Abstract
Although Glomerella glycines, Colletotrichum magnum and C. orchidearum are known as causal agents of anthracnose of soybean, Cucurbitaceae and Orchidaceae, respectively, their taxonomy remains unresolved. In preliminary analyses based on ITS, strains of these species appear basal in Colletotrichum phylogenies, clustering close to C. cliviae, C. brevisporum and other recently described species from tropical or subtropical regions. Phylogenetic analyses (ITS, GAPDH, CHS-1, HIS3, ACT, TUB2) of 102 strains previously identified as Ga. glycines, C. magnum and C. orchidearum as well as other related strains from different culture collections and studies placed these taxa in three species complexes, and distinguished at least 24 species, including 11 new species. In this study, C. magnum, C. orchidearum and C. piperis were epitypified and their taxonomy resolved, while C. cliviicola was proposed as a new name for C. cliviae. Furthermore, a sexual morph was observed for C. yunnanense, while C. brevisporum, C. cliviicola and C. tropicicola were reported from new hosts or countries. Regarding their conidial morphology, species in the C. dracaenophilum, C. magnum and C. orchidearum species complexes are reminiscent of C. gloeosporioides or C. boninense s. lat., and were likely to be confused with them in the past.
Key words: Anthracnose, Ascomycota, Colletotrichum, Gloeosporium, Glomerella, Phylogeny, Systematics
Taxonomic novelties: New name: Colletotrichum cliviicola Damm & Crous for C. cliviae Yan L. Yang et al.
New species: C. cacao Damm; C. cattleyicola Damm & Toy. Sato; C. coelogynes Damm; C. lobatum Damm; C. merremiae Damm; C. musicola Damm; C. okinawense Damm & Toy. Sato; C. panamense Damm; C. plurivorum Damm, Alizadeh & Toy. Sato; C. sojae Damm & Alizadeh; C. vittalense Damm
Epitypifications (basionyms): Glomerella magna S.F. Jenkins & Winstead, C. orchidearum Allesch., C. piperis Petch
Introduction
During a systematic study of Colletotrichum species, strains were detected that were reminiscent of C. gloeosporioides regarding conidial morphology, but did not belong to any of the well-studied species complexes (Cannon et al. 2012). Several of these strains were previously identified as Glomerella glycines, Ga. magna and C. orchidearum.
Glomerella glycines is known as the causal agent of anthracnose of soybean. It was described by Lehman & Wolf (1926) from soybean stems as the sexual morph of Colletotrichum glycines (Hemmi 1920). The Compendium of Soybean Diseases (Sinclair 1982) lists two species on soybean, the first being C. dematium var. truncatum (syn. of C. truncatum, Damm et al. 2009) represented by the line drawing of C. glycines by Hemmi (1920), which was apparently regarded as a synonym of C. truncatum, and the second being Ga. glycines. Further confusion was caused by connecting C. destructivum to Ga. glycines (Tiffany and Gilman, 1954, Manandhar et al., 1986). However, a recent molecular study has shown that C. destructivum belongs to the C. destructivum species complex, while isolates from the study of Manandhar et al. (1986) were not closely related to C. destructivum, belonging to a different species complex (Damm et al. 2014). In contrast, von Arx & Müller (1954) treated Ga. glycines as a form of Ga. cingulata with large ascospores. Based on these records, Ga. glycines was previously thus connected to at least three different species complexes.
Glomerella magna (syn. C. magnum) was described on watermelon (Citrullus lanatus) in the USA and is heterothallic (Jenkins & Winsteat 1964). According to Jenkins & Winsteat (1964), Ga. magna is pathogenic to many species of Cucurbitaceae, including watermelon, cantaloup (Cucumis melo), squash and pumpkin (Cucurbita spp.), and sometimes cucumber (Cucumis sativus). Grand (1985) lists this species as a pathogen of Citrullus lanatus, Cucumis melo, Cucumis sativus, Cucurbita pepo, Cucurbita sp. and Trichosanthes anguina in North Carolina, USA. Recently, Ga. magna was also reported from Cucumis sativus, Lagenaria siceraria and Luffa cylindrica (Cucurbitaceae) in Taiwan (Tsay et al. 2010), as an anthracnose pathogen of papaya (Carica papaya) in Brazil, Mexico and Costa Rica (Nascimento et al., 2010, Tapia-Tussell et al., 2016, Molina-Chaves et al., 2017) and from Lobelia chinensis in China (Li et al. 2013). Wasilwa et al. (1993) found that strains of this species were less aggressive compared with those of C. orbiculare. Freeman and Rodriguez, 1992, Freeman and Rodriguez, 1993 and Redman et al. (1999) studied the lifestyle of this fungus, and were able to disrupt pathogenicity in C. magnum using an ultraviolet radiation treatment, transforming it to a non-pathogenic, endophytic mutualist. In the ITS and MAT1-2 phylogenies by Du et al. (2005), strain L2.5 (CBS 519.07) grouped outside any known species complex. The sequence of the mating type protein (MAT1-2) gene is also included in Marcelino et al. (2008), confirming this result. The species was however never included in any of the recent multilocus studies of the genus (Cannon et al., 2012, Marin-Felix et al., 2017), and its close relatives are unknown.
Colletotrichum orchidearum is known as a causal agent of anthracnose of Orchidaceae. The species was described by Allescher (1902) with three formae on three different Orchidaceae species from the Munich Botanical Garden. Recently, strains from different Orchidaceae in China were included in a multilocus analysis of Orchidaceae plants from China and identified as C. orchidearum (Yang et al. 2011). Farr & Rossman (2017) list C. orchidearum from numerous Orchidaceae hosts from Asian, African and Latin American countries. Xu et al. (2016) also reported this species from Arctium lappa in China. However, except for Yang et al. (2011) and Xu et al. (2016), these reports originate from checklists or other reports from the pre-molecular era. There are sequences of strains called C. orchidearum from two further studies in GenBank, one of them displayed a strain as “type strain of C. orchidearum” (Z. Zhang et al., unpubl. data). However, there are no strains available from the original publication. The species was lectotypified recently (Damm et al. 2012a), but still awaits epitypification to fix the genetic application of the name.
In preliminary analyses based on ITS sequence data, strains of Ga. glycines, C. magnum and C. orchidearum appear basal in Colletotrichum phylogenies close to C. cliviae, C. brevisporum and other recently described species from mainly tropical regions (Yang et al., 2009, Noireung et al., 2012), and indicating that more than one species complex was involved. The aim of this study was therefore to clarify the systematic position of Ga. glycines, C. magnum and C. orchidearum and related species, resolve the respective species complexes and characterise the species in these complexes morphologically and by means of multilocus sequence analyses.
Materials and methods
Isolates
A total of 102 strains was studied, previously identified as C. cliviae, C. orchidearum, C. brevisporum, Ga. glycines, C. magnum and C. dracaenophilum, as well as other related strains from the culture collections of the Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands (CBS), the Genetic Resources Center, National Agriculture and Food Research Organization, Tsukuba, Ibaraki, Japan (MAFF), the University of Tehran, University College of Agriculture and Natural Resources, Tehran, Iran (UTFC) and CABI Europe UK Centre, Egham, UK (IMI) as well as from recent studies in the literature. The Iranian strains were part of recent collections by A. Alizadeh and O. Atghia from cultivated and wild plants in Iran. Type material (holotypes, lectotypes and epitypes) of the species studied are located in the fungaria of the CBS, the US National Fungus Collections, Beltsville, Maryland, USA (BPI), the fungaria based in the Royal Botanic Gardens, Kew, UK (IMI and K(M)), the Botanische Staatssammlung München (M), Germany and the Herbarium Hamburgense (HBG), Germany. All descriptions are based on the ex-holotype or ex-epitype cultures, if not stated otherwise. Features of other strains are added if deviant. Subcultures of the ex-holotypes and ex-epitypes, respectively, as well as all other isolates used for morphological and sequence analyses are maintained in the culture collections of CBS, IMI, MAFF and UTFC (Table 1).
Table 1.
Strains of Colletotrichum spp. studied, with collection details and GenBank accession numbers.
| Species | Accession no.1 | Host | Country | GenBank No.2 |
|||||
|---|---|---|---|---|---|---|---|---|---|
| ITS | GAPDH | CHS-1 | HIS3 | ACT | TUB2 | ||||
| C. brevisporum | CBS 129957 | Anthurium sp. | Thailand | MG600762 | MG600822 | MG600869 | MG600908 | MG600966 | MG601029 |
| CBS 129958 | Anthurium sp. | Thailand | MG600763 | MG600823 | MG600870 | MG600909 | MG600967 | MG601030 | |
| CBS 512.75 | Carica papaya | Australia | MG600761 | MG600821 | MG600868 | MG600907 | MG600965 | MG601028 | |
| BCC 38876* | Neoregalia sp. | Thailand | JN050238 | JN050227 | — | — | JN050216 | JN050244 | |
| MFLUCC100182 | Pandanus pygmaeus | Thailand | JN050239 | JN050228 | — | — | JN050217 | JN050245 | |
| MAFF 305751 | Passiflora edulis | Japan | MG600764 | MG600824 | MG600871 | — | MG600968 | MG601031 | |
| C. cacao | CBS 119297* | Theobroma cacao | Costa Rica | MG600772 | MG600832 | MG600878 | MG600916 | MG600976 | MG601039 |
| C. cattleyicola | CBS 170.49* | Cattleya sp. | Belgium | MG600758 | MG600819 | MG600866 | MG600905 | MG600963 | MG601025 |
| MAFF 238321 | Cattleya sp. | Japan | MG600759 | — | — | — | — | MG601026 | |
| C. cliviicola | CBS 125375* | Clivia miniata | China | MG600733 | MG600795 | MG600850 | MG600892 | MG600939 | MG601000 |
| CSSS2 | Clivia miniata | China | GU109480 | GU085868 | GU085866 | — | GU085862 | GU085870 | |
| CBS 133705 | Clivia sp. | South Africa | MG600732 | MG600794 | MG600849 | MG600891 | MG600938 | MG600999 | |
| C. coelogynes | CBS 132504* | Coelogyne sp. | Germany | MG600713 | MG600776 | MG600836 | MG600882 | MG600920 | MG600980 |
| CBS 132515 | Coelogyne sp. | Germany | MG600714 | MG600777 | MG600837 | MG600883 | MG600921 | MG600981 | |
| C. dracaenophilum | CBS 121453 | Dracaena sanderana | Bulgaria | MG600712 | MG600775 | MG600835 | MG600881 | MG600919 | MG600979 |
| CBS 119360 | Dracaena sanderana | China | MG600711 | MG600774 | MG600834 | MG600880 | MG600918 | MG600978 | |
| CBS 118200 | Dracaena sanderana | China | MG600710 | MG600773 | MG600833 | MG600879 | MG600917 | MG600977 | |
| CBS 118199* | Dracaena sanderana | China | JX519222 | JX546707 | JX519230 | JX546756 | JX519238 | JX519247 | |
| C. excelsum-altitudinum | CGMCC 3.15130* | Bletilla ochracea | China | HM751815 | KC843502 | — | — | KC843548 | JX625211 |
| CGMCC 3.15131 | Bletilla ochracea | China | JX625182 | KC843503 | — | — | KC843549 | JX625212 | |
| C. gloeosporioides | CBS 112999* | Citrus sinensis | Italy | JQ005152 | JQ005239 | JQ005326 | JQ005413 | JQ005500 | JQ005587 |
| C. liaoningense 1 | CGMCC 3.17616, CAUOS2* | Capsicum annuum | China | KP890104 | KP890135 | KP890127 | — | KP890097 | KP890111 |
| C. liaoningense 1 | CAUOS6 | Capsicum annuum | China | — | — | KP890131 | — | — | KP890115 |
| C. liaoningense 2 | CAUOS3 | Capsicum annuum | China | KP890105 | KP890136 | KP890128 | — | — | KP890112 |
| C. liaoningense 2 | CAUOS4 | Capsicum annuum | China | KP890106 | KP890137 | KP890129 | — | KP890099 | KP890113 |
| C. lobatum | IMI 79736* | Piper catalpaefolium | Trinidad and Tobago | MG600768 | MG600828 | MG600874 | MG600912 | MG600972 | MG601035 |
| C. magnum | CBS 519.97* | Citrullus lanatus | USA | MG600769 | MG600829 | MG600875 | MG600913 | MG600973 | MG601036 |
| IMI 391662 | Citrullus lanatus | USA | MG600771 | MG600831 | MG600877 | MG600915 | MG600975 | MG601038 | |
| CBS 575.97 | Citrullus lanatus | USA | MG600770 | MG600830 | MG600876 | MG600914 | MG600974 | MG601037 | |
| C. merremiae | CBS 124955* | Merremia umbellata | Panama | MG600765 | MG600825 | MG600872 | MG600910 | MG600969 | MG601032 |
| C. musicola | CBS 132885* | Musa sp. | Mexico | MG600736 | MG600798 | MG600853 | MG600895 | MG600942 | MG601003 |
| CBS 127557 | Musa sp. | Mexico | MG600737 | MG600799 | MG600854 | MG600896 | MG600943 | MG601004 | |
| C. okinawense | MAFF 240517 | Carica papaya | Japan | MG600767 | MG600827 | — | — | MG600971 | MG601034 |
| C. orchidearum | CORCX6 | Cattleya sp. | China | HM585403 | HM585393 | HM582027 | — | HM581997 | HM585419 |
| UTFC 262 | Cordyline terminalis | Iran | MG600742 | MG600804 | — | — | MG600948 | MG601009 | |
| CORCG3 | Cymbidium hookerianum | China | HM585402 | HM585392 | HM582026 | — | HM581996 | HM585416 | |
| CBS 135131* | Dendrobium nobile | Netherlands | MG600738 | MG600800 | MG600855 | MG600897 | MG600944 | MG601005 | |
| CBS 136877 | Dendrobium nobile | Netherlands | MG600739 | MG600801 | MG600856 | MG600898 | MG600945 | MG601006 | |
| MAFF 239931 | Dendrobium phalaenopsis | Japan | MG600745 | MG600807 | — | — | MG600951 | MG601012 | |
| MAFF 240480 | Dendrobium phalaenopsis | Japan | MG600746 | MG600808 | MG600858 | — | MG600952 | MG601013 | |
| UTFC 266 | Epipremnum aureum | Iran | MG600741 | MG600803 | — | — | MG600947 | MG601008 | |
| MFLUCC 12-0531* (HT of C. hymenocallidicola) | Hymenocallis sp. | Thailand | KT290264 | KT290263 | KT290262 | — | — | — | |
| CGMCC 3.14982* (HT of C. aracearum) | Monstera deliciosa | China | KX853166 | KX893585 | — | — | KX893577 | KX893581 | |
| CORCX11 | Oncidium flexuosum | China | HM585404 | HM585394 | HM582028 | — | HM581998 | HM585417 | |
| MAFF 238499 | Oncidium sp. | Japan | MG600743 | MG600805 | — | — | MG600949 | MG601010 | |
| MAFF 238779 | Oncidium sp. | Japan | MG600744 | MG600806 | MG600857 | — | MG600950 | MG601011 | |
| MAFF 306084 | Phalaenopsis sp. | Japan | MG600747 | — | — | — | — | MG601014 | |
| CGMCC 3.14983 | Philodendron selloum | China | KX853167 | KX893586 | — | — | KX893578 | KX893582 | |
| UTFC 265 | Philodendron sp. | Iran | MG600740 | MG600802 | — | — | MG600946 | MG601007 | |
| MAFF 240504 | Vanda sp. | Japan | MG600748 | MG600809 | MG600859 | — | MG600953 | MG601015 | |
| C. panamense | CBS 125386* | Merremia umbellata | Panama | MG600766 | MG600826 | MG600873 | MG600911 | MG600970 | MG601033 |
| C. piperis | IMI 71397, CPC 21195* | Piper nigrum | Malaysia | MG600760 | MG600820 | MG600867 | MG600906 | MG600964 | MG601027 |
| C. plurivorum | MAFF 306008 | Abelmoschus esculentus | Japan | MG600727 | MG600790 | — | — | MG600933 | MG600994 |
| MAFF 243073 | Amorphophallus rivieri | Japan | MG600730 | MG600793 | MG600847 | — | MG600936 | MG600997 | |
| CORCX9 | Arundina graminifolia | China | HM585398 | HM585381 | HM582025 | — | HM581986 | HM585423 | |
| CGMCC 3.17358 | Camellia sinensis | China | KJ955215 | KJ954916 | — | — | KJ954483 | KJ955361 | |
| LJTJ30* (HT of C. sichuanensis) | Capsicum annuum | China | KP748221 | KP823800 | — | — | KP823741 | KP823853 | |
| LJTJ3 | Capsicum annuum | China | KP748193 | KP823773 | — | — | KP823738 | KP823850 | |
| LJTJ16 | Capsicum annuum | China | KP748207 | KP823786 | — | — | KP823739 | KP823851 | |
| LJTJ22 | Capsicum annuum | China | KP748213 | KP823792 | — | — | KP823740 | KP823852 | |
| MAFF 238697 | Carica papaya | Japan | MG600724 | MG600787 | — | — | MG600931 | MG600991 | |
| CBS 125474* | Coffea sp. | Vietnam | MG600718 | MG600781 | MG600841 | MG600887 | MG600925 | MG600985 | |
| CBS 125473 | Coffea sp. | Vietnam | MG600717 | MG600780 | MG600840 | MG600886 | MG600924 | MG600984 | |
| CORCG2 | Cymbidium hookerianum | China | HM585397 | HM585380 | HM582024 | — | HM581985 | HM585422 | |
| MAFF 238875 | Glycine max | Japan | MG600725 | MG600788 | — | — | — | MG600992 | |
| LFN0008 | Glycine max. | Brazil | KT696336 | KT696289 | — | KT696311 | KT696275 | KT696282 | |
| CBS 132443 | Gossypium sp. | Brazil | MG600719 | MG600782 | MG600842 | MG600888 | MG600926 | MG600986 | |
| CBS 132444 | Gossypium sp. | Brazil | MG600720 | MG600783 | MG600843 | MG600889 | MG600927 | MG600987 | |
| MAFF 306007 | Lycopersicon esculentum | Japan | MG600728 | MG600791 | MG600846 | — | MG600934 | MG600995 | |
| CMM 3742 | Mangifera indica | Brazil | KC702980 | KC702941 | KC598100 | — | KC702908 | KC992327 | |
| CMM 3746 | Mangifera indica | Brazil | KC702981 | KC702942 | KC598101 | — | KC702909 | KC992328 | |
| MAFF 305790 | Musa sp. | Japan | MG600726 | MG600789 | MG600845 | — | MG600932 | MG600993 | |
| MAFF 238315 | Oncidium sp. | Japan | MG600729 | MG600792 | — | — | MG600935 | MG600996 | |
| MAFF 305974 | Passiflora edulis | Japan | MG600731 | — | MG600848 | — | MG600937 | MG600998 | |
| CBS 903.69 | Phaseolus lunatus | Benin | MG600721 | MG600784 | MG600844 | MG600890 | MG600928 | MG600988 | |
| UTFC 261 | Phaseolus vulgaris | Iran | MG600722 | MG600785 | — | — | MG600929 | MG600989 | |
| UTFC 260 | Spathiphyllum wallisii | Iran | MG600723 | MG600786 | — | — | MG600930 | MG600990 | |
| C. sojae | SAUCC 1407 | Arctium lappa | China | KT362184 | KT362188 | KT362187 | — | KT362189 | KT362185 |
| CGMCC 3.15171 | Bletilla ochracea | China | HM751813 | KC843501 | — | — | KC843550 | KC244161 | |
| CAUOS5 | Capsicum sp. | China | KP890107 | KP890138 | KP890130 | — | — | KP890114 | |
| LFN0009 | Glycine max | Brazil | KT696354 | KT696295 | — | KT696318 | KT696281 | KT696288 | |
| UTFC 288 | Glycine max | Iran | MG600755 | MG600816 | — | — | MG600960 | MG601022 | |
| CBS 134.87 | Glycine max | Italy | MG600752 | MG600813 | MG600863 | MG600902 | MG600957 | MG601019 | |
| CBS 181.81 | Glycine max | Serbia | MG600753 | MG600814 | MG600864 | MG600903 | MG600958 | MG601020 | |
| CBS 182.81 | Glycine max | Serbia | MG600754 | MG600815 | MG600865 | MG600904 | MG600959 | MG601021 | |
| ATCC 62257* | Glycine max | USA | MG600749 | MG600810 | MG600860 | MG600899 | MG600954 | MG601016 | |
| IL18A | Glycine max | USA | KC110792 | KC110810 | — | KC110801 | KC110828 | KC110819 | |
| IL26A | Glycine max | USA | KC110793 | KC110811 | — | KC110802 | KC110829 | KC110820 | |
| ATCC 11871 | Medicago sativa | USA | MG600750 | MG600811 | MG600861 | MG600900 | MG600955 | MG601017 | |
| CBS 128510 | Medicago sativa | USA | MG600751 | MG600812 | MG600862 | MG600901 | MG600956 | MG601018 | |
| UTFC 301 | Phaseolus vulgaris | Iran | MG600756 | MG600817 | — | — | MG600961 | MG601023 | |
| UTFC 303 | Vigna unguiculata | Iran | MG600757 | MG600818 | — | — | MG600962 | MG601024 | |
| C. tropicicola 1 | BCC 38877, MFLUCC 110114* | Citrus maxima | Thailand | JN050240 | JN050229 | — | — | JN050218 | JN050246 |
| C. tropicicola 1 | CBS 127555, CPC 15927 | Citrus sp. | Mexico | MG600715 | MG600778 | MG600838 | MG600884 | MG600922 | MG600982 |
| C. tropicicola 1 | CBS 133174, CPC 15924 | Citrus sp. | Mexico | MG600716 | MG600779 | MG600839 | MG600885 | MG600923 | MG600983 |
| C. tropicicula 2 | MFLUCC100167 | Paphiopedilum bellatolum | Thailand | JN050241 | JN050230 | — | — | JN050219 | JN050247 |
| C. vittalense | GUFCC 15503 | Calamus thwaitesii | India | JN390935 | KC790759 | KF451996 | — | KC790646 | KC790892 |
| CBS 126.25 | orchid | unknown | MG600735 | MG600797 | MG600852 | MG600894 | MG600941 | MG601002 | |
| CBS 181.82* | Theobroma cacao | India | MG600734 | MG600796 | MG600851 | MG600893 | MG600940 | MG601001 | |
| C. yunnanense | CBS 132135, AS3.9617* | Buxus sp. | China | JX546804 | JX546706 | JX519231 | JX546755 | JX519239 | JX519248 |
| AS3.9616 | Buxus sp. | China | EF369491 | — | — | — | — | — | |
| Colletotrichum sp. | GZAAS5 09545 | Citrus medica | China | JQ247623 | JQ247599 | — | — | JQ247647 | JQ247635 |
| COUFAL7300 | Sechium edule | Brazil | KT285378 | KT285381 | KT285380 | — | KT285378 | KT285383 | |
* Ex-holotype or ex-epitype cultures.
ATCC: American Type Culture Collection, Virginia, USA; BCC: BIOTEC culture collection, Bangkok, Thailand; CBS: Culture collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands; CGMCC: China General Microbiological Culture Collection Center, Beijing, China; CMM: Culture Collection of Phytopathogenic Fungi Prof. Maria Menezes, Federal Rural University of Pernambuco, Brazil; CPC: Culture collection of Pedro Crous, housed at CBS; COUFAL: Coleção de Culturas de Fungos Fitopatogênicos da Universidade Federal de Alagoas, Brazil; GUFCC: Goa University, Fungal Culture Collection, Taleigao Plateau, Goa, India; GZAAS: Guizhou Academy of Agricultural Sciences, Guiyang, China; IMI: Culture collection of CABI Europe UK Centre, Egham, UK; LARS: Culture collection of Long Ashton Research Station, Bristol, UK (no longer existing); MAFF: MAFF Genebank Project, Ministry of Agriculture, Forestry and Fisheries, Tsukuba, Japan; MFLUCC: Culture collection of the Mae Fah Luang University, Chiang Rai, Thailand ; SAUCC: Culture collection of the Department of Plant Pathology, College of Plant Protection, Shenyang Agricultural University, China; STE-U: Culture collection of the Department of Plant Pathology, University of Stellenbosch, South Africa; UTFC: Culture collection of the University of Tehran, University College of Agriculture and Natural Resources, University of Tehran, Karaj, Iran.
ITS: internal transcribed spacers and intervening 5.8S nrDNA; GAPDH: partial glyceraldehyde-3-phosphate dehydrogenase gene; CHS-1: partial chitin synthase-1 gene; HIS: partial histone H3 gene; ACT: partial actin gene; TUB2: partial beta-tubulin gene. Sequences generated in this study are emphasised in bold face.
Morphological analysis
To enhance sporulation, autoclaved filter paper and double-autoclaved stems of Anthriscus sylvestris were placed onto the surface of synthetic nutrient-poor agar medium (SNA; Nirenberg 1976). SNA and OA (oatmeal agar; Crous et al. 2009) cultures were incubated at 20 °C under near UV light with a 12 h photoperiod for 10 d. Measurements and photographs of characteristic structures were made according to Damm et al. (2007). Appressoria on hyphae were observed on the reverse side of SNA plates or on slide cultures (Damm et al. 2013). Microscopic preparations were made in clear lactic acid, with 30 measurements per structure and observed with a Nikon SMZ1000 dissecting microscope (DM), or with a Nikon Eclipse 80i microscope using differential interference contrast (DIC) illumination. Colony characters and pigment production on SNA and OA cultures incubated at 20 °C under near UV light with 12 h photoperiod were determined after 10 d. Colony colours were rated according to Rayner (1970). Growth rates were measured after 7 and 10 d.
Phylogenetic analysis
Genomic DNA of the isolates was extracted using the method of Damm et al. (2008). The 5.8S nuclear ribosomal RNA gene with the two flanking internal transcribed spacers (ITS), a 200-bp intron of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH), partial sequences of the chitin synthase 1 (CHS-1), histone H3 (HIS3), actin (ACT) and beta-tubulin (TUB2) genes were amplified and sequenced using the primer pairs ITS-1F (Gardes & Bruns 1993) + ITS-4 (White et al. 1990), GDF1 + GDR1 (Guerber et al. 2003), CHS-354R + CHS-79F (Carbone & Kohn 1999), CYLH3F + CYLH3R (Crous et al. 2004b), ACT-512F + ACT-783R (Carbone & Kohn 1999) and T1 (O'Donnell & Cigelnik 1997) + Bt-2b (Glass & Donaldson 1995) or T1 + BT4R (Woudenberg et al. 2009), respectively. The PCRs were performed in a 2720 Thermal Cycler (Applied Biosystems, Foster City, California) in a total volume of 12.5 μL. The GAPDH, CHS-1, HIS3, ACT and TUB2 PCR mixture contained 1 μL 20× diluted genomic DNA, 0.2 μM of each primer, 1x PCR buffer (Bioline, Luckenwalde, Germany), 2 mM MgCl2, 20 μM of each dNTP, 0.7 μL DMSO and 0.25 U Taq DNA polymerase (Bioline). Conditions for PCR of these genes constituted an initial denaturation step of 5 min at 94 °C, followed by 40 cycles of 30 s at 94 °C, 30 s at 52 °C and 30 s at 72 °C, and a final denaturation step of 7 min at 72 °C, while the ITS PCR was performed as described by Woudenberg et al. (2009). The DNA sequences generated with forward and reverse primers were used to obtain consensus sequences using Bionumerics v. 4.60 (Applied Maths, St-Marthens-Lathem, Belgium), and the alignment assembled and manually adjusted using Sequence Alignment Editor v. 2.0a11 (Rambaut 2002).
To determine whether the six sequence datasets were congruent and combinable, tree topologies of 70 % reciprocal Neighbour-Joining bootstrap with Maximum Likelihood distances (10 000 replicates) with substitution models determined separately for each partition using MrModeltest v. 2.3 (Nylander 2004) were compared visually (Mason-Gamer & Kellogg 1996). Maximum parsimony analyses were performed on the multilocus alignment (ITS, GAPDH, CHS-1, HIS3, ACT, TUB2) as well as for each gene separately with PAUP (Phylogenetic Analysis Using Parsimony) v. 4.0b10 (Swofford 2003) using the heuristic search option with 100 random sequence additions and tree bisection and reconstruction (TBR) as the branch-swapping algorithm. Alignment gaps were treated as missing and all characters were unordered and of equal weight. No more than 10 trees of score (length) greater than or equal to 10 were saved in each replicate. Tree length, consistency index (CI), retention index (RI), rescaled consistency index (RC) and homoplasy index (HI) were calculated for the resulting tree. The robustness of the trees obtained was evaluated by 10 000 bootstrap replications using the Fast-stepwise addition algorithm (Hillis & Bull 1993). A Markov Chain Monte Carlo (MCMC) algorithm was used to generate phylogenetic trees with Bayesian probabilities using MrBayes v. 3.2.6 (Ronquist & Huelsenbeck 2003) for the combined sequence datasets. Models of nucleotide substitution for each gene determined by MrModeltest v. 2.3 were included for each gene partition. The analyses of two MCMC chains were run from random trees for 10 00 000 generations and sampled every 100 generations. The likelihood score of the two runs were 2 630 and 2 600 and therefore, the first 2 615 (the average of both) trees were discarded as the burn-in phase of the analysis and posterior probabilities determined from the remaining trees. For additional comparison, a Neighbour-Joining analysis was performed on the multigene alignment using PAUP and 1 000 bootstrap replications. Sequences derived in this study have been lodged at GenBank, the alignment and trees in TreeBASE (www.treebase.org/treebase-web/home.html), and taxonomic novelties in MycoBank (Crous et al. 2004a).
Results
Phylogeny
The six individual datasets did not show any conflicts in tree topology of the 70 % reciprocal bootstrap trees, which allowed us to combine them. In the multigene analyses (gene boundaries of ITS: 1–558, GAPDH: 569–900, CHS-1: 911–1 211, HIS3: 1 222–1 642, ACT: 1 653–1 940, TUB2: 1 951–2 498) of 102 strains previously identified as C. cliviae, C. orchidearum, C. brevisporum, Ga. glycines, C. magnum, C. dracaenophilum as well as other related strains, including the outgroup (C. gloeosporioides strain CBS 112999), 2 500 characters including the alignment gaps were processed, of which 613 characters were parsimony-informative, 158 parsimony-uninformative and 1 729 constant. After a heuristic search using PAUP, the maximum of 1 000 equally most parsimonious trees were retained (length = 1 470 steps, CI = 0.723, RI = 0.942, RC = 0.681, HI = 0.277), of which one is shown in Fig. 1. The topology of the 1 000 trees was similar, which was verified for a large selection of trees. They differed only in the position of taxa within the subclades and in the position of some of the subclades within the main clades. For the Bayesian analyses, a GTR+G model was selected for ITS, a GTR+I+G model for CHS-1, a HKY+G model for GAPDH, TUB2 and HIS3 and a HKY+I model for ACT, and incorporated in the analysis. The consensus tree obtained from Bayesian analyses confirmed the tree topology obtained with parsimony. Bayesian posterior probability values agreed with bootstrap support values (Fig. 1).
Fig. 1.
The first of 1 000 equally most parsimonious trees obtained from a heuristic search of the combined ITS, GAPDH, CHS-1, HIS3, ACT and TUB2 sequence alignment of the Colletotrichum dracaenophilum, C. magnum and C. orchidearum species complexes. Bootstrap support values above 70 % (bold) and Bayesian posterior probability values above 0.90 are shown at the nodes. Colletotrichum gloeosporioides strain CBS 112999 is used as outgroup. Numbers of ex-holotype and ex-epitype isolates are emphasised with an asterisk. Strain numbers are followed by substrate (host genus) and country of origin, NL = Netherlands, Trin Tobago = Trinidad and Tobago. Species complexes are indicated by blue lines. Branches that are crossed by diagonal lines are shortened by 50 %.
The analyses resulted in detection of three main clades and 24 subclades, presumably representing different Colletotrichum species. The first clade (orchidearum, 99 % bootstrap support) consists of eight subclades, of which three subclades include a large number of strains from several host genera, and five subclades with one to three strains of either the same host genus or the same country. The two subclades on the top of the phylogeny are short-branched with bootstrap support values of 80 % and 94 %, respectively. Except for the single-strain clade of C. piperis, all other subclades are longer-branched with bootstrap support values ranging between 82 and 100 %. The second clade (magnum, 99 %) consists of 10 subclades; half of them are single-strain clades. Two further subclades containing five and three strains are on short branches. The remaining three subclades consist of strains from three studies, the sequences of which were downloaded from GenBank; the four strains of C. liaoningense form two subclades. There are six subclades in the third clade (dracaenophilum, 86 %); three of them are very long-branched, while the other three subclades are closely related. There was only the ITS sequence available of the second strain of C. yunnanense; the bootstrap support value of this long branch is therefore only 87 %. One strain previously identified as C. tropicicola (MFLUCC 10-0167) formed a separate lineage between the C. tropicicola clade containing the ex-holotype of that species and C. excelsum-altitudinum. All six subclades contain strains from only one host genus each.
The three subclades, further referred to as C. dracaenophilum, C. magnum and C. orchidearum species complexes, respectively, are all well supported in the multilocus phylogeny, however, the clades are not supported with some of the single-locus phylogenies (not shown). Isolates can be best assigned to one of these species complexes by using TUB2 sequence data.
Taxonomy
Based on DNA sequence data and morphology, the 102 strains studied (Table 1) are assigned to 24 species, of which 8, 10 and 6 species, respectively, belong to the Colletotrichum dracaenophilum, C. magnum and C. orchidearum species complexes, including 11 species that proved to be new to science and are described. Five species formed sexual morphs in vitro. All species studied in culture are characterised below.
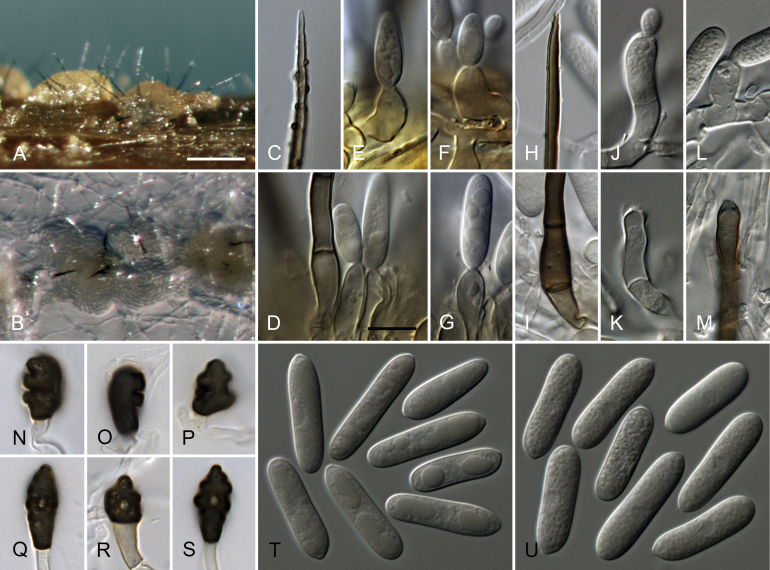
Colletotrichum brevisporum Noireung et al., Cryptog. Mycol. 33: 350. 2012. Fig. 2.
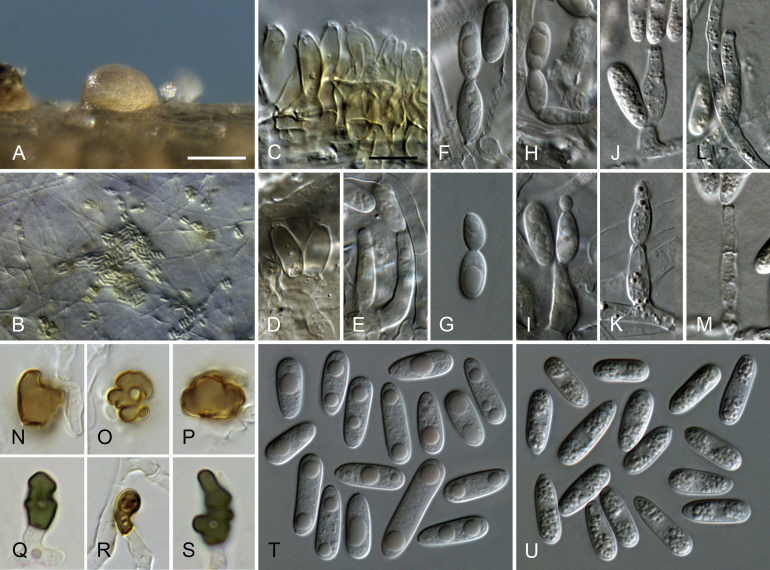
Fig. 2.
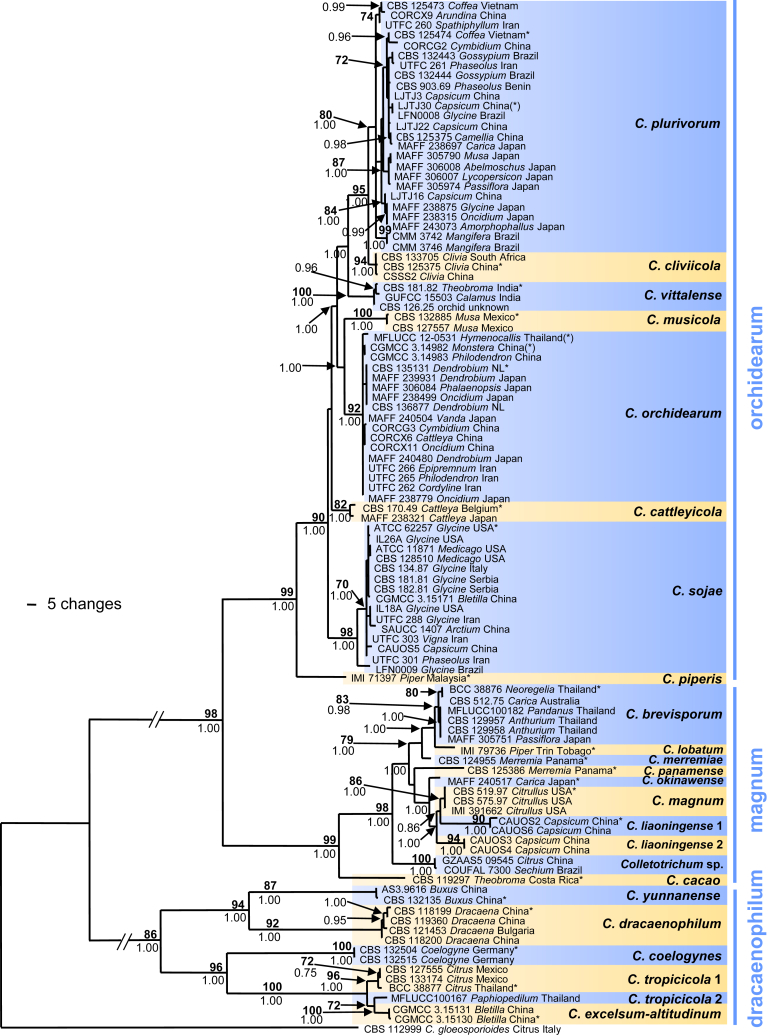
Colletotrichum brevisporum (A–N, U–V. from culture CBS 129957. O–T. from culture CBS 512.75). A–B. Conidiomata. C, I. Tips of setae. D, J. Bases of setae. E–H, K–N. Conidiophores. O–T. Appressoria. U–V. Conidia. A, C–H, U. from Anthriscus stem. B, I–T, V. from SNA. A–B. Dissecting microscope (DM). C–V. Differential interference contrast illumination (DIC). Scale bars: A = 100 μm, E = 10 μm. Scale bar of A applies to A–B. Scale bar of E applies to C–V.
A description of the type specimen is provided by Noireung et al. (2012). The description below is based on strains from Anthurium sp. in Thailand and Carica papaya in Australia.
Sexual morph not observed. Asexual morph on SNA (CBS 129957). Vegetative hyphae 2–8 μm diam, hyaline to pale brown, smooth-walled, septate, branched. Chlamydospores not observed. Conidiomata, conidiophores and setae formed directly on hyphae. Setae medium brown, verrucose, 60–110 μm long, 2–4-septate, base pale brown, cylindrical, sometimes slightly inflated, 4–7 μm diam, tip rounded to ± acute. Conidiophores hyaline to pale brown, smooth-walled, septate, branched, to 40 μm long. Conidiogenous cells (hyaline to) pale brown, smooth-walled, cylindrical to ampulliform, 12–24 × 3–6 μm, opening 1–2 μm diam, collarette 0.5 μm long, periclinal thickening distinct. Conidia hyaline, smooth-walled, aseptate, the apex rounded, the base rounded to truncate, (14–)16.5–19(–19.5) × (4–)4.5–5(–5.5) μm, mean ± SD = 17.7 ± 1.3 × 4.8 ± 0.4 μm, L/W ratio = 3.7, conidia of strain CBS 512.75 shorter, measuring (10.5–)12.5–14.5(–15.5) × (3.5–)4–5(–6) μm, mean ± SD = 13.4 ± 1.1 × 4.6 ± 0.6 μm, L/W ratio = 2.9. Appressoria not formed by CBS 129957 after > 2 wk, appressoria of strain CBS 129958 single, dark brown, smooth-walled, irregularly roundish outline, with an undulate to lobate margin, (5.5–)8–13(–16) × (4–)6–10(–12.5) μm, mean ± SD = 10.4 ± 2.5 × 8.0 ± 2.2 μm, L/W ratio = 1.3.
Asexual morph on Anthriscus stem (CBS 129957). Conidiomata, conidiophores and setae formed on pale brown, angular cells, 3–8 μm diam. Setae medium brown, verrucose, 50–120(–160) μm long, 1–3-septate, base cylindrical to slightly inflated, 5–6.5 μm diam, tip ± acute to ± rounded. Conidiophores pale brown, smooth-walled. Conidiogenous cells pale brown, smooth-walled, ellipsoidal to cylindrical, 8–22 × 4–6 μm, opening 1–2 μm diam, conidiogenous cells of strain CBS 512.75 often bent and inflated in the upper part and sometimes extending to form new conidiogenous loci, collarette 0.5 μm long, periclinal thickening visible or distinct. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical, the apex rounded, the base rounded to truncate, (17–)18–20.5(–22) × (4–)4.5–5(–5.5) μm, mean ± SD = 19.3 ± 1.2 × 4.6 ± 0.3 μm, L/W ratio = 4.2, conidia of strain CBS 512.75 shorter and wider, measuring (11–)12.5–16(–16) × (4.5–)5–5.5(–6) μm, mean ± SD = 13.6 ± 1.3 × 5.3 ± 0.4 μm, L/W ratio = 2.6.
Cultural characteristics (CBS 129957): Colonies on SNA flat with entire margin, medium hyaline to pale ochreous, filter paper partly olivaceous grey, Anthriscus stem, filter paper and medium partly covered with olivaceous grey, iron grey or salmon acervuli and felty whitish aerial mycelium, reverse same colours; 25–27.5 mm in 7 d (≥40 mm in 10 d). Colonies on OA flat with entire margin, surface buff to grey olivaceous, covered with pale olivaceous grey, iron grey or salmon acervuli and very short, felty whitish aerial mycelium, reverse buff, pale to olivaceous grey to olivaceous grey; 26–27 mm in 7 d (≥40 mm in 10 d). Conidial mass salmon.
Materials examined: Australia, Victoria, Melbourne, from stem rot of Carica papaya, collection date and collector unknown (deposited in CBS collection Oct. 1975 by D.G. Parbery), CBS H-21067, CBS 512.75 = INB 13412. Thailand, Chiang Mai, Mesapok waterfall, from leaf spot of Anthurium sp., 5 Oct. 2010, P.W. Crous, CBS H-21064, culture CBS 129957; Chiang Mai, Mesapok waterfall, from leaf spot of Anthurium sp., 5 Oct. 2010, P.W. Crous, culture CBS 129958.
Notes: Colletotrichum brevisporum was described as a causal agent of leaf anthracnose of Neoregelia sp. and Pandanus pygmaeus in Thailand (Noireung et al. 2012), and has been reported as endophytes and pathogens of several host plants in tropical regions, including Capsicum chinense in Brazil (De Almeida et al. 2017), Capsicum annuum in China (Liu et al. 2016), Carica papaya in Australia (Shivas et al. 2016) and Brazil (Vieira et al. 2013), Passiflora edulis in Australia (Shivas et al. 2016), Citrus medica in China (Peng et al. 2012), Lycium chinense in Korea (Paul et al. 2014) and Sechium edule in Brazil (Bezerra et al. 2016).
Although there are only ITS sequences available of the strains from papaya and passion fruit in the paper of Shivas et al. (2016), which is not absolute proof of their identity as C. brevisporum, different strains included in this study (CBS 512.75, MAFF 305751) confirm the occurrence of this species on these hosts. However, we also included a strain from papaya in Japan (MAFF 240517), that represents a different species in the C. magnum species complex (see C. okinawense). We also report here C. brevisporum on Anthurium in Thailand (Fig. 2A–N, U–V). A different strain from Anthurium in Thailand was identified as C. karstii (CBS 129927) by Damm et al. (2012b), belonging to the C. boninense species complex.
The ITS and ACT sequences of C. brevisporum are the same as those of C. merremiae and C. lobatum; the ITS sequence is also identical with that of C. okinawense. Colletotrichum brevisporum can be identified based on its GAPDH sequence, but two groups are resolved. The TUB2 sequences have only one difference from C. lobatum. There are no CHS-1 and HIS3 sequences of the ex-type strain available, but based on the strains included in this study, there is one additional nucleotide difference from C. lobatum and C. panamaense, and from C. lobatum and C. merremiae, respectively.
In a blastn search on NCBI GenBank, the TUB2 sequence of the ex-type strain BCC 38876 is 100 % identical with the two strains of the original paper (Noireung et al. 2012), and with the sequence of C. brevisporum strain CCCM12 from Cucurbita moschata in China (KY797630, Liu et al. 2018). The only ACT and GAPDH sequences that are 100 % identical with the ex-type strain are those from the ex-type strain itself. The ACT sequence of the strain from Pandanus from the original paper as well as C. brevisporum strains CCCM12 from Cucurbita moschata in China (KY797629, Liu et al. 2018), strain IRA93 from Capsicum in Brazil (KU315567, De Almeida et al. 2017), strains CRI-L1 and CRI-N2 (KT185055, KT185056, L. Huang, unpubl. data) and four strains from Carica papaya in Brazil (Vieira et al. 2013) are 99 % identical (1 or 2 nucleotides difference). The GAPDH sequence of the strain from Pandanus is 99 % identical (2 nucleotides difference). The ITS sequences of a large number of unidentified strains and strains identified as C. brevisporum, C. magnum, Ga. magna and Ga. cingulata var. brevispora are 100 % identical with the ITS sequence of the ex-type strain of C. brevispora.
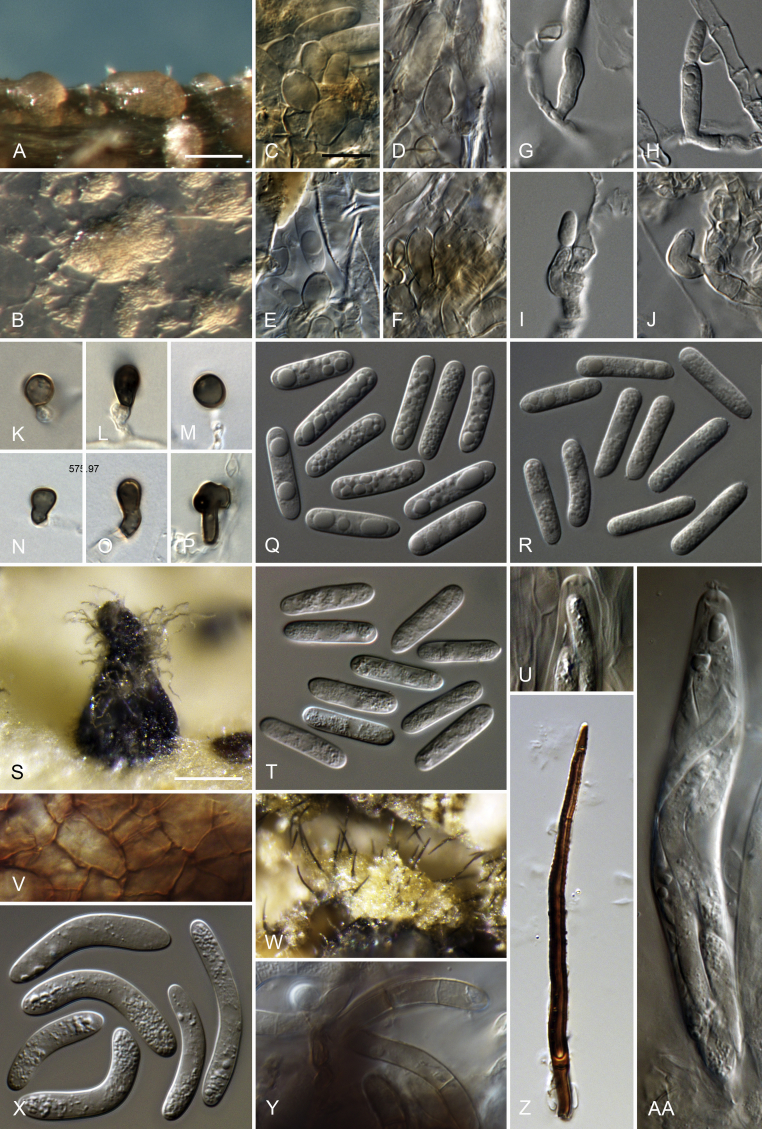
Colletotrichum cacao Damm, sp. nov. MycoBank MB824219. Fig. 3.
Fig. 3.
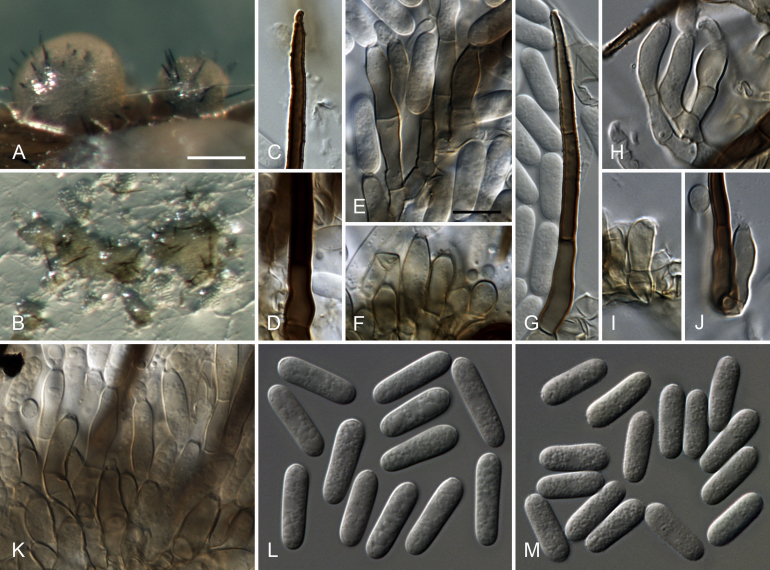
Colletotrichum cacao (from ex-holotype culture CBS 119297). A–B. Conidiomata. C. Tip of a seta. D. Base of a seta. E–F, H–K. Conidiophores. G. Seta. L–M. Conidia. A, C–F, K–L. from Anthriscus stem. B, G–J, M. from SNA. A–B. DM. C–M. DIC. Scale bars: A = 100 μm, E = 10 μm. Scale bar of A applies to A–B. Scale bar of E applies to C–M.
Etymology: The species epithet is derived from the host plant, Theobroma cacao.
Sexual morph not observed. Asexual morph on SNA. Vegetative hyphae 1.5–5.5 μm diam, hyaline, smooth-walled, septate, branched. Chlamydospores not observed. Conidiomata, conidiophores and setae formed directly on hyphae. Setae pale to medium brown, verrucose, 35–85 μm long, 1–2(–4)-septate, base cylindrical to somewhat inflated, 4–7.5 μm diam, tip ± acute. Conidiophores pale brown, smooth-walled, septate, sometimes branched, to 45 μm long. Conidiogenous cells pale brown, smooth-walled, ellipsoidal, obpyriform to clavate, 9–22 × (3–)4–6 μm, opening 1.5–2 μm diam, collarette 0.5–1 μm long, periclinal thickening visible, sometimes distinct. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical, the apex and base rounded, (13–)14–15.5(–17) × 5(–5.5) μm, mean ± SD = 14.7 ± 0.8 × 5.1 ± 0.2 μm, L/W ratio = 2.9. Appressoria not formed.
Asexual morph on Anthriscus stem. Conidiomata, conidiophores and setae formed directly on hyphae, no basal cells observed, but also formed in medium brown, closed conidiomata. Setae medium brown, smooth-walled, verruculose towards the tip, 50–130 μm long, 1–5-septate, base cylindrical, conical to slightly inflated, 4.5–6.5 μm diam, tip ± acute to ± rounded or functioning as conidiogenous locus. Conidiophores pale brown, smooth-walled to verrucose, septate, branched, to 60 μm long. Conidiogenous cells pale brown, smooth-walled to verrucose, cylindrical to clavate, 9–24 × 3.5–6 μm, opening 1.5–2 μm diam, collarette 0.5–1 μm long, periclinal thickening visible to distinct. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical, the apex and base rounded, (15.5–)16.5–18.5(–20.5) × 4.5–5.5 μm, mean ± SD = 17.4 ± 1.1 × 5.0 ± 0.3 μm, L/W ratio = 3.5.
Culture characteristics: Colonies on SNA flat with entire margin, agar medium hyaline to pale cinnamon, filter paper, Anthriscus stem and agar medium partly covered with isabelline acervuli, aerial mycelium lacking, reverse same colours; 20–25 mm in 7 d (30–34 mm in 10 d). Colonies on OA flat with entire margin, surface buff, almost entirely covered with small iron grey acervuli, with rosy buff to salmon spore masses towards the centre, whitish towards the margin, aerial mycelium lacking, reverse olivaceous buff to olivaceous grey; 22.5–23 mm in 7 d (34–34.5 mm in 10 d). Conidial mass rosy buff to salmon.
Material examined: Costa Rica, endophyte of Theobroma cacao, collection date and collector unknown (deposited in CBS collection by A. Rossman, INB 13412) (CBS H-21068 holotype, culture ex-type CBS 119297 = MCA 2773).
Notes: There are many Colletotrichum species occurring on Theobroma cacao. For example, C. ignotum, C. theobromicola and C. tropicale were recognised as endophytes of T. cacao by Rojas et al. (2010) and described or epitypified in that study; they belong to the C. gloeosporioides species complex, that was reviewed recently by Weir et al. (2012). One species from the C. acutatum species complex, C. sloanei, was described from Theobroma in Malaysia, belonging to the C. acutatum species complex (Damm et al. 2012a). A leaf endophyte of T. cacao in Panama from the study of Rojas et al. (2010) was re-identified as C. karstii (CBS 124951) by Damm et al. (2012b), belonging to the C. boninense species complex. In contrast, the species described here, C. cacao, belongs to the C. magnum species complex. Rojas et al. (2010) noted several further unidentified taxa amongst their collections on T. cacao from Panama.
Additional Colletotrichum species that have been described on T. cacao (see notes under C. sloanei in Damm et al. 2012a) include C. brachytrichum and C. theobromae from leaves of T. cacao in Trinidad and fruits of T. cacao in Cameroon that form conidia that are smaller than those of C. cacao, measuring 10–13.5 × 3–3.7 μm and 9–12 × 3–5 μm, respectively (Saccardo 1906). Colletotrichum cradwickii, described from branches of T. cacao in Jamaica, forms conidia that are elongate, constricted in the middle, measuring 14–17 × 5 μm; while C. luxificum was collected from branches, buds and fruits of T. cacao in Surinam and Demerara (now Guyana) forming ovoid-oblong conidia with both ends rounded, that are sometimes slightly constricted in the centre, measuring 13–19 × 4–5 μm (Saccardo & Trotter 1913). The conidial dimensions are similar to those of C. cacao. However, in both descriptions a constriction in the centre of the conidia is mentioned that was not observed in C. cacao.
The formation of setae that function as conidiogenous loci and conidia in closed conidiomata was not observed in any of the other species of the three species complexes studied here. Closed conidiomata are known from the C. boninense and C. gloeosporioides species complexes (Damm et al. 2012b, B. Weir, unpubl. data), and setae forming conidia is only known from C. theobromicola (syn. C. fragariae) in the C. gloeosporioides species complex (Villanueva-Arce et al. 2005).
Colletotrichum cacao can be identified with all loci studied. There is no TUB2 sequence in GenBank that is > 96 % identical, no ACT sequence > 97 % identical, no HIS3 sequence > 91 % identical, no CHS-1 sequence > 98 % identical and no GAPDH sequence > 96 % identical to the respective sequences of C. cacao. The ITS sequence of strain CBS 119297 is 100 % identical to that of Colletotrichum sp. MCA 2773 (DQ286217), the same strain sequenced by Farr et al. (2006), Colletotrichum sp. FH2 (FJ919388) from bitter gourd, probably from India (V. Jayakumar et al., unpubl. data), fungal endophyte STRI:ICBG-Panama:TK766 from a tropical woody plant (KF436361, Higginbotham et al. 2013) and C. gloeosporioides isolates S166, S170, S183 and S193 from tissue of Aristolochia triangularis in Brazil (MF076612–MF076615, A.K. Stuart et al., unpubl. data).
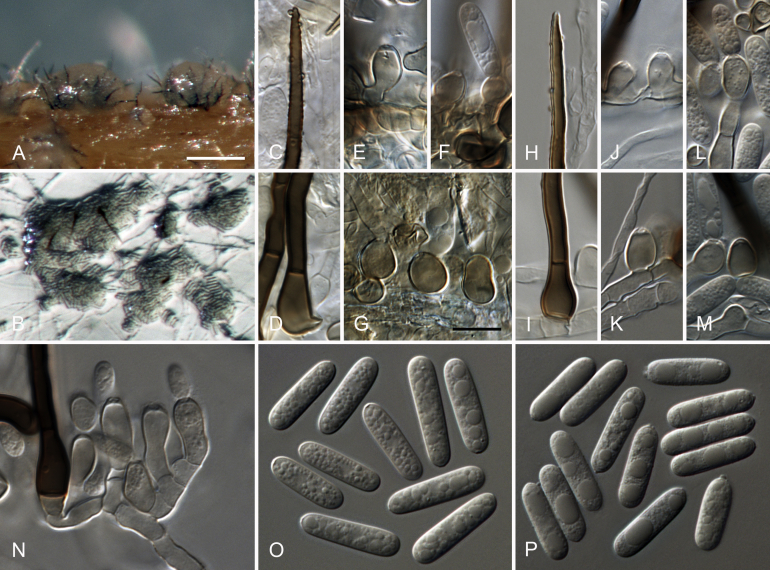
Colletotrichum cattleyicola Damm & Toy. Sato, sp. nov. MycoBank MB824220. Fig. 4.
Fig. 4.
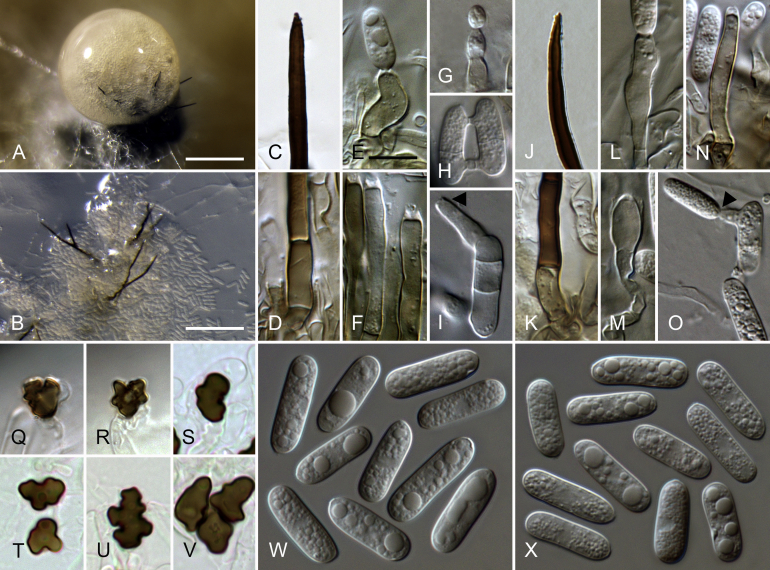
Colletotrichum cattleyicola (from ex-holotype culture CBS 170.49). A–B. Conidiomata. C, H. Tips of setae. D, I. Bases of setae. E–G, J–M. Conidiophores. N–S. Appressoria. T–U. Conidia. A, C–G, T. from Anthriscus stem. B, H–S, U. from SNA. A–B. DM. C–U. DIC. Scale bars: A = 100 μm, E = 10 μm. Scale bar of A applies to A–B. Scale bar of E applies to C–U.
Etymology: The species epithet is derived from the host plant, Cattleya.
Sexual morph not observed. Asexual morph on SNA. Vegetative hyphae 1–8 μm diam, hyaline, smooth-walled, septate, branched. Chlamydospores not observed. Conidiomata, conidiophores and setae formed directly on hyphae. Setae medium brown, verruculose to verrucose, 50–80 μm long, 2–3-septate, base cylindrical, slightly inflated to conical, 4.5–6 μm diam, tip ± acute to ± rounded, setae of strain MAFF 238321 up to 280 μm long. Conidiophores hyaline to pale brown, smooth-walled, septate, branched, to 35 μm long. Conidiogenous cells hyaline to pale brown, smooth-walled, cylindrical to ellipsoidal, 9–35 × 3.5–5.5 μm, opening 1.5–2 μm diam, collarette 0.5–1 μm long, periclinal thickening visible. Conidia hyaline, smooth-walled, aseptate, straight, sometimes very slightly curved, cylindrical, the apex and base rounded, (10.5–)14.5–19(–20) × (4–)5–5.5(–6) μm, mean ± SD = 16.9 ± 2.1 × 5.3 ± 0.4 μm, L/W ratio = 3.2, conidia of strain MAFF 238321 longer, measuring (14–)16.5–20,5(–22) × 5–6(–6.5) μm, mean ± SD = 18,7 ± 2.0 × 5.5 ± 0.3 μm, L/W ratio = 3.4. Appressoria single, pale to dark brown, smooth-walled, elongate rectangular, clavate to irregular in outline, with an undulate to lobate margin, (6–)8.5–13.5(–16) × (3.5–)4–6.5(–8) μm, mean ± SD = 11.0 ± 2.5 × 5.1 ± 1.3 μm, L/W ratio = 2.2, appressoria of strain MAFF 238321 wider and more variable in size, measuring (5–)6–16.5(–29) × (4.5–)5.5–11.5(–16.5) μm, mean ± SD = 11.3 ± 5.3 × 8.4 ± 2.9 μm, L/W ratio = 1.3.
Asexual morph on Anthriscus stem. Conidiomata, conidiophores and setae formed on pale brown, angular cells, 3–7.5 μm diam. Setae pale to medium brown, verrucose, 50–100 μm long, 1–3-septate, base conical, to ± inflated, 4.5–7.5 μm diam, tip ± rounded, setae of strain MAFF 238321 up to 330 μm long. Conidiophores pale brown, smooth-walled to verruculose, simple or septate and branched, to 50 μm long. Conidiogenous cells pale brown, smooth-walled to verruculose, doliiform to cylindrical, 7–18 × 4.5–5 μm, opening 1–1.5 μm diam, collarette 0.5–1 μm long, periclinal thickening visible, sometimes distinct. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical, the apex and base rounded, (13–)16–19(–20.5) × (5–)5.5–6(–6.5) μm, mean ± SD = 17.6 ± 1.4 × 5.9 ± 0.3 μm, L/W ratio = 3.0, conidia of strain MAFF 238321 longer, measuring (16–)18.5–21.5(–24.5) × (4.5–)5–6(–6.5) μm, mean ± SD = 20.0 ± 1.7 × 5.6 ± 0.4 μm, L/W ratio = 3.6.
Culture characteristics: Colonies on SNA flat with entire margin, hyaline to pale honey, agar medium partly covered with very short whitish aerial mycelium, reverse same colours; growth 30–32.5 mm in 7 d (≥ 40 mm in 10 d). Colonies on OA flat with entire margin; olivaceous buff to primrose, covered with very short whitish aerial mycelium, reverse same colours, growth 31.5–34 mm in 7 d (≥ 40 mm in 10 d). Conidial mass whitish to very pale salmon.
Materials examined: Belgium, from a root of Cattleya sp., collection date and collector unknown (isolated by J. van Holder, identified by A.C. Stolk, deposited in the CBS collection Feb. 1949) (CBS H-21502 holotype, culture ex-holotype CBS 170.49). Japan, Mie Prefecture, from a lesion on a stem sheath of Cattleya sp., unknown collection date, T. Kobayashi (isolated by T. Kobayashi, Jul. 2000), GLM-F111629, culture MAFF 238321 = CBS 143245 = GLMC 1836.
Notes: Colletotrichum cattleyicola belongs to the C. orchidearum species complex. Based on our study, C. cattleyicola is only known from a root of Cattleya in Belgium and from lesions on a stem sheath of Cattleya sp. in Japan. The strain from Belgium was originally identified as C. orchidearum, which is however epitypified in this study and belongs to the C. dracaenophilum species complex. The strain from Japan was originally identified as C. gloeosporioides (Sato et al. 2012) and later re-identified as C. orchidearum as well (T. Sato, unpubl. data).
Numerous Colletotrichum/Gloeosporium species were described on Orchidaceae, among them four species that were described on Cattleya. Colletotrichum cattleyae Verpl. was described from dead leaves of Brasso-Cattleya (Brassavola × Cattleya) hybrid 'Woluwe' in Belgium with oblong to ovoid conidia that measure 7–10 × 3.5–7 μm, mean ± SD = 9.56 ± 0.18 × 6.22 ± 0.12 μm, L/W ratio = 3.2 (Verplancke 1935b); they are shorter and have a different shape than conidia of C. cattleyicola that are cylindrical. Gloeosporium cattleyae Henn., Hedwigia 48: 16 (1908) [Nom. illegit., Art. 53.1] was described from Cattleya leopoldii in Sao Paulo, Brazil, with ellipsoidal conidia with both ends rounded, that are larger than those of C. cattleyicola, measuring 15–22 × 7–11 μm, while Gl. cattleyae Henn. var. macrospora Verpl. forms conidia that are even larger, measuring 21–31 × 7–10 μm (Verplancke 1935a). The earlier homonym Gl. cattleyae Sacc. & D. Sacc. (Saccardo 1906) was described from dead leaves of Cattleya mossia in Paris, France, with elongate conidia with both ends rounded, measuring 15–20 × 4–6 μm. They regard this fungus as the asexual morph of Physalospora cattleyae Maubl. & Lasnier (Saccardo 1905) that forms ascospores that measure 20–25 × 5–7 μm. The conidial size is similar to that of C. cattleyicola; however, we were not able to locate the type specimen in order to verify the morphology of this species. Another species was described fom Cattleya sp. in Italy, C. servazzii (Gallucci-Rangone 1955); conidia are larger than those of C. cattleyicola, measuring 26 × 7 μm.
Appressoria of C. cattleyicola are narrow and very different in shape (mostly clavate or elongate cylindrical) compared to other species in the C. orchidearum complex. Colletotrichum cattleyicola can be identified based on its unique ITS, HIS3 and TUB2 sequences, while the ACT sequence is the same as that of C. vittalense and the GAPDH sequence is the same as those of C. sojae and C. orchidearum. The ITS of strain CBS 170.49 is identical to that of Colletotrichum strains ITCC 5213 from Cattleya in India (JN390844, Sharma et al. 2013b) and Colletotrichum sp. strain GLB3 from damaged roots of Vanilla planifolia in Mexico (KX953436, M.C.C. Gonzalez-Chavez et al., unpubl. data), indicating possible further occurrences of this species. The closest match with the TUB2 sequence of strain CBS 170.49 was with 99 % identity (5 nucleotides difference) Colletotrichum sp. MST 6-3 from leaves of Coffea arabica in Puerto Rico (KJ883603, M.C.C. Gonzalez-Chavez et al., unpubl. data). There is only one nucleotide difference between the GAPDH sequence of strain CBS 170.49 and those of three unidentified Colletotrichum isolates, C08116, C07004 and C07010 (GU935864–GU935866), probably from Korea (Choi et al. 2011). The ACT sequence of C. cattleyicola is 100 % identical with that of C. cliviae strain GUFCC15503 from Calamus thwaitesii in India (KC790646, Sharma et al. 2013a) that is re-identified as C. vittalense in this study.
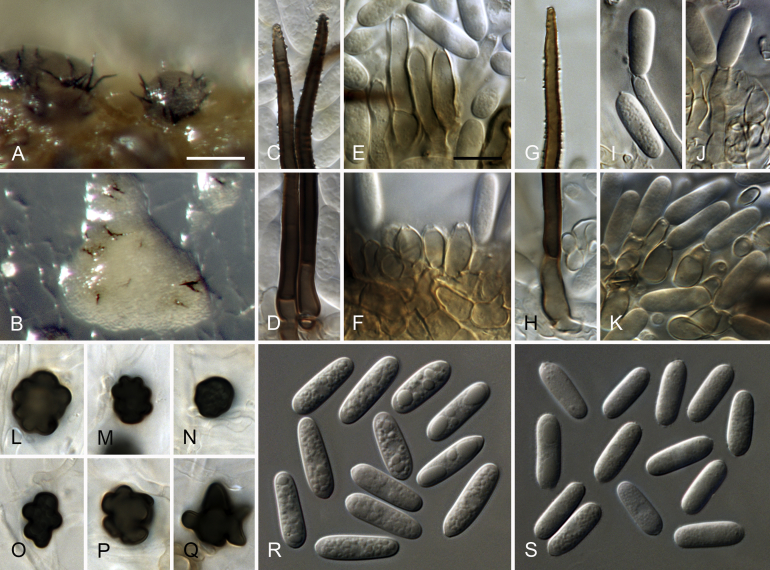
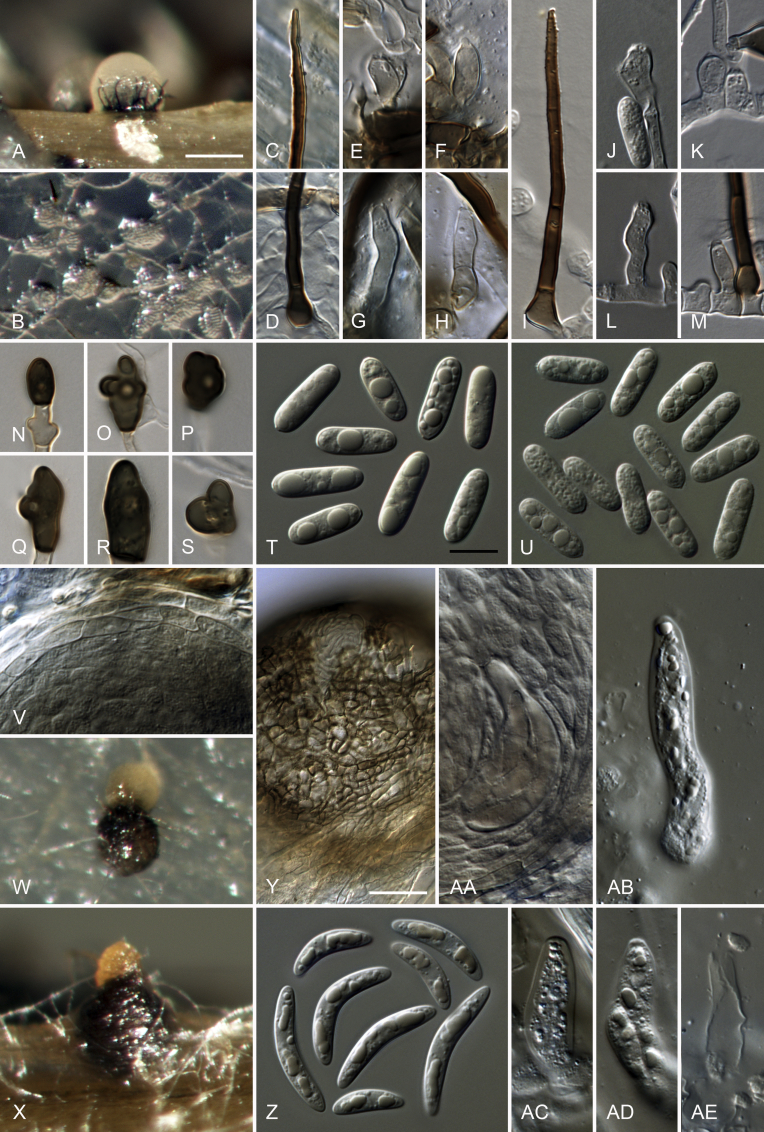
Colletotrichum cliviicola Damm & Crous, nom. nov. MycoBank MB824221. Fig. 5.
Fig. 5.
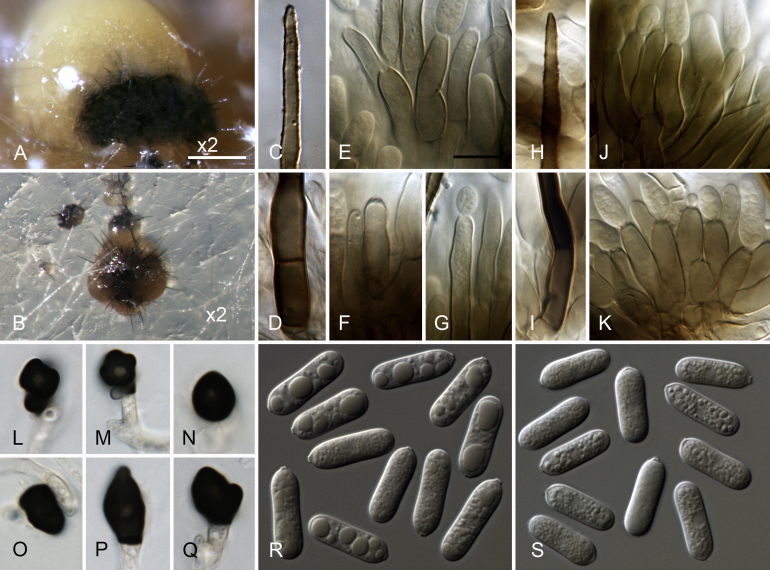
Colletotrichum cliviicola (A–O, W–X. from ex-holotype culture CBS 125375). A–B. Conidiomata. C, J. Tips of setae. D, K. Bases of setae. E–G, L–N. Conidiophores. H. Two 1-septate conidia forming anastomoses. I, O. Aging conidia forming conidiogenous loci (arrow heads). Q–V. Appressoria. W–X. Conidia. A, C–I, W. from Anthriscus stem. B, J–V, X. from SNA. A–B. Dissecting microscope (DM). C–X. Differential interference contrast illumination (DIC). Scale bars: A = 200 μm, B = 100 μm, E = 10 μm. Scale bar of E applies to C–X.
Basionym: Colletotrichum cliviae Yan L. Yang et al., Fungal Diversity 39: 133. 2009, nom. illeg. [ICN (Melbourne) Art. 53.1], non Chaetostroma cliviae Oudem., Verslagen van de Gewone Vergaderingen der Wis- en Natuurkundige Afdeeling: 226. 1896. Colletotrichum cliviae (Oudem.) Arnaud, Bulletin de la Société de pathologie végétale de France I: 37. 1914. Colletotrichum cliviae (Oudem.) Petr., Sydowia 1 (1–3): 82 (1947), nom. illeg. [ICN (Melbourne) Art. 53.1].
Etymology: The species epithet is derived from the host plant, Clivia.
Sexual morph not observed. Asexual morph on SNA. Vegetative hyphae 1–10 μm diam, hyaline, smooth-walled, septate, branched. Chlamydospores not observed. Conidiomata, conidiophores formed directly on hyphae. Setae (only formed after > 2 wk) pale to dark brown (opaque), sometimes with a pale brown to white tip, smooth-walled to verruculose, 50–130 μm long, 2–4-septate, base cylindrical or slightly inflated, 4.5–5.5 μm diam, tip ± acute. Conidiophores hyaline, smooth-walled, septate, branched, to 30 μm long, after 3 wk turning pale brown and elongating up to 70 μm. Conidiogenous cells hyaline, smooth-walled, cylindrical to doliiform, often ± flexuous, upper part sometimes surrounded by a mucous sheath, 7.5–23 × 4.5–7.5 μm, opening 1.5–2 μm diam, collarette 0.5–1 μm long, periclinal thickening distinct. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical, the apex and base rounded, (11–)15.5–20.5(–26.5) × (4–)5.5–6.5(–7) μm, mean ± SD = 17.9 ± 2.5 × 5.9 ± 0.6 μm, L/W ratio = 3.0, after 3 wk often 1-septate, forming anastomosis or secondary conidia (microcyclic conidiation) from short phialides formed from conidia or anastomosis tubes. Appressoria single, medium to dark brown, smooth-walled, elliptical, subcircular or irregular in outline, with an undulate to lobate margin, (7–)8.5–11.5(–12.5) × (4.5–)6.5–8.5(–9.5) μm, mean ± SD = 10.0 ± 1.7 × 7.4 ± 1.1 μm, L/W ratio = 1.3., appressoria of strain CBS 133705 differ in shape and are longer, clavate to navicular in outline, with an undulate to lobate margin, (5.5–)12–21.5(–24) × (4.5–)5–8(–10.5) μm, mean ± SD = 16.6 ± 4.8 × 6.4 ± 1.5 μm, L/W ratio = 2.6.
Asexual morph on Anthriscus stem. Conidiomata, conidiophores formed on pale brown, angular cells, 3.5–8.5 μm diam. Setae (only formed after >2 wk) pale to dark brown (opaque), smooth-walled to verruculose, 110–220 μm long, 2–7-septate, base cylindrical or slightly inflated, 3.5–7.5 μm diam, tip ± acute. Conidiophores hyaline to pale brown, smooth-walled, septate, to 30 μm long, after 3 wk elongating up to 80 μm. Conidiogenous cells hyaline to pale brown, smooth-walled, cylindrical to doliiform, often ± flexuous, occasionally extending to form new conidiogenous loci, 7–19 × 4.5–7.5 μm, after 3 wk elongating up to 40 μm, opening 1.5–2 μm diam, collarette 0.5–1 μm long, periclinal thickening distinct. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical, the apex and base rounded, (13–)16.5–20.5(–22.5) × (5–)6–6.5(–7) μm, mean ± SD = 18.4 ± 1.9 × 6.2 ± 0.4 μm, L/W ratio = 3.0, after 3 wk often 1–3-septate, forming anastomosis or secondary conidia (microcyclic conidiation) from short phialides formed from conidia or anastomosis tubes.
Culture characteristics: Colonies on SNA flat with entire margin, hyaline to pale saffron, agar medium and Anthriscus stems partly covered with whitish to pale grey aerial mycelium, Anthriscus stems also covered with grey to black conidiomata, reverse hyaline to pale saffron, partial smoke grey to greyish sepia; growth 35–36 mm in 7 d (≥ 40 mm in 10 d). Colonies on OA flat with entire margin; grey olivaceous to olivaceous black, partly covered with floccose whitish to pale grey aerial mycelium, reverse pale olivaceous grey to olivaceous grey, growth 38–40 mm in 7 d (≥ 40 mm in 10 d). Conidial mass whitish to very pale salmon.
Materials examined: China, Yunnan Province, Kunming, on leaf of Clivia miniata, 10 Aug. 2008, Y.L. Yang (GZAAS 080005 holotype [not seen], GLM-F112434 isotype, culture ex-holotype CBS 125375 = CSSK4 = GLMC 1861). South Africa, Western Cape Province, Stellenbosch, Botanical Garden, on Clivia sp., 18. Jul. 2012, P.W. Crous, culture CBS 133705 = CPC 21079.
Notes: Oudemans (1896) described a fungus on leaves of Clivia nobilis (Amaryllidaceae) in the Netherlands as Chaetostroma cliviae. Arnaud (1914) combined this species in the genus Colletotrichum; Petrak (1947) did the same, apparently unaware of the publication by Arnaud (1914). Both combinations of Chaetostroma cliviae are not listed in Index Fungorum and MycoBank, which was apparently the reason for Yang et al. (2009) to overlook them and describe C. cliviae as a new species from leaves of Clivia miniata in China. Colletotrichum cliviae (Oud.) Arnaud has priority over the two later homonyms C. cliviae (Oud.) Petr. and C. cliviae Yan L. Yang et al.; both are therefore illegitimate names (Art. 53.1). In order to replace the latter name, the new name C. cliviicola nom. nov. is provided.
Conidia of Chaetostroma cliviae are cylindrical with both ends rounded, measuring 23–28 × 5–7 μm (Saccardo and Sydow, 1899), that means they are longer than those of C. cliviicola, measuring (11–)15.5–20, 5(–26.5) × (4–)5.5–6.5(–7) μm on SNA and (13–)16.5–20.5(–22.5) × (5–)6–6.5(–7) μm on Anthriscus stems. Another species described on Clivia miniata in greenhouses in Turnau (today Turnov, Czech Republik), C. himantophylli Bubák & Kabát 1907, was regarded as a synonym of C. cliviae (Oud.) Arnaud by Arnaud (1914). However, its conidia are narrower than this species, and C. cliviicola, measuring 14–24 × 4–4.5 μm (Bubák & Kabát 1907). A later homonym, C. himantophylli Verpl. & Claess. 1934 (Nom. illegit., Art. 53.1), described from Clivia nobilis in Belgium, forms cylindrical but narrower conidia, measuring 15–23 × 4–4.5 μm (Trotter & Cash 1972).
Colletotrichum cliviae Yan L. Yang et al. was reported as an anthracnose pathogen of Arundina graminifolia, Capsicum sp., Clivia miniata, Cymbidium hookerianum and Zamioculas zamiifolia in China (Diao et al., 2017, Yang et al., 2009, Yang et al., 2011, Zhou and Li, 2017) and of soybean, lima bean and grapevine in Brazil (Santos et al., 2018, Sousa et al., 2018, Barbieri et al., 2017), on Cattleya sp., Calamus thwaitesii, Phaseolus sp. and Saccharum sp. in India (Sharma et al., 2013b, Chowdappa et al., 2014), Myrianthus arboreus in Cameroon, Citrus limon in Vietnam (Douanla-Meli et al. 2018) and as an endophyte on Camellia sinensis and Mangifera indica in Brazil and China, respectively (Vieira et al., 2014, Liu et al., 2015). However, in this study, only strains from Clivia grouped with the ex-holotype strain of C. cliviicola, while all “C. cliviae” strains from hosts other than Clivia (Yang et al., 2011, Vieira et al., 2014, Liu et al., 2015, Barbieri et al., 2017), including strains from the MAFF culture collection that were included in our study, were revealed to be mostly C. plurivorum, a species closely related to C. cliviicola, C. sojae or C. vittalense.
In a study of Douanla-Meli et al. (2018), C. plurivorum (as C. sichuanensis) was regarded as a synonym of C. cliviicola (as C. cliviae), although both species formed well supported clades in the phylogeny based on a multilocus data set and there was no indication of disconcordance between the gene trees. Based on this study, C. cliviicola is a distinct species. The C. cliviae clade in Douanla-Meli et al. (2018) also included strains from an undetermined ornamental plant in India that could also be Clivia.
Strains from the studies of Douanla-Meli et al., 2018, Liu et al., 2015 and Yang et al. (2011) previously identified as C. cliviae that were re-identified as C. plurivorum in this study, formed a sexual morph. However, no sexual morph was observed in strains from Clivia, identified as C. cliviicola in this study (Yang et al., 2009, Yang et al., 2011, this study). Moreover, microcyclic conidiation and the formation of anastomoses were observed in the ex-type strain of C. cliviicola, but not in C. plurivorum. This species is also the fastest growing species compared to all species treated in this study.
Colletotrichum cliviicola belongs to the C. orchidearum species complex and differs from the closely related C. plurivorum in its TUB2, HIS and GAPDH sequences, while their CHS-1 sequences are identical. In the ITS and ACT trees (not shown), C. cliviicola was also separated from C. plurivorum, but forms a subgroup within the C. plurivorum clade.
In a pathogenicity test by Yang et al. (2011), this species caused symptoms on Clivia spp. and Bletilla striata (Orchidaceae), but on none of the nine other test plants belonging to Amaryllidaceae and other plant families.
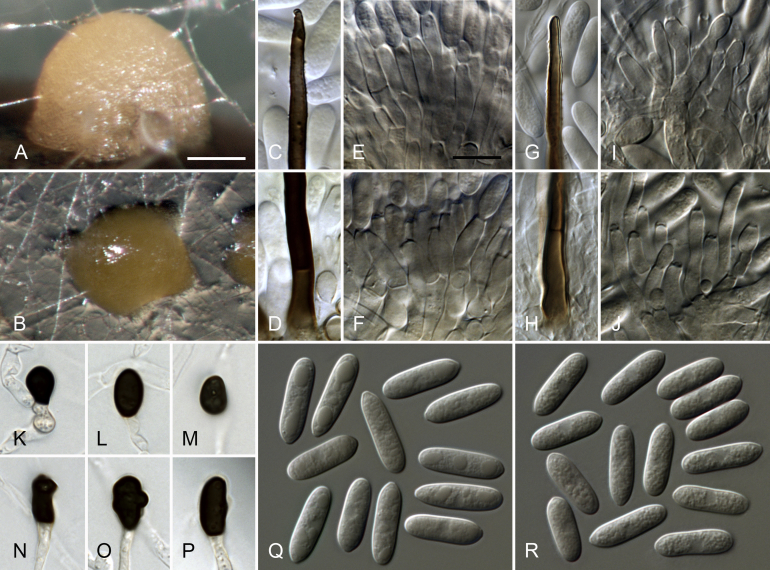
Colletotrichum coelogynes Damm, sp. nov. MycoBank MB824222. Fig. 6.
Fig. 6.
Colletotrichum coelogynes (from ex-holotype culture CBS 132504). A–B. Conidiomata. C, H. Tips of setae. D, I. Bases of setae. E–G, J–K. Conidiophores. L–Q. Appressoria. R–S. Conidia. A, C–G, R. from Anthriscus stem. B, H–Q, S. from SNA. A–B. DM. C–S. DIC. Scale bars: A = 200 μm, E = 10 μm. Scale bar of A applies to A–B. Scale bar of E applies to C–S.
Etymology: Named after the host plant, Coelogyne.
Sexual morph not observed. Asexual morph on SNA. Vegetative hyphae 1.5–11.5 μm diam, hyaline, smooth-walled, septate, branched. Chlamydospores not observed. Conidiomata, conidiophores and setae formed directly on hyphae. Setae medium brown, smooth-walled, often verruculose towards the tip, 50–190 μm long, 2–8-septate, base cylindrical, 3.5–5.5 μm diam, tip ± acute. Conidiophores pale to medium brown, smooth-walled, septate, branched, to 80 μm long. Conidiogenous cells pale to medium brown, smooth-walled, cylindrical, 12–24 × 3.5–5.5 μm, the upper part often surrounded by a gelatinous sheath, opening 1.5–2.5 μm diam, collarette 0.5 μm long, periclinal thickening sometimes visible. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical, the apex and base rounded, often with a prominent scar and slightly constricted in the middle, (15–)15.5–18(–20) × (4.5–)5–6(–6.5) μm, mean ± SD = 16.7 ± 1.2 × 5.5 ± 0.4 μm, L/W ratio = 3.0. Appressoria single or in loose groups, (pale) medium to dark brown, smooth-walled, navicular, ± circular or irregular in outline, with an undulate or entire margin, (8–)10.5–16.5(–20.5) × (6.5–)8–12(–16) μm, mean ± SD = 13.4 ± 3.1 × 9.8 ± 2.1 μm, L/W ratio = 1.4.
Asexual morph on Anthriscus stem. Conidiomata, conidiophores and setae formed on pale to medium brown, angular cells, 4–9.5 μm diam. Setae medium brown, verruculose to verrucose towards the tip, 70–210 μm long, 2–7-septate, base cylindrical to conical, 5–10 μm diam, tip ± acute, often with a constriction close to the tip. Conidiophores pale to medium brown, smooth-walled, septate, branched, to 60 μm long. Conidiogenous cells pale to medium brown, smooth-walled, cylindrical, 16–25 × 3–5 μm, opening 1–1.5 μm diam, collarette 0.5 μm long, periclinal thickening visible, sometimes distinct. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical, the apex and base rounded, often with a prominent scar and slightly constricted in the middle, 16–19(–23.5) × 5–6 μm, mean ± SD = 17.5 ± 1.6 × 5.5 ± 0.3 μm, L/W ratio = 3.2.
Culture characteristics: Colonies on SNA flat with entire margin, medium buff to pale honey, chervil stem, filter paper and SNA medium partly covered with greyish acervuli and short whitish aerial mycelium, reverse buff to pale honey with the grey acervuli shining through; 26–27.5 mm in 7 d (32.5–35 mm in 10 d). Colonies on OA flat with entire margin, surface salmon, partly covered with dark grey to orange acervuli and floccose whitish to pale grey aerial mycelium, reverse vinaceous buff, rosy buff to purplish grey; 27.5–32.5 mm in 7 d (≥ 40 mm in 10 d). Conidial mass orange.
Materials examined: Germany, Munich, glasshouse, from leaves of Coelogyne sp., 20 Nov. 2010, U. Damm (CBS H-21069 holotype, culture ex-holotype CBS 132504); Munich, glasshouse, from leaves of Coelogyne sp., 20 Nov. 2010, U. Damm, culture CBS 132515.
Notes: Typical for C. coelogynes are the large conidiomata and the comparatively high growth rate; C. coelogynes is the fastest growing species in the C. dracaenophilum complex. In contrast to C. orchidearum, C. coelogynes forms conidia that often have a prominent scar and are slightly constricted in the middle, as well as have longer setae. See C. orchidacearum for other species described and reported from Orchidaceae.
Colletotrichum coelogynes can be identified with all loci studied. Closest matches with the ITS sequence of C. coelogynes strain CBS 132504 in GenBank were with 99 % identity (1 and 4 nucleotides difference) endophytic Colletotrichum isolates, probably both from Dendrobium spp. from China (FJ042517, Yuan et al. 2009 and FJ544250, C. Gao & S.X. Guo, unpubl. data). Closest matches with the TUB2 and the GAPDH sequences of strain CBS 132504 were with 92 % and 84 % identity, respectively, the two C. tropicicola strains from Noireung et al. (2012). Closest matches with the ACT and HIS3 sequences were, both with 92 % identity, C. tropicicola strain MFLUCC 11-0114 and the two C. excelsum-altitudinum strains (Noireung et al., 2012, Tao et al., 2013) and C. pseudomajus strain CBS 571.88 and C. radicis strain CBS 529.93 (KF687864, KF687847, C. gigasporum complex, Liu et al. 2014), respectively. Closest match with the CHS-1 sequence of strain CBS 132504 with 95 % identity were C. yunnanense strain CBS 132135 (JX519231, Cannon et al. 2012) as well as sequences of several species belonging to the C. gloeosporioides und C. gigasporum complexes.
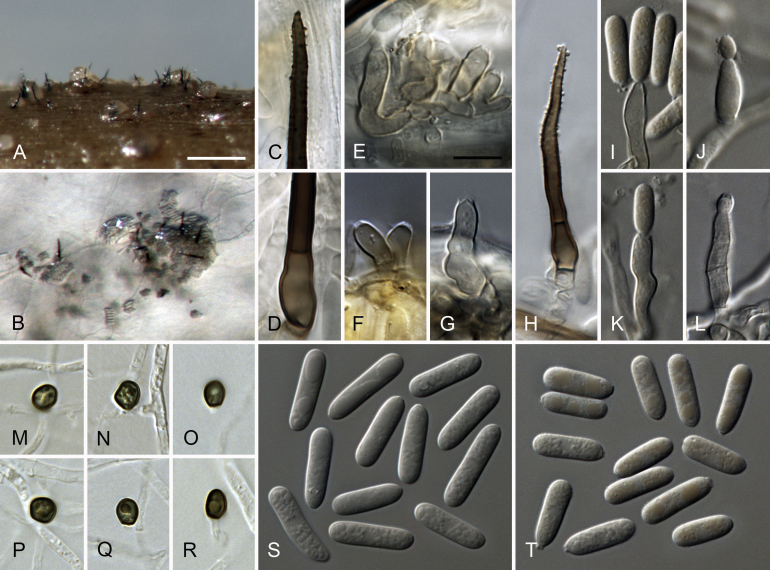
Colletotrichum dracaenophilum D.F. Farr & M.E. Palm, Mycol. Res. 110: 1401. 2006. Fig. 7.
Fig. 7.
C. dracaenophilum (from ex-holotype culture CBS 118199). A–B. Conidiomata. C, H. Tips of setae. D. Base of a setae and conidiophores. I. Base of a setae. E–G, J–M. Conidiophores. M. Seta ending in a conidiogenous opening. N–S. Appressoria. T–U. Conidia. A, C–G, T. from Anthriscus stem. B, H–S, U. from SNA. A–B. DM. C–U. DIC. Scale bars: A = 100 μm, D = 10 μm. Scale bar of A applies to A–B. Scale bar of D applies to C–U.
Sexual morph not observed. Asexual morph on SNA. Vegetative hyphae 1.5–5.5 μm diam, hyaline, smooth-walled, septate, branched. Chlamydospores not observed. Conidiomata, conidiophores and setae formed directly on hyphae. Setae medium brown, smooth-walled, verruculose towards the tip, 50–130 μm long, 2–5-septate, base cylindrical, sometimes slightly inflated, 5–8 μm diam, tip ± acute, often ending in a conidiogenous opening. Conidiophores hyaline (setae ending in a conidiogenous opening medium brown), smooth-walled, septate, branched. Conidiogenous cells hyaline (on setae ending in a conidiogenous opening medium brown), smooth-walled, cylindrical, the upper part sometimes surrounded by a gelatinous sheath, 10–16 × 5.5–6.5 μm, opening 1.5–2 μm diam, collarette 0.5 μm long, periclinal thickening distinct. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical, the apex rounded, the base rounded or truncate, sometimes tapering to the base and sometimes slightly curved, (15.5–)20–24.5(–28.5) × (5.5–)6.5–7 μm, mean ± SD = 22.2 ± 2.3 × 6.6 ± 0.3 μm, L/W ratio = 3.4, conidia of strain CBS 121453 shorter, measuring (15.5–)17–21.5(–25.5) × 6–7(–8) μm, mean ± SD = 19.3 ± 2.4 × 6.7 ± 0.4 μm, L/W ratio = 2.9. Appressoria single or in loose groups, dark brown, smooth-walled, navicular, cigar- to bullet-shaped in outline, with an lobate, crenate or undulate margin, (7–)10–18(–22.5) × (5.5–)6–8.5(–10.5) μm, mean ± SD = 13.9 ± 4.0 × 7.4 ± 1.3 μm, L/W ratio = 1.9, appressoria of strain CBS 121453 wider, measuring (5–)8–19(–27) × (4.5–)6.5–11.5(–15.5) μm, mean ± SD = 13.5 ± 5.5 × 9.1 ± 2.6 μm, L/W ratio = 1.5.
Asexual morph on Anthriscus stem. Conidiomata, no basal cells were found, on which conidiophores and setae are formed. Setae pale to medium brown, smooth-walled, verrucose towards the tip, 60–190 μm long, 4–6-septate, base cylindrical to ± inflated, 4–7 μm diam, very thin towards the tip, tip ± rounded. Conidiophores pale brown, smooth-walled. Conidiogenous cells pale brown, smooth-walled, cylindrical to ampulliform, 10–16 × 5–7 μm, opening 1–1.5 μm diam, collarette 0.5 μm long, rarely observed, periclinal thickening distinct. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical, the apex rounded, the base rounded or truncate, sometimes with a prominent scar, sometimes tapering towards the base, (17.5–)20–25(–28) × 5.5–6.5 μm, mean ± SD = 22.4 ± 2.6 × 6.1 ± 0.4 μm, L/W ratio = 3.7.
Culture characteristics: Colonies on SNA flat with entire margin, hyaline to pale ochreous, filter paper partly pale ochreous, chervil stem, filter paper and SNA medium partly covered with short white aerial mycelium, reverse same colours; 16.5–17 mm in 7 d (24–25.5 mm in 10 d). Colonies on OA flat with entire margin, surface moist, saffron to orange due to sporulation, or with some buff sectors with grey spots, aerial mycelium lacking, reverse saffron to buff; 14.5–15.5 mm in 7 d (25–26.5 mm in 10 d). Conidial mass saffron to orange.
Materials examined: Bulgaria, Plovdiv, from plants of Dracaena sanderiana, bought from the market, collection date unknown, S.G. Bobev, CBS 121453 = AR 4406 = No. 1688/1. China, intercepted in San Francisco, California, USA, from dying stems of Dracaena sanderiana, 25 Sep. 2002, J.R. Nelson (BPI 871498 holotype [not seen], culture ex-holotype CBS 118199 = MEP 1532).
Notes: Colletotrichum dracaenophilum was described as a stem pathogen of Dracaena sanderiana; it is based on Dracaena stems that were intercepted in California, USA, but originated from China (Farr et al. 2006). The species is only known from Dracaena, from D. sanderiana in Australia, Bulgaria, China, Egypt and Florida (USA), and on D. braunii in Brazil (Farr et al., 2006, Bobev et al., 2008, Sharma et al., 2014, Macedo and Barreto, 2016, Morsy and Elshahawy, 2016, Shivas et al., 2016). The reports from Bulgaria and Florida (USA) could be debated, as the plants were imported from other countries, probably from Asia (Bobev et al. 2008) and could have been infected by the fungus prior to import. The strain from Bobev et al. (2008) is included in this study.
Another species on Dracaena, C. petchii, was found on D. marginata and Dracaena sp. in Italy and the Netherlands, respectively, and on D. sanderiana in China and Australia, and belongs to the C. boninense species complex (Damm et al., 2012b, Shivas et al., 2016). Both species, C. dracaenophilum and C. petchii, are only known from Dracaena spp.
Colletotrichum dracaenophilum can be identified with all loci studied. The closest matches with the sequences of all loci of the ex-type strain CBS 118199 in GenBank are all those of the C. dracaenophilum strains included in this study or mentioned above. This species belongs to the C. dracaenophilum species complex. Conidia of C. dracaenophilum are larger than those of all other species treated in this study.
Colletotrichum excelsum-altitudinum G. Tao et al. [as ‘excelsum-altitudum’], Fungal Diversity 61: 152. 2013.
Description: See Tao et al. (2013).
Notes: Colletotrichum excelsum-altitudinum was described from healthy leaves of Bletilla ochracea (Orchidaceae) in Guizhou, China, and reported from this host together with 16 other endophytic species, including a further six new Colletotrichum species from the same host (Tao et al. 2013).
Colletotrichum excelsum-altitudinum belongs to the C. dracaenophilum species complex. This species forms shorter conidia than the other species in this complex and can be identified with sequences of all loci available (ITS, GAPDH, ACT, TUB2), best with GAPDH and TUB2. The ITS sequence of the ex-type strain, CGMCC 3.15130, is 100 % identical with the two sequences of C. excelsum-altitudinum (Tao et al. 2013) and with that from C. excelsum-altitudinum isolate OBitC1 from Momordica charantia (Cucurbitaceae) in India (KU239167, P. Chowdappa et al., unpubl. data). The GAPDH sequence of the ex-type strain, CGMCC 3.15130, is 100 % identical with the two sequences of C. excelsum-altitudinum (Tao et al. 2013); the sequences of all other species are ≤ 96 % identical. The ACT sequence of strain CGMCC 3.15130 is 100 % identical with the two sequences of C. excelsum-altitudinum (Tao et al. 2013) and 99 % identical (1 and 2 nucleotides difference) with those of the C. tropicicola strains from Citrus and Paphiopedilum (Noireung et al. 2012), while the TUB2 sequence is 100 % and 99 % identical (1 nucleotides difference) with those of the C. excelsum-altitudinum strains and 99 % identical (10 nucleotides difference) with those of both C. tropicicola strains.
Colletotrichum liaoningense Y.Z. Diao et al., Persoonia 38: 34. 2017.
Description: See Diao et al. (2017).
Notes: This species belongs to the C. magnum species complex and is so far only known from Capsicum in China (Diao et al. 2017).
There are five strains from Capsicum sp. in China cited in Diao et al. (2017) that belong to the species complexes treated in this paper; four strains were described as C. liaoningense, while one strain was identified as C. cliviae. The C. cliviae strain CAUOS5 was re-identified as C. sojae (C. orchidearum species complex) in this study. However, there are several irregularities related to the sequence data from Diao et al. (2017). We suspect, for example, that the ACT sequence KP890098 is actually from CAUOS5 as well and not from CAUOS3, as it is identical with that of several strains of C. sojae; there is no ACT sequence of CAUOS5 listed in the paper. The number of this strain in the strain table is given as CAUOS6. The sequences of the four C. liaoningense strains deposited by Diao et al. (2017) also all differ from one another. For example, the ITS sequence of strain CAUOS2, the ex-type strain of C. liaoningense differs in numerous positions from those of the other three strains of this species, while the TUB2 sequences are identical with C. magnum (see notes of this species).
The four strains of C. liaoningense form two clades in the phylogeny of this study, suggesting that their identification requires verification.
Colletotrichum lobatum Damm, sp. nov. MycoBank MB824223. Fig. 8.
Fig. 8.
Colletotrichum lobatum (from ex-holotype culture IMI 79736). A–B. Conidiomata. C, G. Tips of setae. D, H. Bases of setae. E–F, I–K. Conidiophores. L–Q. Appressoria. R–S. Conidia. A, C–F, R. from Anthriscus stem. B, G–Q, S. from SNA. A–B. DM. C–S. DIC. Scale bars: A = 100 μm, E = 10 μm. Scale bar of A applies to A–B. Scale bar of E applies to C–S.
Etymology: The species epithet is derived from the lobate edge of the appressoria.
Sexual morph not observed.
Asexual morph on SNA. Vegetative hyphae 1–6.5 μm diam, hyaline, smooth-walled, septate, branched. Chlamydospores not observed. Conidiomata, conidiophores and setae formed directly on hyphae. Setae pale to medium brown, verrucose, 35–90 μm long, 1–3-septate, base cylindrical to slightly inflated, 4.5–7 μm diam, tip ± rounded to ± acute. Conidiophores hyaline to pale brown, smooth-walled, septate, branched, to 45 μm long. Conidiogenous cells hyaline to pale brown, smooth-walled, cylindrical, 5–30 × 3–5 μm, opening 1.5–2 μm diam, collarette 0.5–1 μm long, distinct, periclinal thickening distinct. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical, the apex and base rounded, (10.5–)12.5–14.5(–16) × (4–)4.5–5 μm, mean ± SD = 13.7 ± 1.0 × 4.7 ± 0.3 μm, L/W ratio = 2.9. Appressoria not formed on the backside of the SNA plate, but in slide culture, single, medium brown, smooth-walled, subglobose, elliptical to irregular in outline, with an lobate or undulate margin, (7–)7.5–15.5(–25) × (5.5–)6.5–11(–15) μm, mean ± SD = 11.6 ± 4.0 × 8.9 ± 2.3 μm, L/W ratio = 1.3.
Asexual morph on Anthriscus stem. Conidiomata, conidiophores and setae formed on pale brown, angular cells, 3.5–7 μm diam. Setae medium brown, verrucose, 45–90 μm long, 1–3-septate, base cylindrical, sometimes slightly inflated, 4–6.5 μm diam, tip ± acute to ± rounded. Conidiophores hyaline to pale brown, smooth-walled to verruculose, simple or septate, branched, to 30 μm long. Conidiogenous cells pale brown, smooth-walled to verruculose, cylindrical to doliiform, 7.5–15 × 3.5–5.5 μm, opening 1.5–2 μm diam, collarette 0.5 μm long, periclinal thickening distinct. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical, the apex and base rounded, (13.5–)14.5–17(–18) × (4–)4.5–5(–5.5) μm, mean ± SD = 15.6 ± 1.2 × 4.9 ± 0.3 μm, L/W ratio = 3.2.
Culture characteristics: Colonies on SNA flat with entire margin, hyaline, honey to pale cinnamon, agar medium, filter paper and Anthriscus stem partly covered with tiny grey to salmon acervuli, aerial mycelium lacking, reverse same colours, growth 23–24.5 mm in 7 d (33.5–35 mm in 10 d). Colonies on OA flat with entire margin; cinnamon, entirely covered by tiny grey dots formed by conidiomata, aerial mycelium lacking, reverse buff to vinaceous buff, growth 25.5–27 mm in 7 d (35.5–37.5 mm in 10 d). Conidial mass salmon.
Material examined: Trinidad and Tobago, from Piper catalpaefolium, collection date and collector unknown (IMI 79736 holotype, CBS H-21506 isotype, culture ex-holotype IMI 79736 = CPC 21196).
Notes: There are two species on Piper spp. treated in this study, C. lobatum and C. piperis. Colletotrichum lobatum belongs to the C. magnum species complex, while C. piperis belongs to the C. orchidearum complex. The conidial sizes and shapes are very similar. In contrast to C. piperis, C. lobatum forms lobate appressoria and setae that are unbranched, and colonies that grow faster. Moreover, C. lobatum strain IMI 79736 originates from Latin America, while C. piperis is only known from Asia (Malaysia, Sri Lanka). See C. piperis for other species described and reported from Piper spp.
The mostly regularly lobed roundish appressoria are also different from the irregular appressoria of the closely related C. brevisporum that also has faster growing colonies.
The species can be identified best by its unique GAPDH sequence. There is respectively one nucleotide difference in the TUB2, CHS-1 and HIS3 sequences to those of C. brevisporum, while ITS and ACT sequences of the two species are identical. The closest matches in a blastn search with the GAPDH sequence in GenBank were with 97 % C. liaoningense strains CAUOS3 and CAUOS4 (KP890136, KP890137, Diao et al. 2017), while the closest matches with the TUB2 sequence were with 99 % identity (1 nucleotide difference) C. brevisporum strains L57/LC0600 (ex-holotype strain) and BTL23/LC0870 (JN050244 and JN050245, Noireung et al. 2012) and strain CCCM12 from Cucurbita moschata (KY797630, Liu et al. 2018). The result of the blastn search with the ITS sequence was the same as that for C. brevisporum.
Colletotrichum magnum (S.F. Jenkins & Winstead) Rossman & W.C. Allen, IMA Fungus 7: 4. 2016. Fig. 9.
Fig. 9.
Colletotrichum magnum (A–J, Q–R. from ex-epitype culture CBS 519.97. K–P. from culture CBS 575.97. S–AA. from holotype BPI 596678). A–B, W. Conidiomata. C–J. Conidiophores. K–P. Appressoria. Q–R, T. Conidia. S. Ascoma. V. Outer surface of peridium. X–Y. Ascospores. Z. Seta. U, AA. Asci. A, C–F, Q. from Anthriscus stem; B, G–P, R. from SNA. S–AA. from filter paper. A–B, S, W. DM. C–R, T–V. X–AA. DIC. Scale bars: A, S = 100 μm, C = 10 μm. A applies to A–B. S applies to S, W. C applies to C–R, T–V, X–AA.
Basionym: Glomerella magna S.F. Jenkins & Winstead, Phytopathology 54: 453. 1964.
Sexual morph on filter paper (only observed on the type specimen BPI 596678). Ascomata perithecia, solitary, superficial or immersed, non-stromatic, medium to dark brown, subglobose to pyriform, 355–500 × 200–355 μm, ostiolate, with a neck. Peridium composed of medium brown flattened textura angularis with cells 8–17 μm diam. Ascogenous hyphae and interascal tissue not observed. Asci unitunicate, probably 8-spored, but the number of spores per ascus could not be seen, cylindrical, tapering to apex and base, smooth-walled, 71–122 × 12–15.5 μm. Ascospores initially hyaline and aseptate but can become pale brown and septate with age, smooth-walled, allantoid, curved most in the middle, with rounded ends, (23–)27–37(–43.5) × (4.5–)5–6.5(–7) μm, mean ± SD = 32.1 ± 5.0 × 5.6 ± 0.7 μm, L/W ratio = 5.7.
Asexual morph on filter paper (type specimen BPI 596678). Conidiomata, conidiophores (disintegrated) and setae formed on pale brown, angular cells. Setae medium brown, verruculose to verrucose, 50–70 μm long, 1–3-septate, base cylindrical, 3–4 μm diam, tip rounded. Conidiophores and conidiogenous cells not observed. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical, the apex and base rounded, (17–)18–20(–20.5) × 4.5–5(–5.5) μm, mean ± SD = 19.1 ± 1.0 × 4.9 ± 0.3 μm, L/W ratio = 3.9.
Asexual morph on SNA (ex-epitype strain CBS 519.97). Vegetative hyphae 1.5–11 μm diam, hyaline, smooth-walled, septate, branched. Chlamydospores not observed. Conidiomata, conidiophores formed directly on hyphae. Setae not observed. Conidiophores hyaline, smooth-walled, septate, not branched, to 20 μm long, conidiophores of strain CBS 575.97 pale to medium brown, smooth-walled to verruculose, septate, branched, to 50 μm long. Conidiogenous cells hyaline, smooth-walled, cylindrical to ellipsoidal, often ± flexuous, 13–20 × 4–5 μm, conidiogenous cells of strain CBS 575.97 pale to medium brown, smooth-walled to verruculose, cylindrical to clavate, opening 1.5–2 μm diam, collarette 0.5 μm long, periclinal thickening visible. Conidia hyaline, smooth-walled, aseptate, straight, sometimes very slightly curved, cylindrical, the apex and base rounded, (11–)15.5–19(–20.5) × 4–4.5 μm, mean ± SD = 17.4 ± 1.8 × 4.2 ± 0.2 μm, L/W ratio = 4.1. Appressoria single or in loose groups, medium to dark brown, smooth-walled, ± circular, elliptical, clavate, spathulate or irregular in outline, with an entire or undulate margin, (5–)6.5–12.5(–16) × (3.5–)4.5–7.5(–10) μm, mean ± SD = 9.4 ± 2.9 × 5.9 ± 1.5 μm, L/W ratio = 1.6.
Asexual morph on Anthriscus stem (ex-epitype strain CBS 519.97). Conidiomata, conidiophores formed directly on hyphae or on pale brown, angular cells, 3–8.5 μm diam. Setae not observed. Conidiophores pale brown, smooth-walled, sometimes septate, rarely branched, to 20 μm long. Conidiogenous cells pale brown, smooth-walled, subsphaerical to broadly ellipsoidal, sometimes ampulliform, 5.5–12 × 4–7 μm, opening 1.5–2 μm diam, collarette not observed, periclinal thickening visible. Conidia hyaline, smooth-walled, aseptate, sometimes very slightly curved, cylindrical, the apex and base rounded, (17–)18–20(–21.5) × 4.5–5(–5.5) μm, mean ± SD = 19.2 ± 1.1 × 4.8 ± 0.3 μm, L/W ratio = 4.0, conidia of strain CBS 575.97 narrower, measuring (17–)19–22(–24) × (3.5–)4–4.5(–5) μm, mean ± SD = 20.5 ± 1.4 × 4.3 ± 0.3 μm, L/W ratio = 4.8.
Culture characteristics: Colonies on SNA flat with entire margin, hyaline to pale cinnamon, aerial mycelium lacking, in strain CBS 575.97 Anthriscus stem and medium partly covered with felty white aerial mycelium, reverse same colours; 24.5–25.5 mm in 7 d (36–36.5 mm in 10 d), strain CBS 575.97 faster growing, 28.5–31.5 mm in 7 d (≥ 40 mm in 10 d). Colonies on OA flat with entire margin, surface isabelline to citrine, towards the margin straw, aerial mycelium lacking, reverse olivaceous buff to olivaceous, 24.5–26 mm in 7 d (36.5 mm in 10 d), strain CBS 575.97 differing in colour, mycelium formation and growth rate: dark olivaceous, olivaceous to olivaceous black, partly covered with short felty whitish aerial mycelium, reverse olivaceous buff, olivaceous grey to iron grey, 25.5–27.5 mm in 7 d (36–≥ 40 mm in 10 d). Conidial mass saffron.
Materials examined: USA, from Citrullus lanatus, collection date and collector unknown, [BPI 596678 (S120 A × S120 F, dried culture on filter paper) holotype]; from Citrullus lanatus, collection date and collector unknown (CBS H-21063 epitype, here designated MBT380418, culture ex-epitype CBS 519.97 = L2.5 = LARS 688); from Citrullus lanatus, collection date and collector unknown, culture CBS 575.97 = DXD = LARS 687; from Citrullus lanatus, collection date and collector unknown (deposited in IMI collection by D.S. Freeman), culture IMI 391662 = L2 5 = CPC 19388.
Notes: An anthracnose disease of cucurbits caused by Ga. magna was reported from the USA (Jenkins & Winstead 1964). The causal agent was isolated from Citrullus lanatus and described as a heterothallic species with very large asci and ascospores that were formed by strains crossed in the laboratory. The species was combined into Colletotrichum by Rossman et al. (2016).
Compatible strains 120A (=ATCC 15015 = S120A) and 120F (=ATCC 15016 = S120F) that were crossed in the laboratory to produce the sexual morph were deposited in ATCC by Jenkins & Winstead (1964). In the resulting publication, the authors state that they deposited herbarium material in BPI as S 120A and S 120F, apparently two specimens and not indicating it as type material. That would mean the species is not validly published, as it was published after 1 Jan. 1958 without designating a type (Art. 40.1). However, the label on BPI 596678 says “possible type”; further information on the specimen: “S.F.J. & N.N.W. No. S120 A × S120 F, Comm. N.N. Winstead May 10, 1963” clearly reveals that the authors deposited only one specimen, the result of the crossing of the two strains and not two, and that the details of this specimen agree with the information in the original description. BPI 596678 is therefore regarded as the holotype.
The two strains 120A and 120F that were crossed in order to form the sexual morph have no type status, and there is no ex-holotype strain retained. In a subsequent PhD thesis at Cornell University, Ithaca, NY, USA, that was not available to us, S. Brown conducted additional crosses of the two original strains. Strains CBS 519.97 (=LARS 688 = L2.5) and CBS 575.97 (=LARS 687 = DXD) are derived from them (B.G. Turgeon, in litt.). One of these strains, CBS 519.97, was selected as the basis of the epitype.
Conidia of the holotype specimen BPI 596678 measured in this study are smaller than those listed in the original description by Jenkins & Winstead [1964; (24–)28(–40) × (4–)4.5(–6) μm], measuring (17–)18–20(–20.5) × 4.5–5(–5.5) μm]. Conidial measurements from the ex-epitype strain CBS 519.97 agree with those from the holotype. Other features observed on the holotype do not diverge from the original description.
Two further species were described on Citrullus in Russia and the USA, respectively. They are possible Colletotrichum spp. However, none of them seems to be conspecific with C. magnum: Gloeosporium lagenaria var. citrulli forms shorter conidia (14 × 5 μm, Potebnia 1907) than C. magnum, while conidia of Volutella citrulli are elliptical or clavate, sometimes slightly curved, measuring 15–20 × 3–4 μm (Stoneman 1898). Conidia of C. magnum are wider and not clavate.
Although Jenkins & Winstead (1964) proved the pathogenicity of Ga. magna to many species of Cucurbitaceae, especially watermelon, squash, pumpkin and cantaloup, strains and specimens from that study are all from watermelon. Glomerella magna is reported from other Cucurbitaceae in the USA, China and Taiwan (Grand, 1985, Tsay et al., 2010), from Carica papaya in Brazil, Costa Rica and Mexico (Nascimento et al., 2010, Tapia-Tussell et al., 2016, Molina-Chaves et al., 2017) and Lobelia chinensis in China (Li et al. 2013), however, most of them either lack molecular data or have ITS data only. By means of sequence comparison, most of the respective strains could be confirmed as belonging to the C. magnum species complex. However, there is no species identification possible in this complex based on ITS data.
In contrast, further reports of Colletotrichum species on Citrullus lanatus include C. truncatum (as C. capsici) in India, C. gloeosporioides and C. gloeosporioides f. sp. cucurbitae in Brazil, C. lagenarium in the USA, Libya and Zimbabwe as well as C. orbiculare in several countries worldwide (Farr & Rossman 2017). Probably none of these reports was based on molecular data and some could refer to C. magnum. The only strain of a different species from Citrullus identified based on molecular data is strain CBS 128524 from Citrullus lanatus in New Zealand that was identified as C. karstii, belonging to the C. boninense species complex (Damm et al. 2012b).
Colletotrichum magnum is difficult to differentiate based on sequence data. Of the loci included, only the GAPDH and possibly also the HIS3 sequences are unique. However, HIS3 sequences are not available of all species that are included in the multilocus phylogeny in this study. The other loci are identical with strains of other species, mainly C. liaoningense (Diao et al. 2017). Conidia of C. magnum are sometimes slightly curved on both media, having a different shape than those of the closely related C. okinawense, and are longer than those of the also closely related C. liaoningense (71–122 × 12–15.5 μm, Diao et al. 2017). The ascospores are curved and larger than those of any other Colletotrichum species treated in this study and only exceeded in length by C. gigasporum (Rakotoniriana et al. 2013) and C. taiwanense (Sivanesan & Hsieh 1993), the former belonging to the C. gigasporum complex, while the systematic position of the latter is dubious (Liu et al. 2014). Both species have filiform, 0–1- or 3–8-septate, respectively, ascospores that are less strongly curved than those of C. magnum.
Closest matches with the GAPDH sequence of strain CBS 519.97 were with 99 % identity (1 nucleotide difference) Ga. magna strain AK7 probably from watermelon in the USA (DQ792850, Liu et al. 2007a) and C. brevisporum isolate LJTJ59 (KP943513) from Capsicum in China (F. Liu & G. Gong, unpubl. data). Based on blastn searches, the ITS sequence of the ex-type strain CBS 519.97 is 100 % identical e.g. to those of Ga. magna and Colletotrichum sp. strains from Carica papaya in Brazil (Nascimento et al. 2010), Mexico (E.T. Arechiga-Carvajal et al., unpubl. data) and Malaysia (J.H. Sim et al., unpubl. data). However, the ITS of C. magnum is identical with several other species in the C. magnum complex.
Colletotrichum magnum was the basis for a number of molecular, morphological and pathogenicity studies on appressorium formation, pathogenic and symbiotic lifestyles of fungi in plants (Bhairi et al., 1990, Freeman and Rodriguez, 1993, Redman et al., 1999, Rodriguez et al., 2004). For example, mutation of pathogenic C. magnum strains resulted in the loss of a virulence factor and transformation into an endophytic fungus (Freeman & Rodriguez 1993).
Colletotrichum merremiae Damm, sp. nov. MycoBank MB824224. Fig. 10.
Fig. 10.
Colletotrichum merremiae (from ex-holotype culture CBS 124955). A–B. Conidiomata. C. Tip of a seta. D. Base of a seta. H. Seta. E–G, I–L. Conidiophores. M–R. Appressoria. S–T. Conidia. A, C–G, S. from Anthriscus stem. B, H–R, T. from SNA. A–B. DM. C–T. DIC. Scale bars: A = 100 μm, E = 10 μm. Scale bar of A applies to A–B. Scale bar of E applies to C–T.
Etymology: The species epithet is derived from the host plant, Merremia.
Sexual morph not observed. Asexual morph on SNA. Vegetative hyphae 1.5–7 μm diam, hyaline to pale brown, smooth-walled, septate, branched. Chlamydospores not observed. Conidiomata, conidiophores and setae formed directly on hyphae. Setae (pale to) medium brown, smooth-walled, verrucose towards the tip, 30–65 μm long, 1–2-septate, base cylindrical or ± inflated, 4–5.5 μm diam, tip ± acute. Conidiophores pale brown, smooth-walled to verruculose, septate, branched, to 40 μm long. Conidiogenous cells pale brown, smooth-walled to verruculose, cylindrical, sometimes flexuous, 10–21 × 3–5 μm, opening 1.5–2 μm diam, collarette 0.5 μm long, periclinal thickening visible, sometimes distinct. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical, the apex rounded, the base rounded to truncate, (12.5–)14–15.5(–16.5) × (4–)4.5–5(–5.5) μm, mean ± SD = 14.8 ± 0.9 × 4.6 ± 0.3 μm, L/W ratio = 3.2. Appressoria single, medium brown, smooth-walled, ± circular to elliptical in outline, with an entire or undulate margin, (4.5–)5–7.5(–10) × (3–)3.5–5(–5.5) μm, mean ± SD =6.4 ± 1.3 × 4.3 ± 0.7 μm, L/W ratio = 1.5.
Asexual morph on Anthriscus stem. Conidiomata, conidiophores and setae formed directly on hyphae, no basal cells observed. Setae medium brown, verruculose, 45–80 μm long, 1–2-septate, base cylindrical, conical to ± inflated, 4–7.5 μm diam, tip ± acute. Conidiophores pale brown, smooth-walled, septate, rarely branched. Conidiogenous cells pale brown, smooth-walled, cylindrical to ellipsoidal, flexuous and with constrictions, 8–12 × 4.5–6.5 μm, opening 1.5–2 μm diam, collarette 0.5 μm long, periclinal thickening visible. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical, the apex rounded, the base rounded to truncate, (14–)15–16.5(–19) × (4–)4.5–5 μm, mean ± SD = 15.8 ± 0.9 × 4.6 ± 0.2 μm, L/W ratio = 3.4.
Culture characteristics: Colonies on SNA flat with entire margin, hyaline to pale cinnamon, aerial mycelium lacking, reverse same colours, 21.5–22.5 mm in 7 d (31.5–35 mm in 10 d). Colonies on OA flat with entire margin, olivaceous, towards the margin honey to buff, partly covered with short felty whitish aerial mycelium, reverse olivaceous buff to olivaceous grey, 24–24.5 mm in 7 d (35–35.5 mm in 10 d). Conidial mass rosy buff to saffron.
Material examined: Panama, Gamboa, wet lowland forest, leaf endophyte of Merremia umbellata, Nov. 2004, S. VanBael & Z. Maynard, D3-1 (CBS H-21065 holotype, culture ex-type CBS 124955 = Q855).
Notes: Merremia is a genus of flowering plants in the Convolvulaceae; M. umbellata is commonly known as hogvine (www.wikipedia.org). No Colletotrichum species was previously described from Merremia and no strain from this host genus was included in any of the recently studied species complexes. The USDA fungal databases (Farr & Rossman 2017) only list Rojas et al. (2010), the publication where the two strains, CBS 124955 and CBS 125386, described as two different species in this study, C. merremiae and C. panamense, were derived from. Both species belong to the C. magnum species complex.
The conidiogenous cells of C. merremiae are usually cylindrical and often flexuous or with constrictions, while those of C. panamense are mostly subglobose to ellipsoidal. Conidia of C. merremiae are on both media on average at least 2 μm shorter than those of C. panamense. Typical are also the small roundish appressoria, that are very different from the compact appressoria of the closely related C. brevisporum and C. lobatum. According to the measurements in Diao et al. (2017), the appressoria of C. liaoningense are even smaller (3.5–5 × 2.5–4.5 μm).
Colletotrichum merremiae can be identified based on its unique GAPDH, HIS3 and TUB2 sequences. HIS3 differs in one, GAPDH and TUB2 each in four nucleotides from C. brevisporum. The ITS and ACT sequences are identical with those of C. brevisporum and C. lobatum, that of ITS also with C. okinawense. The ITS, GAPDH, CHS-1, HIS3, ACT and TUB2 sequences of the two species from Merremia differ in 3, 8, 5, 12, 9 and 5 nucleotides, respectively.
Closest match in a blastn search with the TUB2 sequence of C. merremiae strain CBS 124955 is with 99 % identity (3 nucleotides difference) C. brevisporum strain CCCM12 from Cucurbita moschata in China (KY797630, Liu et al. 2018) and with the GAPDH sequence with 98 % identity the two C. brevisporum strains from the paper of Noireung et al. (2012). No HIS3 sequence in GenBank is more than 91 % identical to that of C. merremiae. A blastn search with the ITS sequence of C. merremiae resulted in more than 50 identical sequences from strains that were identified as C. brevisporum/Ga. cingulata var. brevisporum, C. magnum/Ga. magna and unidentified Colletotrichum strains including that of the same species sequenced by Rojas et al. (2010, GU994392).
Colletotrichum musicola Damm, sp. nov. MycoBank MB824225. Fig. 11.
Fig. 11.
Colletotrichum musicola (A–C, I–N, V–AF from ex-holotype culture CBS 132885. D–H, O–U. from culture CBS 127557). A–C. Conidiomata. D, M. Tips of setae. E, N. Bases of setae. F–L. Conidiophores. O–T. Appressoria. U–V. Conidia. W, Z, AA. Ascomata. X. Outer surface of peridium. Y. Peridium in cross section. AB. Ascospores. AC. Paraphyses. AD–AF. Asci. D–E, U, Y–AC, AF. from Anthriscus stem. A–C, F–T, V, W–X, AD. from SNA. AE. from OA. A–C, W, Z, AA. DM. D–V, X–Y, AB–AF. DIC. Scale bars: A = 100 μm, E = 10 μm. A applies to A–C, W, Z–AA. E applies to D–V, X–Y, AB–AF.
Etymology: The species epithet is derived from the host plant, Musa.
Sexual morph on Anthriscus stem (only observed in strain CBS 132885). Ascomata perithecia, formed after 4 wk, solitary, superficial, non-stromatic, pyriform, ostiolate, glabrous, medium to dark brown, 200–320 × 130–220 μm diam, ostiolate, glabrous; Peridium 5–10 μm thick, composed of 2–4 layers of medium to dark brown flattened textura angularis with cells 5–16.5 μm diam. Ascogenous hyphae hyaline, smooth-walled, delicate, rarely visible. Interascal tissue formed of paraphyses, hyaline, smooth-walled, cylindrical, with a round tip, disintegrating quickly, septate, apically free, 40–55 μm long, at the basis 2–6 μm wide. Asci unitunicate, 8-spored, cylindrical to clavate, tapering to apex and base, smooth-walled, 76–91.5 × 9.5–12.5 μm. Ascospores uni- or biseriately arranged, aseptate, hyaline, sometimes pale brown, smooth-walled, lunate to fusiform (12.5–)16–21(–25.5) × (4.5–)5.5–6.5(–7) μm, mean ± SD = 18.4 ± 2.4 × 6.0 ± 0.5 μm, L/W ratio = 3.1.
Sexual morph on SNA (only observed in strain CBS 132885). Ascomata pyriform to subspherical, 100–370 × 90–300 μm. Peridium 5–10 μm thick, composed of 2–4 layers of pale to medium brown flattened textura angularis with cells 4.5–15.5 μm diam. Ascogenous hyphae hyaline, smooth, delicate, rarely visible. Interascal tissue formed of paraphyses, hyaline, smooth-walled, cylindrical, disintegrating quickly, rarely observed. Asci unitunicate, 8-spored, narrowly clavate, cylindrical to flask-shaped, fasciculate, 35–90.5 × 10–13.5 μm. Ascospores uni- or biseriately arranged, aseptate, hyaline, sometimes pale brown, smooth-walled, lunate to fusiform, (15–)17.5–21.5(–24.5) × (5–)5.5–6.5(–8) μm, mean ± SD = 19.5 ± 2.2 × 6.0 ± 0.6 μm, L/W ratio = 3.2.
Asexual morph on SNA. Vegetative hyphae 1–10.5 μm diam, hyaline, smooth-walled, septate, branched. Chlamydospores not observed. Conidiomata, conidiophores and setae formed directly on hyphae. Setae pale to medium brown, verruculose to verrucose, 65–95 μm long, 1–2-septate, base cylindrical to ± inflated, 4.5–6.5 μm diam, tip ± rounded. Conidiophores hyaline to pale brown, smooth-walled, single or septate and branched, to 30 μm long. Conidiogenous cells hyaline to pale brown, smooth-walled, cylindrical to conical, 7.5–14.5 × 4–6 μm, opening 1–1.5 μm diam, collarette ≤ 0.5 μm long, periclinal thickening observed, collarette of strain CBS 127557 0.5–1 μm long and periclinal thickening distinct. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical to ellipsoidal, the apex and base rounded, (10.5–)12.5–16.5(–19) × 4.5–5.5(–6) μm, mean ± SD = 14.5 ± 2.1 × 5.2 ± 0.5 μm, L/W ratio = 2.8, conidia of strain CBS 127557 slightly larger, measuring (13.5–)14.5–17(–19) × 5–5.5(–6) μm, mean ± SD = 15.6 ± 1.2 × 5.3 ± 0.3 μm, L/W ratio = 2.9. Appressoria single or in loose groups, medium to dark brown, smooth-walled, elliptical, navicular, bullet-shaped or irregular outline, with undulate or lobate margin, (7.5–)9.5–15(–18.5) × (4.5–)5–7.5(–10) μm, mean ± SD = 12.3 ± 2.8 × 6.4 ± 1.3 μm, L/W ratio = 1.9, appressoria of strain CBS 127557 slightly larger, measuring (9–)11–16.5(–20) × (5–)6–11(–13) μm, mean ± SD = 13.7 ± 2.8 × 8.3 ± 2.5 μm, L/W ratio = 1.6.
Asexual morph on Anthriscus stem not observed in strain CBS 132885, but few conidia and one seta observed in strain CBS 127557. Seta medium brown, verruculose, 62 μm long, 2-septate, base slightly inflated, 5.5 μm diam, tip ± acute. Conidiophores and conidiogenous cells not observed. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical to ellipsoidal, the apex and base rounded, (14–)14.5–17(–17.5) × (5–)5.5–6 μm, mean ± SD = 15.5 ± 1.2 × 5.6 ± 0.3 μm, L/W ratio = 2.8.
Culture characteristics: Colonies on SNA flat with entire margin, hyaline to honey, agar medium, filter paper and Anthriscus stem partly covered with felty whitish aerial mycelium and grey to black conidiomata/ascomata, reverse same colours; growth 27.5–29.5 mm in 7 d (≥ 40 mm in 10 d). Colonies on OA flat with entire margin; olivaceous buff, grey olivaceous to olivaceous, partly covered with short felty whitish aerial mycelium and grey conidiomata/ascomata, reverse olivaceous buff to olivaceous grey, growth 28–30 mm in 7 d (≥ 40 mm in 10 d). Conidial mass pale salmon.
Materials examined: Mexico, from Musa sp., 16 Dec. 2008, M. de J. Yanez Morales (CBS H-21500 holotype, culture ex-holotype CBS 132885 = CPC 16328); Tapachula, Chiapas, from Musa sp., 16 Dec. 2008, M. de J. Yanez Morales, CBS H-21501, culture CBS 127557 = CPC 16329.
Notes: Colletotrichum musicola can be identified with all loci studied. Closest matches with the ITS sequence of isolate CBS 132885 were with 99 % identity (1 nucleotide difference) “C. gloeosporioides” isolates CG0305 from Glycine max in Taiwan (FJ172224, Chen et al. 2006) and AC4-M-Mexico from post-harvest fruit of Musa acuminata in Mexico (M. Espinoza-Ortega et al., unpubl. data), possibly a similar strain or even one of the strains studied here. Closest matches to the TUB2 sequence of CBS 132885 are with seven nucleotides difference (99 % identity) C. orchidearum strains CORCG3 and CORCX6 from Cymbidium and Cattleya, respectively, in China (HM585418 and HM585416, Yang et al. 2011). Closest match with the GAPDH sequence of isolate CBS 132885 was with 99 % identity (2 nucleotides difference) “C. cliviae” strains LC0551 and LC1238, isolated as endophytes from Pennisetum purpureum in Thailand (KC835389, KC835390, Manamgoda et al. 2013). There is no ACT and no CHS-1 sequence with > 98 % identity and no HIS3 sequence with > 96 % identity to those of C. musicola available in GenBank.
This species belongs to the C. orchidearum species complex. Several Colletotrichum species are known from Musa, for example C. aotearoa, C. chrysophilum, C. musae, C. siamense, C. theobromicola and C. tropicale belonging to the C. gloeosporioides complex (Weir et al., 2012, Sharma et al., 2015, Vieira et al., 2018), C. karstii, belonging to the C. boninense complex (Damm et al. 2012b), C. paxtonii and C. scovillei belonging to the C. acutatum complex (Damm et al., 2012a, Zhou et al., 2017) and C. gigasporum belonging to the C. gigasporum complex (Liu et al. 2014). Further Colletotrichum species were described on Musa spp. without sequence data (see notes under C. paxtonii in Damm et al. 2012a). We could not locate the type material of these species to confirm their taxonomic positions.
One sexual morph, Ga. musarum, was described by Petch (1917) on Musa paradisiaca in Ceylon (Sri Lanka) as associated with Gloeosporium musarum Cooke & Massee; its ascospores are hyaline, aseptate, cymbiform (boat-shaped, navicular), straight or curved, obtuse and measure 14–18 × 3.5–4 μm. Ascospores of C. musicola have a different shape and are larger, measuring on average 19.5 × 6 μm (SNA) or 18.4 × 6 μm (Anthriscus stem). Ascospore shape and size of strains called Ga. musae by Rodrigues & Owen (1992) were again different and cannot be linked either. The sexual morph of the latter species was obtained by means of laboratory crossings, while C. musicola formed a sexual morph in pure culture; it is apparently homothallic. Compared to other sexual morphs developed in the C. orchidearum complex, C. musicola forms ascospores that are similar in length to those of C. sojae, however much wider in the middle and with a different shape (fusiform).
Colletotrichum okinawense Damm & Toy. Sato, sp. nov. MycoBank MB824226. Fig. 12.
Fig. 12.
Colletotrichum okinawense (from ex-holotype culture MAFF 240517). A–B. Conidiomata. C–E, J–M. Conidiophores. F–I. Microcyclic conidiation. N–S. Appressoria. T–U. Conidia. A, C–I, T. from Anthriscus stem. B, J–S, U. from SNA. A–B. DM. C–U. DIC. Scale bars: A = 100 μm, C = 10 μm. Scale bar of A applies to A–B. Scale bar of C applies to C–U.
Etymology: The species epithet is derived from the Japanese island Okinawa, where the species was collected.
Sexual morph not observed. Asexual morph on SNA. Vegetative hyphae 2–8 μm diam, hyaline, smooth-walled, septate, branched. Chlamydospores not observed. Conidiomata, conidiophores formed directly on hyphae. Setae not observed. Conidiophores hyaline, smooth-walled, septate, branched. Conidiogenous cells hyaline, smooth-walled, cylindrical to doliiform, sometimes lacking a basal septum, 8–25 × 3–6 μm, opening 1–2 μm diam, collarette 0.5–1 μm long, periclinal thickening observed. Conidia hyaline, smooth-walled, aseptate, straight, clavate to cylindrical, the apex rounded, the base rounded or truncate, (10.5–)12–14.5(–16.5) × (3.5–)4.5–5.5 μm, mean ± SD = 13.4 ± 1.3 × 5.0 ± 0.4 μm, L/W ratio = 2.7, few conidia up to 22 μm long were observed. Appressoria single or in loose groups, pale to medium brown, smooth-walled, elliptical, navicular, bullet-shaped or irregular in outline, with undulate or lobate margin, (5.5–)7–15(–23) × (2.5–)5–10.5(–13) μm, mean ± SD = 11.1 ± 3.9 × 7.7 ± 2.6 μm, L/W ratio = 1.4.
Asexual morph on Anthriscus stem. Conidiomata, conidiophores formed on pale to medium brown, angular cells, 3–7 μm diam. Setae not observed. Conidiophores hyaline to very pale brown, smooth-walled, septate, branched, to 25 μm long. Conidiogenous cells hyaline, smooth-walled, cylindrical to conical, 4.5–21 × 3.5–5.5 μm, opening 1–2 μm diam, collarette 0.5 μm long, periclinal thickening observed. Microcyclic conidiation observed, sometimes from conidia still attached to the conidiogenous cell. Conidia hyaline, smooth-walled, aseptate, straight, clavate to cylindrical, the apex rounded, the base rounded or truncate, (12–)12.5–15(–16.5) × (4.5–)5–5.5 μm, mean ± SD = 13.9 ± 1.3 × 5.1 ± 0.3 μm, L/W ratio = 2.7.
Culture characteristics: Colonies on SNA flat with entire margin, medium hyaline, aerial mycelium lacking, reverse same colours; 31.5–34 mm in 7 d (≥ 40 mm in 10 d). Colonies on OA flat with entire margin, surface saffron to salmon, aerial mycelium lacking, reverse salmon; 31.5–34 mm in 7 d (≥ 40 mm in 10 d). Conidial mass saffron.
Materials examined: Brazil, Minas Gerais, Uberaba, on stems of Carica papaya, Jun. 1892, E. Ule n. 1947 (H holotype of Gloeosporium papayae). Japan, Okinawa prefecture, Miyakojima Island, from a petiole of Carica papaya, 28 Sep. 2007, S. Sato (isolated Sep. 2007 by T. Sato) (GLM-F 111630 holotype of C. okinawense, culture ex-holotype MAFF 240517 = NBRC 104626 = CBS 143246 = GLMC 1837).
Notes: This species belongs to the C. magnum species complex. Strain MAFF 240517 was originally identified as C. gloeosporioides by J. Moriwaki and recently re-identified as C. brevisporum by T. Sato. Based on the multilocus phylogeny in this study this species is closely related to C. brevisporum, but represents a distinct species.
Many Colletotrichum species have been reported or were described on papaya (Carica papaya) (Farr & Rossman 2017); a number of them were studied based on molecular data. For example, C. acutatum and C. simmondsii were described on Carica papaya in Australia. They belong to the C. acutatum species complex; both species occur on many other host plants as well (Damm et al. 2012a). A strain from Carica papaya in Brazil was identified as C. karstii (CBS 106.91); this species belongs to the C. boninense species complex and also occurs on many host plants (Damm et al. 2012b). Colletotrichum queenslandicum, originally described as C. gloeosporioides var. minus from Carica papaya in Australia, and C. siamense strain ICMP 18739 from Carica papaya in South Africa belong to the C. gloeosporioides species complex (Weir et al. 2012). There are also two other strains from papaya included in this study, one from Japan (MAFF 238697) that is re-identified as C. plurivorum (C. orchidearum complex), and one from Australia (CBS 512.75) that was re-identified as C. brevisporum that belongs to the C. magnum complex, too.
Two species were described by Hennings, 1895, Hennings, 1908 from papaya in Brazil: C. papayae Henn. 1908 and Gloeosporium papayae Henn. 1895. They are not synonyms as listed in Saccardo et al., 1931, Petrak, 1953 combined the latter in Colletotrichum as C. papayae (P. Henn.) Petr. 1953, apparently not being aware of the older homonym (nom. illegit., Art. 53.1). Conidia of C. papayae Henn. are cylindrical, straight to curved, measuring 12–20 × 5–7 μm, while those of Gl. papayae Henn. are cylindrical to subclavate, obtuse, straight, measuring 11–14 × 5–6 μm according to Hennings (1895) and (12.5–)13–15(–15.5) × 4.5–6 μm, mean ± SD = 14 ± 1.2 × 5.3 ± 0.5 μm, L/W ratio = 2.7 according to our measurements. The conidia size of strain MAFF 240517 is similar to that of Gl. papayae. However, the shape of the conidia observed on the type specimen of Gl. papayae was cylindrical with parallel walls or even a constriction at the middle, not distinctly attenuated to the base as those of C. okinawense.
GAPDH, ACT and TUB2 sequences of C. okinawense differ from all other species treated in this study (no HIS3 sequence available), while the ITS is identical to that of C. brevisporum, C. merremiae and C. lobatum, and the CHS-1 sequence to that of C. brevisporum. The ACT sequence is 100 % identical to those of C. brevisporum isolates CMM1672, CMM1702, CMM1822 and CMM2005 from Carica papaya in Brazil (KC702903–KC702906, Vieira et al. 2013) that possibly belong to the same species. The closest matches with the TUB2 sequence are with 99 % identity (6 nucleotides difference) to the four C. liaoningense strains from Capsicum in China (KP890111–KP890113, KP890115, Diao et al. 2017). The closest matches with the GAPDH sequence are with 97 % identity to the C. liaoningense strains CAUOS 3 and CAUOS 4 (KP890136, KP890137, Diao et al. 2017). The ITS sequence is 100 % identical to several unidentified isolates and isolates identified as C. brevisporum, C. magnum, Ga. magna and C. cingulata var. brevispora.
In contrast to all other species in the C. magnum complex, conidia of C. okinawense are predominantly clavate; it is also the fastest growing species in this complex. Microcyclic conidiation has only rarely been observed in the genus Colletotrichum. Only one other species with this feature is known to us, C. cliviicola, that belongs to the C. orchidearum complex. Secondary conidia of C. okinawense are formed directly from conidiogenous openings on conidia, and sometimes already when the mother cell itself is still attached to its conidiogenous cell, while secondary conidia of C. cliviicola are usually formed on short pegs or phialides that appear on older conidia that are sometimes already septate.
Colletotrichum orchidearum Allesch., Rabenh. Krypt.-Fl., Edn 2 (Leipzig) 1(7): 563. 1902 (1903). Fig. 13.
Fig. 13.
C. orchidearum (A–W, Y–Z. from ex-epitype culture CBS 133131, X, AA. from culture CBS 136877). A–B. Conidiomata, arrow head in A: Conidioma with seta. C, G. Tips of setae. D, F, H. Bases of setae. E, I–L. Conidiophores. M–R. Appressoria. S–T. Conidia. U. Ascoma. V. Peridium in cross section. W. Outer surface of peridium. X. Paraphyses. Y. Ascospores. Z–AA. Asci. A, C–F, S. from Anthriscus stem. B, G–R, T–AA. from SNA. A–B, U. DM. C–T, V–AA. DIC. Scale bars: A = 100 μm, E = 10 μm. A applies to A–B. E applies to C–T, V–AA.
Synonyms: Colletotrichum hymenocallidicola Chethana et al., Fungal Diversity 75: 160. 2015.
Colletotrichum aracearum L.W. Hou & L. Cai, Mycosphere 7: 1115. 2016.
Sexual morph on SNA. Ascomata perithecia, formed after 4 wk, solitary, non-stromatic, subglobose, ostiolate, 200–300 × 175–250 μm. Peridium 6–15 μm thick, composed of 2–3 layers of medium brown flattened textura angularis with cells 5–22 μm diam. Ascogenous hyphae hyaline, smooth, delicate. Interascal tissue formed of paraphyses, hyaline, smooth-walled, mostly cylindrical but tapering towards the round tip, disintegrating quickly, septate, constricted at the septa, branched, apically free, 40–70 μm long and up to 6–8.5 μm diam. Asci unitunicate, 8-spored, fasciculate, smooth-walled, cylindrical to slightly clavate, tapering to apex and base, round tip, no apical apparatus observed, 70–100 × 11–15 μm. Ascospores uni- or biseriately arranged, aseptate, hyaline, smooth-walled, allantoid, with round ends, (13.5–)16–20(–22) × 5–5.5(–6) μm, mean ± SD = 18.0 ± 1.9 × 5.2 ± 0.3 μm, L/W ratio = 3.5.
Sexual morph on Anthriscus stem. Ascomata perithecia, formed after 4 wk, solitary, non-stromatic, subglobose to ovoidal, dark brown, ostiolate; outer wall composed of medium brown flattened textura angularis with cells 17–22 μm diam. Asci 8-spored, cylindrical to clavate, unitunicate, fasciculate, thin-walled, 56–77 × 9–11.5 μm. Ascospores uni- or biseriately arranged, aseptate, hyaline, smooth-walled, allantoid, with round ends, (13–)15.5–19(–21) × 4.5–5.5(–6) μm, mean ± SD = 17.4 ± 1.8 × 5.1 ± 0.4 μm, L/W ratio = 3.4.
Asexual morph on SNA. Vegetative hyphae 1–11 μm diam, hyaline to pale brown, smooth-walled to verruculose, septate, branched. Chlamydospores not observed. Conidiomata, conidiophores and setae formed directly on hyphae. Setae medium brown, verruculose to verrucose, 55–110 μm long, 1–2(–4)-septate, base ± inflated, 5.5–6 μm diam, tip rounded. Conidiophores hyaline to pale brown, smooth-walled to verruculose, septate, branched. Conidiogenous cells hyaline to pale brown, smooth-walled to verruculose, cylindrical to doliiform, 8–40 × 4–5,5 μm, opening 0.5–2 μm diam, collarette 0.5–1 μm long, periclinal thickening sometimes observed. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical, the apex rounded, the base truncate or rounded, (13–)14–20(–28) × 4.5–5.5(–6.5) μm, mean ± SD = 17.0 ± 3.2 × 5.1 ± 0.5 μm, L/W ratio = 3.4, CBS 136877 forms shorter and wider conidia, measuring (12.5–)13–15.5(–17) × 5–6.5(–7) μm, mean ± SD = 14.2 ± 1.2 × 5.7 ± 0.7 μm, L/W ratio = 2.5. Appressoria single, medium to dark brown, smooth-walled, clavate, navicular, elliptical or circular in outline, with a lobate or undulate margin, (9–)10.5–17.5(–22.7) × (5–)6–10.5(–13) μm, mean ± SD = 14.0 ± 3.3 × 8.4 ± 2.3 μm, L/W ratio = 1.7.
Asexual morph on Anthriscus stem. Conidiomata, conidiophores and setae formed on pale brown, angular cells, 4.5–8 μm diam. Setae (few observed) medium to dark brown, verruculose, 75–130 μm long, 1–5-septate, base cylindrical, conical to ± inflated, 4–8 μm diam, tip acute to ± rounded. Conidiophores hyaline to pale brown, smooth-walled. Conidiogenous cells hyaline to pale brown, smooth-walled, cylindrical to doliiform, 7–10 × 4.5–6 μm, opening 1 μm diam, collarette 0.1–1 μm long, periclinal thickening not observed. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical, the apex rounded, the base truncate or rounded, (14.5–)15.5–17(–17.5) × (4.5–)5–5.5(–6) μm, mean ± SD = 16.2 ± 0.9 × 5.2 ± 0.3 μm, L/W ratio = 3.1.
Culture characteristics: Colonies on SNA flat with entire margin, hyaline to pale honey, agar medium, filter paper and Anthriscus stem partly covered with grey to black fruiting bodies and sparse aerial mycelium, reverse same colours; growth 30–31.5 mm in 7 d (≥ 40 mm in 10 d). Colonies on OA flat with entire margin; buff, honey, grey olivaceous to olivaceous grey, partly covered with short floccose white aerial mycelium, reverse buff to olivaceous grey, growth 30–32 mm in 7 d (≥ 40 mm in 10 d). Conidial mass whitish to pale yellow.
Materials examined: Germany, Munich, glasshouses of Botanical Garden, on dead and dying leaves of Eria javanica (syn. E. stellata), Apr. 1895, J.E. Weiss [M-0140831 lectotype of C. orchidearum (named as forma eriae)]; Iran, Golestan province, Gorgan, from leaves of Epipremnum aureum (syn. Scindapsus aureus), Oct. 2013, A. Alizadeh, strain UTFC 266 = 249C. Netherlands, Utrecht (indoor plant), from anthracnose on leaf of Dendrobium nobile, Apr. 2013, I. Benoit-Gelber (CBS H-21910 epitype, here designated MBT380419, culture ex-epitype CBS 135131); ibid., culture CBS 136877.
Notes: The type specimens of the three forms of C. orchidearum, C. orchidearum f. cymbidii, C. orchidearum f. eriae and C. orchidearum f. physosiphonis (Allescher 1902) were recently investigated by Yang et al. (2011), and strains from different orchid genera from China clustering in the multilocus phylogeny of that study were identified as C. orchidearum based on morphological resemblance. As C. orchidearum was described in Europe, the authors were however reluctant to select an epitype from their Asian collection. Damm et al. (2012a) studied the specimens as well and lectotypified C. orchidearum with the specimen M-0140831 from leaves of Eria javanica.
Different collections of Colletotrichum species from Orchidaceae in Europe were treated in this study, including strains of the species studied by Yang et al. (2011), that were isolated from a leaf of Dendrobium nobile in the Netherlands cultivated as an indoor plant, confirming the occurrence of this species in Europe. Conidia from the lectotype specimen were measured in Damm et al. (2012a): (13.5–)15.5–19.5 × 5–6 μm, mean ± SD = 17.2 ± 1.6 × 5.6 ± 0.3 μm, L/W ratio = 3.1. Conidia from one of the strains from the Netherlands, CBS 135131, agree in conidial dimensions, measuring on average (13–)14–20(–28) × 4.5–5.5(–6.5) μm on SNA and (14.5–)15.5–17(–17.5) × (4.5–)5–5.5(–6) μm on Anthriscus stems. Moreover, conidiomata were minute as observed on the lectotype specimen and the length of the setae was similar (setae from lectotype: 56–120 × 4.5–6 μm). Strain CBS 135131 was therefore used as basis of the epitype.
Our collection from Munich, Germany, had similar conidial dimensions as C. orchidearum; however, conidia often develop a prominent scar and are slightly constricted in the middle. Moreover, the conidiomata of this collection were very large, while those of C. orchidearum were small and longer setae were formed. The species collected in Munich belongs to the C. dracaenophilum complex and was described as C. coelogynes in this study. Another species from orchids described in this study is C. cattleyicola that also belongs to the C. orchidearum species complex.
There are numerous other Colletotrichum species that were described on Orchidaceae, including C. orchidophilum, belonging to the C. acutatum species complex (Damm et al. 2012a), C. cymbidiicola, C. oncidii and C. karstii, belonging to the C. boninense species complex (Yang et al., 2011, Damm et al., 2012b), C. arxii belonging to the C. gigasporum complex (Liu et al. 2014), C. ochracea, C. caudasporum, C. duyunensis, C. endophytum, C. guizhounensis and C. bletillum, belonging to the C. caudatum, C. graminicola and C. spaethianum species complexes, respectively (Tao et al. 2013), C. coelogynes and C. excelsum-altitudinum in the C. dracaenophilum species complex (Tao et al. 2013, this study).
There are sequences of strains called C. orchidearum from three studies in GenBank. Strains from different Orchidaceae hosts from the study of Yang et al. (2011) and isolate SAUCC 1407 from Arctium lappa (Xu et al. 2016) both from China were included in our phylogeny. The strains from Orchidaceae were confirmed as C. orchidearum, while the isolate from Arctium lappa was re-identified as C. sojae. One of two ITS sequences of “C. orchidearum” isolate NW248 from an unpublished and unnamed study was displayed as “type strain of C. orchidearum” (EU520211, EU732727; Z. Zhang et al., unpubl. data); it was revealed to be no Colletotrichum sp. at all but a species of Cytospora (Diaporthales).
The description of C. hymenocallidicola (Ariyawansa et al. 2015) is based on one strain of which five loci were sequenced (ITS, GAPDH, CHS-1, ACT, TUB2). Blastn searches on NCBI GenBank with all sequences of this species revealed that the ACT and TUB2 sequences do not conform with the placement of the species based on ITS, GAPDH and CHS-1 sequences. While the closest matches of the ITS, GAPDH and CHS-1 sequences were with 98–99 % identity strains identified as C. cliviae, placing the species in the C. orchidearum complex, the ACT sequence (KT290260) was 93 % identical with those of C. pseudomajus and C. vietnamense (both C. gigasporum complex) and only 83 % identical with strains identified as C. cliviae, and the TUB2 sequence (KT290261) was 100 % identical with the ex-epitype strain of C. truncatum (Damm et al. 2009). The ACT and TUB2 sequences were possibly mixed-up with those of other strains studied by the authors. Further, the beginning part of the GAPDH sequence apparently includes artefacts as it is completely different from all related species. Therefore, C. hymenocallidicola and C. aracearum were not in the same clade in a recent phylogeny of the genus Colletotrichum (Marin-Felix et al. 2017). The ACT and TUB2 sequences of C. hymenocallidicola as well as the first 22 nucleotides of GAPDH were excluded from the alignment in this study. It is highly recommended to re-sequence the ex-type strain of this species, in order to confirm our results with the other loci and establish a solid basis of this apparently common species.
Colletotrichum hymenocallidicola and C. aracearum were described recently as pathogens of Hymenocallis sp. in Thailand (Ariyawansa et al. 2015) and Monstera deliciosa and Philodendron selloum in China (Hou et al. 2016), respectively, and shown to be synonyms of C. orchidearum in this study. Based on this study, C. orchidearum is known from monocots such as Hymenocallis (Amaryllidaceae), Monstera, Philodendron, Scindapus (Araceae), Cordyline (Asparagaceae) as well as many Orchidaceae, including Cattleya, Cymbidium, Dendrobium, Oncidium, Phalaenopsis and Vanda in Asia (China, Iran, Japan, Thailand) and Europe (as indoor plant in the Netherlands).
Colletotrichum orchidearum can be differentiated with sequences of all loci included, except for GAPDH; sequences of CHS-1 and HIS3 are only available of part of the strains included. GAPDH sequences of C. orchidearum and C. sojae are identical. In a blastn search in GenBank, the TUB2 sequence of strain CBS 135131 was 100 % identical with those of the strains from Orchidaceae in China and 99 % identical (one nucleotide difference) with those of the two C. aracearum strains, while the two C. cliviicola strains CSSS1 and CSSS1 (Yang et al. 2009) differ in eight nucleotides; all are included in this study. In a blastn search with the ACT sequence, the two C. aracearum strains were identical and the strains from Orchidaceae in China differed in one nucleotide; while the C. cliviicola strains differed in four nucleotides. The ITS sequence of strain CBS 135131 was identical with those of the two C. aracearum (strains, “C. gloeosporioides” isolate C10 from Cymbidium in China (Yao et al. 2013), Colletotrichum sp. strain AR3750 and Glomerella sp. strain AR3749 from Dendrobium and Cattleya, respectively, in Thailand (Farr et al. 2006) and differed in one nucleotide from “C. gloeosporioides” isolate C14 from Cymbidium in China (?) (J.A. Yao et al., unpubl. data) and the ex-type strain of C. hymenocallidicola, while the C. cliviicola strains differed in seven nucleotides.
No sexual morph was observed in the strain from Hymenocallis (Ariyawansa et al. 2015), but in those from Monstera deliciosa (Hou et al. 2016) and from Orchidaceae (Yang et al. 2011, this study). In pathogenicity tests by Yang et al. (2011), strain CORCX6 from Cattleya sp. caused lesions on fruits of peppers, apple and tomato by wound/drop inoculation, but none on unwounded fruits.
Colletotrichum panamense Damm, sp. nov. MycoBank MB824227. Fig. 14.
Fig. 14.
Colletotrichum panamense (from ex-holotype culture CBS 125386). A–B. Conidiomata. C, H. Tips of setae. D, I. Bases of setae. E–G, J–N. Conidiophores. O–P. Conidia. A, C–G, O. from Anthriscus stem. B, H–N, P. from SNA. A–B. DM. C–P. DIC. Scale bars: A = 100 μm, G = 10 μm. Scale bar of A applies to A–B. Scale bar of G applies to C–P.
Etymology: The species epithet is derived from the country where the species was collected, Panama.
Sexual morph not observed. Asexual morph on SNA. Vegetative hyphae 1.5–7.5 μm diam, hyaline, smooth-walled, septate, branched. Chlamydospores not observed. Conidiomata, conidiophores and setae formed directly on hyphae. Setae medium brown, smooth-walled, verrucose towards the tip, 40–100 μm long, 2(–3)-septate, base inflated, 6–7 μm diam, tip ± acute. Conidiophores pale brown, smooth-walled, septate or aseptate. Conidiogenous cells pale brown, smooth-walled, broadly ellipsoidal, doliiform to cylindrical, 7–16 × 5.5–7 μm, often intercalary, opening 1.5–2 μm diam, collarette 0.5–1 μm long, periclinal thickening distinct. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical, the apex and base rounded, (14.5–)15.5–18(–19.5) × (4.5–)5–5.5 μm, mean ± SD = 16.9 ± 1.2 × 5.0 ± 0.3 μm, L/W ratio = 3.4. Appressoria not formed.
Asexual morph on Anthriscus stem. Conidiomata, conidiophores and setae formed directly on hyphae, no basal cells observed. Setae medium brown, smooth-walled, verrucose towards the tip, 60–100 μm long, 2–3-septate, base inflated, 5.5–7.5 μm diam, tip ± acute to ± rounded. Conidiophores pale brown, smooth-walled, septate, sometimes branched, to 30 μm long. Conidiogenous cells pale brown, smooth-walled, ovoidal, broadly ellipsoidal to subsphaerical, 6–11.5 × 4.5–7.5 μm, opening 1.5–2 μm diam, collarette not observed, periclinal thickening sometimes visible or distinct. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical, the apex and base rounded, (14–)16–20(–22) × 5–5.5 μm, mean ± SD = 18 ± 2.1 × 5.3 ± 0.2 μm, L/W ratio = 3.4.
Culture characteristics: Colonies on SNA flat with entire margin, medium hyaline to pale ochreous to pale luteous, filter paper partly pale luteous, aerial mycelium lacking, reverse same colours; 18.5–20 mm in 7 d (27.5–30 mm in 10 d). Colonies on OA flat with entire margin, surface rosy buff to saffron with small grey dots due to sporulation, aerial mycelium lacking, reverse rosy buff to buff; 22.5–25.5 mm in 7 d (34–33.5 mm in 10 d). Conidial mass saffron.
Material examined: Panama, Gamboa, wet lowland forest, leaf endophyte of Merremia umbellata, Nov. 2004, S. Van Bael & Z. Maynard, D2-13 (CBS H-21066 holotype, culture ex-type CBS 125386 = Q882).
Notes: Colletotrichum panamense belongs to the C. magnum species complex like C. merremiae that is also described from Merremia umbellata in Panama in this study. In contrast to C. merremiae, the conidiogenous cells of C. panamense are often subglobose to ellipsoidal, on SNA often also intercalary. Conidia of C. panamense are on both media on average at least 2 μm longer.
Colletotrichum panamense differs with all loci studied from all other species of the genus. The ITS sequence of strain CBS 125386 is 100 % identical to Colletotrichum sp. Q882 (GU994392, Rojas et al. 2010), which is the same strain. Other sequences have ≥ 3 bp differences. Closest matches with the TUB2 sequence of the ex-type strain of C. panamense are with 99 % identity (5 nucleotides difference) the four C. liaoningense strains that are included in our study (Diao et al. 2017). Closest matches with the CHS-1 sequence are with 99 % identity (1 nucleotide difference) to those of the ex-type strain of C. brevisporum sequenced by Liu et al. (2014, KF687760) and C. brevisporum strain CCCM12 from Cucurbita moschata in China (KY797631, Liu et al. 2018). No ACT sequence in GenBank is closer than 97 % identical and no HIS3 sequence closer than 90 % identical to those of C. panamense.
Colletotrichum piperis Petch, Ann. R. bot. Gdns Peradeniya 6(3): 239. 1917. Fig. 15.
Fig. 15.
Colletotrichum piperis (from ex-epitype culture IMI 71397). A–B. Conidiomata. C, H. Tips of setae. D, I. Bases of setae. E–G, J–K. Conidiophores. L–Q. Appressoria. R–S. Conidia. A, C–G, R. from Anthriscus stem. B, H–Q, S. from SNA. A–B. DM. C–S. DIC. Scale bars: A = 100 μm, E = 10 μm. Scale bar of A applies to A–B. Scale bar of E applies to C–S.
Sexual morph not observed. Asexual morph on SNA. Vegetative hyphae 1–8 μm diam, hyaline, smooth-walled, septate, branched. Chlamydospores not observed. Conidiomata, conidiophores and setae formed directly on hyphae. Setae pale to medium brown, verruculose to verrucose, 30–95 μm long, 1–3-septate, base cylindrical, 4.5–7 μm diam, tip ± acute to ± rounded. Conidiophores hyaline to pale brown, smooth-walled to verruculose, septate, branched, to 65 μm long. Conidiogenous cells hyaline to pale brown, smooth-walled to verruculose, cylindrical, 10–25 × 3.5–5.5 μm, opening 2–2.5 μm diam, collarette 1–1.5 μm long, periclinal thickening distinct. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical to obclavate, the apex rounded, the base rounded to truncate, (11.5–)13–18.5(–28) × 4–5(–6) μm, mean 15.7 ± 2.7 × 4.7 ± 0.4 μm, L/W ratio = 3.4. Appressoria not formed at the reverse side of SNA plates, but few observed on slide culture after 1 month, single, medium brown, smooth-walled, elliptical, bullet-shaped to rectagular in outline, with an entire margin, (3.5–)6–10(–13) × (3–)4–6(–8.5) μm, mean ± SD =7.9 ± 2.1 × 5.0 ± 1.1 μm, L/W ratio = 1.6.
Asexual morph on Anthriscus stem. Conidiomata, conidiophores and setae formed on pale brown, angular cells. Setae pale to medium brown, verruculose to verrucose, often branched, 50–120 μm long, 1–3-septate, base cylindrical, sometimes slightly inflated, 4.5–7 μm diam, tip ± acute to ± rounded. Conidiophores pale brown, smooth-walled, septate, sometimes branched, to 35 μm long. Conidiogenous cells pale brown, smooth-walled, cylindrical to doliiform, 8–22 × 4–6.5 μm, opening 1–1.5 μm diam, collarette 0.5–1 μm long, periclinal thickening observed. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical, the apex rounded, the base rounded to truncate, (14–)15.5–17.5(–19.5) × (4–)4.5–5(–5.5) μm, mean ± SD = 16.6 ± 1.0 × 4.7 ± 0.3 μm, L/W ratio = 3.6.
Culture characteristics: Colonies on SNA flat with entire margin, hyaline, cinnamon to isabelline in the centre, agar medium, filter paper and Anthriscus stem partly covered with grey to salmon acervuli, aerial mycelium lacking, reverse same colours; growth 15.5–16.5 mm in 7 d (26–27.5 mm in 10 d). Colonies on OA flat with entire margin; grey olivaceous, with a buff margin, covered with translucent salmon conidia masses and partly covered with very short aerial mycelium, reverse olivaceous grey to iron-grey, growth 17.5–19 mm in 7 d (26–29 mm in 10 d). Conidial mass salmon.
Materials examined: Malaysia, from Piper nigrum, unknown collection date and collector (IMI history: 1957 P. Holliday HS 134, 1958 CABI) (IMI 71397 epitype of C. piperis Petch, here designated, MBT380420, CBS H-21503 isoepitype, culture ex-epitype IMI 71397). Puerto Rico, Caguas, from Piper umbellatum, 9 Feb. 1913, FL Stevens, BPI 399521 possible paratype of C. piperis Stevens. Sri Lanka, Medulsima, from leaf of Piper betle, T. Petch 4544, 17 Mar. 1915 (K(M) 235609 B, part of holotype of C. piperis Petch). Taiwan, Tainan, from Piper betle, 29 Oct. 1907, K. Sawada, BPI 397531 possible holotype of C. geniculatum.
Notes: There were several species described on Piper spp. including C. dasturii, C. geniculatum, C. necator, C. piperis Petch and C. piperis Stevens. The two species named C. piperis were described in the same year (1917), C. piperis Petch from leaves of Piper betle and P. nigrum in Sri Lanka (Ceylon) and C. piperis Stevens from Piper umbellatum in Puerto Rico, respectively (Petch, 1917, Stevens, 1917). According to Roy (1948), who investigated the publication dates, the name C. piperis Stevens is illegitimate (nom. illegit., Art. 53.1) as it was published between 10th July and 18th August 1917 and a homonym of the earlier C. piperis Petch (published 4th July 1917) and was therefore replaced by C. stevensii.
Conidia of C. piperis Petch measure 12–19 × 3.5–4.5 μm according to Petch (1917) and 10–16 × 4–6 μm and (13.5–)14–16(–17.5) × (4.5–)4.5–5.5(–6) μm, mean ± SD = 15.1 ± 1.0 × 5.1 ± 0.4 μm, L/W ratio = 3.0 according to measurements from the type specimen by Roy (1948) and the present study. The conidial size and shape of strain IMI 71397 agree with these measurements and our observations (not shown), respectively. This strain originated from P. nigrum in Malaysia, the same region of Asia than the holotype of C. piperis Petch (Sri Lanka). Strain IMI 71397 is therefore used as the basis for the epitype of this species. In contrast, conidia of C. stevensii (syn. C. piperis Stevens) are larger than C. piperis Petch, measuring 17–27 × 7 μm (Stevens 1917); which was confirmed by measurements of conidia of a possible paratype specimen of C. piperis Stevens in this study: (16.5–)19–23.5(–25.5) × (5–)5.5–7.5(–8.5) μm, mean ± SD = 21.2 ± 2.3 × 6.4 ± 0.9 μm, L/W ratio = 3.3. Colletotrichum necator that was described from fruit of Piper sp. in Singapore by Massee (1912) also forms larger conidia (20–23 × 5–7 μm) than C. piperis Petch. Sawada (1959) described C. geniculatum from P. betle in Taiwan. Conidia from the type specimen measure 12.5–14(–15) × (5–)5.5–6.5(–7) μm, mean ± SD = 13.3 ± 0.8 × 6.2 ± 0.6 μm, L/W ratio = 2.2; conidiophores were not observed, setae measure 35–57 × 4–5.5 μm. The conidia size of C. geniculatum is similar to that of C. piperis Petch. However, the conidia are shorter and wider, resulting in a smaller L/W ratio and a different shape. Roy (1948) described C. dasturii from P. betle in India, Bengal; its conidia are curved, while C. piperis Petch has straight conidia.
Sequence comparison of Colletotrichum spp. from Piper spp. in GenBank suggests that species from several species complexes can be found on this host genus. For example, strains from a study of leaf endophytes in Piper hispidum in Brazil identified by ITS belong to the C. boninense and C. gloeosporioides species complexes (Orlandelli et al. 2012). A strain from Piper nigrum (IMI 324991, unknown location) was recently identified as C. fioriniae belonging to the C. acutatum species complex (Damm et al. 2012a), while a C. siamense strain from a leaf lesion of P. nigrum in Australia (James et al. 2014) and two C. truncatum strains (CBS 127.57, IMI 63597) from Peperomia magnoliifolia (Piperaceae) in India (Damm et al. 2009) belong to the C. gloeosporioides and C. truncatum species complexes, respectively. Farr & Rossman (2017) further list C. capsici (syn. of C. truncatum), C. dematium and C. gloeosporioides. However, the respective sources are from the pre-sequence era and therefore need confirmation.
Another species on Piper, C. lobatum, is described in this study from Piper catalpaefolium in Trinidad and Tobago with very similar conidial shape and size, but very different appressoria that are more complex than those of C. piperis Petch. Colletotrichum lobatum belongs to the C. magnum species complex, while C. piperis belongs to the C. orchidearum complex. Apart from the rather simple appressoria with entire edge, C. piperis sometimes forms branched setae, a feature rarely observed in the genus Colletotrichum. Moreover, C. piperis is the slowest growing species in the C. orchidearum complex.
Colletotrichum piperis can be identified with all loci included in this study. Closest matches in a blastn search with the ITS sequence of C. piperis strain IMI 71397 on GenBank were with 99 % identity (5 nucleotides difference) several strains identified as C. gloeosporioides, fungal sp., C. magnum, C. orchidearum, fungal endophyte, C. trifolii, Glomerella glycines, Colletotrichum sp., C. aracearum and Glomerella sp. There is no GAPDH sequence in GenBank with ≥ 96 % identity and no TUB2 sequence with ≥ 97 % identity to that of strain IMI 71397. The closest match with the ACT sequence was with 99 % identity (3 nucleotides difference) C. cliviae strain GUFCC15503 from Calamus thwaitesii in India (KC790646, Sharma et al. 2013a).
Colletotrichum plurivorum Damm, Alizadeh & Toy. Sato, sp. nov. MycoBank MB824228. Fig. 16.
Fig. 16.
Colletotrichum plurivorum (A, C–H, O–T, V–AD. from ex-holotype culture CBS 125474. B, I–N, U. from culture CBS 903.69). A–B. Conidiomata. C, I. Tips of setae. D, J. Bases of setae. E–H, K–N. Conidiophores. O–S. Appressoria. T–U. Conidia. V–W. Ascomata. X. Paraphyses. Y. Outer surface of peridium. Z. Peridium in cross section. AA–AC. Asci. AD. Ascospores. A, C–H,T, V, X, AA, AD. from Anthriscus stem. B, I–S, U, W, Y–Z, AB–AC. from SNA. A–B, V–W. DM. C–U, X–AD. DIC. Scale bars: A = 100 μm, T = 10 μm. A applies to A–B, V–W. T applies to C–U, X–AD.
Synonym: Colletotrichum sichuanensis G.S. Gong & F.L. Liu, Scientific Reports 6(32761): 6. 2016. nom. inval., Art. 40.1 (Melbourne).
Etymology: The species epithet is based on the large host range of this species.
Sexual morph on Anthriscus stem (observed in strains CBS 125474 and UTFC 260). Ascomata perithecia, formed after 4 wk, solitary, superficial or immersed, non-stromatic, globose to obpyriform, ostiolate, glabrous or covered by sparse white aerial mycelium, medium to dark brown, 100–230 × 95–160 μm, surrounded by pale brown, smooth-walled to verruculouse hyphae. Peridium 12–18 μm thick, composed of 3–5 layers of pale brown flattened textura angularis with cells 5–18 μm diam. Ascogenous hyphae hyaline, smooth, delicate, rarely visible. Interascal tissue formed of paraphyses, hyaline, smooth-walled, cylindrical, with a rounded tip, disintegrating quickly, septate, apically free, 50–70 μm long, base 4.5–7.5 μm wide, branched at the base. Asci unitunicate, 8-spored, cylindrical, smooth-walled, 50–65.5 × 10.5–12.5 μm, the base broadly truncate, asci of strain UTFC 260 larger, measuring 70–90 × 9–13.5 μm. Ascospores uni- or biseriately arranged, aseptate, in strain UTFC 260 also septate ascospores observed, initially hyaline, turning pale brown with age, smooth-walled, allantoid to fusiform, with both ends rounded, (13–)14.5–18(–22) × (4–)5–6(–7) μm, mean ± SD = 16.4 ± 1.8 × 5.5 ± 0.5 μm, L/W ratio = 3.0.
Sexual morph on SNA (observed in strains CBS 125474, UTFC 260 and UTFC 261). Ascomata perithecia, globose to obpyriform, 180–340 × 180–230 μm, ostiolate, medium to dark brown, glabrous. Peridium 12–18 μm thick, composed of 4–6 layers of pale brown flattened textura angularis with cells 5–19 μm diam. Interascal tissue formed of paraphyses, hyaline, smooth-walled, cylindrical with a rounded tip, disintegrating quickly, septate, apically free, 40–70 μm long, base 4.5–6 μm wide, branched at the base. Asci unitunicate, 8-spored, cylindrical to clavate, fasciculate, 80.5–93 × 8.5–12 μm. Ascospores uni- or biseriately arranged, aseptate, initially hyaline, turning pale brown with age, smooth-walled, allantoid to fusiform, with both ends rounded, (13–)15–19(–21) × (5–)5.5–6(–7) μm, mean ± SD = 17.0 ± 2.1 × 5.7 ± 0.4 μm, L/W ratio = 3.0.
Asexual morph on SNA (mostly based on strain CBS 903.69, UTFC 260 and UTFC 261, only few conidia observed in strain CBS 125474). Vegetative hyphae 1–8 μm diam, hyaline to pale brown, smooth-walled, septate, branched. Chlamydospores not observed. Conidiomata, conidiophores and setae formed directly on hyphae. Setae not observed in strain CBS 125474, setae of strain CBS 903.69 medium brown, verruculose to verrucose, 40–130 μm long, 1–(2)-septate, base cylindrical to slightly inflated, 3–6 μm diam, tip ± rounded to ± acute, setae of strain UTFC 260 2–3-septate. Conidiophores and conidiogenous cells not observed in strain CBS 125474, conidiophores of strain CBS 903.69 hyaline to pale brown, smooth-walled, septate, branched, to 40 μm long. Conidiogenous cells of strain CBS 903.69 hyaline to pale brown, smooth-walled, cylindrical clavate to doliiform, 6.5–19 × 3.5–4.5 μm, often integrated, some polyphialides observed, opening 1–2 μm diam, collarette 0.5–1 μm long, periclinal thickening distinct. Conidia (only three observed in strain CBS 125474) hyaline, smooth-walled, aseptate, straight, cylindrical, the apex and base rounded, 15–17 × 5.5 μm, mean ± SD = 16.0 ± 0.9 × 5.6 ± 0.1 μm, L/W ratio = 2.9, conidia of strain CBS 903.69 shorter, measuring (9.5–)11.5–14(–15) × (4.5–)5–6 μm, mean ± SD = 12.7 ± 1.3 × 5.4 ± 0.4 μm, L/W ratio = 2.4, conidia of strain UTFC 261 shorter and narrower, measuring (10–)10.5–11.5(–12.5) × (3.5–)4–4.5(–5) μm, mean ± SD = 11 ± 1.1 × 4.5 ± 0.5 μm, L/W ratio = 2.6, conidia of strains UTFC 260 and UTFC 261 sometimes slightly curved. Appressoria single, pale, medium to dark brown, smooth-walled, navicular to bullet-shaped or irregular in outline, with an undulate, crenate to strongly lobate margin, (9–)12.5–18.5(–22.5) × (4.5–)6.5–11.5(–15.5) μm, mean ± SD = 15.4 ± 3.0 × 8.8 ± 2.6 μm, L/W ratio = 1.7, appressoria of strain CBS 903.69 smaller, measuring (5–)8.5–15.5(–24.5) × (5–)6.5–9(–10) μm, mean ± SD = 11.9 ± 3.4 × 7.6 ± 1.3 μm, L/W ratio = 1.6.
Asexual morph on Anthriscus stem. Conidiomata, conidiophores and setae formed on pale brown, angular cells, 2.5–6.5 μm diam. Setae medium brown, verruculose to verrucose, 40–95 μm long, 1–2-septate, base conical, slightly inflated, 4.5–8 μm diam, tip ± acute. Conidiophores pale brown, smooth-walled, simple or septate and branched, to 30 μm long. Conidiogenous cells pale brown, smooth-walled, cylindrical, clavate to doliiform, 7–19 × 4–5.5 μm, opening 1–1.5 μm diam, collarette 0.5–1 μm long, periclinal thickening visible, sometimes distinct. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical, sometimes slightly clavate, the apex and base rounded, (15–)16.5–20(–22.5) × (5–)5.5–6.5(–8) μm, mean ± SD = 18.4 ± 1.8 × 5.9 ± 0.5 μm, L/W ratio = 3.1, conidia of strain CBS 903.69 shorter, measuring (12.5–)13.5–16.5(–16.5) × (5–)5.5–6 μm, mean ± SD = 14.6 ± 1.1 × 5.6 ± 0.2 μm, L/W ratio = 2.6.
Culture characteristics: Colonies on SNA flat with entire margin, hyaline to pale olivaceous grey, agar medium, filter paper and Anthriscus stem partly covered with pale olivaceous grey aerial mycelium and grey conidiomata/ascomata, reverse same colours; growth 27.5–31 mm in 7 d (≥ 40 mm in 10 d). Colonies on OA flat with entire margin; olivaceous buff, grey olivaceous to olivaceous, partly covered with short felty whitish aerial mycelium and pale purplish grey to pale olivaceous grey conidiomata/ascomata, reverse pale olivaceous grey to olivaceous grey, growth 28.5–31.5 mm in 7 d (≥ 40 mm in 10 d). Conidial mass greyish white to very pale salmon, in strain UTFC 260 pale buff to orange.
Materials examined: Benin, Dahomey, Porto Novo, from leaf spot of Phaseolus lunatus, collection date and collector unknown (isolated by G. Weststeijn and deposited in CBS collection Oct. 1969 by G. Weststeijn), CBS H-21497, culture CBS 903.69. Brazil, from Gossypium sp., collection date and collector unknown, culture CBS 132443 = CPC 18211; from Gossypium sp., collection date and collector unknown, culture CBS 132444 = CPC 18212. Iran, Golestan province, Gorgan, from leaves of Spathiphyllum wallisii, May 2013, A. Alizadeh, strain UTFC 260 = 176C; Guilan province, Koochesfehan area, from pods of Phaseolus vulgaris, Oct. 2013, O. Atghia & A. Alizadeh, strain UTFC 261 = 513C. Vietnam, Da Lat-Lam Dong, from anthracnose on leaf of Coffea sp., collection date unknown, P. Nguyen & E. Liljeroth (CBS H-21496 holotype, culture ex-holotype CBS 125474 = LD11(L3)); Da Lat-Lam Dong, from anthracnose on leaf of Coffea sp., collection date unknown, P. Nguyen & E. Liljeroth, culture CBS 125473 = LD 33(L1).
Notes: Colletotrichum plurivorum was first described as C. sichuanensis from Capsicum annuum in the Sichuan Province of China (Liu et al. 2016). The species is based on four “holotype living cultures”, one of which was designated as holotype; no type specimen has been deposited in a fungarium, and the name is therefore invalid (Art. 40.1). This species is described as a new species based on a strain from Coffea in Vietnam (Nguyen et al. 2010). In a study of Douanla-Meli et al. (2018), C. sichuanensis was regarded as a synonym of C. cliviicola (as C. cliviae), although both species formed well supported clades in the phylogeny of that study based on a multilocus data set and there was no indication of disconcordance between the gene trees. Based on the data of our study confirming these clades, C. cliviicola and C. plurivorum are regarded as distinct species.
Colletotrichum plurivorum belongs to the C. orchidearum species complex. It has a large host range, including Anacardiaceae (Mangifera), Araceae (Amorphophallus, Spathiphyllum), Caricaceae (Carica), Fabaceae (Glycine, Phaseolus), Malvaceae (Abelmoschus, Gossypium), Musaceae (Musa), Orchidaceae (Arundina, Cymbidium, Oncidium), Passifloraceae (Passiflora), Rubiaceae (Coffea), Solanaceae (Capsicum, Lycopersicon) and Theaceae (Camellia). Based on DNA sequence comparisons, strains from Vitis spp. (Vitaceae) in Brazil (Santos et al. 2018) and China (Lei et al. 2016), Dioscorea (Dioscoreaceae) in Puerto Rico (S.E. Fuentes Aponte et al., unpubl. data), Myrianthus arboreus (Urticaceae) in Cameroon and Citrus limon (Rutaceae) in Vietnam (Douanla-Meli et al. 2018), previously identified as C. cliviae or Colletrichum sp. were revealed to be C. plurivorum as well. Strains from studies on lima beans (Phaseolus lunatus) in Brazil (Sousa et al., 2018, Cavalcante et al., 2018) probably also belong to this species, based on sequence comparisons of GAPDH and ACT sequences (TUB2 was mixed-up with a C. truncatum strain) or a multilocus phylogeny, respectively. Many species have been reported or were described from these hosts (Farr & Rossman 2017, this study). It is likely that this species has been described previously in the pre-molecular era based on morphology. Due to the small differences in morphology (e.g. conidial size and shape) between the species of the C. orchidearum complex on the one hand and the large interspecific variation on the other, it is hardly possible to link such old species.
Colletotrichum plurivorum formed a sexual morph in culture. However, some strains predominantly formed a sexual morph, while the asexual morph was hardly or not observed; other strains predominantly formed an asexual morph, while the sexual morph was lacking or both morphs were formed. Similar observations were made in other studies. For example, strain CGMCC 3.17358 from Camellia in China (Liu et al. 2015) was sterile on SNA and PDA, but developed the sexual morph on Anthriscus stems; no asexual morph was observed. In contrast, strains from Orchidaceae in Yang et al. (2011) formed both a sexual and an asexual morph. In the study of Douanla-Meli et al.(2018) isolates of C. plurivorum (as C. cliviae) from Citrus limon in Vietnam only produced the sexual morph, while isolates from Myrianthus arboreus in Cameroon produced both the asexual and the sexual morph.
In pathogenicity tests by Liu et al. (2016) strains described as C. sichuanensis from Capsicum annuum in China were able to infect fruits of Capsicum annuum and Pyrus pyrifolia. The pathogenicity of strain UTFC 261 from Phaseolus vulgaris in Iran was confirmed by Atghia (2015).
Colletotrichum plurivorum can be identified by GAPDH, HIS3, and TUB2 sequences with few nucleotides difference in each gene to the closely related C. cliviicola. The asexual morph of the two species is similar. However, C. plurivorum is slower growing on SNA and OA than C. cliviicola. Morover, microcyclic conidiation and the formation of anastomoses were observed in the ex-type strain of C. cliviicola, but not in C. plurivorum, while no sexual morph is so far known in C. cliviicola.
In a blastn search in GenBank with the TUB2 sequence of the ex-type strain CBS 125474 the sequences of several strains were found to be 100 % identical: C. cliviae strains AH1B6, AH1B5 (Wang et al. 2016b) and LF774 (Liu et al. 2015) from tea plants on China, of which the latter was included in this study (as CBS 125375), Colletotrichum sp. strain FJ074 from grapevine in China (Lei et al. 2016) as well as Colletotrichum isolates LJTJ3, LJTJ22 and LJTJ30 that were described by Liu et al. (2016) as C. sichuanensis and included in this study. The closest matches with the HIS3 sequence of the ex-type strain are with 99 % identity (1 nucleotide difference) six C. cliviae strains from soybeans in Brazil (Barbieri et al. 2017), of which one, strain LFN0008, is included in this study. The GAPDH sequence of the ex-type strain of C. plurivorum is 100 % identical to several strains that were previously identified as C. cliviae, including G61B, G14B, F85241C from Dioscorea alata in Puerto Rico (S.E. Fuentes Aponte et al., unpubl. data), JQS from Zamioculcas zamiifolia in China (Zhou & Li 2017), AV1 from grapevine in Brazil (Santos et al. 2018), AH1A2 and AH1B5 from tea plants on China (Wang et al. 2016b), HP157 from papaya in Mexico (I. Marquez-Zequera, unpubl. data), CORCG2 from Cymbidium in China (Yang et al. 2011) that is included in this study, and the six strains from soybeans in Brazil (Barbieri et al. 2017). The blastn search with the ITS sequence of strain CBS 125474 resulted in ≥ 40 sequences with 100 % identity. Most, if not all of these strains probably represent C. plurivorum as well, but this needs to be confirmed based on GAPDH, HIS3, or TUB2 sequences.
The clades representing C. plurivorum and C. sojae in the phylogeny in this study include strains from Phaseolus spp. in Iran and Benin. The well-known anthracnose pathogen of common beans, C. lindemuthianum had been reported to form sexual morphs in culture that were called Ga. lindemuthiana (Shear & Wood 1913) or Ga. cingulata f. sp. phaseoli (Kimati & Galli 1970). However, in an extensive study on C. lindemuthianum that included the epitypification of this species, several methods were unsuccessfully applied to induce the formation of a sexual morph of this species (Liu et al. 2013). Liu et al. (2013) assumed the sexual morphs not to be conspecific with C. lindemuthianum.
The species studied by Shear & Wood (1913) formed conidia that measure 10.5–16.5 × 4.5–5 μm on corn meal agar and 12–16.5 × 4.5–6 μm on host tissue and curved ascospores that measure 15–22.5 × 4.5–6 μm; it could well be a species of the C. orchidearum complex. The ascospores of C. sojae studied by us are narrower, while the ascospores of C. plurivorum observed in this study and the studies of Liu et al. (2015) and Yang et al. (2011) have a different shape, being only very slightly curved. However, ascospores reported in Liu et al. (2016) from Capsicum in China have a similar shape, and are similarly arranged in the ascus. It is therefore possible that C. plurivorum is the same fungus as “Ga. lindemuthiana”. In contrast, ascospores of Ga. lindemuthiana shown by Rodríguez-Guerra et al. (2005) are broader than both species, measuring 15.5–29 × 4.5–7 μm; the size differs from both. Kimati & Galli (1970) observed two types of asci and ascospores of Ga. cingulata f. sp. phaseoli: (1–)4(–8)-spored asci with mean ascospores dimensions, 20 × 6.5 μm, and 8-spored asci, with ellipsoidal ascospores, measuring on average 10 × 4 μm, that also do not match with any of the species treated in this study.
While no sequence data were included in these papers, few studies showed that strains from Phaseolus vulgaris with sexual morphs and those forming asexual morphs belong to different RAPD groups (Talamini et al. 2006) or clades in phylogenies based on ITS and HMG sequence data (Barcelos et al., 2011, Barcelos et al., 2014), one of which clustered with Ga. magna strains (Barcelos et al. 2014). A blast search with a sequence of strain UFLAG07-3 (KF604738) from that study was 99 % identical (4 nucleotides difference) with Ga. glycines strain LFN0009 from soybean in Brazil (Barbieri et al. 2017), that is included in the C. orchidearum subclade in this study; based on that it is probably none of the species treated in this study. More loci need to be sequenced to resolve the position of this species.
There are also two strains from Gossypium in Brazil that were identified as C. plurivorum. However, the common causal agents of cotton anthracnose and ramulose, C. gossypii var. gossypii and C. gossypii var. cephalosporioides, respectively, belong to the C. gloeosporioides complex (Salustiano et al. 2014, U. Damm, unpubl. data).
Colletotrichum sojae Damm & Alizadeh sp. nov. MycoBank MB824229. Fig. 17.
Fig. 17.
Colletotrichum sojae (B–D, I–V, Y–AE. from ex-holotype culture ATCC 62257. A, E–H. from culture ATCC 11871. W–X. from culture CBS 134.87). A–B. Conidiomata. C. Tip of a seta. D. Base of a seta. I. Seta. E–H, J–M. Conidiophores. N–S. Appressoria. T–U. Conidia. V. Peridium in cross section. W–X. Ascomata. Y. Outer surface of peridium. Z. Ascospores. AA–AD. Immature asci. AE. Disintegrating apex of an ascus apparently after ascospore release. A, C–H, T, V, X–AE. from Anthriscus stem. B, I–S, U, W. from SNA. A–B, W–X. DM. C–V, Y–AE. DIC. Scale bars: A = 100 μm, T = 10 μm, Y = 40 μm. A applies to A–B, W–X. T applies to C–U, V, Y–AE.
Non Vermicularia truncata Schwein., Trans. Am. phil. Soc. 4(2): 230. 1832. Glomerella glycines Lehman & F.A. Wolf, J. Agric. Res., Washington 33(4): 381. 1926. [=Colletotrichum truncatum (Schwein.) Andrus & W.D. Moore].
Etymology: The species epithet is derived from the host plant Glycine max, soybean.
Sexual morph on Anthriscus stem (observed in strains ATCC 62257, CBS 134.87, UTFC 288, UTFC 301 and UTFC 303). Ascomata perithecia, solitary, superficial, non-stromatic, globose to subglobose, ostiolate, pale to medium brown, 80–150 μm diam, glabrous or covered with few stiff hairs, 10–70 μm long. Peridium 10–15 μm thick, composed of 3 layers of pale brown flattened textura angularis. Ascogenous hyphae hyaline, smooth-walled, delicate, rarely visible. Interascal tissue not observed. Asci mostly immature asci of strain ATCC 62257 observed, mature asci apparently disintegrating quickly, unitunicate, 8-spored, cylindrical to clavate, smooth-walled, the base broadly truncate, mature asci of strain UTFC 288 thin-walled, clavate, measuring 77–80 × 11–11.5 μm. Ascospores aseptate, hyaline, smooth-walled, fusiform, curved, sometimes additionally slightly flexed in the middle, with rounded ends, (14–)15.5–23(–33.5) × (3.5–)4–5 μm, mean ± SD = 19.3 ± 3.7 × 4.5 ± 0.3 μm, L/W ratio = 4.3, ascospores of strain UTFC 288 wider, measuring 14.5–20 × 5–6.5 μm, mean ± SD = 17 ± 1.49 × 5.5 ± 0.4, L/W ratio = 3.1.
Sexual morph on SNA (only observed in strain CBS 134.87). Ascomata perithecia, solitary or in clusters, superficial or immersed in the agar medium, non-stromatic, ± globose, ostiolate, pale to medium brown, glabrous. Asci not observed. Ascospores aseptate, hyaline, smooth-walled, fusiform, curved, with rounded ends, (18.5–)20–27(–37) × (4–)4.5–5 μm, mean ± SD = 27.7 ± 3.5 × 4.6 ± 0.3 μm, L/W ratio = 5.2.
Asexual morph on SNA. Vegetative hyphae 1–7.5 μm diam, hyaline to pale brown, smooth-walled, septate, branched. Chlamydospores not observed. Conidiomata, conidiophores and setae formed directly on hyphae. Setae medium brown, smooth-walled to verruculose, 30–75 μm long, 2–3-septate, base cylindrical, conical to slightly inflated, 3.5–6 μm diam, tip ± rounded to slightly acute. Conidiophores hyaline, smooth-walled, simple, sometimes septate, to 25 μm long. Conidiogenous cells hyaline, smooth-walled, cylindrical to ovoidal, often integrated and conical or clavate, 7–22.5 × 3–6.5(–8) μm, opening 1–2 μm diam, collarette 0.5–1 μm long, periclinal thickening distinct. Conidia hyaline, smooth-walled, initially aseptate, becoming septate with age, straight, the apex and base rounded, (11–)14–17(–17.5) × (4.5–)5–6(–6.5) μm, 15.7 ± 1.4 × 5.4 ± 0.4 μm, L/W ratio = 2.9, conidia of strain ATCC 11871 narrower, measuring (14–)15–18(–19.5) × (4–)4.5–5(–5.5) μm, mean ± SD = 16.5 ± 1.5 × 4.9 ± 0.3 μm, L/W ratio = 3.4. Appressoria (only few observed) single, medium brown, smooth-walled, navicular, bullet-shaped or irregular in outline, with an undulate to lobate margin, (9–)12–19(–24) × (6–)8–11.5(–13) μm, mean ± SD = 15.3 ± 3.6 × 9.7 ± 1.8 μm, L/W ratio = 1.6, appressoria of strain UTFC 288 smaller, measuring 11–21 × 5–9.5 μm, mean ± SD = 14.5 ± 3.1 × 7.5 ± 1.4 μm, L/W ratio = 1.9.
Asexual morph on Anthriscus stem. Sporulation sparse. Conidiomata not observed in strain ATCC 62257, conidiophores and setae formed by strain ATCC 11871 on pale brown angular cells, 3–8.5 μm diam. Setae (few observed) medium to dark brown, smooth-walled or verruculose, 50–130 μm long, 1–12-septate, base cylindrical to conical, or ± inflated, 3–8.5 μm diam, tip ± acute to ± rounded. Conidiophores and conidiogenous cells not observed in strain ATCC 62257. Conidiophores in strain ATCC 11871 hyaline to pale brown, smooth-walled, septate, branched, to 40 μm long. Conidiogenous cells in strain ATCC 11871 hyaline to pale brown, smooth-walled, cylindrical, 8–25 × 4–5.5 μm, opening 1–2 μm diam, collarette 0.5 μm long, rarely seen, periclinal thickening observed. Conidia hyaline, smooth-walled, aseptate, straight, the apex and base rounded, (12.5–)14–17.5(–18.5) × 5–6(–6.5) μm, mean ± SD = 15.8 ± 1.5 × 5.5 ± 0.4 μm, L/W ratio = 2.9.
Culture characteristics: Colonies on SNA flat, with entire margin, hyaline to honey, covered by thin, floccose, whitish to pale grey, aerial mycelium, reverse same colours; 28–29 mm in 7 d (38.5–≥ 40 mm in 10 d). Colonies on OA flat with entire margin, buff, olivaceous buff to olivaceous, partly covered with thin, floccose, whitish aerial mycelium, reverse buff to olivaceous grey, 30–32.5 mm in 7 d (≥ 40 mm in 10 d). Conidial mass not observed in strain ATCC 62257, those of strain UTFC 288 pale yellow.
Materials examined: Iran, Golestan province, Tooskestan area, isolated from leaves of Glycine max, Oct. 2013, A. Alizadeh & O. Atghia, strain UTFC 288 = 442C; Guilan province, Roodsar area, from leaves of Phaseolus vulgaris, Oct. 2013, A. Alizadeh & O. Atghia, strain UTFC 301 = 507C; Guilan province, Loolman-Astaneh ashrafie road, from leaves of Vigna unguiculata, Oct. 2013, A. Alizadeh & O. Atghia, strain UTFC 303 = 524C. Italy, Bologna, from Glycine max, unknown collection date and collector (sent to CBS Jan. 1987 for identification by P. Giunchi, Osservatorio Malattie Delle Piante, Bologna, Italy, identification by H.A. van der Aa, No. 9944), CBS H-12958, culture CBS 134.87. Serbia and Montenegro, Novi Sad, from Glycine max, unknown collection date, Z. Klocokar-Smit (isolated 1980 by Z. Klocokar-Smit and deposited in CBS collection Jan. 1981), CBS H-12907, culture CBS 181.81; Novi Sad, from Glycine max, unknown collection date and collector (isolated 1980 by Z. Klocokar-Smit and deposited in CBS collection Jan. 1981), culture CBS 182.81. USA, Illinois, from Glycine max, unknown collection date, J.B. Manandhar (received from R. O'Connell as Ga. glycines, before from J.B. Sinclair) (CBS H-21495 holotype of C. sojae, culture ex-holotype ATCC 62257 = CPC 19367); Iowa, from stem of Medicago sativa, unknown collection date and collector (received from R. O'Connell as Ga. glycines, before from L.H. Tiffany), culture ATCC 11871 = CPC 18948; Iowa, from stem of Medicago sativa unknown collection date and collector (received from R. O'Connell as C. destructivum, before from A. Alfaro, originally from L.H. Tiffany), culture CBS 128510 = LARS 204 = CECT 2873 = Alfaro 212 = ATCC 11871 = DSM 1167 (apparently the same strain as above); North Carolina, Raleigh, from anthracnose of Glycine max, unknown collection date, S.G. Lehman & F.A. Wolf [BPI 596658 (dried culture) holotype of Ga. glycines]; North Carolina, Raleigh, from anthracnose of Glycine soja (=G. max), unknown collection date and collector (isolated by S.G. Lehman as strain 1936, deposited in CBS collection as Ga. glycines in May 1932 by J.A. Stevenson), (strain CBS 195.32 ex-holotype strain of Ga. glycines).
Notes: Soybean (Glycine max) anthracnose was first reported in 1917 from Korea, where it was attributed to C. glycines (Hemmi 1920). Anthracnose occurs in all soybean producing areas and can cause yield losses of up to 100 % (Sinclair 1982). Lehman & Wolf (1926) studied soybean anthracnose in the USA and noticed an asexual morph they identified as C. glycines and a sexual morph they regarded as the sexual morph of this species, hence naming it Ga. glycines.
The only specimen located that could directly be linked to Lehman & Wolf is BPI 596658. A handwritten note with that specimen reads “ATCC #1936, G. glycines (Hori) n.n. (n.n. was later crossed out and replaced by “Lehman & Wolf”), S.G. Lehmann and Frederick A. Wolf, Jour. Agric. Research 33: 381–390, Aug. 15, 1926”, referring to the publication of that species. We therefore regard this specimen as the holotype of Ga. glycines.
Specimen BPI 596658 contains two dried cultures of Ga. glycines strain ATCC 1936 (Label on tubes: Glomerella glycines, A.T.C.C. # 1936, 11-19-1929, Vaxter agar on soybean). No conidia or ascospores were observed on these cultures. The same number (1936) appeared with the strain information of strain CBS 195.32 that was isolated by S.G. Lehman from Glycine max in Raleigh, North Carolina, USA, originally identified as Ga. glycines and deposited to the CBS collection in May 1932 by J.A. Stevenson. This means CBS 195.32 is apparently the same strain as ATCC 1936 and therefore regarded as the ex-holotype strain of Ga. glycines. The ATCC collection lists a strain ATCC 1936, but recorded as “unknown”. According to a request at ATCC (A. Khashnobish, in lit.), no strain ATCC 1936 exists anymore.
However, strain CBS 195.32 was identified as C. truncatum by Damm et al. (2009). Drawings and description of the asexual morph of Ga. glycines observed by Lehman & Wolf (1926) agree with this. Colletotrichum truncatum is a common pathogen of soybean (Ramos et al., 2013, Yang et al., 2014, Rogério et al., 2016). However, it is dubious that this soybean pathogen formed a sexual morph in the study of Lehmann & Wolf, as a sexual morph was never observed for any strain of C. truncatum, neither in the extensive study of Damm et al. (2009) nor in any other study on this pathogen (e.g. Shenoy et al., 2007, Than et al., 2008, Rogério et al., 2016).
Lehman & Wolf (1926) stated that they made isolations “from diseased stems and pods bearing the conidial stage, from infected seed, and from old, decaying stems bearing the ascogenous stage” and further: “No efforts have been made to make single-spore cultures from either conidia or ascospores...” There is no indication that the authors would have found conidia and ascospores on the same plant parts. They transferred blocks of agar containing several spores each. Although the authors did not observe cultural differences, which was obviously the reason for them to conclude they were studying one species, it is likely that two species were isolated and even mixed cultures were prepared: “Some strains from conidia have born conidia alone, even after repeated transfer and cultivation on a variety of media; other strains from ascospores, however, have consistently yielded both the conidial and ascospore stages when cultivated upon the same kind of media” (Lehman & Wolf 1926). So, it is likely that the sexual morph and the asexual morph observed by them are different species. Yang et al. (2014) reported two species to be common pathogens of soybean in the USA, namely “Ga. glycines” and C. truncatum. Apparently, Lehman & Wolf never noticed the asexual morph that really belongs to the observed sexual morph.
According to Tiffany & Gilman (1954) and Lin & Wu (1966), the asexual morph of Ga. glycines is not C. glycines, but a species similar to C. destructivum. Manandhar et al. (1986) claimed C. destructivum to be the asexual morph of Ga. glycines by morphological comparison of strains from Glycine forming a sexual morph as described for Ga. glycines (Lehman & Wolf 1926) that were therefore considered to be Ga. glycines, and strains from Medicago that were considered to be C. destructivum and gave a detailed description of both morphs. Strains from Manandhar et al. (1986) from both hosts (ATCC 62257 from Glycine max and ATCC 11871 from Medicago), were included in this study and confirmed to be conspecific. However, these strains are not closely related to C. destructivum that was epitypified in a recent study and belongs to the C. destructivum complex (Damm et al. 2014).
One of the two strains from Manandhar et al. (1986) easily formed the very typical sexual morph in culture that agrees with the pictures and the description of Lehman & Wolf (1926): perithecia 220–340 μm diam, asci 70–106 × 9.5–13.5 μm, mean = 80 × 12 μm; ascospores in culture were (13.12–)18.75–28.12(–43.35) μm long. The ascospores of C. plurivorum, also occurring on soybeans and also described in this study, have a different shape (see Fig. 16).
The name Ga. glycines was applied by Lehman & Wolf (1926) based on the asexual morph that was identified by them as C. glycines and the simultaneously observed sexual morph that was apparently wrongly connected to the asexual morph. However, the type specimen of Ga. glycines only includes the asexual morph that was re-identified as C. truncatum by Damm et al. (2009). Colletotrichum truncatum is based on Vermicularia truncata (Schweinitz 1832), which has priority over Ga. glycines. There is no name available for the second species that forms the sexual morph. Consequently, this species is described here as a new species, C. sojae.
According to von Arx (1957), C. glycines Hori ex Hemmi is a synonym of C. dematium f. truncata, which is a synonym of C. truncatum (Damm et al. 2009). However, von Arx neither saw the type specimen nor a strain of C. glycines. In the Compendium of Soybean Diseases, the two species on soybean are treated as C. truncatum (= C. glycines) and Ga. glycines (Sinclair 1982). It is possible that C. glycines and C. truncatum are synonyms as both types represent species with curved conidia. However, Yang et al. (2014) collected three Colletotrichum species with curved conidia on soybeans in the USA, C. incanum, C. chlorophyti and C. truncatum. Colletotrichum glycines Hori ex Hemmi could be a synonym of any of these species, or even a different species. We did not study the type of C. glycines and there is no ex-type strain of C. glycines available for molecular analysis. In order to clarify its systematic position, C. glycines would need to be epitypified.
Another species named C. glycines, C. glycines Gonz. Frag. (González Fragoso 1924) was not described from soybean, but from Wisteria sinensis (syn.: Glycine sinensis) in Portugal. The name is illegitimate, according to Art. 53.1.
Within the strains of this species there is considerable variation in the formation of sexual and asexual morphs. There are strains that form the asexual morph only and strains that predominantly form the sexual morph. It is very likely the latter case is the reason for Lehman & Wolf (1926) to overlook the asexual morph of this species.
Colletotrichum sojae occurs mainly on Fabaceae (Glycine, Medicago, Phaseolus, Vigna), but also on Amaranthaceae (Amaranthus), Asteraceae (Arctium), Solanaceae (Capsicum) and Orchidaceae (Bletilla). There is another species occurring on Fabaceae (including Glycine and Phaseolus) treated in this study (see C. plurivorum).
Colletotrichum sojae can be identified by HIS3, ACT and TUB2 sequences, while the ITS is identical with that of C. vittalense. Closest matches in blastn searches with the TUB2 sequence of the ex-type strain of C. sojae, ATCC 62257, resulted in 100 % matches with Ga. glycines strain ATCC 62257 (Yang et al. 2014), Colletotrichum strain CGMCC 3.15171 from Bletilla in China (Tao et al. 2013), C. cliviae strain CAUOS5 from Capsicum in China (Diao et al. 2017) and with 99 % identity (1 nucleotide difference) Ga. glycines strains IL18A and IL26A (Yang et al. 2014) and C. orchidearum isolate SAUCC 1407 (Xu et al. 2016), both included in this study. The ACT sequence of strain ATCC 62257 resulted in 100 % identity with the three Ga. glycines strains of Yang et al. (2014), Colletotrichum strains CGMCC 3.15171 from Bletilla (Tao et al. 2013) and CAUGOS3 from Capsicum in China (Diao et al. 2017), all included in this study, as well as Colletotrichum sp. SKH-2010 isolates C07010 and C08116 (GU944760, GU935805, Choi et al. 2011) and with 99 % identity (1 and 2 nucleotides difference), C. orchidearum isolate SAUCC 1407 (Xu et al. 2016) and Ga. glycines strain LFN0009 (Barbieri et al. 2017), both included in this study. The GAPDH of strain ATCC 62257 is 100 % identical with Ga. glycines strains IL26A, ATCC 62257, IL18A (Yang et al. 2014) and LFN0009 (Barbieri et al. 2017) and C. aracearum strains LC1033 and LC1041 (Hou et al. 2016), all included in this paper, as well as Colletotrichum isolates C08116, C07004 and C07010 (GU935864, GU935865, GU935866, Choi et al. 2011). Closest matches in blastn searches with the ITS sequence of the ex-type strain resulted in a large number of identical sequences from several host plants in different countries.
Colletotrichum orchidearum isolate SAUCC 1407 from Arctium lappa in China (Xu et al. 2016), C. cliviae strains LFN0009 (as Ga. glycines in GenBank) from Glycine max in Brazil (Barbieri et al. 2017) and CAUOS5 from Capsicum in China (Diao et al. 2017), Colletotrichum sp. from Bletilla in China (CGMCC 3.15171, Tao et al. 2013) as well as Ga. glycines isolates IL18A, IL26A from soybean in the USA (Yang et al. 2014) could be re-identified as C. sojae based on a sequence comparison in this study. We suspect the ACT sequence KP890098 is actually from CAUOS5 as well, and not from CAUOS3. There is no ACT sequence of CAUOS5 listed in the paper (Diao et al. 2017). Colletotrichum sp. SKH-2010 isolates C07010, C07004 and C08116 were also identified as C. sojae. These strains were cited from Choi et al. (2011) in GenBank, but not included in that paper. As there is no strain information, we also did not include the strains in our phylogeny. In contrast, strain MAFF 238875 from Glycine max in Japan previously identified as C. cliviae was revealed to be C. plurivorum. Moreover, the ITS of “Ga. glycines” isolate IFO 7384 from an unknown host (GenBank AB057435, Moriwaki et al. 2002) is 100 % identical with that of CBS 127604, the ex-holotype strain of C. lentis (Damm et al. 2014).
Colletotrichum tropicicola Phouliv. et al., Cryptog. Mycol. 33(3): 353. 2012. Fig. 18.
Fig. 18.
Colletotrichum tropicicola (A–B, E–R. from culture CBS 127555. C–D from culture CBS 133174). A–B. Conidiomata. C, G. Tips of setae. D–H. Bases of setae. E–F, I–J. Conidiophores. K–P. Appressoria. Q–R. Conidia. A, C–F, Q. from Anthriscus stem. B, G–P, R. from SNA. A–B. DM. C–R. DIC. Scale bars: A = 100 μm, E = 10 μm. A applies to A–B. E applies to C–R.
A description of the type specimen is provided by Noireung et al. (2012). The description below is based on strains from Citrus sp. collected in Mexico.
Sexual morph not observed.
Asexual morph on SNA (CBS 127555). Vegetative hyphae 1.5–10.5 μm diam, hyaline, smooth-walled, septate, branched. Chlamydospores not observed. Conidiomata, conidiophores and setae formed directly on hyphae. Seta (only one observed) medium brown, verruculose, 81 μm long, 3-septate, base cylindrical, 5.5 μm diam, tip ± rounded. Conidiophores hyaline, smooth-walled, septate, branched, to 40 μm long. Conidiogenous cells hyaline, smooth-walled, cylindrical, (3–)5–18 × (2–)3–4.5 μm, often extending to form new conidiogenous loci, opening 1.5–2 μm diam, collarette 1 μm long, periclinal thickening distinct. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical, the apex rounded, sometimes tapering to the rounded or truncate base, (13.5–)14–17.5(–22) × 4.5–5(–5.5) μm, mean ± SD = 15.7 ± 1.7 × 4.8 ± 0.3 μm, L/W ratio = 3.3, conidia of strain CBS 133174 wider, measuring (14–)15–17(–18.5) × (5–)5.5–6 μm, mean ± SD = 16.2 ± 1.0 × 5.5 ± 0.3 μm, L/W ratio = 2.9. Appressoria single or in loose groups, medium to dark brown, smooth-walled, ellipsoidal to bullet-shaped in outline, with an entire or undulate margin, (7–)8–11(–14.5) × (4–)5.5–7(–7.5) μm, mean ± SD = 9.5 ± 1.5 × 6.2 ± 0.7 μm, L/W ratio = 1.6, appressoria of strain CBS 133174 larger, only few observed, measuring 11.5–20(–24.5) × 5–8(–10) μm, mean ± SD = 15.8 ± 4.1 × 6.7 ± 1.6 μm, L/W ratio = 2.4.
Asexual morph on Anthriscus stem (CBS 127555). Conidiomata, conidiophores formed directly on hyphae. Setae not observed. Conidiophores hyaline, smooth-walled, septate, branched, to 30 μm long. Conidiogenous cells hyaline, smooth-walled, cylindrical to ± inflated, 8–25 × 3.5–4.5(–6) μm, opening 1.5–2 μm diam, collarette 0.5 μm long, periclinal thickening distinct. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical, the apex rounded, sometimes tapering to the rounded or truncate base, (13.5–)15–17(–18) × 4.5–5(–5.5) μm, mean ± SD = 15.9 ± 1.0 × 5.0 ± 0.2 μm, L/W ratio = 3.2, conidia of strain CBS 133174 larger, measuring (15.5–)17–19.5(–20) × 5–5.5 μm, mean ± SD = 18.2 ± 1.0 × 5.4 ± 0.2 μm, L/W ratio = 3.4.
Cultural characteristics (CBS 127555). Colonies on SNA flat with entire margin, medium buff to pale honey, Anthriscus stem, filter paper and SNA medium partly covered with greyish to orange acervuli and whitish to grey aerial mycelium, reverse buff to honey with grey to orange acervuli shining through; 15–18 mm in 7 d (28.5–32 mm in 10 d). Colonies on OA flat with entire margin, surface buff, pale olivaceous grey to olivaceous grey, partly covered with floccose to felty pale olivaceous grey to olivaceous grey aerial mycelium and grey to orange acervuli, reverse buff, rosy buff, pale olivaceous grey to olivaceous grey; 16–17.5 mm in 7 d (30–32.5 mm in 10 d). Conidial mass orange.
Materials examined: Mexico, Tamaulipas, from Citrus sp., 16 Oct. 2008, M. de Jesus Yanez-Morales, CBS H-21070, culture CBS 127555 = CPC 15927; Tamaulipas, from Citrus sp., 16 Oct. 2008, M. de Jesus Yanez-Morales, culture CBS 133174 = CPC 15924.
Notes: This species belongs to the C. dracaenophilum species complex. Noireung et al. (2012) described C. tropicicola from leaves of Citrus maxima and Paphiopedilum bellatolum in Thailand. Further strains from Citrus sp. in Mexico could be identified as C. tropicicola in this study based on the multilocus phylogeny. While these two strains formed a well-supported clade with the ex-type strain of C. tropicicola in our phylogeny, the strain from Paphiopedilum formed a sister clade to C. excelsum-altitudinum and could represent a further species (referred to as “C. tropicicola 2” in the phylogeny). Thus C. tropicicola s. str. only occurs on Citrus spp., according to our data.
Colletotrichum tropicicola can be identified with sequences of all loci available (ITS, GAPDH, ACT, TUB2), but best with GAPDH and TUB2. The ITS sequence of the ex-type strain, MFLUCC 11-0114, is 99 % identical (2, 7 and 6 nucleotides difference) to those of the apparently wrongly identified C. cordylinicola strain LC0886 (KC790944, Sharma et al. 2013a), the two C. excelsum-altitudinum isolates (Tao et al. 2013) and MFLUCC 10-0167, the C. tropicicola strain from Paphiopedilum, respectively. The GAPDH sequence of the ex-type strain is 99 % identical (4 nucleotides difference) to that of the C. tropicicola strain from Paphiopedilum and 96 % identical with the two C. excelsum-altitudinum isolates. The ACT sequence of the ex-type strain is 99 % identical (3 and 11 nucleotides difference) to that of the C. tropicicola strain from Paphiopedilum and the two C. excelsum-altitudinum isolates, respectively. The TUB2 sequence of the ex-type strain is 98 % identical (10 and 8 nucleotides difference) with the two original C. excelsum-altitudinum isolates and that of the C. tropicicola strain from Paphiopedilum, respectively.
Colletotrichum vittalense Damm, sp. nov. MycoBank MB824230. Fig. 19.
Fig. 19.
Colletotrichum vittalense (from ex-holotype culture CBS 181.82). A–B. Conidiomata. C, G. Tips of setae. D–H. Bases of setae. E–F, I–K. Conidiophores. L–Q. Appressoria. R–S. Conidia. T. Outer surface of peridium. U. Peridium in cross section. V. Paraphyses. W. Ascoma. X. Ascospores. Y–Z. Asci. A, C–F, R, U, W–Z. from Anthriscus stem. B, G–Q, S–T, V. from SNA. A–B, W. DM. C–V, X–Z. DIC. Scale bars: A = 100 μm, E = 10 μm. A applies to A–B, W. E applies to C–V, X–Z.
Etymology: The species epithet is derived from Vittal, the place in Karnataka, India, where the type was collected.
Sexual morph on SNA (only observed in strain CBS 181.82). Ascomata perithecia, formed after 4 wk, solitary, superficial or immersed in the agar medium, non-stromatic, globose to obpyriform, ostiolate, glabrous, dark brown, 250–300 × 200–250 μm. Peridium 10–15 μm thick, composed of 3–5 layers of medium brown flattened textura angularis with cells 5.5–15 μm diam. Ascogenous hyphae hyaline, smooth, delicate, rarely visible. Interascal tissue formed of paraphyses, hyaline, smooth-walled, cylindrical, disintegrating quickly, septate, apically free. Asci unitunicate, 8-spored, cylindrical to clavate, tapering to apex and base, smooth-walled, 53–83 × 9–11 μm, the base broadly truncate. Ascospores uni- or biseriately arranged, aseptate, hyaline to pale brown, smooth-walled, allantoid, fusiform to ellipsoidal, with both ends rounded, curved or straight, (11.5–)14.5–19(–23.5) × (4–)4.5–5.5(–6) μm, mean ± SD = 16.8 ± 2.3 × 5.1 ± 0.4 μm, L/W ratio = 3.3.
Sexual morph on Anthriscus stem (only observed in strain CBS 181.82). Ascomata perithecia, formed after 4 wk, solitary, superficial, non-stromatic, globose to obpyriform, ostiolate, glabrous, dark brown, 150–320 × 130–200 μm diam. Peridium 11–13 μm thick, composed of 4–6 layers of medium brown flattened textura angularis with cells 5–15.5 μm diam. Paraphyses not observed. Asci 8-spored, unitunicate, cylindrical to clavate, 53–83 × 9–11 μm. Ascospores uni- or biseriately arranged, aseptate, hyaline to pale brown, smooth-walled, allantoid, fusiform to ellipsoidal, with both ends rounded, curved or straight, (12–)14–16.5(–17.5) × (4.5–)5–5.5(–6) μm, mean ± SD = 15.1 ± 1.3 × 5.3 ± 0.4 μm, L/W ratio = 2.8.
Asexual morph on SNA. Vegetative hyphae 1.5–6.5 μm diam, hyaline to pale brown, smooth-walled, septate, branched. Chlamydospores not observed. Conidiomata, conidiophores and setae formed directly on hyphae. Setae medium to dark brown, verruculose to verrucose, 50–120 μm long, 1–3-septate, base cylindrical to conical, 3.5–5.5 μm diam, tip ± acute. Conidiophores pale brown, smooth-walled to verruculose, septate, branched, to 60 μm long. Conidiogenous cells pale brown, smooth-walled to verruculose, cylindrical to clavate, 7–22.5 × 2.5–6.5 μm, opening 1.5–2 μm diam, collarette 0.5–1 μm long, periclinal thickening distinct, conidiogenous cells of strain CBS 126.25 sometimes integrated. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical, sometimes slightly clavate with the widest part at the base, the apex and base rounded, (11.5–)13.5–16(–18.5) × 5–5.5(–7) μm, mean ± SD = 14.8 ± 1.4 × 5.4 ± 0.4 μm, L/W ratio = 2.8, conidia of strain CBS 126.25 slightly longer, measuring (11–)12.5–15(–18) × 5–5.5 μm, mean ± SD = 13.6 ± 1.2 × 5.2 ± 0.2 μm, L/W ratio = 2.6. Appressoria single or in loose groups, medium to dark brown, smooth-walled, navicular, bullet-shaped, elliptical or irregular in outline, with a lobate or undulate margin, (6–)9.5–16.5(–22.5) × (5–)7.5–11(–14.5) μm, mean ± SD = 13.1 ± 3.5 × 9.1 ± 1.7 μm, L/W ratio = 1.4, appressoria of strain CBS 126.25 smaller, measuring (7–)8.5–14.5(–18.5) × (5–)5.5–8(–9.5) μm, mean ± SD = 11.3 ± 3.0 × 6.8 ± 1.1 μm, L/W ratio = 1.7.
Asexual morph on Anthriscus stem. Conidiomata, conidiophores and setae formed on medium brown, angular cells, 3–7 μm diam. Setae medium brown, verruculose to verrucose, 55–120 μm long, 1–2(–3)-septate, base cylindrical to conical, 4.5–7.5 μm diam, tip ± acute. Conidiophores pale brown, smooth-walled. Conidiogenous cells pale brown, smooth-walled, cylindrical to doliiform, 7.5–10.5 × 3.5–5.5 μm, opening 1–2 μm diam, collarette ± 0.5 μm long, periclinal thickening distinct. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical, the apex and base rounded, 15–17(–18.5) × 5–5.5(–6) μm, mean ± SD = 16.1 ± 0.9 × 5.3 ± 0.3 μm, L/W ratio = 3.0, conidia of strain CBS 126.25 shorter, measuring (12.5–)13.5–15.5(–17.5) × 5–5.5 μm, mean ± SD = 14.5 ± 1.0 × 5.2 ± 0.2 μm, L/W ratio = 2.8.
Culture characteristics: Colonies on SNA flat with entire margin, hyaline to pale olivaceous grey, agar medium, filter paper and Anthriscus stem partly covered with felty white to pale olivaceous grey aerial mycelium and grey to black conidiomata/ascomata, reverse same colours; growth 29–30.5 mm in 7 d (≥ 40 mm in 10 d), strain CBS 126.25 grows more slowly: 22.5–24 mm in 7 d (31–32.5 mm in 10 d). Colonies on OA flat with entire margin; olivaceous buff, grey olivaceous to olivaceous, partly covered with short felty whitish aerial mycelium and pale grey conidiomata/ascomata, reverse olivaceous buff, pale olivaceous grey to olivaceous grey, growth 28.5–30 mm in 7 d (≥ 40 mm in 10 d), strain CBS 126.25 grows more slowly: 24–26 mm in 7 d (28.5–34 mm in 10 d). Conidial mass greyish white.
Materials examined: India, Karnataka, Vittal, from Theobroma cacao (pathogenic), collection date and collector unknown (isolated and deposited in CBS collection by F.H. Beyma, May 1928) (CBS H-21498 holotype, culture ex-holotype CBS 181.82). Unknown, from an unknown Orchidaceae plant, collection date and collector unknown (isolated and deposited in CBS collection by K.N. Murthy, Mar. 1928), CBS H-21499, culture CBS 126.25.
Notes: Colletotrichum vittalense belongs to the C. orchidearum species complex and is only known as pathogen of Theobroma cacao in India and from an unknown Orchidaceae plant.
Several species were described on Theobroma cacao, including C. cacao in the C. magnum complex that is also described in this study. Numerous species were described on Orchidaceae hosts, including C. cattleyicola, also belonging to the C. orchidearum species complex and C. orchidearum belonging to the C. orchidearum species complex described and epitypified, respectively, in this study. See notes under C. cacao, C. cattleyicola and C. orchidearum for further species and reports on these hosts. The two C. vittalense strains from the CBS collection were previously identified as Ga. cingulata var. cingulata and C. gloeosporioides, Ga. lagenaria and Gl. affine, the latter is a species that was described from Hoya (Apocynaceae) and Vanilla (Orchidaceae) from the botanical garden in Padua, Italy, with similar conidial shape (cylindrical with both ends rounded) and size (14–20 × 4–6 μm) as C. vittalense (Saccardo 1878). However, we could not locate the type material of this species to confirm its taxonomic position.
A typical feature of C. vittalense are the very short ascospores with round ends that vary between straight to strongly curved. This species can be identified by its unique GAPDH, CHS-1, HIS3 and TUB2 sequences. In a blastn search on GenBank with the GAPDH sequence of the ex-type strain of C. vittalense, CBS 181.82, the only 100 % identical sequence was C. cliviae strain GUFCC15503 from Calamus thwaitesii in India (KC790759, Sharma et al. 2013a) that was included in the phylogeny of this study and re-identified as C. vittalense. Strain GUFCC15503 originates from Netravali in Goa, also in the south-west of India like Vittal. The GAPDH sequence of the ex-type strain of C. cliviicola differs in 2 nucleotides from that of the ex-type strain of C. vittalense. Strain GUFCC15503 (KF451996, Sharma et al. 2015) was also the closest match in a blastn search with the ACT, CHS-1 and TUB2 sequences of strain CBS 181.82 with none, one or two nucleotides difference, respectively. The ITS sequence of strain CBS 181.82 is 100 % identical with that of strains from various hosts in different countries from different studies, including C. sojae strains from Glycine max in the USA (Yang et al. 2014), that were included in this study. The ITS of the ex-type strain of C. cliviicola differs in 3 nucleotides.
Colletotrichum yunnanense Xiao Ying Liu & W.P. Wu, Mycotaxon 100: 139. 2007. Fig. 20.
Fig. 20.
Colletotrichum yunnanense (from ex-holotype culture CBS 132135). A–B. Conidiomata. C, G. Tips of setae. D–H. Bases of setae. E–F, I–K. Conidiophores. L–Q. Appressoria. R–S. Conidia. T. Ascoma. U. Peridium in cross section. V. Outer surface of peridium. W–X. Asci. Y. Tip of an ascus. Z. Paraphyses. AA. Ascospores. A, C–F, R. from Anthriscus stem. B, G–Q, S–AA. from SNA. A–B, T. DM. C–S, U–AA. DIC. Scale bars: A = 100 μm, F = 10 μm. A applies to A–B, T. F applies to C–S, U–AA.
Sexual morph on SNA. Ascomata perithecia, solitary, superficial or immersed in the agar medium, non-stromatic, subglobose, ostiolate, medium to dark brown, 150–200 × 140–260 μm. Peridium 10–13 μm thick, composed of 4–6 layers of pale brown textura angularis. Ascogenous hyphae hyaline, smooth-walled, delicate. Interascal tissue formed of paraphyses, hyaline, smooth-walled, cylindrical, apically free, tapering towards the round tip, disintegrating quickly, septate, branched, 50–65 μm long, base 2–2.5 μm diam. Asci unitunicate, 8-spored, cylindrical to clavate, tapering to apex and base, smooth-walled, 57–65.5 × 10.5–11.5 μm, the base broadly truncate. Ascospores uni- or biseriately arranged, aseptate, hyaline, smooth-walled, fusiform with acute ends, slightly curved, (13.5–)14.5–17.5(–19.5) × 5–5.5(–6) μm, mean ± SD = 16.1 ± 1.7 × 5.4 ± 0.3 μm, L/W ratio = 3.0.
Asexual morph on SNA. Vegetative hyphae 1–9 μm diam, hyaline, smooth-walled, septate, branched. Chlamydospores not observed. Conidiomata, conidiophores and setae formed directly on hyphae. Setae medium brown, verrucose, warts up to 2.5 μm diam, 70–130 μm long, 2–5-septate, base slightly inflated, 4–6.5 μm diam, tip ± acute. Conidiophores pale brown, smooth-walled, septate, branched, to 40 μm long. Conidiogenous cells pale brown, smooth-walled, clavate, 8–28 × 5–7 μm, the upper part often surrounded by a gelatinous sheath, opening 1.5–2 μm diam, collarette 0.5 μm long, periclinal thickening observed. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical, the apex and base rounded, with a prominent scar, (13–)15.5–18(–19.5) × 4.5–5(–5.5) μm, rarely up to 29 μm long, mean ± SD = 16.7 ± 1.2 × 5.0 ± 0.2 μm, L/W ratio = 3.4. Appressoria in loose groups, medium to dark brown, smooth-walled, subglobose to ellipsoidal in outline, with an entire or lobate margin, (3.5–)5.5–8(–10) × (3–)4.5–7(–7.5) μm, mean ± SD = 6.8 ± 1.3 × 5.8 ± 1.1 μm, L/W ratio = 1.2.
Asexual morph on Anthriscus stem. Conidiomata, conidiophores and setae formed on pale brown, angular cells, 3.5–7.5 μm diam. Setae medium brown, verrucose, 75–120 μm long, 2–3-septate, base cylindrical, conical to slightly inflated, 5–8 μm diam, tip ± rounded. Conidiophores (hyaline to) pale brown, smooth-walled, septate, branched, to 35 μm long. Conidiogenous cells (hyaline to) pale brown, smooth-walled, cylindrical, ellipsoidal to doliiform, 9.5–26 × 3.5–6 μm, the upper part often surrounded by a gelatinous sheath, opening 1–1.5 μm diam, collarette 0.5 μm long, periclinal thickening visible, sometimes distinct. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical, the apex and base rounded, with a prominent scar, (16–)17–18.5(–20.5) × 5–5.5 μm, mean ± SD = 17.7 ± 0.9 × 5.2 ± 0.2 μm, L/W ratio = 3.4.
Culture characteristics: Colonies on SNA flat with entire margin, hyaline, filter paper partly straw, agar medium, filter paper and Anthriscus stem partly covered with grey acervuli and thin whitish aerial mycelium, reverse same colours; growth 10–11.5 mm in 7 d (17–17.5 mm in 10 d). Colonies on OA flat with entire margin; buff to grey olivaceous, partly covered with short whitish to grey aerial mycelium and salmon to grey acervuli, reverse buff, honey, olivaceous to olivaceous-grey, growth 11–12.5 mm in 7 d (18–19.5 mm in 10 d). Conidial mass salmon.
Material examined: China, Yunnan, Kunming Botanical Garden, from healthy leaves of Buxus sp., 5 Nov. 2004, W.P. Wu (WU47182 holotype [not seen], culture ex-holotype CBS 132135 = AS 3.9617 = LC1526).
Notes: Colletotrichum yunnanense was described from healthy leaves of Buxus sp. in Yunnan (China); the species was isolated as an endophyte; no sexual morph was observed by incubating it on PDA at 25 °C in the dark (Holotype WUWP 47182, Liu et al. 2007b). The ex-holotype strain was re-examined on SNA and OA in this study, and the formation of a sexual morph was observed on SNA. Colletotrichum yunnanense is the only species in the C. dracaenophilum complex for which a sexual morph is known. It is the species in this complex with the smallest appressoria; because of their often roundish outline, these appressoria also have the lowest L/W ratio. The growth rate of C. yunnanense is the lowest of all species treated in this study.
There are no reports of this species other than that published by Liu et al. (2007b). Moreover, only one other Colletotrichum species was reported on Buxus: C. theobromicola on Buxus microphylla var. japonica in the USA (Singh et al. 2015).
Colletotrichum yunnanense can be identified with all loci studied. The closest matches with the sequences of all loci of the ex-type strain CBS 132135 in GenBank are all those of this strain itself and of the ITS sequence of strain AS 3.9616, respectively. One further ITS sequence, LN552210, of the endophytic Colletotrichum sp. BS4 from leaves of Buxus sinica in Guangzhou, China (Wang et al. 2016a) was 99 % identical (6 nucleotides difference) to that of strain CBS 132135. The differences in the sequences are likely to be artefacts, as they are in the beginning of the sequence, where there are several Ns. We assume that this strain is also representative of C. yunnanense. The authors discovered three new antibacterial azaphilones, colletotrichones A−C (1−3) and one known compound, chermesinone B (4a). This isolate tested positive against environmental and human pathogenic clinical bacterial strains (Wang et al. 2016a). The boxwood plant, Buxus sinica, is used in traditional Chinese medicine.
Colletotrichum sp. strains GZAAS5.09545 and COUFAL7300
Notes: Based on our multilocus phylogeny, two strains, GZAAS5.09545 from citron leaves (Citrus medica, Rutaceae) in Puer City, Yunnan, China (Peng et al. 2012) and COUFAL7300 from anthracnose of chayote fruits (Sechium edule, Cucurbitaceae) in Antonio Calos, Santa Catarina State, Brazil (Bezerra et al. 2016) formed a clade with high bootstrap support. This clade could represent a further unnamed species in the C. magnum species complex. Both strains were originally identified as C. brevisporum. The latter strain caused lesions on fruits of Sechium edule and Cucumis melo (Bezerra et al. 2016).
Discussion
All three species complexes treated in this study, namely of C. dracaenophilum, C. magnum and C. orchidearum, form straight conidia, similar to those in the C. gloeosporioides species complex (Weir et al. 2012). They could have been regarded as C. gloeosporioides in the past. This is corroborated by records in Farr & Rossman (2017), and previous identifications linked to sequences deposited in GenBank.
Except for C. dracaenophilum that forms larger conidia, the conidial sizes in all three species complexes differ more within the species (provided that more than one strain was available/measured) than between them. Differences were mainly observed among appressoria and ascospore sizes and shapes. But these structures were not always available as they were not formed by all species and cultures. In this study, nine previously described species were assigned to one of the three species complexes treated in this study. Together with the C. orbiculare complex, these species representing the three species complexes constitute the most basal lineages of the genus Colletotrichum (Marin-Felix et al. 2017).
Many species in the three species complexes are possibly host specific — apparently mainly specialised to monocots, while others occur on many hosts. Most of the species were predominately isolated from plants in tropical or subtropical regions of the world, mainly from Asia and Latin America. The rare collections from the temperate climate in Central-Europe are from ornamental plants.
One of the plant families often found as host plants in the three species complexes is the Orchidaceae. This family is often colonised by Colletotrichum species. Farr & Rossman (2017) list 34 Colletotrichum species on Orchidaceae. A number of Colletotrichum species were described on Orchidaceae, including C. orchidophilum in the C. acutatum complex (Damm et al. 2012a), C. cymbidiicola and C. oncidii in the C. boninense complex (Damm et al. 2012b) and C. arxii in the C. gigasporum complex (Liu et al. 2014). There are also several species on Orchidaceae treated in this study: C. excelsum-altitudinum (Tao et al. 2013) and C. coelogynes and a possible further Colletotrichum species on Paphiopedilum in Thailand in the C. dracaenophilum species complex, and C. cattleyicola, C. orchidearum, C. plurivorum, C. sojae and C. vittalense in the C. orchidearum species complex.
The Colletotrichum dracaenophilum species complex contains a few apparently host-specific species. Based on the few strains available, these species seem to be uncommon. Most of the species are rather distantly related; it is not a species complex in the strict sense. All species form cylindrical conidia with round ends, some with sometimes truncate bases or with a basal scar. In some species, we also observed conidiogenous cells that are extending to form new conidiogenous loci or mucous layers in the upper parts of the conidiogenous cells, similar to the C. boninense complex (Damm et al. 2012b). A sexual morph is only known in C. yunnanense.
The Colletotrichum magnum species complex consists of nine closely related species. Except for C. brevisporum and an undescribed species from two independent publications, they are known only from one host species each. However, for some of these species only a single strain is available. A sexual morph is only known from Ga. magna that is heterothallic; the sexual morph was the result of a laboratory crossing (Jenkins & Winstead 1964). The ascospores are curved and very large, larger than those of any other Colletotrichum species treated in this study and probably only exceeded in length by those of C. gigasporum (Rakotoniriana et al. 2013) and C. taiwanense (Sivanesan & Hsieh 1993), the former belonging to the C. gigasporum complex, while the systematic position of the latter is dubious (Liu et al. 2014).
The Colletotrichum orchidearum species complex currently consists of eight closely related species, including three species (C. orchidearum, C. plurivorum, C. sojae) that are very common and occur on many host species and a number of less common species that seem to be either host-specific (C. cliviicola, C. musicola, C. cattleyicola, C. piperis) or restricted to a specific country and region (C. vittalense). Sexual morphs were observed in cultures of many of the species of the C. orchidearum complex; we assume them to be homothallic. However, this needs to be confirmed.
In some species in the C. orchidearum complex we observed different strains that were only/predominantly producing the sexual or the asexual morph, respectively. A dominating sexual morph of one Colletotrichum species and another Colletotrichum species forming only the asexual morph co-occurring on the same host, is one of the reasons for wrong connections of sexual and asexual morphs of Colletotrichum species. This was clearly the case with Ga. glycines by Lehman & Wolf (1926). A similar scenario is possible with the sexual morphs ascribed to C. lindemuthianum that were called Ga. lindemuthiana or Ga. cingulata f. sp. phaseoli (Shear and Wood, 1913, Kimati and Galli, 1970). It is also possible that the asexual morphs were just assumed to be C. lindemuthianum based on the host plants (common beans) or C. gloeosporioides based on conidial morphology, respectively. Epitypifications of the respective species would clarify this situation.
Nothing is known about the infection strategies of the individual species in the three species complexes treated here, and little is known about their distribution, host specificity and their impact on the host plants. The high number of sequences from unidentified or basically only tentatively identified strains in GenBank suggests a larger distribution and probably more species in these complexes than included in the present study.
Acknowledgements
We thank the curators of the CBS, MAFF, UTFC and CABI culture collections as well as Dr Richard O'Connell, UMR1290 BIOGER-CPP, INRA-AgroParisTech, 78850 Thiverval-Grignon, France, Dr Isabelle Benoit-Gelber, Department of Biology, Concordia University, Montreal, Canada, and Omid Atghita, Department of Plant Protection, College of Agriculture and Natural Resources, University of Tehran, Karaj, Iran, for kindly supplying isolates or collecting plant material for this study. We kindly thank the curators of the fungaria at the Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands, the Herbarium Hamburgense, Hamburg, Germany, the Botanische Staatssammlung München, Germany, the Royal Botanic Gardens in Kew, UK, and the US National Fungus Collections, Beltsville, Maryland, USA, for providing access to important herbarium specimens. Prof. Dr Eva H. Stukenbrock, Department of Environmental Genomics, Botanical Institute, Christian-Albrechts University of Kiel, Germany, is gratefully acknowledged for financial support in sequencing the UTFC strains. Prof. Dr Uwe Braun, Institut für Geobotanik und Botanischer Garten, Martin-Luther-Universität Halle-Wittenberg, Germany, is thanked for verifying the Latin names and Dr Jens Wesenberg, Department of Botany, Senckenberg Museum of Natural History Görlitz, Germany, for the identification of an orchid host plant. This research was also supported by the Dutch Ministry of Agriculture, Nature and Food Quality through an endowment of the FES programme “Versterking infrastructuur Plantgezondheid”.
Footnotes
Peer review under responsibility of Westerdijk Fungal Biodiversity Institute.
References
- Allescher A. 2nd edn. 1(7) Rabenhorst's Kryptogamen-Flora von Deutschland; Oesterreich und der Schweiz: 1902. pp. 385–704. (Fungi Imperfecti). [Google Scholar]
- Ariyawansa H.A., Hyde K.D., Jayasiri S.C. Fungal diversity notes 111–252-taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity. 2015;75:27–274. [Google Scholar]
- Arnaud G. Une maladie des Clivia en serre. Bulletin de la Société de pathologie végétale de France. 1914;1:36–37. [Google Scholar]
- Atghia O. University of Tehran; Iran: 2015. Etiology of anthracnose on common bean (Phaseolus vulgaris) in northern and west northern provinces of Iran; p. 204. M.Sc. thesis. [Google Scholar]
- Barbieri M.C.G., Ciampi-Guillardi M., Moraes S.R.G., Bonaldo S.M., Rogerio F., Linhares S.M., Massola N.S., Jr. First report of Colletotrichum cliviae causing anthracnose of soybean in Brazil. Plant Disease. 2017;101:1677. [Google Scholar]
- Barcelos Q.L., de Souza E.A., Vaillancourt L. Morphological and phylogenetic analysis of Glomerella and C. lindemuthianum strains isolated from common bean anthracnose lesions. Annual Report of the Bean Improvement Cooperative. 2011;54:110–111. [Google Scholar]
- Barcelos Q.L., Pinto J.M., Vaillancourt L.J. Characterization of Glomerella strains recovered from anthracnose lesions on common bean plants in Brazil. PLoS One. 2014;9(3):E90910. doi: 10.1371/journal.pone.0090910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezerra J.P., Ferreira P.V., da F Barbosa L. First report of anthracnose on chayote fruits (Sechium edule) caused by Colletotrichum brevisporum. Plant Disease. 2016;100:217. [Google Scholar]
- Bhairi S., Buckley E.H., Staples R.C. Protein synthesis and gene expression during appressorium formation in Glomerella magna. Experimental Mycology. 1990;14:207–217. [Google Scholar]
- Bobev S.G., Castlebury L.A., Rossman A.Y. First Report of Colletotrichum dracaenophilum on Dracaena sanderiana in Bulgaria. Plant Disease. 2008;92:173. doi: 10.1094/PDIS-92-1-0173A. [DOI] [PubMed] [Google Scholar]
- Bubák F., Kabát J.E. Mykologische Beiträge. Hedwigia. 1907;46:288–298. [Google Scholar]
- Cannon P.F., Damm U., Johnston P.R. Colletotrichum – current status and future directions. Studies in Mycology. 2012;73:181–213. doi: 10.3114/sim0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone I., Kohn L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91:553–556. [Google Scholar]
- Cavalcante G.R.S., Vieira W.A.S., Michereff S.J. First report of anthracnose caused by Colletotrichum sichuanensis on Phaseolus lunatus in Brazil. Plant Disease. 2018;102:680. [Google Scholar]
- Chen L.S., Chu C., Liu C.D. PCR-based detection and differentiation of anthracnose pathogens, Colletotrichum gloeosporioides and C. truncatum, from vegetable soybean in Taiwan. Journal of Phytopathology. 2006;154:654–662. [Google Scholar]
- Choi K.J., Kim W.G., Kim H.G. Morphology, molecular phylogeny and pathogenicity of Colletotrichum panacicola causing anthracnose of Korean ginseng. The Plant Pathology Journal. 2011;27:1–7. [Google Scholar]
- Chowdappa P., Chethana C.S., Pant R.P. Multilocus gene phylogeny reveals occurrence of Colletotrichum cymbidiicola and C. cliviae on orchids in North East India. Journal of Plant Pathology. 2014;96:327–334. [Google Scholar]
- Crous P.W., Gams W., Stalpers J.A. MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology. 2004;50:19–22. [Google Scholar]
- Crous P.W., Groenewald J.Z., Risede J.M. Calonectria species and their Cylindrocladium anamorphs: species with sphaeropedunculate vesicles. Studies in Mycology. 2004;50:415–430. doi: 10.3114/sim.55.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P.W., Verkleij G.J.M., Groenewald J.Z. Centraalbureau voor Schimmelcultures; Utrecht, Netherlands: 2009. Fungal biodiversity. CBS laboratory manual series 1. [Google Scholar]
- Damm U., Cannon P.F., Liu F. The Colletotrichum orbiculare species complex: important plant pathogens and mycoherbicides. Fungal Diversity. 2013;61:29–59. [Google Scholar]
- Damm U., Cannon P.F., Woudenberg J.H.C. The Colletotrichum acutatum species complex. Studies in Mycology. 2012;73:37–113. doi: 10.3114/sim0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm U., Cannon P.F., Woudenberg J.H.C. The Colletotrichum boninense species complex. Studies in Mycology. 2012;73:1–36. doi: 10.3114/sim0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm U., Crous P.W., Fourie P.H. Botryosphaeriaceae as potential pathogens of Prunus species in South Africa, with descriptions of Diplodia africana and Lasiodiplodia plurivora spp. nov. Mycologia. 2007;99:664–680. doi: 10.3852/mycologia.99.5.664. [DOI] [PubMed] [Google Scholar]
- Damm U., Mostert L., Crous P.W. Novel Phaeoacremonium species associated with necrotic wood of Prunus trees. Persoonia. 2008;20:87–102. doi: 10.3767/003158508X324227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm U., O’Connell R.J., Crous P.W. The Colletotrichum destructivum species complex – hemibiotrophic pathogens of forage and field crops. Studies in Mycology. 2014;79:49–84. doi: 10.1016/j.simyco.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm U., Woudenberg J.H.C., Cannon P.F., Crous P.W. Colletotrichum species with curved conidia from herbaceous hosts. Fungal Diversity. 2009;39:45–87. [Google Scholar]
- De Almeida L.B., Matos K.S., Assis L.A.G. First report of anthracnose of Capsicum chinense in Brazil caused by Colletotrichum brevisporum. Plant Disease. 2017;101:1035–1036. [Google Scholar]
- Diao Y.Z., Zhang C., Liu F. Colletotrichum species causing anthracnose disease of chili in China. Persoonia. 2017;38:20–37. doi: 10.3767/003158517X692788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douanla-Meli C., Unger J.-G., Langer E. Multi-approach analysis of the diversity in Colletotrichum cliviae sensu lato. Antonie van Leeuwenhoek. 2018;111:423–435. doi: 10.1007/s10482-017-0965-9. [DOI] [PubMed] [Google Scholar]
- Du M., Schardl C.L., Nuckles E.M., Vaillancourt L.J. Using mating-type gene sequences for improved phylogenetic resolution of Colletotrichum species complexes. Mycologia. 2005;97:641–658. doi: 10.3852/mycologia.97.3.641. [DOI] [PubMed] [Google Scholar]
- Farr D.F., Aime M.C., Rossman A.Y. Species of Colletotrichum on Agavaceae. Mycological Research. 2006;110:1395–1408. doi: 10.1016/j.mycres.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Farr D.F., Rossman A.Y. U.S. National Fungus Collections, ARS, USDA; 2017. Fungal Databases.https://nt.ars-grin.gov/fungaldatabases/ Retrieved June 14, 2017, from. [Google Scholar]
- Freeman S., Rodriguez R.J. A rapid, reliable bioassay for pathogenicity of Colletotrichum magna on cucurbits and the isolation of non–pathogenic mutants. Plant Disease. 1992;76:901–905. [Google Scholar]
- Freeman S., Rodriguez R.J. Genetic conversion of a fungal plant pathogen to a nonpathogenic, endophytic mutualist. Science. 1993;260:75–78. doi: 10.1126/science.260.5104.75. [DOI] [PubMed] [Google Scholar]
- Gallucci-Rangone M.M. Una nuova specie di Colletotrichum rinvenuta su Catteya sp. Annali della sperimentazione agraria, N. S. 1955;9(Suppl):11–15. [Google Scholar]
- Gardes M., Bruns T.D. ITS primers with enhanced specificity for basidiomycetes – application to the identification of mycorrhizae and rusts. Molecular Ecology. 1993;2:113–118. doi: 10.1111/j.1365-294x.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- Glass N.L., Donaldson G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology. 1995;61:1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González Fragoso R. Contribución a la flora micológica lusitánica. Boletím da Sociedade Broteriana. 1924;2(Sér. 2):3–83. [Google Scholar]
- Grand L.F. North Carolina Plant Disease Index North Carolina Agricultural Research Service. Technical Bulletin. 1985;240:1–157. [Google Scholar]
- Guerber J.C., Liu B., Correll J.C. Characterization of diversity in Colletotrichum acutatum sensu lato by sequence analysis of two gene introns, mtDNA and intron RFLPs, and mating compatibility. Mycologia. 2003;95:872–895. [PubMed] [Google Scholar]
- Hemmi T. Beiträge zur Kenntnis der Morphologie und Physiologie der Japanischen Gloeosporien. Journal of the College of Agriculture, Hokkaido Imperial University. 1920;9:1–159. [Google Scholar]
- Hennings P.C. Fungi Goyazenses. Hedwigia. 1895;34:88–116. [Google Scholar]
- Hennings P.C. Fungi S. Paulenses IV. a cl. Puttemans collecti. Hedwigia. 1908;48:1–20. [Google Scholar]
- Higginbotham S.J., Arnold A.E., Ibanez A. Bioactivity of fungal endophytes as a function of endophyte taxonomy and the taxonomy and distribution of their host plants. PLoS One. 2013;8(9):E73192. doi: 10.1371/journal.pone.0073192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillis D.M., Bull J.J. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Systematic Biology. 1993;42:182–192. [Google Scholar]
- Hou L.W., Liu F., Duan W.J. Colletotrichum aracearum and C. camelliae-japonicae, two holomorphic new species from China and Japan. Mycosphere. 2016;7:1111–1123. [Google Scholar]
- James R.S., Ray J., Tan Y.P. Colletotrichum siamense, C. theobromicola, and C. queenslandicum from several plant species and the identification of C. asianum in the Northern Territory, Australia. Australasian Plant Disease Notes. 2014;9:138. [Google Scholar]
- Jenkins S.F., Winstead N.N. Glomerella magna, cause of a new anthracnose of cucurbits. Phytopathology. 1964;54:452–454. [Google Scholar]
- Kimati H., Galli F. Glomerella cingulata (Stonem.) Spauld. Et v. Scherenk. f. sp. phaseoli n. f., fase ascógena do agente causal da antracnose do feijoeiro. Anais da Escola Superior de Agricultura “Luiz de Queiroz”, Piracicaba. 1970;27:411–437. [Google Scholar]
- Lehman S.G., Wolf F.A. Soy-bean anthracnose. Journal of agricultural Research. 1926;33:381–390. [Google Scholar]
- Lei Y., Tang X.B., Jayawardena R.S. Identification and characertization of Colletotrichum species causing grape rot in southern China. Mycosphere. 2016;7:1177–1191. [Google Scholar]
- Li Q.L., Mo J.Y., Huang S.P. First report of leaf spot disease caused by Glomerella magna on Lobelia chinensis in China. Plant Disease. 2013;97:1383. doi: 10.1094/PDIS-03-13-0346-PDN. [DOI] [PubMed] [Google Scholar]
- Lin Y.-S., Wu L.C. Seed-borne diseases of soybean in Taiwan. III. Anthracnose. Plant Protection Bulletin (Taiwan) 1966;8:305–317. [Google Scholar]
- Liu B., Wasilwa L.A., Morelock T.E. Comparison of Colletotrichum orbiculare and several allied Colletotrichum spp. for mtDNA RFLPs, intron RFLP and sequence variation, vegetative compatibility and host specificity. Phytopathology. 2007;97:1305–1314. doi: 10.1094/PHYTO-97-10-1305. [DOI] [PubMed] [Google Scholar]
- Liu F., Cai L., Crous P.W. Circumscription of the anthracnose pathogens Colletotrichum lindemuthianum and C. nigrum. Mycologia. 2013;105:844–860. doi: 10.3852/12-315. [DOI] [PubMed] [Google Scholar]
- Liu F., Cai L., Crous P.W. The Colletotrichum gigasporum species complex. Persoonia. 2014;33:83–97. doi: 10.3767/003158514X684447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Tang G., Zheng X. Molecular and phenotypic characterization of Colletotrichum species associated with anthracnose disease in peppers from Sichuan Province, China. Scientific Reports. 2016;6:32761. doi: 10.1038/srep32761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Weir B.S., Damm U. Unravelling Colletotrichum species associated with Camellia: employing ApMat and GS loci to resolve species in the C. gloeosporioides complex. Persoonia. 2015;35:63–86. doi: 10.3767/003158515X687597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Zhang L., Qiu P. First report of Colletotrichum brevisporum causing anthracnose on pumpkin in China. Plant Disease. 2018;102:1038. [Google Scholar]
- Liu X., Xie X., Duan J. Colletotrichum yunnanense sp. nov., a new endophytic species from Buxus sp. Mycotaxon. 2007;100:137–144. [Google Scholar]
- Macedo D.M., Barreto R.W. Colletotrichum dracaenophilum causes anthracnose on Dracaena braunii in Brazil. Australasian Plant Disease Notes. 2016;11:5. [Google Scholar]
- Manamgoda D.S., Udayanga D., Cai L. Endophytic Colletotrichum from tropical grasses with a new species C. endophytica. Fungal Diversity. 2013;61:107–115. [Google Scholar]
- Manandhar J.B., Hartman G.L., Sinclair J.B. Colletotrichum destructivum, the anamorph of Glomerella glycines. Phytopathology. 1986;76:282–285. [Google Scholar]
- Marcelino J., Giordano R., Gouli S. Colletotrichum acutatum var. fioriniae (teleomorph: Glomerella acutata var. fioriniae var. nov.) infection of a scale insect. Mycologia. 2008;100:353–374. doi: 10.3852/07-174r. [DOI] [PubMed] [Google Scholar]
- Marin-Felix Y., Groenewald J.Z., Cai L. Genera of phytopathogenic fungi: GOPHY 1. Studies in Mycology. 2017;86:99–216. doi: 10.1016/j.simyco.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason-Gamer R.J., Kellogg E.A. Testing for phylogenetic conflict among molecular data sets in the tribe Triticeae (Gramineae) Systematic Biology. 1996;45:524–545. [Google Scholar]
- Massee G.E. Fungi exotici. XIII. Bulletin of Miscellaneous Information. Royal Botanic Gardens, Kew. 1912;1912:189–191. [Google Scholar]
- Molina-Chaves A., Gómez-Alpízar L., Umaña-Rojas G. Identificación de especies del género Colletotrichum asociadas a la antracnosis en papaya (Carica papaya L.) en Costa Rica [Identification of Colletotrichum species associated with anthracnose in papaya (Carica papaya L.) in Costa Rica.] Agronomía Costarricense. 2017;41:69–80. [Google Scholar]
- Moriwaki J., Tsukiboshi T., Sato T. Grouping of Colletotrichum species in Japan based on rDNA sequences. Journal of General Plant Pathology. 2002;68:307–320. [Google Scholar]
- Morsy A.A., Elshahawy I.E. Anthracnose of lucky bamboo Dracaena sanderiana caused by the fungus Colletotrichum dracaenophilum in Egypt. Journal of Advanced Research. 2016;7:327–335. doi: 10.1016/j.jare.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento R.J., Mizubuti E.S.G., Câmara M.P.S. First report of papaya fruit rot caused by Colletotrichum magna in Brazil. Plant Disease. 2010;94:1506. doi: 10.1094/PDIS-08-10-0555. [DOI] [PubMed] [Google Scholar]
- Nguyen P.T.H., Vinnere Pettersson O., Olsson P. Identification of Colletotrichum species associated with anthracnose disease of coffee in Vietnam. European Journal of Plant Pathology. 2010;127:73–87. [Google Scholar]
- Nirenberg H.I. Untersuchungen über die morphologische und biologische Differenzierung in der Fusarium-Sektion Liseola. Mitteilungen aus der Biologischen Bundesanstalt für Land- und Forstwirtschaft Berlin-Dahlem. 1976;169:1–117. [Google Scholar]
- Noireung P., Phoulivong, Liu F. Novel species of Colletotrichum revealed by morphology and molecular analysis. Cryptogamie, Mycologie. 2012;33:347–362. [Google Scholar]
- Nylander J.A.A. Evolutionary Biology Centre, Uppsala University; 2004. MrModeltest v2. Program distributed by the author. [Google Scholar]
- O'Donnell K., Cigelnik E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution. 1997;7:103–116. doi: 10.1006/mpev.1996.0376. [DOI] [PubMed] [Google Scholar]
- Orlandelli R.C., Alberto R.N., Rubin Filho C.J. Diversity of endophytic fungal community associated with Piper hispidum (Piperaceae) leaves. Genetics and Molecular Research. 2012;11:1575–1585. doi: 10.4238/2012.May.22.7. [DOI] [PubMed] [Google Scholar]
- Oudemans C.A.J.A. Notice sur quelques champignons nouveaux. Verslagen van de Gewone Vergaderingen der Wis- en Natuurkundige Afdeeling. 1896;5:224–233. [Google Scholar]
- Paul N.C., Lee H.B., Lee J.H. Endophytic fungi from Lycium chinense Mill and characterization of two new Korean records of Colletotrichum. International Journal of Molecular Sciences. 2014;15:15272–15286. doi: 10.3390/ijms150915272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Yang Y., Hyde K.D. Colletotrichum species on Citrus leaves in Guizhou and Yunnan provinces, China. Cryptogamie, Mycologie. 2012;33:267–283. [Google Scholar]
- Petch T. Additions to Ceylon fungi. Annals of the Royal Botanic Gardens of Peradeniya. 1917;6:195–256. [Google Scholar]
- Petrak F. Über einige wenig bekannte, durch Colletotrichum cliviae (Oud.) Petr. verursachte Blattkrankheit der Clivien. Sydowia. 1947;1(1–3):80–82. [Google Scholar]
- Petrak F. Beiträge zur Pilzflora von Hawaii. Sydowia. 1953;7:381–409. [Google Scholar]
- Potebnia A. Mycologische Studien. Annales Mycologici. 1907;5(1):1–28. [Google Scholar]
- Rambaut A. University of Oxford; Oxford, UK: 2002. Sequence alignment editor. Version 2.0. [Google Scholar]
- Rakotoniriana E.F., Scauflaire J., Rabemanantsoa C. Colletotrichum gigasporum sp. nov., a new species of Colletotrichum producing long straight conidia. Mycological Progress. 2013;12:403–412. [Google Scholar]
- Ramos A.M., Tadic L.F., Cinto I. Molecular characterization of Colletotrichum species causing soybean anthracnose in Argentina. Mycotaxon. 2013;123:457–465. [Google Scholar]
- Rayner R.W. Commonwealth Mycological Institute; Kew, UK: 1970. A mycological colour chart. [Google Scholar]
- Redman R.S., Ranson J., Rodriguez R.J. Conversion of the pathogenic fungus Colletotrichum magna to a nonpathogenic endophytic mutualist by gene disruption. Molecular Plant Microbe Interactions. 1999;12:969–975. [Google Scholar]
- Rodrigues R.J., Owen J.L. Isolation of Glomerella musae[teleomorph of Colletotrichum musae (Berk. & Curt.) Arx.] and segregation analysis of ascospore progeny. Experimental Mycology. 1992;16:291–301. [Google Scholar]
- Rodrígues-Guerra R., Ramírez-Rueda M.T., Cabral-Enciso M. Heterothallic mating observed between Mexican isolates of Glomerella lindemuthiana. Mycologia. 2005;97:793–803. doi: 10.3852/mycologia.97.4.793. [DOI] [PubMed] [Google Scholar]
- Rodriguez R.J., Redman R.S., Henson J.M. The role of fungal symbioses in the adaptation of plants to high stress environments. Mitigation and Adaptation Strategies for Global Change. 2004;9:261–272. [Google Scholar]
- Rogério F., Ciampi-Guillardi M., Barbieri M.C.G. Phylogeny and variability of Colletotrichum truncatum associated with soybean anthracnose in Brazil. Journal of Applied Microbiology. 2016;122:402–415. doi: 10.1111/jam.13346. [DOI] [PubMed] [Google Scholar]
- Rojas E.I., Rehner S.A., Samuels G.J. Colletotrichum gloeosporioides s.l. associated with Theobroma cacao and other plants in Panama: multilocus phylogenies distinguish pathogen and endophyte clades. Mycologia. 2010;102:1318–1338. doi: 10.3852/09-244. [DOI] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rossman A.Y., Allen W.C., Castlebury L.A. New combinations of plant-associated fungi resulting from the change to one name for fungi. IMA Fungus. 2016;7:1–7. doi: 10.5598/imafungus.2016.07.01.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy T.C. Anthracnose of Piper betle L. caused by Colletotrichum dasturii Boy sp. nov. in Bengal. Journal of the Indian Botanical Society. 1948;27:96–102. [Google Scholar]
- Saccardo P.A. Fungi novi ex herbarium professoris Doct. P. Magnus Berolinensis. Michelia. 1878;1(2):117–132. [Google Scholar]
- Saccardo P.A. Vol. 17. 1905. pp. 1–991. (Sylloge Fungorum). Italy, Patavii. [Google Scholar]
- Saccardo P.A. Vol. 18. 1906. pp. 1–839. (Sylloge Fungorum). Italy, Patavii. [Google Scholar]
- Saccardo P.A., Saccardo D., Traverso G.B. Vol. 25. 1931. pp. 1–1093. (Sylloge Fungorum). Italy, Abellini; Pergola, Saccardo Estate. [Google Scholar]
- Saccardo P.A., Sydow P. 14(2) 1899. pp. 725–1316. (Sylloge Fungorum). Italy, Patavii; P.A. Saccardo. [Google Scholar]
- Saccardo P.A., Trotter A. Vol. 22. 1913. pp. 1–1612. (Sylloge Fungorum). Italy, Patavii; P.A. Saccardo. [Google Scholar]
- Salustiano M.E., Rondon M.N., Abreu L.M. The etiological agent of cotton ramulosis represents a single phylogenetic lineage within the Colletotrichum gloeosporioides species complex. Tropical Plant Pathology. 2014;39:357–367. [Google Scholar]
- Santos R.F., Ciampi-Guillardi M., Amorim L. Aetiology of anthracnose on grapevine shoots in Brazil. Plant Pathology. 2018;67:692–706. [Google Scholar]
- Sato T., Moriwaki J., Uzuhashi S. Molecular phylogenetic analyses and morphological re-examination of strains belonging to three rare Colletotrichum species in Japan. Microbiology and Culture Collections. 2012;28:121–134. [Google Scholar]
- Sawada K. Descriptive catalogue of Taiwan (Formosan) Fungi. Part XI. Special Publication of the College of Agriculture, National Taiwan University. 1959;8:1–268. [Google Scholar]
- Sharma G., Kumar N., Weir B.S., Hyde K.D., Shenoy B.D. The ApMat marker can resolve Colletotrichum species: a case study with Mangifera indica. Fungal Diversity. 2013;61:117–138. [Google Scholar]
- Sharma G., Kumar Pinnaka A., Shenoy B.D. Resolving the Colletotrichum siamense species complex using ApMat marker. Fungal Diversity. 2015;71:247–264. [Google Scholar]
- Sharma G., Pinnaka A.K., Shenoy B.D. ITS-based diversity of Colletotrichum from India. Current Research in Environmental & Applied Mycology. 2013;3:194–220. [Google Scholar]
- Sharma K., Merritt J., Palmateer A. Isolation, characterization, and management of Colletotrichum spp. causing anthracnose on lucky bamboo (Dracaena sanderiana) Hortscience. 2014;49:453–459. [Google Scholar]
- Shear C., Wood A. Studies of fungous parasites belonging to the genus Glomerella. U.S. Department of Agriculture. 1913;252:1–110. [Google Scholar]
- Shenoy B.D., Jeewon R., Lam W.H. Morpho-molecular characterisation and epitypification of Colletotrichum capsici (Glomerellaceae, Sordariomycetes), the causative agent of anthracnose in chilli. Fungal Diversity. 2007;27:197–211. [Google Scholar]
- Shivas R.G., Tan Y.P., Edwards J. Colletotrichum species in Australia. Australasian Plant Pathology. 2016;45:447–464. [Google Scholar]
- Sinclair J.B. 2nd edn. American Phytopathological Society; Saint Paul, USA: 1982. Compendium of Soybean Diseases. [Google Scholar]
- Singh R., Graney L., Williamson M. First report of boxwood dieback caused by Colletotrichum theobromicola in the United States. Plant Disease. 2015;99:1274. [Google Scholar]
- Sivanesan A., Hsieh W.H. A new ascomycete, Glomerella septospora sp. nov. and its coelomycete anamorph, Colletotrichum taiwanense sp. nov. from Taiwan. Mycological Research. 1993;97:1523–1529. [Google Scholar]
- Sousa E.S., Silva J.R.A., Assunção I.P. Colletotrichum species causing anthracnose on lima bean in Brazil. Tropical Plant Pathology. 2018;43:78–84. [Google Scholar]
- Stevens F.L. Porto Rican fungi, old and new. Transactions of the Illinois State Academy of Science. 1917;10:162–218. [Google Scholar]
- Stoneman B. A comparative study of the development of some anthracnoses. Botanical Gazette. 1898;26:69–120. [Google Scholar]
- Swofford D.L. Sinauer Associates; Sunderland, Massachusetts: 2003. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. [Google Scholar]
- Talamini V., Souza E.A., Pozza E.A. Genetic divergence among and within Colletotrichum lindemuthianum races assessed by RAPD. Fitopatologia Brasileira. 2006;31:545–550. [Google Scholar]
- Tao G., Liu Z.Y., Liu F. Endophytic Colletotrichum species from Bletilla ochracea (Orchidaceae), with descriptions of seven new species. Fungal Diversity. 2013;61:139–164. [Google Scholar]
- Tapia-Tussell R., Cortés-Velázquez A., Valencia-Yah T. First report of Colletotrichum magnum causing anthracnose in papaya in Mexico. Plant Disease. 2016;100:2323. [Google Scholar]
- Than P.P., Jeewon R., Hyde K.D. Characterization and pathogenicity of Colletotrichum species associated with anthracnose on chilli (Capsicum spp.) in Thailand. Plant Pathology. 2008;57:562–572. [Google Scholar]
- Tiffany L.H., Gilman J.C. Species of Colletotrichum from Legumes. Mycologia. 1954;46:52–75. [Google Scholar]
- Trotter A., Cash E.K. Vol. 26. Johnson Reprint Corporation; USA, New York & UK, London: 1972. pp. 1–1563. (Sylloge Fungorum). [Google Scholar]
- Tsay J.G., Chen R.S., Wang W.L. First report of anthracnose on cucurbitaceous crops caused by Glomerella magna in Taiwan. Plant Disease. 2010;94:787. doi: 10.1094/PDIS-94-6-0787A. [DOI] [PubMed] [Google Scholar]
- Verplancke G. Bijdrage tot de flora der woekerzwammen van België. V. Studie der zwammen gevonden op Orchidaceae. Mededeelingen der Landbouwhoogeschool en der Opzoekingsstations van der Staat, te Gent. 1935;III(1):3–33. [Google Scholar]
- Verplancke G. Bijdrage tot de flora der woekerzwammen van België. V. Studie der zwammen gevonden op Orchidaceae. Mededeelingen der Landbouwhoogeschool en der Opzoekingsstations van der Staat, te Gent. 1935;III(2):81–112. [Google Scholar]
- Vieira W.A.S., Lima W.G., Nascimento E.S. The impact of phenotypic and molecular data on the inference of Colletotrichum diversity associated with Musa. Mycologia. 2018;109:912–934. doi: 10.1080/00275514.2017.1418577. [DOI] [PubMed] [Google Scholar]
- Vieira W.A.S., Michereff S.M., Morais Endophytic species of Colletotrichum associated with mango in northeastern Brazil. Fungal Diversity. 2014;67:181–202. [Google Scholar]
- Vieira W.A.S., Nascimento R.J., Michereff S.J. First report of papaya fruit anthracnose caused by Colletotrichum brevisporum in Brazil. Plant Disease. 2013;97:1659. doi: 10.1094/PDIS-05-13-0520-PDN. [DOI] [PubMed] [Google Scholar]
- Villanueva-Arce R., Hernández-Anguiano A.M., de Yáñez-Morales M. Caracterización e identificación de Colletotrichum fragariae en frutos de cherimoya (Characterization and identification of Colletotrichum fragariae on cherimoya fruits) Agrociencia. 2005;39:93–106. [Google Scholar]
- Von Arx J.A. Die Arten der Gattung Colletotrichum Cda. Phytopathologische Zeitschrift. 1957;29:413–468. [Google Scholar]
- Von Arx J.A., Müller E. Die Gattungen der amerosporen Pyrenomyceten. Beiträge zur Kryptogamenflora der Schweiz. 1954;11(1):1–434. [Google Scholar]
- Von Schweinitz L.D. Synopsis fungorum in America boreali media degentium. Transactions of the American Philosophical Society of Philadelphia New series. 1832;4:141–316. [Google Scholar]
- Wang W.-X., Kusari S., Laatsch H. Antibacterial azaphilones from an endophytic fungus Colletotrichum sp. BS4. Journal of natural products. 2016;79:704–710. doi: 10.1021/acs.jnatprod.5b00436. [DOI] [PubMed] [Google Scholar]
- Wang Y.C., Hao X.Y., Wang L. Diverse Colletotrichum species cause anthracnose of tea plants (Camellia sinensis (L.) O. Kuntze) in China. Scientific Repeports. 2016;6:35287. doi: 10.1038/srep35287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasilwa L.A., Correll J.C., Morelock T.E., McNew R.E. Reexamination of races of the cucurbit anthracnose pathogen Colletotrichum orbiculare. Phytopathology. 1993;83:1190–1198. [Google Scholar]
- Weir B., Damm U., Johnston P.R. The Colletotrichum gloeosporioides species complex. Studies in Mycology. 2012;73:115–180. doi: 10.3114/sim0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T.J., Bruns T., Lee S. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M.A., Gelfand D.H., Sninsky J.J., White T.J., editors. PCR protocols: a guide to methods and applications. Academic Press; San Diego, USA: 1990. pp. 315–322. [Google Scholar]
- Woudenberg J.H.C., Aveskamp M.M., de Gruyter J. Multiple Didymella teleomorphs are linked to the Phoma clematidina morphotype. Persoonia. 2009;22:56–62. doi: 10.3767/003158509X427808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H.J., Zhou R.J., Fu J.F. Morphological and molecular identification of anthracnose on Arctium lappa caused by C. simmondsii in China. Plant Disease. 2016;100:1010. [Google Scholar]
- Yang H.-C., Haundenshield J.S., Hartman G.L. Colletotrichum incanum sp. nov., a curved-conidial species causing soybean anthracnose in USA. Mycologia. 2014;106:32–42. doi: 10.3852/13-013. [DOI] [PubMed] [Google Scholar]
- Yang Y.L., Cai L., Yu Z.N. Colletotrichum species on Orchidaceae in southwest China. Cryptogamie Mycologie. 2011;32:229–253. [Google Scholar]
- Yang Y.L., Liu Z.Y., Cai L. Colletotrichum anthracnose of Amaryllidaceae. Fungal Diversity. 2009;39:123–146. [Google Scholar]
- Yao J.A., Yu D.Y., Chen F. Sequence analysis of ITS and PCR rapid-detection of Colletotrichum gloeosporioides from Cymbidium ensifolium. Acta Phytophylacica Sinica. 2013;40:249–254. [Google Scholar]
- Yuan Z.L., Chen Y.C., Yang Y. Diverse non-mycorrhizal fungal endophytes inhabiting an epiphytic medicinal orchid (Dendrobium nobile): estimation and characterization. World Journal of Microbiology & Biotechnology. 2009;25:295–303. [Google Scholar]
- Zhou Y., Huang J.S., Yang L.Y. First report of banana anthracnose caused by Colletotrichum scovillei in China. Plant Disease. 2017;101:381–382. [Google Scholar]
- Zhou Z., Li Y.L. First report of Colletotrichum cliviae causing anthracnose on Zamioculcas zamiifolia in Henan Province, China. Plant Disease. 2017;101:838. [Google Scholar]