Abstract
High-risk human papillomavirus-associated cancers express viral oncoproteins (e.g., E6 and E7) that induce and maintain the malignant phenotype. The viral origin of these proteins makes them attractive targets for development of a therapeutic vaccine. Camelid-derived single-domain antibody fragments (nanobodies or VHHs) that recognize cell surface proteins on antigen-presenting cells (APCs) can serve as targeted delivery vehicles for antigens attached to them. Such VHHs were shown to induce CD4+ and CD8+ T-cell responses against model antigens conjugated to them via sortase, but antitumor responses had not yet been investigated. Here, we tested the ability of an anti-CD11b VHH (VHHCD11b) to target APCs and serve as the basis for a therapeutic vaccine to induce CD8+ T cell responses against HPV+ tumors. Mice immunized with VHHCD11b conjugated to an H-2Db-restricted immunodominant E7 epitope (E749–57) had more E7-specific CD8+ T cells compared to those immunized with E749–57 peptide alone. These CD8+ T cells acted prophylactically and conferred protection against a subsequent challenge with HPV E7-expressing tumor cells. In a therapeutic setting, VHHCD11b-E749–57 vaccination resulted in greater numbers of CD8+ tumor-infiltrating lymphocytes compared to mice receiving E749–57 peptide alone in HPV+ tumor-bearing mice, as measured by in vivo noninvasive VHH-based immune-positron emission tomography (immunoPET), which correlated with tumor regression and survival outcome. Together, these results demonstrate that VHHs can serve as a therapeutic cancer vaccine platform for HPV-induced cancers.
Introduction
Prophylactic vaccination against human papillomavirus (HPV) using capsid protein-based virus-like particles (VLPs) elicits protective humoral immunity against subsequent viral exposure and can prevent HPV-induced malignancies (1). However, for economic, political, and cultural reasons, not all individuals at risk for HPV infection receive the vaccine (2, 3). Consequently, transmission of HPV and the attendant risk of malignancies remain major global concerns (4). HPV-induced malignancies are of viral origin, and as such are excellent immunological targets because viral antigens (Ags) are distinct from self-Ags. HPV type 16 (HPV16) is the most common oncogenic high-risk HPV strain (5), with its primary drivers of oncogenesis, the early genes E6 and E7, constitutively expressed in HPV-induced tumors (6), making them major targets of antitumor vaccination strategies (7). Although prophylactic vaccination with VLP-based vaccines is protective prior to HPV exposure, established HPV+ tumors will require a vaccination approach that elicits early gene product-specific cytotoxic CD8+ T cells to eradicate tumors. Targeted delivery of Ags to professional antigen-presenting cells (APCs) enhances immunity against the delivered Ags through the generation of Ag-specific CD8+ T cells (8, 9). Here we explored single-domain antibody fragments (also known as nanobodies or VHHs) as a therapeutic vaccine platform to deliver and induce cellular immunity against such VHH-conjugated antigens in an HPV cancer model.
VHHs are derived from the variable region of heavy chain-only antibodies naturally present in the serum of camelids (10). They are the smallest antibody fragments that retain Ag specificity (11). Their small size confers a number of desirable properties, such as stability, solubility, ease of production and modification, short circulatory half-lives, and similarity to human immunoglobulin V region sequences (12, 13). We demonstrated that VHHs specific for immune cell surface proteins (i.e. MHC class II, CD36, and CD11b) can deliver conjugated Ags to APCs and elicit an immune response to them (14). An Ag conjugated to the anti-CD11b VHH (VHHCD11b or DC13) raised the strongest CD8+ T-cell response against a model peptide antigen derived from ovalbumin. CD11b is the α subunit of integrin αMβ2 (Mac-1), expressed on neutrophils, macrophages, myeloid dendritic cells (DCs), NK cells, and subsets of CD8+ T cells and B cells (15). To conjugate an Ag to a specific site on the VHH, we installed a sortase A recognition motif near the C-terminus of the VHH, and used an enzymatic transacylation reaction to attach a triglycine-equipped peptide (G3-peptide) containing the Ag of interest (16, 17). We thus conjugated an immunodominant HPV type 16 E7 Ag (HPV16 E749–57) to VHHCD11b via a sortase A-catalyzed reaction (Fig. 1). This VHH-Ag conjugate was successful as a therapeutic vaccine in an HPV cancer model in mice.
Figure 1.
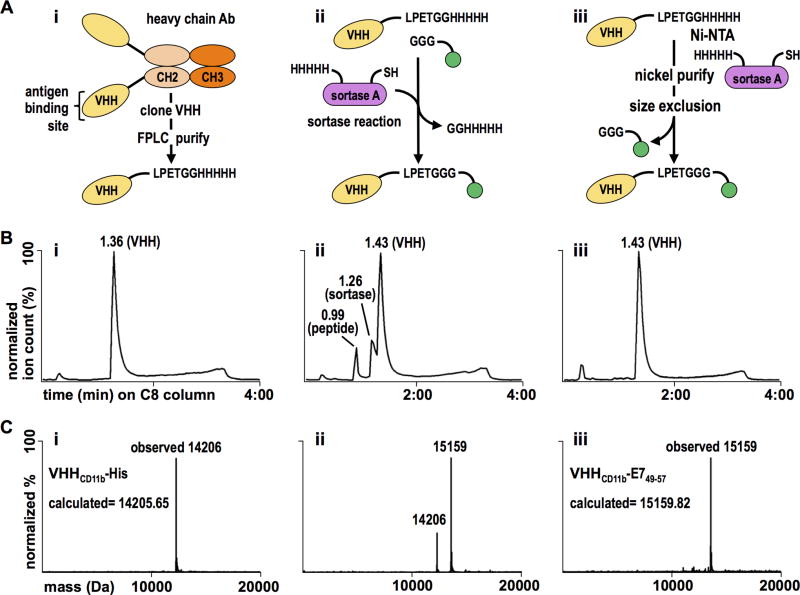
VHHCD11b-E749–57 conjugate design, purification, and validation. A) i. VHHCD11b (yellow) was cloned from the VHH genes of an immunized alpaca, expressed with a C-terminal sortase-recognition motif (LPETG), and purified via FPLC. ii. The E749–57 antigen (green) was synthesized with a G3 motif, and was site-specifically conjugated to VHHCD11b via a sortase A (purple)-mediated reaction. iii. The reaction mixture was purified in two steps. First, Ni-NTA beads were used to remove His-tag containing sortase A and unreacted VHHCD11b. Second, size exclusion was used to remove unreacted E749–57 antigen. B) Chromatograms showing the retention times of different species on a C8 HPLC column of (i) the purified sortase-ready VHH, (ii) the complete reaction mixture, and (iii) the final product following two-step purification. C i–iii) The calculated and observed mass(es) of the primary peaks (at 1.36, 1.43, and 1.43 min in the panels above, respectively) of the corresponding HPLC chromatograms.
Methods
All procedures were performed in accordance with institutional guidelines and approved by the Boston Children’s Hospital Institutional Animal Care and Use Committee (IACUC protocol number 16-12-3328).
Antibodies and reagents
Rat anti-CD11b-PE (clone M1/70) was purchased from eBioscience (San Diego, CA). Anti-mouse H-2Db, H-2Kb, and CD80 were purchased from BioLegend (San Diego, CA). Anti-mouse CD40 (clone 1C10) was purchased from Southern Biotech (Birmingham, AL). polyinosinic-polycytidylic acid (Poly(I:C)) and lipopolysaccharide (LPS) from Escherichia coli (E.coli) were purchased from Sigma Aldrich (St. Louis, MO).
Mice and cell culture
Specific pathogen-free female 6–8-week-old C57BL/6 mice were purchased from Jackson Laboratory (000664), and OT-I Rag-deficient (Rag−/−) mice were purchased from Taconic Biosciences (2334-F). Tumor challenge studies were performed with the C3.43 HPV16-transformed murine cell line (18, 19). C3.43 cells were a gift from W Martin Kast, PhD and were maintained in Iscove's modified Dulbecco's medium (IMDM) supplemented with 10% heat-inactivated fetal bovine serum (IFS), 50 µM 2-mercaptoethanol, and 100 U/mL penicillin/streptomycin. Cells used for tumor challenges were cultured no longer than 2 weeks from original seed stocks before in vivo injection. The murine dendritic DC2.4 and B cell lymphoma A20 cell lines were obtained from the ATCC and maintained in RPMI 1640 supplemented with 10% IFS, 2 mM glutamine, MEM nonessential amino acids, 1 mM sodium pyruvate, 50 µM 2-mercaptoethanol, and 100 U/mL penicillin/streptomycin. DC2.4 cells were characterized as CD11b+H-2Db+H-2Kb+CD80+ as assessed by cytofluorimetry (Fig. 2A and 3A). A20 cells did not express CD11b (Fig. 2A). DC2.4 and A20 cells routinely test positive for known DC and B cell markers (i.e. DEC-205 and CD19, respectively) as examined by FACS and are passaged no more than 5 times from cryopreserved stocks. All cell lines were found to be mouse pathogen free (and Mycoplasma pulmonis free) by IDEXX BioResearch pathogen testing.
Figure 2.
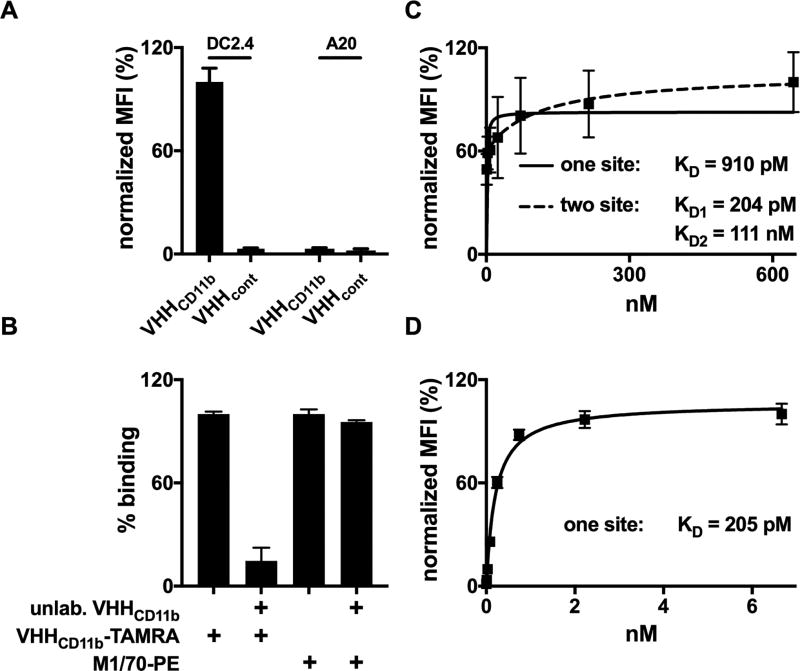
Binding of VHHCD11b to CD11b+ cells. A) CD11b+ DC2.4 cells or CD11b− A20 cells were incubated with TAMRA-labeled VHHCD11b or VHHcont (anti-GFP control VHH). The mean fluorescence intensity (MFI) as assessed by cytofluorimetry was normalized to the average maximum observed MFI of VHHCD11b-TAMRA on DC2.4 cells. B) DC2.4 cells were pre-incubated with unlabeled VHHCD11b (10 µg/mL) for 30 min at 4°C, and then stained with VHHCD11b-TAMRA or M1/70-PE for 30 min at 4°C. MFI was assessed via cytofluorimetry and binding was normalized to those observed with VHHCD11b-TAMRA (left 2 columns) or M1/70-PE (right 2 columns). C–D) DC2.4 cells were incubated with different concentrations of VHHCD11b-TAMRA (C) or M170-PE (D) for and 30 min at 4°, and the MFI as assessed by cytofluorimetry was normalized to the maximum observed MFI for VHHCD11b-TAMRA or M170-PE, respectively. One-site (solid) or two-site (dashed) hyperbolic binding models were then used to calculate relative KD values. All data shown are representatives of at least two independent experiments performed in triplicate (mean ± SD).
Figure 3.
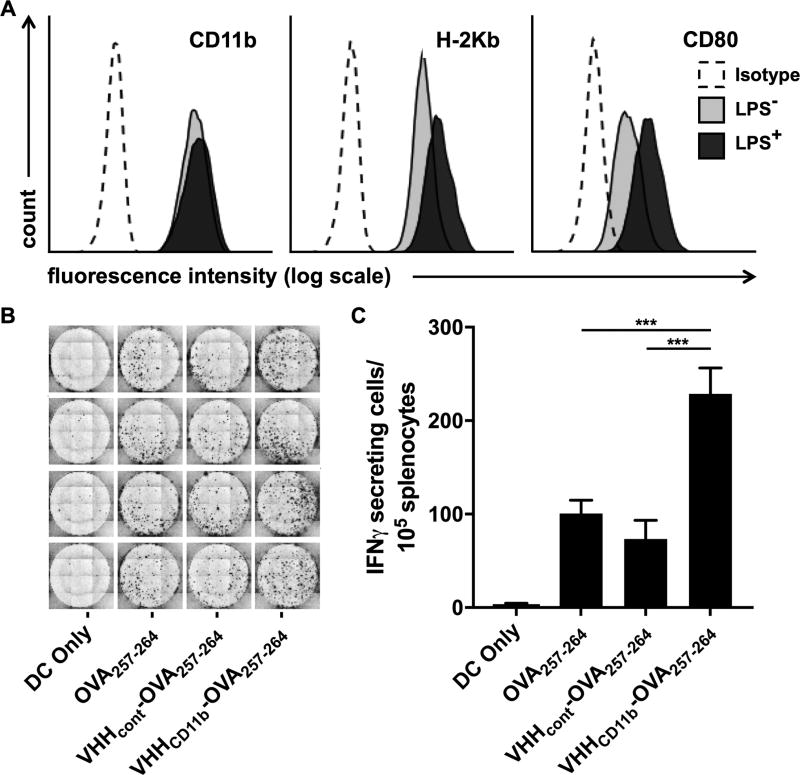
Antigen presentation is increased with VHHCD11b. A) DC2.4 cells express CD11b, H-2Kb (MHCI known to present OVA257–264), and CD80 (costimulatory molecule) as assessed via cytofluorimetry (light gray histograms). Isotype controls (Isotype) are shown as empty dashed histograms. CD80 and H-2Kb expression were increased following 24-hr incubation with LPS (dark gray histograms). B-C) DC2.4 cells were treated as indicated before LPS stimulation, followed by coculture with splenocytes from a RAG−/− OT-I mouse (1 DC2.4 cell: 20 OT-I splenocytes), and the number of IFNγ-secreting cells was measured via ELISpot assay. The spots shown in B are quantified in C (means ± SD are shown; ***P < 0.001) of an experiment performed in quadruplicate, and is a representative to three independent experiments.
Recombinant protein expression, peptide synthesis, and sortase reactions
Recombinant VHHs containing a sortase recognition motif and 5× His-tag for purification were expressed in WK6 E.coli overnight in Terrific Broth at 30°C after isopropyl β-D-thiogalactopyranoside induction (1 mM) at an OD600 of ~0.6 (sequences available in Supplemental Table 1). VHHs were harvested from the periplasm by osmotic shock and purified by adsorption to Ni-NTA beads (MCLab, San Francisco, CA), followed by size-exclusion chromatography on a Superdex 75 16/600 column (GE Healthcare, Piscataway NJ) in PBS or Tris-HCl (50 mM, pH 7.5). VHHs were then concentrated using centrifugal filters with a 10 kDa molecular weight cut-off (Millipore Sigma, UFC901024). G3-TAMRA and G3-RRAAYRAHYNIVTF were synthesized as described (20). G3-AAYSIINFEKL was synthesized at the MIT Biopolymers & Proteomics Core Facility (Cambridge, MA). Heptamutant Ca2+-independent sortase A from Staphylococcus aureus (SrtAstaph7M) was expressed with a 5× his-tag and purified following published procedures (20, 21). All sortase reactions were performed in 50 mM Tris-HCl (pH 7.5) for 2 h at 20–25°C as described (20). The reagent concentrations were VHH (2 mg/mL or ~140 µM), 300 µM nucleophile (G3-RRAAYRAHYNIVTF, etc.), and 10 µM SrtAstaph7M. Residual unconjugated VHH and SrtAstaph7M were removed by adsorption to Ni-NTA beads (MCLab). Excess nucleophile was removed via centrifugal filters with a 10-kDa molecular weight cut-off (Millipore Sigma, UFC901024). Identity of the final products was confirmed by liquid chromatography-mass spectrometry (LC-MS) (Fig. 1).
In vitro experiments
For binding experiments, CD11b+ DC2.4 cells or CD11b− A20 cells were incubated with different concentrations of TAMRA-labeled VHHCD11b or VHHcont (anti-GFP VHH) (22), or PE-labeled anti-CD11b mAb (M1/70-PE) for 30 min at 4°C, and washed. Mean fluorescence intensity (MFI) was assessed by cytofluorimetry. For competition binding experiments, DC2.4 cells were pre-incubated with unlabeled VHHCD11b (10 µg/mL) for 30 min at 4°C, and then incubated with VHHCD11b-TAMRA (1 µg/mL) or M1/70-PE for 30 min at 4°C. MFI was then assessed via cytofluorimetry after washing. For in vitro presentation assays, DC2.4 cells were treated with 100 nM OVA257–264 (SIINFEKL), VHHcont-OVA257–264, or VHHCD11b-OVA257–264 for 1 h in complete medium at 37°C and then washed with PBS. Cells were then stimulated with LPS (1 µg/mL) in complete medium overnight at 37°C. The DC2.4 cells were then collected and seeded onto pre-coated 96-well enzyme-linked immunosorbent spot (ELISpot) plates (see ELISpot assay below), to which 105 splenocytes from a RAG−/− OT-I mouse were added and incubated for 24 h at 37°C (1 DC2.4 cell: 20 OT-I splenocyte). DC2.4 cells and OT-I splenocytes are both C57BL/6-derived. The number of OVA257–264-specific interferon-γ (IFNγ)-secreting cells was then measured via IFNγ ELISpot assay (see below).
In vivo experiments
To evaluate the induction of HPV16 E7-specific T cells, wild-type C57BL/6 mice were injected intraperitoneally (IP) with 6 nmol VHHCD11b, E749–57, or VHHCD11b-E749–57 plus adjuvant (adjuvant: 50 µg Poly(I:C) + 50 µg agonistic anti-CD40), or adjuvant only. 14-days post-vaccination, spleens were harvested and anti-E749–57-specific CD8+ T cells were enumerated by IFNγ ELISpot assay. In a similar experiment, mice were injected IP with 6 nmol VHHCD11b-E749–57 or VHHcont (anti-GFP VHH)-E749–57 plus adjuvant, or adjuvant only 14 days prior to IFNγ ELISpot assay. To evaluate the protective effects of the induced CD8+ T cells, C57BL/6 mice were injected IP with adjuvant alone; adjuvant plus 6 nmol E749–57 or VHHCD11b-E749–57 on days −21 and −7; or left untreated (n = 5 per group). Serum was collected on days −14 and −7 to assess anti-VHH responses (Supplemental Fig. 1). On day 0, mice were challenged with 3 × 105 C3.43 cells subcutaneously (SC). Tumor growth and overall survival were then monitored. For therapeutic studies, C57BL/6 mice were challenged with 3 × 105 C3.43 cells on day 0, resulting in palpable tumors by day 8, and then randomized. On day 8 and 15, mice received an IP injection with adjuvant alone; adjuvant plus 6 nmol E749–57 or VHHCD11b-E749–57; or left untreated (n = 8 per group for the E749–57 and VHHCD11b-E749–57 groups, and n = 4 for the untreated group). CD8+ tumor-infiltrating lymphocytes (TILs) were evaluated via immunoPET on day 8. To evaluate immunological memory, mice with complete tumor regression from the E749–57 and VHHCD11b-E749–57 groups in the therapeutic study (n = 3 per group) were challenged with 3×105 C3.43 cells contralaterally, and then tumor growth and overall survival were monitored.
ELISpot assay
IFNγ ELISpot assays were performed following published procedures (23). Briefly, 96-well ELISpot plates (BD ELISPOT Mouse IFNγ ELISPOT Set, BD Biosciences, San Jose, CA) were coated with an IFNγ capture antibody (BD Biosciences) in PBS overnight at 4°C and plates were blocked with complete medium for 2 h at room temperature (RT). For in vitro presentation experiments, DC2.4 cell/OT-I splenocyte cocultures were set-up in quadruplicate as described above. Negative control wells contained only DC2.4 cells. For in vivo experiments, splenocytes from vaccinated mice were added in triplicate to wells containing the H-2Db-restricted E749–57 peptide (2 µg/mL) in complete medium or medium alone as a negative control. Positive control wells for all experiments contained 1× Cell Stimulation Cocktail (Invitrogen, Carlsbad, CA). Post splenocyte plating, ELISpot plates were incubated overnight at 37°C. After 24 hrs, plates were washed and incubated with a biotinylated IFNγ detection antibody (BD Biosciences) for 2 h, followed by streptavidin-horse radish peroxidase (BD Biosciences) for 1 h at RT. The plates were developed with 3-amino-9-ethyl-carbazole substrate (BD ELISPOT AEC Substrate Set) for 5 min and dried for at least 24 h. Spots were enumerated using the KS ELISpot analysis system (Carl Zeiss, Thornwood, NY), and the number of spots in negative control wells was subtracted from experimental wells to determine the number of spots above background.
ImmunoPET imaging and analysis
Radiolabeling of the anti-CD8 VHH with 89Zr (VHHCD8-89Zr) was performed as described (Fig. 5D) (24). Specifically, VHHCD8 was conjugated to G3-desferrioxamine (DFO)-azide via SrtAstaph7M. The azide group was used to install 20 kDa polyethylene glycol (PEG) moiety (as DBCO-PEG), which improves the quality of the PET signal seen with VHHCD8 in vivo (24). A volume of 89Zr4+ stock solution (1.0 M oxalic acid adjusted to pH 6.8–7.5 using 2.0 M Na2CO3) corresponding to 1.0−1.5 mCi was added to 200 µL VHHCD8-DFO solution (~2.0 mg of chelexed PEGylated-VHHCD8-DFO in 0.5 M HEPES buffer, pH 7.5) in a 2 mL microcentrifuge tube, and the total volume was adjusted to 300 µL using 1.0 M oxalic acid. The reaction mixture was incubated for 60 min at RT with agitation, loaded onto a PD-10 size-exclusion column (GE Healthcare), and eluted with PBS. Radiolabeling of VHHCD11b with 64Cu (VHHCD11b-64Cu) was performed as described (25). In brief, VHHCD11b was conjugated to G3-NOTA via SrtAstaph7M. 400 µL VHHCD11b-NOTA (20 µM in 200 mM NH4OAc buffer; pH 6.5) was mixed with 75 µL 64CuCl2 (~6 mCi in 200 mM NH4OAc buffer; pH 6.5). The reaction mixture was incubated for 20 min at RT with agitation, loaded onto a PD-10 size-exclusion column (GE Healthcare), and eluted with PBS. PET-CT was performed following published procedures (25). Briefly, mice were anaesthetized using isoflurane, injected with 20–50 µCi radiolabeled VHH via the tail vain, and imaged by PET-CT 24 h later using a G8 PET-CT small animal scanner (PerkinElmer, Norwalk, CT). PET and CT acquisition was ~10 min and 1.5 min, respectively, per animal. Images were reconstructed using the manufacturer’s software. Data were further analyzed and quantified using VivoQuant software (inviCRO Imaging Service and Software, Boston, MA). PET signal values were converted to units of percentage of injected dose per gram by using as input the total radioactivity at the time of measurement with the preprocessing tool. CT scans overlaid with PET signals were used to generate 3D regions of interest (ROIs) to represent certain regions within each mouse (i.e. tumor or lymph node). Once all ROIs were generated, a table was exported containing statistical information, such as mean PET signal or variation for each ROI. TIL and lymph node (LN) intensity was defined as the PET signal observed from 89Zr-labeled VHHCD8 at the ROI over background PET signal observed in muscle tissue ROI.
Figure 5.
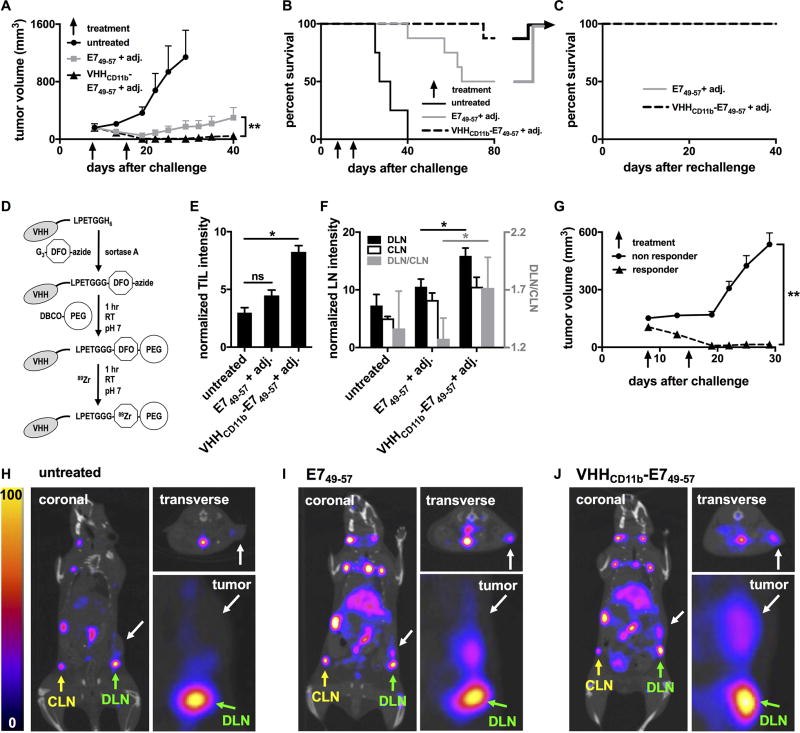
VHHCD11b-E749–57 vaccine induced responses in tumor-bearing mice in vivo. A–B) Wild-type C57BL/6 mice were challenged with 3×105 C3.43 cells on day 0 resulting in palpable tumors (~200 mm3) on day 8. Mice were then injected IP with E749–57 (n = 8) or VHHCD11b-E749–57 (n = 8) plus adj. on days 8 and 15, or left untreated (n = 4). Tumor growth (A) and survival were monitored (means ± SEM are shown; **P < 0.01 as assessed by an unpaired t test). C) Mice with complete tumor regression from E749–57 or VHHCD11b-E749–57 groups in (B) were rechallenged 80 days after the initial challenge with 3 × 105 C3.43 cells contralaterally and survival was monitored (n = 3 per group). D) Schematic of 89Zr-labeled VHHs preparation for immunoPET imaging (DFO: desferoxamine; PEG: polyethylene glycol). E–F) All tumor-bearing mice from A were injected via the tail vein with VHHCD8-89Zr on day 15 and CD8+ T cells were imaged in vivo on day 16 by PET-CT on the same day. TIL, draining lymph node (DLN), and contralateral lymph node (CLN) CD8+ T-cell intensities from the PET images were quantified (means ± SEM are shown; *P < 0.05). The ratios of the intensities in the DLN to CLN shown in F (gray bars) are plotted according to the right y-axis (shown in gray). G) Tumor growth was stratified by TIL intensity (responder TIL intensity >5 (n = 9); nonresponder <5 (n = 11); means ± SEM are shown; **P < 0.001). H–J) Representative PET-CT images from the untreated group (H), E749–57 + adj. group (I), and VHHCD11b-E749–57 + adj. group (J) from one of two independent experiments. White arrows in the coronal (left), insets from the coronal (lower right), and transverse (upper right) images indicate the sites of the tumors. Green and yellow arrows indicate the sites of the DLN and CLN, respectively.
Statistical analyses
All statistical analyses were performed on GraphPad Prism version 7.0 (GraphPad Software Inc., San Diego, CA). One- and two-site hyperbolic binding curves were used to estimate the equilibrium dissociation constants (KD values) with R-square values used as a metric to evaluate the quality of fit of the model to the data. Statistical significance for ELISpot assays, tumor growth curves, and CD8+ TILs were assessed with two-tailed unpaired t-tests or ANOVA. Survival curves were assessed via the Kaplan-Meier method, with statistical significance determined by the log-rank (Mantel-Cox) test. Alternatively, two-tailed Fisher’s exact tests were used to assess differences between outcomes among groups. Significance was defined as P < 0.05 for all experiments.
Results
Design of VHH-antigen constructs
Mouse models of E6/E7-positive tumors show that HPV Ag-specific T cells are few and comparatively poor at homing to and infiltrating tumor tissue, unless the tumor-bearing animal is vaccinated against agents containing or encoding the immunodominant E7-derived peptide, E749–57 (RAHYNIVTF) (26, 27). This peptide is recognized by CD8+ T cells in H-2Db-restricted fashion (18). Therefore, we synthesized the E749–57 peptide as a sortase A-ready nucleophile for conjugation to an anti-CD11b VHH (14). In between the G3 motif at the N-terminus and the E749–57 epitope, two arginines (RR) were included to increase solubility; an AAY linker was added to enhance cross-presentation upon internalization by target APCs (28). The resultant sequence was GGGRRAAYRAHYNIVTF. Although standard genetic means are an obvious alternative to create the requisite fusions, the ability to multiplex enzymatic conjugations and extend this approach to multiple peptide or protein Ags affords flexibility, including the site-specific installation of post-translational modifications (20, 21) and radioisotopes for positron emission tomography (PET) (25). A genetic fusion would have to be purified from an appropriate source, where appendage of a C-terminal tag (e.g. a His tag) would be deployed for affinity enrichment. However, the presence of such C-terminal extensions would require further processing steps that would likely limit the generation of the relevant peptide epitope for MHC binding, as ER peptidases primarily trim N-terminal residues (29). Purification to homogeneity of the desired fusion in the absence of a tag may not always be possible. Because of the efficiency and specificity of sortase A-catalyzed reactions, we therefore applied the enzymatic approach, which yields the desired fusion in yield and purity. Accordingly, VHHCD11b was expressed with a C-terminal sortase-recognition motif (LPETG) and 5× His-tag for purification (Fig. 1 Ai). The E7 Ag nucleophile (G3-RRAAYRAHYNIVTF) was site-specifically conjugated using sortase A and purified (Fig. 1 Aii–Aiii) as verified by liquid chromatography-mass spectrometry (LC-MS) (Fig. 1B–C). The purified VHH-Ag construct (VHHCD11b-E749–57) was used in subsequent in vivo vaccination experiments.
VHHCD11b specifically binds to CD11b+ cells with high affinity
VHHCD11b binds to the splenocytes of wild-type but not CD11b−/− mice (14). Here we show that VHHCD11b binds to CD11b+ DC2.4 cells, but not to CD11b− A20 cells (Fig. 2A). Competition assays with unlabeled VHHCD11b showed that the fluorophore attached to VHHCD11b did not affect its binding to DC2.4, and that VHHCD11b binds an epitope different from that recognized by the well-established anti-mouse CD11b mAb M1/70 (Fig. 2B). Data from our binding studies fit substantially better to a two-site binding model compared to a one-site model (R2 = 0.9895 and 0.6182, respectively), notwithstanding monovalency of VHHCD11b. Paradoxically, M1/70 binding was well-modelled using a one-site binding model (R2 = 0.9949) (Fig. 2C–D). As CD11b is a member of the integrin family capable of undergoing dramatic conformational changes, these results may indicate that VHHCD11b recognizes two distinct conformations of CD11b, one with high affinity (KD1 of 0.204 ± 0.090 nM) and one with moderate affinity (KD2 = 110 ± 47 nM), whereas M1/70 binds the different conformers with equal affinity.
Cross-presentation of OVA257–264 is increased when conjugated to VHHCD11b
The efficacy of therapeutic vaccines against HPV-driven cancers lies in their ability to elicit strong CD8+ T-cell responses against E6 and E7 proteins, which in the case of most peptide-based vaccines requires internalization, processing, and cross-presentation of the Ags on class I MHC (MHCI) (7). To explore the ability of VHHCD11b to enhance Ag presentation, DCs were treated with an H-2Kb–restricted ovalbumin peptide (OVA257–264) or an equivalent molar amount of peptide conjugated to VHHCD11b (VHHCD11b-OVA257–264). OVA257–264 was chosen for these experiments because no TCR transgenic mice are available that recognize HPV antigens to serve as a source of adequate numbers of E7-specific CD8+ T cells. We measured the expression of relevant surface markers on the treated DCs after activation with LPS (Fig. 3A) and the corresponding CD8+ T-cell responses following coculture of the differentially activated DCs with splenocytes from an OT-I OVA257–26–specific T-cell receptor (TCR) transgenic mouse (30). Treatment with the VHHCD11b-OVA257–264 conjugate resulted in a greater than two-fold increase in the number of IFNγ-secreting effector CD8+ T cells compared to peptide alone or peptide conjugated to a nontargeting control VHH (VHHcont) (Fig. 3B–C). While exogenously added peptides can bind directly to cell surface MHCI, proteins when added at equimolar concentrations obviously still require processing and cross-presentation (31). These results show that VHHCD11b-OVA257–264 efficiently entered the cross-presentation pathway.
VHHCD11b-E749–57 vaccination elicits stronger T cell responses than E749–57 alone
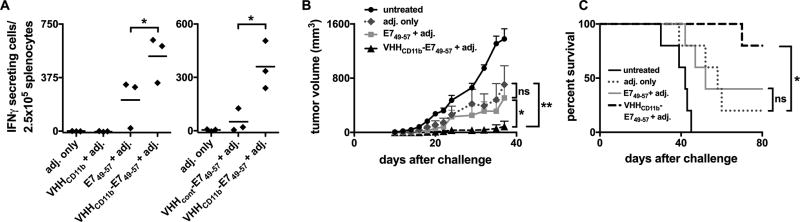
Vaccination with 30 nmol E749–57 peptide with the adjuvant poly(I:C) and an agonistic mAb to CD40 results in robust HPV-specific T-cell responses (32). Therefore, to assess the ability of VHHCD11b-E749–57 to enhance HPV-specific T-cell responses in vivo, the dose of E749–57 peptide and VHHCD11b-E749–57 was reduced to just 6 nmol, corresponding to ~100 µg of VHHCD11b-E749–57 injected per mouse. Mice were injected IP with VHHCD11b, E749–57, or VHHCD11b-E749–57 plus adjuvant (poly(I:C) and anti-CD40), or adjuvant only. Two weeks after vaccination, spleens were harvested and anti-E749–57 specific CD8+ T cells were enumerated via IFNγ ELISpot assay. A single vaccination with VHHCD11b-E749–57 resulted in a two-fold increase in the number of IFNγ-secreting effector CD8+ T cells compared to peptide alone (Fig. 4A left), highlighting the enhanced CD8+ T-cell response due to nanobody targeting via CD11b. In a similar experiment, mice were injected IP with VHHCD11b-E749–57 or E749–57 conjugated to a control VHH (anti-GFP: VHHcont) with adjuvant, or with adjuvant only. The control VHH-peptide adduct did not elicit a robust anti-E749–57 response, indicating that conjugation of the peptide to a nontargeting VHH does not enhance the antigen-specific response (Fig. 4A right).
Figure 4.
VHHCD11b-E749–57 vaccine-induced HPV-specific T-cell responses and VHHCD11b-E749–57 vaccine-induced protection against C3.43 tumor growth in vivo. A) Wild-type C57BL/6 mice were injected IP with VHHCD11b, E749–57, or VHHCD11b-E749–57 plus adjuvant (adj. = 50 µg Poly(I:C) + 50 µg agonistic anti-CD40 Ab), or adj. only (left panel). 14-days post-vaccination, spleens were harvested and anti-E749–57 specific CD8+ T cells were enumerated via IFNγ ELISpot. In a separate experiment, mice were injected IP with 6 nmol VHHCD11b-E749–57 or VHHcont (anti-GFP VHH)-E749–57 plus adj., or adj. only 14 days prior to IFNγ ELISpot assay (right panel). Horizontal bars indicate the average number of spots per group from one of two experiments (n = 3 per group per experiment; *P < 0.05). B–C) Wild-type C57BL/6 mice were injected IP with VHHCD11b, E749–57, or VHHCD11b-E749–57 plus adj., or adj. only on days −21 and −7 or left untreated (n = 5 per group). On day 0, mice were challenged with 3×105 C3.43 cells. Tumor growth (C) and survival (D) were monitored, and shown is the average group means from one of two independent experiments (group means ± SEM are shown; *P < 0.05, **P < 0.01).
Prophylactic vaccination with VHHCD11b-E749–57 confers tumor-growth protection
We next evaluated whether vaccination with VHHCD11b, E749–57 conferred protection against a subsequent tumor challenge in vivo, using the cell line C3.43 as the HPV cancer model. C3.43 cells are derived from the C3 line by transformation with a complete HPV16 genome-containing pRSVneo-derived plasmid (18, 19). C3.43 cells retain HPV16 E6 and E7 expression, and peptides from them are presented on MHC class I. By day 40, only one of five mice primed (day −21) and boosted (day −7) with VHHCD11b-E749–57 and adjuvant prior to C3.43 challenge (day 0) had developed a tumor. There was therefore a significantly lower tumor burden in this group compared to that of mice receiving E749–57 plus adjuvant (three mice developed tumors) or adjuvant only (four mice developed tumors) (P = 0.018 and 0.008, respectively; Fig. 4B). The single tumor in the VHHCD11b-E749–57 group grew more slowly than tumors of the E749–57 plus adjuvant and adjuvant only groups, resulting in survival until day 70 (Fig. 4C). VHHCD11b-E749–57 conferred a significant overall survival benefit compared to that of the adjuvant-only group (P = 0.03; P = 0.1 between VHHCD11b-E749–57 and E749–57 groups), whereas no difference in survival was observed between the E749–57 plus adjuvant and adjuvant-only groups (P = 0.8; Fig. 4C).
Therapeutic vaccination with VHHCD11b-E749–57 induces tumor regression in vivo
We next evaluated the therapeutic potential of Ag conjugation to VHHCD11b. Tumor-bearing mice were treated with VHHCD11b-E749–57 and adjuvant, or E749–57 and adjuvant on days 8 and 15 after a subcutaneous challenge with C3.43 cells. Tumor growth and overall survival were monitored. The VHHCD11b-E749–57 plus adjuvant group had a significantly lower tumor burden (7/8 mice showed complete tumor regression) than that of the E749–57 plus adjuvant group (4/8 mice had complete regression) (P = 0.002; Fig. 5A), and the survival benefit of VHHCD11b-E749–57 over E749–57 trended towards significance (P = 0.08, Fig. 5B). No mice in the untreated group exhibited tumor regression (0/4) and all died within 40 days of challenge; thus, both the VHHCD11b-E749–57 and E749–57 treatment groups provided significant survival benefits over untreated controls (P < 0.01). After day 40, the tumor in the one tumor-bearing mouse from the VHHCD11b-E749–57 plus adjuvant group grew more slowly than those of tumor-bearing mice in the E749–57 plus adjuvant group, resulting in survival until day 75 (Fig. 5B). Mice with complete tumor regression from both the E749–57 and VHHCD11b-E749–57 groups were rechallenged on the opposite flank with C3.43 tumor cells 80 days after the initial tumor challenge. None of these rechallenged mice developed tumors, suggesting that the level of immunological memory was no different between the two groups (Fig. 5C).
We have developed methods to noninvasively image immune cell subsets in vivo using nanobody-based immuno-positron emission tomography (immunoPET; Fig. 5D) (24, 25). We used immunoPET to monitor CD8+ tumor-infiltrating lymphocytes (TILs) in all tumor-bearing mice 15 days after tumor challenge, which was 7 days after the first vaccination for the VHHCD11b-E749–57 and E749–57 groups (representative PET images are shown in Fig. 5H–5J). We saw greater TIL intensity (see Methods) in the tumors of VHHCD11b-E749–57-treated mice compared to those in untreated mice (Fig. 5E). No obvious difference was seen when comparing the E749–57 and untreated groups. When stratified by TIL intensity as measured by PET (responder >5; nonresponder <5), there was a significantly lower tumor burden in responders than in nonresponders (Fig. 5G), and TIL intensity was significantly correlated with survival (P = 0.01), indicating the prognostic value of TILs visualized noninvasively by immunoPET. Similarly, we measured the CD8+ T-cell signal intensities in the draining and contralateral lymph nodes (DLN and CLN, respectively) via immunoPET. The CD8+ T-cell signal intensities in both the DLN and CLN were increased in the VHHCD11b-E749–57 group compared to the other groups, with a significant difference observed between the DLN signal intensities of the VHHCD11b-E749–57 and E749–57 groups (Fig. 5F). We hypothesized that the better induction of antigen-specific CD8+ T cells in the VHHCD11b-E749–57 group as seen by ELISpot (Fig. 4A) would yield a higher CD8+ T-cell signal in the DLN of E7+ tumors, where antigen-specific T cells are expected to accumulate during an antitumor response. Indeed, we found that the ratio of CD8+ T-cell signal intensity in the DLN over that in the CLN was highest in the VHHCD11b-E749–57 group (Fig. 5F gray bars). We suggest this is due to the presence of a greater number of antigen-specific T cells.
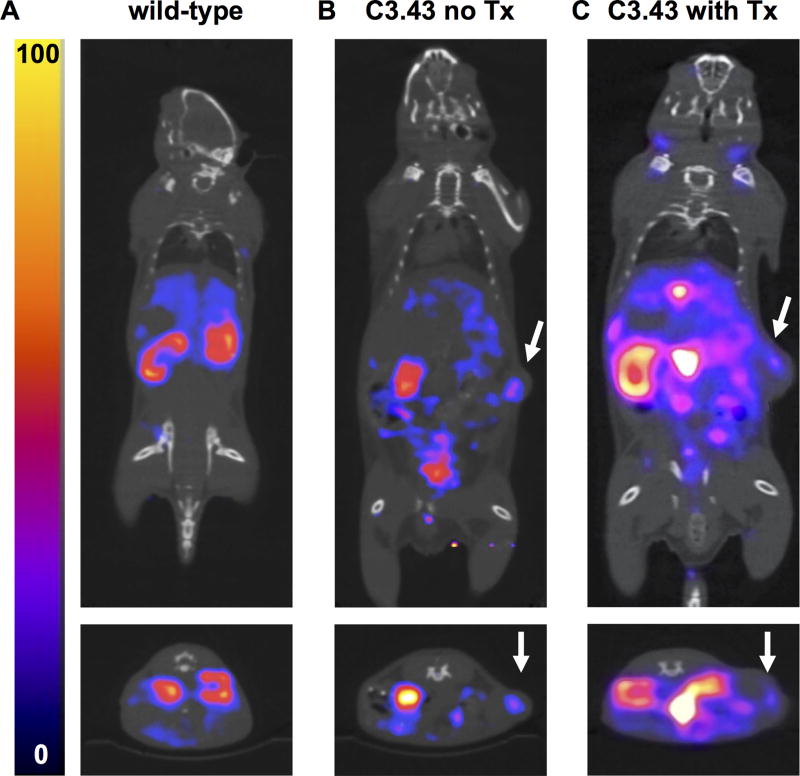
CD11b+ cells are found in C3.43 tumors in vivo
CD11b+ cells have been observed in the skin of transgenic K14E7 mice that constitutively express E7 in squamous epithelium (33). Therefore, we utilized VHHCD11b as an immunoPET imaging agent in C3.43 tumor-bearing mice to test for the presence of intratumoral CD11b+ cells. We labeled VHHCD11b with 64Cu and injected it into wild-type mice or mice harboring ~200 mm3 (16 days after challenge) C3.43 tumors with or without VHHCD11b-E749–57 treatment (mice were treated on day 8). C3.43 tumors were found to be positive for VHHCD11b-64Cu staining (Fig 6B–C). Although we do not know where exactly induction of the anti-E7 response occurs in VHHCD11b-E749–57 vaccinated mice, a pool of Ag delivered to the tumor site may allow resident APCs to acquire it. If these APCs were to migrate to the draining lymph node, further E7-specific T cells may be recruited from that lymph node and participate in the antitumor response. Such localized delivery may be especially important when antigen levels are limiting. This imaging experiment further highlights the utility of sortase A to conjugate payloads to VHHs, as we used the identical sortase-ready VHH as a delivery vehicle for the E7 antigen in one setting, and as an immunoPET imaging agent in another.
Figure 6.
ImmunoPET analysis of CD11b+ cells in vivo. Wild-type mice or mice challenged with 3×105 C3.43 cells 16 days prior (with or without VHHCD11b-E749–57 treatment (Tx) on day 8) were injected via the tail vein with VHHCD11b-64Cu and imaged by PET-CT (n = 4 per group). Representative PET-CT images from A) wild-type, B) C3.43 tumor-bearing without Tx, and C) C3.43 tumor-bearing with Tx animals are shown from two independent experiments. Images from A and B were collected on the same day, and C was acquired on a separate day. White arrows in the coronal (top), and transverse (bottom) images indicate the sites of the tumors.
Discussion
As HPV DNA testing becomes more widely implemented as a primary screening method for HPV infection (34), more women will test positive for high-risk HPV infection, including those with normal cytology. More than 15% of women infected with high-risk HPV strains do not initiate effective immune responses against the virus, leaving them at risk for the development of low-grade cervical intraepithelial neoplastic (CIN1) lesions (35). Although CIN1 is often eliminated by the host’s immune system, regression of high-grade lesions (CIN2/3) is far less common. Progression is thus correlated with limited immune responses, increasing the risk for developing invasive cancers (36, 37). A therapeutic HPV vaccine that can stimulate the immune system to clear established HPV infections is needed, especially for patients with consecutive HPV+ DNA tests or with CIN1 lesions, for which the only treatment option is watchful waiting (38).
We explored whether an HPV vaccine based on anti-CD11b VHH-targeted delivery of an HPV antigen-derived peptide would elicit stronger antitumor CD8+ T cell responses than vaccination with peptide alone. Our data support that hypothesis. Specifically, MHC class I presentation of an ovalbumin-derived peptide was highest when conjugated to VHHCD11b, compared to equimolar amounts of peptide delivered via a control VHH or as peptide alone. Other conjugates comprising VHHCD11b and a different peptide should be handled similarly. The VHHCD11b-E749–57 construct elicited a strong CD8+ T-cell response in vivo, which afforded prophylaxis against C3.43 tumor growth. VHHCD11b-E749–57 therapeutic vaccination induced TILs as seen in live mice by immunoPET and resulted in tumor regression and increased survival. Considerably less Ag was needed than previously reported to achieve strong E749–57-specific responses (i.e. 6 nmol vs. 30 nmol) when conjugated to VHHCD11b (32). The approach reported here can be extended to the inclusion of multiple epitopes. Collectively, these data demonstrate that targeted delivery of HPV Ags to CD11b+ cells via VHH conjugates is a viable strategy to elicit effective cellular immune responses against HPV-induced cancers. An extension to other tumor models would require the availability of a similarly immunodominant peptide, but currently such sequences are not known for other commonly studied preclinical cancer models, such as murine B16-F10 melanoma or MC-38 colon adenocarcinoma.
Other studies have shown that targeted delivery of an Ag to CD11b+ cells can induce strong CD8+ T-cell responses. Specifically, a detoxified form of Bordetella pertussis adenylate cyclase toxin (CyaA), a ligand of CD11b, has been used to deliver Ags as CyaA fusion products to elicit MHC class I– and MHC class II–restricted T-cell responses, both in vitro and in vivo (39, 40). Such CyaA-E7 fusions elicit HPV-specific CD8+ T-cell responses and corresponding antitumor responses in vivo (41). However, the molecular mass of these CyaA fusions is ~180 kDa, which may limit access to tissues harboring appropriate CD11b+ populations of APCs. In contrast, the VHH-Ag constructs are ~15 kDa, providing superior tissue penetration, even compared to conventional antibodies (~150 kDa) (42, 43). Our previous work already showed that a conventional full-size Ag-modified antibody was less efficient in eliciting Ag-specific CD4+ T-cell responses than a VHH conjugated covalently to that same Ag (14). As to the use of CyaA as an alternative delivery vehicle, its bacterial origin entails the added risk of immunogenicity and the generation of CyaA-neutralizing antibodies, which may compromise efficacy when a CyaA-based vaccine requires repeated administration. VHHs are comparatively poorly immunogenic as well (Supplementary Fig. 1), and can be humanized if necessary (44). VHHCD11b binds more tightly to CD11b+ cells (KD value 0.204 ± 0.090 nM on DC2.4 cells) than CyaA (KD 9.2 ± 4.5 and 3.2 ± 1.9 nM depending on cell type). Our previous (14) and current work shows that VHHCD11b-Ag constructs have their payloads presented on MHCI. Their ease of expression, excellent tissue penetration, high affinity, and interchangeability of VHHs and Ags in VHH-Ag constructs thus create a flexible vaccine platform.
E7 expression in the skin of mice results in expansion of CD11b+ cells (33). In particular, more CD11b+Gr1+ cells were present in the skin of transgenic K14E7 mice (33), which constitutively express HPV16 E7 in squamous epithelium (45). CD11b+Gr1+ cells are defined as myeloid-derived suppressor cells (MDSCs) that can show intratumoral immunosuppressive activity (46). If sites of HPV infection and/or HPV-induced cancers are at the same time foci of CD11b expression, then delivering immunodominant HPV Ags to these locations under stimulatory conditions may activate local APCs, which may also pick up other HPV Ags in addition to those delivered, thus allowing for epitope spreading. This might occur despite the immunosuppression associated with increased CD11b+Gr1+ cells, but if and how a balance between the action of stimulatory APCs and suppressive MDSCs is established in our vaccination protocol will require further study. We readily visualized the presence of CD11b+ cells in E7-expressing C3.43 tumors in vivo via immunoPET, but it is not yet known whether these cells are associated with the observed anti-E7 immune responses in VHHCD11b-E749–57 vaccinated mice.
Spatial resolution and imaging depth by PET is superior to bioluminescence-based in vivo noninvasive imaging methods such as the current in vivo imaging systems (IVIS), a technique not easily translatable to a clinical setting (47). Moreover, PET provides quantitative information on the distribution of label (i.e. 89Zr) throughout the entire animal, with a spatial resolution of ~1 mm. Although two-photon microscopy provides better cellular and temporal resolution, it is an invasive method that requires surgical intervention, for example to expose lymph nodes for imaging, or to implant a window that allows inspection of the underlying tissue (48, 49). Multi-photon microscopy does not provide information on the distribution of label over the entire animal. We combined the spatial resolution of PET with the specificity of an anti-CD8 VHH to noninvasively visualize TILs in vivo. We found that TILs as seen via immunoPET were prognostic for tumor regression, in agreement with our previous work (24). These techniques could be applied to any number of cancer immunotherapy animal models and could potentially be translated to human cancers to not only provide prognostic information about treatment outcomes, but inform physicians whether patients are responding to immunotherapeutic interventions during a given course of treatment.
Supplementary Material
Acknowledgments
JGS was supported by the Keck School of Medicine/USC Graduate School PhD Fellowship. ELISpot plate reading was performed at the USC Immune Monitoring Core Facility supported in part by the National Cancer Institute Cancer Center Support Grant award P30CA014089. The authors also thank Jessica Ingram for providing VHHCD11b.
Financial Support: AWW is supported by the Arnold O. Beckman Postdoctoral Fellowship. RWC is supported in part by funding from the Cancer Research Institute Irvington Postdoctoral Fellowship. WMK holds the Walter A. Richter Cancer Research Chair and is supported by NIH R01 CA074397. HLP is supported by the Lustgarten Foundation (award ID 388167).
Footnotes
Disclosure of Potential Conflicts of Interest
AWW, RWC, JJL, MR, SCK, MM, JND, JB, JGS, DMD, WMK, and HLP have no conflicts of interest to disclose.
References
- 1.Wang JW, Roden RB. Virus-like particles for the prevention of human papillomavirus-associated malignancies. Expert Rev Vaccines. 2013;12(2):129–41. doi: 10.1586/erv.12.151. PubMed PMID: 23414405; PMCID: PMC3835148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stokley S, Jeyarajah J, Yankey D, Cano M, Gee J, Roark J, Curtis RC, Markowitz L. Human papillomavirus vaccination coverage among adolescents, 2007–2013, and postlicensure vaccine safety monitoring, 2006–2014--United States. MMWR Morbidity and mortality weekly report. 2014;63(29):620–4. Epub 2014/07/24. PubMed PMID: 25055185. [PMC free article] [PubMed] [Google Scholar]
- 3.Brawner BM, Baker JL, Voytek CD, Leader A, Cashman RR, Silverman R, Peter N, Buchner BJ, Barnes CA, Jemmott LS, Frank I. The Development of a Culturally Relevant, Theoretically Driven HPV Prevention Intervention for Urban Adolescent Females and Their Parents/Guardians. Health Promot Pract. 2012 doi: 10.1177/1524839912462389. Epub 2012/10/27. PubMed PMID: 23099659. [DOI] [PubMed] [Google Scholar]
- 4.Haedicke J, Iftner T. Human papillomaviruses and cancer. Radiother Oncol. 2013 doi: 10.1016/j.radonc.2013.06.004. Epub 2013/07/09. PubMed PMID: 23830197. [DOI] [PubMed] [Google Scholar]
- 5.Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, Snijders PJ, Peto J, Meijer CJ, Munoz N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. JPathol. 1999;189(1):12–9. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. PubMed PMID: 1. [DOI] [PubMed] [Google Scholar]
- 6.Stanley MA, Pett MR, Coleman N. HPV: from infection to cancer. Biochem Soc Trans. 2007;35(Pt 6):1456–60. doi: 10.1042/BST0351456. Epub 2007/11/23. PubMed PMID: 18031245. [DOI] [PubMed] [Google Scholar]
- 7.Skeate JG, Woodham AW, Einstein MH, Da Silva DM, Kast WM. Current therapeutic vaccination and immunotherapy strategies for HPV-related diseases. Hum Vaccin Immunother. 2016;12(6):1418–29. doi: 10.1080/21645515.2015.1136039. PubMed PMID: 26835746; PMCID: PMC4964648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonifaz LC, Bonnyay DP, Charalambous A, Darguste DI, Fujii S, Soares H, Brimnes MK, Moltedo B, Moran TM, Steinman RM. In vivo targeting of antigens to maturing dendritic cells via the DEC-205 receptor improves T cell vaccination. J Exp Med. 2004;199(6):815–24. doi: 10.1084/jem.20032220. PubMed PMID: 15024047; PMCID: PMC2212731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fayolle C, Osickova A, Osicka R, Henry T, Rojas MJ, Saron MF, Sebo P, Leclerc C. Delivery of multiple epitopes by recombinant detoxified adenylate cyclase of Bordetella pertussis induces protective antiviral immunity. J Virol. 2001;75(16):7330–8. doi: 10.1128/JVI.75.16.7330-7338.2001. PubMed PMID: 11462005; PMCID: PMC114968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, Songa EB, Bendahman N, Hamers R. Naturally occurring antibodies devoid of light chains. Nature. 1993;363(6428):446–8. doi: 10.1038/363446a0. PubMed PMID: 8502296. [DOI] [PubMed] [Google Scholar]
- 11.Arbabi Ghahroudi M, Desmyter A, Wyns L, Hamers R, Muyldermans S. Selection and identification of single domain antibody fragments from camel heavy-chain antibodies. FEBS Lett. 1997;414(3):521–6. doi: 10.1016/s0014-5793(97)01062-4. PubMed PMID: 9323027. [DOI] [PubMed] [Google Scholar]
- 12.Muyldermans S. Nanobodies: natural single-domain antibodies. Annu Rev Biochem. 2013;82:775–97. doi: 10.1146/annurev-biochem-063011-092449. PubMed PMID: 23495938. [DOI] [PubMed] [Google Scholar]
- 13.Helma J, Cardoso MC, Muyldermans S, Leonhardt H. Nanobodies and recombinant binders in cell biology. J Cell Biol. 2015;209(5):633–44. doi: 10.1083/jcb.201409074. PubMed PMID: 26056137; PMCID: PMC4460151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duarte JN, Cragnolini JJ, Swee LK, Bilate AM, Bader J, Ingram JR, Rashidfarrokhi A, Fang T, Schiepers A, Hanke L, Ploegh HL. Generation of Immunity against Pathogens via Single-Domain Antibody-Antigen Constructs. J Immunol. 2016;197(12):4838–47. doi: 10.4049/jimmunol.1600692. PubMed PMID: 27821668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arnaout MA. Structure and function of the leukocyte adhesion molecules CD11/CD18. Blood. 1990;75(5):1037–50. PubMed PMID: 1968349. [PubMed] [Google Scholar]
- 16.Popp MW, Ploegh HL. Making and breaking peptide bonds: protein engineering using sortase. Angew Chem Int Ed Engl. 2011;50(22):5024–32. doi: 10.1002/anie.201008267. PubMed PMID: 21538739. [DOI] [PubMed] [Google Scholar]
- 17.Popp MW, Antos JM, Grotenbreg GM, Spooner E, Ploegh HL. Sortagging: a versatile method for protein labeling. Nat Chem Biol. 2007;3(11):707–8. doi: 10.1038/nchembio.2007.31. PubMed PMID: 17891153. [DOI] [PubMed] [Google Scholar]
- 18.Feltkamp MC, Smits HL, Vierboom MP, Minnaar RP, de Jongh BM, Drijfhout JW, ter Schegget J, Melief CJ, Kast WM. Vaccination with cytotoxic T lymphocyte epitope-containing peptide protects against a tumor induced by human papillomavirus type 16-transformed cells. Eur J Immunol. 1993;23(9):2242–9. doi: 10.1002/eji.1830230929. PubMed PMID: 7690326. [DOI] [PubMed] [Google Scholar]
- 19.Smith KA, Meisenburg BL, Tam VL, Pagarigan RR, Wong R, Joea DK, Lantzy L, Carrillo MA, Gross TM, Malyankar UM, Chiang CS, Da Silva DM, Kundig TM, Kast WM, Qiu Z, Bot A. Lymph node-targeted immunotherapy mediates potent immunity resulting in regression of isolated or metastatic human papillomavirus-transformed tumors. Clin Cancer Res. 2009;15(19):6167–76. doi: 10.1158/1078-0432.CCR-09-0645. PubMed PMID: 19789304; PMCID: PMC2756704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guimaraes CP, Witte MD, Theile CS, Bozkurt G, Kundrat L, Blom AE, Ploegh HL. Site-specific C-terminal and internal loop labeling of proteins using sortase-mediated reactions. Nat Protoc. 2013;8(9):1787–99. doi: 10.1038/nprot.2013.101. PubMed PMID: 23989673; PMCID: PMC3943461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Theile CS, Witte MD, Blom AE, Kundrat L, Ploegh HL, Guimaraes CP. Site-specific N-terminal labeling of proteins using sortase-mediated reactions. Nat Protoc. 2013;8(9):1800–7. doi: 10.1038/nprot.2013.102. Epub 2013/08/31. PubMed PMID: 23989674; PMCID: 3941705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirchhofer A, Helma J, Schmidthals K, Frauer C, Cui S, Karcher A, Pellis M, Muyldermans S, Casas-Delucchi CS, Cardoso MC, Leonhardt H, Hopfner KP, Rothbauer U. Modulation of protein properties in living cells using nanobodies. Nat Struct Mol Biol. 2010;17(1):133–8. doi: 10.1038/nsmb.1727. PubMed PMID: 20010839. [DOI] [PubMed] [Google Scholar]
- 23.Yan L, Woodham AW, Da Silva DM, Kast WM. Functional analysis of HPV-like particle-activated Langerhans cells in vitro. Methods Mol Biol. 2015;1249:333–50. doi: 10.1007/978-1-4939-2013-6_25. Epub 2014/10/29. PubMed PMID: 25348318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rashidian M, Ingram JR, Dougan M, Dongre A, Whang KA, LeGall C, Cragnolini JJ, Bierie B, Gostissa M, Gorman J, Grotenbreg GM, Bhan A, Weinberg RA, Ploegh HL. Predicting the response to CTLA-4 blockade by longitudinal noninvasive monitoring of CD8 T cells. J Exp Med. 2017;214(8):2243–55. doi: 10.1084/jem.20161950. PubMed PMID: 28666979; PMCID: PMC5551571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rashidian M, Keliher EJ, Bilate AM, Duarte JN, Wojtkiewicz GR, Jacobsen JT, Cragnolini J, Swee LK, Victora GD, Weissleder R, Ploegh HL. Noninvasive imaging of immune responses. Proc Natl Acad Sci U S A. 2015;112(19):6146–51. doi: 10.1073/pnas.1502609112. Epub 2015/04/23. PubMed PMID: 25902531; PMCID: 4434737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feltkamp MC, Vreugdenhil GR, Vierboom MP, Ras E, van der Burg SH, ter Schegget J, Melief CJ, Kast WM. Cytotoxic T lymphocytes raised against a subdominant epitope offered as a synthetic peptide eradicate human papillomavirus type 16-induced tumors. Eur J Immunol. 1995;25(9):2638–42. doi: 10.1002/eji.1830250935. PubMed PMID: 7589138. [DOI] [PubMed] [Google Scholar]
- 27.Kanodia S, Da Silva DM, Karamanukyan T, Bogaert L, Fu YX, Kast WM. Expression of LIGHT/TNFSF14 combined with vaccination against human papillomavirus Type 16 E7 induces significant tumor regression. Cancer Research. 2010;70(10):3955–64. doi: 10.1158/0008-5472.CAN-09-3773. Epub 2010/05/13. PubMed PMID: 20460520; PMCID: 2873073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Velders MP, Weijzen S, Eiben GL, Elmishad AG, Kloetzel PM, Higgins T, Ciccarelli RB, Evans M, Man S, Smith L, Kast WM. Defined flanking spacers and enhanced proteolysis is essential for eradication of established tumors by an epitope string DNA vaccine. J Immunol. 2001;166(9):5366–73. doi: 10.4049/jimmunol.166.9.5366. PubMed PMID: 11313372. [DOI] [PubMed] [Google Scholar]
- 29.Blum JS, Wearsch PA, Cresswell P. Pathways of antigen processing. Annu Rev Immunol. 2013;31:443–73. doi: 10.1146/annurev-immunol-032712-095910. PubMed PMID: 23298205; PMCID: PMC4026165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clarke SR, Barnden M, Kurts C, Carbone FR, Miller JF, Heath WR. Characterization of the ovalbumin-specific TCR transgenic line OT-I: MHC elements for positive and negative selection. Immunol Cell Biol. 2000;78(2):110–7. doi: 10.1046/j.1440-1711.2000.00889.x. PubMed PMID: 10762410. [DOI] [PubMed] [Google Scholar]
- 31.Joffre OP, Segura E, Savina A, Amigorena S. Cross-presentation by dendritic cells. Nat Rev Immunol. 2012;12(8):557–69. doi: 10.1038/nri3254. PubMed PMID: 22790179. [DOI] [PubMed] [Google Scholar]
- 32.Barrios K, Celis E. TriVax-HPV: an improved peptide-based therapeutic vaccination strategy against human papillomavirus-induced cancers. Cancer Immunol Immunother. 2012;61(8):1307–17. doi: 10.1007/s00262-012-1259-8. PubMed PMID: 22527249; PMCID: PMC3446251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Damian-Morales G, Serafin-Higuera N, Moreno-Eutimio MA, Cortes-Malagon EM, Bonilla-Delgado J, Rodriguez-Uribe G, Ocadiz-Delgado R, Lambert PF, Gariglio P. The HPV16 E7 Oncoprotein Disrupts Dendritic Cell Function and Induces the Systemic Expansion of CD11b(+)Gr1(+) Cells in a Transgenic Mouse Model. Biomed Res Int. 2016;2016:8091353. doi: 10.1155/2016/8091353. PubMed PMID: 27478837; PMCID: PMC4958469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abraham J, Stenger M. Cobas HPV test for first-line screening for cervical cancer. J Community Support Oncol. 2014;12(5):156–7. doi: 10.12788/jcso.0039. Epub 2014/06/28. PubMed PMID: 24971425. [DOI] [PubMed] [Google Scholar]
- 35.Giuliano AR, Harris R, Sedjo RL, Baldwin S, Roe D, Papenfuss MR, Abrahamsen M, Inserra P, Olvera S, Hatch K. Incidence, prevalence, and clearance of type-specific human papillomavirus infections: The Young Women's Health Study. J Infect Dis. 2002;186(4):462–9. doi: 10.1086/341782. Epub 2002/08/27. PubMed PMID: 12195372. [DOI] [PubMed] [Google Scholar]
- 36.Rodriguez AC, Schiffman M, Herrero R, Wacholder S, Hildesheim A, Castle PE, Solomon D, Burk R Proyecto Epidemiologico Guanacaste G. Rapid clearance of human papillomavirus and implications for clinical focus on persistent infections. J Natl Cancer Inst. 2008;100(7):513–7. doi: 10.1093/jnci/djn044. PubMed PMID: 18364507; PMCID: PMC3705579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walker JL, Wang SS, Schiffman M, Solomon D. Predicting absolute risk of CIN3 during post-colposcopic follow-up: results from the ASCUS-LSIL Triage Study (ALTS) Am J Obstet Gynecol. 2006;195(2):341–8. doi: 10.1016/j.ajog.2006.02.047. Epub 2006/08/08. PubMed PMID: 16890545. [DOI] [PubMed] [Google Scholar]
- 38.Massad LS, Einstein MH, Huh WK, Katki HA, Kinney WK, Schiffman M, Solomon D, Wentzensen N, Lawson HW. 2012 updated consensus guidelines for the management of abnormal cervical cancer screening tests and cancer precursors. Obstet Gynecol. 2013;121(4):829–46. doi: 10.1097/AOG.0b013e3182883a34. Epub 2013/05/03. PubMed PMID: 23635684. [DOI] [PubMed] [Google Scholar]
- 39.Guermonprez P, Khelef N, Blouin E, Rieu P, Ricciardi-Castagnoli P, Guiso N, Ladant D, Leclerc C. The adenylate cyclase toxin of Bordetella pertussis binds to target cells via the alpha(M)beta(2) integrin (CD11b/CD18) J Exp Med. 2001;193(9):1035–44. doi: 10.1084/jem.193.9.1035. PubMed PMID: 11342588; PMCID: PMC2193436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loucka J, Schlecht G, Vodolanova J, Leclerc C, Sebo P. Delivery of a MalE CD4(+)-T-cell epitope into the major histocompatibility complex class II antigen presentation pathway by Bordetella pertussis adenylate cyclase. Infect Immun. 2002;70(2):1002–5. doi: 10.1128/iai.70.2.1002-1005.2002. PubMed PMID: 11796640; PMCID: PMC127677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Esquerre M, Bouillette-Marussig M, Goubier A, Momot M, Gonindard C, Keller H, Navarro A, Bissery MC. GTL001, a bivalent therapeutic vaccine against human papillomavirus 16 and 18, induces antigen-specific CD8+ T cell responses leading to tumor regression. PLoS One. 2017;12(3):e0174038. doi: 10.1371/journal.pone.0174038. PubMed PMID: 28301611; PMCID: PMC5354464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smolarek D, Bertrand O, Czerwinski M. Variable fragments of heavy chain antibodies (VHHs): a new magic bullet molecule of medicine? Postepy Hig Med Dosw (Online) 2012;66:348–58. doi: 10.5604/17322693.1000334. Epub 2012/06/19. PubMed PMID: 22706121. [DOI] [PubMed] [Google Scholar]
- 43.Hassanzadeh-Ghassabeh G, Devoogdt N, De Pauw P, Vincke C, Muyldermans S. Nanobodies and their potential applications. Nanomedicine (Lond) 2013;8(6):1013–26. doi: 10.2217/nnm.13.86. PubMed PMID: 23730699. [DOI] [PubMed] [Google Scholar]
- 44.Vincke C, Loris R, Saerens D, Martinez-Rodriguez S, Muyldermans S, Conrath K. General strategy to humanize a camelid single-domain antibody and identification of a universal humanized nanobody scaffold. J Biol Chem. 2009;284(5):3273–84. doi: 10.1074/jbc.M806889200. PubMed PMID: 19010777. [DOI] [PubMed] [Google Scholar]
- 45.Herber R, Liem A, Pitot H, Lambert PF. Squamous epithelial hyperplasia and carcinoma in mice transgenic for the human papillomavirus type 16 E7 oncogene. J Virol. 1996;70(3):1873–81. doi: 10.1128/jvi.70.3.1873-1881.1996. PubMed PMID: 8627712; PMCID: PMC190015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9(3):162–74. doi: 10.1038/nri2506. PubMed PMID: 19197294; PMCID: PMC2828349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zinn KR, Chaudhuri TR, Szafran AA, O'Quinn D, Weaver C, Dugger K, Lamar D, Kesterson RA, Wang X, Frank SJ. Noninvasive bioluminescence imaging in small animals. ILAR J. 2008;49(1):103–15. doi: 10.1093/ilar.49.1.103. PubMed PMID: 18172337; PMCID: PMC2614121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dzhagalov IL, Melichar HJ, Ross JO, Herzmark P, Robey EA. Two-photon imaging of the immune system. Curr Protoc Cytom. 2012;Chapter 12(Unit12):26. doi: 10.1002/0471142956.cy1226s60. PubMed PMID: 22470153; PMCID: PMC3662370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Helmchen F, Denk W. Deep tissue two-photon microscopy. Nat Methods. 2005;2(12):932–40. doi: 10.1038/nmeth818. PubMed PMID: 16299478. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.