Abstract
Point mutations in the seed sequence of miR-142-3p are present in a subset of AML and in several subtypes of B cell lymphoma. Here we show that mutations associated with AML result both in loss of miR-142-3p function and in decreased miR-142-5p expression. Mir142 loss altered the hematopoietic differentiation of multipotent hematopoietic progenitors, enhancing their myeloid potential while suppressing their lymphoid potential. During hematopoietic maturation, loss of Mir142 increased Ash1l protein expression and consequently resulted in the aberrant maintenance of Hoxa gene expression in myeloid-committed hematopoietic progenitors. Mir142 loss also enhanced the disease-initiating activity of IDH2 mutant hematopoietic cells in mice. Together these data suggest a novel model in which miR-142, through repression of ASH1L activity, plays a key role in suppressing HOXA9/A10 expression during normal myeloid differentiation. AML-associated loss-of-function mutations of MIR142 disrupt this negative signaling pathway, resulting in sustained HOXA9/A10 expression in myeloid progenitors/myeloblasts and ultimately contributing to leukemic transformation.
INTRODUCTION
MicroRNAs (miRNAs) are non-coding RNAs, which inhibit the expression of messenger RNAs (mRNAs) (1). MiRNA genes are transcribed as primary miRNAs, which are processed by the Drosha microprocessor complex into precursor miRNAs. Precursor miRNAs are processed by Dicer, generating a duplex, which contains two distinct mature miRNA species, referred to as 5p or 3p depending on whether they are derived from the 5′ or 3′ end of the primary transcript. Only one strand is loaded into the RNA induced silencing complex (RISC); the other is discarded. The incorporated strand guides the RISC to semi-complementary mRNA targets and inhibits their expression by either directly repressing translation or promoting mRNA decay (2). A miRNA can target hundreds of mRNAs based on sequence complementarity.
There is evidence that the dysregulation of certain miRNAs contributes to leukemogenesis. For example, MIR125 overexpression, due to the rare chromosomal translocation t(2:11)(p21;p23) or through epigenetic mechanisms, promotes leukemogenesis in mouse models (3). The genetic loss or epigenetic silencing of miRNAs also has been implicated in AML pathogenesis. For example, the loss of MIR145 and MIR146A in myeloid neoplasms carrying 5q chromosomal deletions has been shown to contribute to AML (4). In contrast, genetic alterations solely targeting miRNAs are uncommon in AML (5). In the TCGA cohort of 200 AML patients, MIR142 was the only recurrently mutated miRNA (6). MIR142 point mutations were identified in 4 cases, with one case bi-allelic; all mutations localized to the miR-142-3p seed sequence. A subsequent study produced similar results (7). MIR142 is also mutated in follicular and diffuse large B cell lymphoma at reported frequencies of 10–25% and 12–20% respectively (8–11). These mutations also often localize to the miR-142-3p seed sequence.
MIR142 is highly-conserved and broadly expressed in mature hematopoietic cells (12,13). It is expressed at lower levels in hematopoietic stem/progenitor cells (HSPCs). In mice, Mir142 loss results in splenomegaly from an expansion of myeloid and CD19+ B cells (14,15). Mir142 loss is also associated with the impaired development and function of multiple hematopoietic lineages, including B1 B cells (14), T cells (16,17), CD4+ dendritic cells (15), mast cells (18), megakaryocytes (19), and erythroid precursors (20,21). MiR-142’s role in regulating HSPCs is less well defined, although inhibition of miR-142-3p in zebrafish results in reduced HSPC activity (22).
Here, we show that AML-associated MIR142 mutations disrupt miR-142-3p function and result in decreased miR-142-5p expression. In mice, Mir142 loss enhances the myeloid potential, while suppressing the lymphoid potential, of multipotent HSPCs. Moreover, Mir142 loss enhances the disease initiating capacity of IDH2R172K mutant hematopoietic cells. We provide evidence that these phenotypes are due, in part, to derepression of Ash1l (absent, small, or homeotic 1-like), a histone methyltransferase, which positively regulates Hoxa gene expression. These data suggest a novel model in which MIR142 mutations, through derepression of ASH1L, cause sustained HOXA9/A10 expression during myeloid differentiation, ultimately contributing to leukemic transformation.
MATERIALS AND METHODS
Patient Studies
Bone marrow samples from patients with AML were collected as part of a single-institution banking protocol approved by the Washington University Human Studies Committee (IRB#01-1014). Healthy donor bone marrow samples were obtained following written informed consent as approved by the Washington University Institutional Review Board. All patient studies were performed in accordance with the Declaration of Helsinki.
Mouse models
Mice were maintained under standard pathogen-free conditions with animal studies conducted in accordance with IACUC-approved Washington University protocols. Mice on a C57Bl/6 background with homozygous deletion of Mir142 (Mir142−/−) were generated and described by Sun et al. (17). Mice on a C57Bl/6 background with a gene trap insertion cassette causing >90% reduction of full length Ash1l transcript (Ash1lGT/GT) were generated and described by Jones et al. (23). Experimental and control mice were age (8–12 weeks) and sex-matched.
Cell lines and cell culture
HEK293T cells were obtained from ATCC and cultured in a humidified incubator at 37°C with 5% CO2. They were maintained in Dulbecco’s Modified Eagle Medium (Gibco) with 10% fetal bovine serum (Atlas), penicillin (100 Units/ml) (Gibco) and streptomycin (100 μg/ml) (Gibco). Cells were passaged upon reaching a confluency of 75–80% with Trypsin-EDTA (0.25%) (Gibson). HEK293T cells were not authenticated or tested for Mycoplasma and were used within 3–5 passages after initial expansion.
RNA expression in murine hematopoietic populations
The NucleoSpin RNA kit (Macherey-Nagel) was used to isolate RNA from CD150+CD48−c-kit+Sca1+lineage− cells. For other populations, RNA was isolated with Trizol LS (Thermo Fisher). RNA expression was assessed with two-step RT-qPCR using iScript reverse transcriptase (Bio-Rad) and iTaq DNA polymerase (Bio-Rad).
Protein extraction
To isolate histones, bone marrow c-kit+-selected cells were isolated with CD117 MicroBeads on the autoMACS Pro Separator (Miltenyi). They were washed twice with PBS and lysed in a PBS solution with 0.5% Triton X-100 (Sigma), 2 mM PMSF (Sigma), and 0.02% sodium azide (Sigma). After 10 minutes on ice, samples were centrifuged at 2,000 rpm for 10 minutes at 4°C. The pellets were resuspended in 0.2N HCl and histones extracted at 4°C overnight. Finally, samples were centrifuged at 2,000 rpm for 10 minutes at 4°C and the histone-containing supernatant collected.
To isolate ASH1L, 2x106 bone marrow cells were washed twice with PBS and lysed in a buffer consisting of 7M urea (Sigma), 2M thiourea (Sigma), 4%CHAPS (Sigma), 30 mM Tris (Sigma), and a protease inhibitor cocktail (Thermo Fisher). To prepare for Western blotting, the solution was frozen and thawed three times on dry ice, dissolved in Laemmli buffer, heated at 99°C for 5 minutes, and sonicated.
Viral transduction and bone marrow transplantation
For competitive transplantation, bone marrow cells from Mir142+/+ or Mir142−/− Ly5.2 mice were mixed with competitor wild type bone marrow cells expressing Ly5.1 and retro-orbitally transplanted into lethally irradiated (1000 cGy) Ly5.1/5.2 recipient mice.
For transplantation of phenotypic stem cells, 20–25 CD150+CD48−c-kit+Sca1+ lineage− cells from Mir142+/+ or Mir142−/− Ly5.2 mice were sorted into individual wells containing 2.5–3x105 support bone marrow cells (Ly5.1 or Ly5.1/Ly5.2) and injected retro-orbitally into lethally irradiated (1000 cGy) recipient mice (Ly5.1 or Ly5.1/Ly5.2). The lineage output of the transplanted HSCs was determined 24 weeks post-transplantation in the peripheral blood of mice with identifiable Ly5.2 engraftment in both myeloid (Gr1+ or CD115+) and lymphoid (B220+ or CD3+) lineages.
Human IDH2R172K over-expression was achieved using an MSCV-IRES-mcherry vector (Addgene #52114). HEK293T cells were transfected with the retroviral vector and an Ecopak plasmid (Addgene #12371) using the Calcium Phosphate Transfection Kit (Thermo Fisher). Viral supernatant collected 48 hours post-transfection was added to bone marrow c-kit+ cells that were isolated using CD117 MicroBeads on the autoMACS Pro Separator and cultured in MEMα (Gibco) with 15% fetal bovine serum (Atlas) and a cytokine cocktail (TPO 10 ng/mL, IL-3 10 ng/mL, SCF 100 ng/mL, and Flt3-L 50 ng/mL; Peprotech). Cells were centrifuged at 966xg for 90 minutes at 30°C and incubated at 37°C overnight. Viral transduction was repeated the following day. 2–3x105 cells were injected retro-orbitally into lethally irradiated (1000 cGy) mice. Mice were followed and sacrificed if they became symptomatic from either hematologic or non-hematologic etiologies, had a hemoglobin <5 g/dl, or reached the end of the follow-up period (>1 year). Mice were censored if they were sacrificed for non-hematologic reasons or reached the end of the follow-up period.
For secondary transplantation, one million splenocytes from sacrificed mice previously transplanted with single mutant IDH2R172K or double mutant Mir142−/−IDH2R172K cells were injected retro-orbitally into sublethally irradiated (650 cGy) mice.
Statistical Analyses
Data is represented as the mean ± standard error of the mean with relevant statistical tests noted in the figure legends.
Data availability
Small RNA sequencing data from AML samples were submitted to dbGaP (accession number phs000159.v9). All other relevant data are included in the article or supplementary files, or available from the authors upon request.
RESULTS
MIR142 mutations attenuate miR-142-3p and miR-142-5p function
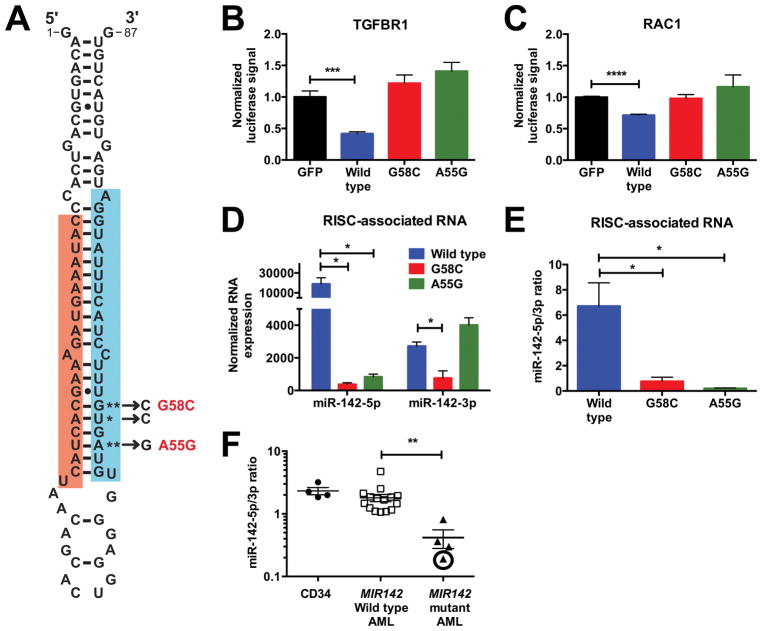
All MIR142 mutations in the TCGA AML cohort localized to the miR-142-3p seed sequence (Fig 1A). MiRNA target prediction programs (mirdb.org) suggested that all three mutations would cause a loss of most 3p targets with no commonly gained targets shared amongst all three mutants. As expected, both the A55G and G58C MIR142 mutants were unable to inhibit translation in a luciferase assay using the 3′ untranslated regions (UTRs) of the known miR-142-3p targets RAC1 (24,25) and TGFBR1 (26) (Fig 1B–C), indicating that AML-associated MIR142 mutations disrupt its 3p function.
Figure 1. MIR142 mutations attenuate miR-142-3p and miR-142-5p functions.
a, MIR142 hairpin. Red: MiR-142-5p. Blue: MiR-142-3p. Mutations in the TCGA cohort are starred with G58C and A55G recurrently observed. b–c, Luciferase assays measuring the activity of wild type, G58C mutant, or A55G mutant MIR142 hairpins against the 3′ UTRs of TGFBR1 (b) (n=4–5) and RAC1 (c) (n=6). d–e, Total levels (d) and ratio (e) of RISC-associated miR-142-5p and miR-142-3p after overexpression of the wild type MIR142 hairpin (n=3) or of its G58C (n=3) and A55G (n=3) mutants. For d, normalized expression is defined as (number of reads mapped to a gene/total number of reads) x106. f, Ratio of miR-142-5p and miR-142-3p in normal donor CD34+ cells, AML cells without MIR142 mutations, and MIR142 mutant AML cells. The circled data point was derived from an AML with bi-allelic MIR142 mutations. Significance was determined with an unpaired t test. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.
Interestingly, overexpression of the A55G and G58C mutant MIR142 hairpins resulted in significantly reduced levels of mature 5p compared to wild type, despite these mutations being localized to 3p (Suppl. Fig 1A). In contrast, levels of the mutant 3p strand derived from these hairpins were similar or increased when compared to wild type, resulting in a reduced 5p:3p ratio (Suppl. Fig 1B). Expression of the mutant MIR142 precursors was similar to that of wild type (Suppl. Fig 1C). Thermodynamic stability at the 5′ end of miRNAs within the small RNA duplex influences miRNA loading into the RISC; the strand with decreased stability towards its 5′ end is preferentially loaded (27). Theoretically, AML-associated MIR142 mutations should decrease stability at the 5′ end of 3p, causing its preferential loading at 5p’s expense. To test this, we assessed the levels of RISC-associated miR-142-5p and miR-142-3p. Indeed overexpression of the A55G and G58C mutant MIR142 hairpins resulted in decreased RISC-associated miR-142-5p compared to wild type and a reduced 5p:3p ratio (Fig 1D–E).
We then assessed miR-142-5p and miR-142-3p expression in AML with or without MIR142 mutations. Consistent with prior reports (28), compared with healthy donor CD34+ cells, miR-142-5p and 3p expression were on average 16 and 31-fold higher respectively in AML (Suppl. Fig 1D). In both AML with wild type MIR142 and in normal CD34+ cells, 5p had slightly higher expression than 3p (Fig 1F & Suppl. Fig 1E). However, in the four MIR142 mutated AMLs, we observed significantly decreased expression of 5p relative to 3p. This was most striking in the AML with bi-allelic MIR142 mutations where 5p levels were markedly reduced. Collectively, these data show that AML-associated MIR142 mutations cause both loss of miR-142-3p function and reduced RISC incorporation and stability of miR-142-5p.
Mir142 loss alters hematopoietic differentiation
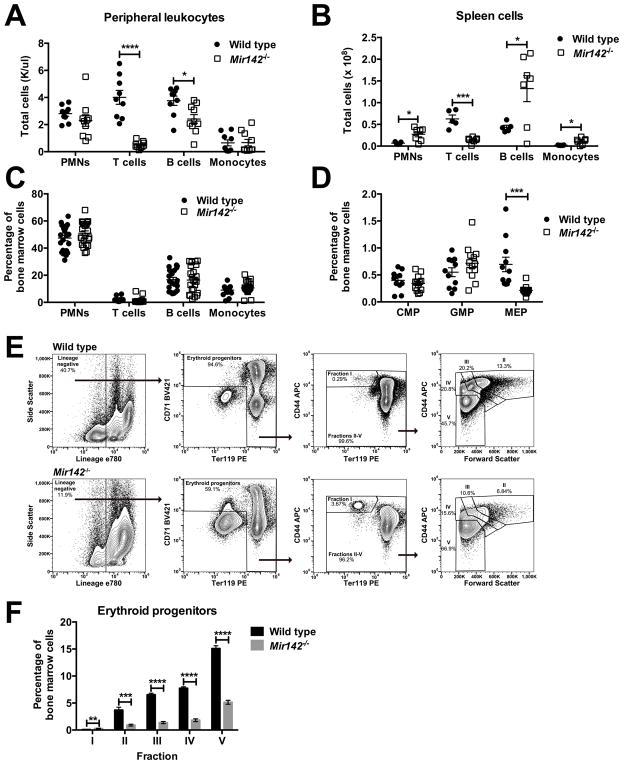
Since AML-associated MIR142 mutations cause a functional loss of both miR-142-3p and 5p, we assessed hematopoiesis in Mir142−/− mice. As reported previously, these mice were leukopenic (19,29), primarily due to a loss of T cells and to a lesser extent of B cells (Fig 2A & Suppl. Fig 2A–B). They also exhibited splenomegaly, with an increased number of mature granulocytes and B cells and a reduced number of T cells (Fig 2B & Suppl. Fig 2C–D) (14,15). In contrast, the bone marrow cellularity of Mir142−/− mice was similar to wild type, with a similar percentage of neutrophils, monocytes, and B cells (Fig 2C & Suppl. Fig 2E–2F). Although at baseline Mir142−/− mice had normal red blood cell and platelet parameters (Suppl. Fig 2G–I), they exhibited a significant decrease in both bone marrow megakaryocyte-erythrocyte progenitors (MEPs) and erythroid progenitors, consistent with a recent report (Fig 2D–F) (20). In contrast, the percentage of granulocyte-monocyte progenitors (GMPs) and common myeloid progenitors (CMPs) in the bone marrow of Mir142−/− mice was similar to wild type (Fig 2D).
Figure 2. Mir142 loss alters differentiation in mature hematopoietic populations.
a–b, Number of cells for the indicated hematopoietic lineage in peripheral blood (a) and spleen (b). c, Percentage of mature hematopoietic cells in the bone marrow. PMNs: neutrophils. d, Percentage of CMPs, GMPs, or MEPs in the bone marrow. e, Representative flow plots of bone marrow erythroid progenitor analysis. f, Percentage of erythroid progenitors in Mir142−/− (n=5) bone marrow compared to control (n=5). Significance was determined with an unpaired t test. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.
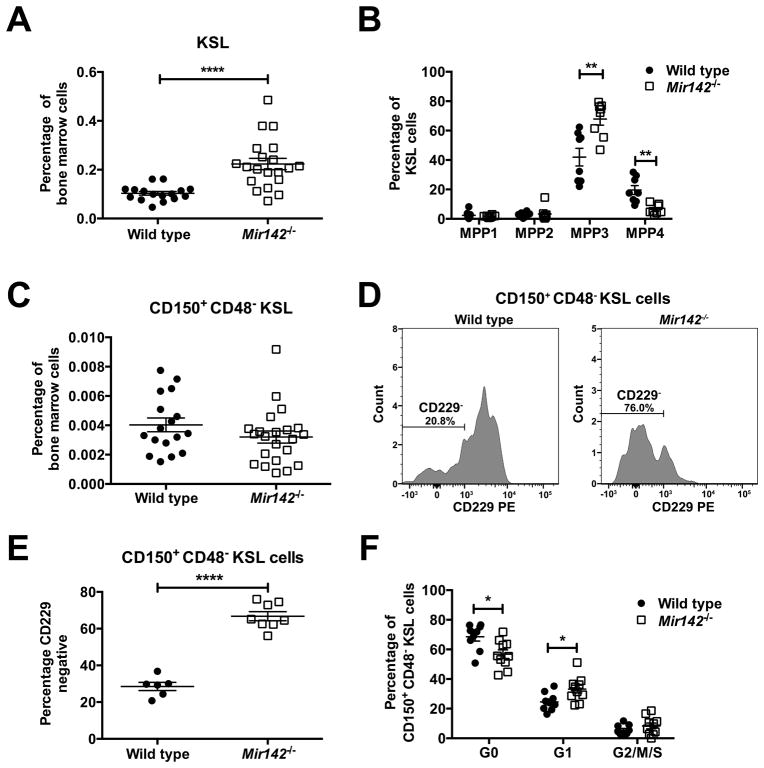
We asked whether Mir142 loss alters multipotent hematopoietic progenitors (MPPs). We observed a modest increase in Kit+Sca+lineage− (KSL) cells in Mir142−/− mice (Fig 3A). As previously described (30), MPPs can be analyzed based on CD34, CD48, CD135, and CD150 expression (Suppl. Fig 3A). We found that the KSL expansion in Mir142−/− mice was due to an increase in myeloid-biased MPP3 (CD150−CD48+CD34+CD135−) cells (Fig 3B). The percentage of phenotypic HSCs (CD150+CD48−KSL cells) in the bone marrow was similar in Mir142−/− and wild type mice (Fig 3C). CD229 surface expression discriminates between lymphoid-biased and more myeloid-biased HSCs, with most HSCs being lymphoid-biased and expressing CD229 in young mice (31). However, in Mir142−/− mice, most phenotypic HSCs had low CD229 expression, suggesting a loss of lymphoid potential (Fig 3D–E). Interestingly, a modest decrease in HSC quiescence was observed in Mir142−/− mice (Fig 3F). These data suggest that Mir142 loss not only alters differentiation in mature hematopoietic populations but also in immature HSPC populations, resulting in a loss of erythroid and lymphoid potential while maintaining myeloid potential.
Figure 3. Mir142 loss alters differentiation in MPPs and HSCs.
a, Percentage of KSL cells in the bone marrow. b, Percentage of multipotent progenitors (MPP1, MPP2, MPP3, and MPP4 cells) within the KSL gate. c, Percentage of LT-HSCs (CD150+CD48−KSL cells) in the bone marrow. d, Representative histograms showing CD229 surface expression on LT-HSCs. e, CD229− cells as a percentage of bone marrow LT-HSCs. f, Cell cycle distribution of LT-HSCs. Significance was determined with an unpaired t test. *P<0.05; **P<0.01; ****P<0.0001.
Mir142−/− bone marrow exhibits reduced repopulating activity with a loss of HSC lymphoid potential
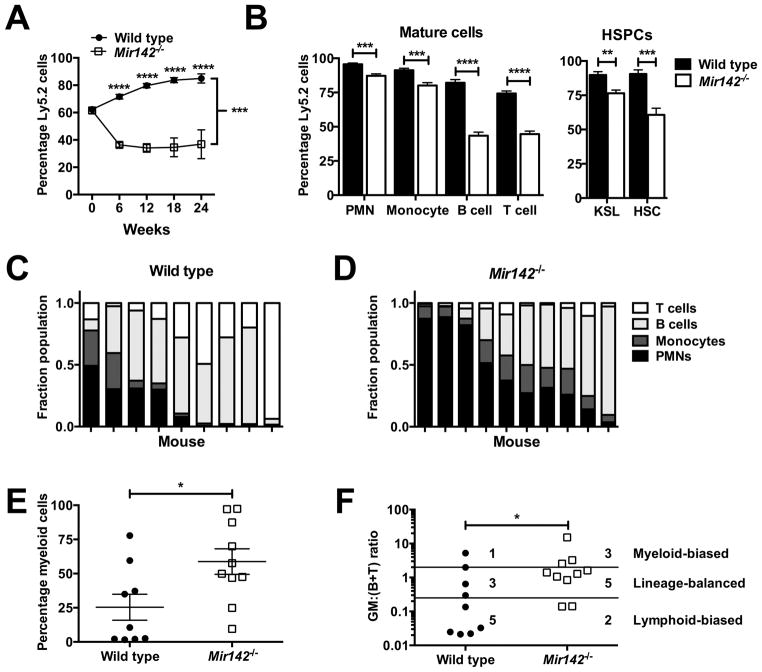
We next assessed HSC function through transplantation experiments. Whole bone marrow competitive repopulation assays showed a modest repopulating defect with Mir142−/− bone marrow (Fig 4A). Post-transplantation, Mir142−/− cells had reduced contribution to the bone marrow myeloid, lymphoid, and HSPC populations (Fig 4B). Consistent with the phenotype of Mir142−/− mice, the reduction was more pronounced in lymphoid versus myeloid cells, suggesting that Mir142−/− HSCs may be deficient in lymphoid potential. To address this, we performed transplantation experiments with a limited number of CD150+CD48−KSL cells. As expected, since young (8–10 week-old) donor mice were used, the majority of wild type HSCs primarily contributed to the lymphoid lineages (Fig 4C). In contrast, Mir142−/− HSCs contributed more significantly to the myeloid lineages (Fig 4D–E). As Mir142 is essential for proper T cell development, we repeated this analysis excluding T cells; again, Mir142−/− HSCs contributed more significantly to the myeloid lineages (Suppl. Fig 4A–C). Using the previously described GM:(B+T) ratio (32), wild type HSCs were more likely to be lymphoid-biased, with Mir142−/− HSCs more likely to be myeloid-biased or balanced (Fig 4F). These data show that Mir142 loss results in a modest decrease in total bone marrow repopulating activity with a loss of HSC lymphoid potential.
Figure 4. Mir142−/− bone marrow exhibits reduced repopulating activity with a loss of HSC lymphoid potential.
a, Percentage donor (Ly5.2) chimerism in the peripheral blood following competitive transplantation of wild type or Mir142−/− bone marrow (week 0: input chimerism). Two independent experiments were performed with a combined total of 9–10 recipients per group. b, Percentage bone marrow donor chimerism 12 weeks post-transplantation (n = 5). KSL: Kit+Sca+Lin− cells. HSC: CD150+CD48−KSL cells. c–d, Sorted donor CD150+CD48−KSL cells (20–25 per recipient) were transplanted into recipients in two independent experiments with peripheral blood donor chimerism assessed 24 weeks later. Shown is the contribution of the indicated lineage to the total donor cell pool in mice transplanted with wild type (c) or Mir142−/− (d) HSCs. Each column represents a mouse. e, Percentage of cells in c or d that were myeloid (Gr1+ or CD115+). f, Quotient of the percentage donor contribution to the overall myeloid lineage (GM; Gr1+ or CD115+) over the sum of the percentage donor contributions to the overall B and T cell lineages (B+T) for individual mice. Myeloid-biased mice had a ratio >2, lineage balanced mice between 0.25 and 2, and lymphoid-biased mice < 0.25. For a, significance was determined with a mixed-model analysis of covariance using input chimerism as a covariate. Pairwise differences were examined with Tukey-Kramer adjusted tests. Significance was determined with a two-way anova for b, with an unpaired t test for e, and with a Mann-Whitney test for f. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.
Mir142 loss increases Hoxa9/a10 expression through derepression of Ash1l
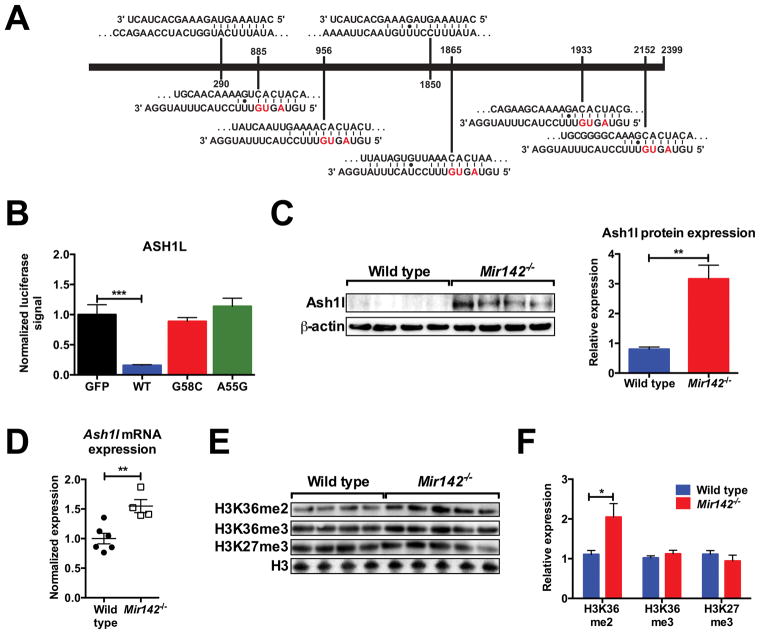
ASH1L was identified as a miR-142 target in thyroid neoplasias (33), with two putative miR-142-5p and five putative miR-142-3p binding sites in its 3′ UTR (Fig. 5A). ASH1L is a member of the trithorax family of histone methyltransferases and is linked to the trimethylation of histone 3 lysine 4 (H3K4me3) (34) and the dimethylation of histone 3 lysine 36 (H3K36me2) (35). In hematopoiesis, ASH1L is essential for maintenance of HSC self-renewal (23) and plays a key role in MLL-induced leukemogenesis (36). In hematopoietic cell lines, ASH1L knockdown promotes erythropoiesis while inhibiting granulopoiesis (37). These observations suggest that loss of miR-142-mediated suppression of Ash1l activity in Mir142−/− mice may contribute to their hematopoietic phenotype.
Figure 5. Mir142 loss results in increased Ash1l activity.
a, Predicted miR-142-5p (top) and miR-142-3p (bottom) target sites in the ASH1L 3′ UTR. Red: mutated in the TCGA cohort. b, Activity of wild type, G58C mutant, or A55G mutant MIR142 hairpins in luciferase assays against the ASH1L 3′ UTR (n=5). c, Western blot measuring Ash1l protein in wild type or Mir142−/− bone marrow cells. Quantification is to the right. d, Ash1l mRNA expression in c-kit+-selected bone marrow cells normalized to wild type. e, Western blot measuring H3K36me2, H3K36me3, and H3K27me3 levels in c-kit+-selected bone marrow cells. f, Quantification of e. Significance was determined with an unpaired t test. *P<0.05; **P<0.01; ***P<0.001.
To test this hypothesis, we first assessed the ability of MIR142 hairpins to suppress protein expression in a luciferase assay using ASH1L’s 3′ UTR (Fig. 5B). Wild type MIR142 decreased the luciferase signal by >80%. In contrast, the A55G and G58C mutant hairpins didn’t suppress luciferase activity. Ash1l protein levels were increased ~three-fold in total bone marrow cells from Mir142−/− mice compared to wild type, confirming miR-142’s importance in suppressing Ash1l protein expression in vivo (Fig. 5C). Interestingly, Ash1l mRNA expression was increased in Mir142−/− c-kit+-selected bone marrow cells, suggesting that miR-142 also regulates Ash1l mRNA levels by promoting its decay (Fig. 5D). To determine the consequences of Ash1l overexpression, we interrogated global histone modification levels in c-kit+-selected bone marrow cells. Consistent with Ash1l’s function as an H3K36 dimethyltransferase, global H3K36me2 levels were increased two-fold in Mir142−/− cells compared to wild type; other histone modification marks (H3K36me3 and H3K27me3) were unchanged (Fig 5E–F).
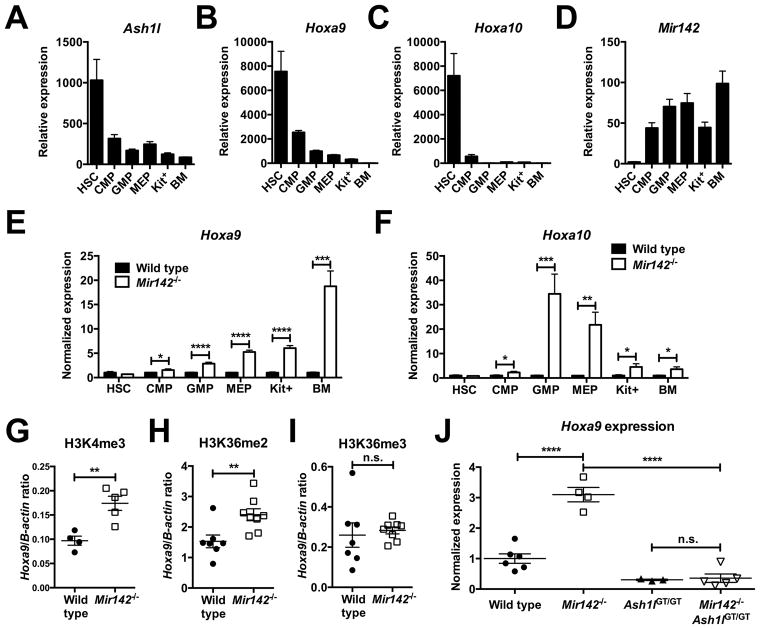
ASH1L is an important positive regulator of HOX gene expression (34,35,37). During murine hematopoietic differentiation, Ash1l and Hoxa9/a10 exhibit a similar pattern of mRNA expression, with high expression in HSCs and a progressive decline with differentiation (Fig 6A–C). In contrast, Mir142 is expressed at lower levels in HSCs, increasing in expression in more mature populations (Fig 6D). As miR-142 suppresses Ash1l activity in hematopoietic cells, we hypothesized that it may contribute to the decrease in Hoxa9/a10 mRNA expression during hematopoietic differentiation. To test this, we measured Hoxa9/a10 expression in sorted Mir142−/− hematopoietic populations. As expected, Mir142 loss did not affect Hoxa9/a10 expression in HSCs. However, we observed a significant increase in Hoxa9/a10 expression in both Mir142−/− myeloid committed progenitors and total bone marrow cells (Fig 6E–F).
Figure 6. Mir142 loss increases Hoxa9/a10 expression through the derepression of Ash1l.
a–d, RNA expression of Ash1l (a), Hoxa9 (b), Hoxa10 (c), and Mir142-5p (d) in the indicated hematopoietic cell populations in wild type mice (n=4–7). Kit+: purified c-kit+ bone marrow cells; BM: unselected bone marrow cells. For a–c, expression is relative to Actb. For d, expression is relative to Sno202. e–f, RNA expression of Hoxa9 (e) and Hoxa10 (f) in the indicated hematopoietic cell populations (n=4–7). Data are normalized to wild type. g–i, Ratio of cross-linked DNA at the Hoxa9 promoter versus at the β-actin promoter pulled down with antibodies against H3K4me3 (g), H3K36me2 (h), or H3K36me3 (i). j, Hoxa9 expression in total bone marrow cells. Data is normalized to wild type. Significance was determined with an unpaired t test. n.s. not significant. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.
We next measured histone modifications at the Hoxa9 promoter. We focused on H3K4 trimethylation and H3K36 dimethylation as Ash1l is implicated in the regulation of these histone modifications. H3K4me3 and H3K36me2 (but not H3K36me3) occupancy at the Hoxa9 promoter was increased in Mir142−/− c-kit+ bone marrow cells when compared to wild type (Figure 6G–I). Collectively, these data suggest that Mir142 loss results in increased Hoxa9 expression through enhanced Ash1l activity. To test this, we crossed Mir142−/− mice with Ash1l deficient mice (Ash1lGT/GT). Ash1lGT/GT mice have a gene trap insertion cassette following exon 1 of Ash1l, causing a >90% reduction in its full-length transcript (23). We then assessed Hoxa9 expression in total bone marrow cells (Fig 6J). As previously demonstrated, Mir142−/− mice had significantly increased Hoxa9 expression compared to wild type. As reported previously, Ash1lGT/GT mice had slightly decreased Hoxa9 expression. In double mutant Mir142−/−Ash1lGT/GT mice, the effect of Mir142 loss on Hoxa9 expression was abrogated, showing that miR-142 regulates Hoxa9 expression primarily by repressing Ash1l activity.
Mir142 loss enhances IDH2R172K disease-initiating capacity
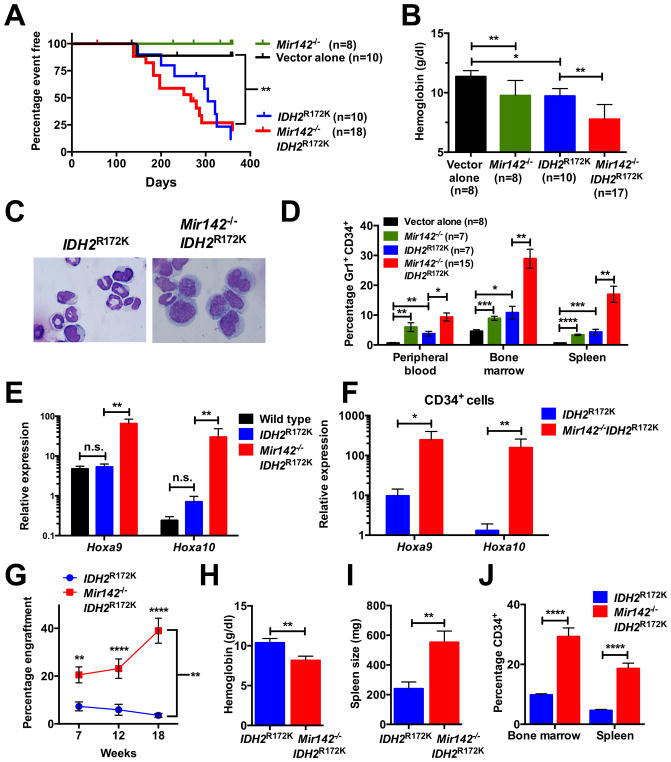
In the TCGA AML cohort, all MIR142 mutant patients harbored mutations in IDH1 or IDH2 (6). To determine whether MIR142 loss cooperates with IDH mutations to induce AML, we reconstituted lethally irradiated mice with wild type or Mir142−/− c-kit+-selected cells transduced with a retrovirus expressing human IDH2R172K. As reported previously (38,39), mice transplanted with wild type cells expressing IDH2R172K developed a fatal myelodysplastic-like phenotype characterized by anemia, thrombocytopenia, splenomegaly, bone marrow hypocellularity, and a variable white blood cell count (Fig 7A, Suppl. Fig 5A–E). Concomitant loss of Mir142 did not affect the latency or penetrance of this disorder (Fig. 7A). However, consistent with the Mir142−/− phenotype, double mutant mice had a significantly more severe anemia (Fig 7B). They also exhibited increased hematopoietic immaturity with a higher percentage of myeloid blasts in the bone marrow, blood, and spleen (Fig 7C–D, Suppl. Fig 5F). In fact, 11/15 (73.3%) of double mutant mice exceeded the 20% myeloblasts diagnostic threshold for AML, compared to only 1/7 (14.3%) mice expressing IDH2R172K alone (P<0.05 by Fisher’s Exact Test). Mice transplanted with wild type or Mir142−/− cells lacking IDH2R172K expression did not develop a progressive hematologic disorder.
Figure 7. Mir142 loss enhances IDH2R172K disease initiating capacity.
a–f, Mice were transplanted with wild type or Mir142−/− c-kit+ cells transduced with empty retrovirus (vector alone) or retrovirus expressing IDH2R172K. a, Mouse survival. Mice were sacrificed if distressed or if their hemoglobin was < 5 g/dl. Mice sacrificed for non-hematologic reasons were censored. b, Hemoglobin twenty weeks post-transplantation. c, Representative bone marrow cytospins. d, Percentage of Gr-1+CD34+ cells in the peripheral blood, bone marrow, or spleen upon sacrifice. e–f, Hoxa9/a10 RNA expression in total bone marrow (e) or sorted CD34+ (f) cells upon sacrifice (n=4–6). Expression is relative to Actb. g–j, 1x106 splenocytes from IDH2R172K or Mir142−/−IDH2R172K primary recipient mice were transplanted into sublethally irradiated secondary recipients. Four IDH2R172K and five Mir142−/−IDH2R172K donors were separately transplanted into 3–7 recipients, with 20 and 25 recipients respectively analyzed. Shown are the donor chimerism in peripheral blood (g), hemoglobin 18 weeks post-transplantation (h), and the spleen size (i) and percentage of CD34+ cells in the bone marrow and spleen (j) upon sacrifice. Significance was determined with a Mantel-Cox log rank test for a, with an unpaired t test for b, d, and h–j, and with a Mann-Whitney test for e–f. For g, significance was determined with a mixed-model analysis of variance using log (percent engraftment) as the dependent variable. Pairwise differences were examined with Tukey-Kramer adjusted tests. n.s. not significant. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.
Previous studies showed that Hoxa9 overexpression cooperates with Idh mutations to induce a myeloproliferative-like disorder in mice (38,40,41). Since Mir142 loss is associated with increased Hoxa9/a10 expression, we measured expression of these two genes in total bone marrow cells from IDH2R172K or IDH2R172KMir142−/− mice. IDH2R172K expression alone did not cause increased Hoxa9/a10 expression (Fig 7E). In contrast, both Hoxa9 and Hoxa10 expression were significantly increased in IDH2R172KMir142−/− cells. Increased Hoxa9/a10 expression also was observed in flow-sorted CD34+ cells from IDH2R172KMir142−/− mice compared to IDH2R172K mice (Fig 7F).
Increased Hoxa9/a10 expression has been linked to enhanced self-renewal in hematopoietic cells (42,43). Thus, we asked whether Mir142 loss increased the ability of IDH2R172K mutant hematopoietic cells to initiate disease following transplantation. We transplanted one million splenocytes from single (IDH2R172K alone) or double mutant mice exhibiting the previously described phenotype into sub-lethally irradiated recipients. Whereas cells from most single mutant donors failed to engraft, cells from all double mutant donor mice engrafted at varying efficiency (Fig 7G). Engrafted mice developed a similar phenotype to their donors, exhibiting anemia, splenomegaly, and hematopoietic immaturity (Fig 7H–J). Collectively, these data suggest that Mir142 loss increases the disease-initiating activity of IDH2R172K mutant hematopoietic cells.
DISCUSSION
In this study, we show that AML-associated MIR142 mutations disrupt miR-142-3p’s ability to inhibit expression of its target genes. Moreover, by altering the preference for which miRNA strand is incorporated into the RISC, they also decrease miR-142-5p levels. This may be applicable to other miRNA mutations. Any mutation disrupting Watson Crick base pairing at the ends of the miRNA duplex may influence the preferential loading of one miRNA strand versus the other. In the TCGA sequencing of solid malignancies, somatic mutations were identified in the 5p and 3p seed sequence of multiple miRNAs (44). In a significant fraction of cases, the more highly expressed miRNA strand was the non-mutated strand (http://www.mirbase.org). In these cases, the primary effect of these mutations may be to reduce expression of the opposite strand.
To model the effect of miR-142-3p seed sequence mutations, we characterized hematopoiesis in Mir142−/− mice. We confirmed prior studies showing that Mir142 loss results in multiple hematopoietic alterations, including myeloid expansion, impaired erythropoiesis, and T cell lymphopenia. However, we also show that Mir142 plays a role in the function and lineage determination of multipotent HSPCs. In Mir142−/− mice, total bone marrow repopulating activity and HSC quiescence is modestly reduced. Among multipotent progenitors, there is an expansion of myeloid-biased MPP3 cells. Transplantation studies using a small number of phenotypic HSCs provide evidence that Mir142−/− HSCs have reduced lymphoid potential. Thus, although Mir142 is expressed at lower levels in HSPCs, it contributes to their function.
Our data show miR-142 to be a key post-transcriptional regulator of Ash1l in hematopoietic cells, with increased Ash1l expression and activity in Mir142−/− mice. Ashl1 is a key positive regulator of Hoxa gene family expression, whose loss results in a decline of functional HSCs (23). Our data suggest that Mir142 limits Hoxa gene expression during hematopoietic differentiation by suppressing Ash1l activity. Mir142 loss had no effect on Hoxa9/a10 expression in HSCs but caused a significant increase in Hoxa9/a10 expression in myeloid committed and total bone marrow cells. Of note, enforced expression of Hoxa9 or Hoxa10 phenocopies some of the hematopoietic alterations in Mir142−/− mice, including myeloid expansion and impaired lymphopoiesis and erythropoiesis (43,45). However, not all hematopoietic abnormalities in Mir142−/− mice can be explained by Ash1l overexpression. For example, Ash1l loss results in a marked reduction in HSCs (23). Increased Ash1l expression in HSPCs would not explain the modest loss of Mir142−/− total bone marrow repopulating activity. Mir142 target genes contributing to the altered functionality and reduced lymphoid potential of Mir142−/− HSCs require further investigation.
Hoxa gene family expression is essential for normal HSC function. HSCs lacking specific Hoxa genes have an impaired ability to respond to stress and reduced repopulating ability (42). Conversely, enforced expression of specific Hoxa genes results in HSC expansion and a transplantable myeloproliferative phenotype (43,46). Certain oncogenes (e.g. MLL translocations) directly induce HOX expression (47). In most AMLs, the cause of elevated HOX expression remains unclear. Many AML subtypes have a similar HOX expression pattern to normal CD34+ cells, suggesting that, in these AMLs, HOX genes aren’t actively dysregulated but instead “captured” in their normal stem cell state (48). The mechanisms through which stem-cell-like HOX expression patterns are maintained in myeloblasts are largely unknown. We provide evidence that MIR142 loss-of-function mutations, by derepressing ASH1L, represent a novel mechanism of increased HOXA gene expression in AML.
We show that MIR142 loss isn’t sufficient to induce leukemia, indicating that additional leukemia-initiating events are required. All four MIR142 mutant AMLs in the TCGA cohort carried mutations in IDH1 or IDH2. Our data suggest that concomitant Mir142 loss enhances the disease-initiating activity of IDH2R172K mutant hematopoietic cells. In mice, Hoxa9 overexpression cooperates with IDH1/2 mutations and their oncogenic metabolite (R)-2-hydroxyglutarate to promote leukemogenesis Our data suggest that MIR142 loss enhances the disease-initiating activity of IDH2R172K mutant hematopoietic cells through derepression of ASH1L and increased HOXA gene expression. Unlike other leukemia drivers, IDH mutations are rarely observed in clonal hematopoiesis and haven’t been shown to provide a fitness advantage upon transplantation. It is possible that IDH mutations do not by themselves provide a self-renewal advantage and require the maintenance of HOX expression for leukemic transformation. This is consistent with data showing that mutant IDH1/2 expression in mice doesn’t result in enhanced HSC self-renewal (38,41) and with our data showing that the MDS-like disorder induced by retroviral overexpression of IDH2R172K is not highly transplantable.
MIR142 mutations are rare in AML, occurring in 0.5–2% of patients in several cohorts (6,7) and not identified in other series (49). Consistent with our hypothesis that loss of MIR142 results in sustained HOXA9/A10 expression, all three MIR142-mutated AMLs in the TCGA cohort for whom RNA sequencing was available had HOXA9 expression >1.0 by FPKM (6). However, high HOXA9/A10 expression was observed in most IDH1/2-mutated AMLs, regardless of MIR142 mutation status. As our data suggests that IDH mutations do not independently promote HOX expression, other mechanisms likely contribute to sustained HOXA9/10 expression in these cases.
In summary, we show that AML-associated MIR142 mutations cause functional loss of miR-142-3p and miR-142-5p. In mice, Mir142 loss results in altered HSC function with reduced lymphoid potential and enhances the disease initiating capacity of IDH2 mutant hematopoietic cells. Mir142 loss causes derepression of Ash1l, resulting in the inappropriate maintenance of Hoxa9/a10 gene expression during hematopoietic differentiation. Thus, AML-associated MIR142 mutations may contribute to leukemogenesis by increasing HOXA gene expression in myeloid-committed progenitors. Given the importance of HOXA expression in maintaining self-renewal, targeting these pathways (e.g. by inhibiting ASH1L methyltransferase activity) may have therapeutic benefit against AML.
Supplementary Material
Significance.
These findings provide mechanistic insights into the role of miRNAs in leukemogenesis and hematopoietic stem cell function.
Acknowledgments
This work was supported by National Institutes of Health, National Cancer Institute grants PO1 CA101937 (D.C. Link & T.J. Ley), P50 CA171963 (D.C. Link), K08 CA197369 (T.N. Wong), T32 CA113275 (J.C. Yao), and F32 CA200253 (M.M. Berrien-Elliott), by National Institutes of Health, National Heart, Lung, and Blood Institute grants T32 HL7088-37 (M.C. Trissal) and K12 HL087107 (T.N. Wong), by National Institutes of Health, National Institute on Aging grant R01 AG050509 (I.P. Maillard), by National Institutes of Health, National Institute of Allergy and Infectious Diseases grant R01 AI102924 (T.A. Fehniger), and by the University of Michigan Organogenesis Training Program grant T32 HD007505 (G. Jih). Technical support was provided by the Alvin J. Siteman Cancer Center Tissue Procurement Core and the High-Speed Cell Sorting Core at Washington University School of Medicine, which are supported by National Institutes of Health, National Cancer Institute grant P30 CA91842.
The authors thank Amy P. Schmidt for technical assistance and Jackie Tucker-Davis for animal care.
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
References
- 1.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–85. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 2.Iwakawa HO, Tomari Y. The Functions of MicroRNAs: mRNA Decay and Translational Repression. Trends Cell Biol. 2015;25:651–65. doi: 10.1016/j.tcb.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 3.Shaham L, Binder V, Gefen N, Borkhardt A, Izraeli S. MiR-125 in normal and malignant hematopoiesis. Leukemia. 2012;26:2011–8. doi: 10.1038/leu.2012.90. [DOI] [PubMed] [Google Scholar]
- 4.Starczynowski DT, Kuchenbauer F, Argiropoulos B, Sung S, Morin R, Muranyi A, et al. Identification of miR-145 and miR-146a as mediators of the 5q- syndrome phenotype. Nat Med. 2010;16:49–58. doi: 10.1038/nm.2054. [DOI] [PubMed] [Google Scholar]
- 5.Ramsingh G, Jacoby MA, Shao J, De Jesus Pizzaro RE, Shen D, Trissal M, et al. Acquired copy number alterations of miRNA genes in acute myeloid leukemia are uncommon. Blood. 2013;122:e44. doi: 10.1182/blood-2013-03-488007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research N. Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–74. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thol F, Scherr M, Kirchner A, Shahswar R, Battmer K, Kade S, et al. Clinical and functional implications of microRNA mutations in a cohort of 935 patients with myelodysplastic syndromes and acute myeloid leukemia. Haematologica. 2015;100:e122–4. doi: 10.3324/haematol.2014.120345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouska A, Zhang W, Gong Q, Iqbal J, Scuto A, Vose J, et al. Combined copy number and mutation analysis identifies oncogenic pathways associated with transformation of follicular lymphoma. Leukemia. 2017;31:83–91. doi: 10.1038/leu.2016.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hezaveh K, Kloetgen A, Bernhart SH, Mahapatra KD, Lenze D, Richter J, et al. Alterations of microRNA and microRNA-regulated messenger RNA expression in germinal center B-cell lymphomas determined by integrative sequencing analysis. Haematologica. 2016;101:1380–9. doi: 10.3324/haematol.2016.143891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwanhian W, Lenze D, Alles J, Motsch N, Barth S, Doll C, et al. MicroRNA-142 is mutated in about 20% of diffuse large B-cell lymphoma. Cancer Med. 2012;1:141–55. doi: 10.1002/cam4.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morin RD, Assouline S, Alcaide M, Mohajeri A, Johnston RL, Chong L, et al. Genetic Landscapes of Relapsed and Refractory Diffuse Large B-Cell Lymphomas. Clin Cancer Res. 2016;22:2290–300. doi: 10.1158/1078-0432.CCR-15-2123. [DOI] [PubMed] [Google Scholar]
- 12.Merkerova M, Belickova M, Bruchova H. Differential expression of microRNAs in hematopoietic cell lineages. Eur J Haematol. 2008;81:304–10. doi: 10.1111/j.1600-0609.2008.01111.x. [DOI] [PubMed] [Google Scholar]
- 13.Petriv OI, Kuchenbauer F, Delaney AD, Lecault V, White A, Kent D, et al. Comprehensive microRNA expression profiling of the hematopoietic hierarchy. Proc Natl Acad Sci U S A. 2010;107:15443–8. doi: 10.1073/pnas.1009320107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kramer NJ, Wang WL, Reyes EY, Kumar B, Chen CC, Ramakrishna C, et al. Altered lymphopoiesis and immunodeficiency in miR-142 null mice. Blood. 2015;125:3720–30. doi: 10.1182/blood-2014-10-603951. [DOI] [PubMed] [Google Scholar]
- 15.Mildner A, Chapnik E, Manor O, Yona S, Kim KW, Aychek T, et al. Mononuclear phagocyte miRNome analysis identifies miR-142 as critical regulator of murine dendritic cell homeostasis. Blood. 2013;121:1016–27. doi: 10.1182/blood-2012-07-445999. [DOI] [PubMed] [Google Scholar]
- 16.Mildner A, Chapnik E, Varol D, Aychek T, Lampl N, Rivkin N, et al. MicroRNA-142 controls thymocyte proliferation. Eur J Immunol. 2017;47:1142–52. doi: 10.1002/eji.201746987. [DOI] [PubMed] [Google Scholar]
- 17.Sun Y, Oravecz-Wilson K, Mathewson N, Wang Y, McEachin R, Liu C, et al. Mature T cell responses are controlled by microRNA-142. J Clin Invest. 2015;125:2825–40. doi: 10.1172/JCI78753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamada Y, Kosaka K, Miyazawa T, Kurata-Miura K, Yoshida T. miR-142-3p enhances FcepsilonRI-mediated degranulation in mast cells. Biochem Biophys Res Commun. 2014;443:980–6. doi: 10.1016/j.bbrc.2013.12.078. [DOI] [PubMed] [Google Scholar]
- 19.Chapnik E, Rivkin N, Mildner A, Beck G, Pasvolsky R, Metzl-Raz E, et al. miR-142 orchestrates a network of actin cytoskeleton regulators during megakaryopoiesis. Elife. 2014;3:e01964. doi: 10.7554/eLife.01964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rivkin N, Chapnik E, Birger Y, Yanowski E, Curato C, Mildner A, et al. Rac1 functions downstream of miR-142 in regulation of erythropoiesis. Haematologica. 2017 doi: 10.3324/haematol.2017.171736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rivkin N, Chapnik E, Mildner A, Barshtein G, Porat Z, Kartvelishvily E, et al. Erythrocyte survival is controlled by microRNA-142. Haematologica. 2017;102:676–85. doi: 10.3324/haematol.2016.156109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu X, Li X, He Q, Gao J, Gao Y, Liu B, et al. miR-142-3p regulates the formation and differentiation of hematopoietic stem cells in vertebrates. Cell Res. 2013;23:1356–68. doi: 10.1038/cr.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones M, Chase J, Brinkmeier M, Xu J, Weinberg DN, Schira J, et al. Ash1l controls quiescence and self-renewal potential in hematopoietic stem cells. J Clin Invest. 2015;125:2007–20. doi: 10.1172/JCI78124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu L, Cai C, Wang X, Liu M, Li X, Tang H. MicroRNA-142-3p, a new regulator of RAC1, suppresses the migration and invasion of hepatocellular carcinoma cells. FEBS Lett. 2011;585:1322–30. doi: 10.1016/j.febslet.2011.03.067. [DOI] [PubMed] [Google Scholar]
- 25.Zheng Z, Ding M, Ni J, Song D, Huang J, Wang J. MiR-142 acts as a tumor suppressor in osteosarcoma cell lines by targeting Rac1. Oncol Rep. 2015;33:1291–9. doi: 10.3892/or.2014.3687. [DOI] [PubMed] [Google Scholar]
- 26.Lei Z, Xu G, Wang L, Yang H, Liu X, Zhao J, et al. MiR-142-3p represses TGF-beta-induced growth inhibition through repression of TGFbetaR1 in non-small cell lung cancer. FASEB J. 2014;28:2696–704. doi: 10.1096/fj.13-247288. [DOI] [PubMed] [Google Scholar]
- 27.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–16. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 28.Dahlhaus M, Roolf C, Ruck S, Lange S, Freund M, Junghanss C. Expression and prognostic significance of hsa-miR-142-3p in acute leukemias. Neoplasma. 2013;60:432–8. doi: 10.4149/neo_2013_056. [DOI] [PubMed] [Google Scholar]
- 29.Shrestha A, Carraro G, El Agha E, Mukhametshina R, Chao CM, Rizvanov A, et al. Generation and Validation of miR-142 Knock Out Mice. PLoS One. 2015;10:e0136913. doi: 10.1371/journal.pone.0136913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cabezas-Wallscheid N, Klimmeck D, Hansson J, Lipka DB, Reyes A, Wang Q, et al. Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell. 2014;15:507–22. doi: 10.1016/j.stem.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 31.Oguro H, Ding L, Morrison SJ. SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell Stem Cell. 2013;13:102–16. doi: 10.1016/j.stem.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dykstra B, Kent D, Bowie M, McCaffrey L, Hamilton M, Lyons K, et al. Long-term propagation of distinct hematopoietic differentiation programs in vivo. Cell Stem Cell. 2007;1:218–29. doi: 10.1016/j.stem.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 33.Colamaio M, Puca F, Ragozzino E, Gemei M, Decaussin-Petrucci M, Aiello C, et al. miR-142-3p down-regulation contributes to thyroid follicular tumorigenesis by targeting ASH1L and MLL1. J Clin Endocrinol Metab. 2015;100:E59–69. doi: 10.1210/jc.2014-2280. [DOI] [PubMed] [Google Scholar]
- 34.Gregory GD, Vakoc CR, Rozovskaia T, Zheng X, Patel S, Nakamura T, et al. Mammalian ASH1L is a histone methyltransferase that occupies the transcribed region of active genes. Mol Cell Biol. 2007;27:8466–79. doi: 10.1128/MCB.00993-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanaka Y, Katagiri Z, Kawahashi K, Kioussis D, Kitajima S. Trithorax-group protein ASH1 methylates histone H3 lysine 36. Gene. 2007;397:161–8. doi: 10.1016/j.gene.2007.04.027. [DOI] [PubMed] [Google Scholar]
- 36.Zhu L, Li Q, Wong SH, Huang M, Klein BJ, Shen J, et al. ASH1L Links Histone H3 Lysine 36 Dimethylation to MLL Leukemia. Cancer Discov. 2016;6:770–83. doi: 10.1158/2159-8290.CD-16-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanaka Y, Kawahashi K, Katagiri Z, Nakayama Y, Mahajan M, Kioussis D. Dual function of histone H3 lysine 36 methyltransferase ASH1 in regulation of Hox gene expression. PLoS One. 2011;6:e28171. doi: 10.1371/journal.pone.0028171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kats LM, Reschke M, Taulli R, Pozdnyakova O, Burgess K, Bhargava P, et al. Proto-oncogenic role of mutant IDH2 in leukemia initiation and maintenance. Cell Stem Cell. 2014;14:329–41. doi: 10.1016/j.stem.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sasaki M, Knobbe CB, Munger JC, Lind EF, Brenner D, Brustle A, et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. 2012;488:656–9. doi: 10.1038/nature11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chaturvedi A, Araujo Cruz MM, Jyotsana N, Sharma A, Goparaju R, Schwarzer A, et al. Enantiomer-specific and paracrine leukemogenicity of mutant IDH metabolite 2-hydroxyglutarate. Leukemia. 2016;30:1708–15. doi: 10.1038/leu.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chaturvedi A, Araujo Cruz MM, Jyotsana N, Sharma A, Yun H, Gorlich K, et al. Mutant IDH1 promotes leukemogenesis in vivo and can be specifically targeted in human AML. Blood. 2013;122:2877–87. doi: 10.1182/blood-2013-03-491571. [DOI] [PubMed] [Google Scholar]
- 42.Lawrence HJ, Christensen J, Fong S, Hu YL, Weissman I, Sauvageau G, et al. Loss of expression of the Hoxa-9 homeobox gene impairs the proliferation and repopulating ability of hematopoietic stem cells. Blood. 2005;106:3988–94. doi: 10.1182/blood-2005-05-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thorsteinsdottir U, Mamo A, Kroon E, Jerome L, Bijl J, Lawrence HJ, et al. Overexpression of the myeloid leukemia-associated Hoxa9 gene in bone marrow cells induces stem cell expansion. Blood. 2002;99:121–9. doi: 10.1182/blood.v99.1.121. [DOI] [PubMed] [Google Scholar]
- 44.Bhattacharya A, Cui Y. SomamiR 2. 0: a database of cancer somatic mutations altering microRNA-ceRNA interactions. Nucleic Acids Res. 2016;44:D1005–10. doi: 10.1093/nar/gkv1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buske C, Feuring-Buske M, Antonchuk J, Rosten P, Hogge DE, Eaves CJ, et al. Overexpression of HOXA10 perturbs human lymphomyelopoiesis in vitro and in vivo. Blood. 2001;97:2286–92. doi: 10.1182/blood.v97.8.2286. [DOI] [PubMed] [Google Scholar]
- 46.Thorsteinsdottir U, Sauvageau G, Hough MR, Dragowska W, Lansdorp PM, Lawrence HJ, et al. Overexpression of HOXA10 in murine hematopoietic cells perturbs both myeloid and lymphoid differentiation and leads to acute myeloid leukemia. Mol Cell Biol. 1997;17:495–505. doi: 10.1128/mcb.17.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–17. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 48.Spencer DH, Young MA, Lamprecht TL, Helton NM, Fulton R, O’Laughlin M, et al. Epigenomic analysis of the HOX gene loci reveals mechanisms that may control canonical expression patterns in AML and normal hematopoietic cells. Leukemia. 2015;29:1279–89. doi: 10.1038/leu.2015.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen FF, Lou YJ, Chen J, Jin J, Zhou JN, Yin XF, et al. Absence of miR-142 mutation in Chinese patients with acute myeloid leukemia. Leuk Lymphoma. 2014;55:2961–2. doi: 10.3109/10428194.2014.907892. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Small RNA sequencing data from AML samples were submitted to dbGaP (accession number phs000159.v9). All other relevant data are included in the article or supplementary files, or available from the authors upon request.