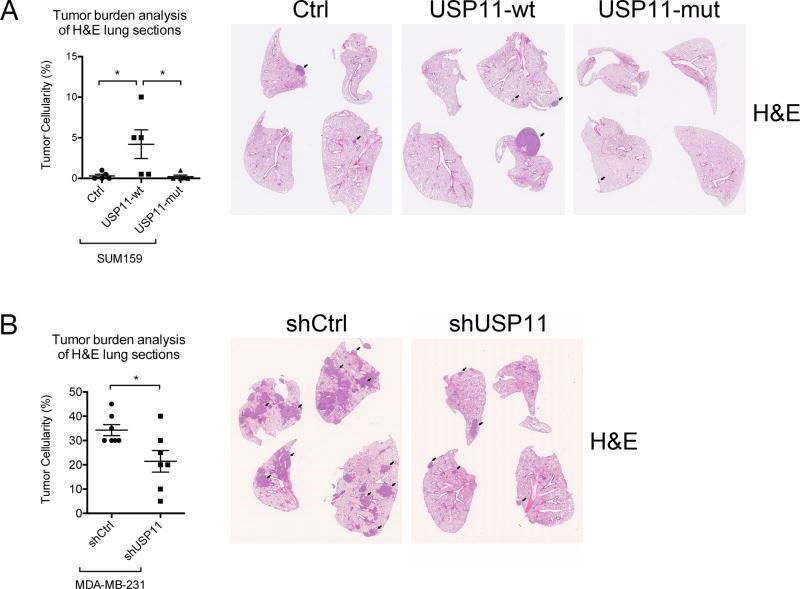
On the basis of the results from our in vitro experiments described above, we hypothesized that modulating USP11 expression in human breast cancer cell lines would affect their behavior in vivo. To directly test this hypothesis, we used an experimental tail vein metastasis assay. Indeed, we found that SUM159 cells overexpressing USP11-wt, but not USP11-mut, colonized lung tissue as metastases 10-fold more than control cells (Fig. 6A). Alternatively, when USP11 expression was knocked down with stable shRNA expression, MDA-MB-231 cells colonized lung tissue approximately 40% less than control cells (Fig. 6B). These results suggest that the effect of USP11 expression on in vitro behaviors of cancer cell lines can be recapitulated with a more stringent in vivo test of metastatic capacity.
Figure 6. USP11 regulates experimental metastasis of human breast cancer cells in mice.
A, B, Either 1.5×106 SUM159 cells (A) or 1×106 MDA-MB-231 cells (B) were resuspended in PBS and injected via tail vein into NSG mice at a volume of 100 µl. After 6–8 weeks, mice were sacrificed and lungs were analyzed for metastatic colonization by fixation, paraffin embedding. H&E-stained sections were examined for percentage of tumor cells amongst normal cells (tumor cellularity). Each data point represents the average tumor cellularity of three step sections (100 µm apart) from one mouse. The black arrows indicate examples of metastatic tumors within lung tissue.
USP11 regulates the stability of TGFBR2 and downstream TGFβ signaling in human breast cancer cells
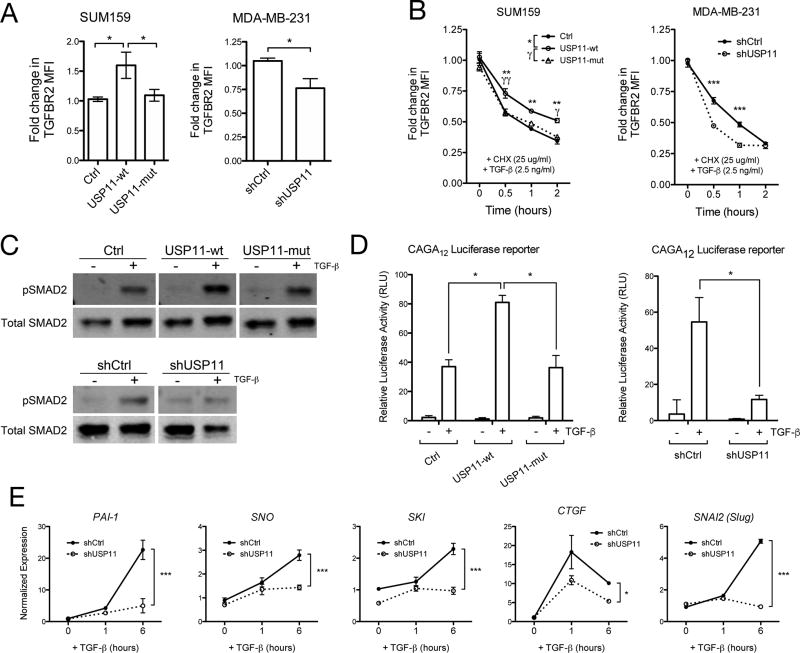
Given the known relationship between TGFβ receptors and DUBs (14,16,25,26), and that the USP11-dependent regulation of transwell migration we observed is TGFβ-dependent (Supp. Fig. 5), we hypothesized that USP11 was affecting TGFβ receptor stability in human breast cancer cells. To determine the mechanism of USP11’s effects on cancer cell behavior, we measured the level of TGFβ type II receptor (TGFBR2) using flow cytometry. Overexpression of USP11-wt, but not USP11-mut, caused a 50% increase in TGFBR2 levels in SUM159 cells maintained in normal culture conditions (Fig. 7A). Conversely, USP11 depletion caused a 25% decrease in TGFBR2 levels in MDA-MB-231 cells maintained in normal culture conditions (Fig. 7A). To investigate the stability and degradation of TGFBR2 in cancer cell lines, we performed a cycloheximide timecourse with simultaneous TGFβ treatment in cells that were serum-starved overnight prior to treatment. While TGFBR2 was degraded over time in all cell lines, TGFBR2 exhibited increased stability in cells overexpressing USP11-wt, but not USP11-mut (Fig. 7B). Conversely, TGFBR2 was degraded more quickly in cells lacking USP11 (Fig. 7B). These results show that USP11 regulates the stability of TGFBR2 during TGFβ stimulation.
To examine how USP11 affects TGFβ signaling, we performed western blots to measure the level of TGFβ pathway activation. We found that USP11 modulates TGFβ signaling over short timescales in human breast cancer cells, as differences in phospho-SMAD2 levels were evident following 30 minutes of TGFβ treatment (Fig. 7C). TGFβ transcriptional activity is also affected by USP11, as the CAGA12 SMAD-dependent luciferase reporter (14) was differentially activated when USP11 expression was altered in SUM159 cells (Fig. 7D). Finally, we found that downstream targets of TGFβ are affected by USP11, as USP11 depletion in MDA-MB-231 cells prevented the expression of PAI-1, SNO, SKI, CTGF, and SNAI2 to the same extent as control cells (Fig. 7E). These effects are recapitulated in the induction of EMT markers by TGFβ at the protein level in SUM159 and MDA-MB-231 cells (Supp. Fig. 6). Together, these results confirm that USP11 regulates TGFβ signaling and is essential for TGFβ-dependent responses in human breast cancer cells.
Figure 7. USP11 regulates TGFBR2 stability and signaling in human breast cancer cells.
A, Flow cytometry analysis of TGFBR2 median fluorescence intensity (MFI) in SUM159 cells overexpressing GFP (Ctrl), USP11-wt, or USP11-mut and MDA-MB-231 cells expressing shCtrl or shUSP11. B, Flow cytometry analysis of TGFBR2 MFI over time after CHX and TGFβ treatment in SUM159 and MDA-MB-231 cells. C, Western blot analysis of phosho-SMAD2 (pSMAD2) in SUM159 cells overexpressing GFP (Ctrl), USP11-wt, or USP11-mut and MDA-MB-231 cells expressing shCtrl or shUSP11 before and after TGFβ treatment for 30 minutes. All blot images are from the same membrane with intervening irrelevant samples removed for clarity. D, Effect of USP11-wt or USP11-mut overexpression and shRNA knockdown on CAGA12-luciferase transcriptional response induced by TGFβ in SUM159 cells. E, RT-qPCR analysis of mRNA levels of TGFβ target genes in MDA-MB-231 cells expressing shCtrl or shUSP11.
Discussion
While EMT has been studied extensively and clearly plays a role in metastasis, novel druggable regulators of EMT have yet to be uncovered and introduced into the clinic. Since the majority of cancer mortalities are due to metastasis, anti-EMT drugs are an important deficiency in the current arsenal of cancer therapies. In this study, we confirm the role of USP11 in EMT using a robust EMT cell culture model. Furthermore, we establish for the first time USP11’s importance in human breast cancer cell migration, self-renewal, and metastasis.
In search of novel regulators of EMT within the ubiquitin-proteasome pathway, we mined previously published microarray data and found USP11, USP13, and UCHL1 to be upregulated during EMT. Using human mammary epithelial cells (HMLE), a robust in vitro model for EMT, we show that overexpression of these DUBs alone does not change their phenotype. However, upon EMT induction with TGFβ, only USP11 overexpression enhanced the EMT phenotype. Further, modulation of USP11 expression affects the development of mesenchymal characteristics in HMLE cells, namely mesenchymal marker expression and self-renewal ability. These experiments are in line with previous work that established a role for USP11, but not USP13 or UCHL1, in TGFβ signaling and regulation of TGFBR1 and TGFBR2 stability (16,26). While these studies use mouse or non-mammary cell lines, the current study provides for the first time a thorough phenotypic and functional characterization using a human model of EMT in cells derived from the mammary epithelium.
Our study is also the first to show the dependence of human breast cancer cells on USP11 for cell migration, self-renewal, and metastasis. We showed that shRNA knockdown of USP11 slowed scratch wound closure and decreased transwell migration, while the opposite effect was shown in a previous study in 786-O cells, a human renal cell adenocarcinoma cell line (27). In that same study, shRNA knockdown of USP11 in 786-O cells increased cell proliferation, which contrasts with our observation that overexpression or shRNA knockdown of USP11 in SUM159 and MDA-MB-231 cells, respectively, had no effect on cell proliferation in a 2D growth assay. However, we did find that USP11 knockdown resulted in smaller T47D spheres at the secondary passage, raising the possibility that USP11 may regulate cell proliferation. Moreover, the tail vein metastasis assay, which tends to assess parameters related to survival and outgrowth, demonstrated alterations in lung colonization with USP11 overexpression or knockdown. Thus, it remains unclear whether USP11 directly regulates cell proliferation, but it appears that USP11’s role may be context-specific with different targets for deubiquitination in different cell and tissue types.
In terms of self-renewal, modulating USP11 expression had a significant effect in HMLE normal epithelial cells and T47D human breast cancer cells. In general, we found that USP11 is required for proper sphere formation and sufficient to increase sphere formation, specifically at the secondary passage and independent of apoptosis. These results suggest that USP11 is responsible for the maintenance of stem cell characteristics in stem-like human breast cancer cells. Unexpectedly, we see USP11-dependent effects on sphere size in T47D cells. Sphere size is not well studied, but it is thought that sphere size indicates the nature of the founder clone cell as being more stem-like in terms of being able to produce more progenitor cells (28). Our results show for the first time USP11’s role in regulating stem-like behavior of human breast cancer cells in vitro. While sphere number and size are not necessarily a definitive readout of the in vivo presence of cancer stem cells, in vivo models of metastasis are rigorous tests of these characteristics. Our study shows that, in this case, the in vivo experimental metastasis model does faithfully recapitulate the in vitro behaviors of human breast cancer cells.
Tumor-intrinsic subtype analysis of patient survival outcomes provided information about which patient subpopulations are more at risk for USP11-associated disease progression and metastasis. In these analyses, we found high USP11 expression in Luminal-type tumors (Luminal A/B, ER+), but not Basal-like tumors, was associated with decreased survival and metastasis. Luminal-type tumors tend to have more epithelial characteristics, and we speculate that those tumor types are more sensitive to EMT-inducing signals, especially those enhanced by USP11. Since Basal-like tumors have already adopted a mesenchymal phenotype, USP11 may not be predictive of survival in those patients. By contrast, we observed an effect of USP11 in regulating the behavior of Basal-like human breast cancer cells in our xenograft studies. This apparent lack of correlation represents a limitation of the study, but raises the possibility that characteristics beyond tumor subtype may influence whether or not USP11 can influence growth, survival, and metastasis.
Mechanistically, we show that USP11 enhances TGFBR2 stability, thereby enhancing TGFβ signaling and metastasis. USP11 has been shown to target both TGFBR1 and TGFBR2 for deubiquitination (16,26) and our study confirms this pathway for the first time in human breast cancer cells. It has not escaped our notice that USP11 likely deubiquitinates TGFBR1 in human breast cancer cells in order to affect EMT and metastasis. Since TGFβ has been linked to EMT and metastasis-promoting functions, it is likely that USP11 regulates the metastatic cascade at multiple points (invasion, intravasation, extravasation, colonization, any step that depends on TGFβ signaling). Thus, our current study suggests a role for USP11 in the complex process of metastasis and implicates USP11 as a potential therapeutic target in breast cancer
Supplementary Material
Implications.
Implications: USP11 regulates TGFβ-induced epithelial-mesenchymal plasticity and human breast cancer metastasis and may be a potential therapeutic target for breast cancer.
Acknowledgments
Supported by NIH grants (DK093507 and OD008469 to J.T.C. and CA168689, CA174869, CA206880 to J.Y.). J.T.C. is a Howard Hughes Medical Institute Physician-Scientist Early Career Awardee and a V Foundation for Cancer Research V Scholar Awardee. D.A.G. is a Howard Hughes Medical Institute Gilliam Fellow. The Tissue Technology Shared Resource is supported by a National Cancer Institute Cancer Center Support Grant (CCSG Grant CA23100). We thank Dwayne Stupack (UC San Diego), Gen-Sheng Feng (UC San Diego), and the members of the Chang and Bui laboratory for helpful discussions.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–64. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 2.Weigelt B, Peterse JL, van Veer LJ. Breast cancer metastasis: markers and models. Nature reviews Cancer. 2005;5:591–602. doi: 10.1038/nrc1670. [DOI] [PubMed] [Google Scholar]
- 3.Yeung KT, Yang J. Epithelial-mesenchymal transition in tumor metastasis. Molecular oncology. 2017;11:28–39. doi: 10.1002/1878-0261.12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Padua D, Massagué J. Roles of TGFβ in metastasis. Cell Research. 2008;19:89–102. doi: 10.1038/cr.2008.316. [DOI] [PubMed] [Google Scholar]
- 5.Bhola NE, Balko JM, Dugger TC, Kuba M, Sánchez V, Sanders M, Stanford J, Cook RS, Arteaga CL. TGF-β inhibition enhances chemotherapy action against triple-negative breast cancer. Journal of Clinical Investigation. 2013;123:1348–58. doi: 10.1172/JCI65416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katsuno Y, Lamouille S, Derynck R. TGF-β signaling and epithelial-mesenchymal transition in cancer progression. Current opinion in oncology. 2013;25:76–84. doi: 10.1097/CCO.0b013e32835b6371. [DOI] [PubMed] [Google Scholar]
- 7.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–96. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banno A, Garcia DA, van Baarsel ED, Metz PJ, Fisch K, Widjaja CE, Kim SH, Lopez J, Chang AN, Geurink PP, Florea BI, Overkleeft HS, Ovaa H, Bui JD, Yang J, Chang JT. Downregulation of 26S proteasome catalytic activity promotes epithelial-mesenchymal transition. Oncotarget. 2016;7:21527–41. doi: 10.18632/oncotarget.7596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Voutsadakis IA. Proteasome expression and activity in cancer and cancer stem cells. Tumor Biology. 2017;39 doi: 10.1177/1010428317692248. [DOI] [PubMed] [Google Scholar]
- 10.Eichhorn PJA, Rodón L, Gonzàlez-Juncà A, Dirac A, Gili M, Martínez-Sáez E, Aura C, Barba I, Peg V, Prat A, Cuartas I, Jimenez J, García-Dorado D, Sahuquillo J, Bernards R, Baselga J, Seoane J. USP15 stabilizes TGF-β receptor I and promotes oncogenesis through the activation of TGF-β signaling in glioblastoma. Nature Medicine. 2012;18:429–35. doi: 10.1038/nm.2619. [DOI] [PubMed] [Google Scholar]
- 11.Wu Y, Wang Y, Lin Y, Liu Y, Wang Y, Jia J, Singh P, Chi Y-II, Wang C, Dong C, Li W, Tao M, Napier D, Shi Q, Deng J, Evers BM, Zhou BP. Dub3 inhibition suppresses breast cancer invasion and metastasis by promoting Snail1 degradation. Nature communications. 2017;8:14228. doi: 10.1038/ncomms14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu Y, Wang Y, Yang XH, Kang T, Zhao Y, Wang C, Evers BM, Zhou BP. The deubiquitinase USP28 stabilizes LSD1 and confers stem-cell-like traits to breast cancer cells. Cell reports. 2013;5:224–36. doi: 10.1016/j.celrep.2013.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J, Zhang P, Wei Y, Piao H-LL, Wang W, Maddika S, Wang M, Chen D, Sun Y, Hung M-CC, Chen J, Ma L. Deubiquitylation and stabilization of PTEN by USP13. Nature cell biology. 2013;15:1486–94. doi: 10.1038/ncb2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L, Zhou F, Drabsch Y, Gao R, Snaar-Jagalska EB, Mickanin C, Huang H, Sheppard K-A, Porter JA, Lu CX, ten Dijke P. USP4 is regulated by AKT phosphorylation and directly deubiquitylates TGF-β type I receptor. Nature Cell Biology. 2012;14:717–26. doi: 10.1038/ncb2522. [DOI] [PubMed] [Google Scholar]
- 15.Zhou Z, Zhang P, Hu X, Kim J, Yao F, Xiao Z, Zeng L, Chang L, Sun Y, Ma L. USP51 promotes deubiquitination and stabilization of ZEB1. American journal of cancer research. 2017;7:2020–31. [PMC free article] [PubMed] [Google Scholar]
- 16.Jacko AM, Nan L, Li S, Tan J, Zhao J, Kass DJ, Zhao Y. De-ubiquitinating enzyme, USP11, promotes transforming growth factor β-1 signaling through stabilization of transforming growth factor β receptor II. Cell death & disease. 2016;7 doi: 10.1038/cddis.2016.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang H, Meng F, Liu G, Zhang B, Zhu J, Wu F, Ethier SP, Miller F, Wu G. Forkhead transcription factor foxq1 promotes epithelial-mesenchymal transition and breast cancer metastasis. Cancer Res. 2011;71:1292–301. doi: 10.1158/0008-5472.CAN-10-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meerbrey KL, Hu G, Kessler JD, Roarty K, Li MZ, Fang JE, Herschkowitz JI, Burrows AE, Ciccia A, Sun T, Schmitt EM, Bernardi RJ, Fu X, Bland CS, Cooper TA, Schiff R, Rosen JM, Westbrook TF, Elledge SJ. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3665–70. doi: 10.1073/pnas.1019736108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, Yang J, Hartwell K, Onder TT, Gupta PB, Evans KW, Hollier BG, Ram PT, Lander ES, Rosen JM, Weinberg RA, Mani SA. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:15449–54. doi: 10.1073/pnas.1004900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Györffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Research and Treatment. 2010;123:725–31. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 22.Geback T, Schulz MM, Koumoutsakos P, Detmar M. TScratch: a novel and simple software tool for automated analysis of monolayer wound healing assays. Biotechniques. 2009;46:265–74. doi: 10.2144/000113083. [DOI] [PubMed] [Google Scholar]
- 23.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–15. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bayraktar S, Gutierrez Barrera AM, Liu D, Pusztai L, Litton J, Valero V, Hunt K, Hortobagyi GN, Wu Y, Symmans F, Arun B. USP-11 as a predictive and prognostic factor following neoadjuvant therapy in women with breast cancer. Cancer J. 2013;19:10–7. doi: 10.1097/PPO.0b013e3182801b3a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aggarwal K, Massagué J. Ubiquitin removal in the TGF-β pathway. Nature Cell Biology. 2012;14:656–7. doi: 10.1038/ncb2534. [DOI] [PubMed] [Google Scholar]
- 26.Al-Salihi MA, Herhaus L, Macartney T, Sapkota GP. USP11 augments TGFβ signalling by deubiquitylating ALK5. Open biology. 2012;2:120063. doi: 10.1098/rsob.120063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang E, Shen B, Mu X, Qin Y, Zhang F, Liu Y, Xiao J, Zhang P, Wang C, Tan M, Fan Y. Ubiquitin-specific protease 11 (USP11) functions as a tumor suppressor through deubiquitinating and stabilizing VGLL4 protein. American journal of cancer research. 2016;6:2901–9. [PMC free article] [PubMed] [Google Scholar]
- 28.Pastrana E, Silva-Vargas V, Doetsch F. Eyes wide open: a critical review of sphere-formation as an assay for stem cells. Cell stem cell. 2011;8:486–981. doi: 10.1016/j.stem.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.