ABSTRACT
Energy conservation via hydrogen cycling, which generates proton motive force by intracellular H2 production coupled to extracellular consumption, has been controversial since it was first proposed in 1981. It was hypothesized that the methanogenic archaeon Methanosarcina barkeri is capable of energy conservation via H2 cycling, based on genetic data that suggest that H2 is a preferred, but nonessential, intermediate in the electron transport chain of this organism. Here, we characterize a series of hydrogenase mutants to provide direct evidence of H2 cycling. M. barkeri produces H2 during growth on methanol, a phenotype that is lost upon mutation of the cytoplasmic hydrogenase encoded by frhADGB, although low levels of H2, attributable to the Ech hydrogenase, accumulate during stationary phase. In contrast, mutations that conditionally inactivate the extracellular Vht hydrogenase are lethal when expression of the vhtGACD operon is repressed. Under these conditions, H2 accumulates, with concomitant cessation of methane production and subsequent cell lysis, suggesting that the inability to recapture extracellular H2 is responsible for the lethal phenotype. Consistent with this interpretation, double mutants that lack both Vht and Frh are viable. Thus, when intracellular hydrogen production is abrogated, loss of extracellular H2 consumption is no longer lethal. The common occurrence of both intracellular and extracellular hydrogenases in anaerobic microorganisms suggests that this unusual mechanism of energy conservation may be widespread in nature.
KEYWORDS: Methanosarcina, energy conservation, hydrogenase, methanogenesis
IMPORTANCE
ATP is required by all living organisms to facilitate essential endergonic reactions required for growth and maintenance. Although synthesis of ATP by substrate-level phosphorylation is widespread and significant, most ATP is made via the enzyme ATP synthase, which is energized by transmembrane chemiosmotic gradients. Therefore, establishing this gradient across the membrane is of central importance to sustaining life. Experimental validation of H2 cycling adds to a short list of mechanisms for generating a transmembrane electrochemical gradient that is likely to be widespread, especially among anaerobic microorganisms.
INTRODUCTION
An essential requirement for life is the ability to couple exergonic metabolism to the endergonic synthesis of ATP. While some ATP is made by direct phosphorylation of ADP using “high-energy” metabolites such as phosphoenolpyruvate or 1,3-diphosphoglycerate, the vast majority is produced via the enzyme ATP synthase using energy stored in a transmembrane proton (or sodium) gradient. These electrochemical gradients are typically established during the process of electron transport by membrane proteins that couple exergonic redox reactions to generation of an ion motive force by one of three general mechanisms: (i) vectorial proton pumping; (ii) scalar movement of protons across the membrane, as in the Q-cycle or Q-loop; or (iii) coupled reactions that consume protons within the cell and produce protons on the outside (1, 2). Given the importance of this process, it is not surprising that this central aspect of living systems has been the subject of intense study (and at least three Nobel Prizes). Indeed, we now possess a detailed, molecular-level understanding of chemiosmotic energy conservation as it applies to photosynthesis and aerobic respiration in a wide variety of organisms, including eukaryotes, bacteria, and archaea. Nevertheless, unique and sometimes surprising mechanisms for generation of chemiosmotic gradients continue to be found, including sodium-pumping methyltransferases in methanogenic archaea (3), electrogenic formate:oxalate antiporters in bacteria (4, 5), and light-driven, proton-pumping rhodopsins (6).
A controversial, and as yet unproven, mechanism for creating transmembrane proton gradients called H2 cycling was proposed by Odom and Peck in 1981 to explain ATP synthesis in sulfate-reducing bacteria (7). In this proposed energy-conserving process, protons in the cytosol are reduced to molecular H2 by enzymes known as hydrogenases. The H2 so produced then diffuses across the membrane where it is reoxidized by extracellular hydrogenases, releasing protons that contribute to a transmembrane proton gradient that can be used to make ATP. The electrons produced by this reaction are returned to the cytoplasm via a membrane-bound electron transport chain, completing the redox process.
Although H2 cycling has been suggested to occur in a number of anaerobic organisms (7–11), the hydrogen cycling hypothesis has not been widely accepted. A key argument against the idea is based on the high diffusion rate of molecular hydrogen. Thus, unless extracellular recapture is exceptionally efficient, hydrogen produced in the cytoplasm would be easily lost, resulting in redox imbalance and presumably cell death. Nevertheless, experimental demonstration of simultaneous production and consumption of H2 by Desulfovibrio vulgaris supports the model (12), as does metabolic modeling (13). However, other data are inconsistent with the idea, including the ability of hydrogenase mutants to grow on lactate (14) and the inability of high external H2 pressures to inhibit substrate catabolism (15). Thus, the H2 cycling model for energy conservation remains unproven.
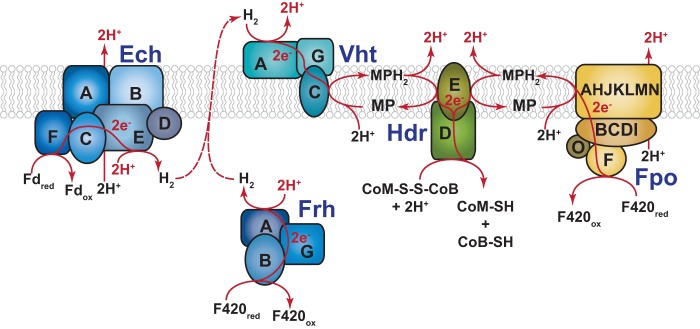
On the basis of a series of genetic experiments, we proposed that the methanogenic archaeon Methanosarcina barkeri employs H2 cycling during growth on one-carbon (C1) substrates and acetate (16, 17). During growth on C1 compounds such as methanol, the putative cycling pathway would produce H2 using the cytoplasmic F420-dependent (Frh) and energy-converting ferredoxin-dependent (Ech) hydrogenases, while H2 production during growth on acetate would be mediated solely by Ech. Both pathways would converge on the methanophenazine-dependent hydrogenase (Vht), which is thought to have an active site on the outer face of the cell membrane (18), to consume extracellular H2 and deliver electrons to the membrane-bound electron transport chain, where they serve to reduce the coenzyme M-coenzyme B heterodisulfide (CoM-S-S-CoB) produced during the production of methane (Fig. 1). However, these genetic studies remain incomplete because neither the role of Vht nor the production and consumption of hydrogen were examined. Here we explicitly test both, providing strong experimental support for the role of H2 cycling in energy conservation in M. barkeri.
FIG 1 .
Putative H2 cycling electron transport chain of M. barkeri. Growth on C1 substrates generates reduced cofactor F420 (F420red), which is a hydride carrying cofactor analogous to NADH, and the reduced form of the small electron-carrying protein ferredoxin (Fdred). During aceticlastic methanogenesis, only Fdred is produced. These reduced electron carriers are reoxidized in the cytoplasm by the Frh and Ech hydrogenases, respectively, with concomitant consumption of protons to produce molecular H2. H2 subsequently diffuses out of the cell where it is reoxidized by the Vht hydrogenase, which has an active site located on the outer face of the cell membrane. This reaction releases protons on the outside of the cell and produces reduced methanophenazine (MPH2), a membrane-bound electron carrier analogous to ubiquinone. MPH2 subsequently delivers electrons to the enzyme heterodisulfide reductase (Hdr), which serves as the terminal step in the Methanosarcina electron transport chain. This final reaction regenerates coenzyme B (CoB-SH) and coenzyme M (CoM-SH) from the mixed disulfide (CoM-S-S-CoB), which is produced from the free thiol cofactors during methanogenic metabolism. Electron (e−) flow and scalar protons (H+) are shown in red. It should be noted that M. barkeri can also reoxidize F420red using the membrane-bound, proton-pumping F420-dehydrogenase (Fpo). Thus, the cell has a branched electron transport chain, and therefore, it is not dependent on H2 cycling during growth on methylotrophic substrates (16); however, both pathways for electron transport from F420 have identical levels of energy conservation: namely, 4 H+/2e−. It should also be noted that the Ech hydrogenase acts as a proton pump in addition to its role in H2 cycling, thus electron transport from Fdred during methylotrophic and aceticlastic methanogenesis conserves 6H+/2e−. Individual subunits of the various enzymes are indicated by capital letters (e.g., A, B, C…).
RESULTS AND DISCUSSION
Hydrogenases of M. barkeri.
Three distinct types of hydrogenases are encoded by Methanosarcina barkeri Fusaro (see Fig. S1 in the supplemental material) (19). The F420-reducing hydrogenase (Frh) is a cytoplasmic, three-subunit (α, β, and γ) enzyme encoded by the frhADGB operon, which also includes a maturation protease, FrhD (20). This enzyme couples the oxidation/reduction of the deazaflavin cofactor F420 with production/consumption of H2. The membrane-bound Vht hydrogenase utilizes the quinone-like electron carrier, methanophenazine, as a cofactor (21). Like Frh, Vht is a three-subunit enzyme encoded by a four-gene operon (vhtGACD) that includes a maturation protease, VhtD (19). M. barkeri also contains genes that encode homologs of both the frh and vht operons (the freAEGB and vhxGAC operons, respectively); however, multiple lines of evidence suggest that these genes are incapable of producing active hydrogenases (16, 22). Thus, the presence of these genes has no bearing on the results presented herein. The final hydrogenase encoded by M. barkeri is a membrane-bound, energy-converting hydrogenase (Ech), which couples the oxidation/reduction of ferredoxin and H2 to the production/consumption of a proton motive force (23, 24). Thus, the enzyme can use proton motive force to drive the endergonic reduction of ferredoxin by H2, which is required for CO2 reduction during hydrogenotrophic methanogenesis and for biosynthesis during growth by H2-dependent reduction of C1 compounds (methyl-reducing methanogenesis). During both methylotrophic and aceticlastic methanogenesis, Ech is believed to couple oxidation of reduced ferredoxin to production of proton motive force and H2. The hydrogen thus produced would need to be recaptured by Vht in a putative H2 cycling process that contributes to proton motive force (Fig. 1) (17).
Hydrogenase operons in Methanosarcina barkeri. Three distinct types of hydrogenase are encoded by M. barkeri Fusaro. The frh and fre operons encode putative F420-reducing hydrogenases, while the vht and vhx operons encode putative methanophenazine-reducing hydrogenases. Genetic and biochemical data show that neither the fre nor the vhx operon is capable of producing an active hydrogenase under any growth condition yet examined (T. D. Mand, G. Kulkarni, and W. W. Metcalf, 2018 bioRxiv, https://doi.org/10.1101/334656). The ech operon encodes a ferredoxin-dependent energy-conserving hydrogenase. The locus tags are shown below each gene, with the prefix “Mbar_” omitted to save space (indicated by an asterisk) in some cases. Download FIG S1, PDF file, 0.7 MB (696.1KB, pdf) .
Copyright © 2018 Kulkarni et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The cytoplasmic Frh hydrogenase is responsible for production of H2 during growth on methanol.
A number of studies have shown that assorted Methanosarcina strains produce H2 during growth on methylotrophic and aceticlastic substrates (9, 25–30); however, to our knowledge, this has never been assessed in M. barkeri strain Fusaro. To test this, we quantified the accumulation of CH4 and H2 during growth on methanol-containing medium (Fig. 2). Consistent with the hydrogen cycling hypothesis, we observed significant H2 production, which reached a maximum partial pressure of ca. 20 Pa near the end of exponential growth. As expected, the culture also produced substantial levels of methane. As previously observed (16), a mutant lacking Frh (strain WWM115 [Table S1]) grew at a lower rate than its isogenic parent and produced somewhat smaller amounts of methane. Very little H2 (<4 Pa) was produced during growth of the Δfrh mutant; however, after growth ceased, the H2 concentration slowly rose, reaching a maximum level of 7 Pa. Thus, Frh is responsible for most hydrogen production during growth of M. barkeri Fusaro on methanol, although some hydrogen is still produced in the Δfrh mutant. As will be shown below, Ech is probably responsible for the low levels of H2 seen in the Δfrh mutant.
FIG 2 .
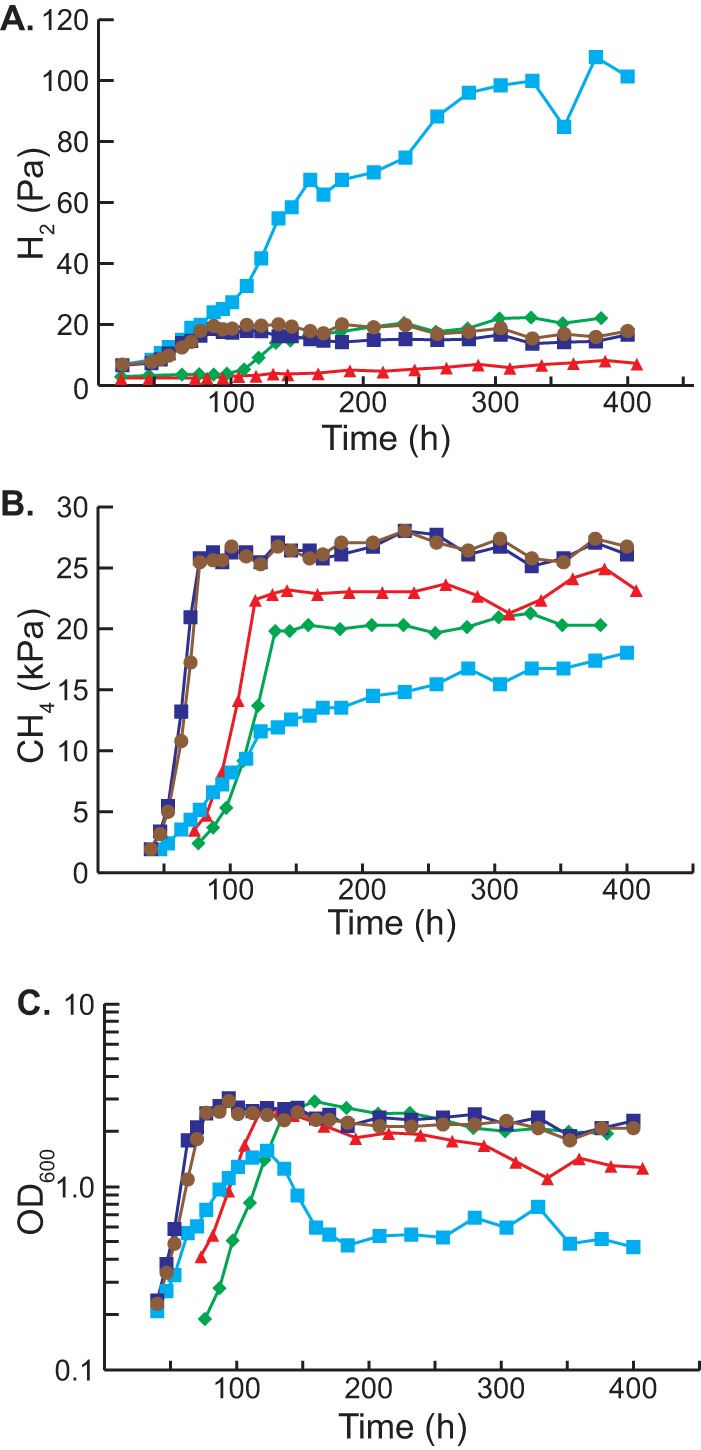
Hydrogen and methane production during methylotrophic growth. (A to C) The partial pressures of H2 (A) and methane (B) were monitored during the course of growth (as indicated by optical density [C]) in methanol-containing medium for various M. barkeri strains. Strains used were M. barkeri isogenic parental strain (WWM85 [brown circles]), tetracycline-regulated Ptet::vht mutant (WWM157) with tetracycline (dark blue squares) and without tetracycline (light blue squares), Δfrh mutant (WWM115 [red triangles]), and Δfrh Δvht double mutant (WWM351 [green diamonds]). Measurements were performed in triplicates as described in Materials and Methods. Complete strain genotypes can be found in Table S1 in the supplemental material.
Methanosarcina barkeri Fusaro strains used in this study. Download TABLE S1, DOC file, 0.04 MB (46KB, doc) .
Copyright © 2018 Kulkarni et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Vht activity is required for viability of M. barkeri.
To investigate the role of Vht during growth of M. barkeri, we attempted to delete the vhtGACD operon via homologous gene replacement (31, 32). However, despite numerous attempts, including selection on a variety of media, with and without supplementation of potential biosynthetic intermediates, no mutant colonies were obtained. We also attempted to delete the vht operon using the markerless deletion method of genetic exchange (33). This method relies on construction of a merodiploid strain with both mutant and wild-type alleles. Upon segregation of the merodiploid, 50% of the recombinants are expected to be mutants if there is no selective pressure against the mutant allele. However, if the mutation causes a reduction in growth rate (with lethality being the most extreme case), the probability of obtaining recombinants with the mutant allele is severely reduced. We tested 101 haploid recombinants obtained from a vhtGACD+/ΔvhtGACD merodiploid; all carried the wild-type vht allele. Taken together, these data suggest that the vhtGACD operon is critical for normal growth of M. barkeri.
To test whether Vht is essential, we constructed a mutant in which the vht operon was placed under control of a tightly regulated, tetracycline-dependent promoter (34). We then examined the viability of the mutant and its isogenic parent by spotting serial dilutions on a variety of media, with and without tetracycline. As shown in Fig. 3, the Ptet::vht mutant is unable to grow in the absence of the inducer but grew well when tetracycline was added, whereas the isogenic parent grew with or without the addition of tetracycline. These phenotypes were observed on a variety of media, including media containing (i) methanol, (ii) methanol plus H2, (iii) H2/CO2, and (iv) acetate, which were chosen because they encompass growth conditions that require each of the four known methanogenic pathways used by M. barkeri (Fig. 4). It should be stressed that the Ptet::vht mutant used in this experiment was pregrown in the presence of inducer. Thus, at the start of the experiment, all cells have active Vht. However, during cultivation in the absence of tetracycline, preexisting Vht is depleted by protein turnover and cell division, thereby allowing characterization of the Vht-deficient phenotype. The absence of growth of the diluted cultures in all media shows that Vht is essential for growth via the methylotrophic (methanol), methyl-reducing (methanol plus H2), hydrogenotrophic (H2/CO2), and aceticlastic (acetate) methanogenic pathways.
FIG 3 .
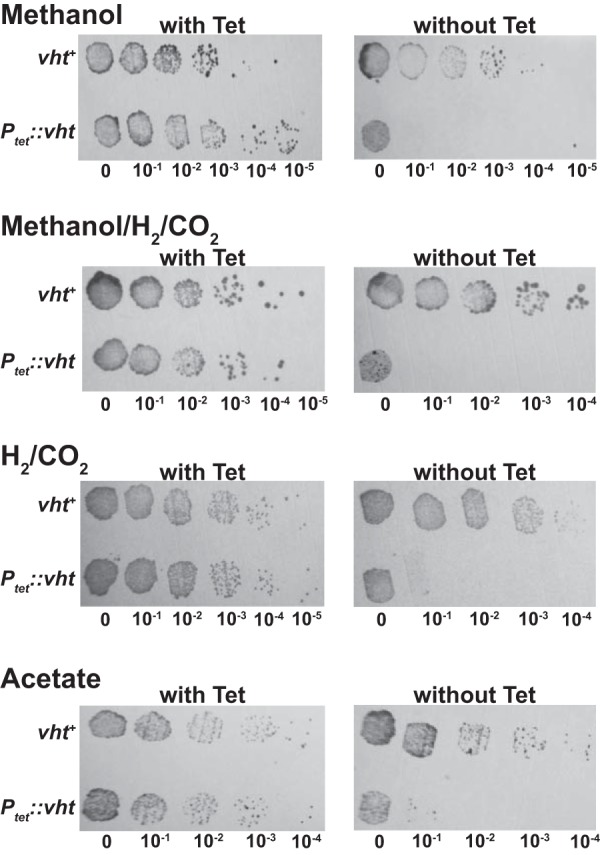
Essentiality of the Vht hydrogenase in M. barkeri. Cultures of the Ptet::vht mutant (WWM157) and its isogenic parent (WWM154) were adapted to four different substrates of interest (and in the presence of tetracycline for Ptet::vht), then washed, serially diluted, and incubated with each substrate with and without tetracycline (Tet). The media used indicate the ability to grow via each of the four known methanogenic pathways: (i) methylotrophic (methanol), (ii) methyl reduction (methanol/H2/CO2), (iii) hydrogenotrophic (H2/CO2), and (iv) aceticlastic (acetate).
FIG 4 .
Role of H2 cycling in the four methanogenic pathways of M. barkeri. M. barkeri utilizes four distinct methanogenic pathways to allow growth on a variety of substrates. In the hydrogenotrophic pathway (shown in red), CO2 is reduced to methane using electrons derived from H2, while in the methyl-reducing pathway (shown in orange), H2 is used to reduce C1 compounds, such as methanol, directly to CH4. During methylotrophic methanogenesis (shown in green), C1 compounds are disproportionated to CO2 and methane, with one molecule of the C1 compound oxidized to provide electrons for reduction of three additional molecules to methane. Finally, in the aceticlastic pathway (shown in blue), acetate is split into a methyl group and an enzyme-bound carbonyl moiety. The latter is oxidized to CO2 to provide electrons required for reduction of the methyl group to methane. The steps catalyzed by Fpo, Frh, Vht, Ech, and Hdr proteins are indicated. Steps involving H2 cycling are shown by labeled dashed arrows. An alternate, H2-independent electron transport pathway is shown in brown. Experimental data support the function of this alternate pathway during methylotrophic methanogenesis, but not in hydrogenotrophic or methyl-reducing methanogenesis (as indicated by the dashed brown box). Abbreviations; Fpo, F420 dehydrogenase; Frh, F420-reducing hydrogenase; Vht, methanophenazine-dependent hydrogenase; Ech, energy-converting ferredoxin-dependent hydrogenase; Hdr, heterodisulfide reductase; CoM, coenzyme M; CoB, coenzyme B; CoB-CoM, mixed disulfide of CoB and CoM; MP/MPH2, oxidized and reduced methanophenazine; F420ox/F420red, oxidized and reduced cofactor F420, respectively; Fdox/Fdred, oxidized and reduced ferredoxin, respectively; CHO-MF, formyl-methanofuran; H4SPT, tetrahydrosarcinapterin; CHO-H4SPT, formyl-H4SPT; CH≡H4SPT, methenyl-H4SPT; CH2=H4SPT, methylene-H4SPT; CH3-H4SPT, methyl-H4SPT; CH3-CoM, methyl-coenzyme M; CoA, coenzyme A; CH3CO-CoA, acetyl-coenzyme A; [CO], enzyme-bound carbonyl moiety.
Depletion of Vht results in H2 accumulation and cell lysis.
To help understand why Vht is essential, we quantified production of H2 and CH4 in cultures of the Ptet::vht strain with and without tetracycline (Fig. 2). When the strains were grown in methanol-containing medium in the presence of tetracycline, the accumulation of H2 and CH4 was essentially identical to that of the isogenic parent. Cultures in which vht is not expressed (i.e., without tetracycline) grew initially but growth rapidly slowed and reached an optical density that was less than half of that obtained when vht was expressed. The optical density subsequently dropped, suggesting cell death and lysis. Similarly, methane accumulation in cultures not expressing vht was much slower than in induced cultures and only reached half of that seen under inducing conditions. In contrast, H2 accumulation was much higher in the absence of Vht, with final levels nearly sixfold higher than those seen in cultures that express Vht. These data clearly show that Vht is required for efficient recapture of H2 produced by Frh and Ech. Moreover, they suggest that H2 loss is responsible for the lethal consequences of vht repression.
Vht is not essential in Δfrh mutants.
If the inability to recapture H2 is responsible for the essentiality of Vht, then it should be possible to delete the vht operon in strains that do not produce hydrogen. As described above, Frh is responsible for the majority of H2 production during growth. Thus, we attempted to introduce a Δvht allele into the Δfrh host. In contrast to our prior unsuccessful attempts to create a Δvht single mutant, the Δvht Δfrh double mutant was isolated in the first attempt. Therefore, Vht is not required when Frh is absent. Like the Δfrh single mutant, the Δvht Δfrh double mutant grows slowly on methanol and produces lower levels of methane (Fig. 2). Significantly, the double mutant does not produce the excessive level of H2 seen in the uninduced Ptet::vht strain, instead accumulating H2 at levels similar to those of the parental strain (ca. 20 Pa). Because Ech is the only active hydrogenase remaining in the Δvht Δfrh double mutant, it must be responsible for H2 production in this strain. This begs the question of why H2 accumulation stops at 20 Pa in the double mutant, while the uninduced Ptet::vht strain produces much higher levels. We suggest that the coupling of Ech activity to generation of proton motive force thermodynamically restrains excessive H2 production, even in the absence of H2 uptake by Vht. This would also explain the viability of the Δvht Δfrh double mutant. This situation is in stark contrast to that seen in the vht-depleted strain, where the F420-dependent Frh is responsible for most of the H2 production (see above). Accordingly, at the low H2 partial pressures observed in our experiments, reduction of protons with F420 is strongly exergonic, allowing excessive hydrogen accumulation. This is also consistent with the observation that the redox state of F420 is in rapid equilibrium with H2 (35). Interestingly, the smaller amount of H2 accumulation in the Δfrh mutant relative to that seen in the Δvht Δfrh double mutant shows that Vht also consumes H2 produced by Ech. This supports previous studies indicating potential energy conservation via Ech/Vht H2 cycling during acetate metabolism (17, 23).
M. barkeri has a bifurcated electron transport chain with H2-dependent and -independent branches.
We previously showed that M. barkeri has a branched electron transport chain, with Frh- and F420 dehydrogenase (Fpo)-dependent branches (16). The data reported here extend our understanding of the Frh-dependent branch and are fully consistent with the model depicted in Fig. 1. Thus, during growth on methylotrophic substrates such as methanol, reduced F420 is preferentially oxidized via an energy-conserving, H2 cycling electron transport chain that requires Frh. However, in the absence of Frh, reduced F420 is channeled into the Fpo-dependent electron transport chain, which supports growth at a significantly lower rate (Fig. 1 and 2). This alternate pathway accounts for the viability of the Δfrh mutant, which is lost when both frh and fpo are deleted (16). Similar but less severe phenotypes have been observed in fpo and frh mutants of Methanosarcina mazei, thus it seems likely that H2 cycling also occurs in this closely related species (36). However, many Methanosarcina species, especially those that inhabit marine environments, are devoid of hydrogenase activity, despite the presence of hydrogenase-encoding genes. We, and others, have interpreted this to be an adaptation to the marine environment, where H2-utilizing sulfate reducers are likely to disrupt H2 cycling due to the superior thermodynamics of H2 oxidation coupled to sulfate reduction (19, 37).
A similar branched electron transport chain may also explain the contradictory evidence regarding H2 cycling in Desulfovibrio species. Thus, the viability of Desulfovibrio hydrogenase mutants and the inability of excess H2 to suppress substrate catabolism can both be explained by the presence of alternative electron transport mechanisms. Indeed, metabolic modeling of Desulfovibrio vulgaris strongly supports this interpretation (13). Thus, it is critical that experiments designed to test the H2 cycling mechanism be interpreted within a framework that includes the possibility of branched electron transport chains. With this in mind, it seems likely that many anaerobic organisms might use H2 cycling for energy conservation. Indeed, since it was originally proposed, H2 cycling has been suggested to occur in the acetogen Acetobacterium woodii (10) and in the Fe(III) respiring Geobacter sulfurreducens (8).
Why are Vht mutants inviable during growth on methanol/H2 or H2/CO2?
Although the data presented here strongly support the H2 cycling model, they raise additional questions regarding H2-dependent methanogenesis that are not easily explained. In particular, it is not readily apparent why the uninduced Ptet::vht mutants are inviable during hydrogenotrophic or methyl-reducing growth. As shown in Fig. 4, it should be possible to channel electrons from H2 oxidation into the electron transport chain via Frh and Fpo. Indeed, Thauer et al. have proposed that this alternate pathway is functional in Methanosarcina (38). Nevertheless, the Ptet::vht mutant does not grow under repressing conditions on either H2/CO2 or methanol plus H2. It should be stressed that we use high concentrations of hydrogen during growth on these substrates. Thus, it is expected that reduction of F420 via Frh should be exergonic in our experiments, which would favor this pathway. (This is in contrast to the methylotrophic or aceticlastic growth conditions described above, under which the reverse reaction [i.e., hydrogen production] is favored.) Thus, a thermodynamic argument cannot easily explain the results. Further, based on available evidence (16, 39, 40), energy conservation via the Vht-dependent pathway should be identical to that of the alternate Frh/Fpo-dependent pathway. Thus, an energy conservation argument also cannot explain the phenomenon. One might argue that faster kinetics of the Vht-dependent pathway could be responsible, but in our opinion, the growth (albeit slower than wild type) of the Δfrh and Δvht Δfrh mutants during methylotrophic growth, which depends on Fpo, argues against this explanation. Therefore, as yet unknown regulatory and/or biochemical constraints on hydrogen metabolism in Methanosarcina await discovery.
MATERIALS AND METHODS
Strains, media, and growth conditions.
The construction and genotypes of all Methanosarcina strains are presented in Table S1 in the supplemental material. Methanosarcina strains were grown as single cells (41) at 37°C in high-salt (HS) broth medium (42) or on agar-solidified medium as described previously (43). Growth substrates provided were methanol (125 mM in broth medium and 50 mM in agar-solidified medium) or sodium acetate (120 mM) under a headspace of either N2/CO2 (80/20%) at 50 kPa over ambient pressure or H2/CO2 (80/20%) at 300 kPa over ambient pressure. Cultures were supplemented as indicated with 0.1% yeast extract, 0.1% Casamino Acids, 10 mM sodium acetate, or 10 mM pyruvate. Puromycin (CalBioChem, San Diego, CA) was added at 2 µg/ml for selection of the puromycin transacetylase (pac) gene (33). 8-Aza-2,6-diaminopurine (8-ADP) (Sigma, St. Louis, MO) was added at 20 µg/ml for selection against the presence of hpt (33). Tetracycline was added at 100 µg/ml to induce the tetracycline-regulated PmcrB(tetO3) promoter (34). Standard conditions were used for growth of Escherichia coli strains (44) DH5α/λ-pir (45) and DH10B (Stratagene, La Jolla, CA), which were used as hosts for plasmid constructions.
DNA methods and plasmid construction.
Standard methods were used for plasmid DNA isolation and manipulation using E. coli hosts (46). Liposome-mediated transformation was used for Methanosarcina as described previously (47). Genomic DNA isolation and DNA hybridization were performed as described previously (32, 42, 43). DNA sequences were determined from double-stranded templates by the W. M. Keck Center for Comparative and Functional Genomics, University of Illinois. Plasmid constructions are described in the supporting information (Tables S2 and S3).
Plasmids used in this study. Download TABLE S2, DOC file, 0.05 MB (49KB, doc) .
Copyright © 2018 Kulkarni et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download TABLE S3, DOC file, 0.03 MB (34.5KB, doc) .
Copyright © 2018 Kulkarni et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Construction of the Δfrh and Δvht Δfrh mutants.
The markerless genetic exchange method (33) using plasmid pGK4 was employed to delete frhADGB (Δfrh) in the Δhpt background of M. barkeri Fusaro (Tables S1, S2, and S3) using methanol/H2/CO2 as the growth substrate. The Δvht Δfrh mutant was constructed by deleting vhtGACD in the Δfrh markerless mutant by the homologous recombination-mediated gene replacement method (32). To do this, the 5.6-kb XhoI/NotI fragment of pGK82B was used to transform the Δfrh mutant to puromycin resistance on methanol-containing medium. The mutants were confirmed by PCR and DNA hybridization (data not shown).
Construction of the tetracycline-regulated vht mutant (Ptet::vht).
The tetracycline-regulated PmcrB(tetO3) promoter was employed to drive conditional expression of the vht operon in M. barkeri WWM157 (34). This strain was constructed by transforming strain WWM154 to puromycin resistance using the 7-kb NcoI/SpeI fragment of pGK61A (Tables S1, S2, and S3). The transformants were selected on methanol plus H2/CO2 medium in the presence of puromycin and tetracycline. The Ptet::vht strain was confirmed by DNA hybridization (data not shown). To ensure that the native vht promoter (Pvht) did not interfere with expression from PmcrB(tetO3), 382 bp upstream of vhtG were deleted in Ptet::vht. This left 1,038 bp intact for the expression of the hyp operon, which is upstream of the vht operon and expressed in the opposite direction.
Determination of Vht essentiality during growth on all substrate types.
Growth of strains WWM157 (Ptet::vht) and WWM154 (isogenic parent) on methanol, methanol/H2/CO2, H2/CO2, and acetate were analyzed by the spot-plate method (48). Cultures were first adapted for at least 15 generations to the substrate of interest; tetracycline was added to each medium for growth of strain WWM157. Upon reaching stationary phase, 10 ml of culture was washed three times and resuspended in 5 ml HS medium that lacked growth substrate. Subsequently, 10 µl of 10-fold serial dilutions was spotted onto the following: three layers of GB004 paper (Whatman, NJ), two layers of GB002 paper (Schleicher & Schuell BioScience, NH), one layer of 3 MM paper (Whatman, NJ), and a 0.22 mM nylon membrane (GE Water and Process Technologies, PA) soaked in 43 ml of HS medium containing the substrate of interest with and without tetracycline. The plates were sealed and incubated at 37°C for at least 2 weeks in an intrachamber anoxic incubator (49). Growth on acetate and methanol was tested under an atmosphere of N2/CO2/H2S (80/19.9/0.1 ratio), while growth on methanol/H2/CO2 or H2/CO2 was tested under an atmosphere of H2/CO2/H2S (80/19.9/0.1 ratio).
Measurement of H2, CH4, and OD600 during growth on methanol.
M. barkeri WWM85 (isogenic parent), WWM157 (Ptet::vht; grown in the presence of tetracycline), WWM115 (Δfrh), and WWM351 (Δvht Δfrh) were grown on methanol until mid-exponential phase (optical density at 600 nm [OD600] of ca. 0.5) and then 1 ml (WWM85 and WWM157) or 5 ml (WWM115 and WWM351) was inoculated into 100 ml HS-methanol in a 500-ml serum bottle. For WWM157, the culture was washed once prior to inoculation with or without tetracycline. To measure H2 and CH4, ca. 1-ml or 2-ml headspace sample was withdrawn aseptically from the culture at various time points with a syringe that had been flushed with sterile, anaerobic N2. The gas sample was then diluted into 70 ml helium. A gas-tight syringe flushed with helium was subsequently used to withdraw 3 ml of the diluted sample, which was then injected into an SRI gas chromatograph, equipped with a reduction gas detector (RGD) and a thermal conductivity detector (TCD) at 52°C. The RGD column was a three-foot-long 13× molecule sieve, whereas the TCD column was a six-foot HayeSep D porous polymer column. The RGD column was used to detect H2 by peak height, and the TCD column was used to detect CH4 by peak area. Helium was used as the carrier gas. OD600 was also measured during the growth curve.
ACKNOWLEDGMENTS
We thank Rob Sanford for providing assistance and facilities for measurement of low hydrogen partial pressures.
We acknowledge the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the U.S. Department of Energy through grant DE-FG02-02ER15296 for funding this work.
Footnotes
Citation Kulkarni G, Mand TD, Metcalf WW. 2018. Energy conservation via hydrogen cycling in the methanogenic archaeon Methanosarcina barkeri. mBio 9:e01256-18. https://doi.org/10.1128/mBio.01256-18.
REFERENCES
- 1.White D. 2000. Electron transport, p 103–131. In The physiology and biochemistry of prokaryotes, 2nd ed. Oxford University Press, New York, NY. [Google Scholar]
- 2.White D. 2000. Bioenergetics in the cytosol, p 165–179. In The physiology and biochemistry of prokaryotes, 2nd ed. Oxford University Press, New York, NY. [Google Scholar]
- 3.Gottschalk G, Thauer RK. 2001. The Na+-translocating methyltransferase complex from methanogenic archaea. Biochim Biophys Acta 1505:28–36. doi: 10.1016/S0005-2728(00)00274-7. [DOI] [PubMed] [Google Scholar]
- 4.Anantharam V, Allison MJ, Maloney PC. 1989. Oxalate:formate exchange. The basis for energy coupling in Oxalobacter. J Biol Chem 264:7244–7250. [PubMed] [Google Scholar]
- 5.Kuhner CH, Hartman PA, Allison MJ. 1996. Generation of a proton motive force by the anaerobic oxalate-degrading bacterium Oxalobacter formigenes. Appl Environ Microbiol 62:2494–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Béjà O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP, Jovanovich SB, Gates CM, Feldman RA, Spudich JL, Spudich EN, DeLong EF. 2000. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289:1902–1906. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- 7.Odom JM, Peck HD. 1981. Hydrogen cycling as a general mechanism for energy coupling in the sulfate-reducing bacteria, Desulfovibrio sp. FEMS Microbiol Lett 12:47–50. doi: 10.1111/j.1574-6968.1981.tb07609.x. [DOI] [Google Scholar]
- 8.Coppi MV. 2005. The hydrogenases of Geobacter sulfurreducens: a comparative genomic perspective. Microbiology 151:1239–1254. doi: 10.1099/mic.0.27535-0. [DOI] [PubMed] [Google Scholar]
- 9.Lovley DR, Ferry JG. 1985. Production and consumption of H2 during growth of Methanosarcina spp. on acetate. Appl Environ Microbiol 49:247–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Odom JM, Peck HD. 1984. Hydrogenase, electron-transfer proteins, and energy coupling in the sulfate-reducing bacteria Desulfovibrio. Annu Rev Microbiol 38:551–592. doi: 10.1146/annurev.mi.38.100184.003003. [DOI] [PubMed] [Google Scholar]
- 11.Lupa B, Hendrickson EL, Leigh JA, Whitman WB. 2008. Formate-dependent H2 production by the mesophilic methanogen Methanococcus maripaludis. Appl Environ Microbiol 74:6584–6590. doi: 10.1128/AEM.01455-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peck HD Jr, LeGall J, Lespinat PA, Berlier Y, Fauque G. 1987. A direct demonstration of hydrogen cycling by Desulfovibrio vulgaris employing membrane-inlet mass spectrometry. FEMS Microbiol Lett 40:295–299. doi: 10.1111/j.1574-6968.1987.tb02042.x. [DOI] [Google Scholar]
- 13.Noguera DR, Brusseau GA, Rittmann BE, Stahl DA. 1998. A unified model describing the role of hydrogen in the growth of Desulfovibrio vulgaris under different environmental conditions. Biotechnol Bioeng 59:732–746. doi:. [DOI] [PubMed] [Google Scholar]
- 14.Odom JM, Wall JD. 1987. Properties of a hydrogen-inhibited mutant of Desulfovibrio desulfuricans ATCC 27774. J Bacteriol 169:1335–1337. doi: 10.1128/jb.169.3.1335-1337.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pankhania IP, Gow LA, Hamilton WA. 1986. The effect of hydrogen on the growth of Desulfovibrio vulgaris (Hildenborough) on lactate. Microbiology 132:3349–3356. doi: 10.1099/00221287-132-12-3349. [DOI] [Google Scholar]
- 16.Kulkarni G, Kridelbaugh DM, Guss AM, Metcalf WW. 2009. Hydrogen is a preferred intermediate in the energy-conserving electron transport chain of Methanosarcina barkeri. Proc Natl Acad Sci U S A 106:15915–15920. doi: 10.1073/pnas.0905914106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meuer J, Kuettner HC, Zhang JK, Hedderich R, Metcalf WW. 2002. Genetic analysis of the archaeon Methanosarcina barkeri Fusaro reveals a central role for Ech hydrogenase and ferredoxin in methanogenesis and carbon fixation. Proc Natl Acad Sci U S A 99:5632–5637. doi: 10.1073/pnas.072615499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deppenmeier U, Blaut M, Lentes S, Herzberg C, Gottschalk G. 1995. Analysis of the vhoGAC and vhtGAC operons from Methanosarcina mazei strain Gö1, both encoding a membrane-bound hydrogenase and a cytochrome b. Eur J Biochem 227:261–269. doi: 10.1111/j.1432-1033.1995.tb20383.x. [DOI] [PubMed] [Google Scholar]
- 19.Guss AM, Kulkarni G, Metcalf WW. 2009. Differences in hydrogenase gene expression between Methanosarcina acetivorans and Methanosarcina barkeri. J Bacteriol 191:2826–2833. doi: 10.1128/JB.00563-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaupel M, Thauer RK. 1998. Two F420-reducing hydrogenases in Methanosarcina barkeri. Arch Microbiol 169:201–205. doi: 10.1007/s002030050561. [DOI] [PubMed] [Google Scholar]
- 21.Brodersen J, Bäumer S, Abken HJ, Gottschalk G, Deppenmeier U. 1999. Inhibition of membrane-bound electron transport of the methanogenic archaeon Methanosarcina mazei Gö1 by diphenyleneiodonium. Eur J Biochem 259:218–224. doi: 10.1046/j.1432-1327.1999.00017.x. [DOI] [PubMed] [Google Scholar]
- 22.Mand TD, Kulkarni G, Metcalf WW. 2018. Genetic, biochemical, and molecular characterization of Methanosarcina barkeri mutants lacking three distinct classes of hydrogenase. bioRxiv doi: 10.1101/334656. [DOI] [PMC free article] [PubMed]
- 23.Meuer J, Bartoschek S, Koch J, Künkel A, Hedderich R. 1999. Purification and catalytic properties of Ech hydrogenase from Methanosarcina barkeri. Eur J Biochem 265:325–335. doi: 10.1046/j.1432-1327.1999.00738.x. [DOI] [PubMed] [Google Scholar]
- 24.Peters JW, Schut GJ, Boyd ES, Mulder DW, Shepard EM, Broderick JB, King PW, Adams MWW. 2015. [FeFe]- and [NiFe]-hydrogenase diversity, mechanism, and maturation. Biochim Biophys Acta 1853:1350–1369. doi: 10.1016/j.bbamcr.2014.11.021. [DOI] [PubMed] [Google Scholar]
- 25.Phelps TJ, Conrad R, Zeikus JG. 1985. Sulfate-dependent interspecies H2 transfer between Methanosarcina barkeri and Desulfovibrio vulgaris during coculture metabolism of acetate or methanol. Appl Environ Microbiol 50:589–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhatnagar L, Krzycki JA, Zeikus JG. 1987. Analysis of hydrogen metabolism in Methanosarcina barkeri: regulation of hydrogenase and role of CO-dehydrogenase in H2 production. FEMS Microbiol Lett 41:337–343. doi: 10.1111/j.1574-6968.1987.tb02223.x. [DOI] [Google Scholar]
- 27.Boone DR, Menaia JAGF, Boone JE, Mah RA. 1987. Effects of hydrogen pressure during growth and effects of pregrowth with hydrogen on acetate degradation by Methanosarcina species. Appl Environ Microbiol 53:83–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krzycki JA, Morgan JB, Conrad R, Zeikus JG. 1987. Hydrogen metabolism during methanogenesis from acetate by Methanosarcina barkeri. FEMS Microbiol Lett 40:193–198. doi: 10.1111/j.1574-6968.1987.tb02024.x. [DOI] [Google Scholar]
- 29.Ahring BK, Westermann P, Mah RA. 1991. Hydrogen inhibition of acetate metabolism and kinetics of hydrogen consumption by Methanosarcina thermophila TM-1. Arch Microbiol 157:38–42. doi: 10.1007/BF00245332. [DOI] [Google Scholar]
- 30.Zinder SH, Anguish T. 1992. Carbon monoxide, hydrogen, and formate metabolism during methanogenesis from acetate by thermophilic cultures of Methanosarcina and Methanothrix strains. Appl Environ Microbiol 58:3323–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maeder DL, Anderson I, Brettin TS, Bruce DC, Gilna P, Han CS, Lapidus A, Metcalf WW, Saunders E, Tapia R, Sowers KR. 2006. The Methanosarcina barkeri genome: comparative analysis with Methanosarcina acetivorans and Methanosarcina mazei reveals extensive rearrangement within methanosarcinal genomes. J Bacteriol 188:7922–7931. doi: 10.1128/JB.00810-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang JK, White AK, Kuettner HC, Boccazzi P, Metcalf WW. 2002. Directed mutagenesis and plasmid-based complementation in the methanogenic archaeon Methanosarcina acetivorans C2A demonstrated by genetic analysis of proline biosynthesis. J Bacteriol 184:1449–1454. doi: 10.1128/JB.184.5.1449-1454.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pritchett MA, Zhang JK, Metcalf WW. 2004. Development of a markerless genetic exchange method for Methanosarcina acetivorans C2A and its use in construction of new genetic tools for methanogenic archaea. Appl Environ Microbiol 70:1425–1433. doi: 10.1128/AEM.70.3.1425-1433.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guss AM, Rother M, Zhang JK, Kulkarni G, Metcalf WW. 2008. New methods for tightly regulated gene expression and highly efficient chromosomal integration of cloned genes for Methanosarcina species. Archaea 2:193–203. doi: 10.1155/2008/534081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Poorter LM, Geerts WJ, Keltjens JT. 2005. Hydrogen concentrations in methane-forming cells probed by the ratios of reduced and oxidized coenzyme F420. Microbiology 151:1697–1705. doi: 10.1099/mic.0.27679-0. [DOI] [PubMed] [Google Scholar]
- 36.Welte C, Deppenmeier U. 2011. Re-evaluation of the function of the F420 dehydrogenase in electron transport of Methanosarcina mazei. FEBS J 278:1277–1287. doi: 10.1111/j.1742-4658.2011.08048.x. [DOI] [PubMed] [Google Scholar]
- 37.Deppenmeier U. 2004. The membrane-bound electron transport system of Methanosarcina species. J Bioenerg Biomembr 36:55–64. doi: 10.1023/B:JOBB.0000019598.64642.97. [DOI] [PubMed] [Google Scholar]
- 38.Thauer RK, Kaster A-K, Seedorf H, Buckel W, Hedderich R. 2008. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat Rev Microbiol 6:579–591. doi: 10.1038/nrmicro1931. [DOI] [PubMed] [Google Scholar]
- 39.Ide T, Bäumer S, Deppenmeier U. 1999. Energy conservation by the H2:heterodisulfide oxidoreductase from Methanosarcina mazei Gö1: identification of two proton-translocating segments. J Bacteriol 181:4076–4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baumer S, Ide T, Jacobi C, Johann A, Gottschalk G, Deppenmeier U. 2000. The F420H2 dehydrogenase from Methanosarcina mazei is a redox-driven proton pump closely related to NADH dehydrogenases. J Biol Chem 275:17968–17973. doi: 10.1074/jbc.M000650200. [DOI] [PubMed] [Google Scholar]
- 41.Sowers KR, Boone JE, Gunsalus RP. 1993. Disaggregation of Methanosarcina spp. and growth as single cells at elevated osmolarity. Appl Environ Microbiol 59:3832–3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Metcalf WW, Zhang JK, Shi X, Wolfe RS. 1996. Molecular, genetic, and biochemical characterization of the serC gene of Methanosarcina barkeri Fusaro. J Bacteriol 178:5797–5802. doi: 10.1128/jb.178.19.5797-5802.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boccazzi P, Zhang JK, Metcalf WW. 2000. Generation of dominant selectable markers for resistance to pseudomonic acid by cloning and mutagenesis of the ileS gene from the archaeon Methanosarcina barkeri Fusaro. J Bacteriol 182:2611–2618. doi: 10.1128/JB.182.9.2611-2618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wanner BL. 1986. Novel regulatory mutants of the phosphate regulon in Escherichia coli K-12. J Mol Biol 191:39–58. doi: 10.1016/0022-2836(86)90421-3. [DOI] [PubMed] [Google Scholar]
- 45.Miller VL, Mekalanos JJ. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol 170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. 1992. Current protocols in molecular biology. Wiley & Sons, New York, NY. [Google Scholar]
- 47.Metcalf WW, Zhang JK, Apolinario E, Sowers KR, Wolfe RS. 1997. A genetic system for Archaea of the genus Methanosarcina: liposome-mediated transformation and construction of shuttle vectors. Proc Natl Acad Sci U S A 94:2626–2631. doi: 10.1073/pnas.94.6.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buan N, Kulkarni G, Metcalf W. 2011. Genetic methods for Methanosarcina species. Methods Enzymol 494:23–42. doi: 10.1016/B978-0-12-385112-3.00002-0. [DOI] [PubMed] [Google Scholar]
- 49.Metcalf WW, Zhang JK, Wolfe RS. 1998. An anaerobic, intrachamber incubator for growth of Methanosarcina spp. on methanol-containing solid media. Appl Environ Microbiol 64:768–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Hydrogenase operons in Methanosarcina barkeri. Three distinct types of hydrogenase are encoded by M. barkeri Fusaro. The frh and fre operons encode putative F420-reducing hydrogenases, while the vht and vhx operons encode putative methanophenazine-reducing hydrogenases. Genetic and biochemical data show that neither the fre nor the vhx operon is capable of producing an active hydrogenase under any growth condition yet examined (T. D. Mand, G. Kulkarni, and W. W. Metcalf, 2018 bioRxiv, https://doi.org/10.1101/334656). The ech operon encodes a ferredoxin-dependent energy-conserving hydrogenase. The locus tags are shown below each gene, with the prefix “Mbar_” omitted to save space (indicated by an asterisk) in some cases. Download FIG S1, PDF file, 0.7 MB (696.1KB, pdf) .
Copyright © 2018 Kulkarni et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Methanosarcina barkeri Fusaro strains used in this study. Download TABLE S1, DOC file, 0.04 MB (46KB, doc) .
Copyright © 2018 Kulkarni et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plasmids used in this study. Download TABLE S2, DOC file, 0.05 MB (49KB, doc) .
Copyright © 2018 Kulkarni et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download TABLE S3, DOC file, 0.03 MB (34.5KB, doc) .
Copyright © 2018 Kulkarni et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.