ABSTRACT
The recent emergence of a transferable colistin resistance mechanism, MCR-1, has gained global attention because of its threat to clinical treatment of infections caused by multidrug-resistant Gram-negative bacteria. However, the possible transmission route of mcr-1 among Enterobacteriaceae species in clinical settings is largely unknown. Here, we present a comprehensive genomic analysis of Escherichia coli isolates collected in a hospital in Hangzhou, China. We found that mcr-1-carrying isolates from clinical infections and feces of inpatients and healthy volunteers were genetically diverse and were not closely related phylogenetically, suggesting that clonal expansion is not involved in the spread of mcr-1. The mcr-1 gene was found on either chromosomes or plasmids, but in most of the E. coli isolates, mcr-1 was carried on plasmids. The genetic context of the plasmids showed considerable diversity as evidenced by the different functional insertion sequence (IS) elements, toxin-antitoxin (TA) systems, heavy metal resistance determinants, and Rep proteins of broad-host-range plasmids. Additionally, the genomic analysis revealed nosocomial transmission of mcr-1 and the coexistence of mcr-1 with other genes encoding β-lactamases and fluoroquinolone resistance in the E. coli isolates. These findings indicate that mcr-1 is heterogeneously disseminated in both commensal and pathogenic strains of E. coli, suggest the high flexibility of this gene in its association with diverse genetic backgrounds of the hosts, and provide new insights into the genome epidemiology of mcr-1 among hospital-associated E. coli strains.
KEYWORDS: E. coli, genetic diversity, mcr-1, population genomics, transmission
IMPORTANCE
Colistin represents one of the very few available drugs for treating infections caused by extensively multidrug-resistant Gram-negative bacteria. The recently emergent mcr-1 colistin resistance gene threatens the clinical utility of colistin and has gained global attention. How mcr-1 spreads in hospital settings remains unknown and was investigated by whole-genome sequencing of mcr-1-carrying Escherichia coli in this study. The findings revealed extraordinary flexibility of mcr-1 in its spread among genetically diverse E. coli hosts and plasmids, nosocomial transmission of mcr-1-carrying E. coli, and the continuous emergence of novel Inc types of plasmids carrying mcr-1 and new mcr-1 variants. Additionally, mcr-1 was found to be frequently associated with other genes encoding β-lactams and fluoroquinolone resistance. These findings provide important information on the transmission and epidemiology of mcr-1 and are of significant public health importance as the information is expected to facilitate the control of this significant antibiotic resistance threat.
INTRODUCTION
The relentless increase in the populations of multidrug-resistant (MDR) and extensively drug-resistant (XDR) Gram-negative bacterial strains is worrying, not least because we apparently have no new clinical options. Moreover, the antibiotic pipeline is bereft of novel entities to potentially cover MDR and XDR Gram-negative infections (1–4). Recent global attention has focused on the plight of our entry into the “postantibiotic era” in the face of the rapid dissemination of carbapenem-resistant mechanisms (NDM-1, KPC, and OXA-48/181/232) and the realization of the very limited number of antibiotics, e.g., colistin, that we have left to treat serious infections (5–7). Until recently, colistin resistance was observed to be mediated by chromosomal mutations only and commonly occurred in Klebsiella pneumoniae and Pseudomonas aeruginosa but rarely in Escherichia coli (8). However, the first transferable colistin resistance mechanism, termed mcr-1, was recently reported in Enterobacteriaceae from both food-producing animals and human origins, in particular, E. coli (9). Perhaps of greater concern is the coexistence of mcr-1 and carbapenem resistance genes, such as blaNDM-5/9 and blaKPC-2, recently identified in E. coli from human infections and poultry production (10–13), as the common occurrence of the mcr-1 gene in carbapenem-resistant Enterobacteriaceae (CRE) would seriously compromise current treatment options not just in China but globally.
The unprecedented global increase in the populations of CRE, and now of mcr-1-positive Enterobacteriaceae (MCRPE), has placed further pressure on drug discovery programs to produce novel antimicrobials. It is still unclear what drives CRE and MCRPE, and although the increase in antibiotic consumption (e.g., the use of carbapenems) has been attributed to this increase, the remarkable plasticity and fluidity of DNA structures in Gram-negative bacteria have made a significant contribution. In Enterobacteriaceae, this horizontal gene transfer is fueled by a potent cocktail of plasmids, transposons, insertion sequence (IS) elements, and insertion sequence common region (ISCR) elements (14, 15). For example, the blaNDM-1 gene can be found in considerably greater numbers of bacteria than its KPC counterpart—in part, as a result of the diversity of plasmids it is associated with (16). The immediate genetic context surrounding the blaNDM-1 gene is also remarkably heterogeneous and has contributed to its translocation between chromosome and plasmid, and vice versa, and between plasmids (16).
Due to its global significance, many studies have been reported on investigating the prevalence of mcr-1 and have characterized its genetic environments in Enterobacteriaceae. To date, more than 70 completed sequences of plasmids carrying mcr-1 have been deposited into the GenBank database and their data show that they are relatively narrow in range and contain few other antibiotic resistance genes (11, 17–27). Thus far, many whole-genome sequences have been retrospectively searched for mcr-1; however, the majority of the reports focus only on the gene or plasmid and few have analyzed its associated bacterial hosts (18, 28). Moreover, data on the possible transmission routes of mcr-1 among MCRPE are largely lacking.
In order to aid understanding of the prevalence and outcomes of the presence of MCRPE in patients as well as in healthy adults, two studies were recently published on mcr-1-positive isolates and their impacts on nosocomial infections (29, 30). Statistical data suggest that mcr-1-positive E. coli (MCRPEC) infections were found to be associated with male sex, immunosuppression, and antibiotic usage (30). Multiple studies have also shown that E. coli is the most frequently observed Enterobacteriaceae species carrying the mcr-1 gene and that the gene can be transferred to other Enterobacteriaceae species from MCRPEC at high frequencies (9, 29, 30). However, despite those studies, there is a marked paucity of genetic data on MCRPE in hospitals that can help researchers to understand their circulation and thus their potential impact on infection control policies. Here, we present an extensive whole-genome analysis of 80 E. coli strains isolated from both clinical samples and fecal samples of patients and healthy human volunteers in one hospital in Hangzhou, Zhejiang Province, in China.
RESULTS
Overview of mcr-1-positive E. coli.
During our previous epidemiological and clinical study, we obtained a considerable number of MCRPEC isolates from patients and healthy adults in Hangzhou, Zhejiang, China (30). Here, we performed whole-genome analysis of 80 MCRPEC strains to address the possible dissemination of the mcr-1 gene (see Table S1 in the supplemental material). Briefly, 36 MCRPEC isolates were derived from nosocomial infections (urinary tract, surgical wounds, respiratory tract, etc.) of inpatients (n = 2,577), while 27 were from fecal samples of inpatients (n = 1,028) and 17 were from fecal samples of healthy volunteers (n = 2,909) collected in 2015. Detailed clinical information on these 80 mcr-1-carrying E. coli isolates is presented in Table S2.
The E. coli isolates in this study and their information from sequencing analysis. Download TABLE S1, XLSX file, 0.04 MB (38.1KB, xlsx) .
Copyright © 2018 Shen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Clinical information on 80 E. coli isolates with the mcr-1 gene and the MICs of multiple antimicrobial agents. Download TABLE S2, XLSX file, 0.05 MB (48.8KB, xlsx) .
Copyright © 2018 Shen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genome sequencing of the 80 mcr-1-positive E. coli isolates.
At least 100× coverage of raw reads from Illumina sequencing was obtained for each isolate. The draft genomes were assembled de novo using CLC Genomics Workbench (version 8.5). The number of contigs ranged from 51 to 232, while the N50 of contigs ranged from 43 kb to 521 kb (Table S1) for the isolates assembled by CLC Genomics Workbench (version 8.5). Since the majority of the mcr-1 genes were located on the plasmids, we further used the plasmidSPAdes program to optimize the assembly of plasmids. The mcr-1-containing contigs for each isolate were extracted from the two assemblies, and the longer contig was used to determine the genetic context of mcr-1. The lengths of mcr-1-carrying contigs generated by Illumina sequencing ranged from 2,388 to 141,207 bp (Table S1). Due to the shortness of the reads generated by Illumina sequencing and the high number of insertion elements, the assembled mcr-1-carrying contigs for nine isolates were short (2.3 to 3.8 kb). Thus, these isolates were resequenced by single-molecule real-time (SMRT) sequencing to generate complete chromosomes and plasmids (Table S1 and S3). A further six isolates were also subjected to SMRT sequencing.
The assembly stats for the PacBio genomes. Download TABLE S3, XLSX file, 0.03 MB (33.1KB, xlsx) .
Copyright © 2018 Shen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genomic epidemiology of the mcr-1-carrying E. coli isolates.
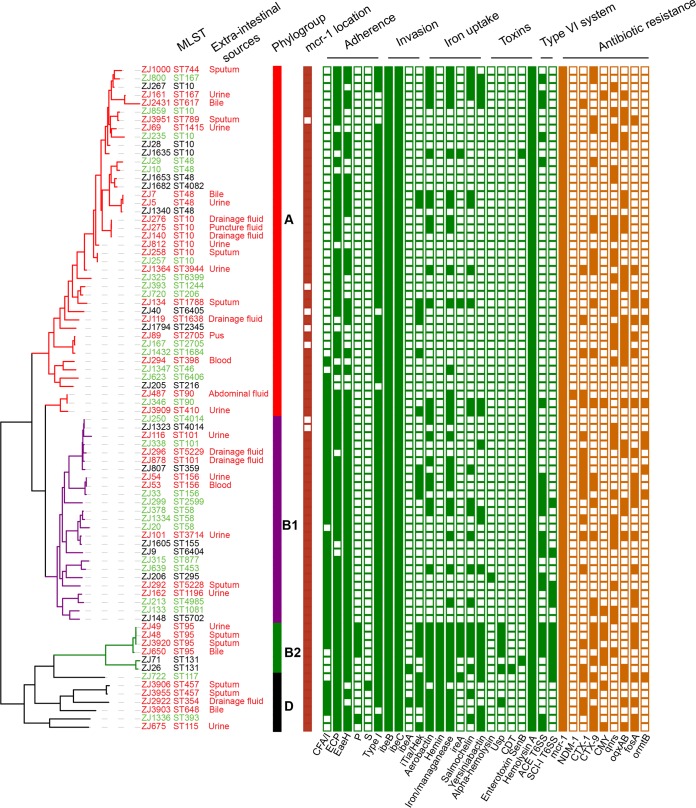
The genomic and epidemiological relationships among all MCRPEC isolates were investigated (Fig. 1). Classification of strains to phylogenetic subgroups (31, 32) and of sequence types using multilocus sequence typing (MLST) (33) was performed through in silico analysis (Fig. 1). The core genome phylogenetic tree indicates 3 main clusters, but MLST analysis revealed that the 80 MCRPEC isolates were significantly diverse: 76 isolates were assigned to 46 known MLST types, and 4 other isolates (ST6399, ST6404, ST6405, and ST6406) possessed novel STs (Fig. 1). The most prevalent ST of isolates harboring mcr-1 gene was ST10, accounting for 12.5% of all isolates. This ST is also commonly observed in extended-spectrum-β-lactamase (ESBL)-carrying E. coli isolates of both human and animal origins (34, 35). The classification of phylogenetic subgroups demonstrated that the 80 isolates were distributed throughout the four phylogroups (A, B1, B2, and D) (32), but the majority fell within the A and B1 groups, with 51.3% (n = 41) in the A group and 33.8% (n = 27) in the B1 group. Only 6.3% (n = 5), and 8.8% (n = 7) of the strains belonged to group B2 and group D, respectively (Fig. 1). Analyzing the etiology of group A and B1 isolates, we found that intestinal and extraintestinal isolates were extensively disseminated throughout each phylogenetic group, without significant enrichment in either of the phylogroups (P > 0.05 [Fisher’s exact test]). These results indicated that no clear phylogenomic division exists between intestinal strains and extraintestinal strains as observed in the disparate sequence types (such as ST10, ST48, ST156, etc.) (Fig. 1). Similar distributions were observed in groups B2 and D, although each group contained only a limited number of isolates.
FIG 1 .
Genomic analysis of mcr-1-carrying E. coli isolates in a single hospital system in Zhejiang Province, China. A maximum-likelihood phylogenetic tree was constructed using the core genome SNPs and midpoint rooted. Sources of the isolates are indicated by different colors for strain identification (ID) plus MLST (red, infectious sample of inpatient; green, feces of inpatient; black, feces of healthy volunteer). E. coli phylogroups are denoted by colored strips, and the branches of the tree are colored in correspondence to the coloring of the strips. The location of mcr-1 on a plasmid or chromosome (dark red) and the presence or absence of virulence genes (green) and antibiotic resistance genes (orange) are denoted by filled and empty squares, respectively. Only categories of the virulence genes and 9 clinically important antibiotic resistance genes are shown. Details of the genes in each category are given in Table S4 and S5.
VFs and antibiotic resistance profiles of mcr-1-carrying isolates.
The virulence factors (VFs) in E. coli play an important role in conferring selective advantages and defining pathogenicity profiles. We therefore cataloged the known VFs, including genes associated with adherence, autotransporters, invasion, iron uptake, toxins, and secretion systems, as well as the antibiotic resistance phenotypes. The 12 isolates from phylogroups B2 and D possessed the greatest number of VFs, while 68 isolates of phylogroup A and B strains displayed low prevalence of VFs (Fig. 1; see also Table S4). However, no significant difference was observed in the frequency of VFs between the intestinal and extraintestinal isolates. Furthermore, no VFs related to intestinal pathogenic E. coli could be found, which is consistent with the clinical observation of the patients. Several genes encoding fimbriae were commonly present among the isolates, including those encoding E. coli common pilus (ECP), EaeH, and type 1 fimbriae; however, colonization factor antigen I (CFA/I) fimbriae were less frequently associated with phylogroup A. Types P and S fimbriae, related to extraintestinal infection, were found in phylogroups B2 and D (Fig. 1). Antibiotic resistance profiling showed that all strains were resistant to multiple categories of drugs, including colistin. Two isolates, ZJ134 (from sputum of an inpatient) and ZJ33 (from feces of an inpatient), were resistant to 11 classes of antimicrobials (Table S2); however, the resistance profiles of the isolates were not specific to any phylogenetic group. A total of 58.8% (47/80) of the isolates carried both mcr-1 and ESBL genes (Table S5). Of particular note, one isolate, ZJ487 (ST90), from intra-abdominal fluid contained not only mcr-1 but also blaCTX-M-1, blaNDM-1, blaCTX-M-55, blaOXA-1, and blaSHV-12. E. coli ST131 is the most prevalent sequence type associated with extraintestinal infections (36–38), and we identified two mcr-1-positive ST131 isolates of the serotype of H4:O25. One ST131 isolate, ZJ26, from the fecal sample of a healthy volunteer, carried VF genes iroN, iha, gad, iss, cma, tsh, and fimH22 and antibiotic resistance genes blaCTX-M-55 and mcr-1 and the oqxAB plasmid-mediated quinolone resistance gene. The other ST131 isolate, ZJ71, from the fecal sample of a healthy volunteer, possessed fluoroquinolone resistance alleles gyrA1AB and parC1aAB, blaCTX-M-14, blaTEM-1B, tet(A), and VF genes sat, iss, iha, and senB; the fimH30-gyrA1AB-parC1aAB allelic profile of that isolate was considered to show that it was the more pandemic of the two ST131 isolates (39).
Characterization of virulence genes in 80 mcr-1-carrying E. coli isolates. Download TABLE S4, XLSX file, 0.1 MB (104.2KB, xlsx) .
Copyright © 2018 Shen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Characterization of resistance genes in 80 mcr-1-carrying E. coli isolates. Download TABLE S5, XLSX file, 0.1 MB (57.7KB, xlsx) .
Copyright © 2018 Shen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
mcr-1 is mainly plasmid mediated.
To classify the locations of mcr-1 in the genomes, mcr-1-containing contigs were extracted from the complete genomes of 15 isolates subjected to SMRT sequencing and the draft genomes of 65 isolates sequenced by Illumina. The results of analysis of the complete genomes of 15 isolates suggested that mcr-1 is located on the chromosome in 6 isolates and on plasmids in 9 isolates (Fig. 2; see also Table S1 and S3). Interestingly, isolates ZJ1432 and ZJ859 contain two copies of mcr-1, which were located on distinct plasmids (Fig. 2). mcr-1 was carried on an ~34-kb IncX4 plasmid and an ~62-kb IncI2 plasmid in ZJ1432, while mcr-1 was carried on an ~68-kb IncI2 plasmid and an ~82-kb IncFIB plasmid in ZJ859. To determine the locations of mcr-1 in the rest of the 65 isolates sequenced by Illumina, mcr-1-containing contigs from the 65 isolates were searched against the PlasmidFinder database (40) to define their plasmid types, and the whole draft genomes of each of the isolates were subjected to a BLAST search against the complete mcr-1-carrying plasmids deposited in GenBank to find the closest reference. PlasmidFinder detected that 54 of the 65 mcr-1-positive contigs also carried plasmid replicons, suggesting that their mcr-1 genes are plasmid-borne genes. The other 11 isolates did not possess any known plasmid replicons within the mcr-1-positive contigs. When the complete set of contigs for each isolate was mapped to reference plasmids, 10 of 11 isolates mapped more than 95% to their closest reference plasmids (Fig. 2; see also Table S1). Replicons of the 10 isolates were reexamined using the complete set of contigs of each genome. The results showed that all 10 isolates contain the same replicons as the reference plasmids (see Fig. S1 to S4 at https://doi.org/10.6084/m9.figshare.6281579.v1), but the replicons were separated from mcr-1-containing contigs due to the incomplete assembly. The only exception was the mcr-1 contig (~19.7 kb) from isolate ZJ3951, which did not contain any plasmid replicons and did not map sufficiently with any known plasmids. However, that contig is highly similar to homologous regions of the E. coli chromosome available in the GenBank database (e.g., CP018962.1 and CP018103.1), indicating that mcr-1 in ZJ3951 is located on the chromosome (see Fig. S5 at https://doi.org/10.6084/m9.figshare.6281579.v1). As expected, the locations of mcr-1 based on the whole-genome sequence analysis were highly consistent with the S1 pulsed-field gel electrophoresis (PFGE) results (see Fig. S6 at https://doi.org/10.6084/m9.figshare.6281579.v1), represented as chromosome-borne or plasmid-borne. In total, mcr-1 was found to be chromosome-borne in 7 isolates and plasmid-borne in 73 isolates. As indicated in the NCBI database, IncI2 (n = 29, 38.7%) and IncX4 (n = 28, 37.3%) were the two main plasmid types carrying mcr-1 gene (Fig. 2). A 97-kb p0111 plasmid was first found to carry the mcr-1 gene by SMRT sequencing in two isolates, ZJ275 and ZJ276.
FIG 2 .
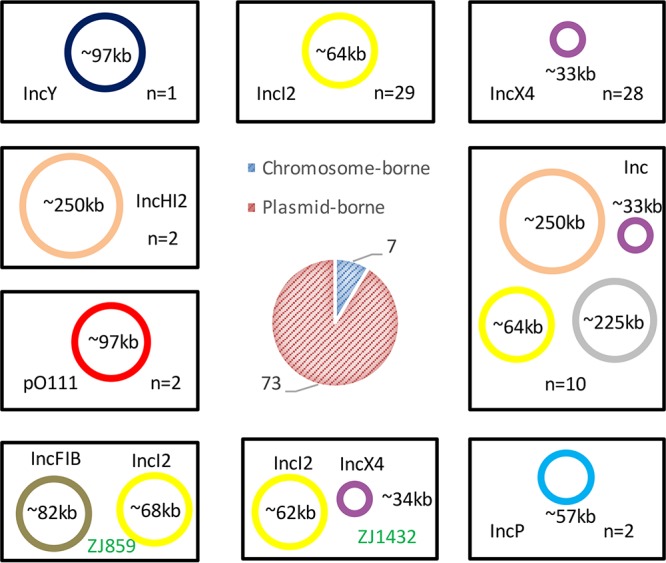
Schematic depiction of the prevalence of mcr-1 in various types of plasmids or on the chromosomes. The types of plasmids potentially carrying the mcr-1 gene were derived from the mcr-1-carrying contigs of each isolate. A colored circle represents a reference mcr-1-carrying plasmid of each type or the complete plasmid determined by SMRT sequencing in this study, and the size of the circle is proportional to that of the reference plasmid. Ten mcr-1-carrying plasmids whose replicons were not in the same contigs as mcr-1 are grouped in one box (Inc). mcr-1 is chromosome-borne in 7 isolates and plasmid-borne in 73 isolates (central pie chart). IncI2 and IncX4 are the major types of mcr-1-carrying plasmids. Isolates ZJ859 and ZJ1432 contained two mcr-1-carrying plasmids each. Thus, the total number of mcr-1-carrying plasmids is 75 in this study.
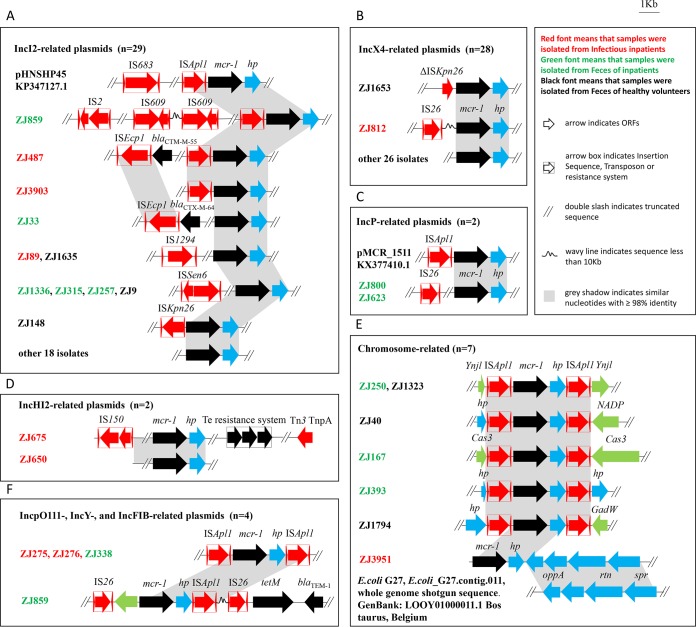
Genetic context of mcr-1 in the 80 E. coli isolates.
To characterize the mobile genetic elements and antibiotic resistance genes in the immediate proximity of mcr-1, we extracted the mcr-1-bearing contigs from 65 draft genomes and 15 complete genomes. Of the 65 contigs from draft genomes, 63 ranged from 12.1 kb to 141.2 kb in size; the remaining two were ~6.7 kb and ~8.3 kb in ZJ1682 and ZJ878, respectively (Table S1). We classified the genetic context into the following nine groups: an IncI2-related group (n = 29), an IncX4-related group (n = 28), an IncHI2-related group (n = 2), an IncP-related group (n = 2), an Incp0111-related group (n = 2), an IncY-related group (n = 1), an IncFIB-related group (n = 1), an non-Inc-related group (n = 10), and an chromosome-related group (n = 7). Overall, considerable diversity was found in the immediate environment of mcr-1. Compared to the previous reports, several novel genetic environments of mcr-1 were found in this study, especially among those located on the chromosomes (Fig. 3).
FIG 3 .
The diverse genetic environments of mcr-1 in 80 E. coli isolates. In panels A to G, red fonts indicate isolates from inpatients with infections; green fonts denote isolates from feces of inpatients; and black fonts depict isolates from feces of healthy volunteers. APEC, avian pathogenic E. coli; ORF, open reading frame.
IncI2-related contigs.
The contigs from 29 isolates were IncI2-related (Fig. 2; see also Fig. S7 at https://doi.org/10.6084/m9.figshare.6281579.v1). The flanking sequences of mcr-1 in all 29 isolates were very similar to sequences in plasmid pHNSHP45 (IncI2; KP347127.1), the first reported mcr-1-carrying plasmid in E. coli of swine origin in China. The ISApl1 insertion sequence is located immediately upstream of mcr-1 in pHNSHP45. However, only three of our isolates (ZJ487, ZJ859, and ZJ3903) contained ISApl1 upstream of mcr-1, while the other 26 isolates showed no evidence of ISApl1 adjunct to mcr-1 (Fig. 3A; see also Fig. S7 at https://doi.org/10.6084/m9.figshare.6281579.v1). Among the 26 isolates, 18 did not contain any insertion sequence and 8 contained IS elements other than ISApl1 (Fig. 3A). ISSen6, initially found in Salmonella enterica (NC_006511), was present in mcr-1-carrying contigs in four isolates (ZJ1336, ZJ315, ZJ9, and ZJ257). ISKpn26, initially found in K. pneumoniae (NC_016845), was detected in mcr-1-positive contigs in ZJ148, while ISEcp1 and IS1294, initially found in E. coli, were identified in mcr-1-positive contigs in ZJ33, ZJ89, and ZJ1635. Even though multiple ESBL genes were detected in isolate ZJ487, only blaCTX-M-1 was found to be located on the mcr-1-carrying plasmid shown by SMRT sequencing. blaNDM-1 and blaSHV-12 were found to be located on an ~54-kb plasmid, and blaCTX-M-55 and blaOXA-1 were found to be located on another, ~129-kb plasmid. In isolate ZJ33, blaCTX-M-64 flanked by ISEcp1 was shown to be located in the same mcr-1-carrying contig. Isolate ZJ859 harbored two IS609 copies and one IS2 copy in a mcr-1-carrying contig.
IncX4-related contigs.
Twenty-eight contigs were associated with IncX4 (Fig. 2; see also Fig. S8 at https://doi.org/10.6084/m9.figshare.6281579.v1). The overall genetic contexts of these contigs showed similarity to plasmid pECGD-8-33 (IncX4; KX254343.1), and 26 of them did not show any evidence of insertion sequences (Fig. 3B; see also Fig. S8 at https://doi.org/10.6084/m9.figshare.6281579.v1). However, a truncated insertion sequence, ISKpn26, was found upstream of mcr-1 in ZJ1653, and IS26 was found ~2 kb upstream of mcr-1 in ZJ812. It is known that IS26 is commonly associated with resistance genes in E. coli and other species (Fig. 3B) (41). However, no resistance genes were found on the mcr-1-carrying contigs of ZJ1653 or on the other IncX4-related contigs.
IncHI2-related contigs.
Two mcr-1-positive contigs from isolates ZJ675 and ZJ650 were found to be IncHI2-related (Fig. 2; see also Fig. S9 at https://doi.org/10.6084/m9.figshare.6281579.v1), and the genetic context was found to be similar to that of pHNSHP45-2 (IncHI2; KU341381.1). The mcr-1-carrying contig in ZJ650 was 107,130 bp and did not harbor any IS elements or other antibiotic resistance genes. The mcr-1-positive contig in ZJ675 (141,207 bp) contains tellurium resistance system genes, i.e., a ter (tellurium resistance [Ter]) operon (terZABCDEF), conferring resistance to tellurite (42). In addition, IS150 and Tn3 were identified in the mcr-1-positive contig of ZJ675 (Fig. 3D; see also Fig. S9 at https://doi.org/10.6084/m9.figshare.6281579.v1).
IncP-related contigs.
The genetic contexts of mcr-1-carrying contigs from isolates ZJ800 and ZJ623 are IncP-related and resemble DNA regions from plasmid pMCR_1511 (IncP; KX377410.1) (Fig. 2; see also Fig. S10 at https://doi.org/10.6084/m9.figshare.6281579.v1). ISApl1 is missing in the two mcr-1-carrying contigs (Fig. 3C); instead, IS26 is distally present with mcr-1. No other antibiotic resistance genes were found in these two contigs.
Incp0111-, IncY-, and IncFIB-related contigs.
The complete mcr-1 plasmids in ZJ275 and ZJ276 possess the Incp0111 replicon (see Fig. S11 at https://doi.org/10.6084/m9.figshare.6281579.v1), while the complete mcr-1-positive plasmid in ZJ338 contains the IncY replicon (see Fig. S12 at https://doi.org/10.6084/m9.figshare.6281579.v1). The genetic contexts of Incp0111 and IncY are very similar, and mcr-1 was flanked by ISApl1 at both ends, represented as ISApl1–mcr-1–hp–ISApl1 (Fig. 3F; see also Fig. S11 at https://doi.org/10.6084/m9.figshare.6281579.v1), with the two ISApl1 elements in the same orientation. Inverse PCR using primers located within mcr-1 was performed to detect the small circular form of this region in ZJ275 and ZJ276. PCR produced a 3,696-bp amplicon containing mcr-1, the region between mcr-1 and two ISApl1 genes, and one intact ISApl1 element (data not shown), suggesting that recombination between the two ISApl1 copies may occur and may facilitate the transmission of mcr-1. One complete plasmid (81,972 bp) of ZJ859 possesses the IncFIB replicon (see Fig. S13 at https://doi.org/10.6084/m9.figshare.6281579.v1), in which mcr-1 is flanked by ISApl1 and IS26 upstream and IS26 downstream (Fig. 3; see also Fig. S14 at https://doi.org/10.6084/m9.figshare.6281579.v1). Furthermore, genes corresponding to Tn3, IS1A, another two IS26 elements, and a type IV secretion system were present in the plasmid (Fig. 3F). This plasmid also contains two additional antibiotic resistance genes, blaTEM-1 and tetM, which were associated with Tn3 and proximal to mcr-1.
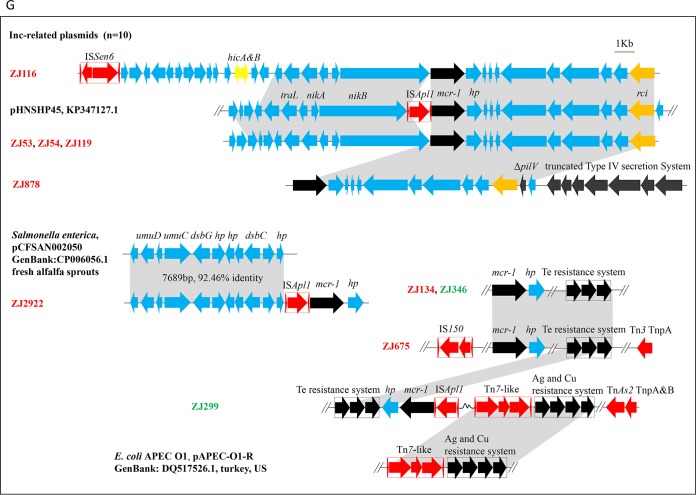
Non-Inc-related contigs.
Ten mcr-1-positive contigs did not have replicon genes due to incomplete assembly (Fig. 2). As described previously, the possible Inc types of these contigs carrying plasmids were predicted through mapping the contigs to the reference plasmids (see Fig. S1 to S4 at https://doi.org/10.6084/m9.figshare.6281579.v1). The mcr-1-positive contigs of ZJ53, ZJ54, JZ116, ZJ119, and ZJ878 were similar to those of pHNSHP45 (IncI2) (see Fig. S1 at https://doi.org/10.6084/m9.figshare.6281579.v1); the mcr-1-positive contig of ZJ116 contains a copy of ISSen6. In isolate ZJ2922, ISApl1–mcr-1–hp was located downstream of a 7,689-bp segment that exhibited 92.5% nucleotide identity to the corresponding region of plasmid pCFSAN002050 from Salmonella enterica (GenBank accession number CP006056.1) (Fig. 3G). In isolates ZJ134, ZJ346, ZJ299, and ZJ675, the mcr-1-positive contigs contain the tellurium resistance system, which is commonly carried by IncHI2 or InHII plasmids (43). Moreover, a Tn7-like transposon element and two resistance systems for heavy metals, the pco (COPR) operon (pcoEABCDRSE) for copper resistance and the sil (silR) operon (silESRCBAP) for silver resistance, were identified upstream of ISApl1–mcr-1–hp in isolate ZJ299, which is similar to the corresponding region of IncHI2 plasmid pAPEC-O1-R (DQ517526) from E. coli isolated from turkey originating from United States (44) (Fig. 3G).
Chromosome-borne mcr-1.
mcr-1 was located on the chromosome in seven isolates; six of the isolates were subjected to SMRT sequencing, and the remaining isolate, ZJ3951, was subjected to Illumina sequencing (Fig. 3E; see also Fig. S15 at https://doi.org/10.6084/m9.figshare.6281579.v1). The mcr-1-carrying contig of ZJ3951 is ~19.7 kb in size and similar to the chromosome of E. coli G27 from Belgium (accession number LOOY01000011.1) (Fig. 3E). In six isolates, ZJ40, ZJ167, ZJ250, ZJ393, ZJ1323, and ZJ1794, mcr-1 was flanked by two ISApl1 elements, forming a unit of ISApl1–mcr-1–hp–ISApl1 as observed in plasmids from ZJ275 and ZJ276. Interestingly, mcr-1 was inserted in the same location in only two isolates, ZJ1323 and ZJ250, which belong to ST4014, and was located in the quite different regions in the other five isolates. All five isolates belong to different ST clades, and the integration sites of mcr-1 on the chromosome were distinct (Fig. 3E), indicating that the mobile genetic unit of “ISApl1–mcr-1–hp–ISApl1” has been recently disseminated among E. coli isolates.
mcr-1 allelic variations.
Here, 10 MCR-1 variants (mcr-1.2 to mcr-1.11) were deposited in NCBI, and in this study novel mcr-1 variants were detected in six isolates (ZJ10, ZJ2431, ZJ299, ZJ3920, ZJ346, and ZJ1432) from various sources (Table S6). The allelic variants from isolates ZJ10, ZJ2431, and ZJ346 exhibited synonymous mutations at positions 18 (T→C), 27 (C→T), and 552 (C→T), respectively. Moreover, ZJ299 and ZJ3920 each harbored a single nucleotide mutation in mcr-1, resulting in nonsynonymous mutations at positions 145 (G→A, Gly→Ser), and 1423 (G→A, Val→Ile), respectively. Finally, isolate ZJ1432, which had two mcr-1 copies in two different plasmids, contained a synonymous mutation at site 27 (C→T) in the mcr-1 copy on the IncX4 plasmid. All the mutations were confirmed by Sanger sequencing (data not shown).
Allelic variations of seven mcr-1-carrying isolates. Download TABLE S6, XLSX file, 0.03 MB (34.3KB, xlsx) .
Copyright © 2018 Shen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
In this study, 80 mcr-1-positive E. coli isolates of intestinal or extraintestinal sources from a single hospital in China were analyzed by a combination of Illumina and SMRT sequencing. We observed significant diversity not only in the genetic context of mcr-1 but particularly in the host E. coli. The mcr-1-positive E. coli isolates are distributed throughout the four phylogroups (A, B1, B2, and D) (32) and represented by 50 ST clades, indicating a lack of clonal spread and implying no outbreak of mcr-1-positive E. coli in this hospital (Fig. 1). In a recent study conducted by another Chinese group, MCRPEC isolates of fecal sources were also found to be genetically diverse (45). However, a case of possible nosocomial transmission of mcr-1-positive E. coli was observed. The complete genomes of ZJ275 and ZJ276 revealed that the two isolates contain identical chromosomes and plasmids, with only 32 single nucleotide variants (SNVs) (4 single nucleotide polymorphisms [SNPs] and 28 indels) in the chromosome and 2 SNVs in the mcr-1-positive plasmid. It should be noted that the mcr-1 gene was located on p0111 plasmids, which were specific to these two strains. Furthermore, ZJ275 and ZJ276 were isolated from the puncture fluid of a 76-year-old man and from the drainage fluid of a 38-year-old woman. The patients were kept in the same surgical intensive care unit during the same period and therefore, the possibility exists of nosocomial transmission of mcr-1-positive E. coli, highlighting the importance of hospital-acquired infections and hospital infection control programs (46).
Previous epidemiological studies indicated that groups A and B1 are usually composed of commensal strains or intestinal pathogenic strains (47) and groups B2 and D are associated with extraintestinal pathogenic strains (36, 48). However, in this study, no clear phylogenomic divisions were observed between intestinal strains and extraintestinal strains. Interestingly, examination of 14 extraintestinal isolates from bloodstream (n = 2) and urinary tract (n = 12) showed that only two isolates belonged to groups B2 and D (Fig. 1). Furthermore, analysis of virulence factors did not show any correlation between intestinal isolates and extraintestinal isolates (Fig. 1; see also Table S4 in the supplemental material). Although the data set is limited, this observation suggests that the intestinal strains may potentially cause systemic infections and serve as a source of nosocomial infections. In hospital settings, many patients are immunocompromised, have indwelling urinary catheters, and are exposed to numerous antimicrobials (Table S2), which might promote extraintestinal infection by antibiotic-resistant E. coli strains that are not normally considered extraintestinal pathogenic E. coli strains.
Acquisition of antibiotic resistance determinants through horizontal gene transfer is highly problematic. The presence of antibiotic resistance gene-linked transferable elements facilitates their spread among different clones and different bacterial species (49). mcr-1 can be found in various plasmids and in different locations on the chromosomes of E. coli. The plasmids were classified into 8 Inc types by bioinformatics analysis, and the IncI2 (38.7%) and IncX4 (37.3%) plasmids were found to be the most prevalent types among the isolates, consistent with previous reports (21, 50, 51). IncI2 mcr-1-carrying plasmids were previously found to be dominant in E. coli-associated bloodstream infections (29). However, our study data indicate that neither IncI2 plasmids nor IncX4 plasmids were predominantly associated with intestinal or extraintestinal sources, patients (6/27 IncI2, 11/27 IncX4), or volunteers (8/17 IncI2, 5/17 IncX4). Interestingly, the genetic contexts of mcr-1 were found to be highly divergent, and yet the majority of mcr-1 genes were not adjacent to IS elements (17 were flanked with IS elements [Fig. 3]). ISApl1 was the most common IS element and was found to be adjacent to mcr-1 at one end or both ends in 15 isolates. The presence of ISApl1 at the both ends of mcr-1 makes the ISApl1–mcr-1–hp–ISApl1 unit potentially active, as was evidenced by the circular form shown by inverse PCR, which can integrate into various locations on the chromosome and/or plasmids in a manner consistent with previous reports (52, 53). Furthermore, ISKpn26 was adjacent to mcr-1 in isolates ZJ148 and ZJ1653, and several other IS elements such as IS2, IS609, IS683, IS1294, IS150, and ISSen6 were found in the mcr-1-positive contigs but not in locations immediately proximal to mcr-1. Whether these IS elements could contribute to the mobilization of mcr-1 remains unknown, but their presence would enhance the transmission of mcr-1 and facilitate its close association with other antibiotic resistance genes as seen in isolates ZJ134, ZJ33, ZJ487, ZJ26, and ZJ71 (41, 54, 55).
In this study, a strategy combining Illumina sequencing and SMRT sequencing was applied to decipher the genetic diversity of mcr-1-positive E. coli isolates. Although we successfully decoded the phylogenetic relationships, virulence factors, antibiotic resistance genes, and genetic contexts of mcr-1 of 80 isolates, we could not definitively determine how many antibiotic resistance genes coexist with mcr-1 on the same plasmids due to some incomplete assemblies (generated by short reads of Illumina sequencing). SMRT sequencing offers a definitive solution for deciphering individual bacterial isolates by virtue of its long reads, but the significantly higher cost limits its application for large numbers of samples. We therefore subjected 15 isolates to SMRT sequencing to complement the data set generated by Illumina sequencing. This combined-sequencing method revealed that mcr-1 was located on the chromosome of 7 isolates and in the plasmids of 73 isolates. New sequencing technologies (such as nanopore sequencing) that generate long sequence reads may be used in future studies to decipher mcr-1-carrying MDR plasmids (56), which would facilitate our understanding of the cotransfer of mcr-1 with other antimicrobial resistance genes in bacterial plasmids.
MATERIALS AND METHODS
Ethical approval.
Ethical approval of the study was granted by the Second Affiliated Hospital of Zhejiang University.
Genome sequencing and assembly.
Genomic DNA of all mcr-1-positive isolates was extracted using a Wizard genomic DNA purification kit (Promega, Beijing, China), following the manufacturer’s instructions. Indexed Illumina sequencing libraries were prepared using a TruSeq DNA PCR-free sample preparation kit (Illumina Inc., San Diego, CA) following the standard protocol and were sequenced on an Illumina HiSeq 2500 platform according to the manufacturer’s protocols; the sequencing produced 250-bp paired-end reads (Bionova, Beijing, China). The draft genomes were assembled using the SPAdes algorithm (57) and CLC Genomics workbench 8.5 (CLC Bio, Aarhus, Denmark). To optimize the assembly of plasmids, Illumina reads of each isolates were reassembled by plasmidSPAdes, a program to assemble plasmids from whole-genome sequencing data (58). All the assemblies were further corrected by Pilon (59). The mcr-1-containing contigs for each isolate were extracted from the two assemblies, and the longer one was used for analysis of the genetic context of mcr-1.
Fifteen isolates (9 isolates that generated only short mcr-1-containing contigs in Illumina sequencing and 6 isolates that were able to cotransfer other antibiotic resistance genes with mcr-1) were further sequenced by the use of SMRT sequencing (60). Genomic DNA was sheared to 10 to 17 kb using Covaris g-tubes (Covaris) and converted into SMRTbell template libraries. The libraries were subsequently subjected to DNA size selection using a BluePippin instrument (Sage Science) to select the longest DNA fragments (lower size cutoff value of ~5 kb). Sequencing was performed on a PacBio RSII system using P6 polymerase binding and C4 sequencing kits with magnetic bead loading and 120-min acquisition (Sinobiocore, Beijing, China). Genome assemblies were performed using HGAP and Quiver as part of SMRTAnalysis version 2.3 by the use of the HGAP3 protocol and corrected using Pilon. Shotgun sequences of all 80 isolates have been deposited in the NCBI database (see below).
Molecular epidemiology and analysis of virulence and antibiotic resistance genes.
Assembled genomes from Illumina and Pacbio sequencing were aligned, and a core-genome phylogenetic tree was generated by Parsnp in the Harvest package (61). MLST analysis and examinations of known virulence-associated genes and antibiotic resistance genes were carried out using pipeline SRST2, which takes Illumina reads as the input (62). Reference sequences of virulence genes and antibiotic resistance genes were from databases VFDB (63) and ARG-ANNOT (64), respectively. The tree and the molecular features of each isolate were visualized by the use of the online tool iTOL (65). Classification of strains to phylogenetic groups was performed by using assembled contigs according to a scheme described previously (32, 66).
Analysis of mcr-1 location.
The plasmid or chromosome location of the mcr-1 gene in 80 E. coli isolates was first determined by S1-nuclease digestion, pulsed-field gel electrophoresis, and probing with the mcr-1 DNA fragment as described previously (9). The location of mcr-1 was further confirmed by sequence analysis. mcr-1-containing contigs generated by Illumina and Pacbio sequencing were examined for Inc types by PlasmidFinder (40). The mcr-1-carrying contigs encoding plasmid replicons were considered to come from plasmids, and all the contigs of the genome of each corresponding isolate were mapped to the reference plasmids by BLASTN (67). A closest reference plasmid was selected based on the mapping coverage to scaffold the mcr-1-carrying plasmid from each isolate.
Analysis of the genetic context of mcr-1.
The mcr-1-carrying contigs were annotated by the use of the RAST annotation server (68). The insertion sequences were identified by ISfinder (69). For contigs in which mcr-1 was immediately surrounded by IS elements or only the mcr-1-hp core region was observed, an inverse PCR assay using primers F-primer (TATTCTGTGCCGTGTATGTT) and R-primer (TATCAGGCTTGGTTGCTT) (annealing temperature, 55°C) located within the mcr-1 gene was performed to determine the possible existence of a free circular form containing IS-flanked sequence.
Additional material.
Supplemental figures can be found at figshare (https://doi.org/10.6084/m9.figshare.6281579.v1). These show the genetic contexts of mcr-1 in the E. coli isolates from one hospital.
Accession number(s).
Shotgun sequences of all 80 isolates have been deposited in the NCBI database (BioProject accession no. PRJNA331013 and BioSample accession no. SAMN05437795 to SAMN05437874), while 15 SMRT sequences have been deposited under other accession numbers (BioProject accession no. PRJNA380845 and BioSample accession no. SAMN06649969 to SAMN06649983).
ACKNOWLEDGMENTS
This work was supported in part by the National Natural Science Foundation of China (grants 31672604 and 31422055) and the Natural Science Foundation of Zhejiang Province (2013C33G2010505), Higher Education Funding Council and European Commission, United Kingdom.
Footnotes
Citation Shen Y, Wu Z, Wang Y, Zhang R, Zhou H-W, Wang S, Lei L, Li M, Cai J, Tyrrell J, Tian G-B, Wu C, Zhang Q, Shen J, Walsh TR, Shen Z. 2018. Heterogeneous and flexible transmission of mcr-1 in hospital-associated Escherichia coli. mBio 9:e00943-18. https://doi.org/10.1128/mBio.00943-18.
REFERENCES
- 1.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. 2007. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Discov 6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 2.Alanis AJ. 2005. Resistance to antibiotics: are we in the post-antibiotic era? Arch Med Res 36:697–705. doi: 10.1016/j.arcmed.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 3.Fears R, ter Meulen V. 2014. What do we need to do to tackle antimicrobial resistance? Lancet Glob Health 2:e11–e12. doi: 10.1016/S2214-109X(13)70086-X. [DOI] [PubMed] [Google Scholar]
- 4.Sprenger M, Fukuda K. 2016. Antimicrobial resistance. New mechanisms, new worries. Science 351:1263–1264. doi: 10.1126/science.aad9450. [DOI] [PubMed] [Google Scholar]
- 5.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, Kumarasamy K, Livermore DM, Maya JJ, Nordmann P, Patel JB, Paterson DL, Pitout J, Villegas MV, Wang H, Woodford N, Quinn JP. 2013. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis 13:785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, Chaudhary U, Doumith M, Giske CG, Irfan S, Krishnan P, Kumar AV, Maharjan S, Mushtaq S, Noorie T, Paterson DL, Pearson A, Perry C, Pike R, Rao B, Ray U, Sarma JB, Sharma M, Sheridan E, Thirunarayan MA, Turton J, Upadhyay S, Warner M, Welfare W, Livermore DM, Woodford N. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis 10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans BA, Amyes SG. 2014. OXA beta-lactamases. Clin Microbiol Rev 27:241–263. doi: 10.1128/CMR.00117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bialvaei AZ, Samadi Kafil H. 2015. Colistin, mechanisms and prevalence of resistance. Curr Med Res Opin 31:707–721. doi: 10.1185/03007995.2015.1018989. [DOI] [PubMed] [Google Scholar]
- 9.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. [DOI] [PubMed] [Google Scholar]
- 10.Falgenhauer L, Waezsada SE, Yao Y, Imirzalioglu C, Käsbohrer A, Roesler U, Michael GB, Schwarz S, Werner G, Kreienbrock L, Chakraborty T; RESET Consortium . 2016. Colistin resistance gene mcr-1 in extended-spectrum β-lactamase-producing and carbapenemase-producing Gram-negative bacteria in Germany. Lancet Infect Dis 16:282–283. doi: 10.1016/S1473-3099(16)00009-8. [DOI] [PubMed] [Google Scholar]
- 11.Li A, Yang Y, Miao M, Chavda KD, Mediavilla JR, Xie X, Feng P, Tang YW, Kreiswirth BN, Chen L, Du H. 2016. Complete sequences of mcr-1-harboring plasmids from extended-spectrum-beta-lactamase- and carbapenemase-producing Enterobacteriaceae. Antimicrob Agents Chemother 60:4351–4354. doi: 10.1128/AAC.00550-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yao X, Doi Y, Zeng L, Lv L, Liu JH. 2016. Carbapenem-resistant and colistin-resistant Escherichia coli co-producing NDM-9 and MCR-1. Lancet Infect Dis 16:288–289. doi: 10.1016/S1473-3099(16)00057-8. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y, Zhang R, Li J, Wu Z, Yin W, Schwarz S, Tyrrell JM, Zheng Y, Wang S, Shen Z, Liu Z, Liu J, Lei L, Li M, Zhang Q, Wu C, Zhang Q, Wu Y, Walsh TR, Shen J. 2017. Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat Microbiol 2:16260. doi: 10.1038/nmicrobiol.2016.260. [DOI] [PubMed] [Google Scholar]
- 14.Ochman H, Lawrence JG, Groisman EA. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 15.Toleman MA, Bennett PM, Walsh TR. 2006. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol Mol Biol Rev 70:296–316. doi: 10.1128/MMBR.00048-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poirel L, Dortet L, Bernabeu S, Nordmann P. 2011. Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrob Agents Chemother 55:5403–5407. doi: 10.1128/AAC.00585-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGann P, Snesrud E, Maybank R, Corey B, Ong AC, Clifford R, Hinkle M, Whitman T, Lesho E, Schaecher KE. 2016. Escherichia coli harboring mcr-1 and blaCTX-M on a novel IncF plasmid: first report of mcr-1 in the United States. Antimicrob Agents Chemother 60:4420–4421. doi: 10.1128/AAC.01103-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pham Thanh D, Thanh Tuyen H, Nguyen Thi Nguyen T, Chung The H, Wick RR, Thwaites GE, Baker S, Holt KE. 2016. Inducible colistin resistance via a disrupted plasmid-borne mcr-1 gene in a 2008 Vietnamese Shigella sonnei isolate. J Antimicrob Chemother 71:2314–2317. doi: 10.1093/jac/dkw173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poirel L, Kieffer N, Brink A, Coetze J, Jayol A, Nordmann P. 2016. Genetic features of MCR-1-producing colistin-resistant Escherichia coli isolates in South Africa. Antimicrob Agents Chemother 60:4394–4397. doi: 10.1128/AAC.00444-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xavier BB, Lammens C, Butaye P, Goossens H, Malhotra-Kumar S. 2016. Complete sequence of an IncFII plasmid harbouring the colistin resistance gene mcr-1 isolated from Belgian pig farms. J Antimicrob Chemother 71:2342–2344. doi: 10.1093/jac/dkw191. [DOI] [PubMed] [Google Scholar]
- 21.Sun J, Li XP, Yang RS, Fang LX, Huo W, Li SM, Jiang P, Liao XP, Liu YH. 2016. Complete nucleotide sequence of an IncI2 plasmid coharboring blaCTX-M-55 and mcr-1. Antimicrob Agents Chemother 60:5014–5017. doi: 10.1128/AAC.00774-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao R, Wang Q, Li P, Li Z, Feng Y. 2016. Genome sequence and characteristics of plasmid pWH12, a variant of the mcr-1-harbouring plasmid pHNSHP45, from the multi-drug resistant E. coli. Virulence 7:732–735. doi: 10.1080/21505594.2016.1193279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhi C, Lv L, Yu LF, Doi Y, Liu JH. 2016. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 16:292–293. [DOI] [PubMed] [Google Scholar]
- 24.Li R, Xie M, Zhang J, Yang Z, Liu L, Liu X, Zheng Z, Chan EW, Chen S. 2017. Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J Antimicrob Chemother 72:393–401. doi: 10.1093/jac/dkw411. [DOI] [PubMed] [Google Scholar]
- 25.Yu CY, Ang GY, Chong TM, Chin PS, Ngeow YF, Yin WF, Chan KG. 2017. Complete genome sequencing revealed novel genetic contexts of the mcr-1 gene in Escherichia coli strains. J Antimicrob Chemother 72:1253–1255. doi: 10.1093/jac/dkw541. [DOI] [PubMed] [Google Scholar]
- 26.Li R, Xie M, Lv J, Wai-Chi Chan E, Chen S. 2017. Complete genetic analysis of plasmids carrying mcr-1 and other resistance genes in an Escherichia coli isolate of animal origin. J Antimicrob Chemother 72:696–699. doi: 10.1093/jac/dkw509. [DOI] [PubMed] [Google Scholar]
- 27.Sun J, Fang LX, Wu Z, Deng H, Yang RS, Li XP, Li SM, Liao XP, Feng Y, Liu YH. 2017. Genetic analysis of the IncX4 plasmids: implications for a unique pattern in the mcr-1 acquisition. Sci Rep 7:424. doi: 10.1038/s41598-017-00095-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hasman H, Hammerum AM, Hansen F, Hendriksen RS, Olesen B, Agersø Y, Zankari E, Leekitcharoenphon P, Stegger M, Kaas RS, Cavaco LM, Hansen DS, Aarestrup FM, Skov RL. 2015. Detection of mcr-1 encoding plasmid-mediated colistin-resistant Escherichia coli isolates from human bloodstream infection and imported chicken meat, Denmark 2015. Euro Surveill 20. doi: 10.2807/1560-7917.ES.2015.20.49.30085. [DOI] [PubMed] [Google Scholar]
- 29.Quan J, Li X, Chen Y, Jiang Y, Zhou Z, Zhang H, Sun L, Ruan Z, Feng Y, Akova M, Yu Y. 2017. Prevalence of mcr-1 in Escherichia coli and Klebsiella pneumoniae recovered from bloodstream infections in China: a multicentre longitudinal study. Lancet Infect Dis 17:400–410. doi: 10.1016/S1473-3099(16)30528-X. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y, Tian GB, Zhang R, Shen Y, Tyrrell JM, Huang X, Zhou H, Lei L, Li HY, Doi Y, Fang Y, Ren H, Zhong LL, Shen Z, Zeng KJ, Wang S, Liu JH, Wu C, Walsh TR, Shen J. 2017. Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1-positive Enterobacteriaceae in patients and healthy adults from China: an epidemiological and clinical study. Lancet Infect Dis 17:390–399. doi: 10.1016/S1473-3099(16)30527-8. [DOI] [PubMed] [Google Scholar]
- 31.Escobar-Páramo P, Le Menac’h A, Le Gall T, Amorin C, Gouriou S, Picard B, Skurnik D, Denamur E. 2006. Identification of forces shaping the commensal Escherichia coli genetic structure by comparing animal and human isolates. Environ Microbiol 8:1975–1984. doi: 10.1111/j.1462-2920.2006.01077.x. [DOI] [PubMed] [Google Scholar]
- 32.Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol 66:4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tartof SY, Solberg OD, Manges AR, Riley LW. 2005. Analysis of a uropathogenic Escherichia coli clonal group by multilocus sequence typing. J Clin Microbiol 43:5860–5864. doi: 10.1128/JCM.43.12.5860-5864.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oteo J, Diestra K, Juan C, Bautista V, Novais A, Pérez-Vázquez M, Moyá B, Miró E, Coque TM, Oliver A, Cantón R, Navarro F, Campos J; Spanish Network in Infectious Pathology Project (REIPI) . 2009. Extended-spectrum beta-lactamase-producing Escherichia coli in Spain belong to a large variety of multilocus sequence typing types, including ST10 complex/A, ST23 complex/A and ST131/B2. Int J Antimicrob Agents 34:173–176. doi: 10.1016/j.ijantimicag.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 35.Cortés P, Blanc V, Mora A, Dahbi G, Blanco JE, Blanco M, López C, Andreu A, Navarro F, Alonso MP, Bou G, Blanco J, Llagostera M. 2010. Isolation and characterization of potentially pathogenic antimicrobial-resistant Escherichia coli strains from chicken and pig farms in Spain. Appl Environ Microbiol 76:2799–2805. doi: 10.1128/AEM.02421-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salipante SJ, Roach DJ, Kitzman JO, Snyder MW, Stackhouse B, Butler-Wu SM, Lee C, Cookson BT, Shendure J. 2015. Large-scale genomic sequencing of extraintestinal pathogenic Escherichia coli strains. Genome Res 25:119–128. doi: 10.1101/gr.180190.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adams-Sapper S, Diep BA, Perdreau-Remington F, Riley LW. 2013. Clonal composition and community clustering of drug-susceptible and -resistant Escherichia coli isolates from bloodstream infections. Antimicrob Agents Chemother 57:490–497. doi: 10.1128/AAC.01025-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gibreel TM, Dodgson AR, Cheesbrough J, Fox AJ, Bolton FJ, Upton M. 2012. Population structure, virulence potential and antibiotic susceptibility of uropathogenic Escherichia coli from northwest England. J Antimicrob Chemother 67:346–356. doi: 10.1093/jac/dkr451. [DOI] [PubMed] [Google Scholar]
- 39.Petty NK, Ben Zakour NL, Stanton-Cook M, Skippington E, Totsika M, Forde BM, Phan MD, Gomes Moriel D, Peters KM, Davies M, Rogers BA, Dougan G, Rodriguez-Baño J, Pascual A, Pitout JD, Upton M, Paterson DL, Walsh TR, Schembri MA, Beatson SA. 2014. Global dissemination of a multidrug resistant Escherichia coli clone. Proc Natl Acad Sci U S A 111:5694–5699. doi: 10.1073/pnas.1322678111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harmer CJ, Hall RM. 2016. IS26-mediated formation of transposons carrying antibiotic resistance genes. mSphere 1:e00038-16. doi: 10.1128/mSphere.00038-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whelan KF, Sherburne RK, Taylor DE. 1997. Characterization of a region of the IncHI2 plasmid R478 which protects Escherichia coli from toxic effects specified by components of the tellurite, phage, and colicin resistance cluster. J Bacteriol 179:63–71. doi: 10.1128/jb.179.1.63-71.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor DE. 1999. Bacterial tellurite resistance. Trends Microbiol 7:111–115. doi: 10.1016/S0966-842X(99)01454-7. [DOI] [PubMed] [Google Scholar]
- 44.Johnson TJ, Wannemeuhler YM, Scaccianoce JA, Johnson SJ, Nolan LK. 2006. Complete DNA sequence, comparative genomics, and prevalence of an IncHI2 plasmid occurring among extraintestinal pathogenic Escherichia coli isolates. Antimicrob Agents Chemother 50:3929–3933. doi: 10.1128/AAC.00569-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhong LL, Phan HTT, Shen C, Doris-Vihta K, Sheppard AE, Huang X, Zeng KJ, Li HY, Zhang XF, Patil S, Crook DW, Walker AS, Xing Y, Lin JL, Feng LQ, Doi Y, Xia Y, Stoesser N, Tian GB. 2018. High rates of human fecal carriage of mcr-1-positive multi-drug resistant Enterobacteriaceae isolates emerge in China in association with successful plasmid families. Clin Infect Dis 66:676–685. doi: 10.1093/cid/cix885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peleg AY, Hooper DC. 2010. Hospital-acquired infections due to gram-negative bacteria. N Engl J Med 362:1804–1813. doi: 10.1056/NEJMra0904124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.von Mentzer A, Connor TR, Wieler LH, Semmler T, Iguchi A, Thomson NR, Rasko DA, Joffre E, Corander J, Pickard D, Wiklund G, Svennerholm AM, Sjöling Å, Dougan G. 2014. Identification of enterotoxigenic Escherichia coli (ETEC) clades with long-term global distribution. Nat Genet 46:1321–1326. doi: 10.1038/ng.3145. [DOI] [PubMed] [Google Scholar]
- 48.Picard B, Garcia JS, Gouriou S, Duriez P, Brahimi N, Bingen E, Elion J, Denamur E. 1999. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect Immun 67:546–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martínez JL, Coque TM, Lanza VF, de la Cruz F, Baquero F. 2017. Genomic and metagenomic technologies to explore the antibiotic resistance mobilome. Ann N Y Acad Sci 1388:26–41. doi: 10.1111/nyas.13282. [DOI] [PubMed] [Google Scholar]
- 50.Fernandes MR, McCulloch JA, Vianello MA, Moura Q, Pérez-Chaparro PJ, Esposito F, Sartori L, Dropa M, Matté MH, Lira DP, Mamizuka EM, Lincopan N. 2016. First report of the globally disseminated IncX4 plasmid carrying the mcr-1 gene in a colistin-resistant Escherichia coli sequence type 101 isolate from a human infection in Brazil. Antimicrob Agents Chemother 60:6415–6417. doi: 10.1128/AAC.01325-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Falgenhauer L, Waezsada SE, Gwozdzinski K, Ghosh H, Doijad S, Bunk B, Spröer C, Imirzalioglu C, Seifert H, Irrgang A, Fischer J, Guerra B, Käsbohrer A, Overmann J, Goesmann A, Chakraborty T. 2016. Chromosomal locations of mcr-1 and blaCTX-M-15 in fluoroquinolone-resistant Escherichia coli ST410. Emerg Infect Dis 22:1689–1691. doi: 10.3201/eid2209.160692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Snesrud E, Ong AC, Corey B, Kwak YI, Clifford R, Gleeson T, Wood S, Whitman TJ, Lesho EP, Hinkle M, McGann P. 2017. Analysis of serial isolates of mcr-1-positive Escherichia coli reveals a highly active ISApl1 transposon. Antimicrob Agents Chemother 61:e00056-17. doi: 10.1128/AAC.00056-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Snesrud E, He S, Chandler M, Dekker JP, Hickman AB, McGann P, Dyda F. 2016. A model for transposition of the colistin resistance gene mcr-1 by ISApl1. Antimicrob Agents Chemother 60:6973–6976. doi: 10.1128/AAC.01457-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haas M, Rak B. 2002. Escherichia coli insertion sequence IS150: transposition via circular and linear intermediates. J Bacteriol 184:5833–5841. doi: 10.1128/JB.184.21.5833-5841.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gonçalves GA, Oliveira PH, Gomes AG, Prather KL, Lewis LA, Prazeres DM, Monteiro GA. 2014. Evidence that the insertion events of IS2 transposition are biased towards abrupt compositional shifts in target DNA and modulated by a diverse set of culture parameters. Appl Microbiol Biotechnol 98:6609–6619. doi: 10.1007/s00253-014-5695-6. [DOI] [PubMed] [Google Scholar]
- 56.Li R, Xie M, Dong N, Lin D, Yang X, Wong MHY, Chan EW, Chen S. 2018. Efficient generation of complete sequences of MDR-encoding plasmids by rapid assembly of MinION barcoding sequencing data. Gigascience 7:1–9. doi: 10.1093/gigascience/gix132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Antipov D, Hartwick N, Shen M, Raiko M, Lapidus A, Pevzner PA. 2016. plasmidSPAdes: assembling plasmids from whole genome sequencing data. Bioinformatics 32:3380–3387. doi: 10.1093/bioinformatics/btw493. [DOI] [PubMed] [Google Scholar]
- 59.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, Bibillo A, Bjornson K, Chaudhuri B, Christians F, Cicero R, Clark S, Dalal R, Dewinter A, Dixon J, Foquet M, Gaertner A, Hardenbol P, Heiner C, Hester K, Holden D, Kearns G, Kong X, Kuse R, Lacroix Y, Lin S, Lundquist P, Ma C, Marks P, Maxham M, Murphy D, Park I, Pham T, Phillips M, Roy J, Sebra R, Shen G, Sorenson J, Tomaney A, Travers K, Trulson M, Vieceli J, Wegener J, Wu D, Yang A, Zaccarin D, et al. 2009. Real-time DNA sequencing from single polymerase molecules. Science 323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 61.Treangen TJ, Ondov BD, Koren S, Phillippy AM. 2014. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 15:524. doi: 10.1186/PREACCEPT-2573980311437212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Inouye M, Dashnow H, Raven LA, Schultz MB, Pope BJ, Tomita T, Zobel J, Holt KE. 2014. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med 6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen L, Zheng D, Liu B, Yang J, Jin Q. 2016. VFDB 2016: hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res 44:D694–D697. doi: 10.1093/nar/gkv1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, Landraud L, Rolain JM. 2014. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother 58:212–220. doi: 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. Blast+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomason JA III, Stevens R, Vonstein V, Wattam AR, Xia F. 2015. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 5:8365. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The E. coli isolates in this study and their information from sequencing analysis. Download TABLE S1, XLSX file, 0.04 MB (38.1KB, xlsx) .
Copyright © 2018 Shen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Clinical information on 80 E. coli isolates with the mcr-1 gene and the MICs of multiple antimicrobial agents. Download TABLE S2, XLSX file, 0.05 MB (48.8KB, xlsx) .
Copyright © 2018 Shen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The assembly stats for the PacBio genomes. Download TABLE S3, XLSX file, 0.03 MB (33.1KB, xlsx) .
Copyright © 2018 Shen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Characterization of virulence genes in 80 mcr-1-carrying E. coli isolates. Download TABLE S4, XLSX file, 0.1 MB (104.2KB, xlsx) .
Copyright © 2018 Shen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Characterization of resistance genes in 80 mcr-1-carrying E. coli isolates. Download TABLE S5, XLSX file, 0.1 MB (57.7KB, xlsx) .
Copyright © 2018 Shen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Allelic variations of seven mcr-1-carrying isolates. Download TABLE S6, XLSX file, 0.03 MB (34.3KB, xlsx) .
Copyright © 2018 Shen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.