Abstract
Objective
To characterize otitis media–associated structures affixed to the mucosal surface of the tympanic membrane (TM) in vivo and in surgically recovered in vitro samples.
Study Design
Prospective case series without comparison.
Setting
Outpatient surgical care center.
Subjects and Methods
Forty pediatric subjects scheduled for tympanostomy tube placement surgery were imaged intraoperatively under general anesthesia. Postmyringotomy, a portable optical coherence tomography (OCT) imaging system assessed for the presence of any biofilm affixed to the mucosal surface of the TM. Samples of suspected microbial infection–related structures were collected through the myringotomy incision. The sampled site was subsequently reimaged with OCT to confirm collection from the original image site on the TM. In vitro analysis based on confocal laser scanning microscope (CLSM) images of fluorescence in situ hybridization–tagged samples and polymerase chain reaction (PCR) provided microbiological characterization and verification of biofilm activity.
Results
OCT imaging was achieved for 38 of 40 subjects (95%). Images from 38 of 38 (100%) of subjects observed with OCT showed the presence of additional microbial infection–related structures. Thirty-four samples were collected from these 38 subjects. CLSM images provided evidence of clustered bacteria in 32 of 33 (97%) of samples. PCR detected the presence of active bacterial DNA signatures in 20 of 31 (65%) of samples.
Conclusion
PCR and CLSM analysis of fluorescence in situ hybridization–stained samples validates the presence of active bacteria that have formed into a middle ear biofilm that extends across the mucosal layer of the TM. OCT can rapidly and noninvasively identify middle ear biofilms in subjects with severe and persistent cases of otitis media.
Keywords: otitis media, biofilm, bacteria, middle ear, tympanic membrane, fluorescence in situ hybridization, PCR, optical coherence tomography
Otitis media (OM) occurs in >80% of children before the age of 2 years,1 with severe or persistent cases of OM—including recurrent acute OM (RAOM) and chronic OM with effusion (COME)—having an impact on speech, language, and learning development. With a high prevalence among children, repeated medical visits, and surgical intervention for severe cases, the overall treatment of OM entails significant costs.2,3 Once specific criteria are met,4 children with COME or RAOM (with effusion) are often treated with the surgical placement of tympanostomy tubes (TTs) into the tympanic membrane (TM)5,6 to maintain an aerated middle ear space and to help restore normal hearing.
Biofilms are a source of recurrent and persistent infection,7 especially in the respiratory tract,8,9 and mounting evidence indicates that RAOM is a biofilm-associated infec-tion.10–15 Biofilms are collections of bacteria encapsulated in a self-generated matrix of extracellular polymeric substance, which provides a protective microenvironment where bacteria can develop increased resistance to host defense mechanisms16,17 and antibiotic treatments.18 Studies have characterized biofilms on the middle ear mucosa (MEM) in vivo in animals19 and on the MEM of pediatric subjects with RAOM.10 Currently, there is a lack of practical noninvasive diagnostic techniques to determine biofilm presence and provide quantitative metrics for evidence-based treatment decisions. Optical coherence tomography (OCT) is one possible technology that can identify middle ear biofilms in patients with OM. OCT is a noninvasive medical imaging technology20 similar to ultrasound imaging, which detects reflections of light rather than sound. OCT provides real-time micron-scale cross-sectional images of the TM and adjacent middle ear cavity (MEC) with low-power near-infrared light.
Through numerous previous clinical OCT studies,20–27 our group has imaged subjects receiving treatment for acute OM, RAOM, and COME, as compared with control (healthy) subjects. Additional microbial infection–related structures thought to be middle ear biofilms affixed to the MEM of the TM have been identified with OCT in patients with RAOM and COME. Normative OCT image–based features from a normal ear and in RAOM are provided in Figure 1.
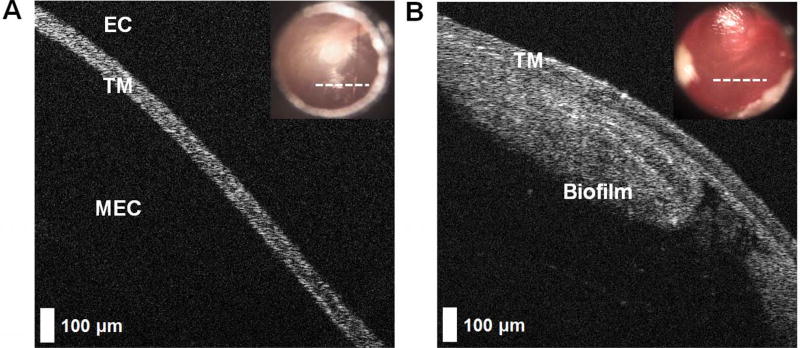
Figure 1.
Optical coherence tomography images demonstrating optical and microstructural differences of a normal ear and one with recurrent acute otitis media. (A) In cross section, a normal tympanic membrane (TM) is a thin, highly scattering ribbon of tissue approximately 100 µm thick. Near the light reflex, no other structures (eg, ossicles) appear in the middle ear cavity (MEC) behind the TM, and no signal is observed from the air-filled ear canal (EC). (B) This is in contrast to the TM from a subject with eustachian tube dysfunction and recurrent acute otitis media. A microbial infection–related structure is found adhered to the medial mucosal surface of the TM and within the MEC, having a thickness of ~350 µm. Digital otoscopy images are inset in each panel. White dashed lines indicate the physical location on the TM where the optical coherence tomography scan was taken.
Past studies based on our OCT systems with handheld probes identified and characterized infection states in vivo24,25,28,29 and the physical and functional properties of the TM with pneumatic-enabled OCT.26 In a recent OCT study,27 longitudinal effects of TT surgery were associated with elimination of biofilms from the TM. However, no validation or biological characterization of these OCT-observed biofilms has been performed to date.
In this work, we imaged, identified, and characterized suspected middle ear biofilms in vivo with intraoperative OCT and in vitro with polymerase chain reaction (PCR) and confocal laser scanning microscopy (CLSM) images of fluorescence in situ hybridization (FISH)–tagged surgically recovered samples. This study determined that structures adhered to the TM in subjects with severe and persistent OM and observed with OCT are consistent with a middle ear biofilm. Furthermore, this validates the feasibility of OCT to rapidly and noninvasively assess the TM and middle ear for the presence of biofilms.
Methods
In this study, 40 pediatric subjects previously diagnosed with RAOM and/or COME and scheduled for surgery (myringotomy and TT placement) were recruited from Urbana-Champaign, Illinois, receiving care in the Department of Otolaryngology at Carle Foundation Hospital. All subjects provided informed consent and assent in accordance with protocols approved by the Institutional Review Boards of Carle Foundation Hospital and the University of Illinois at Urbana-Champaign. In this study, standard-of-care treatment followed established definitions and guidelines for acute OM,1 OM with effusion,4 and RAOM.5 Subjects were diagnosed with RAOM if multiple infections occurred over at least 3 to 6 months with resolution of symptoms between episodes, alongside concerns of developmental delays and hearing loss. Subjects with COME additionally had a persistent middle ear effusion (MEE) identified for >3 months. No subjects were excluded according to ethnicity, sex, or race, or recruited per the presence or absence of any type of effusion.
Imaging and Sample Collection
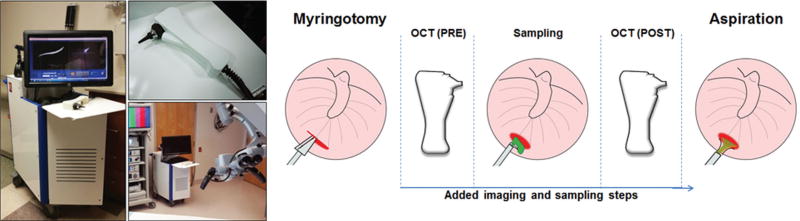
Immediately after making a surgical incision in the TM (myringotomy), a handheld OCT probe was used to assess both TMs for the presence of a middle ear biofilm. Cross-sectional OCT images, ~5 mm (transverse) × 3 mm (depth), were acquired at 30 frames per second, with a depth resolution of 2.4 µm in air. The imaging beam was positioned near the incision via real-time video otoscopy images from a color camera integrated in the handheld probe. Further system details are available in a prior publication.28 Any blood that obscured the TM was aspirated per standard of care. However, the MEC was not aspirated before sampling, to prevent disruption of any biofilm structure adhered to the TM. A digital video otoscope (Welch Allyn, Skaneateles Falls, New York) was used to record color surface images of each TM. A 90° gross curette was inserted through the myringotomy incision of each ear to collect samples of middle ear content from the imaging site (mucosal surface of TM). The sampled site was subsequently reimaged with OCT to confirm sample collection from the original imaging site on the TM. Multiple stacks of 40 previously visualized scans were saved during pre-and postsampling time points for later analysis. All subsequent steps in the surgical procedure were performed following standard of care. Figure 2 shows the portable OCT system and handheld probe, and visually presents the imaging and sampling protocol. All OCT imaging was performed immediately postmyringotomy and pre-TT placement to avoid structural tissue deformation that may occur from the myringotomy, which would have otherwise complicated direct correlation and visualization of biofilm sampling in OCT images. No more than 5 additional minutes (on average) of surgery and anesthesia time was added when imaging each ear.
Figure 2.
(Left) Portable optical coherence tomography (OCT) system and handheld probe. For scale comparison, the system is shown in the operating theater alongside standard visualization equipment. (Right) Imaging and sampling protocol.
OCT imaging and sample collection were successful in a majority of subjects, while unsuccessful sample collection was likely due to the limited grip of the curette on the amorphous microbial structures. Collected samples were immediately placed into 4% paraformaldehyde, stored at 4°C overnight, and transferred to a 50/50 phosphate-buffered saline (PBS) and ethanol solution for longer-term storage. In vitro FISH and PCR analysis provided microbiological characterization.
OCT Image Analysis
Representative OCT images were extracted from image stacks to compare structures present on the TM at pre- and postsampling time points. With previously developed OCT image processing protocols, images were collected27 and analyzed24 by readers experienced with OCT and middle ear imaging, although there was no specific training for this study. Presampling OCT images showed the presence of a biofilm adhered to the TM. Postsampling OCT images from the same site provided evidence of biofilm sampling from the mucosal surface of the TM and were used to correlate with PCR and CLSM/FISH data. OCT image interpretation was blinded from any clinical or surgical reports, and physicians were blinded to OCT imaging results.
FISH and PCR Processing
Samples were analyzed for the 3 most common microorganisms responsible for OM30—specifically, Moraxella catarrhalis, nontypeable Haemophilus influenzae, and Streptococcus pneumoniae—in addition to a universal domain bacteria probe (EUB335) that detected all bacterial strains. Samples were rinsed of storage media in PBS and divided for PCR and FISH processing.
Half of each sample was embedded for cryosectioning. Six-micrometer sections were prepared and detected by FISH with bacterial 16s rRNA probes as described previously.10 Briefly, slides were washed sequentially with PBS, PBS:ethanol (1:1), 80% ethanol, and 100% ethanol and then treated with 10 mg/mL of lysozyme (Sigma-Aldrich) in 0.1M Tris–0.05M EDTA at 37°C for 1 hour and washed with ultrapure water. Slides were then blocked with nonspecific DNA (human Cot-1 DNA; Life Technologies, Carlsbad, California) at 37°C for 6 hours. Specimens were stained with a 16s rRNA probe mixture of universal P-Eub335 (cy3-GCTGCCTCCCGTAGGAGT) paired with P-Hinf (GCCATGATGAGCCCAAGTGG-C3-fluorecein, H influenzae) and P-Spn (Cy5-GTGATGCAAGT GCACCTT, S pneumoniae) paired with P-Mcat (TGAAAG GGGGCTTTTAGCTC-Cal-fluor orange 560, M catarrhalis). Specimens were mounted with SlowFade Gold antifade reagent with DAPI (Life Technologies) and examined with CLSM (LSM 510; Carl Zeiss, Oberkochen, Germany) and software (LSM Image Browser; Carl Zeiss).
For PCR processing, bacterial DNA was extracted from biofilm samples with a QIAamp UCP Pathogen Mini Kit (Qiagen, Hilden, Germany) per the manufacturer’s protocol. Fragments from 16S rRNA of the 3 bacteria (H influenzae, S pneumoniae, and M catarrhalis) were amplified in a 25-µL reaction with 30 to 300 ng of the isolated DNA as template. A no-template negative control and a species-specific positive control were included. The assay was performed on an MJ Mini Thermal Cycler (Bio-Rad, Hercules, California). The PCR primers and conditions used in the assay were as previously described.10
Results
Forty subjects participated in this study, which concluded without any adverse events. A brief description of the samples analyzed is provided in Table 1 . OCT imaging was performed in 38 of 40 subjects. One subject had a collapsed inaccessible ear canal, preventing proper insertion of the handheld probe speculum. In the other subject, due to delays unrelated to this study, there were concerns about overextending anesthesia time, so only sample collection was performed (no OCT imaging). Analysis of OCT images identified biofilms in 100% (38 of 38) of subjects observed. A total of 34 small (~1 mm3) biological samples were successfully collected from the interior (medial) mucosal surface of the TM. Samples were divided for analysis for CLSM (33 of 34) and PCR (31 of 34). One of the 34 samples had poor quality FISH staining; thus, no CLSM data were obtained from this sample. Three of the 34 samples were too small for analysis by CLSM and PCR processing and, as such, were analyzed only with CLSM/FISH.
Table 1.
Study Results of Middle Ear Biofilm Detection and Validation With OCT, FISH/CLSM, and PCR Analysis of Samples.
| Analysis | Samples, n |
|---|---|
| OCT | |
| Diagnosed with COME/RAOM and observed intraoperatively | 40 of 40 |
| OCT imaging achieved | 38 of 40 |
| OCT identified biofilm | 38 of 38 |
| CLSM (FISH labeled) | |
| Universal probe EUB335 | 32 of 33 |
| Universal and at least 1 probe | 28 of 33 |
| Polymicrobial population | 24 of 33 |
| Haemophilus influenzae | 19 of 24 |
| Streptococcus pneumoniae | 21 of 24 |
| Moraxella catarrhalis | 18 of 24 |
| PCR | |
| Insufficient DNA for identification | 11 of 31 |
| OM-related bacteria identified | 20 of 31 |
| H influenzae | 14 of 20 |
| S pneumoniae | 7 of 20 |
| M catarrhalis | 8 of 20 |
Abbreviations: CLSM, confocal laser scanning microscopy; COME, chronic otitis media with effusion; FISH, fluorescence in situ hybridization; OCT, optical coherence tomography; OM, otitis media; PCR, polymerase chain reaction; RAOM, recurrent acute otitis media.
Table 2 presents data related to each sample that was collected and analyzed, detailing patient history from the physician’s report, intraoperative observations from the surgical microscope, the identified presence of a biofilm with OCT, and results from FISH and PCR. Analysis of CLSM images identified active bacterial biofilms in 32 of 33 samples with the universal domain probe and in 28 of 33 samples with the universal domain probe and at least 1 other probe, while 24 of 33 contained polymicrobial populations. Of 31 samples, 20 yielded sufficient DNA for PCR analysis, although 11 of 31 samples were negative for specific genetic bacteria markers. Overall, 100% of samples (34 of 34) had bacteria positively detected by either PCR or FISH.
Table 2.
Data from All Samples Collected and Processed in This Study.a
| No. | Patient History | Surgical Findings | OCT | EUB335 | NTHi | SP | Mcat | PM | Notes | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ETD, RAOM, COME | MPE | Biofilm | + | + | Δ | + | + | |||||
| 2 | ETD, CMOM | MPE | Biofilm | + | + | Δ | |||||||
| 3 | ETD, CMOM | Bulging TM, MPE | Biofilm | + | Δ | Δ | Δ | Δ | |||||
| 4 | ETD, CMOM | Injected, dull TM, MPE | Biofilm | + | Δ | + | + | Δ | + | Δ | |||
| 5 | ETD, CMOM | Bulging, injected TM, MPE | Biofilm | + | Δ | ||||||||
| 6 | ETD, CMOM | Bulging, injected TM, ME | Biofilm | + | + | Δ | Δ | Δ | |||||
| 7 | ETD, RAOM | Retracted (mild) TM, SE | Biofilm | + | + | + | Δ | + | |||||
| 8 | ETD, CMOM | Injected, dull TM, thick MPE | Biofilm | + | + | + | + | + | PCR (NEG) | ||||
| 9 | ETD, CMOM | Injected, dull TM, thick MPE | Biofilm | + | + | Δ | + | + | |||||
| 10 | ETD, RAOM, past TT (R) | ME | Biofilm | Δ | Poor-quality FISH | ||||||||
| 11 | ETD, CMOM | Retracted (mild), injected TM, thick ME | Biofilm | + | + | + | + | No PCR | |||||
| 12 | ETD, CMOM, past TT (Ex) | Retracted (mild) TM, SE | Biofilm | + | PCR (NEG) | ||||||||
| 13 | ETD, RAOM, COME | ME | No OCT | + | + | Δ | |||||||
| 14 | ETD, CMOM | Retracted (mild), injected TM, thick ME | Biofilm | + | + | + | + | PCR (NEG) | |||||
| 15 | ETD, CMOM | Retracted (mild), injected TM, thick ME | Biofilm | + | + | Δ | + | Δ | + | Δ | + | Δ | |
| 16 | RAOM, OME, SL | ME | Biofilm | + | Δ | Δ | Δ | ||||||
| 17 | ETD, RAOM | ME | Biofilm | + | + | Δ | + | + | + | ||||
| 18 | ETD, OME, past TT (R) | ME, tonsillectomy + adenoidectomy | Biofilm | + | + | + | + | + | PCR (NEG) | ||||
| 19 | ETD, COME | ME | Biofilm | + | + | PCR (NEG) | |||||||
| 20 | Chronic ETD, COME, torticollis | ME | No OCT | + | + | + | + | No PCR | |||||
| 21 | Chronic ETD, COME, torticollis | ME | Biofilm | + | + | + | + | Δ | + | ||||
| 22 | ETD, COME, SL | ME | Biofilm | + | + | + | Δ | + | Δ | + | Δ | ||
| 23 | ETD, COME, SL | ME | Biofilm | + | + | + | Δ | + | Δ | + | Δ | ||
| 24 | ETD, RAOM | Injected, “full” TM, glue-like effusion | Biofilm | + | + | + | + | PCR (NEG) | |||||
| 25 | ETD, RAOM | Injected, “full” TM, glue-like effusion | Biofilm | + | + | Δ | + | + | |||||
| 26 | ETD, CMOM, frequent OM, (DS) | Retracted (mild) TM, thick ME | Biofilm | + | + | PCR (NEG) | |||||||
| 27 | ETD, CMOM, conductive hearing loss, SL | Thick ME | Biofilm | + | + | + | + | PCR (NEG) | |||||
| 28 | ETD, CMOM, conductive hearing loss, SL | Thick ME | Biofilm | Δ | + | + | + | ||||||
| 29 | ETD, CMOM, persistent OME | Injected, dull TM, thick ME | Biofilm | + | + | + | + | PCR (NEG) | |||||
| 30 | ETD, CMOM, persistent OME | Injected, dull TM, thick ME | Biofilm | + | + | + | + | + | PCR (NEG) | ||||
| 31 | ETD, CMOM, persistent OME | Injected, dull TM, thick ME | Biofilm | + | Δ | + | + | + | |||||
| 32 | ETD, CMOM, persistent OME | Injected, dull TM, thick ME | Biofilm | + | + | Δ | + | + | |||||
| 33 | ETD, CMOM, past TT (Ex) | Retracted (severe), thin TM, thick ME | Biofilm | + | + | + | + | + | PCR (NEG) | ||||
| 34 | ETD, CSOM, persistent OME, OSA tonsillar and adenoid hypertrophy (3+) | Thick SE | Biofilm | + | + | + | + | + | No PCR | ||||
Abbreviations: CMOM, chronic mucoid otitis media; COME, chronic otitis media with effusion; CSOM, chronic serous otitis media; DS, Down syndrome; ETD, eustachian tube dysfunction; Ex, extruded; FISH, fluorescence in situ hybridization; Mcat, Moraxella catarrhalis; ME, mucoid effusion; MPE, mucopurulent effusion; NEG, negative; NTHi, Haemophilus influenzae; OCT, optical coherence tomography; OME, otitis media with effusion; OSA, obstructive sleep apnea; PCR, polymerase chain reaction; PM, polymicrobial; R, resolved (extruded and healed); RAOM, recurrent acute otitis media; SE, serous effusion; SL, hearing or speech and language delay concerns; SP, Streptococcus pneumoniae; TM, tympanic membrane; TT, tympanostomy tube.
Positive identification of a biofilm or bacterial strain as OCT is marked as “biofilm,” FISH as +, and PCR as Δ for each genetic probe.
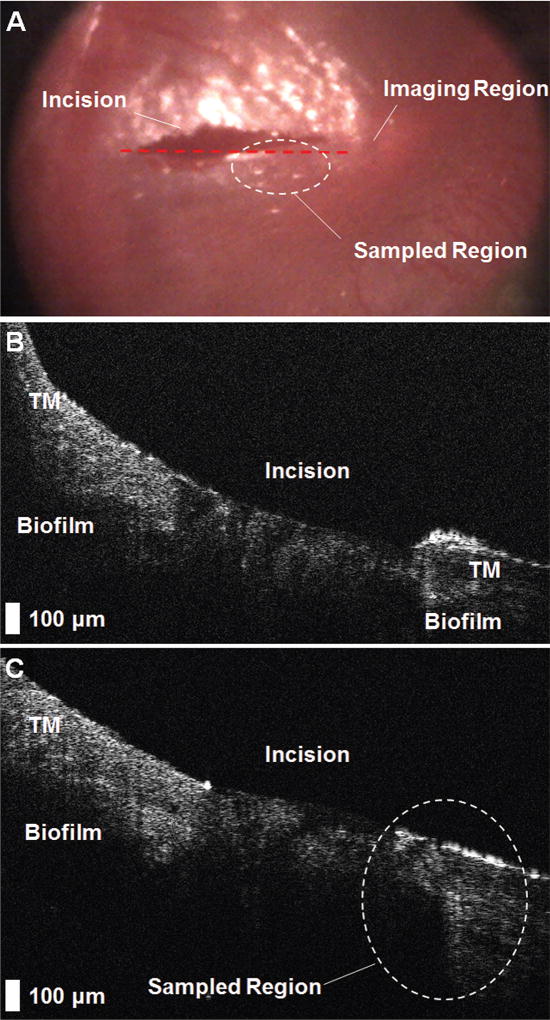
Figure 3 shows representative imaging data. This subject was diagnosed with chronic ETD and COME and scheduled for surgery. Sample 12 was collected from this ear.
Figure 3.
Representative results from the imaging and sampling protocol. (A) Digital otoscopy image of the tympanic membrane (TM) immediately after myringotomy, which identifies the imaging region (red dashed line) and the sampling region (white dashed circle). (B) A presampling optical coherence tomography image of the TM. (C) The postsampling image demonstrates microstructural changes to the sampled region (white dashed circle) and confirms that sampling was performed near the original imaging site.
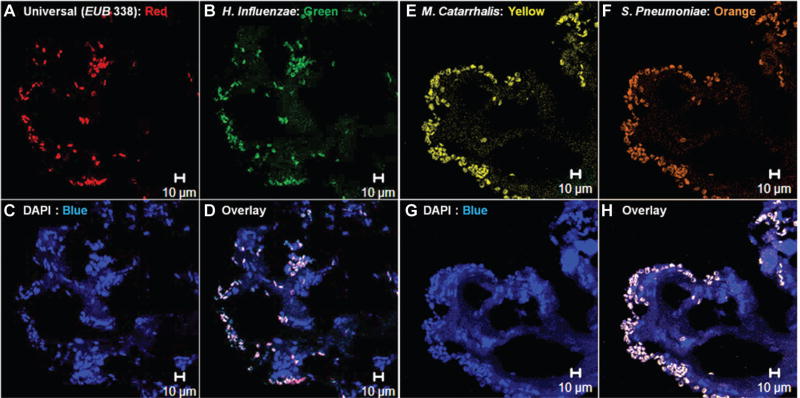
CLSM images were evaluated for bacterial clustering and compared with known morphology.10,31,32 Images that showed evidence of biofilm ultrastructure demonstrated bacterial presence with the universal bacterial domain probe or colocalization with species-specific probes. Figure 4 presents representative CLSM images from sample 21. Figure 4D and 4H illustrate the colocalized presence of bacteria within a biofilm-like ultrastructure.
Figure 4.
Representative confocal laser scanning microscope images from fluorescence in situ hybridization–tagged sample 21. (A) Components identified with the EUB335 domain probe, which colocalizes with (B) the Haemophilus influenzae probe. (C) A nuclei stain (DAPI) detects other unrelated and unknown genetic components in the sample, likely originating from white blood cells, cell fragments, genetic components from host/immune cells, or bacterial populations outside the selected fluorescence in situ hybridization probes. (D) An overlay of these channels reveals the presence of bacteria dispersed throughout the sample, with little background noise. (E–H) Similar colocalized fluorescence from Moraxella catarrhalis and Streptococcus pneumoniae fluorescence in situ hybridization probes, as well as DAPI acquired from an adjacent histologic section.
Discussion
Collectively, OCT, CLSM, and PCR results provided compelling evidence for the presence of a biofilm affixed to the mucosal surface of the TM. Past characterization of the TM and MEC with OCT identified and established optical and image-based features for controls and subjects diagnosed with acute and RAOM.24 The microbial infection–related structures identified in this study with OCT were similar to those consistently identified in past subjects with severe cases of RAOM. OCT can noninvasively identify the presence of additional microbial structures based on their inherent optical scattering properties and without the use of any exogenous dyes or stains. OCT can simultaneously and quantitatively measure the thickness of these structures and the TM, which was shown to be statistically different among normal ears, ears with acute OM, and ears with a biofilm.24 However, OCT does not provide information related to the microbiological content, as the contrast mechanism in OCT is sensitive only to optical refractive index differences.33 A previous study integrated low-coherence interferometry (single-point OCT) and Raman spectroscopy to correlate structural and biochemical properties of the middle ear.34 This system is currently under further development.
PCR and CLSM/FISH images were used to provide biochemical and morphologic characterization of sampled biofilm structures to validate OCT findings and demonstrate that the observed structures were indeed biofilms. CLSM/FISH images provided highly specific visualization of the spatial distribution of bacteria,15 where other dyes may non-specifically adsorb or absorb to other biological components present in MEEs and biofilms. While PCR can identify bacteria with sufficient available genetic material,35,36 PCR does not categorize structural morphology. Additionally, the technically challenging and lengthy sample preparation for PCR or FISH cannot be performed for rapid point-of-care diagnosis or in vivo, and repeated invasive sampling of patients for monitoring OM is impractical. In the future, it may be possible to identify, quantify, and longitudinally track in vivo dynamics of these biofilms based on OCT image features.
While these results are promising, the clinical utility of detecting middle ear biofilms during OM remains unclear. Large-scale clinical trials are needed to define a clinical management strategy following detection of a middle ear biofilm. Comparing OM with other biofilm-mediated diseases may provide insight into expectations and potential treatment regimens.37–41 Typically, single- and multispecies biofilms act as reservoirs for reseeding infections in recurrent cases. As biofilms mature and expand, encapsulated bacteria multiply, are protected from the host immune system,42,43 and develop antibiotic resistance.44 Eventually a “critical mass” is reached, and bacteria are dispersed. Recent research verified that biofilm dispersal mechanisms are directly related to proliferation of infection, as demonstrated in a mouse and indwelling catheter model.45 Another study found evidence of biofilms within the MEE of patients with COME,46 likely caused by biofilm dispersal. Consequently, episodes of recurrent OM are probably the result of a biofilm within the MEC.
More effective methods of treating severe and persistent cases of OM and any biofilm would perhaps include disruption of its signaling, formation capability, or structural integrity, thereby exposing pathogens to the host immune system and possibly to concurrently utilized antibiotics.47–49 Novel treatments that specifically target biofilms are an active area of development, including hydrogel-mediated transtympanic delivery of antibiotics,50 techniques for photo irradiation,51 acoustic disruption,52 cold plasma–based irradiation,53 ionic liquid–based penetration for enhanced antimicrobial activity,54 and even bacteriophage therapy.55 Noninvasively assessing the presence and characteristics of middle ear biofilms with OCT offers an opportunity to readily perform in vivo human studies and trials as compared to animal studies with ex vivo histologic endpoints or invasive surgical sampling studies in humans.
During this study, there were no instances of confounding ear pathology, such as tympanosclerosis, cholesteatoma, dimeric TMs, or retraction pockets that would affect the assessment of OCT images for the presence or absence of a middle ear biofilm. These conditions arise from separate physiologic processes and have distinct OCT image–based features that distinguish them from middle ear biofilms, as previously demonstrated.56–58
There are several limitations in this study. First, there was no control group. No TM mucosa samples were collected for analysis from healthy pediatric subjects undergoing non-OM-related surgeries. However, it was previously demonstrated that normal ears have no biofilms on the MEM.10 Other studies similarly reported that normal ears lack biofilm-related structures, as shown in a rat model with a combination of OCT and histology21 and in normal adult20 and pediatric24 ears with OCT.
Prior to sample collection, the MEC was not aspirated to remove any effusion, and samples were not washed before being placed in fixative. Given the numerous FISH processing steps, it is unlikely that an effusion had any significant effect on these results. Moreover, positive CLSM images were evaluated by consistent and repeated fluorescent signal embedded within the biofilm matrix, not from the exterior of the structure. Aspiration of any MEE before imaging and sampling may also inadvertently remove biofilm material and confound sample collection.
It is possible that some samples, once divided for PCR and FISH/CLSM, did not have active bacterial populations. However, it is likely that in other samples, the amount of genetic material for analysis was simply limited. Some recovered samples were small (~1 mm3), and no additional culturing to expand bacterial concentration was performed. While FISH results were able to identify single bacteria, PCR requires a minimum amount of genetic material,36 which may explain why some samples had no identifiable bacteria. Furthermore, our study analyzed the 3 most common bacterial species known to cause OM,59 although many other bacterial strains have been identified.60 In aggregate, these factors may explain why some samples did not confirm our hypothesis with combined PCR and CLSM/FISH imaging results. However, when sufficient genetic material was present for 1 or both techniques, the resulting measurements were not degraded by the heterogeneous composition of these samples, which can include white and red blood cells, MEE fluid, other bacteria, and cell and biofilm fragments.
The OCT system provided an imaging depth up to ~2 mm into tissue, even semitransparent or highly scattering tissues such as the TM. This capability allows cross-sectional depth-resolved visualization and quantification of the TM and any adjacent structure in the MEC. Since the MEM is known to support biofilms,10 our group is developing a swept-source OCT system to provide visualization of deeper structures within the MEC, up to a centimeter or more,61 including the ossicles and the MEM.
Conclusion
Based on the direct observation, sampling, and analysis of structures that extend across the mucosal surface of the TM, this study confirmed that OCT image–based findings of microbial infection–related structures in this cohort of subjects with RAOM and/or COME are indeed middle ear biofilms. Furthermore, results demonstrated that OCT provides a means to quickly and noninvasively assess the middle ear and TM for the presence of these biofilms. In the future, OCT could be used to rapidly and quantitatively assess for the presence of a middle ear biofilm without invasive sampling, as in the primary care office. This capability allows for the longitudinal tracking of middle ear biofilms, specifically their formation and resolution at different stages of OM and when exposed to existing or newly developed pharmacologic or surgical treatment strategies.
Acknowledgments
We acknowledge the logistical assistance from Marina Marjanovic, PhD; the research staff at Carle Foundation Hospital, specifically Deveine Toney and Alexandra Almasov; and the nursing staff at the Carle Ambulatory Surgery Center for their assistance during subject examination and surgery. Additional information can be found at http://biophotonics.illinois.edu.
Funding source: National Institutes of Health (1R01EB013723 to S.A.B.).
Footnotes
This article was presented at the 2017 AAO-HNSF Annual Meeting & OTO Experience; September 10–13, 2017; Chicago, Illinois.
Author Contributions
Guillermo L. Monroy, substantial contributions to the conception, design, and/or execution of the work, drafting, final approval, and accountability for the work; Wenzhou Hong, substantial contributions to the conception, design, and/or execution of the work, drafting, final approval, and accountability for the work; Pawjai Khampang, substantial contributions to the conception, design, and/or execution of the work, drafting, final approval, and accountability for the work; Ryan G. Porter, substantial contributions to the conception, design, and/or execution of the work, drafting, final approval, and accountability for the work; Michael A. Novak, substantial contributions to the conception, design, and/or execution of the work, drafting, final approval, and accountability for the work; Darold R. Spillman, substantial contributions to the conception, design, and/or execution of the work, drafting, final approval, and accountability for the work; Ronit Barkalifa, substantial contributions to the conception, design, and/or execution of the work, drafting, final approval, and accountability for the work; Eric J. Chaney, substantial contributions to the conception, design, and/or execution of the work, drafting, final approval, and accountability for the work; Joseph E. Kerschner, substantial contributions to the conception, design, and/or execution of the work, drafting, final approval, and accountability for the work; Stephen A. Boppart, substantial contributions to the conception, design, and/or execution of the work, drafting, final approval, and accountability for the work.
Disclosures
Competing interests: Michael Novak—has equity interest in and serves on the clinical advisory board of PhotoniCare, Inc. Stephen A. Boppart—cofounder and chief medical officer of PhotoniCare, Inc; received royalties from patents licensed by MIT related to OCT.
Sponsorships: None.
References
- 1.Lieberthal AS, Carroll AE, Chonmaitree T, et al. The diagnosis and management of acute otitis media. Pediatrics. 2013;131:964–999. doi: 10.1542/peds.2012-3488. [DOI] [PubMed] [Google Scholar]
- 2.Sjogren PP, Gale C, Henrichsen J, et al. Variation in costs among surgeons and hospitals in pediatric tympanostomy tube placement. Laryngoscope. 2016;126:1935–1939. doi: 10.1002/lary.25775. [DOI] [PubMed] [Google Scholar]
- 3.Keyhani S, Kleinman LC, Rothschild M, Bernstein JM, Anderson R, Chassin M. Overuse of tympanostomy tubes in New York metropolitan area: evidence from five hospital cohort. BMJ. 2008;337:a1607. doi: 10.1136/bmj.a1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenfeld RM, Shin JJ, Schwartz SR, et al. Clinical practice guideline: otitis media with effusion (update) Otolaryngol Head Neck Surg. 2016;154(1):S1–S41. doi: 10.1177/0194599815623467. [DOI] [PubMed] [Google Scholar]
- 5.Rosenfeld RM, Schwartz SR, Pynnonen MA, et al. Clinical practice guideline: tympanostomy tubes in children—executive summary. Otolaryngol Head Neck Surg. 2013;149:8–16. doi: 10.1177/0194599813490141. [DOI] [PubMed] [Google Scholar]
- 6.Cullen KA, Hall MJ, Golosinskiy A. Ambulatory surgery in the United States. Natl Health Stat Report. 2009;(11):1–25. [PubMed] [Google Scholar]
- 7.Joo H-S, Otto M. Molecular basis of in-vivo biofilm formation by bacterial pathogens. Chem Biol. 2012;19:1503–1513. doi: 10.1016/j.chembiol.2012.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tóth L, Csomor P, Sziklai I, Karosi T. Biofilm detection in chronic rhinosinusitis by combined application of hematoxylineosin and gram staining. Eur Arch Otorhinolaryngol. 2011;268:1455–1462. doi: 10.1007/s00405-011-1623-x. [DOI] [PubMed] [Google Scholar]
- 9.Nistico L, Kreft R, Gieseke A, et al. Adenoid reservoir for pathogenic biofilm bacteria. J Clin Microbiol. 2011;49:1411–1420. doi: 10.1128/JCM.00756-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall-Stoodley L, Hu FZ, Gieseke A, et al. Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. JAMA. 2006;296:202–211. doi: 10.1001/jama.296.2.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akyıldız İ, Take G, Uygur K, Kızıl Y, Aydil U. Bacterial biofilm formation in the middle-ear mucosa of chronic otitis media patients. Indian J Otolaryngol Head Neck Surg. 2013;65(suppl 3):557–561. doi: 10.1007/s12070-012-0513-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osgood R, Salamone F, Diaz A, Casey JR, Bajorski P, Pichichero ME. Effect of pH and oxygen on biofilm formation in acute otitis media associated NTHi clinical isolates. Laryngoscope. 2015;125:2204–2208. doi: 10.1002/lary.25102. [DOI] [PubMed] [Google Scholar]
- 13.Coticchia JM, Cohen D, Sachdeva L. Grand challenges in pediatric otolaryngology. Front Pediatr. 2013;1:10. doi: 10.3389/fped.2013.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen RG, Johansen HK, Bjarnsholt T, Eickhardt-Sørensen SR, Homøe P. Recurrent otorrhea in chronic suppurative otitis media: is biofilm the missing link? Eur Arch Otorhinolaryngol. 2017;274:1–7. doi: 10.1007/s00405-017-4586-8. [DOI] [PubMed] [Google Scholar]
- 15.Thornton RB, Rigby PJ, Wiertsema SP, et al. Multi-species bacterial biofilm and intracellular infection in otitis media. BMC Pediatrics. 2011;11:1–10. doi: 10.1186/1471-2431-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thurlow LR, Hanke ML, Fritz T, et al. Staphylococcus aureus biofilms prevent macrophage phagocytosis and attenuate inflammation in vivo. J Immunol. 2011;186:6585–6596. doi: 10.4049/jimmunol.1002794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fedtke I, Gotz F, Peschel A. Bacterial evasion of innate host defenses—the Staphylococcus aureus lesson. Int J Med Microbiol. 2004;294:189–194. doi: 10.1016/j.ijmm.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 18.Savage VJ, Chopra I, O’Neill AJ. Staphylococcus aureus biofilms promote horizontal transfer of antibiotic resistance. Antimicrob Agents Chemother. 2013;57:1968–1970. doi: 10.1128/AAC.02008-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Post JC. Direct evidence of bacterial biofilms in otitis media. Laryngoscope. 2001;111:2083–2094. doi: 10.1097/00005537-200112000-00001. [DOI] [PubMed] [Google Scholar]
- 20.Nguyen CT, Jung W, Kim J, et al. Noninvasive in vivo optical detection of biofilm in the human middle ear. Proc Natl Acad Sci U S A. 2012;109:9529–9535. doi: 10.1073/pnas.1201592109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen CT, Tu H, Chaney EJ, Stewart CN, Boppart SA. Non-invasive optical interferometry for the assessment of biofilm growth in the middle ear. Biomed Opt Express. 2010;1:1104–1116. doi: 10.1364/BOE.1.001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chaney EJ, Nguyen CT, Boppart SA. Novel method for non-invasive induction of a middle-ear biofilm in the rat. Vaccine. 2011;29:1628–1633. doi: 10.1016/j.vaccine.2010.12.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen CT, Robinson SR, Jung W, Novak MA, Boppart SA, Allen JB. Investigation of bacterial biofilm in the human middle ear using optical coherence tomography and acoustic measurements. Hear Res. 2013;301:193–200. doi: 10.1016/j.heares.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monroy GL, Shelton RL, Nolan RM, et al. Noninvasive depth-resolved optical measurements of the tympanic membrane and middle ear for differentiating otitis media. Laryngoscope. 2015;125:E276–E282. doi: 10.1002/lary.25141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monroy GL, Pande P, Shelton RL, et al. Non-invasive optical assessment of viscosity of middle ear effusions in otitis media. J Biophotonics. 2016;10:394–403. doi: 10.1002/jbio.201500313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shelton RL, Nolan RM, Monroy GL, et al. Quantitative pneumatic otoscopy using a light-based ranging technique. J Assoc Res Otolaryngol. 2017;18:555–568. doi: 10.1007/s10162-017-0629-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monroy GL, Pande P, Nolan RM, et al. Noninvasive in vivo optical coherence tomography tracking of chronic otitis media in pediatric subjects after surgical intervention. J Biomed Opt. 2017;22:1–11. doi: 10.1117/1.JBO.22.12.121614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hubler Z, Shemonski ND, Shelton RL, Monroy GL, Nolan RM, Boppart SA. Real-time automated thickness measurement of the in vivo human tympanic membrane using optical coherence tomography. Quant Imaging Med Surg. 2015;5:69–77. doi: 10.3978/j.issn.2223-4292.2014.11.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shelton RL, Jung W, Sayegh SI, McCormick DT, Kim J, Boppart SA. Optical coherence tomography for advanced screening in the primary care office. J Biophotonics. 2014;7:525–533. doi: 10.1002/jbio.201200243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leibovitz E, Broides A, Greenberg D, Newman N. Current management of pediatric acute otitis media. Expert Rev Anti infect Ther. 2010;8:151–161. doi: 10.1586/eri.09.112. [DOI] [PubMed] [Google Scholar]
- 31.Kerschner JE, Hong W, Khampang P, Johnston N. Differential response of gel-forming mucins to pathogenic middle ear bacteria. Int J Pediatr Otorhinolaryngol. 2014;78:1368–1373. doi: 10.1016/j.ijporl.2014.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bales PM, Renke EM, May SL, Shen Y, Nelson DC. Purification and characterization of biofilm-associated EPS exopolysaccharides from ESKAPE organisms and other pathogens. PLoS ONE. 2013;8:e67950. doi: 10.1371/journal.pone.0067950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Izatt JA, Choma MA. Theory of optical coherence tomography. In: Drexler W, Fujimoto JG, editors. Optical Coherence Tomography. Berlin, Germany: Springer; 2008. pp. 47–72. [Google Scholar]
- 34.Zhao Y, Monroy GL, You S, et al. Rapid diagnosis and differentiation of microbial pathogens in otitis media with a combined Raman spectroscopy and low-coherence interferometry probe: toward in vivo implementation. J Biomed Opt. 2016;21:107005. doi: 10.1117/1.JBO.21.10.107005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aly BH, Hamad MS, Mohey M, Amen S. Polymerase chain reaction (PCR) versus bacterial culture in detection of organisms in otitis media with effusion (OME) in children. Indian J Otolaryngol Head Neck Surg. 2012;64:51–55. doi: 10.1007/s12070-011-0161-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Forootan A, Sjöback R, Björkman J, Sjögreen B, Linz L, Kubista M. Methods to determine limit of detection and limit of quantification in quantitative real-time PCR (qPCR) Biomol Detect Quantif. 2017;12(suppl C):1–6. doi: 10.1016/j.bdq.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levy LL, Jiang N, Smouha E, Richards-Kortum R, Sikora AG. Optical imaging with a high-resolution microendoscope to identify cholesteatoma of the middle ear. Laryngoscope. 2013;123:1016–1020. doi: 10.1002/lary.23710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zijnge V, van Leeuwen MBM, Degener JE, et al. Oral biofilm architecture on natural teeth. PLoS ONE. 2010;5:e9321. doi: 10.1371/journal.pone.0009321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fastenberg JH, Hsueh WD, Mustafa A, Akbar NA, Abuzeid WM. Biofilms in chronic rhinosinusitis: pathophysiology and therapeutic strategies. World J Otorhinolaryngol Head Neck Surg. 2016;2:219–229. doi: 10.1016/j.wjorl.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson A, Gray D, Karakiozis J, Thomas J. Advanced endotracheal tube biofilm stage, not duration of intubation, is related to pneumonia. J Trauma Acute Care Surg. 2012;72:916–923. doi: 10.1097/TA.0b013e3182493a10. [DOI] [PubMed] [Google Scholar]
- 41.Cole SJ, Records AR, Orr MW, Linden SB, Lee VT. Catheter-associated urinary tract infection by Pseudomonas aeruginosa is mediated by exopolysaccharide-independent biofilms. Infect Immun. 2014;82:2048–2058. doi: 10.1128/IAI.01652-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Donlan RM. Biofilms: microbial life on surfaces. Emerg Infect Dis. 2002;8(9):8881–890. doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents. 2010;35:322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 45.Wang R, Khan BA, Cheung GY, et al. Staphylococcus epidermidis surfactant peptides promote biofilm maturation and dissemination of biofilm-associated infection in mice. J Clin Invest. 2011;121:238–248. doi: 10.1172/JCI42520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Hoecke H, De Paepe AS, Lambert E, et al. Haemophilus influenzae biofilm formation in chronic otitis media with effusion. Eur Arch Otorhinolaryngol. 2016;1273:3553–3560. doi: 10.1007/s00405-016-3958-9. [DOI] [PubMed] [Google Scholar]
- 47.Rabin N, Zheng Y, Opoku-Temeng C, Du Y, Bonsu E, Sintim HO. Agents that inhibit bacterial biofilm formation. Future Med Chem. 2015;7:647–671. doi: 10.4155/fmc.15.7. [DOI] [PubMed] [Google Scholar]
- 48.Römling U, Balsalobre C. Biofilm infections, their resilience to therapy and innovative treatment strategies. J Intern Med. 2012;272:541–561. doi: 10.1111/joim.12004. [DOI] [PubMed] [Google Scholar]
- 49.Wu H, Moser C, Wang H-Z, Høiby N, Song Z-J. Strategies for combating bacterial biofilm infections. Int J Oral Sci. 2015;7:1–7. doi: 10.1038/ijos.2014.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang R, Sabharwal V, Okonkwo OS, et al. Treatment of otitis media by transtympanic delivery of antibiotics. Sci Transl Med. 2016;8:356ra120. doi: 10.1126/scitranslmed.aaf4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hughes G, Webber MA. Novel approaches to the treatment of bacterial biofilm infections. British J Pharmacol. 2017;174:2237–2246. doi: 10.1111/bph.13706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gnanadhas DP, Elango M, Janardhanraj S, et al. Successful treatment of biofilm infections using shock waves combined with antibiotic therapy. Sci Rep. 2015;5:17440. doi: 10.1038/srep17440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Delben JA, Zago CE, Tyhovych N, Duarte S, Vergani CE. Effect of atmospheric-pressure cold plasma on pathogenic oral biofilms and in vitro reconstituted oral epithelium. PLoS ONE. 2016;11:e0155427. doi: 10.1371/journal.pone.0155427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pendleton JN, Gilmore BF. The antimicrobial potential of ionic liquids: a source of chemical diversity for infection and biofilm control. Int J Antimicrob Agents. 2015;46:131–139. doi: 10.1016/j.ijantimicag.2015.02.016. [DOI] [PubMed] [Google Scholar]
- 55.Chaudhry WN, Concepción-Acevedo J, Park T, Andleeb S, Bull JJ, Levin BR. Synergy and order effects of antibiotics and phages in killing Pseudomonas aeruginosa biofilms. PLoS ONE. 2017;12:e0168615. doi: 10.1371/journal.pone.0168615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Djalilian HR, Ridgway J, Tam M, Sepehr A, Chen Z, Wong BJ. Imaging the human tympanic membrane using optical coherence tomography in vivo. Otol Neurotol. 2008;29:1091–1094. doi: 10.1097/MAO.0b013e31818a08ce. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van der Jeught S, Dirckx JJ, Aerts JR, Bradu A, Podoleanu AG, Buytaert JA. Full-field thickness distribution of human tympanic membrane obtained with optical coherence tomography. J Assoc Res Otolaryngol. 2013;14:483–494. doi: 10.1007/s10162-013-0394-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Djalilian HR, Rubinstein M, Wu EC, et al. Optical coherence tomography of cholesteatoma. Otol Neurotol. 2010;31:932–935. doi: 10.1097/mao.0b013e3181e711b8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ramakrishnan K, Sparks R, Berryhill W. Diagnosis and treatment of otitis media. Am Fam Physician. 2007;76:1650–1658. [PubMed] [Google Scholar]
- 60.Ngo CC, Massa HM, Thornton RB, Cripps AW. Predominant bacteria detected from the middle ear fluid of children experiencing otitis media: a systematic review. PLoS ONE. 2016;11:e0150949. doi: 10.1371/journal.pone.0150949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.MacDougall D, Farrell J, Brown J, Bance M, Adamson R. Long-range, wide-field swept-source optical coherence tomography with GPU accelerated digital lock-in Doppler vibrography for real-time, in vivo middle ear diagnostics. Biomed Opt Express. 2016;7:4621–4635. doi: 10.1364/BOE.7.004621. [DOI] [PMC free article] [PubMed] [Google Scholar]