Summary paragraph
Ebola virus (EBOV) in humans causes a severe illness with high mortality rates. Several strategies have been developed in the past to treat EBOV infection, including the antibody cocktail ZMappTM that has been shown to be effective in nonhuman primate models of infection1 and has been used under compassionate-treatment protocols in humans2. ZMappTM is a mixture of three chimerized murine monoclonal antibodies (mAbs)3–6 that target EBOV-specific epitopes on the surface glycoprotein (GP)7,8. However, ZMappTM mAbs do not neutralize other species from the Ebolavirus genus, such as Bundibugyo virus (BDBV), Reston virus (RESTV) or Sudan virus (SUDV). Here we describe three naturally-occurring human cross-neutralizing mAbs, from BDBV survivors, that target an antigenic site in the canonical heptad repeat 2 (HR2) region near the membrane proximal external region (MPER) of GP. The identification of a conserved neutralizing antigenic site in the GP suggests that these mAbs could be used to design universal antibody therapeutics against diverse ebolavirus species. Furthermore, we found that immunization with a peptide comprising the HR2/MPER antigenic site elicits neutralizing antibodies in rabbits. Structural features determined by conserved residues in the antigenic site described here could inform an epitope-based vaccine design against infection caused by diverse ebolavirus species.
Main text
In previous work, we described a large panel of neutralizing mAbs using peripheral blood B cells from survivors of the 2007 BDBV outbreak in Uganda9. Three non-clonally related cross-reactive mAbs designated BDBV223, BDBV317 and BDBV340 were isolated from two donors who survived BDBV infection. These three mAbs competed for binding with BDBV-specific neutralizing mAbs that bound to an epitope extending down below the base of GP, a domain that is poorly characterized as an antigen in filovirus GPs9.
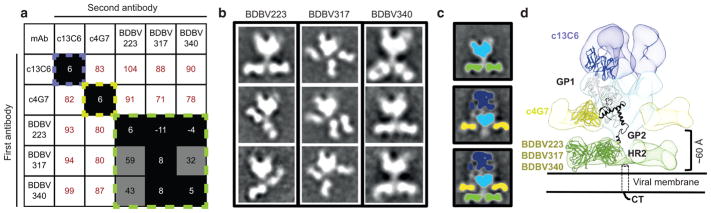
To delineate the binding sites of these cross-reactive mAbs on GP, we performed quantitative competition-binding assays with BDBV223, BDBV317 and BDBV340 as well as previously described ebolavirus-specific mAbs targeting the glycan cap or the GP base (Fig. 1a). As expected, BDBV223, BDBV317 and BDBV340 did not compete with either the glycan cap specific mAb c13C6 or the base region-binding mAb c4G7 (Fig. 1a). To determine the location of the cross-reactive antigenic site on the GP stem region targeted by these human mAbs, we performed negative-stain single-particle electron microscopy (EM) studies. The EM class averages confirmed that BDBV223, BDBV317, and BDBV340 each bind to the bottom of the GP, in the canonical heptad repeat 2 (HR2) domain near the MPER (Fig. 1b,c). To confirm their location on GP, we overlayed a class average of BDBV223 Fab bound to BDBV GP with a class average of c13C6 Fab:c4G7 Fab bound to EBOV GP. This showed that BDBV223, BDBV317, and BDBV340 bound to a region on GP closer to the viral membrane, well below the epitope of the mAb c4G7 site of vulnerability at the GP1/GP2 interface (Fig. 1d)7. Measurements of the distance from the bottom of the GP globular head to the mid-point of the Fab in the class averages showed a distance of ~60 Å, which indicates that Fabs bind to the HR2 region (PDBID 5JQ3)10, which was also previously crystalized as post-fusion GP2 (PDBID 1EBO)11 (Fig. 1d). Three BDBV317 Fabs could be seen in the class average images, indicating that three antibody molecules can bind simultaneously to the HR2/MPER region of GP trimer (Fig. 1b).
Figure 1. Cross-reactive neutralizing antibodies from BDBV survivors bind near the membrane proximal region of GP.
a) Data from competition-binding assays using BDBV223, BDBV317 or BDBV340; antibodies from ZMappTM cocktail (c2G4 and c13C6). Numbers indicate the percent binding of the second mAb in the presence of the first mAb, compared to binding of second mAb alone. MAbs were judged to compete for the same site if maximum binding of the second mAb was reduced to <30% of its un-competed binding (black boxes with white numbers). MAbs were considered non-competing if maximum binding of the second mAb was >70% of its un-competed binding (white boxes with red numbers). Grey boxes with black numbers indicate an intermediate phenotype (between 30 and 70% of un-competed binding). Multiple readings were used to determine the maximum signal for each mAb alone and a single reading of a mAb in combination with each competing antibody was recorded.
b) Representative negative stain class averages of antibodies that bind GP2 exclusively in the HR2/MPER region. Complexes are of BDBV Fabs bound to BDBV GPΔmuc.
c) (Top) A class average of BDBV GPΔmuc bound to BDBV223 demonstrates the location of each component, with the core GP colored blue and the Fabs in green. (Middle) A class average of c13C6 Fab:c4G7 Fab bound to EBOV GPΔTM7 (with c13C6 in dark blue, c4G7 in yellow and GP core in light blue). (Bottom) Overlaying a class average of c13C6 Fab:c4G7 Fab bound to EBOV GPΔTM7 (with c13C6 in dark blue, c4G7 in yellow and GP core in light blue) over a class average of BDBV223 Fab bound to BDBV GPΔmuc (with BDBV223 in green and GP core in light blue), demonstrates that BDBV223 binds significantly lower down on GP, well below the epitope of the c4G7 site of vulnerability at the GP1/GP2 interface.
d) A model of the c13C6 Fab:c4G7 Fab bound to EBOV GPΔTM (EMDB ID-6152, Fab variable regions from PDB 5KEN, EBOV GPΔMuc from PDB 5JQ3) is shown with the relative location of BDBV223/317/340 Fabs (segmented c4G7 Fab from the above map placed in the relative location on GP as indicated by class averages, representative Fab from PDB 3CSY).
As BDBV223, BDBV317 and BDBV340 mAbs reacted with all three GPs from BDBV, EBOV and SUDV9, we next sought to determine whether these mAbs possess cross-neutralizing capacity. All three human mAbs, which were isolated from a BDBV-immune donor, neutralized BDBV as well as the heterologous Mayinga (historical) and Makona-Gueckedou-C07 (contemporary) strains of EBOV (Fig. 2A, Supplementary Figure 1). BDBV317 also neutralized two strains of Reston virus (RESTV). Although these human mAbs recognized SUDV GP in ELISA9, they failed to neutralize SUDV (Fig. 2a). Previously, we identified species-specific neutralizing human mAbs that bound the BDBV GP near the MPER region and competed for binding with cross-reactive mAbs BDBV223, BDBV317 and BDBV340, although their epitopes were not determined9. Recently, another group isolated several EBOV neutralizing mAbs that recognize the GP stalk region12. One HR2/MPER-specific mAb (ADI-16061) from that antibody panel was reported to neutralize vesicular stomatitis viruses bearing EBOV, BDBV, and Taï Forest virus (TAFV) GPs13. Also, a previous study described generation of a rabbit cross-reactive non-neutralizing mAb 15H10 that bound to peptides corresponding to the HR2/MPER region14. Therefore, the HR2/MPER region is a complex antigenic site on GP that could be targeted by species-specific or cross-reactive as well as neutralizing or non-neutralizing antibodies.
Figure 2. Neutralization and protective efficacy of HR2/MPER-specific mAbs.
a) Neutralization activity of BDBV223, BDBV317 or BDBV340 against BDBV, EBOV, RESTV or SUDV. Means ± SD of triplicates or quadruplicates are shown.
b) Post-exposure protection of EBOV-inoculated mice treated with BDBV223, BDBV317, BDBV340 or a control mAb 2D22 one day after virus challenge (n = 5 animals/group). Kaplan-Meier survival curves are shown.
c) Post-exposure protection of EBOV-inoculated guinea pigs treated with BDBV223, BDBV317, or a control mAb 2D22 one day after EBOV challenge (n = 5 animals/group). Kaplan-Meier survival curves are shown.
d) Post-exposure protection of BDBV-inoculated ferrets treated with BDBV223 or a control 2D22 mAb at days 3 and 6 after BDBV challenge (n = 4 animals/group). Kaplan-Meier survival curves are shown.
Separate two-sided log-rank (Mantel-Cox) tests were used for pairwise comparisons of survival curves between each BDBV mAb-treated group and control 2D22-treated mAb group (b-d). P-values were not adjusted for multiple comparisons. P ≤0.05 considered significant; ns – not significant.
To determine the protective capacity of HR2/MPER-specific mAbs, we tested cross-reactive antibodies in mice against a heterologous mouse-adapted EBOV, strain Mayinga. Two mAbs (BDBV223 and BDBV317) provided complete protection from death when delivered 1 day after challenge with heterologous EBOV (Fig. 2b, Supplementary Figures 2). BDBV223 and BDBV317 also conferred partial protection against heterologous EBOV in guinea pigs when delivered 24 hours after the virus challenge (Fig. 2c, Supplementary Figure 2). Finally, we tested BDBV223 in a ferret model of BDBV infection, which is one of only two models described for BDBV challenge (the other is non-human primates)15,16. Animals were treated at day 3 and 6. All animals treated with the control mAb (2D22, Dengue virus-specific) succumbed to death at day 7 following challenge. In contrast, 2 of 4 animals treated with mAb BDBV223 survived the infection (Fig. 2d).
To map the epitope of cross-reactive human mAbs, we used a comprehensive alanine scanning approach and tested the binding of BDBV223, BDBV317, and BDBV340 to individual members of a shotgun mutagenesis alanine scan library of EBOV GP. Consistent with the EM data, we identified critical contact residues in GP2 for BDBV223, BDBV317, and BDBV340 that spanned the C-terminal part of HR2 region and the N-terminal part of the MPER region (Fig. 3a). Single critical residues in the HR2 region were identified for BDBV317 (K633) and BDBV340 (D624). Two critical residues (D624 and D632) were identified for BDBV223 (Fig. 3a, Supplementary Table 1). For comparison we also indicate the location of several critical residues identified previously for c4G7, including D552 in the internal fusion loop and C556, which is part of the HR18.
Figure 3. Structural and functional analysis of GP residues important for mAb cross-reactivity and neutralization.
a) Sequence alignment of GP2 from BDBV, EBOV, RESTV and SUDV. The numbers above the sequence correspond to the amino acid position in GP. Amino acids identical to BDBV are indicated by dots. Color-coded circles indicate the position of residues at which alanine substitutions disrupt mAb binding, as determined by alanine-scanning mutagenesis (ASM). Color-coded triangles indicate the residues at which mutations disrupt the binding to peptides in ELISA (PE) and color-coded squires indicate escape mutations for BDBV223, BDBV317 or BDBV340. BDBV1p, BDBV2p, EBOV2p or SUDV2p peptide sequences analyzed are indicated by grey, black, blue or purple lines, respectively. Vertical lines indicate C-terminal ends of the GP constructs used for ELISA binding assay in Figure 4.
b) Neutralization activity of BDBV223, BDBV317 or BDBV340 against wild-type BDBV (black), BDBV223 (green), BDBV317 (blue) or BDBV340 (purple) escape mutants. Means ± SD of triplicates are shown.
c) Binding of BDBV223, BDBV317 or BDBV340 to BDBV1p (grey), BDBV2p (black), EBOV2p (brown) or SUDV2p (orange) peptides. Means ± SD of quadruplicates are shown.
d) Binding of BDBV223, BDBV317 or BDBV340 to HR2/MPER peptides. The EC50 value for each peptide-mAb combination is shown. EC50 values greater than 20,000 are indicated (>).
We passaged the chimeric filovirus with BDBV GP17 in the presence of HR2/MPER-specific mAbs and generated antibody escape mutant viruses for mAbs BDBV223 (an isolate with a P634H mutation), BDBV317 (an isolate with the K633R mutation), and BDBV340 (an isolate with the D522E mutation). We found that BDBV223 P634H escape mutant resisted neutralization by all three HR2/MPER mAbs. BDBV223 and BDBV340 neutralized the BDBV317 escape mutant virus with about 100-fold lower potency than wild-type virus (Fig. 3b). The BDBV340 escape mutant virus (D522E) was completely resistant to neutralization by BDBV340 and BDBV317 and only weakly neutralized by BDBV223. Interestingly, the D522E mutation is located in the internal fusion loop region outside of the expected epitope for these mAbs and is likely to cause conformational changes affecting binding of HR2/MPER-specific mAbs.
According to several crystallographic studies, the HR2 α-helix ends around residue D63210,11,18, indicating that HR2/MPER peptides might adopt an α-helical conformation that is recognized by the ebolavirus cross-reactive mAbs. To determine whether BDBV223, BDBV317 or BDB340 can recognize a linear conserved epitope in the HR2/MPER region, we synthesized peptides spanning the GP amino acids 620-635 in the HR2/MPER region of three virus species (designated peptides BDBV2, EBOV2 and SUDV2) as well as a control peptide from the N-terminus of HR2 region (amino acids 599-613), designated the BDBV1 peptide. MAbs BDBV223, BDBV317 and BDBV340 recognized the BDBV2 peptide containing critical residues determined by alanine-scanning mutagenesis, but not the negative control BDBV1 peptide from the HR2 region, as expected (Fig. 3c). While BDBV223 bound to BDBV2, EBOV2 and SUDV2 peptides, BDBV317 and BDBV340 recognized only BDBV2 and EBOV2 peptides (Fig. 3c,d). Binding capacity of the three mAbs to the HR2/MPER peptides (Fig. 3c,d) were consistent with their neutralization potencies in vitro (Fig. 2a). Thus, BDBV317 bound equally well to BDBV2 and EBOV2 peptides and neutralized BDBV and EBOV with the same potency. BDBV223 and BDBV340 bound better to the BDBV2 peptide, and the same antibodies neutralized BDBV with higher potency than EBOV. These results suggest that the mAbs recognize a relatively linear peptide epitope in the HR2/MPER region. In a recent study, the HR2 and the N-terminal part of the MPER was reported to be proximal to the lipid bilayer19, which would make the HR2/MPER epitope accessible for antibody binding on the viral surface.
To determine the basis of mAb cross-reactivity to multiple ebolavirus species, we synthesized a panel of chimeric BDBV peptides containing polymorphic residues from EBOV or SUDV. There are two amino acid differences between the BDBV2 and EBOV2 peptides (at residues 631 and 634), located in the C-terminus of the peptide (Fig. 3a,d). To determine which substitutions were responsible for the better binding of BDBV223 to the BDBV2 peptide, we synthesized two chimeric BDBV peptides with single EBOV substitutions at residues 631 or 634 (designated peptides BDBV2A and BDBV2B). While BDBV223 bound equally well to BDBV2 and BDBV2A peptides, it bound relatively weakly to the BDBV2B peptide, suggesting that P634T substitution in BDBV2B peptide is responsible for reduced binding of BDBV223 to the EBOV2 peptide (Fig. 3c,d). We also synthesized chimeric BDBV2C and BDBV2D peptides to introduce two SUDV substitutions at residues 624 and 633. While both BDBV223 and BDBV317 bound equally well to BDBV2D and BDBV2 peptides, they failed to recognize the BDBV2C peptide, suggesting that the N624 residue is critical for binding of SUDV by MPER/HR-specific mAbs (Fig. 3d). To investigate the role of residue D624 in the context of live virus neutralization, we constructed a recombinant BDBV bearing D624N mutation, which completely resisted neutralization with BDBV223 and BDBV340 but was sensitive to BDBV317 (Supplementary Figure 3).
To determine the immunogenicity of HR2/MPER peptides, we set up immunization experiments with the BDBV2 (Fig. 4, animals 1 and 2) and SUDV2 (Fig. 4, animals 3 and 4) peptides conjugated to keyhole limpet hemocyanin (KLH) through a PEG6 linker. Immunization of rabbits with the both peptides elicited a peptide-reactive IgG response in serum (Fig. 4a). We next isolated affinity-purified polyclonal Abs using a BDBV2p- or SUDV2p-coupled resin for capturing antibodies binding to HR2/MPER peptides and filovirus GPs. In ELISA, the immunoaffinity-purified polyclonal Abs displayed broad cross-reactivity, binding to peptides (BDBV2p, EBOV2p, and SUDV2p) and GPs (BDBV, EBOV, and SUDV) from multiple ebolavirus species (Fig. 4b,c). Finally, we tested polyclonal Abs in a neutralization experiment against BDBV, EBOV and SUDV. Immunization with BDBV2p-PEG6-KLH or SUDV2p-PEG6-KLH formulations elicited BDBV- or SUDV-specific neutralizing pAbs (Fig. 4d), suggesting the feasibility of an HR2/MPER peptide-based vaccine approach to elicit ebolavirus-specific neutralizing Abs. We also detected weak neutralization against heterologous ebolavirus species at the highest Ab concentrations tested in animals 1, 2 and 4. Animal 3 immunized with SUDV2p-PEG6-KLH formulation, developed cross-neutralizing response to three ebolavirus species: BDBV, EBOV, and SUDV. Future studies are needed to determine the optimal conformation of HR2/MPER peptide and formulation of peptide immunogen to elicit potent ebolavirus cross-neutralizing Ab response.
Figure 4. Immunization with HR2/MPER peptide elicits peptide and protein antigen-reactive and neutralizing antibody response.
a) Pre-immune serum (black) or immune serum (grey) samples from animals were tested in ELISA for binding to BDBV2p (animals 1 and 2) or SUDV2p (animals 3 and 4) peptides. Means ± SD of quadruplicates are shown.
b) Binding of rabbit polyclonal Abs to BDBV2p (black), EBOV3p (brown), SUDV2p (orange), or BDBV1p (grey) peptides. Means ± SD of quadruplicates are shown.
c) Binding of rabbit polyclonal Abs to BDBV (black), EBOV (brown), SUDV (orange), or MARV (grey) glycoproteins. Means ± SD of quadruplicates are shown.
d) Neutralization activity of polyclonal Abs against BDBV (black), EBOV (brown), or SUDV (orange). Means ± SD of triplicates are shown.
Here we describe human cross-reactive, neutralizing, and protective monoclonal antibodies that target an antigenic region near the MPER region of EBOV GP. These antibodies recognize a distinct antigenic site, and, therefore, do not compete for binding with therapeutic antibodies used in the past to treat EBOV infection. This finding suggests that MPER-specific mAbs could be used to design a new antibody therapeutic cocktail with broader potency against multiple ebolaviruses. Increased breadth and potency can be achieved by combining HR2/MPER-specific mAbs with previously described potent cross-reactive mAbs that target other conserved epitopes in glycan cap9,12,13, GP base12,13, and internal fusion loop20. Alternatively, the mAbs described here might be beneficial to include in existing experimental therapeutic antibody cocktails to increase the breadth of those cocktails.
The MPER region in the GP of enveloped viruses is increasingly recognized as an important region for recognition by broad and potent human mAbs. Several mAbs that neutralize a broad range of HIV21,22 isolates are known to bind conserved epitopes in the HIV gp41 MPER region. The identification of potent HIV MPER-specific mAbs has spurred important progress in the effort to design HIV vaccines rationally using antigens designed to induce such mAbs23. Neutralizing mAbs that recognize the influenza hemagglutinin surface protein stem region24–26 also have spurred research into the possibility of a universal influenza vaccine. Here we report the structural and function features of conserved epitopes in the MPER region of EBOV GP that could inform the design of an MPER-based universal vaccine effective against multiple filoviruses.
Materials and Methods
Ethics Statement
The animal protocols for testing of mAbs in mice, guinea pigs and ferrets were approved by the Institutional Animal Care and Use Committee of the University of Texas Medical Branch (UTMB) in compliance with the Animal Welfare Act and other applicable federal statutes and regulations relating to animals and experiments involving animals. Challenge studies were conducted under maximum containment in an animal biosafety level 4 (ABSL-4) facility of the Galveston National Laboratory, UTMB.
Cell lines
Vero-E6 cell line was obtained from the American Type Culture Collection and cultured in Minimal Essential Medium (MEM) (ThermoFisher Scientific) supplemented with 10% fetal bovine serum (HyClone) and 1% penicillin-streptomycin at 5% CO2, 37°C. FreeStyle 293F cell line was purchased from Thermo Fisher Scientific and cultured according to the manufacturer’s protocol. Drosophila Schneider S2 cells for protein expression were grown in Insect-XPRESSTM protein-free medium with L-glutamine (Lonza) supplemented with puromycin (6 μg/mL) (InvivoGen) at 27˚C. All cell lines were tested on a monthly basis for Mycoplasma and found to be negative in all cases.
Human mAb and Fab expression and purification
Human hybridoma cell lines were expanded in post-fusion medium, as previously described27. HiTrap Protein G or HiTrap MabSelectSure columns (GE Healthcare) were used to purify antibodies from filtered culture supernatants. Fab fragments were generated by papain digestion, as described previously 27.
Expression and purification of filovirus GPs
The ectodomains of EBOV GPΔTM (residues 1-636; strain Makona; GenBank: KM233070), BDBV GPΔTM (residues 1-643; strain 200706291 Uganda; GenBank: NC_014373), and SUDV GPΔTM (residues 1-637, strain Gulu; GenBank: NC_006432) were expressed transiently in Expi293F cells with a C-terminus strep II tag using the pcDNA3 plasmid vector. Secreted proteins were purified using a 5 mL StrepTrap HP column (GE Healthcare) following the manufacturer’s protocol, and then purified further and buffer exchanged into PBS using Superdex200 (GE Healthcare) size exclusion chromatography (SEC). Recombinant BDBV GP ectodomain lacking residues 312–463 of the mucin-like domain (GPΔmuc)28,29 was produced by transfection of Drosophila Schneider 2 (S2) cells with modified pMTpuro vectors, followed by stable selection of transfected cells with 6 μg/mL puromycin. Secreted GP ectodomain expression was induced with 0.5 mM CuSO4 for 4 d. Proteins were engineered with a modified double strep tag at the C terminus (enterokinase cleavage site followed by a strep tag/linker/strep tag) to facilitate purification using Strep-Tactin resin (Qiagen). Proteins were purified further by Superdex 200 (S200) SEC in 10 mM Tris and 150 mM NaCl, pH 7.5 (1× TBS).
Antibody neutralization experiments
All work with filoviruses, including the chimeric filoviruses, was performed in the BSL-4 facility of the Galveston National Laboratory. Antibody neutralization assays were performed against the recombinant EBOV strain Mayinga expressing green fluorescent protein from an added gene30 and its derivatives in which GP was replaced with its counterpart from BDBV (strain 200706291 Uganda) or SUDV (strain 200011676 Gulu)17 to which we refer as EBOV, BDBV and SUDV for simplicity. In some antibody neutralization experiments, biological isolates of additional viruses were used: EBOV strain Mayinga (Fig. 2A), EBOV strain Makona-Gueckedou-C07, accession number KJ660347.2 (Supplementary Figure 1), REST strain 119810 Reston RIID Mky 53 (Fig. 2A) and REST strain 200809254 Plum Island (Philippines) (Supplementary Figure 1). The assays were performed in a high-throughput format, as previously described17, or using a standard plaque reduction assay as previously described9.
Generation of recombinant BDBV with the D624N mutation
To introduce the D624N mutation in BDBV GP of the chimeric filovirus used in the study17 the pEBOwtΔBamHI-SbfI,AscI-PspOMI subclone of the full-length clone encoding the viral genome17 was mutagenized using the QuikChange site-directed mutagenesis kit (Stratagene). Then the ApaI-SacI fragment of the subclone, which includes the mutated GP, was used to replace the corresponding fragment of the chimeric filovirus full-length clone. The mutated chimeric virus was recovered as previously described31.
Generation and testing of antibody escape filovirus mutants
To generate escape mutants, 100 PFU of recombinant chimeric EBOV with GP derived from BDBV were combined with 2-fold dilutions of mAbs starting at 200 μg/mL in U-bottom 96-well plates and incubated for 1 hr at 37°C. Mixtures were placed on Vero-E6 cell monolayer cultures in 96-well plates and incubated for 1 hr. Supernatants were removed, fresh mAbs were added at the same concentrations in 200 μL of MEM supplemented with 2% FBS, and plates were incubated for 7 days at 37°C. Viruses that replicated in the presence of the highest concentrations of mAbs, as determined by UV microscopy, were collected. 20 μL aliquots were incubated with 2-fold dilutions of mAbs starting at 200 μg/mL, and viruses were propagated in the presence of mAbs as above. The procedure was repeated once more with mAb dilutions starting at 400 μg/mL. Viruses that replicated at the highest mAb concentrations were amplified in Vero-E6 cell culture monolayers in 24-well plates in the presence of mAbs at 200 μg/mL for 7 days. Cells were used for isolation of RNA using TRIzol reagent, and GP gene ORFs were PCR-amplified and sequenced. To determine susceptibility of the isolated escape mutants to mAbs, 100 PFU of the viruses in MEM supplemented with 2% FBS in triplicate were combined in U-bottom 96-well plates with 8 to 12 two-fold dilutions of mAbs, starting at 200 μg/mL, in total volumes of 50 μL, and incubated for 1 hr at 37°C. The virus/antibody mixtures then were placed in triplicate Vero-E6 cell culture monolayers in 96-well plates, incubated for 1 hr at 37°C, washed with MEM, overlaid with 200 μL of MEM containing 2% FBS and 0.8% methylcellulose, and incubated for 48 hrs at 37°C. Plates were fixed with 10% phosphate-buffered formalin (Fisher) and taken out of the BSL-4 facility according the UTMB BSL-4 standard operating procedures. Plaques were counted using a fluorescence microscope.
Biolayer interferometry competition binding assay
Competition-binding studies using biolayer interferometry and biotinylated EBOV GP (EZ-link® Micro NHS-PEG4-Biotinylation Kit, Thermo Scientific #21955) (5 μg/mL) were performed on an Octet RED biosensor (ForteBio Menlo Park, CA), as described previously27. In brief, the antigen was immobilized onto streptavidin-coated biosensor tips. After a brief washing step, biosensor tips were immersed first into the wells containing primary antibody at a concentration of 100 μg/mL and then into the wells containing competing mAbs at a concentration of 100 μg/mL. The percent binding of the competing mAb in the presence of the first mAb was determined by comparing the maximal signal of competing mAb applied after the first mAb complex to the maximal signal of competing mAb alone.
Electron microscopy
To determine the epitope of HR2/MPER-directed mAbs, BDBV223, 317, or 340 Fabs were generated as described above and added in 10 M excess to BDBV GPΔmuc and allowed to bind overnight at 4°C9. Complexes were subsequently purified by size exclusion chromatography on an S200 Increase column (GE) and stained as previously described7. Particles were visualized using an FEI Tecnai Spirit electron microscope operating at 120kV and images were collected on a TVIPS TemCam-F416 (4k x 4k) CCD camera using Leginon32 with the following settings: magnification of 52,000X that resulted in a pixel size of 2.05Å at the specimen plane, a constant defocus of -1.00 μm and an electron dosage of ~30e−/Å2. Images were processed using the Appion platform33. Particles were picked using DoG Picker34, stacks were created and 2D reference-free class averages were generated using iterative MRA-MSA35. For all complexes, there was a strong bias toward side-views. Further, the region containing the HR2/MPER epitope in our soluble GP constructs is flexible, as indicated by the variety of positions that bound Fabs adopted in the class averages. Therefore, our data was refractory to a reconstruction, although class averages could be compared to previous class averages of known complexes to determine the spatial location of the epitope on GP.
Epitope mapping using an EBOV GP alanine-scan mutation library
Epitope mapping was carried out as described previously8. Comprehensive high-throughput alanine scanning (‘shotgun mutagenesis’) was carried out on a full-length EBOV GP expression construct (based on the Yambuku-Mayinga variant GP sequence), mutagenizing GP residues 33-676 to create a library of clones, each representing an individual point mutant. Residues were changed to alanine (with alanine residues changed to serine). The resulting library, covering 641 of 644 (99.5%) of target residues was arrayed into 384-well plates, one mutant per well, then transfected into HEK-293T cells and allowed to express for 22 hours. Cells, unfixed or fixed in 4% paraformaldehyde, were incubated with primary antibody then with an Alexa Fluor 488-conjugated secondary antibody (Jackson ImmunoResearch Laboratories, Westgrove, PA). After washing, cellular fluorescence was detected using the Intellicyt high throughput flow cytometer (Intellicyt, Albuquerque, NM). MAb reactivity against each mutant EBOV GP clone was calculated relative to wild-type EBOV GP reactivity by subtracting the signal from mock-transfected controls and normalizing to the signal from wild-type GP-transfected controls.
Mutated residues within clones were identified as critical to the mAb epitope if they did not support reactivity of the test mAb but did support reactivity of other control EBOV mAbs. This counter-screen strategy facilitated the exclusion of GP mutants that were misfolded locally or that exhibited an expression defect. The detailed algorithms used to interpret shotgun mutagenesis data were described previously36.
Peptide synthesis and purification
Peptides were synthesized using standard Fmoc solid-phase methods on a Peptide Machines Discovery-4 synthesizer on rink resin37–41. All coupling reactions were performed with ten-fold excess (vs. load capacity of the resin) of activated amino acid (Aapptec or Advanced Chemtech), using FMOC amino acids/HBTU/HoBt/DIEA (1:1:1:2.5) in DMF for 60 minutes. Deprotection of the FMOC group was accomplished in 20% v/v piperidine diluted in DMF for 30 minutes. Peptides used for ELISA experiments were acetylated at the N-terminus by reaction with 1:1:2 v/v acetic anhydride:DIEA:DMF. The peptides were cleaved by exposure to a 90:5:3:2 v/v mixture of trifluoroacetic acid, thioanisole, ethanedithiol, and anisole for two hours. Peptides were precipitated by addition of cold diethyl ether, lyophilized and purified by reverse-phase HPLC (Waters Prep LC 4000) equipped with a Waters 2487 detector and C18 column. Final purified fractions were lyophilized until further use. Isolation of the target peptide was confirmed by MALDI mass spectrometry.
Half maximal effective concentration (EC50) ELISA binding analysis
Soluble forms of the full-length extracellular domain of BDBV, EBOV, SUDV or MARV GPs or corresponding peptides were coated overnight onto 384-well plates at 1 μg/mL. For EC50 binding analysis by ELISA, purified antibodies were applied to the plates at a concentration range of 30 μg/mL to 170 ng/mL, using three-fold serial dilutions. The presence of antibodies bound to the GP was determined using goat anti-human IgG alkaline phosphatase conjugate and p-nitrophenol phosphate substrate tablets, with optical density read at 405 nm after 120 minutes. A non-linear regression analysis was performed on the resulting curves using Prism version 5 (GraphPad) to calculate EC50 values.
Testing of mAbs in vivo
The 7–8-week-old BALB/c female mice (Charles River Laboratories) at five animals per group were injected with 1,000 PFU of the mouse-adapted EBOV, strain Mayinga, by the intraperitoneal route. 24 hours later, animals were injected with individual mAbs at 100 μg per animal by the intraperitoneal route. Animals treated with 2D22 mAb served as controls. Mice were monitored twice daily from day 0 to day 14 post challenge, followed by once daily monitoring from day 15 to the end of the study at day 28. The disease was scored using the following parameters: dyspnea (possible scores 0–5), recumbency (0–5), unresponsiveness (0–5), and bleeding/hemorrhage (0–5). All mice were euthanized at day 28 post EBOV challenge. To test the protective efficacy of mAbs in guinea pigs (strain Hartley), five- to six-week-old female guinea pigs at five animals per group were injected with 1,000 PFU of guinea pig-adapted EBOV, strain Mayinga, by the intraperitoneal route. 24 hours later, animals were injected with individual mAbs at 5 mg per animal by the intraperitoneal route. Animals were monitored and weighed daily for 28 days. After animals became symptomatic, they were examined no less than twice per day. The disease was scored using the same parameters as used for mice. All guinea pigs were euthanized at day 28 post EBOV challenge. The animals were randomly assigned for the experimental groups. UTMB Animal Resource Center veterinary staff was blinded for the antibodies’ administration to experimental groups.
Ferret challenge with BDBV
Groups of 6-month-old male and female animals (Mustela putorius furo, Marshall BioResources) were challenged intramuscularly with 1,000 pfu of BDBV as described previously16. Animals were treated with 20 mg of BDBV223 or control 2D22 antibody on day 3, and the same dose of the antibody on day 6 after challenge. The disease scores were assessed as follows: healthy, 1; developing clinical disease, 2; advanced disease, 3; moribund, 4. Ferrets were monitored for 28 days after infection and then euthanized. Five animals per experimental group were used, based on 100% mortality following infection with filoviruses, which allowed statistical assessment at the 95% confidence level (1-tailed Fisher exact test). The animals were randomly assigned for the experimental groups. UTMB Animal Resource Center veterinary staff was blinded for the antibodies’ administration to experimental groups.
Analysis of viremia by plaque assay
Virus titration was performed on serum samples collected from the guinea pigs and ferrets in Vero-E6 cells by plaque assay as previously described17 with some modifications. Briefly, duplicate 10-fold serial dilutions of sera were adsorbed to Vero-E6 cell monolayers in 96 well plates for 1 hour, covered with 100 μl of 0.9% methylcellulose (Sigma-Aldrich, St. Louis, MO) overlay and incubated at 37°C for 6 days. The overlay was removed, cell monolayers were fixed with formalin for 24 hours, and plates were brought out of BSL-4 as per UTMB BSL-4 standard operating procedures. Fixed cell monolayers were washed three times with phosphate buffered saline, blocked for 1 hour with 5% skim milk in phosphate buffered saline containing 0.1% Tween-20, and plaques were immunostained with rabbit-anti GP primary antibody at 1:5,000 (IBT Bioservices, Rockville, MD) followed by goat-anti rabbit secondary antibody labeled with horseradish peroxidase at 1:1,000 (Southern Biotech, Birmingham, AL), both antibodies diluted in 5% skim milk in phosphate buffered saline containing 0.1% Tween-20. Virus plaques were visualized by staining with 4CN two component peroxidase substrate system (Seracare, Milford, MA).
Polyclonal Ab generation and characterization
New Zealand white rabbits (3–4 month-old) were immunized subcutaneously with 0.2 mg of HR2/MPER peptide-KLH conjugate consisting of BDBV2p peptide (TDKIDQIIHDFIDKPL) or SUDV2 peptide (TDKINQIIHDFIDNPL) conjugated to KLH through C-terminal cysteine attached to Peg6 linker (NH-(PEG)6-CH2CH2COOH) in Freund’s complete adjuvant (GenScript, polyclonal antibody service). Subsequently, the animals were boosted three times subcutaneously with the same conjugates in Freund’s incomplete adjuvant. Filter sterilized pre-immune and test antiserum after the second boost were tested in ELISA for the presence of polyclonal Abs to BDBV2p or SUDV2p peptides. Polyclonal Abs were purified using BDBV2p or SUDV2p peptides conjugated to iodoacetyl resin (GenScript, polyclonal antibody service). Purified pAbs were tested in ELISA for binding to BDBV2p, EBOV2, SUDV2 or BDBV1 peptides or filovirus GPs. The presence of antibodies bound to the antigen was determined using mouse anti-rabbit IgG horseradish peroxidase conjugate (GenScript, A01827-200). Polyclonal Abs were tested in a neutralization assay against the recombinant EBOV expressing green fluorescent protein from an added gene30 and its derivatives in which GP was replaced with its counterpart from BDBV or SUDV.
Data availability
The data that support the findings of this study are available from the corresponding authors upon request.
Supplementary Material
Acknowledgments
This project received support from the Defense Threat Reduction Agency (grant HDTRA1-13-1-0034 to JEC and AB), U.S. NIH grant U19 AI109711 (to JEC and AB), U19 AI109762 (to EOS and ABW), and NIH contract HHSN272201400058C (to BJD), and R01 AI067927 (to EOS). EOS is an Investigator in the Pathogenesis of Infectious Disease of the Burroughs Wellcome Fund. The project was supported by NCRR Grant UL1 RR024975-01, and is now at the National Center for Advancing Translational Sciences, Grant 2 UL1 TR000445-06. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. We thank Stephen Graham and Leah Loerinc at Vanderbilt for help with expression of recombinant antibodies. We thank Dr. Patrick Younan at UTMB for assisting with statistical calculations. We also thank Andrew McNeal, Srikar Reddy, and Rachel Fong for technical help with shotgun mutagenesis epitope mapping. CDM is supported by a National Science Foundation Graduate Research Fellowship.
Footnotes
Author Contributions
A.I.F, A.B., and J.E.C. conceived the study. A.I.F., N.K., C.D.M., C.B., E.D., P.G., P.A.I., X.S., K.H., P.R., H.T., R.L., N.K., H.K., G.S., conducted experiments. C.P.G., M.L.F., D.W.W., and E.O.S. provided reagents. A.I.F., N.K., C.D.M., E.D., P.G., P.A.I., X.S., P.R., B.J.D., T.G.K., A.B.W., A.B., and J.E.C. analyzed data. A.I.F. and J.E.C. wrote the paper. All authors reviewed, edited and approved the paper.
Competing financial interests.
C.B., E.D., and B.J.D. are employees of Integral Molecular. B.J.D. is a shareholder of Integral Molecular. J.E.C. is a consultant for Sanofi, and is on the Scientific Advisory Boards of PaxVax, CompuVax, GigaGen, Meissa Vaccines, is a recipient of previous unrelated research grants from Moderna and Sanofi and is founder of IDBiologics. A.I.F., P.A.I., A.B and J.E.C. are co-inventors on a patent applied for that includes the BDBV223, BDBV317, and BDBV340 antibodies.
References
- 1.Qiu X, et al. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature. 2014;514:47–53. doi: 10.1038/nature13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lyon GM, et al. Clinical care of two patients with Ebola virus disease in the United States. N Engl J Med. 2014;371:2402–2409. doi: 10.1056/NEJMoa1409838. [DOI] [PubMed] [Google Scholar]
- 3.Qiu X, et al. Characterization of Zaire ebolavirus glycoprotein-specific monoclonal antibodies. Clin Immunol. 2011;141:218–227. doi: 10.1016/j.clim.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 4.Qiu X, et al. Successful treatment of ebola virus-infected cynomolgus macaques with monoclonal antibodies. Sci Transl Med. 2012;4:138ra181. doi: 10.1126/scitranslmed.3003876. [DOI] [PubMed] [Google Scholar]
- 5.Olinger GG, Jr, et al. Delayed treatment of Ebola virus infection with plant-derived monoclonal antibodies provides protection in rhesus macaques. Proc Natl Acad Sci U S A. 2012;109:18030–18035. doi: 10.1073/pnas.1213709109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson JA, et al. Epitopes involved in antibody-mediated protection from Ebola virus. Science. 2000;287:1664–1666. doi: 10.1126/science.287.5458.1664. [DOI] [PubMed] [Google Scholar]
- 7.Murin CD, et al. Structures of protective antibodies reveal sites of vulnerability on Ebola virus. Proc Natl Acad Sci U S A. 2014;111:17182–17187. doi: 10.1073/pnas.1414164111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davidson E, et al. Mechanism of Binding to Ebola Virus Glycoprotein by the ZMapp, ZMAb, and MB-003 Cocktail Antibodies. J Virol. 2015;89:10982–10992. doi: 10.1128/JVI.01490-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flyak AI, et al. Cross-Reactive and Potent Neutralizing Antibody Responses in Human Survivors of Natural Ebolavirus Infection. Cell. 2016;164:392–405. doi: 10.1016/j.cell.2015.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Y, et al. Toremifene interacts with and destabilizes the Ebola virus glycoprotein. Nature. 2016;535:169–172. doi: 10.1038/nature18615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weissenhorn W, Carfi A, Lee KH, Skehel JJ, Wiley DC. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol Cell. 1998;2:605–616. doi: 10.1016/s1097-2765(00)80159-8. [DOI] [PubMed] [Google Scholar]
- 12.Bornholdt ZA, et al. Isolation of potent neutralizing antibodies from a survivor of the 2014 Ebola virus outbreak. Science. 2016;351:1078–1083. doi: 10.1126/science.aad5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wec AZ, et al. Antibodies from a Human Survivor Define Sites of Vulnerability for Broad Protection against Ebolaviruses. Cell. 2017;169:878–890. e815. doi: 10.1016/j.cell.2017.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu JS, et al. Detection of Ebola virus envelope using monoclonal and polyclonal antibodies in ELISA, surface plasmon resonance and a quartz crystal microbalance immunosensor. J Virol Methods. 2006;137:219–228. doi: 10.1016/j.jviromet.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 15.Cross RW, et al. The Domestic Ferret (Mustela putorius furo) as a Lethal Infection Model for 3 Species of Ebolavirus. J Infect Dis. 2016;214:565–569. doi: 10.1093/infdis/jiw209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozak R, et al. Ferrets Infected with Bundibugyo Virus or Ebola Virus Recapitulate Important Aspects of Human Filovirus Disease. J Virol. 2016;90:9209–9223. doi: 10.1128/JVI.01033-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ilinykh PA, et al. Chimeric Filoviruses for Identification and Characterization of Monoclonal Antibodies. J Virol. 2016;90:3890–3901. doi: 10.1128/JVI.00101-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malashkevich VN, et al. Core structure of the envelope glycoprotein GP2 from Ebola virus at 1.9-A resolution. Proc Natl Acad Sci U S A. 1999;96:2662–2667. doi: 10.1073/pnas.96.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee J, et al. Structure of the Ebola virus envelope protein MPER/TM domain and its interaction with the fusion loop explains their fusion activity. Proc Natl Acad Sci U S A. 2017;114:E7987–E7996. doi: 10.1073/pnas.1708052114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao X, et al. Immunization-Elicited Broadly Protective Antibody Reveals Ebolavirus Fusion Loop as a Site of Vulnerability. Cell. 2017;169:891–904. e815. doi: 10.1016/j.cell.2017.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muster T, et al. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J Virol. 1993;67:6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zwick MB, et al. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J Virol. 2001;75:10892–10905. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montero M, van Houten NE, Wang X, Scott JK. The membrane-proximal external region of the human immunodeficiency virus type 1 envelope: dominant site of antibody neutralization and target for vaccine design. Microbiol Mol Biol Rev. 2008;72:54–84. doi: 10.1128/MMBR.00020-07. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ekiert DC, et al. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sui J, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corti D, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 27.Flyak AI, et al. Mechanism of human antibody-mediated neutralization of marburg virus. Cell. 2015;160:893–903. doi: 10.1016/j.cell.2015.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee JE, et al. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee JE, et al. Techniques and tactics used in determining the structure of the trimeric ebolavirus glycoprotein. Acta Crystallogr D Biol Crystallogr. 2009;65:1162–1180. doi: 10.1107/S0907444909032314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Towner JS, et al. Generation of eGFP expressing recombinant Zaire ebolavirus for analysis of early pathogenesis events and high-throughput antiviral drug screening. Virology. 2005;332:20–27. doi: 10.1016/j.virol.2004.10.048. [DOI] [PubMed] [Google Scholar]
- 31.Lubaki NM, et al. The lack of maturation of Ebola virus-infected dendritic cells results from the cooperative effect of at least two viral domains. J Virol. 2013;87:7471–7485. doi: 10.1128/JVI.03316-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suloway C, et al. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 33.Lander GC, et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J Struct Biol. 2009;166:205–213. doi: 10.1016/j.jsb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 36.Davidson E, Doranz BJ. A high-throughput shotgun mutagenesis approach to mapping B-cell antibody epitopes. Immunology. 2014;143:13–20. doi: 10.1111/imm.12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bodanszky M, Bodanszky A. The practice of peptide synthesis. 2. Springer-Verlag; 1994. [Google Scholar]
- 38.Grant GA. Synthetic peptides : a user’s guide. W.H. Freeman; 1992. [Google Scholar]
- 39.Fields GB, Noble RL. Solid phase peptide synthesis utilizing 9-fluorenylmethoxycarbonyl amino acids. Int J Pept Protein Res. 1990;35:161–214. doi: 10.1111/j.1399-3011.1990.tb00939.x. [DOI] [PubMed] [Google Scholar]
- 40.Stuber W, Knolle J, Breipohl G. Synthesis of peptide amides by Fmoc-solid-phase peptide synthesis and acid labile anchor groups. Int J Pept Protein Res. 1989;34:215–221. doi: 10.1111/j.1399-3011.1989.tb00233.x. [DOI] [PubMed] [Google Scholar]
- 41.Atherton ECLR, Sheppard RC. Peptide synthesis : Part 10. Use of pentafluorophenyl esters of fluorenylmethoxycarbonylamino acids in solid phase peptide synthesis. Tetrahedron. 1988;44:843–857. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon request.