Abstract
Proper cell-material interactions are critical to remain cell function and thus successful tissue regeneration. Many fabrication processes have been developed to create microenvironments to control cell attachment and organization on a 3D scaffold. However, these approaches often involve heavy engineering and only the surface layer can be patterned. We found that 3D extrusion based printing at high temperature and pressure will result an aligned effect on the polymer molecules, and this molecular arrangement will further induce the cell alignment and different differentiation capacities. In particular, articular cartilage tissue is known to have zonal collagen fiber and cell orientation to support different functions, where collagen fibers and chondrocytes align parallel, randomly, and perpendicular, respectively, to the surface of the joint. Therefore, cell alignment was evaluated in a cartilage model in this study. We used small angle X-ray scattering analysis to substantiate the polymer molecule alignment phenomenon. The cellular response was evaluated both in vitro and in vivo. Seeded mesenchymal stem cells showed different morphology and orientation on scaffolds, as a combined result of polymer molecule alignment and printed scaffold patterns. Gene expression results showed improved superficial zonal chondrogenic marker expression in parallel-aligned group. The cell alignment was successfully maintained in the animal model after 7 days with distinct MSC morphology between the casted and parallel printed scaffolds. This 3D printing induced polymer and cell alignment will have a significant impact on developing scaffold with controlled cell-material interactions for complex tissue engineering while avoiding complicated surface treatment, and therefore provides new concept for effective tissue repairing in future clinical applications.
Keywords: poly(lactic-co-glycolic acid), scaffold, 3D printing, cell alignment, polymer alignment, cell-material interaction
1. Introduction
Recognizing the relatively more flexible fabrication process and reduced labor and cost of acellular scaffolds compared to cell-laden approaches, the tissue engineering field has advanced to develop scaffolds that can better support biological functions for cells introduced in vitro or in vivo. 1 Among many cell types that have been combined with acellular scaffold to form an engineered tissue construct, stem cells are the most commonly used due to the advantages of multipotency and more abundant cell sources. 2–5 Besides widely used chemical cues such as growth factors to control stem cell lineage commitment, material cues including stiffness and geometry have been shown to contribute in stem cell differentiation. 6–9
Therefore, cell-surface interactions with extracellular biomaterials are an important consideration when constructing tissue regenerative devices as such interactions can influence cellular attachment, alignment, proliferation, and differentiation when using multipotent cells such as mesenchymal stem cells (MSCs). The basement membrane in vertebrates is composed of a complex mix of ECM proteins with varied pore and fiber size and orientation serves as the substrata for supporting cellular structures. 10 Although the process is not well understood, it is generally accepted that the surface topography of the basement membrane or synthesized material provides cues to influence cellular function and differentiation, through activation of plasma membrane integrin receptors. 11,12 Surface features at the microscale influence cell adhesion, alignment, morphology, proliferation and differentiation; having an even greater impact at the nanoscale. 13,14 For example, topographically influenced MSCs showed similar efficiency when differentiating into bone lineage as to MSCs treated with osteogenic media. 15 In another study, nanotopography also showed a stronger effect on the upregulation of neuronal markers than biochemical cues alone on unpatterned surfaces. 16 In general, the presence of surface features on the scaffold such as ridges, steps or spaces affects cell attachment and proliferation by as the cells increase their complement of filopodia and microspikes, which are known to be the “sensing” organelles. 17
Articular cartilage tissue exemplifies the impact of cellular organization on tissue function. It is composed of 3 zones: the superficial or tangential zone, the middle or transitional zone, and the deep zone, each with different biological functions. Cartilage in the superficial zone provides high tensile properties by having parallel collagen fibers and chondrocytes to the surface of the cartilage.18 Cartilage in the middle zone provides a transition between the superficial and deep zones. Collagen fibers and chondrocytes in this zone are randomly organized to provide a functional bridge between the superficial and deep zones. The deep zone collagen fibers and cells are arranged perpendicular to calcified cartilage and bone, which allows it to adhere to bone tissue and provide resistance to compression. Zonal differences in matrix organization and structure are due to variations in the cellular activity of each zone. For example, proteoglycan 4 (PRG4), which is a large glycoprotein contributes to the lubricating function of the synovial fluid, is predominantly produced by the superficial zone chondrocytes.
The dysfunction of articular cartilage affects millions of people of all ages. The prevalence of clinical osteoarthritis had grown to nearly 27 million by 2007 with trends indicating this number will continue to increase every year. 19 A limited self-renewal capacity owing to lack of blood vessels and low cellularity makes cartilage one of the most difficult tissues to regenerate. A few of the most frequently employed treatments include microfracture, autologous chondrocyte implantation (ACI), but these strategies have limited success. 20 To significantly reduce the potential immune response from transplanting allogeneic cartilage tissue and the cost and time associated with ACI, the use of biomimetic acellular scaffolds which provide zonal biological and mechanical cues to stimulate seeded stem cell differentiation is an attractive alternative for functional cartilage regeneration.
Our present study investigated the impact of 3D printing induced surface properties on MSC adhesion and chondrogenic differentiation, both at a scaffold pattern (~100 μm) and polymer molecule level (nano scale) so that cell distribution as a population and cell alignment at the single cell level were observed. Unlike the complicated nanofabrication process used in previous studies to achieve desired surface patterns, the polymer molecule and cell alignment we observed was a result from the 3D extrusion without any additional processing. The objective of this study is to demonstrate effect of polymer molecule alignment and induced cell alignment caused by extrusion based 3D printing. The cell alignment was revealed and quantified both in in vitro and in vivo models. Finally, the chondrogenic differentiation of MSCs on scaffolds printed with different patterns was evaluated.
2. Materials and Methods
2.1 Scaffold fabrication
PLGA with LA:GA ratio of 85:15 and molecular weight of 35kD was purchased PolySciTech (West Lafayette, IN). The 3D printed scaffold was fabricated using 3D Bioplotter (EnvisionTEC, Gladbeck, Germany) with direct melt extrusion technique. The scaffold was programmed with inner patterns using the provided EnvisionTEC software. For printing, the material was loaded into the printing cartridge and melted at 165°C and extruded at 9 bar with an average speed of 1.5 mm/s using a 0.2 mm inner diameter needle based on previously established methods 21. Parallel pattern scaffold has fibers diameter of 0.2 mm parallel to each other with 0.2 mm edge-to-edge spacing of two adjacent fibers. For random pattern, the angle to the contour and the spacing of each layer were randomly selected using a random generator package in R software. All scaffolds have a dimension of 4 mm (length) × 4 mm (width) × 1.5 mm (height). The casted PLGA was made by melting the raw PLGA material and shape to the same size as the printed scaffold.
2.2 Small Angle X-Ray Scattering
SAXS has been used to determine the material internal structure as the interference pattern is characteristic to the molecule orientation in the material. Therefore, by recording the scattered pattern or signal distribution on each direction, the intrinsic molecule alignment of the material can be interpreted. SAXS measurements were performed with Xenocs Xeussat system at the X-ray Crystallographic Center, located at Department of Chemistry & Biochemistry, University of Maryland. The system was equipped with 5 Meter system with CuKα sealed 30W tube high brightness micro-focus source at a constant X-ray energy of 10 keV. The samples were taped on a metal holder in the experiment chamber. The exposure time to collect each scattering profile was 600 s. The sample-to-detector distance was set at 2514.72 mm for all samples. The incidence angle between x-rays and the sample surface was fixed at 0.14°. Scattering profiles were recorded on a Pilatus 1M 2-D area detector.
2.3 Cell culture and seeding
Primary hMSCs (P2) were purchased (Lonza, Basel, Switzerland) and expanded in a monolayer in high glucose Dulbecco’s Modified Eagle Medium (DMEM) (Life Technologies, Carlsbad, CA) containing 0.1% penicillin/streptomycin (Life Technologies), 0.1 mM non-essential amino acids (Life Technologies) and 10% fetal bovine serum (Life Technologies, Carlsbad, CA) (MSCs growth media). After reaching the desired amount, cells were lifted with trypsin to form a cell pellet. Approximately 1 million cells were seeding onto each scaffold by dropping 100 μL concentrated cell solution covering the entire scaffold. Before adding fresh MSCs growth media, seeded scaffolds were kept in 37 °C for 4 hours to allow attachment. Cell culture media was changed every the other day during maintenance.
2.4 Live/Dead Staining
Live/Dead assay was performed to show cell viability and morphology. The scaffolds were washed in Hank’s buffered saline solution (HBSS, Life Technologies, Carlsbad, CA) for 5 minutes to remove extra media and other active reagents. The cells on the scaffolds were stained in a 2 μM ethidium homodimer and 4 μM calcein AM (Life Technologies, Carlsbad, CA) combined with HBSS for 30 minutes in the dark.
2.5 Confocal Imaging
Cells were imaged using Zeiss LSM 710 confocal microscope (Carl Zeiss Microscopy, Jena, Germany). Separate channels of lasers were applied, A488 for green, A594 for red, and a DAPI channel. Images were taken at frame scanning mode, with a size of 1024 × 1024 at medium speed to achieve desired resolution. Each laser power was kept in consistent for comparable results.
2.6 Cell Alignment Quantification
The degree of cell alignment was quantified using an Image J plugin Orientation J following a series of steps. First the image taken from Zeiss software was converted to 8 bit in Image J. The region of interest (ROI) was defined by circling cells along their shape. Orientation J automatically measured the alignment angles to the horizontal axis. By extracting the printed fiber orientation, the angle to the printed fiber can be calculated. Images were taken from three different regions to calculate average orientation. All cells on each image was counted to eliminate the biased choices. Single-blinded experiment was conducted to analyze the images.
2.7 Animal Surgery
The scaffolds pre-seeded with rat MSCs were implanted subcutaneously in rats. Three rats were used with 6 implantations per rat, 3 dorsal and 3 ventral. The scaffolds were UV sterilized and seeded with rat MSCs (R&D systems, Minneapolis, MN). The cell-laden scaffold was prepared one day before the implantation and was allowed to culture and maintained at 37 °C until the surgery. After induction and maintenance with isoflurane, a small dorsal incision was made in the skin. Subsequently, a subcutaneous pocket was created cranially and caudally to the incision by blunt dissection and then the scaffolds were placed into the created space. The wound was closed by subcuticular suturing along the incision lines using 4-0 C-14 reverse cutting resorbable sutures. After 7 days of implantation, the rats were euthanized and the implants were collected for analysis. Using both histology and confocal microscopy, cell alignment was examined within the scaffolds. This animal protocol was approved by University of Maryland College Park Institutional Animal Care and Use Committee (IACUC) (Project 955010-1).
2.8 RNA Isolation and qRT-PCR
Toal RNA was isolated from the hMSCs using an RNeasy Plus Mini Kit (Qiagen, Frederick, MD) and then reverse transcribed to complementary DNA (cDNA) using a High Capacity cDNA Archive Kit (Life Technologies, Carlsbad CA). The cDNA solution was then combined with a Universal Master Mix (Life Technologies, Carlsbad CA), as well as oligonucleotide primers and Taqman probes for Sox 9, type II collagen (COL2A1), aggrecan (AGC), and the endogenous gene control glyceraldehyde 3 phosphate dehydrogenase (GAPDH) (Thermo Fisher Scientific, Waltham, MA) to perform quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR). A standard protocol at thermal conditions of 2 min at 50°C, 10 min at 95°C, 40 cycles of 15 s at 95°C, and 1 min at 60°C was applied. The relative gene expression level of each target gene was normalized to the mean of GAPDH in each group then the fold change was determined relative to the first time point baseline gene expression. Fold change was calculated using the ΔΔCT relative comparative method as described previously 22.
2.9 Statistical Analysis
Student t-test was performed to compare cell orientation between two groups. To assess gene expression, data were analyzed using analysis of variance and Tukey’s multiple-comparison test. A significance level of 95% was chosen, and a p-value less than 0.05 was considered to indicate a significant difference between samples.
3. Results
3.1 Polymer Alignment
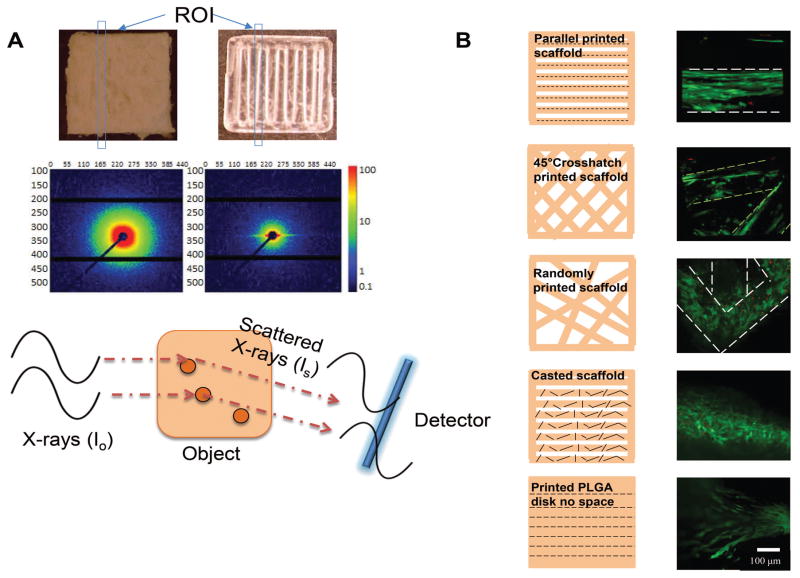
We scanned our printed scaffold and a casted PLGA scaffold using small angle X-ray scattering to investigate the polymer molecule organization between casted and printed scaffolds. Differences in molecular organization of materials results in different scattering maps due to the different paths traveled by the X-ray. Therefore, the distribution of scattering signals on each direction carries structural information of the molecular alignment in each direction. The scattering intensity map here shows homogeneous scattering in each direction for casted PLGA polymer samples, which indicates the polymer molecules were randomly distributed in the material. However, the scattering of printed PLGA was compressed to one direction as a result of constructive interference, which indicates a polymer molecule orientation exists in the printed sample (Figure 1A). Notably, the scaffold was printed in solid construct without spacing and the X-ray beam diameter was smaller than the printing nozzle diameter to avoid structure interference. The intensity was presented as an arbitrary detector unit so that the value itself does not carry any structural information; only the ratio of distribution of the scattered signal in each direction indicates the intrinsic polymer molecules organization.
Figure 1. 3D printing induced polymer molecule alignment and resulted cell alignment.
A. SAXS set up and results comparing casted and printed PLGA scaffolds. Scattered X-ray waves arrive at the detector at different time to form signals as a result of material intrinsic structure. X-ray scattering shows uniform scattering in all directions of the casted scaffold, which indicates random polymer molecule organization. However, scattering shows higher intensity on the horizontal direction of the printed scaffold, which indicates vertically oriented polymer molecule existing after printing. Blue boxes indicate regions of interest (ROI) upon incoming x-ray. B. Cell alignment on scaffolds printed with different patterns. Confocal microscope image showed hMSCs aligned differently on printed patterned scaffolds. Live cells were shown in green and dead cells were shown in red. Short black lines indicates the polymer molecule alignment.
3.2 In Vitro Cell Alignment
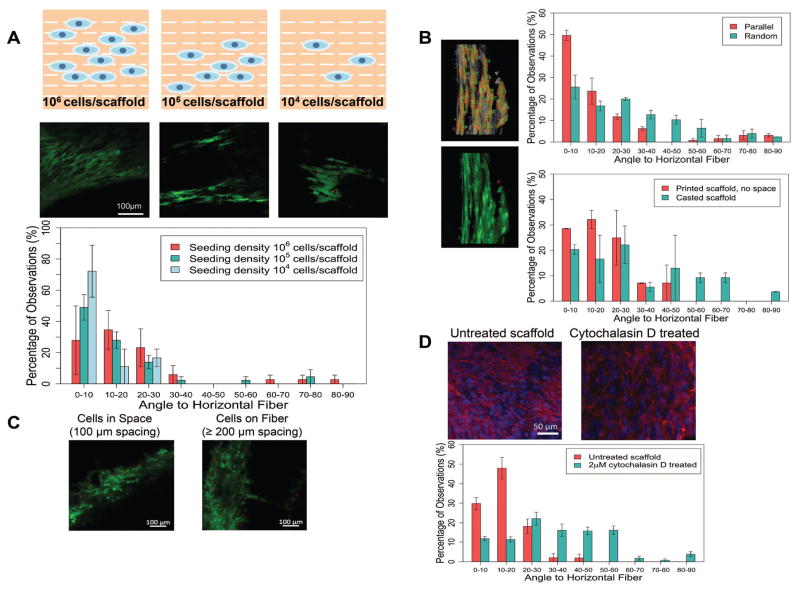
hMSCs were seeded on the 3D printed PLGA scaffolds with different patterns. Confocal microscopy was used to reveal the cell organization followed a quantitative analysis using ImageJ. We observed different cell alignment levels when seeded on scaffolds printed with different patterns (Figure 1B). Since the polymer molecules on each single printed fiber were aligned due to the extrusion processing, cells seeded on scaffolds with parallel fibers showed an induced alignment along the extrusion direction. As a control group, hMSCs were also seeded on casted PLGA scaffold (melt and shaped with parallel pattern), where cells appeared randomly ordered on each fiber. As another control, printed solid scaffold without spacing was seeded with cells to exclude structural cues. Without spacing, cells were still observed to be aligned on the printed direction. For a 45° crosshatch pattern, the printed fibers were less ordered compared with the parallel fibers considering the interactions at the crossed regions. As a result, the cells appeared less ordered than cells seeded on the parallel scaffold. For the random patterned scaffold, with multiple connection points for cells to migrate, cells appeared more randomly aligned compared to the previous two cases. In particular, cells on scaffold corners appeared to be bridging into different directions. In order to test if the cells were encouraged to align due to high concentration, cells were also seeded on the parallel printed scaffold at 10X and 100X diluted concentration. The yield cell attachment efficiency on the scaffold was approximately 15% according to hemacytometer counting. At different seeding concentration, the cells appeared similar degree of alignment, where the majority of the cells attached with an angle less than 30°. Although limited cells can be detected were seen at 104 cells/scaffold seeding density after attachment, the detected cells showed positive alignment along the printed fiber direction (Figure 2A).
Figure 2. Cell alignment quantification and the impact of different factors on cell attachment.
A. Cell alignment and quantification for different seeding densities. Cells seeded at various concentration all showed aligned pattern, with more than 80% of the total population had an angle to the fiber less than 30 degree. B. Cell alignment quantification for scaffolds with different patterned and fabrication methods from Figure 1. Individual cells were selected manually in ImageJ, then the angles were automatically calculated by the built in Plugin OrientationJ. Cells on scaffolds with parallel pattern mostly aligned along the fibers, by displaying an alignment angle to the printed fiber of less than 30°. As a comparison, cells on random printed fibers aligned with less order, displaying more evenly distributed angles. As a control, cells on casted scaffold showed evenly distribution of attachment angles. In a nother control using printed solid disk without any patterns, cells showed directional alignment along the printed fibers. All cells from the captured images were included for analysis. C. Preferred adhesion with different scaffold spacing. With a 200 μm printed fiber, when the width of the spacing is smaller than the fiber, most cells appeared in the gap spacing rather than attached on the surface of the scaffold. However, when the spacing was larger than the diameter of the fiber, cells tended to attach on the scaffold surface instead. D. Phalloidin staining of actin fibers and comparison of cell alignment before and after cytochalasin D treatment. After cytochalasin D treatment, the attached cells completely lost their organized direction due to loss of focal adhesion.
The quantified cell alignment revealed that the cells on parallel patterned scaffolds were more organized. The general trend distribution showed that the majority of cells seeded on parallel scaffolds had a smaller contact angle to the horizontal fiber compared to the random patterned scaffold. 50% of cells on the parallel patterned scaffold showed a contact angle less than 10° while only 26% of cells on the random patterned scaffold had contact angles less than 10° (Figure 2B).
The above results suggested that not only the aligned polymer molecules determines the how the cell would adhere, but also the pattern of the printed fibers contributes to the overall cell distribution. Taking account this effect, we conducted experiment on how the orientation of the fiber influence the cell adhesion. We found that when the spacing was 100 μm or less, cells tend to fill the gaps rather than grow on the printed fibers. On the contrary, for spacing larger than 200 μm, cells would choose to attach on top of PLGA fibers (Figure 2C).
The phalloidin staining revealed aligned actin fibers in the ECM of aligned hMSCs. However, after treating the attached and aligned cells with 2μM cytochalasin D, a known inhibitor of actin polymerization, the cells were disassembled and completely lost their organized direction due to loss of focal adhesion. The quantified resulted showed significant different angle distribution between the untreated group and the cytochalasin D treated group (Figure 2D).
3.3 In Vivo Cell Alignment
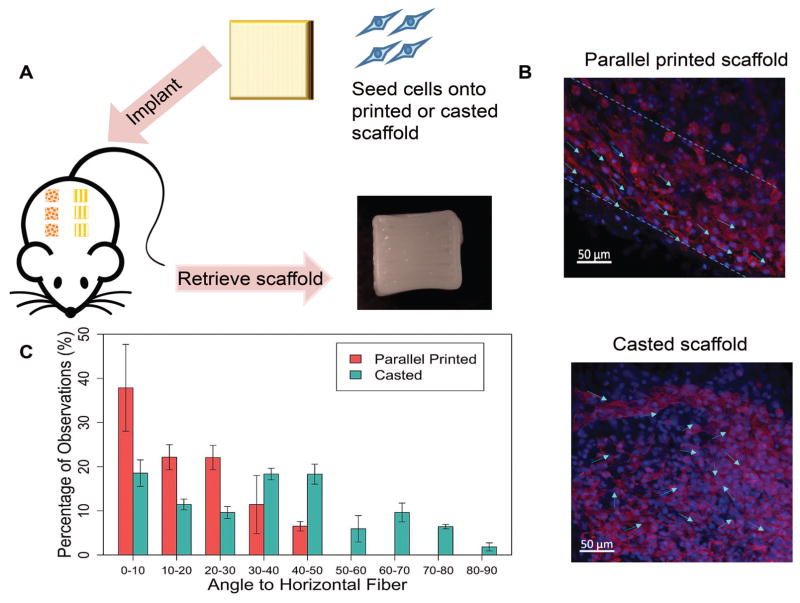
We then conducted an in vivo study using a rat model by subcutaneously implanting the scaffold to see if the cell alignment can be maintained (Figure 3A). After retrieving samples from the rats, the scaffold was visually intact with clear preserved patterns. The whole scaffold was stained and imaged, where cell nuclei were stained blue using DAPI and actin was stained red using phalloidin to show the cell morphology and orientation. While cells appeared randomly distributed on the casted scaffolds, on the printed scaffolds cells were aligned along printed fibers (one fiber marked with dashed line) (Figure 3B). Cell orientation (angle to the fiber) was quantified using ImageJ and the Plugin OrientationJ by manually selecting individual cells and automatically calculating the angles. A total of 178 cells from three images of the casted group and 118 cells from three images of the parallel were included in the single blinded quantification study. Most of the cells on the parallel printed implant displayed an angle less than 50°, with 37% having an angle to the fiber of less than 20°. However, the cells attached on the casted scaffold showed an evenly distributed orientation. In terms of cell morphology, the MSCs on parallel printed scaffolds also appeared more elongated or star shaped compared to those on the casted scaffold.
Figure 3. In vivo cell alignment evaluation and quantification.
A. Experimental flow of the animal study. Rat MSCs were seeded on parallel printed or casted scaffolds, then scaffolds were implanted subcutaneously in rats. After 7 days, retrieved scaffolds showed intact structure and printed patterns. B. Cell alignment was maintained after in vivo implantation. Cells appeared mostly aligned on the parallel patterned scaffold, while cells were more randomly distributed on the casted scaffolds. C. Quantification of cell alignment in vivo. The average number was calculated from three different images.
3.4 Chondrogenesis Response
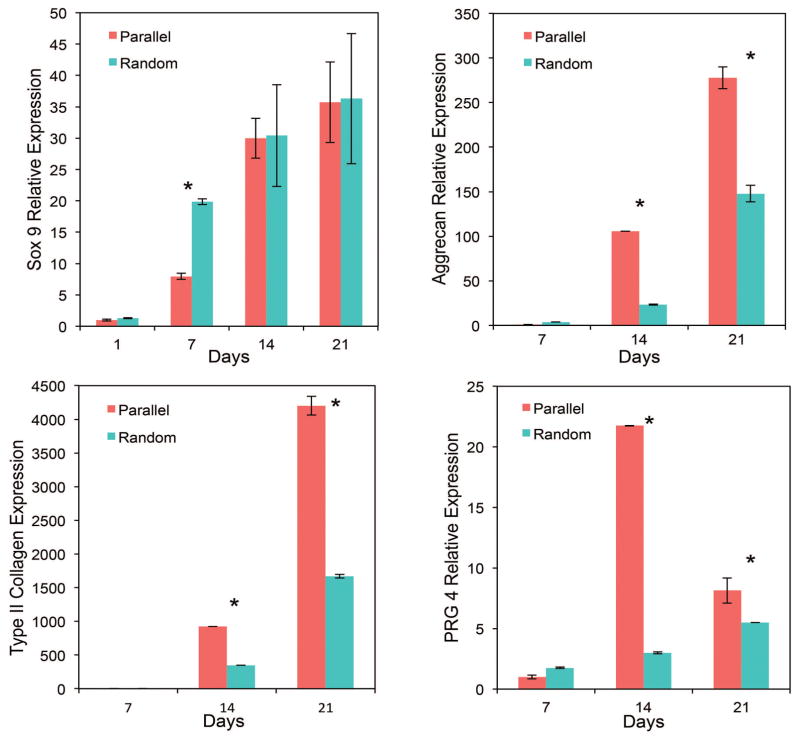
Finally, we investigated the chondrogenesis as a response to different scaffold patterns and the induced cell alignment. Positive markers aggrecan (main proteoglycan in articular cartilage), type II collagen (composes main part of chondrocyte ECM), Sox 9 (transcription marker) and PRG4 (superficial zone marker) were evaluated. Generally, increased expression of positive markers was observed in parallel patterned scaffolds, which indicates a potential to engineer zonal scaffolds with a controlled cellular response. Expression of aggrecan on both day 14 and day 21 was significantly higher for the MSCs seeded on parallel patterned scaffolds. Similarly, approximately 2.5 folders higher expression of type II collagen was observed in the parallel group compared to the random pattern group. In particular, the superficial zonal marker expression was 7 fold increased for cells on parallel aligned scaffolds, and the increased expression was maintained throughout the 21 day study.
4. Discussion
In this study, we investigated the impact of polymer alignment as a result of extrusion-based 3D printing at high pressure and temperature. Most of the reported PLGA printings use solvents or adhesives combining with an ink jet printer. 23,24 Because of the more extreme processing conditions required, only few direct fusion extrusion on PLGA printing has been attempted. As a result, detailed cell-material interactions such as cell attachment or alignment under such fabrication ways have not been studied. In fact, as a result of sheared polymer “solution” under high pressure (9 bar) through a fine needle (0.2 mm), induced MSC alignment as a response to different scaffold fabrication methods and patterns was demonstrated and evaluated both in vitro and in vivo. Our results showed that MSCs exhibited more controlled and organized attachment on printed scaffolds compared to casted scaffolds. The nanoscale polymer molecule alignment is believed to be the major determining factor for cell alignment, although the overall cell distribution could be affected by the micropatterning of the printed scaffold. In one aspect, the aligned polymer molecules caused attached cells to align, where we saw most of the cells aligned along the printed fiber with a pure parallel pattern, with and without a space in between the printed fibers. However, this effect can be compromised by the scaffold patterns that have crossed fibers, so that the level of alignment was weakened as the randomness degree of the polymer fibers increased. These findings will provide useful insight surrounding cell-material interactions, in particular in the context of 3D-printing approaches. The results will help to develop scaffolds with desired cell distribution in complex tissue engineering applications such as engineered zonal cartilage.
SAXS is a useful way to determine the material internal structure since the interference pattern is characteristic to the material structure. The intensity of the interfering waves is a result of distance of the particles and their orientations with respect to the incidence beam, so that the detector can record brightness and darkness based on whether the scattered waves arrive in phase or out of phase. In our case with a solid polymer sample, the orientation of polymer molecules and their degrees can be revealed by the 2D scattering pattern. In SAXS, where the purpose is to reveal the material intrinsic structure properties, only Thomson scattering is concerned as this type of photon collision is strictly synchronous to create coherent scattering that carries structural information. 25 Therefore, only the ratio of intensity distribution on each direction tells the particle or molecule orientation, not the overall intensity of each sample. In the casted samples, the scattering pattern has equal intensities around the incident beam, which indicated randomly oriented polymer molecules. As a contrast, the compressed scatter pattern on the horizontal direction demonstrated a molecule partial orientation along the vertical direction. We hypothesize that this phenomenon is caused by the shear force during high temperature and high pressure printing when polymer undergoes glass transition because similar partial orientation patterns are commonly seen in sheared liquids or spun fibers. 26 In another aspect, such alignment was not observed in our ongoing low temperature and low pressure printing project using the same polymer.
The cell organization on the printed scaffold was investigated. Our results demonstrated that the overall cell distribution combines the effect of aligned polymer molecules and printed fiber orientation. As a major factor during extrusion, sheered polymer “fluid” generated an organized micro-environment of aligned polymer molecules that had an impact on cell alignment. As the polymer molecules and polymer chains align, it may influence the regional wettability chemogradient and thus have an impact on the cell adhesion and organization. 27 However, it was noticed that cells also have an intension to bridge into small gaps (Figure 1B). This bridging feature has been found to be favored by cells during settlement because it reduces the unfavorable energy barriers by increases the tension along the unsupported cell membrane as well as reducing the overall adhesion strength. 28 Therefore, on parallel printed scaffolds, cells appeared highly aligned along each printed fiber because in this case, the molecule alignment is the only dominating factor as all printed fibers were separated from each other. For the crosshatch pattern or random pattern, where fibers crossed each other, the resulted small gaps in crossed regions served as another driving force for cell to settle besides the aligned polymer molecules. As a result, when the degree of randomness of the fiber angle increased (Figure 1B, 2B), attached MSCs were found to be less aligned, due to the effect of alignment caused by polymer molecule alignment was compromised by the gaps between crossed fibers favored by cells to reduce the attachment energy. The later driving force became more and more dominating as there were more connections between fibers present. To exclude the effect that the relatively high cell seeding density (107 cells/ml, equivalent to 106 cells/scaffold, with a yield attachment efficiency of 15%) may have a potential to encourage cells to grow in a certain direction as they were packed, lower seeding densities at 106 cells/ml and 105 cells/ml were performed, where same degree of alignment was demonstrated. Such alignment was based on cell adhesion through ECM. The demonstrated alignment phenomenon is believed to apply to other synthetic polymers going through comparable extrusion process. In fact, similar alignment effect was also achieved using scaffolds printed during our ongoing study using poly (L-lactide-co-caprolactone) (PLCL). After treating with an actin inhibitor cytochalasin D to interrupt the focal adhesion, the aligned pattern was completed lost and resulted random cell organization on the printed scaffold.
Next we expanded the cell alignment study in an in vivo rat model by implanting pre-seeded casted and parallel printed scaffolds subcutaneously. The demonstration of maintaining the desired cell orientation in vivo is an essential step before future pre-clinical investigation, so that such potential was assessed in this study. Besides similar cell orientation observed as the in vitro studies, the anisotropic shape of cells on printed scaffold and the more rounded shape on casted scaffold indicated cellular function difference. These results demonstrated that the cell shape and alignment can be maintained or achieved in a in vivo model, which makes this approach very attractive to develop functional complex tissue with layered or controlled cell behavior such as creating zonal cartilage by printing a scaffold containing different patterns in each zonal.
It was previously discovered that the MSC shape as it adhered to a micro-patterned surface affects differentiation, wherein cellular anisotropy was indicated to have an impact on the lineage commitment of MSCs. 29 Although the full mechanism remains unclear, relevant progress made in the past decade indicated that the attached cellular shape might be a result of cytoskeletal tension. 30–32 In this study, we observed variable cell morphology among different printed patterns and printed vs. casted samples, which we hypothesize to be a result of the initial cellular adhesion response to surface environment (Figure 1B, Figure 3B). The cell shape can further influence differentiation capacity. 33–35 In our evaluation of chondrogenic gene expression, variable levels of chondrogenic differentiation of MSCs was demonstrated in the PCR results (Figure 4). We observed that attached MSCs appeared more elongated and aligned on the parallel patterned scaffolds and had enhanced chondrogenic gene marker expression. In particular, the zonal specific marker PRG4 expression associated with the superficial zone, was significantly higher in the parallel scaffold group where chondrocytes aligned parallel to one another. The recapitulation of articular cartilage zonal structure has been a great challenge in the field. The achievement of zonal marker expression as a result of cell alignment is an important step to develop functional cartilage tissue by engineering the cellular microenvironment. Although future steps of more quantitative analysis and further comprehensive research is needed to understand more about how and why MSC morphology has an impact on differentiation, our demonstrated results will help open a new way to create and control such desired cell organization by using 3D printing techniques. Research in the field will greatly benefit the understanding of cell-cell and cell-material interactions and thus provides new concepts and techniques to develop advanced biomaterials for tissue regeneration.
Figure 4. Comparison of chondrogenic gene expression for cells grown on scaffolds with different patterns.
All chondrogenic markers showed different levels of expression as a response to different scaffold patterns and induced cell alignment. In general, positive chondrogenic markers aggrecan and type II collagen expression were up-regulated for the parallel pattern group. Superficial zonal marker PRG4 expression was significantly higher when hMSCs were parallel aligned.
5. Conclusions
The results indicated that PLGA molecules can be forced to align in the extruded direction at high shear conditions (e.g. high pressure and temperature, small needle). This phenomenon further induced the attached cells to align along the extruded fiber, which we propose could be a result of aligned polymer molecules on scaffold surface involved in cell attachment. However, the overall cell distribution on the scaffold can be influenced by the printed fiber organization as well. With the parallel printed pattern, molecule alignment dominates the cell alignment, where with the random pattern, such alignment can be altered as the cells are bridging the small gaps. The cell orientation was maintained in vivo in a subcutaneous implant, making this approach promising for combining with surgical procedures that release native stem cells for improved cartilage repair. The results on the effect of printed pattern on cell organization and differentiation demonstrated that this approach is promising to create scaffolds with controlled cell and ECM arrangement to achieve desired biological function required for chondrogenesis. Furthermore, the polymer molecule and cell alignment achieved by 3D extrusion will have a significant impact on regenerating biomimetic cartilage tissue in a cost and labor effective way, as well as providing a fundamental understanding of cell-material interactions.
Acknowledgments
This work was supported by National Science Foundation grant CBET 1264517, CBET 1604742, National Institute of Biomedical Imaging and Bioengineering, Center for Engineering Complex Tissues (CECT, P41 EB023833), and University of Maryland Graduate School Fellowships. The quantitative analysis Image J plugin package Orientation J was provided by Dr. Michael Unser’s Biomedical Imaging Group at Swiss federal Institute of Technology in Lausanne (EPFL) for free. The authors thank Dr. Feng Gao for his help on quantitative cell alignment data analysis and programming. We would also like to acknowledge the X-ray Crystallographic Center at University of Maryland for conducting SAXS.
Footnotes
Disclosure Statement
No competing financial interests exist.
References
- 1.Guo T, Lembong J, Zhang L, Fisher J. Three-Dimensional Printing Articular Cartilage: Recapitulating the Complexity of Native Tissue. Tissue Engineering Part B-Reviews. 2017;23(3):225–236. doi: 10.1089/ten.TEB.2016.0316. [DOI] [PubMed] [Google Scholar]
- 2.Discher D, Mooney D, Zandstra P. Growth Factors, Matrices, and Forces Combine and Control Stem Cells. Science. 2009;324(5935):1673–1677. doi: 10.1126/science.1171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Q, Peng J, Guo Q, Huang J, Zhang L, Yao J, Yang F, Wang S, Xu W, Wang A, et al. A cartilage ECM-derived 3-D porous acellular matrix scaffold for in vivo cartilage tissue engineering with PKH26-labeled chondrogenic bone marrow-derived mesenchymal stem cells. Biomaterials. 2008;29(15):2378–2387. doi: 10.1016/j.biomaterials.2008.01.037. [DOI] [PubMed] [Google Scholar]
- 4.Kaur G, Valarmathi M, Potts J, Jabbari E, Sabo-Attwood T, Wang Q. Regulation of osteogenic differentiation of rat bone marrow stromal cells on 2D nanorod substrates. Biomaterials. 2010;31(7):1732–1741. doi: 10.1016/j.biomaterials.2009.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park J, Yang H, Woo D, Jeon S, Park K. The promotion of chondrogenesis, osteogenesis, and adipogenesis of human mesenchymal stem cells by multiple growth factors incorporated into nanosphere-coated microspheres. Biomaterials. 2011;32(1):28–38. doi: 10.1016/j.biomaterials.2010.08.088. [DOI] [PubMed] [Google Scholar]
- 6.Discher D, Janmey P, Wang Y. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310(5751):1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 7.Albrecht D, Underhill G, Wassermann T, Sah R, Bhatia S. Probing the role of multicellular organization in three-dimensional microenvironments. Nature Methods. 2006;3(5):369–375. doi: 10.1038/nmeth873. [DOI] [PubMed] [Google Scholar]
- 8.Reilly G, Engler A. Intrinsic extracellular matrix properties regulate stem cell differentiation. Journal of Biomechanics. 2010;43(1):55–62. doi: 10.1016/j.jbiomech.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 9.Boyan B, Hummert T, Dean D, Schwartz Z. Role of material surfaces in regulating bone and cartilage cell response. Biomaterials. 1996;17(2):137–146. doi: 10.1016/0142-9612(96)85758-9. [DOI] [PubMed] [Google Scholar]
- 10.Flemming R, Murphy C, Abrams G, Goodman S, Nealey P. Effects of synthetic micro- and nano-structured surfaces on cell behavior. Biomaterials. 1999;20(6):573–588. doi: 10.1016/s0142-9612(98)00209-9. [DOI] [PubMed] [Google Scholar]
- 11.JULIANO R, HASKILL S. SIGNAL TRANSDUCTION FROM THE EXTRACELLULAR-MATRIX. Journal of Cell Biology. 1993;120(3):577–585. doi: 10.1083/jcb.120.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mousa S, Cheresh D. Recent advances in cell adhesion molecules and extracellular matrix proteins: Potential clinical implications. Drug Discovery Today. 1997;2(5):187–199. [Google Scholar]
- 13.Curtis A, Wilkinson C. Nantotechniques and approaches in biotechnology. Trends in Biotechnology. 2001;19(3):97–101. doi: 10.1016/s0167-7799(00)01536-5. [DOI] [PubMed] [Google Scholar]
- 14.Lizundia E, Saenz-Perez M, Patrocinio D, Aurrekoetxea I, Vivanco M, Vilas J. Nanopatterned polystyrene-b-poly(acrylic acid) surfaces to modulate cell-material interaction. Materials Science & Engineering C-Materials For Biological Applications. 2017;75:229–236. doi: 10.1016/j.msec.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 15.Dalby M, Gadegaard N, Tare R, Andar A, Riehle M, Herzyk P, Wilkinson C, Oreffo R. The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder. Nature Materials. 2007;6(12):997–1003. doi: 10.1038/nmat2013. [DOI] [PubMed] [Google Scholar]
- 16.Yim E, Pang S, Leong K. Synthetic nanostructures inducing differentiation of human mesenchymal stem cells into neuronal lineage. Experimental Cell Research. 2007;313(9):1820–1829. doi: 10.1016/j.yexcr.2007.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stevens MM, George JH. Exploring and engineering the cell surface interface. Science. 2005;310(5751):1135–8. doi: 10.1126/science.1106587. [DOI] [PubMed] [Google Scholar]
- 18.Klein T, Rizzi S, Reichert J, Georgi N, Malda J, Schuurman W, Crawford R, Hutmacher D. Strategies for Zonal Cartilage Repair using Hydrogels. Macromolecular Bioscience. 2009;9(11):1049–1058. doi: 10.1002/mabi.200900176. [DOI] [PubMed] [Google Scholar]
- 19.Lawrence R, Felson D, Helmick C, Arnold L, Choi H, Deyo R, Gabriel S, Hirsch R, Hochberg M, Hunder G, et al. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States. Arthritis and Rheumatism. 2008;58(1):26–35. doi: 10.1002/art.23176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCormick F, Harris JD, Abrams GD, Frank R, Gupta A, Hussey K, Wilson H, Bach B, Cole B. Trends in the surgical treatment of articular cartilage lesions in the United States: an analysis of a large private-payer database over a period of 8 years. Arthroscopy. 2014;30(2):222–6. doi: 10.1016/j.arthro.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 21.Guo T, Holzberg TR, Lim CG, Gao F, Gargava A, Trachtenberg JE, Mikos AG, Fisher JP. 3D printing PLGA: a quantitative examination of the effects of polymer composition and printing parameters on print resolution. Biofabrication. 2017;9(2):024101. doi: 10.1088/1758-5090/aa6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo T, Yu L, Lim C, Goodley A, Xiao X, Placone J, Ferlin K, Nguyen B, Hsieh A, Fisher J. Effect of Dynamic Culture and Periodic Compression on Human Mesenchymal Stem Cell Proliferation and Chondrogenesis. Annals of Biomedical Engineering. 2016;44(7):2103–2113. doi: 10.1007/s10439-015-1510-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vozzi G, Flaim C, Bianchi F, Ahluwalia A, Bhatia S. Microfabricated PLGA scaffolds: a comparative study for application to tissue engineering. Materials Science & Engineering C-Biomimetic and Supramolecular Systems. 2002;20(1–2):43–47. [Google Scholar]
- 24.Kim S, Utsunomiya H, Koski J, Wu B, Cima M, Sohn J, Mukai K, Griffith L, Vacanti J. Survival and function of hepatocytes on a novel three-dimensional synthetic biodegradable polymer scaffold with an intrinsic network of channels. Annals of Surgery. 1998;228(1):8–13. doi: 10.1097/00000658-199807000-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Putnam C, Hammel M, Hura G, Tainer J. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Quarterly Reviews of Biophysics. 2007;40(3):191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 26.Somani R, Hsiao B, Nogales A, Srinivas S, Tsou A, Sics I, Balta-Calleja F, Ezquerra T. Structure development during shear flow-induced crystallization of i-PP: In-situ small-angle X-ray scattering study. Macromolecules. 2000;33(25):9385–9394. [Google Scholar]
- 27.Khang G, Lee SJ, Lee JH, Kim YS, Lee HB. Interaction of fibroblast cells on poly(lactide-co-glycolide) surface with wettability chemogradient. Biomed Mater Eng. 1999;9(3):179–87. [PubMed] [Google Scholar]
- 28.Carman M, Estes T, Feinberg A, Schumacher J, Wilkerson W, Wilson L, Callow M, Callow J, Brennan A. Engineered antifouling microtopographies - correlating wettability with cell attachment. Biofouling. 2006;22(1):11–21. doi: 10.1080/08927010500484854. [DOI] [PubMed] [Google Scholar]
- 29.Peng R, Yao X, Ding J. Effect of cell anisotropy on differentiation of stem cells on micropatterned surfaces through the controlled single cell adhesion. Biomaterials. 2011;32(32):8048–8057. doi: 10.1016/j.biomaterials.2011.07.035. [DOI] [PubMed] [Google Scholar]
- 30.Kilian K, Bugarija B, Lahn B, Mrksich M. Geometric cues for directing the differentiation of mesenchymal stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(11):4872–4877. doi: 10.1073/pnas.0903269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yim E, Darling E, Kulangara K, Guilak F, Leong K. Nanotopography-induced changes in focal adhesions, cytoskeletal organization, and mechanical properties of human mesenchymal stem cells. Biomaterials. 2010;31(6):1299–1306. doi: 10.1016/j.biomaterials.2009.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thery M, Pepin A, Dressaire E, Chen Y, Bornens M. Cell distribution of stress fibres in response to the geometry of the adhesive environment. Cell Motility and the Cytoskeleton. 2006;63(6):341–355. doi: 10.1002/cm.20126. [DOI] [PubMed] [Google Scholar]
- 33.McBeath R, Pirone D, Nelson C, Bhadriraju K, Chen C. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Developmental Cell. 2004;6(4):483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 34.Ruiz S, Chen C. Emergence of Patterned Stem Cell Differentiation Within Multicellular Structures. Stem Cells. 2008;26(11):2921–2927. doi: 10.1634/stemcells.2008-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao L, McBeath R, Chen C. Stem Cell Shape Regulates a Chondrogenic Versus Myogenic Fate Through Rac1 and N-Cadherin. Stem Cells. 2010;28(3):564–572. doi: 10.1002/stem.308. [DOI] [PMC free article] [PubMed] [Google Scholar]