Abstract
Purpose
To define the genetic landscape of advanced differentiated and anaplastic thyroid cancer and identify genetic alterations of potential diagnostic, prognostic and therapeutic significance.
Experimental design
The genetic profiles of 583 advanced differentiated and 196 anaplastic thyroid cancers (ATC) generated with targeted next-generation sequencing cancer-associated gene panels MSK-IMPACT and FoundationOne were analyzed.
Results
ATC had more genetic alterations per tumor, and pediatric papillary thyroid cancer had fewer genetic alterations per tumor when compared to other thyroid cancer types. DNA mismatch repair deficit and activity of APOBEC cytidine deaminases were identified as mechanisms associated with high mutational burden in a subset of differentiated and anaplastic thyroid cancers. Copy number losses and mutations of CDKN2A and CDKN2B, amplification of CCNE1, amplification of receptor tyrosine kinase genes KDR, KIT and PDGFRA, amplification of immune evasion genes CD274, PDCD1LG2 and JAK2 and activating point mutations in small GTPase RAC1 were associated with ATC. An association of KDR, KIT and PDGFRA amplification with the sensitivity of thyroid cancer cells to lenvatinib was shown in vitro. Three genetically distinct types of ATC are proposed.
Conclusions
This large-scale analysis describes genetic alterations in a cohort of thyroid cancers enriched in advanced cases. Many novel genetic events previously not seen in thyroid cancer were found. Genetic alterations associated with anaplastic transformation were identified. An updated schematic of thyroid cancer genetic evolution is proposed.
Keywords: thyroid cancer, genetic alterations, next generation sequencing, anaplastic transformation, pharmacogenomics
INTRODUCTION
Differentiated thyroid cancer (DTC) is the most common endocrine malignancy, with an estimated 60,000 new cases per year in the United States (1). The incidence of thyroid cancer is growing by an average of 4.5% per year. Although most patients with DTC are cured by surgery with or without radioactive iodine, there is a significant morbidity and mortality associated with distant metastatic disease and anaplastic transformation. Anaplastic thyroid cancer (ATC) is an uncommon but aggressive form of thyroid cancer that is associated with very poor outcomes (2).
At present, systemic therapies for advanced DTC and ATC are given regardless of the tumor’s genetic landscape, in part due to insufficient knowledge of genetic events underlying thyroid cancer progression or anaplastic transformation and lack of validated pharmacogenetic associations. The Cancer Genome Atlas study (TCGA) has defined genetic alterations in papillary thyroid cancer (PTC) with a focus on low-to-intermediate risk tumors(3). Several recently published small studies have begun to explore the genetic landscape of advanced thyroid cancer (4–6).
Targeted next-generation sequencing (NGS) assays, such as MSK-IMPACT™ (7) and FoundationOne® (8), are commonly used for cancer genotyping in clinical practice. We have assembled the largest collection to date of genetic alterations in advanced thyroid cancer by combining data generated with MSK-IMPACT and FoundationOne panels. Analysis of these data identified many novel genetic events in thyroid cancer and helped characterize genetic alterations and gene associations implicated in disease progression and anaplastic transformation. Moreover, several putative predictive biomarkers in thyroid cancer were uncovered, potentially transforming the management of patients with aggressive disease.
METHODS
Data sources
Thyroid cancer genetic data generated by MSK-IMPACT (7) and clinical information on patients’ age, sex, and tumor site were downloaded from the recent publication (9). The subset of anaplastic thyroid cancers from this database was previously analyzed by Landa et al (5). Genetic and clinical data for 630 follicular cell-derived thyroid cancers genotyped by FoundationOne test (8) was provided by Foundation Medicine Inc., Cambridge, MA (Supplementary Table 1). Prior to sequencing MSK-IMPACT (9) and FoundationOne specimens were reviewed by an in-house pathologist for consistency with the previously established diagnosis. The data was generated as part of the clinical care and detailed information on disease stage and clinical course is not available, which is a limitation of this study. All genetic profiles in this study were obtained from unique patients.
Only 6 of the tumors sequenced with Foundation One test were identified as poorly differentiated thyroid carcinoma (data not shown). The comprehensive analysis of poorly differentiated thyroid cancers profiled by MSK-IMPACT was done by Landa et al. (5) and was not repeated in this study.
To maintain the integrity of statistical and machine learning analyses, we have accounted for the differences between older and newer versions of MSK-IMPACT and FoundationOne tests (Supplementary Table 2). Genes with known role in thyroid cancer (BRAF, RAS, RET, ALK etc.) have been tested by all versions of both tests except for PPARG (neither MSK-IMPACT nor FoundationOne sequenced it) and EIF1AX (not tested by FoundationOne). The minority of samples sequenced using older FoundationOne panel 1 were not tested for TERT promoter mutations.
Filtering of germline/non-pathogenic variants and variant annotation
The FoundationOne test does not sequence normal DNA. To remove germline and non-pathogenic variants we employed the following stringent filtering strategy, which is similar to the one used by the American Association for Cancer Research Project GENIE (10):
Variants reported in any of the eight the Exome Aggregation Consortium (11) databases with the frequency of ≥ 0.00001 were removed. However, variants reported as “pathogenic” or “likely pathogenic” by ClinVar (12) were left in the database regardless of the frequency to prevent removal of the relatively frequent pathogenic germline variants causing cancer syndromes.
Germline variants reported by the 1000 Genomes Project (13) were removed.
Unusual point mutations and indels in BRAF, RAS and RET genes that violate mutual exclusivity rule with known pathogenic mutations in any of these genes were removed.
After filtering, the median number of genetic alterations per tumor in MSK-IMPACT and FoundationOne datasets was 3 and 4, respectively. When only genes analyzed by all MSK-IMPACT and FoundationOne panels were included, the median number of genetic alterations per tumor was 2 in both datasets. Despite rigorous filtering, it is possible that a few rare non-pathogenic germline variants remained in the dataset.
Mutations were annotated using ANNOVAR (14). Detailed description of specific annotation tools can be found at the ANNOVAR website http://annovar.openbioinformatics.org/en/latest.
Mutation signatures in thyroid cancer
To assign mutation signatures, we analyzed point mutations, insertions and deletions, but not gene rearrangements or copy number changes. Thyroid cancers with 10 or more mutations (3%, 24 specimens, Supplementary Table 4) were included in the analysis. This threshold was empirically identified as a minimum required to make reliable mutation signature calls. For convenience we labeled this subset of thyroid cancers as “mutation-high”. For each mutation we identified type, substitution class and sequence trinucleotide context and matched mutations against mutation signatures (15), COSMIC, (http://cancer.sanger.ac.uk/cosmic/signatures). Highly recurrent BRAF V600E and TERT promoter mutations were not included in the analysis, because they were not specific to mutation-high cancers and likely occur by a different mechanism.
Mutations were matched to signatures manually by three investigators (NP, LF and SD) and differences in mutation signature calls were reconciled. Two mutation signature groups (APOBEC activity, signatures 2 and 13, and DNA mismatch repair deficit (MMR), signatures 6 and 15) were most prevalent in mutation-high thyroid cancers and have distinct characteristics allowing reliable assignments even with limited genetic data.
Annotation groups of genes
We assigned genes into annotation groups as defined in the Supplementary Table 2. Most of these gene groups were previously described in TCGA (3) and poorly differentiated thyroid cancer/ATC analysis (5).
In vitro drug sensitivity testing
The sensitivity of thyroid cancer cell lines to lenvatinib was tested using CellTiter-Glo 2.0 cell viability assay (Promega, Madison, WI) following manufacturer’s protocol. The viability was measured in quadruplicate after cells were exposed to eight concentrations of lenvatinib (0.64 – 40000 nM) for 3 days. The identity of all cell lines was confirmed with short-tandem repeat profiling (Applied Biosystems AmpFLSTR™ Identifiler™ PCR Amplification Kit). Cell cultures were monitored for Mycoplasma contamination using the Lonza Mycoalert system.
RET, KIT, KDR and PDGFRA expression in thyroid cancer cell lines
The expression of these 4 genes in thyroid cancer cell lines was extracted from transcriptome-wide gene expression profiles generated with Affymetrix Human Genome U133 Plus 2.0 microarrays (unpublished data).
Statistical analysis and machine learning
Kruskal-Wallis test followed by pairwise comparison with Tukey Kramer test was used to compare the number of genetic alterations in thyroid cancer types. χ2 test was used to study associations of genetic alterations and pathways with thyroid cancer subtypes. p-values were adjusted for multiple comparisons using Benjamini-Hotchberg method.
To define ATC classes sharing similar patterns of gene alterations, hierarchical clustering was applied to a binary matrix. The binary similarity metric and Ward aggregation method were used.
Apriori algorithm was used to define associations {gene(s) X} => {gene Y}, which are interpreted as follows: tumors with genetic alterations in gene(s) X are likely to have genetic alteration in gene Y. The genes affected in ≥ 2% of ATCs were included in the analysis (support = 0.02). 70% or more the gene(s) X genetic alterations must follow the rule (confidence = 0.7). Mutations in TERT and TP53 were present in most ATCs and, therefore, were excluded from Apriori analysis.
All statistical and machine learning calculations were performed in R.
RESULTS
MSK-IMPACT and FoundationOne thyroid cancer cohorts
MSK-IMPACT (7) and FoundationOne (8) are hybridization capture–based NGS panels that detect somatic and germline base substitutions, short insertions or deletions (indels), copy number alterations (CNAs), selected promoter mutations and structural rearrangements in a large number of cancer associated genes. Depending on the version, MSK-IMPACT and FoundationOne panels test 287-465 genes (Supplementary Table 2). Of these, 229 genes were included in all versions of the MSK-IMPACT and FoundationOne panels and were tested in all samples.
The combined MSK-IMPACT and FoundationOne database contains genetic data for 779 thyroid cancers (Table 1, Supplementary Table 1) and 394 genes had at least one genetic alteration (Supplementary Table 3). All major DTC subtypes were included: PTC, follicular thyroid cancer (FTC) and Hurthle cell thyroid cancer (HCTC). The largest ATC cohort ever studied, 196 tumors, was analyzed. Fifteen PTC specimens were obtained from pediatric patients (age <21 years).
Table 1.
Thyroid cancer types and tumor sites in combined MSK-IMPACT/FoundationOne cohort.
| MSK-IMPACT (n) | FoundationOne (n) | Total (n) | |
|---|---|---|---|
| Thyroid cancer types
| |||
| PTC | 89 | 379 | 468 |
| Pediatric PTC | 1 | 14 | 15 |
| FTC | 5 | 60 | 65 |
| HCTC | 23 | 12 | 35 |
| ATC | 31 | 165 | 196 |
|
Tumor sites | |||
| Thyroid | 64 | 278 | 342 |
| Lymph Node | 37 | 113 | 150 |
| Head and Neck | 10 | 75 | 85 |
| Mediastinum | 4 | 4 | 8 |
| Skin | 0 | 9 | 9 |
| Soft Tissue | 1 | 42 | 43 |
| Bone | 11 | 22 | 33 |
| Brain | 2 | 12 | 14 |
| Chest Wall | 2 | 6 | 8 |
| Kidney | 0 | 2 | 2 |
| Liver | 3 | 7 | 10 |
| Lung | 14 | 53 | 67 |
| Muscle | 1 | 1 | 2 |
| Other | 0 | 6 | 6 |
|
| |||
| Total | 149 | 630 | 779 |
In general, tumor cohorts tested by high-throughput sequencing of cancer-associated genes are enriched for advanced late-stage cases (9,10). This holds true for the thyroid cancers analyzed here, in which 25% of specimens were obtained from distant metastatic sites (Table 2). By comparison, only 8 (1.7%) samples in TCGA cohort represented distant metastases (3).
Table 2.
Mutation signatures and mechanisms causing high mutation burden in thyroid cancer.
| Mutation signatures and mechanisms | n | Thyroid Cancer Type | Genotype | ||||
|---|---|---|---|---|---|---|---|
| PTC | FTC | HCTC | ATC | BRAF V600E | MMR gene mutations | ||
| Signatures 6 and 15: defective DNA mismatch repair | 11 | 5 | 1 | 5 | 1 | 8‡ | |
| Signatures 2 and 13: APOBEC family of cytidine deaminases activity | 7 | 4 | 3 | 7† | |||
| Signature 1: spontaneous deamination of 5-methylcytosine | 2 | 1 | 1 | ||||
| Unknown | 4 | 4 | 2 | ||||
| Total | 24 | 9 | 2 | 1 | 12* | 10 | 8 |
ATC had disproportionally high number of mutation-high specimens when compared to DTC (χ2, p = 0.009);
mutations in DNA mismatch repair (MMR) genes MLH1, MSH2 and MSH6 were associated with defective DNA mismatch repair mutation signatures (χ2, p = 0.0009);
APOBEC activity mutation signature is exclusively associated with the BRAF V600E genotype (χ2, p = 0.001) in mutation-high thyroid cancers.
Frequency of genetic alterations in thyroid cancers
The median (Md) number of genetic alterations per tumor in the combined cohort was 4 (range 0 to 29), which is consistent with most endocrine-related tumors having low mutational burden (16,17). ATC had significantly more genetic alterations per tumor than any other thyroid cancer subtype (Figure 1, Md = 6, Kruskal-Wallis followed by post-hoc Tukey and Kramer test, p < 0.01). By contrast, pediatric PTC had the fewest genetic alterations per tumor (Md = 2, p < 0.01). The number of genetic alterations per tumor increased with patient age in PTC (Supplementary Figure 1A, ρ = 0.39, p < 2.2e-16) but not in ATC (Supplementary Figure 1B, ρ = 0.02, p = 0.73).
Figure 1. The number of genetic alterations per tumor in thyroid cancer subtypes.
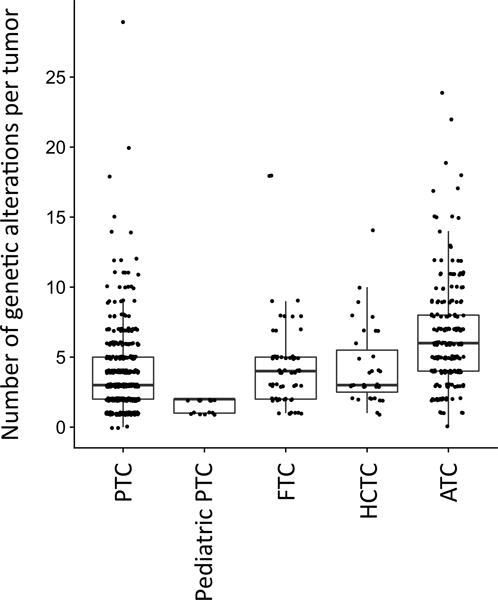
ATC had higher and pediatric PTC had lower number of genetic alterations per tumor, when compared to other thyroid cancer types (Kruskal-Wallis followed by post-hoc Tukey and Kramer test, p<0.01).
Mutation signatures in high mutational burden thyroid cancers
To understand the mechanisms responsible for acquiring relatively large number of mutations by a subset of thyroid cancers, we studied 24 samples (3%) with ≥ 10 mutations per tumor (mutation-high subset, Supplementary Table 4). ATC had significantly more mutation-high tumors than DTC (ATC 12/196 or 6.1% vs DTC 12/583 or 2.1%, χ2, p = 0.009).
Mutation signatures (15) could be assigned to 20 of 24 mutation-high tumors (Table 3, Supplementary Table 4). The deficiency in DNA mismatch repair (MMR) signature (# 6 or 15, http://cancer.sanger.ac.uk/cosmic/signatures) was most frequent in mutation-high thyroid cancers (11/24, 46%). Eight cancers with MMR deficiency signatures had loss-of-function (frameshift, nonsense, splice site) mutations in the MMR genes MLH1, MSH2 or MSH6. None of the tumors with MMR deficiency signature and MMR gene mutation had classic BRAF, RAS or RET thyroid cancer oncogenes. The second most prevalent mutation signature in mutation-high thyroid cancers is associated with increased activity of APOBEC family of cytidine deaminases (signatures # 2 or 13, 7/24, 29%). In contrast to MMR deficiency, APOBEC activity signature was only seen in tumors harboring BRAF V600E mutation. Finally, two mutation-high cancers were assigned signature #1 thought to be caused by spontaneous deamination of 5-methylcytosine.
Table 3.
Pathways and genes more frequently altered in anaplastic thyroid cancer than in differentiated thyroid cancer.
| Gene or group of genes | Prevalence, %
|
p-value* | |
|---|---|---|---|
| DTC | ATC | ||
| Tumor suppressors | 21 | 74 | 1.45e-38 |
| TP53 | 11 | 65 | 2.77e-50 |
| NF2 | 2 | 12 | 4.26e-06 |
| RB1 | 2 | 7 | 0.01 |
| NF1 | 3 | 9 | 0.01 |
|
| |||
| Cell cycle pathway | 13 | 29 | 7.42E-10 |
| CDKN2A | 7 | 22 | 4.29E-06 |
| CDKN2B | 4 | 13 | 0.001 |
| CCNE1 | 0 | 4 | 0.001 |
|
| |||
| PI3K/AKT pathway | 18 | 37 | 9.50E-06 |
| PIK3CA | 5 | 14 | 0.002 |
| PTEN | 4 | 11 | 0.01 |
|
| |||
| SWI/SNF nucleosome modification pathway | 9 | 18 | 0.007 |
| PBRM1 | 1 | 4 | 0.01 |
|
| |||
| Immune evasion | 2 | 5 | 0.07 |
| CD274 | 0 | 3 | 0.03 |
| PDCD1LG2 | 0 | 4 | 0.01 |
| JAK2 | 1 | 4 | 0.03 |
|
| |||
| Hedgehog signaling pathway | 0 | 3 | 0.009 |
|
| |||
| Histone modification | 11 | 19 | 0.03 |
|
| |||
| Mutation-high genotype | 2 | 6 | 0.05 |
|
| |||
| RAC1 | 0 | 4 | 0.004 |
|
| |||
| KIT | 0 | 4 | 0.004 |
| KDR | 0 | 3 | 0.03 |
| PDGFRA | 0 | 3 | 0.03 |
|
| |||
| INPP4B | 0 | 3 | 0.009 |
|
| |||
| NFE2L2 | 0 | 3 | 0.03 |
|
| |||
| CASP8 | 0 | 3 | 0.03 |
|
| |||
| EPHA3 | 1 | 4 | 0.03 |
|
| |||
| NBN | 0 | 3 | 0.03 |
Signaling pathways and groups of genes are highlighted in bold.
- χ2, p-values were adjusted for multiple comparisons using Benjamini-Hotchberg method.
Thyroid cancer subtypes
Papillary thyroid cancer
Four hundred sixty-eight adult (≥ 21 years-old) PTC specimens were analyzed. Seventy-four percent of PTCs had BRAF gene mutations, mostly V600E and BRAF fusions (Supplementary Figure 2), confirming BRAF as the most commonly mutated gene in advanced PTC. Rare BRAF mutations affecting the kinase domain were G469A (reported in lung cancer, (18)), V600_K601>D (melanoma, (19)), V600_S605>D (melanoma, (20)), V600_W604>R (thyroid cancer, (21)) and V600_K601>E (thyroid cancer, (22)). BRAF fusions were mutually exclusive with other BRAF, RAS and RET mutations. Known and novel BRAF fusion partners are listed in Supplementary Table 5.
TERT promoter mutations were the second most frequent genetic alteration in advanced PTC (61%). The prevalence of TERT promoter mutations in PTC from this study was markedly higher than reported by TCGA (9 % (3)) and others (12-23%, (23,24)), reflecting a selection bias towards more aggressive cases in our cohort. This study replicates many previously reported findings such as a higher incidence of C228T than C250T TERT promoter variant, mutual exclusivity of both variants (except for specimen 88W6N5, an ATC with both mutations), lack of TERT promoter mutations in pediatric PTC (25), older age of patients with TERT-mutated cancer (median = 54 and 63 years for wild type and TERT-mutated specimens, respectively, Wilcoxon, p = 4.579e-12 (3)) and an association between BRAF and TERT promoter mutations (χ2, p = 0.002).
RAS gene mutations were found in 42 PTCs (9%). All NRAS and HRAS mutations were hotspots mutations Q61K and Q61R. Conversely, 5 out of 9 KRAS mutations affected glycine-12 (G12C, G12R and G12V).
RET fusions were detected in 34 (7%) tumors, making RET the fifth most frequently altered gene in PTC. In addition, 3 NTRK1, 2 NTRK3 and 2 ALK fusions were observed (Supplementary Table 5).
We found several pathways frequently affected in PTC. Mutations in tumor suppressor genes were seen in 20% of PTC in our cohort (TP53 10%, MEN1 3%, NF1 2%, and NF2 2%), second in frequency only to MAPK pathway genes alterations (84%). PI3K/AKT signaling pathway genes were affected in 18% of PTCs (PIK3CA 6%, PTEN 2%). Histone modification genes were mutated in 11% of PTCs (KMT2C 2%, CREBBP 2%). Genes encoding components of SWI/SNF nucleosome remodeling complexes were altered in 9% of tumors (ARID1A 3%, ARID2 2%, ARID1B 1%) and mutations were mutually exclusive, in agreement with previous reports (5,6). An unexpectedly high percentage of tumors had mutations in DNA repair genes, particularly those belonging to DNA double-strand break repair pathway (9% total; ATM 4%, BRCA2 1%, BRCA1 1%).
Most of the genes altered in PTC in this study were also found to be mutated in the TCGA cohort, however, the rates of mutation were markedly lower in TCGA. For example, tumor suppressors and PI3K/AKT signaling pathway genes were affected in 3.7% and 4.5% of TCGA specimens (3), respectively, compared to 20% and 18% in our cohort. Direct statistical comparison of MSK-IMPACT/FoundationOne and TCGA data cannot be performed due to the differences in the sequencing depth-of-coverage, which results in a lower detection sensitivity for the TCGA analysis. However, it is unlikely that such a dramatic increase in the mutation frequency in PTCs from this cohort is explained solely by methodologic differences. Instead, it more likely reflects selection bias for advanced tumors.
Inactivation of CDKN2A due to copy number losses, gene truncation or loss-of-function mutations was observed in 8% of advanced PTCs. Copy number losses of CDKN2A and CDKN2B genes frequently occurred in the same specimen, which is explained by the co-localization of these genes in the cytogenetic locus 9p21.3.
We found 26 PTCs (7%) with RBM10 mutations. RBM10 has been proposed as a gene associated with PTC virulence (6). In our cohort, most alterations were either frameshift or nonsense mutations causing loss-of-function, consistent with a tumor suppressor role for RBM10 in thyroid cancer. Contrary to prior analysis (6), RBM10 mutations in our cohort frequently co-occurred with BRAF and NRAS mutations but were mutually exclusive with TP53, PIK3CA and ATM mutations. None of the PTCs had a MED12 G44C mutation, reported previously (6).
Pediatric Papillary Thyroid Cancer
Fifteen PTC specimens were from pediatric patients (<21 years old, Supplementary Figure 3). The genetic landscape of pediatric PTC is characterized by fewer genetic alterations, and a high prevalence of RET and ALK gene fusions (9/15 tumors, 60%, Supplementary Table 5). Three out of five ALK gene fusions in the entire cohort were found in pediatric PTC. Oncogene fusion predominance is particularly notable in the youngest patients: all 5 tumors from patients ≤10 years old had RET or ALK fusions. A detailed analysis of pediatric thyroid cancers, including 14 PTC and 2 MTC, has been recently published (26).
Follicular Thyroid Cancer
RAS gene mutations were found in 66% of FTCs (43/65 tumors, Supplementary Figure 4; NRAS 43%, HRAS 18%, KRAS 5%). All NRAS, HRAS, and KRAS mutations in FTC were the hotspot mutations Q61R and Q61K. Five FTCs had BRAF mutations, but only one was BRAF V600E. Three FTC specimens had BRAF K601E mutation and one had previously reported BRAF I592_A598dup mutation (27).
Several other genes were frequently mutated in FTCs. Like other non-pediatric thyroid cancer subtypes, many FTCs had mutations in the TERT promoter (71% of tested specimens). TP53 and RBM10 mutations each affected 12% of FTCs. In comparison with PTC, FTC had significantly more mutations in PTEN (2% and 14% in PTC and FTC, respectively, χ2, p = 0.0001, adjusted for multiple comparisons using Benjamini-Hotchberg method) and RB1 (1% and 9% in PTC and FTC, respectively, χ2, p = 0.0002). Mutations in PTEN, TP53, RB1 and MEN1 co-occurred in a subset of FTCs that had no mutations in RAS or BRAF genes, suggesting that simultaneous loss of multiple tumor suppressors may represent a mechanism of malignant transformation in thyroid cancer. Genetic alterations in GNAS were found in 8% of tumors. The activating GNAS mutation, R201H was found in 3 FTC samples. In one specimen (9RHMJY) it coexisted with NRAS Q61R mutation.
Hurthle Cell Thyroid Cancer
Thirty-five HCTCs were analyzed. Consistent with previous observations (28,29), HCTC has a unique landscape of genetic alterations (Supplementary Figure 5) characterized by few mutations in NRAS (9%) and KRAS (6%), no mutations in BRAF or RET, and relatively frequent mutations of TP53 (20 %). TERT promoter mutations were the most common type of genetic alteration in HCTC (59%), again supporting the association of these mutations and aggressive thyroid cancer in all subtypes. Statistical analysis showed that, in comparison to PTC, HCTC had more mutations in PTEN (2% vs 17%, χ2, p = 0.0002), KEAP1 (0% vs 11%, χ2, p = 1.31E-06) and KMT2C (2% vs 18%, χ2, p = 0.0008). TBX3, GNAS and CDKN1B copy number gains occurred more frequently in HCTC (χ2, p < 0.05). This analysis is limited by a small number of HCTCs.
Anaplastic Thyroid Cancer
One hundred ninety-six ATCs were included in this analysis. The genetic alteration patterns in ATC were distinct from DTC. The two most commonly mutated genes in ATC were TP53 (65%) and TERT (65%, Supplementary Figure 6). A total of 81 ATCs (41%) harbored BRAF gene alterations. RAS genes were mutated in 27% of ATC. Three ATC specimens had mutations affecting RAS glycine-13 (1 NRAS G13V and 2 HRAS G13R), which were not seen in PTC or FTC. One ATC with HRAS G13R mutation also had HRAS Q61K mutation.
The large sample size in this study allowed comparison between mutation frequencies in ATC and DTC, which, thereby, identified genetic events that may contribute to anaplastic transformation. Recapitulating findings from earlier publications (4,5), our ATC cohort was characterized by an increased frequency of mutations in tumor suppressor genes (TP53, NF2, NF1, RB1) and PI3K/AKT pathway genes (PIK3CA, PTEN). The prevalence of genetic alterations and p-values for all genes significantly associated with ATC are listed in Table 3.
Genes associated with histone modification and SNF/SWI nucleosome remodeling were affected in 19% and 18% of ATCs, respectively, which is significantly more often than in DTC. Alterations in cell cycle genes such as CDKN2A and CDKN2B (copy number losses or loss-of-function mutations) and CCNE1 (copy number gains) were found in 29% of ATCs versus 13% of DTCs. Co-amplification of tumor immune evasion genes CD274 (PD-L1), PDCD1LG2 (PD-L2) and JAK2 (cytogenetic locus 9p24.1) described in Hodgkin’s lymphoma (30,31) and non-small cell lung cancer (32) was seen in 5 ATC tumors, but not in other thyroid cancer subtypes (Table 3). KIT amplification was also specific to ATC and coexisted with copy number gains in PDGFRA and KDR (seven ATCs, cytogenetic locus 4q12).
RAC1 encodes a member of the RAS superfamily of small GTPases. Seven ATCs had genetic alterations in this gene including the activating P29S mutation common in melanoma (33), the activating mutation A159V (34), and RAC1 gene amplifications. RAC1 mutations were not mutually exclusive with BRAF and RAS mutations.
Genetic alterations in several other genes, including PI3K/AKT signaling pathway modulator INPP4B, transcription factor NFE2L2, DNA repair gene NBN, caspase CASP8 and a member of ephrin receptor subfamily EPHA3, were infrequent but significantly associated with ATC (Table 3).
Three genetically distinct subtypes of anaplastic thyroid cancer
Theoretically, ATC can evolve from any follicular-derived DTC subtype (PTC, FTC, or HCTC) or directly from normal follicular thyroid cells. Oncogene mutations characterizing DTC subtypes should therefore translate into the genetic heterogeneity of ATC. To test this hypothesis, we applied hierarchical clustering and Apriori machine learning algorithms to define genetically similar subtypes of ATC and find subtype-specific associations of gene alterations. Hierarchical clustering identified four major clusters of ATCs with distinct genetic profiles (Figure 2).
Figure 2. Hierarchical clustering of genetic alterations in ATC.
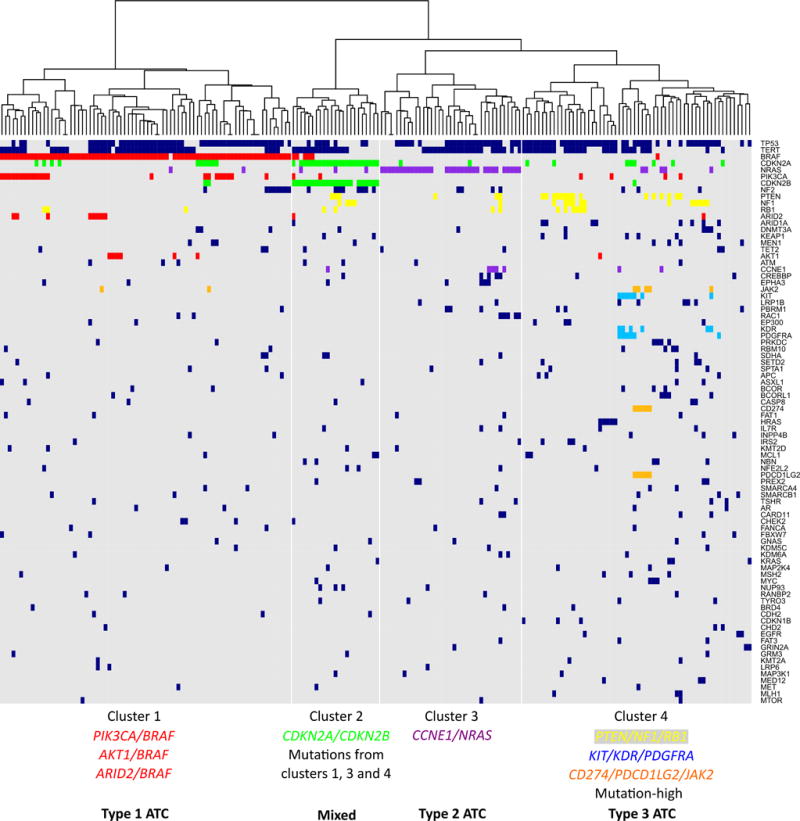
Key associations of genetic alterations identified by Apriori algorithm characterizing each cluster are highlighted by different colors. Four clusters and three ATC types with distinct genetic alteration patterns are proposed.
Cluster 1 consists almost exclusively of tumors with BRAF V600E mutations. The Apriori algorithm identified PIK3CA/BRAF, AKT1/BRAF, and ARID2/BRAF gene mutations associations in ATCs from this cluster (Supplementary Table 6). The PIK3CA/BRAF association has been previously described in ATC (5). The genetic landscape of ATCs in cluster 1 resembles that of PTC, therefore, these anaplastic tumors likely evolved from PTC (Figure 2, type 1 ATC).
Cluster 3 contains ATCs with NRAS mutations. CCNE1 copy number gains were associated with NRAS mutations. These anaplastic thyroid cancers likely originated from NRAS-mutant FTCs (Figure 2, type 2 ATC).
Several ATCs in cluster 4 have oncogenic mutations in RAS genes but most of these cancers do not have mutations in classic thyroid cancer oncogenes. ATCs in this cluster carry PTEN mutations frequently coexisting with NF1 (as it was seen previously in ATC (5)) and RB1 mutations. Other ATCs in this cluster had amplifications of cytogenetic loci 4q12 (KIT/KDR/PDGFR) and 9p24.1 (immune evasion genes CD274/PDCD1LG2/JAK2). Cluster 4 ATCs had overall higher number of genetic alterations and contained mutation-high ATCs with MMR signature and mutations in MSH2 and MLH1 genes. Based on the similarity in genetic profiles, ATCs in cluster 4 most likely originated from HCTC and a subset of RAS-mutant FTC (Figure 2, type 3 ATC).
All tumors in cluster 2 have loss-of-function genetic alterations in the cell cycle regulators CDKN2A and CDKN2B. In addition, tumors in this cluster had genetic features of the three ATC subtypes described above (mutually exclusive BRAF mutations, NRAS mutations and PTEN/NF1/RB1 mutations). Thus, while cluster 2 ATCs may share a common mechanism of aggressiveness and anaplastic transformation, they do not appear to be derived from a single DTC predecessor.
Putative oncogenes and tumor suppressors
To identify genetic alterations of potential significance in thyroid cancer, which are not enriched in a specific thyroid cancer subtype, we reviewed the database for recurrent oncogenic variants and genes with the high proportion of predicted loss-of-function mutations. In addition to classic oncogenic hotspots in BRAF, RAS, and PIK3CA, we found recurrent mutations in AKT1 and AKT2. The mutation E17K was found in 10 instances in AKT1 (5 PTC, 1 FTC and 4 ATC) and 9 instances in AKT2 (7 PTC and 2 ATC); these mutations coexisted with BRAF, HRAS and NRAS mutations, but were mutually exclusive with PTEN alterations. AKT1 E17K was previously reported in advanced thyroid cancer (35), and 2 AKT1 and 1 AKT2 E17K instances were found in TCGA cohort. One FTC had AKT1 E17R mutation, which has not been previously reported in cancer. Recurrent hotspot mutations NUP93 E14K (4 PTC and 2 ATC) and Q15* (5PTC and 1 ATC) have been described in other malignancies (35) but have not been previously reported in thyroid cancer.
Tumor suppressor genes are characterized by mutations causing loss of function (frameshift, nonsense and splice site mutations). To look for putative novel tumor suppressors, we identified genes with the frequencies of loss-of-function mutations exceeding that for TP53 gene (30%, Supplementary Table 7). Using this approach, we found known tumor suppressors in cancer, such as NF1, NF2, RB1, RBM10, MEN1, CDKN2A and PTEN. In addition, we identified several other genes with putative loss-of-function mutations across the coding region, such as transcription regulators CREBBP, ASXL1, BCORL1 and BCOR, components of SWI/SNF nucleosome remodeling complex ARID2 and ARID1A, DNA methyltransferase DNMT3A, methylcytosine deoxygenase TET2, and others. The tumor suppressor role of many of these genes requires experimental validation.
Pharmacogenomics of lenvatinib
Cancer genomic profiles are increasingly used to direct patients towards biomarker-driven clinical trials and prescribe targeted therapies. As a proof-of-principle, we examined whether genetic alterations found in this study are associated with the sensitivity of thyroid cancer cells to lenvatinib, which is approved for the treatment of progressive radioiodine-refractory DTC (36). We tested 30 thyroid cancer cell lines using an in vitro viability assay and identified two cell lines sensitive to lenvatinib: TPC1 and THJ29T (Supplementary Figure 7A). The TPC1 cell line harbors a CCDC6-RET fusion resulting in increased expression of the RET gene (Supplementary Figure 7B), which was shown to result in an increased sensitivity to lenvatinib (37). The THJ29T cell line has an amplification of the 4q21 cytogenetic locus (38) causing overexpression of lenvatinib targets KIT, KDR and PDGFRA (Supplementary Figure 7C-E), which likely explains increased sensitivity to the drug. KIT/KDR/PDGFRA amplification was seen in a subset of ATCs (4%) in our cohort (Figure 2, Supplementary Figure 6). Thus, the sensitivity of thyroid cancer cell lines to lenvatinib in vitro correlated with the presence of genetic alterations in lenvatinib targets.
DISCUSSION
This analysis utilized data obtained with MSK-IMPACT and FoundationOne tests for the in-depth analysis of genetic alterations in a large cohort of 779 advanced DTCs and ATCs. These next-generation sequencing-based tests are not limited to selected hotspot regions and provide detailed genetic information, including data on single nucleotide variants, indels, gene fusions and copy number changes, for hundreds of cancer-related genes. Although not as comprehensive as whole exome or whole genome sequencing, MSK-IMPACT and FoundationOne tests allow for superior sequencing depth of coverage (median of 539× and 613× for the variants in this study, respectively), translating into high sensitivity for detecting genetic alterations with low allele frequency. This is particularly important for ATC, which is heavily contaminated with non-tumor cells (5).
In comparison to the TCGA study (3), our cohort is enriched in advanced thyroid cancers, as evidenced by the large number of specimens obtained from distant metastatic sites as well as the genetic hallmarks of aggressive disease, such as a very high prevalence of TERT promoter mutations. Furthermore, clinicians choosing to sequence large panels of cancer-associated genes, usually reserve this testing for patients with advanced disease, when genetic information can be used to triage patients into biomarker-driven clinical trials or prescribe drugs targeting specific genetic alterations (9,10).
The review of mutation signatures identified two primary mechanisms of acquiring relatively high mutational burden in a subset of thyroid cancers: impaired MMR and activity of APOBEC family of cytidine deaminases. MMR deficiency, when caused by loss-of-function mutations in MMR genes, was independent of mutations in BRAF, RAS and RET oncogenes and may represent novel mechanism of malignant transformation in thyroid cancer. Only one MSH2 mutation was reported by TCGA (3) suggesting that MMR deficiency is associated with advanced DTC (this study) and ATC (this and other studies (4,5)), as opposed to low/intermediate risk PTC. The APOBEC activity signature was previously reported in thyroid cancer (15) and was found in the specimens with highest mutation burden (3), similar to our findings. Interestingly, in our cohort, the APOBEC activity signature was only found in BRAF-mutant PTCs and ATCs. BRAF V600E mutation results from a T>A transversion, which belongs to a mutation class not caused by APOBEC enzymes. Therefore, APOBEC activity is likely a secondary event occurring in BRAF-mutant thyroid cancers, which may contribute to tumor aggressiveness.
This study highlights the importance of cell cycle genes in thyroid cancer pathogenesis. CDKN2A and CDKN2B are negative cell cycle regulators and their loss due to copy number alterations, epigenetic silencing or inactivating mutations is one of the most frequent genetic events encountered in human cancers (39). In thyroid cancer, genetic alterations of CDKN2A/CDKN2B were seen in PTC but were even more frequent in ATC (Table 3), suggesting potential role in anaplastic transformation. CCNE1 amplification was specific to ATC in our cohort. CCNE1 is a regulatory subunit of CDK2 and is required for cell cycle G1/S transition. CCNE1 amplification occurs frequently in non-thyroid cancers (39).
Whereas Q61R and Q61K mutations in RAS genes are common in FTC and were found in PTC, NRAS and HRAS mutations affecting glycine-13 were only seen in ATC. No such mutations were found in TCGA cohort of PTCs (3). HRAS mutations at G13 were previously seen in poorly differentiated thyroid cancer (35), familial non-medullary thyroid carcinoma (40) and medullary thyroid cancer (41). To our best knowledge, the specific NRAS G13V mutation has not been reported previously in thyroid cancer. It is possible that NRAS G13V and HRAS G13R mutations cause an aggressive phenotype with anaplastic transformation and are of prognostic significance.
Cluster analysis uncovered genetic heterogeneity of ATC and three classes of ATC were proposed. These classes have genetic features of three major DTC types, PTC, FTC and HCTC, which supports the current paradigm of anaplastic transformation from differentiated tumors through acquisition of additional oncogenic alterations.
Several genetic alterations described in this study may be important for the personalized management of thyroid cancer. MMR deficiency and CD274/PDCD1LG2/JAK2 amplification are associated with favorable response to immune checkpoint inhibitors, such as pembrolizumab and nivolumab (42,43). Activating RAC1 mutations cause resistance to RAF inhibitors (44). Increased activity of APOBEC enzymes makes cells sensitive to ATR inhibitors resulting in replication catastrophe (45). Finally, we found that the sensitivity of ATC cells to lenvatinib in vitro is associated with 4q21 cytogenetic locus amplification.
We summarized key findings from this study and proposed an updated model of thyroid cancer genetic evolution in Figure 3.
Figure 3. Genetic evolution of thyroid cancer.
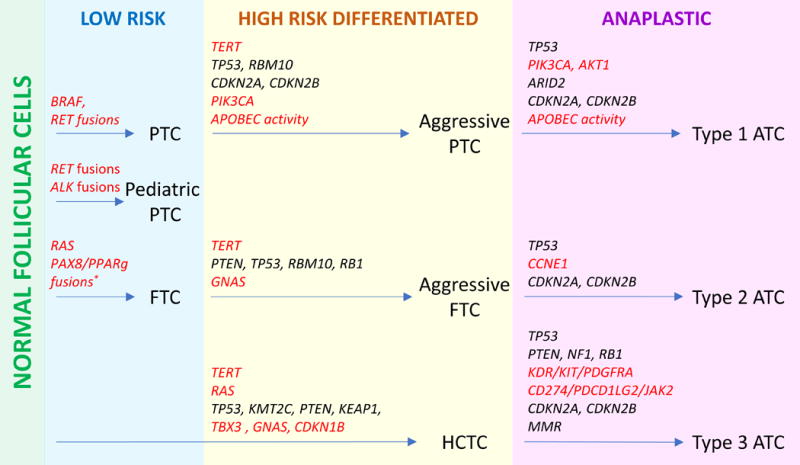
Genetic alterations causing an increase in protein activity (activating point mutations, fusions and gene amplifications) are highlighted in red while loss-of-function mutation and copy number losses are shown in black. * - PAX/PPARg fusions were not tested by MSK-IMPACT and FoundationOne.
This study has some limitations. The data are derived from targeted panels rather than whole exome or genome data. We may have missed important genetical events occurring in advanced thyroid cancer. The outcome analysis and identification of genetic alterations associated with metastatic sites cannot be performed due to limited clinical data. Nevertheless, MSK-IMPACT and FoundationOne panels are the most widely used tests in the clinical setting, making this analysis directly clinically relevant.
In summary, using large combined database of targeted next generation sequencing data, we described known and novel genetic alterations in thyroid cancer, which may be used as prognostic markers and to guide personalized thyroid cancer treatment.
Supplementary Material
STATEMENT OF TRANSLATIONAL RELEVANCE
The knowledge of genetic events associated with advanced differentiated and anaplastic thyroid cancers has prognostic significance and will assist in the development of the next generation of molecular tools for the diagnosis and treatment. Many genetic alterations found in thyroid cancers from this study, such as mutations in DNA mismatch repair genes, amplifications of immune evasion genes and amplifications of receptor tyrosine kinase genes are associated with improved response to checkpoint inhibitors and specific kinase inhibitors and are important for personalized management of advanced thyroid cancer.
Acknowledgments
Financial support. This work has been supported by the American Thyroid Association/Thyroid Cancer Survivors’ Association Research Grant and Paul R. Ohara II Seed Grant to N. Pozdeyev, Mary Rossick Kern and Jerome H Kern Endowment to B.R. Haugen, and NIH Challenge Grant 1RC1CA147371-01 to B.R. Haugen and R.E. Schweppe.
Footnotes
Conflict of interest. Nikita Pozdeyev, Kelsi E. Deaver, Stephanie Davis, Jena D. French, Daniel V. LaBarbera, Aik-Choon Tan, Rebecca E. Schweppe, Lauren Fishbein, Bryan R. Haugen and Daniel W. Bowles declare no conflicts of interest. Laurie M. Gay, Ethan Sokol, Pierre Vanden Borre and Jeffrey S. Ross are employees and shareholders of Foundation Medicine Inc. Ryan Hartmaier was an employee and shareholder of Foundation Medicine Inc. at a time this study was done.
BIBLIOGRAPHY
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Smallridge RC, Copland JA. Anaplastic thyroid carcinoma: pathogenesis and emerging therapies. Clin Oncol (R Coll Radiol) 2010;22:486–97. doi: 10.1016/j.clon.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network. Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159:676–90. doi: 10.1016/j.cell.2014.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kunstman JW, Juhlin CC, Goh G, Brown TC, Stenman A, Healy JM, et al. Characterization of the mutational landscape of anaplastic thyroid cancer via whole-exome sequencing. Hum Mol Genet. 2015;24:2318–29. doi: 10.1093/hmg/ddu749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Landa I, Ibrahimpasic T, Boucai L, Sinha R, Knauf JA, Shah RH, et al. Genomic and transcriptomic hallmarks of poorly differentiated and anaplastic thyroid cancers. J Clin Invest. 2016;126:1052–66. doi: 10.1172/JCI85271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ibrahimpasic T, Xu B, Landa I, Dogan S, Middha S, Seshan V, et al. Genomic Alterations in Fatal Forms of Non-Anaplastic Thyroid Cancer: Identification of MED12 and RBM10 as Novel Thyroid Cancer Genes Associated with Tumor Virulence. Clin Cancer Res. 2017;23:5970–80. doi: 10.1158/1078-0432.CCR-17-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn. 2015;17:251–64. [Google Scholar]
- 8.Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–31. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–13. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.AACR Project GENIE Consortium. AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. 2017;7:818–31. doi: 10.1158/2159-8290.CD-17-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–D8. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. [Google Scholar]
- 14.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fishbein L, Leshchiner I, Walter V, Danilova L, Robertson AG, Johnson AR, et al. Comprehensive Molecular Characterization of Pheochromocytoma and Paraganglioma. Cancer Cell. 2017;31:181–93. doi: 10.1016/j.ccell.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scarpa A, Chang DK, Nones K, Corbo V, Patch AM, Bailey P, et al. Whole-genome landscape of pancreatic neuroendocrine tumours. Nature. 2017;543:65–71. doi: 10.1038/nature21063. [DOI] [PubMed] [Google Scholar]
- 18.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 19.Busser B, Leccia MT, Gras-Combe G, Bricault I, Templier I, Claeys A, et al. Identification of a Novel Complex BRAF Mutation Associated With Major Clinical Response to Vemurafenib in a Patient With Metastatic Melanoma. JAMA Dermatol. 2013;149:1403. doi: 10.1001/jamadermatol.2013.8198. [DOI] [PubMed] [Google Scholar]
- 20.Cruz F, Rubin BP, Wilson D, Town A, Schroeder A, Haley A, et al. Absence of BRAF and NRAS mutations in uveal melanoma. Cancer Res. 2003;63:5761–6. [PubMed] [Google Scholar]
- 21.Barollo S, Pezzani R, Cristiani A, Redaelli M, Zambonin L, Rubin B, et al. Prevalence, Tumorigenic Role, and Biochemical Implications of Rare BRAF Alterations. Thyroid. 2014;24:809–19. doi: 10.1089/thy.2013.0403. [DOI] [PubMed] [Google Scholar]
- 22.Lee Y-W. Peptide nucleic acid clamp polymerase chain reaction reveals a deletion mutation of the BRAF gene in papillary thyroid carcinoma: A case report. Exp Ther Med. 2013;6:1550–2. doi: 10.3892/etm.2013.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu X, Bishop J, Shan Y, Pai S, Liu D, Murugan AK, et al. Highly prevalent TERT promoter mutations in aggressive thyroid cancers. Endocr Relat Cancer. 2013;20:603–10. doi: 10.1530/ERC-13-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landa I, Ganly I, Chan TA, Mitsutake N, Matsuse M, Ibrahimpasic T, et al. Frequent somatic TERT promoter mutations in thyroid cancer: higher prevalence in advanced forms of the disease. J Clin Endocrinol Metab. 2013;98:E1562–6. doi: 10.1210/jc.2013-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ballester LY, Sarabia SF, Sayeed H, Patel N, Baalwa J, Athanassaki I, et al. Integrating Molecular Testing in the Diagnosis and Management of Children with Thyroid Lesions. Pediatr Dev Pathol. 2016;19:94–100. doi: 10.2350/15-05-1638-OA.1. [DOI] [PubMed] [Google Scholar]
- 26.Vanden Borre P, Schrock AB, Anderson PM, Morris JC, 3rd, Heilmann AM, Holmes O, et al. Pediatric, Adolescent, and Young Adult Thyroid Carcinoma Harbors Frequent and Diverse Targetable Genomic Alterations, Including Kinase Fusions. Oncologist. 2017;22:255–63. doi: 10.1634/theoncologist.2016-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Torregrossa L, Viola D, Sensi E, Giordano M, Piaggi P, Romei C, et al. Papillary Thyroid Carcinoma With Rare Exon 15 BRAF Mutation Has Indolent Behavior: A Single-Institution Experience. J Clin Endocrinol Metab. 2016;101:4413–20. doi: 10.1210/jc.2016-1775. [DOI] [PubMed] [Google Scholar]
- 28.Ganly I, Ricarte Filho J, Eng S, Ghossein R, Morris LG, Liang Y, et al. Genomic dissection of Hurthle cell carcinoma reveals a unique class of thyroid malignancy. J Clin Endocrinol Metab. 2013;98:E962–72. doi: 10.1210/jc.2012-3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei S, LiVolsi VA, Montone KT, Morrissette JJ, Baloch ZW. PTEN and TP53 Mutations in Oncocytic Follicular Carcinoma. Endocr Pathol. 2015;26:365–9. doi: 10.1007/s12022-015-9403-6. [DOI] [PubMed] [Google Scholar]
- 30.Green MR, Monti S, Rodig SJ, Juszczynski P, Currie T, O’Donnell E, et al. Integrative analysis reveals selective 9p24.1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma. Blood. 2010;116:3268–77. doi: 10.1182/blood-2010-05-282780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roemer MG, Advani RH, Ligon AH, Natkunam Y, Redd RA, Homer H, et al. PD-L1 and PD-L2 Genetic Alterations Define Classical Hodgkin Lymphoma and Predict Outcome. J Clin Oncol. 2016;34:2690–7. doi: 10.1200/JCO.2016.66.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ikeda S, Okamoto T, Okano S, Umemoto Y, Tagawa T, Morodomi Y, et al. PD-L1 Is Upregulated by Simultaneous Amplification of the PD-L1 and JAK2 Genes in Non-Small Cell Lung Cancer. J Thorac Oncol. 2016;11:62–71. doi: 10.1016/j.jtho.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 33.Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–14. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang MT, Asthana S, Gao SP, Lee BH, Chapman JS, Kandoth C, et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol. 2016;34:155–63. doi: 10.1038/nbt.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ricarte-Filho JC, Ryder M, Chitale DA, Rivera M, Heguy A, Ladanyi M, et al. Mutational profile of advanced primary and metastatic radioactive iodine-refractory thyroid cancers reveals distinct pathogenetic roles for BRAF, PIK3CA, and AKT1. Cancer Res. 2009;69:4885–93. doi: 10.1158/0008-5472.CAN-09-0727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schlumberger M, Tahara M, Wirth LJ, Robinson B, Brose MS, Elisei R, et al. Lenvatinib versus Placebo in Radioiodine-Refractory Thyroid Cancer. N Engl J Med. 2015;372:621–30. doi: 10.1056/NEJMoa1406470. [DOI] [PubMed] [Google Scholar]
- 37.Okamoto K, Kodama K, Takase K, Sugi NH, Yamamoto Y, Iwata M, et al. Antitumor activities of the targeted multi-tyrosine kinase inhibitor lenvatinib (E7080) against RET gene fusion-driven tumor models. Cancer Lett. 2013;340:97–103. doi: 10.1016/j.canlet.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 38.Marlow LA, D’Innocenzi J, Zhang Y, Rohl SD, Cooper SJ, Sebo T, et al. Detailed molecular fingerprinting of four new anaplastic thyroid carcinoma cell lines and their use for verification of RhoB as a molecular therapeutic target. J Clin Endocrinol Metab. 2010;95:5338–47. doi: 10.1210/jc.2010-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cavaco BM, Batista PF, Martins C, Banito A, do Rosario F, Limbert E, et al. Familial non-medullary thyroid carcinoma (FNMTC): analysis of fPTC/PRN, NMTC1, MNG1 and TCO susceptibility loci and identification of somatic BRAF and RAS mutations. Endocr Relat Cancer. 2008;15:207–15. doi: 10.1677/ERC-07-0214. [DOI] [PubMed] [Google Scholar]
- 41.Moura MM, Cavaco BM, Pinto AE, Leite V. High prevalence of RAS mutations in RET-negative sporadic medullary thyroid carcinomas. J Clin Endocrinol Metab. 2011;96:E863–8. doi: 10.1210/jc.2010-1921. [DOI] [PubMed] [Google Scholar]
- 42.Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409–13. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ansell SM, Lesokhin AM, Borrello I, Halwani A, Scott EC, Gutierrez M, et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N Engl J Med. 2015;372:311–9. doi: 10.1056/NEJMoa1411087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watson IR, Li L, Cabeceiras PK, Mahdavi M, Gutschner T, Genovese G, et al. The RAC1 P29S hotspot mutation in melanoma confers resistance to pharmacological inhibition of RAF. Cancer Res. 2014;74:4845–52. doi: 10.1158/0008-5472.CAN-14-1232-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buisson R, Lawrence MS, Benes CH, Zou L. APOBEC3A and APOBEC3B Activities Render Cancer Cells Susceptible to ATR Inhibition. Cancer Res. 2017;77:4567–78. doi: 10.1158/0008-5472.CAN-16-3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


