ABSTRACT
The opportunistic pathogen Streptococcus agalactiae is the major cause of meningitis and sepsis in a newborn’s first week, as well as a considerable cause of pneumonia, urinary tract infections, and sepsis in immunocompromised adults. This pathogen respires aerobically if heme and quinone are available in the environment, and a functional respiratory chain is required for full virulence. Remarkably, it is shown here that the entire respiratory chain of S. agalactiae consists of only two enzymes, a type 2 NADH dehydrogenase (NDH-2) and a cytochrome bd oxygen reductase. There are no respiratory dehydrogenases other than NDH-2 to feed electrons into the respiratory chain, and there is only one respiratory oxygen reductase to reduce oxygen to water. Although S. agalactiae grows well in vitro by fermentative metabolism, it is shown here that the absence of NDH-2 results in attenuated virulence, as observed by reduced colonization in heart and kidney in a mouse model of systemic infection. The lack of NDH-2 in mammalian mitochondria and its important role for virulence suggest this enzyme may be a potential drug target. For this reason, in this study, S. agalactiae NDH-2 was purified and biochemically characterized, and the isolated enzyme was used to screen for inhibitors from libraries of FDA-approved drugs. Zafirlukast was identified to successfully inhibit both NDH-2 activity and aerobic respiration in intact cells. This compound may be useful as a laboratory tool to inhibit respiration in S. agalactiae and, since it has few side effects, it might be considered a lead compound for therapeutics development.
KEYWORDS: bacterial pathogenesis, electron transport chain, NADH dehydrogenase, Streptococcus agalactiae, drug discovery
IMPORTANCE
S. agalactiae is part of the human intestinal microbiota and is present in the vagina of ~30% of healthy women. Although a commensal, it is also the leading cause of septicemia and meningitis in neonates and immunocompromised adults. This organism can aerobically respire, but only using external sources of heme and quinone, required to have a functional electron transport chain. Although bacteria usually have a branched respiratory chain with multiple dehydrogenases and terminal oxygen reductases, here we establish that S. agalactiae utilizes only one type 2 NADH dehydrogenase (NDH-2) and one cytochrome bd oxygen reductase to perform respiration. NADH-dependent respiration plays a critical role in the pathogen in maintaining NADH/NAD+ redox balance in the cell, optimizing ATP production, and tolerating oxygen. In summary, we demonstrate the essential role of NDH-2 in respiration and its contribution to S. agalactiae virulence and propose it as a potential drug target.
INTRODUCTION
Streptococcus agalactiae (group B Streptococcus [GBS]) is a facultative, fermentative commensal bacterium normally living in the gut and urogenital tract of healthy individuals. It belongs to the family Streptococcaceae, many of which are opportunistic pathogens, and is able to transition to invasive niches, causing excessive inflammation, sepsis, and death (1). S. agalactiae is the major cause of meningitis and sepsis in a newborn’s first week of life in the United States, as well as a considerable cause of pneumonia and sepsis in immunocompromised adults (2). In neonates, S. agalactiae is transmitted by the mother via aspiration of fluids during birth. Although most transmission can be prevented by intravenous antibiotic administration during labor, allergies and emerging resistance to such antibiotics are an increasing concern (2). S. agalactiae is also associated with a large fraction of urinary tract infections in the elderly and nursing home residents, including kidney and bladder infections (3).
Despite its capacity for fermentative metabolism, S. agalactiae can perform aerobic respiration in the presence of external sources of heme and quinone. Within the same operon, the genome encodes a cytochrome bd oxygen reductase (cyt bd encoded by cydAB), a putative type 2 NADH dehydrogenase (NDH-2 encoded by ndh), and a 1,4-dihydroxy-2-naphthoate prenyltransferase enzyme (encoded by menA) (4–6). The menA gene is normally involved in the synthesis of demethylmenaquinone (DMK-10). However, genes other than menA that are required to synthesize menaquinone (MK) are not present in S. agalactiae, which is, therefore, not able to synthesize DMK-10 from chorismate (6). Disabling cyt bd (ΔcydA) results in decreased organ colonization and increased survival of neonatal rats compared to wild-type (WT) infection, indicating a link between respiration and virulence (4, 7).
NDH-2 is a homodimeric flavoprotein that catalyzes the oxidation of NADH with the concomitant reduction of quinone. It is a monotopic membrane enzyme that binds at the cytoplasmic surface of the bacterial membrane in order to have access to one of its substrates (quinone) but has no transmembrane domain (8–11). cyt bd is a transmembrane, heme-containing two-subunit enzyme (CydA and CydB) that catalyzes menaquinol:O2 oxidoreductase activity (12). The chemical reaction catalyzed by cyt bd results in the net electrogenic transfer of two protons from the cytoplasm to the extracellular space, contributing to the proton motive force (PMF) (12, 13). Both NDH-2 and cyt bd are absent in mammalian mitochondria, making them plausible drug targets (14). NDH-2, which plays an important role in pathogen survival and virulence, has been pursued as a possible drug target in Mycobacterium tuberculosis (15, 16), Toxoplasma gondii (17), and Plasmodium falciparum (18, 19).
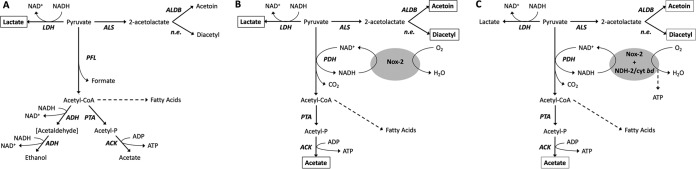
To understand the significance of NDH-2 in S. agalactiae survival and virulence and the consequences of its deficiency, it is important to consider the main metabolic strategies used by this pathogen (Fig. 1). Glycolysis yields 2 eq each of pyruvate and NADH. Growth requires not only ATP production but also a way to recycle NADH to NAD+ to allow glycolysis to proceed. Figure 1 shows alternative pathways for pyruvate catabolism in S. agalactiae that contribute to different degrees, depending on growth conditions. Note that this organism does not have the enzymes required for the tricarboxylic acid (TCA) cycle (20).
FIG 1 .
Pyruvate catabolism in S. agalactiae. One molecule of glucose will produce two of pyruvate via glycolysis, with net reduction of 2 NADH molecules. Fermentation of pyruvate, will allow NADH reoxidation for its recycling and use in a new round of glycolysis. (A) In the absence of oxygen, acetyl coenzyme A (acetyl-CoA) is made from pyruvate by pyruvate formate lyase (PFL), allowing synthesis of fatty acids. The main fermentation product is lactate, but significant amounts of ethanol, acetate, acetoin, and diacetyl are also found (21). Under aerobic conditions, production of acetoin, diacetyl, and acetate is prevalent over that of ethanol. (B) In the presence of oxygen without addition of external heme or quinone sources (respiration nonpermissive condition), lactate is still the main fermentation product, which leads to a significant decrease in pH. (C) Upon addition of external heme and quinone, in the presence of oxygen (respiration permissive condition), the respiratory chain (NDH-2/cyt bd) becomes functional. Metabolism is shifted toward production of acetate, acetoin, and diacetyl, reducing the amount of pyruvate available to be converted into lactate. This results in less acidification of the medium in the stationary phase. Higher growth yield is achieved as a result of enhanced ATP formation via the Pta-Ack pathway and possibly oxidative phosphorylation by the electron transport chain. Boxes indicate the main products for each condition. ACK, acetate kinase; ALS, acetolactate synthase; ALDB, 2-acetolactate decarboxylase; LDH, lactate dehydrogenase; PDH, pyruvate dehydrogenase; PTA, phosphate transacetylase; n.e., nonenzymatic reaction. (Adapted with data from references 21 to 23 and 66.)
Under anaerobic growth conditions, S. agalactiae must grow strictly by fermentation (21–23). Under growth conditions in which sugar metabolism is rapid and there is constant NADH production, NAD+ is regenerated by converting pyruvate to lactate via lactate dehydrogenase (homolactic fermentation) (Fig. 1A). Some growth conditions, however, result in a shift toward mixed-acid fermentation, in which pyruvate can be catabolized by pyruvate dehydrogenase, pyruvate formate lyase, and/or acetolactate synthase, yielding a variety of end products, including formate/CO2, acetate, and acetoin (Fig. 1B). The presence of oxygen has several consequences. First, pyruvate formate lyase is particularly susceptible to inactivation by oxygen. Second, if both heme and quinone can be acquired from the surroundings, S. agalactiae utilizes its respiratory chain to regenerate NAD+, generate a PMF, enhance ATP production via acetate kinase (Ack) and possibly oxidative phosphorylation, and reduce the concentration of O2 (Fig. 1C). Finally, S. agalactiae also has a water-forming NADH oxidase (Nox-2) (23), a soluble enzyme that reduces oxygen to water and regenerates NAD+ but does not contribute to the energy needs of the cell. In the absence of heme and quinone, S. agalactiae relies on Nox-2 for recycling NAD+ to feed glycolysis, to synthesize fatty acids, and for reduction of oxygen to water.
In this study, it is demonstrated that NDH-2 is an essential element for aerobic respiration and that disabling this enzyme reduces virulence in a mouse model of systemic infection. In addition, the S. agalactiae NDH-2 is purified, biochemically characterized, and proven a potential drug target for GBS.
RESULTS
S. agalactiae ndh encodes a highly active NDH-2.
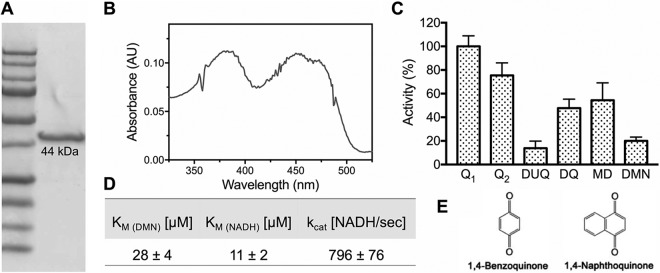
Generally, the low amino acid sequence identity among NDH-2s makes it difficult to distinguish them from other small, soluble flavoenzymes (24–26). The S. agalactiae ndh gene encodes a putative NDH-2 enzyme, showing relatively high identity to Staphylococcus aureus NdhC (42%) (27) and Caldalkalibacillus thermarum NDH-2 (41%) (see Fig. S1 in the supplemental material) (10, 11). The S. agalactiae ndh gene with an N-terminal 8×His tag was cloned and heterologously expressed in E. coli c43. S. agalactiae NDH-2 was purified, and SDS-PAGE analysis shows a single band at the predicted molecular weight of 44 kDa (Fig. 2A). The UV-visible (UV-Vis) spectrum of the air-oxidized protein shows characteristic peaks for flavin adenine dinucleotide (FAD) around 375 and 450 nm, with a shoulder around 470 nm indicating the cofactor is in an apolar environment (28) (Fig. 2B). The FAD cofactor remains in solution after precipitation of the protein with 5% trichloroacetic acid, demonstrating the flavin is noncovalently bound (29). Quantitation of the extracted flavin was calculated to be 0.25 mol FAD/mol protein. Substoichiometric flavin content is often observed with heterologously expressed flavoproteins (27, 30). Since the protein produced in E. coli shows substoichiometric amounts of FAD, assays were performed in the presence of 20 µM FAD. Upon addition of the flavin, a 10-fold increase in activity was observed. All results reported were performed with 20 µM FAD in the assay buffer.
FIG 2 .
The isolated enzyme from S. agalactiae is a highly active NDH-2. (A) SDS-PAGE of ~5 µg of isolated protein. (B) UV-Vis spectrum of the flavin. (C) Enzyme activity was measured with different soluble quinone analogues. Data are expressed as average ± standard deviation (SD) from three independent experiments. Q1, ubiquinone-1; Q2, ubiquinone-2; DUQ, decylubiquinone; DQ, duroquinone; MD, menadione; DMN, 2,3-dimethyl-1,4-naphthoquinone. One hundred percent activity corresponds to 4,190 NADH s−1 observed in the presence of Q1. (D) Turnover number (kcat) and affinity (Km) for the enyzme’s substrates were determined. Data are expressed as average ± SD from three independent experiments. (E) Core structures for benzoquinones (Q1, Q2, DUQ, and DQ) and naphthoquinones (MD and DMN).
S. agalactiae amino acid sequence shows high identity to C. thermarum and S. aureus NDH-2s. Identical residues among the three sequences are shaded in red. The figure was made using ESPript (67). Download FIG S1, TIF file, 1.4 MB (1.4MB, tif) .
Copyright © 2018 Lencina et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Although lacking any transmembrane-spanning elements, NDH-2 is a membrane-bound protein that, upon isolation, requires the presence of detergent to remain in solution. Optimal enzyme activity and stability also required 150 to 300 mM NaCl and pH 7. NADH:quinone oxidoreductase activity was tested in the presence of NADH and different soluble quinone analogues (Fig. 2C). The S. agalactiae endogenous electron acceptor for NDH-2 is DMK-10 (6), a naphthoquinone, but the pure enzyme is also active with ubiquinones (Fig. 2C and E). With 2,3-dimethyl-1,4-naphthoquinone (DMN), the soluble menaquinone analogue most similar to the natural electron acceptor in S. agalactiae membranes, the turnover number is about 800 NADH/s−1 and the Km values for DMN and NADH are 11 µM and 28 µM, respectively. The isolated S. agalactiae NDH-2 does not react directly with O2 in the presence of NADH, unlike the NDH-2 from Corynebacterium glutamicum (31).
NDH-2 is the only entry point for electrons into the respiratory chain of S. agalactiae.
An ndh deletion (Δndh) mutant was constructed to characterize the role of NDH-2 in the S. agalactiae respiratory metabolism. Since both aerobic respiration and Nox-2 are involved in oxygen reduction to water and regeneration of NAD+ (23) (Fig. 1), a Δndh Δnox2 double mutant strain was also constructed to characterize the effect on metabolism and oxygen tolerance. As a control, a previously studied nox2 single-knockout strain (23) was also examined.
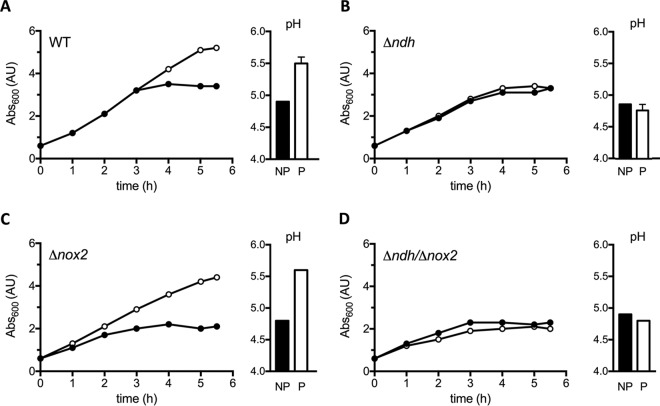
The WT, Δndh, Δnox2, and Δndh Δnox2 strains were grown aerobically in the presence of externally added heme and quinone (referred to as the respiration permissive condition) and in the absence of heme and quinone (referred to as the respiration nonpermissive condition) (Fig. 3). When the WT or Δnox2 mutant strain is grown aerobically in the presence of external heme and quinone, enhanced ATP production results in an improved growth yield (Fig. 3A and C), and there is a metabolic shift favoring mixed-acid fermentation, reduced lactate formation, and hence, a smaller pH drop in the medium (4, 32). In contrast, the Δndh strain does not show any difference in growth rate or pH drop (Fig. 3B), likely due to an increase in consumption of glucose that results in a higher production of lactate (which has a lower pKa) observed under nonpermissive conditions. This is to overcome the lower energetic efficiency derived from homolactic fermentation. The growth behavior and pH of the Δndh strain are restored to that of the WT strain if the deletion is complemented by a plasmid carrying the ndh gene under the control of its own promoter (see Fig. S2 in the supplemental material), indicating the observed effect is in fact due to NDH-2 activity. Similar results are observed for the Δndh Δnox2 strain (Fig. 3D).
FIG 3 .
The NDH-2-deficient strain shows a nonrespiratory phenotype. Growth curves and pH of the media at 8 h of culture are shown for the WT (A) and the Δndh (B), Δnox2 (C), and Δndh Δnox2 (D) mutant strains. Cells were grown under respiration permissive (P [white circles and bars]) and nonpermissive (NP [black circles and bars]) conditions. Growth curves are representative of at least 3 independent experiments. pH data are plotted as the average ± SD from three independent experiments.
The Δndh growth phenotype is complemented by a plasmid-carried copy of ndh. (A and B) Final growth (A) and pH (B) at 8 h of culture under respiration permissive (white bars) or nonpermissive (black bars) conditions for WT and Δndh strains carrying the empty vector (p) or ndh-expressing vector (p-ndh). Data are plotted as the average ± SD from three independent experiments. *, P = 0.0001 via two-way ANOVA with Dunnett’s posttest compared to wild-type nonpermissive. Download FIG S2, TIF file, 0.2 MB (211.6KB, tif) .
Copyright © 2018 Lencina et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
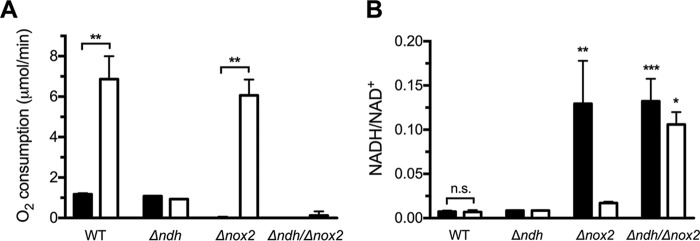
The effect of the deletions on oxygen utilization was tested for cells grown under either respiration permissive or nonpermissive conditions (Fig. 4A). Both respiration (NDH-2/cyt bd) and Nox-2 activity result in oxygen utilization. In the Δndh Δnox2 double deletion strain, no oxygen utilization is observed under any circumstances, suggesting that the elimination of NDH-2 results in the complete absence of respiration.
FIG 4 .
NDH-2 is important for respiration and for maintenance of NADH/NAD + redox balance. Cells were grown under respiration permissive (white bars) or nonpermissive (black bars) conditions and harvested in early stationary phase. (A) Oxygen consumption assays were normalized to an OD600 of 0.3 and started by the addition of 0.2% glucose. (B) The NADH/NAD+ ratio was determined for the WT and isogenic mutant strains. Data are plotted as the average ± SD from three independent experiments. (A) **, P < 0.005 via two-way analysis of variance (ANOVA) with Tukey’s posttest for the indicated comparisons. No significant difference was observed between nonpermissive and permissive conditions for the Δndh or Δndh Δnox2 mutants. (B) n.s., not significant, *, P < 0.05, **, P < 0.005, and ***, P < 0.001, via two-way ANOVA with Dunnett’s posttest compared to wild-type under nonpermissive conditions.
When grown in the absence of heme and quinone, the WT and Δndh strains utilize oxygen at the same rate, since Nox-2 is the only enzyme contributing to the measured depletion of O2. In the presence of heme and quinone, the WT strain has substantially more activity than the Δndh strain, since the WT strain is able to respire, while the Δndh strain cannot. All of the observed oxygen utilization of the Δnox2 strain, grown with heme and quinone, is due to NDH-2/cyt bd respiration. Under the conditions of these experiments and comparing the Δndh and Δnox2 strains, the capacity for O2 utilization by the respiratory chain can be estimated to be about 6-fold higher than that for Nox-2 (Fig. 4A).
Lastly, the importance of aerobic respiration and Nox-2 in NADH reoxidation was evaluated in cells grown under respiration permissive or nonpermissive conditions (Fig. 4B). When grown under permissive conditions, the NADH/NAD+ ratio did not show significant differences in the Δndh and Δnox2 strains. However, both the Δndh Δnox2 strain and the Δnox2 strain, when grown under nonpermissive conditions, show a drastic increase in NADH and the NADH/NAD+ ratio compared to the wild type. This suggests that NDH-2 (respiration) and Nox-2 are the major routes for NAD+ recycling, and these observations support the previously proposed role for respiration in fatty acid biosynthesis (23).
NDH-2 is important for S. agalactiae virulence.
Due to the essential role of NDH-2 in S. agalatiae respiration, we investigated its contribution to the development of invasive disease. The bacterial burdens or fitness of the S. agalactiae WT and mutant strains in different organs were determined after 24, 48, and 72 h postinfection (Fig. 5). Infection assays using the S. agalactiae Δndh strain and Δndh Δnox2 double mutant strain revealed significant attenuation in heart and kidney colonization compared to WT (Fig. 5A and B). The Δnox2 strain is significantly attenuated in heart and spleen compared to the WT (Fig. 5A and C), as reported in previous studies (23). Unexpectedly, no pronounced phenotype is seen in any organ with the Δndh Δnox2 double mutant strain, although this double mutant does not tolerate oxygen well. In an effort to explain this, we investigated the hemolytic activity of these knockout strains (Fig. 6). The cell surface-associated β-hemolysin/cytolysin of S. agalactiae is a nonimmunogenic, oxygen-stable, pore-forming cytolysin and a red polyenic pigment (33, 34). This major virulence factor is encoded by the cylE gene in the cyl locus, a unique 12-gene operon involved in fatty acid biosynthesis (35) that is expressed by most GBS strains. This virulence factor has proapoptotic, proinflammatory, and cytotoxic effects and is necessary for full S. agalactiae virulence in multiple in vivo systems (36). Furthermore, the hemolytic pigment has been shown to protect cells against a panel of stresses (33, 37). Remarkably, increased hemolysis was observed for the double mutant compared to the Δndh strain (Fig. 6), which correlates with the dramatic increase seen for the NADH/NAD+ ratio (Fig. 4B). Thus, this could contribute to the rescue effect observed during infection. Consistent with this interpretation, hemolysis is restored to WT levels upon addition of 0.1% Tween 80 to the cell cultures, effectively providing fatty acids to the cells (Fig. 6).
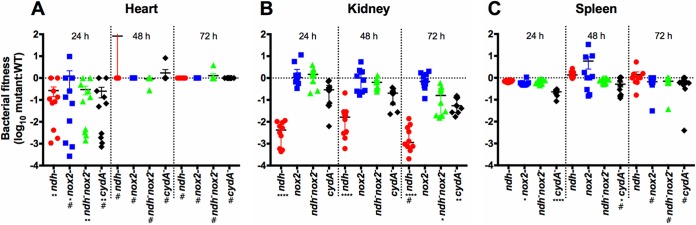
FIG 5 .
The Δndh, Δnox2, and ΔcydA mutant strains are attenuated for virulence in heart, kidney, and spleen. Mice were infected with 2 × 106 CFU of S. agalactiae, and bacteria were counted at 24, 48, and 72 h postinfection in heart (A), kidney (B), and spleen (C). Bacterial fitness of mutants is shown as ratio of the log10 number (CFU/gram) of mutant to WT bacteria. *, P < 0.05, **, P < 0.01, and ****, P < 0.0001 (mutant/WT ratio), by Kruskal-Wallis test followed by Dunn’s multiple comparisons. # denotes a median bacterial load (CFU per milliliter) of zero for the group.
FIG 6 .
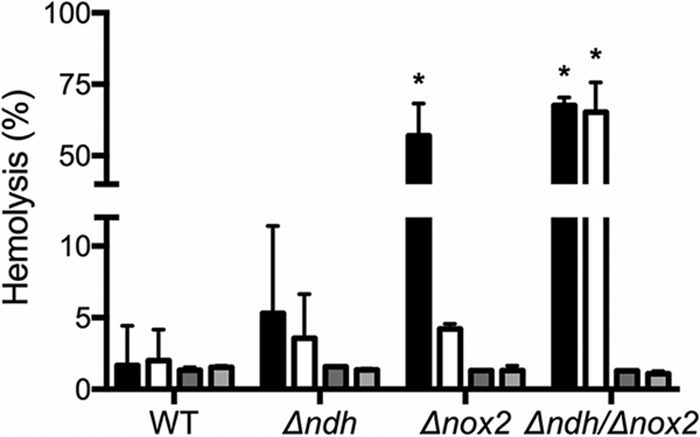
Hemolytic activity correlates with NADH/NAD+ ratios. Cells were grown under respiration permissive (white bars) or nonpermissive (black bars) conditions until the early stationary phase, then washed and incubated with RBCs for 1 h. Dark and light gray bars represent respiration nonpermissive and permissive growth, respectively, with 0.1% Tween 80 added to the media. Data are plotted as the average ± standard error of the mean (SEM) from three independent experiments. *, P < 0.0001 via two-way ANOVA with Dunnet’s posttest compared to the wild type under nonpermissive conditions.
Among the mutants tested, the strain lacking cyt bd (ΔcydA) has been previously shown to result in changes in organ colonization in mice (7). The cydA knockout strain, used here as a control for S. agalactiae attenuation in the model of systemic infection, is significantly attenuated in heart, kidney, and spleen (Fig. 5), as well as liver (see Fig. S3 in the supplemental material), and its attenuation approaches statistical significance in blood (P = 0.08) (Fig. S3), findings consistent with what has been previously observed (7). None of the mutants is significantly attenuated in blood, liver, or brain colonization (Fig. S3A to C). These data support the conclusion that respiration is important for virulence in S. agalactiae (4, 7) and show, furthermore, that NDH-2 contributes to virulence.
The Δndh, Δnox2, and ΔcydA mutant strains are not attenuated for virulence in blood, liver, or brain. Mice were infected with 2 × 106 CFU of S. agalactiae, and bacteria were enumerated at 24, 48, and 72 h postinfection in blood (A), liver (B), and brain (C). Bacterial fitness of mutants is shown as a ratio of the log10 number (CFU/gram) of mutant to WT bacteria. *, P < 0.05 for the mutant/WT ratio by Kruskal-Wallis test followed by Dunn’s multiple comparisons. # denotes a median bacterial load (CFU/milliliter) of zero for the group. Download FIG S3, TIF file, 0.3 MB (269.1KB, tif) .
Copyright © 2018 Lencina et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Inhibitors of S. agalactiae NDH-2 are potential leads against GBS.
Since this work demonstrates that NDH-2 is essential for respiration in S. agalactiae as well as important for virulence, inhibitors of the isolated enzyme were obtained and then tested as inhibitors of respiration in intact S. agalactiae cells. Enzymatic activity was monitored colorimetrically in the presence of NADH and menadione (MD), a commercially available soluble analogue of menaquinone. Two libraries of FDA-approved drugs were screened (about 2,000 compounds) using 25 µM each compound to test for inhibitors of the pure enzyme. Nine compounds inhibited at least 80% of enzymatic activity: alexidine, candesartan, caspofungin, closantel, hexachlorophene, isoquercetin, pramoxine, triclabendazole, and zafirlukast. Except for hexachlorophene, a relatively toxic disinfectant, all other compounds were tested against whole cells growing under respiration permissive conditions, and five of these showed an inhibitory effect on cell growth (Table 1).
TABLE 1 .
FDA-approved compounds that inhibit NDH-2 activity and S. agalactiae growth in vitroa
| Compound | IC50 (µM) | GI50 (µM) | Common use |
|---|---|---|---|
| Alexidine | 16.79 ± 1.26 | 2.70 ± 0.51 | Antiseptic |
| Caspofungin | 15.23 ± 2.33 | 41.03 ± 2.62 | Antifungal |
| Closantel | 5.34 ± 0.65 | 0.25 ± 0.04 | Antihelmintic (veterinary) |
| Triclabendazole | 0.14 ± 0.02 | 15.30 ± 1.92 | Antihelmintic |
| Zafirlukast | 0.83 ± 0.07 | 6.07 ± 0.10 | Asthma treatment |
Cells were grown under respiration permissive conditions.
Although candesartan, isoquercetin, and pramoxine are able to inhibit the purified enzyme in vitro, they show no effect on cell growth. On the other hand, alexidine and closantel inhibit cell growth at concentrations much lower than those required for NDH-2 inhibition, suggesting other targets might be responsible for their effect in whole cells. This is not surprising since both compounds are known to disturb membrane integrity, producing permeabilization (38) and strong uncoupling (39). Finally, caspofungin, triclabendazole, and zafirlukast not only show NDH-2 inhibition in the low micromolar and even nanomolar range but are also able to inhibit cell growth at low micromolar concentrations (Table 1).
Additional assays followed oxygen consumption of WT cells (grown with heme and quinone) in the presence of different concentrations of caspofungin, triclabendazole, or zafirlukast. From these compounds, only zafirlukast shows a dose-dependent linear decline in respiration, with 90% inhibition at 7.5 µM (Fig. 7A). In addition, the Δnox2 strain was grown overnight with and without subinhibitory concentrations of zafirlukast (~50% growth inhibitory concentration [GI50]), and both growth and respiration were examined after overnight incubation. The Δnox2 strain grown in the presence of zafirlukast does not exhibit enhanced growth under respiration permissive conditions compared to nonpermissive conditions. This indicates that the bacterial growth supported by the activity of the electron transport chain is substantially reduced in the presence of zafirlukast (Fig. 7B). Not all of the NDH-2 activity was eliminated since the cells were grown with a concentration of zafirlukast that does not completely inhibit the enzyme. This is shown in Fig. 7C where the oxygen utilization, which must be due to respiration in the Δnox2 strain, is substantially lower than when the cells are grown in the absence of zafirlukast, but not as low as when cells are grown in the absence of heme and quinone. In sum, the data show that zafirlukast inhibits the activity of the isolated S. agalactiae NDH-2 as well as the activity of the enzyme in intact cells.
FIG 7 .
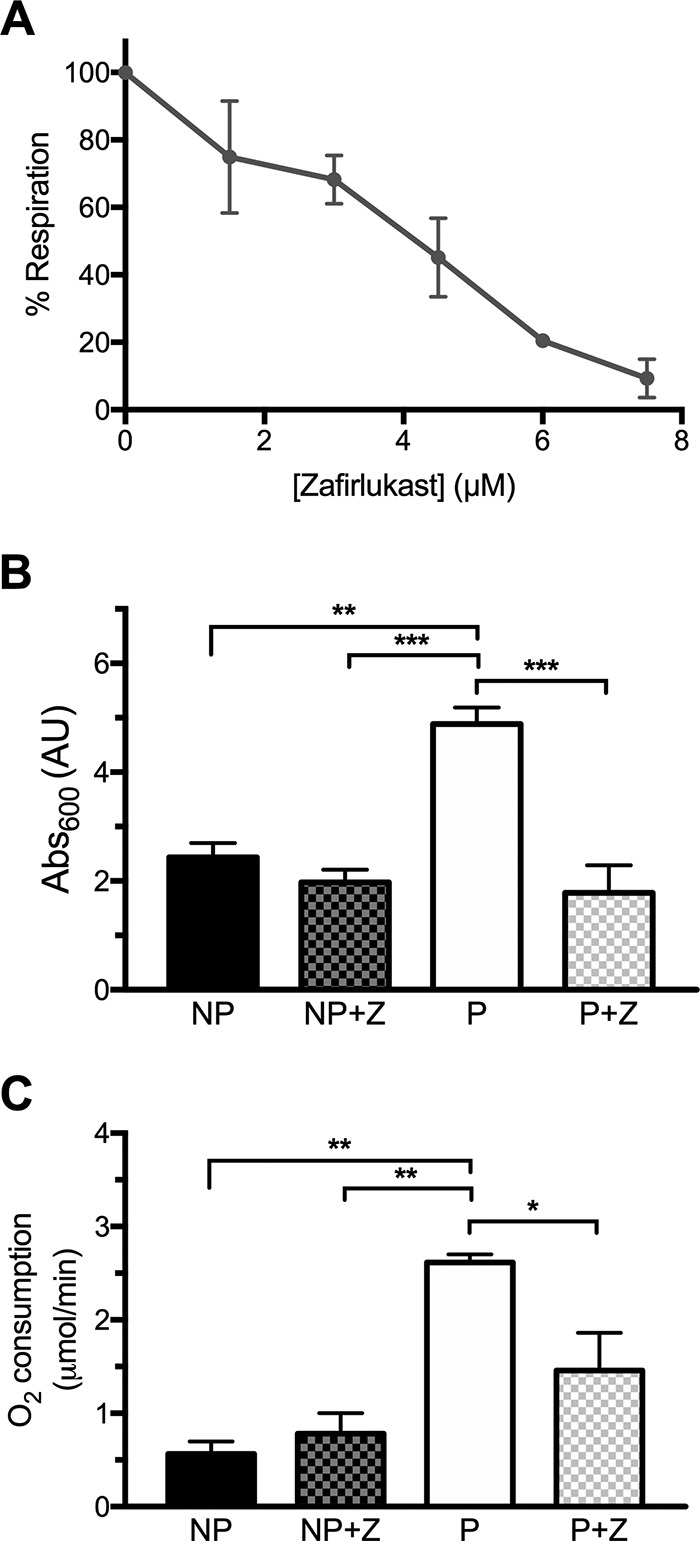
Zafirlukast inhibits S. agalactiae respiration. Cells were grown until the early stationary phase (8 h), and the percentage of respiration was calculated after addition of different concentrations of zafirlukast (A). The percentage of respiration corresponds to the difference in oxygen consumption of cells grown under respiration permissive (P) minus nonpermissive (NP) conditions. (B and C) Overnight growth (B) and oxygen consumption (C) of the Δnox2 mutant strain in the absence or presence of 7.5 µM zafirlukast (Z). Data are plotted as the average ± SD from three independent experiments. In panels B and C, *, P < 0.05, **, P < 0.005, and ***, P < 0.001, via two-way ANOVA with Tukey’s posttest for the indicated comparisons. No significant difference was observed between conditions NP, NP+Z, and P+Z.
DISCUSSION
S. agalactiae can cause invasive disease in newborns leading to meningitis and septicemia and is also associated with urinary tract infections in the elderly population. It is a highly adapted organism capable of metabolizing a wide variety of substrates; however, it is an obligate commensal, auxotrophic for several amino acids and vitamins (1, 20). Although it used to be considered an aerotolerant organism, S. agalactiae was found over 10 years ago to perform aerobic respiration in the presence of external sources of heme and quinone (4). Aerobic respiration consists of the transport of electrons through a series of redox components, from a metabolic reductant (e.g., NADH) and terminating in the reduction of oxygen to water by a terminal oxygen reductase. Importantly, respiration is linked to the generation of PMF and ATP synthesis via Ack and possibly oxidative phosphorylation (Fig. 1C).
S. agalactiae has a two-enzyme respiratory chain.
S. agalactiae has a single oxygen reductase, cyt bd. The first conclusion of the present work is that there is only one enzyme, NDH-2, that provides electrons to the respiratory chain of this organism. Both NDH-2 and cyt bd are encoded by genes (ndh and cydAB, respectively) within the same operon. Although single-subunit respiratory flavoenzymes are often misannotated (40), an exhaustive search of the genome of S. agalactiae shows no other genes for any additional respiratory enzymes, like glycerol-3-P dehydrogenase, pyruvate oxidase, or membrane-bound succinate, lactate, or malate dehydrogenases. A glp operon for glycerol metabolism is present; however, glycerol phosphate dehydrogenase (glpD) is replaced by a glycerol phosphate oxidase (glpO), as in other organisms that do not synthesize heme. The latter enzyme is soluble and reduces oxygen to hydrogen peroxide instead of reducing quinone (41). Also, whereas some bacterial pathogens (e.g., S. aureus) have two different NDH-2 enzymes (27), S. agalactiae has only one, and there is no other type of respiratory NADH dehydrogenase, such as complex I (NDH-1) or the sodium pumping NDH (Nqr) (42). Thus, unlike most other bacteria, the electron transport chain of S. agalactiae is not branched at either end and consists of just two proteins plus DMK-10 (6).
The ndh gene encodes an authentic NDH-2.
The S. agalactiae ndh gene was cloned and expressed in E. coli, and the purified recombinant protein was shown to be an NADH:quinone oxidoreductase. The isolated, recombinant S. agalactiae NDH-2, which has an amino acid sequence identity of 42% with S. aureus NdhC, has a high kcat (~800 NADH s−1) that is similar to that of S. aureus NdhC (~1,500 NADH s−1) (27). These kcat values are substantially higher than those reported for NDH-2s from other organisms, including the NDH-2s from E. coli, with a kcat of ~18 NADH s−1 (43), M. tuberculosis, with a kcat of ~10 NADH s−1 (44), Bacillus pseudofirmus, with a kcat of ~39 NADH s−1 (45), Bacillus subtilis, with a kcat of ~20 NADH s−1 (46), and C. glutamicum, with a kcat of ~213 NADH s−1 (31). The high activities obtained for the enzymes from S. agalactiae and S. aureus are most likely due to the presence of phospholipids and FAD in the reaction buffer of our assays. The S. agalactiae NDH-2 has a lower Km for NADH (11 µM) than those of other previously characterized NDH-2s from bacteria. For example, the apparent Km value for NADH of the NDH-2 from E. coli is 34 µM (43), that from B. subtilis is 60 µM (46), that from B. pseudofirmus is 114 µM (45), and those from S. aureus are 35 µM (NdhC) and 154 µM (NdhF) (27).
Multiple advantages of utilizing aerobic respiration.
S. agalactiae grows well by utilizing a variety of fermentation strategies (Fig. 1), but the operon encoding NDH-2 and cyt bd is expressed constitutively so the cells can very rapidly switch to respiratory growth (4) in the presence of O2 after taking up quinone and heme from the environment (6, 47). Certainly, a major advantage of respiratory growth is a higher bioenergetics capacity via Ack and possibly oxidative phosphorylation. In addition to enhanced growth efficiency, respiration also serves for (i) reduction of the intracellular and local concentrations of O2 and to combat oxygen toxicity, (ii) generation of NAD+ necessary for glycolysis and fatty acid synthesis (23), (iii) generation of the PMF required for powering transporters and essential efflux pumps like PefAB (13, 48), (iv) contribution to resistance against environmental acid stress (5, 49), and (v) production of virulence factors (e.g., respiration is required for nuclease production) (50).
Respiration is linked to virulence and organ colonization.
The different stimuli found in the growth environment, including the carbon source(s), concentration of O2, and the availability of external heme and quinone, determine the metabolic pathways utilized to optimize the growth and survival of S. agalactiae. The growth conditions vary significantly when comparing in vitro batch growth to in vivo growth in a host animal or when comparing the in vivo growth in different organs. The combination of metabolic pathways utilized by S. agalactiae must simultaneously generate ATP, maintain the PMF, reduce levels of O2 if it is present, and maintain the recycling of NADH through NAD+. Since the metabolic strategies used by S. agalactiae can be very different in each organ in an infected animal, it is reasonable that mutations that incapacitate respiration will have different effects on organ colonization, depending on the organ. Based on studies with a cyt bd oxidase deletion mutant, S. agalactiae respiration has been previously linked to virulence and colonization of blood-rich organs (4, 7). It is shown in the present work that the deletion of the gene encoding NDH-2 eliminates in vitro respiration and also results in attenuation of organ colonization in a mouse model for systemic infection. The strongest attenuation from the ndh deletion is in heart and kidney. The apparent significance of respiration for kidney infection may be useful considering the importance of S. agalactiae in urinary tract infections in the elderly (3). Moreover, S. agalactiae is able to synthesize its own DMK-10, starting from 1,4-DHNA (dihydroxy-2-naphthoic acid) or short-chain quinones, such as MK-4 (6), which can be found in high concentrations in the kidney (51). An unexpected finding is that the Δndh Δnox2 double mutant strain is much less attenuated in kidney colonization than the highly attenuated Δndh strain (Fig. 5B). This may be related to the increased hemolysis observed for the double mutant strain compared to the Δndh strain (Fig. 6). The Δndh Δnox2 double mutant strain is highly impaired in NADH oxidation to NAD+ (Fig. 4B), and the deficiency of NAD+ likely results in impaired fatty acid biosynthesis (23). The increased hemolysis observed in the double-knockout strain (Fig. 6) may be the response of S. agalactiae to obtain fatty acids from surrounding host cells (52).
It is noteworthy that the Δndh strain is much more impaired in colonization in the kidney than is the ΔcydA strain, since both of these mutations disrupt respiration. One possible explanation is that the presence of a heme-containing enzyme in the membrane (cyt bd) in the Δndh strain makes the bacterial cells more susceptible to the strong oxidative stress displayed by the kidney during sepsis (53). Alternatively, higher levels of reduced menaquinone in the ΔcydA strain could lead to production of small amounts of intracellular reactive oxygen species (ROS), stimulating a response against oxidative stress through PerR (54, 55), enhancing survival of the strain in the kidney. The precise molecular explanation for the organ-specific effects of the mutants on colonization will require further studies.
Inhibitors of S. agalactiae NDH-2.
Screening for inhibitors against the purified S. agalactiae NDH-2 was done in the presence of lipids to mimic the hydrophobic environment of the enzyme in situ. Although several compounds from libraries of FDA-approved drugs were found to inhibit the enzyme in vitro, only zafirlukast also inhibits respiration in intact cells in a way that suggests NDH-2 is the likely target in vivo. However, it is possible that this compound has other targets in S. agalactiae. Zafirlukast is a cysteinyl leukotriene receptor antagonist, currently used for the prophylaxis and chronic treatment of asthma (56). It is a commercially available generic drug and safe for daily use (57), and it has also been described to have bactericidal activity against M. tuberculosis by inhibiting complex formation between Lsr2 (a nucleoid-associated protein) and DNA (58). Zafirlukast also has been demonstrated to have antibacterial and antibiofilm activity against the nonrespiratory oral pathogens Porphyromonas gingivalis and Streptococcus mutans (59).
It is worth mentioning that despite its high similarity to S. aureus NdhC, S. agalactiae NDH-2 is not inhibited by trifluoperazine or thioridazine, phenothiazines that both show a strong inhibitory effect on S. aureus NdhC (27). These data demonstrate that the NDH-2 enzymes from different organisms, though apparently very similar, can have different inhibition profiles with drugs. These small enzymes, with binding sites for both NADH and quinol, appear to be susceptible to inhibitors that do not bind at the substrate binding sites but, rather, to allosteric sites (44). Although the substrate binding sites may be conserved among different NDH-2s, inhibitor-binding regions may differ greatly. In any event, zafirlukast may be useful as a laboratory tool to inhibit respiration in S. agalactiae and, since it has few side effects (60), might be considered a lead compound for therapeutics.
MATERIALS AND METHODS
Sequence analysis.
Gene sequences encoding S. agalactiae (gbs1788), S. aureus (SAB0708) and C. thermarum (GenBank accession no. ZP_08531709.1) NDH-2s were retrieved from the National Center for Biotechnology Information (NCBI) database. Amino acid sequence alignments were performed using ClustalW (61).
Bacterial strains and growth conditions.
Strains and plasmids used in this work are listed in Table 2. S. agalactiae NEM316, a capsular serotype III strain with a fully sequenced genome, was used as the WT (20, 62). For growth in liquid media, cells were initially grown overnight at 37°C in M17 broth supplemented with 0.2% glucose and 80 µM menaquinone (MK-4), under static microaerobic conditions. These overnight cultures were used to inoculate tubes with M17 broth (1/10 vol/vol ratio) supplemented with 5 µM riboflavin, to an initial optical density at 600 nm (OD600) of ≈0.05. Culture tubes were incubated at 37°C—first statically until they reached an OD600 of ≈0.5 and then transferred to high-aeration conditions (200 rpm). To support respiration, 50 µg ml−1 of hemoglobin (in 0.9% NaCl aqueous solution) and 30 µM MK-4 were added to the cultures before transfer.
TABLE 2 .
Bacterial strains and plasmids used in this study
| Strain or plasmid | Main characteristic(s)a | Reference or source |
|---|---|---|
| Strains | ||
| S. agalactiae | ||
| NEM316 | Serotype III isolated from neonatal blood culture | 20 |
| Δndh mutant | NEM316 with ndh gene deleted | This work |
| Δnox2 mutant | NEM316 nox2 ΩaphA-3 Δnox2 Kmr | 23 |
| Δndh Δnox2 mutant | Δnox2 mutant with ndh gene deleted, Kmr | This work |
| NEMJ01 | NEM316 cydA::aphA-3 Kmr | 4 |
| E. coli | ||
| TG1 | Host strain for sequencing | Lucigen |
| TOP10 | Host strain for molecular cloning | Invitrogen |
| c43(DE3) | Host strain for protein overproduction | Avidis, France |
| Plasmids | ||
| pBR322-pGhost8 | Thermosensitive vector, Tetr | 63 |
| pTCV-lac | Shuttle vector, Eryr Kmr | 64 |
| pTCV-ndhC | ndh complementation plasmid | This work |
| pET-22b | Cloning vector, Ampr | Novagen |
Ampr, Kmr, Tetr, and Eryr indicate resistance to ampicillin, kanamycin, tetracycline, and erythromycin, respectively.
Construction of ndh-deficient mutant and complementation plasmid.
Primers used for constructions are listed in Table 3. Chromosomal DNA from S. galactiae strain NEM316 was used as a template to amplify by PCR a DNA fragment upstream of the gene gbs1788 (ndh) with primer pair gbs1788F and gbs1788intR. A second fragment downstream of gbs1788 was obtained with primer pair gbs1788intF and gbs1788R. Both fragments were fused by PCR with gbs1788F and gbs1788R to give the fragment Δndh, which was further cloned into EcoRI and BamHI sites of the thermosensitive plasmid pBR322-pGhost8 (63) to give the plasmid pBR322-pGhost8-Δndh. The resulting plasmid was established in E. coli strain TG1 for sequencing and then transferred into the S. agalactiae NEM316 WT and Δnox2 mutant strains (23). S. agalactiae transformants were selected at 30°C on brain heart infusion (BHI) plates supplemented with 3 µg/ml tetracycline. The plasmid was integrated in the ndh locus when cells were grown at 37°C (first recombination event), followed by growth at 30°C for excision (second recombination event). Gene deletion was confirmed by PCR with primer pair gbs1788extF and gbs1788extR, designed outside the recombination region. In the Δndh strain, 373 of 402 amino acid residues of NDH-2 protein are absent. For complementation studies, the promoter region of the operon up to downstream of the stop codon of gbs1788 was amplified by PCR with oligonucleotide pair 1788compFor and 1788compRev and chromosomal DNA of a gbs1789 mutant (deletion mutant) and then cloned into EcoRI/BamHI sites of plasmid pTCV-lac (64). The resulting plasmid, pTCV-ndhC, was established in E. coli strain TG1 for sequencing and then in the Δndh mutant. pTCV-lac was used as a control. The plasmids were maintained by addition of kanamycin (Km) at 500 µg ml−1.
TABLE 3 .
Primers used in this study
| Primer designation | Sequence (5′→3′) |
|---|---|
| gbs1788F | GATCGAATTCGTATTTTCTGGCTTGACAATGGG |
| gbs1788intR | GCTAGAAGTTCTTTAAGTCCACCGGCATAACCAGCACCTAAAACTAGG |
| gbs1788intF | CCTAGTTTTAGGTGCTGGTTATGCCGGTGGACTTAAAGAACTTCTAGC |
| gbs1788R | GATCGGATCCGTAAACATCCATAAACCAAGG |
| gbs1788extF | CTGCCTCTTTTATGGATGGG |
| gbs1788extR | GAGCCAAAAACCACTAGC |
| 1788compFor | GCCGGAATTCCGGCTATTTAAAACAAATTGGAGC |
| 1788compRev | CGGCGGATCCCGAAGAGCTAGTTTCCTAGCTTCG |
| FwNdhSNdeIHis | GGAATTCCATATGCATCACCATCACCATCACCATCACAAAGAAATCCTA GTTTTAGGTGC |
| RvNdhSHindIII | CCCAAGCTTTTAATGATATAAATCAAAACGTCCCTTAG |
Oxygen consumption assays and pH determination.
Oxygen consumption by S. agalactiae cells was determined at 37°C using a dual-channel respirometer system (model S782; Strathkelvin Instruments). The concentration of oxygen in the air-saturated buffer at 37°C was assumed to be 237.5 µM. Early-stationary-phase cells (8 h) were washed and resuspended in phosphate-buffered saline (PBS) buffer to an OD of ≈0.3 in a 1-ml chamber; oxygen consumption was monitored after addition of 0.2% glucose. Unless specified otherwise, the effect of different compounds on respiration was determined after 3-min incubations of the cells with the drugs at 37°C before addition of glucose.
The pH of the medium at 8 h of culture was determined using pH strips (MColorphast; Millipore).
Determination of the NADH/NAD+ ratio.
Cells grown under respiration permissive or nonpermissive conditions were collected at early stationary phase (~8 h) by centrifugation at 14,000 × g for 5 min. Pellets were washed once with PBS and resuspended in 400 µl of extraction buffer at an OD of ~30. Samples were homogenized twice in a FastPrep-24 Beadbeater at 6 m/s for 45-s cycles with 2 min of incubation on ice in between. Homogenized samples were centrifuged at 4°C in a microcentrifuge at 14,000 × g for 2 min, and intracellular levels of NADH and NAD+ were measured using the NAD/NADH quantitation kit (MAK037; Sigma-Aldrich, St. Louis, MO) according to the manufacturer’s instructions. NADH and NAD levels were normalized to milligrams of protein.
Hemolysis assay.
Hemolytic activity using whole bacteria was assayed as described by Nizet et al. (65). Briefly, cells grown until the early stationary phase (~8 h), in the presence or absence of 0.1% Tween 80 under respiration permissive or nonpermissive conditions, were pelleted by centrifugation at 14,000 × g, washed once with PBS, and resuspended in 1 ml of PBS with 0.2% glucose to an OD of ~1. In a 96-well conical-bottom microtiter plate, 100 µl of cell suspensions, in triplicate, was incubated with an equal volume of 1% sheep red blood cells (RBCs) in PBS-glucose. The plate was incubated at 37°C for 1 h and then spun down at 640 × g for 5 min to pellet unlysed RBCs and bacteria. The supernatants were transferred to a replica 96-well plate, and hemoglobin release was measured by recording the absorbance at 420 nm. RBCs in PBS-glucose and RBC lysis with 0.1% sodium dodecyl sulfate (SDS) were used as negative and positive controls, respectively.
Animal assays.
Six-week-old female BALB/c mice were purchased from the Animal Resources Centre (Australia). A systemic infection model was used to assess virulence of S. agalactiae. Bacteria used for infection were grown statically in Todd-Hewitt broth (THB) overnight at 37°C. Mice were inoculated through the lateral tail vein with 2 × 106 bacteria in PBS (pH 7.4); inocula were delivered in 200 µl using 27G × 11/4-in. PrecisionGlide needles (BD) connected to 1-ml tuberculin syringes. At various intervals, groups of five mice were sacrificed, and blood was collected by cardiac puncture. Organs were collected, weighed, and homogenized in PBS. The bacterial numbers in blood and organ homogenates were determined by plating on THB agar plates incubated at 37°C. Statistical analysis of bacterial counts was performed using the Kruskal-Wallis test, followed by Dunn’s multiple comparisons. GraphPad Prism (version 7.0b) software was used for all statistical analyses. A P value of <0.05 was considered statistically significant. The animal experiments were repeated twice in independent experiments and were approved by Griffith University Animal Ethics Committee (approval no. MSC/01/15/AEC).
NDH-2 heterologous expression and purification.
The S. agalactiae NDH-2 (ndh [gbs1788]) open reading frame was amplified from NEM316 using primers listed in Table 3 and cloned into the pET22-b expression plasmid, with an N-terminal 8×His tag. The E. coli c43(DE3) strain, carrying the pRARE plasmid and transformed with the resulting plasmid, was grown under aerobic conditions (200 rpm) at 37°C in LB medium supplemented with 50 µg/ml of kanamycin and 100 µg/ml of ampicillin until an OD600 of ≈0.8, and expression was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 4 h. All subsequent steps were performed at 0 to 4°C. Cells were harvested at 14,000 × g for 10 min and resuspended in buffer A (50 mM sodium phosphate buffer [pH 7.5], 300 mM NaCl) plus 5 mM MgSO4, DNase I, and protease inhibitor cocktail (Sigma). These cells were then disrupted by passing three times through a microfluidizer at a pressure of 80,000 lb/in2. The resulting extracts were centrifuged at 14,000 × g for 10 min to remove unbroken cells, and supernatants were subject to ultracentrifugation at 230,000 × g for 4 h to obtain membrane pellets. Membrane fractions were resuspended in buffer A plus the protease inhibitor cocktail and solubilized by addition of a stock solution of 20% dodecyl-β-d-maltoside (DDM) dropwise to a final concentration of 1%. The suspension was incubated at 4°C for 1 h with mild agitation and then cleared by centrifugation at 230,000 × g for 1 h. Solubilized membranes were added to 5 ml of Ni-nitrilotriacetic acid (NTA) resin (Qiagen Sciences, Germantown, MD) preequilibrated with buffer A plus 10 mM imidazole and 0.05% DDM. The protein bound to the resin was washed with buffer A plus increasing concentrations of imidazole (10 to 50 mM) and 0.05% DDM. Protein was eluted with buffer A plus 200 mM imidazole and 0.05% DDM and concentrated by filtration (Millipore concentrator). Imidazole was removed by a series of filtration and washing steps with buffer A plus 0.05% DDM. The purified protein was stored frozen at −80°C after addition of glycerol to a final concentration of 10%.
Biochemical characterization.
Protein concentration was determined using the Pierce bicinchoninic acid (BCA) protein assay kit. Purity of the sample was observed by running SDS-PAGE. The FAD concentration was measured by addition of 5% trichloroacetic acid followed by centrifugation and determination of supernatant absorbance at 450 nm (ε450: 11,300 mM−1 cm−1) (29).
Enzyme activity assay and determination of kinetic parameters.
The rate of NADH oxidation was determined using a UV-Vis spectrophotometer (Agilent Technologies model 8453) by monitoring absorbance at 340 nm (NADH oxidation) upon addition of 100 µM NADH in the presence of 2 nM enzyme and a 100 µM concentration of a quinone soluble analogue. Assays were performed at 37°C in buffer B (50 mM sodium phosphate buffer [pH 7], 150 mM NaCl) plus 125 µg ml−1 of soy asolectin (Sigma) and 20 µM FAD. For determination of kinetic constants, 2 to 200 µM NADH and 2 to 200 µM of DMN were added to the reaction mixture containing 2 nM enzyme. Enzyme rates are expressed as a turnover number (kcat) based on moles of NADH oxidized per second per mole of enzyme.
Inhibitor screening.
Isolated NDH-2 activity was tested against 1,905 compounds from two FDA-approved drug libraries (Selleck Chemicals, with 1,176 compounds, and the NIH Clinical Collection [NCC], with 729 compounds). Enzyme activity was measured in buffer B with 125 µg ml−1 of soy asolectin, 20 µM FAD, and 50 µM MD, in the presence and absence of 25 µM each compound. The reaction was started upon addition of 100 µM NADH, after a 3-min incubation of the reaction mixture at 37°C. The compounds found to inhibit at least 80% of enzyme activity in vitro were further selected to determine their 50% inhibitory concentration (IC50) and GI50. IC50 refers to the concentration that causes 50% inhibition of enzyme activity, while GI50 represents the concentration that causes 50% inhibition of cell growth. The IC50 for each compound was determined by testing enzyme activity after a 3-min incubation in the presence of different concentrations of the inhibitor, before starting the reaction by addition of NADH. GI50 was determined by growing the bacterial cells overnight in the presence of different concentrations of each inhibitor under respiration permissive growth conditions.
ACKNOWLEDGMENTS
We thank members of the Gennis laboratory for help and useful discussions.
This work was supported by grants from the United States National Institutes of Health GM095600 and HL16101 (R.B.G.) and grants from the Australia National Health and Medical Research Council APP1146569 and APP1146820 (G.C.U.).
Footnotes
Citation Lencina AM, Franza T, Sullivan MJ, Ulett GC, Ipe DS, Gaudu P, Gennis RB, Schurig-Briccio LA. 2018. Type 2 NADH dehydrogenase is the only point of entry for electrons into the Streptococcus agalactiae respiratory chain and is a potential drug target. mBio 9:e01034-18. https://doi.org/10.1128/mBio.01034-18.
Contributor Information
Neal D. Hammer, Michigan State University.
Kimberly A. Kline, Nanyang Technological University.
REFERENCES
- 1.Edwards MS, Baker CJ. 2016. Group B streptococcal infections, p 411–456. In Wilson CB, Nizet V, Maldonado Y, Remington JS, Klein JO (ed), Remington and Klein’s infectious disease of the fetus and newborn infant, 8th ed. Elsevier, New York, NY. [Google Scholar]
- 2.National Center for Immunization and Respiratory Diseases, Division of Bacterial Diseases . 2016. Group B Strep (GBS). Centers for Disease Control and Prevention, Atlanta, GA: https://www.cdc.gov/groupbstrep/index.html. [Google Scholar]
- 3.Kline KA, Schwartz DJ, Lewis WG, Hultgren SJ, Lewis AL. 2011. Immune activation and suppression by group B streptococcus in a murine model of urinary tract infection. Infect Immun 79:3588–3595. doi: 10.1128/IAI.00122-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamamoto Y, Poyart C, Trieu-Cuot P, Lamberet G, Gruss A, Gaudu P. 2005. Respiration metabolism of group B Streptococcus is activated by environmental haem and quinone and contributes to virulence. Mol Microbiol 56:525–534. doi: 10.1111/j.1365-2958.2005.04555.x. [DOI] [PubMed] [Google Scholar]
- 5.Rosinski-Chupin I, Sauvage E, Mairey B, Mangenot S, Ma L, Da Cunha V, Rusniok C, Bouchier C, Barbe V, Glaser P. 2013. Reductive evolution in Streptococcus agalactiae and the emergence of a host adapted lineage. BMC Genomics 14:252. doi: 10.1186/1471-2164-14-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franza T, Delavenne E, Derré-Bobillot A, Juillard V, Boulay M, Demey E, Vinh J, Lamberet G, Gaudu P. 2016. A partial metabolic pathway enables group b streptococcus to overcome quinone deficiency in a host bacterial community. Mol Microbiol 102:81–91. doi: 10.1111/mmi.13447. [DOI] [PubMed] [Google Scholar]
- 7.Joubert L, Dagieu JB, Fernandez A, Derré-Bobillot A, Borezée-Durant E, Fleurot I, Gruss A, Lechardeur D. 2017. Visualization of the role of host heme on the virulence of the heme auxotroph Streptococcus agalactiae. Sci Rep 7:40435. doi: 10.1038/srep40435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iwata M, Lee Y, Yamashita T, Yagi T, Iwata S, Cameron AD, Maher MJ. 2012. The structure of the yeast NADH dehydrogenase (Ndi1) reveals overlapping binding sites for water- and lipid-soluble substrates. Proc Natl Acad Sci U S A 109:15247–15252. doi: 10.1073/pnas.1210059109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng Y, Li W, Li J, Wang J, Ge J, Xu D, Liu Y, Wu K, Zeng Q, Wu JW, Tian C, Zhou B, Yang M. 2012. Structural insight into the type-II mitochondrial NADH dehydrogenases. Nature 491:478–482. doi: 10.1038/nature11541. [DOI] [PubMed] [Google Scholar]
- 10.Heikal A, Nakatani Y, Dunn E, Weimar MR, Day CL, Baker EN, Lott JS, Sazanov LA, Cook GM. 2014. Structure of the bacterial type II NADH dehydrogenase: a monotopic membrane protein with an essential role in energy generation. Mol Microbiol 91:950–964. doi: 10.1111/mmi.12507. [DOI] [PubMed] [Google Scholar]
- 11.Nakatani Y, Jiao W, Aragão D, Shimaki Y, Petri J, Parker EJ, Cook GM. 2017. Crystal structure of type II NADH:quinone oxidoreductase from Caldalkalibacillus thermarum with an improved resolution of 2.15 angstrom. Acta Crystallogr Sect F Struct Biol Commun 73:541–549. doi: 10.1107/S2053230X17013073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borisov VB, Gennis RB, Hemp J, Verkhovsky MI. 2011. The cytochrome bd respiratory oxygen reductases. Biochim Biophys Acta 1807:1398–1413. doi: 10.1016/j.bbabio.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brooijmans RJ, Poolman B, Schuurman-Wolters GK, de Vos WM, Hugenholtz J. 2007. Generation of a membrane potential by Lactococcus lactis through aerobic electron transport. J Bacteriol 189:5203–5209. doi: 10.1128/JB.00361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cook GM, Greening C, Hards K, Berney M. 2014. Energetics of pathogenic bacteria and opportunities for drug development. Adv Microb Physiol 65:1–62. doi: 10.1016/bs.ampbs.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 15.Weinstein EA, Yano T, Li LS, Avarbock D, Avarbock A, Helm D, McColm AA, Duncan K, Lonsdale JT, Rubin H. 2005. Inhibitors of type II NADH:menaquinone oxidoreductase represent a class of antitubercular drugs. Proc Natl Acad Sci U S A 102:4548–4553. doi: 10.1073/pnas.0500469102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi L, Sohaskey CD, Kana BD, Dawes S, North RJ, Mizrahi V, Gennaro ML. 2005. Changes in energy metabolism of Mycobacterium tuberculosis in mouse lung and under in vitro conditions affecting aerobic respiration. Proc Natl Acad Sci U S A 102:15629–15634. doi: 10.1073/pnas.0507850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin SS, Gross U, Bohne W. 2009. Type II NADH dehydrogenase inhibitor 1-hydroxy-2-dodecyl-4(1H)quinolone leads to collapse of mitochondrial inner-membrane potential and ATP depletion in Toxoplasma gondii. Eukaryot Cell 8:877–887. doi: 10.1128/EC.00381-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biagini GA, Fisher N, Shone AE, Mubaraki MA, Srivastava A, Hill A, Antoine T, Warman AJ, Davies J, Pidathala C, Amewu RK, Leung SC, Sharma R, Gibbons P, Hong DW, Pacorel B, Lawrenson AS, Charoensutthivarakul S, Taylor L, Berger O, Mbekeani A, Stocks PA, Nixon GL, Chadwick J, Hemingway J, Delves MJ, Sinden RE, Zeeman AM, Kocken CH, Berry NG, O’Neill PM, Ward SA. 2012. Generation of quinolone antimalarials targeting the Plasmodium falciparum mitochondrial respiratory chain for the treatment and prophylaxis of malaria. Proc Natl Acad Sci U S A 109:8298–8303. doi: 10.1073/pnas.1205651109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Y, Yu Y, Li X, Li J, Wu Y, Yu J, Ge J, Huang Z, Jiang L, Rao Y, Yang M. 2017. Target elucidation by cocrystal structures of NADH-ubiquinone oxidoreductase of Plasmodium falciparum (PfNDH2) with small molecule to eliminate drug-resistant malaria. J Med Chem 60:1994–2005. doi: 10.1021/acs.jmedchem.6b01733. [DOI] [PubMed] [Google Scholar]
- 20.Glaser P, Rusniok C, Buchrieser C, Chevalier F, Frangeul L, Msadek T, Zouine M, Couvé E, Lalioui L, Poyart C, Trieu-Cuot P, Kunst F. 2002. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol Microbiol 45:1499–1513. doi: 10.1046/j.1365-2958.2002.03126.x. [DOI] [PubMed] [Google Scholar]
- 21.Garrigues C, Loubiere P, Lindley ND, Cocaign-Bousquet M. 1997. Control of the shift from homolactic acid to mixed-acid fermentation in Lactococcus lactis: predominant role of the NADH/NAD+ ratio. J Bacteriol 179:5282–5287. doi: 10.1128/jb.179.17.5282-5287.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higuchi M, Yamamoto Y, Poole LB, Shimada M, Sato Y, Takahashi N, Kamio Y. 1999. Functions of two types of NADH oxidases in energy metabolism and oxidative stress of Streptococcus mutans. J Bacteriol 181:5940–5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamamoto Y, Pargade V, Lamberet G, Gaudu P, Thomas F, Texereau J, Gruss A, Trieu-Cuot P, Poyart C. 2006. The group B Streptococcus NADH oxidase Nox-2 is involved in fatty acid biosynthesis during aerobic growth and contributes to virulence. Mol Microbiol 62:772–785. doi: 10.1111/j.1365-2958.2006.05406.x. [DOI] [PubMed] [Google Scholar]
- 24.Bandeiras TM, Salgueiro C, Kletzin A, Gomes CM, Teixeira M. 2002. Acidianus ambivalens type-II NADH dehydrogenase: genetic characterisation and identification of the flavin moiety as FMN. FEBS Lett 531:273–277. doi: 10.1016/S0014-5793(02)03514-7. [DOI] [PubMed] [Google Scholar]
- 25.Brito JA, Bandeiras TM, Teixeira M, Vonrhein C, Archer M. 2006. Crystallisation and preliminary structure determination of a NADH: quinone oxidoreductase from the extremophile Acidianus ambivalens. Biochim Biophys Acta 1764:842–845. doi: 10.1016/j.bbapap.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 26.Brito JA, Sousa FL, Stelter M, Bandeiras TM, Vonrhein C, Teixeira M, Pereira MM, Archer M. 2009. Structural and functional insights into sulfide:quinone oxidoreductase. Biochemistry 48:5613–5622. doi: 10.1021/bi9003827. [DOI] [PubMed] [Google Scholar]
- 27.Schurig-Briccio LA, Yano T, Rubin H, Gennis RB. 2014. Characterization of the type 2 NADH:menaquinone oxidoreductases from Staphylococcus aureus and the bactericidal action of phenothiazines. Biochim Biophys Acta 1837:954–963. doi: 10.1016/j.bbabio.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghisla S, Massey V. 1986. New flavins for old: artificial flavins as active site probes of flavoproteins. Biochem J 239:1–12. doi: 10.1042/bj2390001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore EG, Cardemil E, Massey V. 1978. Production of a covalent flavin linkage in lipoamide dehydrogenase. Reaction with 8-Cl−FAD. J Biol Chem 253:6413–6422. [PubMed] [Google Scholar]
- 30.Venkatakrishnan P, Lencina AM, Schurig-Briccio LA, Gennis RB. 2013. Alternate pathways for NADH oxidation in Thermus thermophilus using type 2 NADH dehydrogenases. Biol Chem 394:667–676. doi: 10.1515/hsz-2012-0333. [DOI] [PubMed] [Google Scholar]
- 31.Nantapong N, Otofuji A, Migita CT, Adachi O, Toyama H, Matsushita K. 2005. Electron transfer ability from NADH to menaquinone and from NADPH to oxygen of type II NADH dehydrogenase of Corynebacterium glutamicum. Biosci Biotechnol Biochem 69:149–159. doi: 10.1271/bbb.69.149. [DOI] [PubMed] [Google Scholar]
- 32.Pedersen MB, Gaudu P, Lechardeur D, Petit MA, Gruss A. 2012. Aerobic respiration metabolism in lactic acid bacteria and uses in biotechnology. Annu Rev Food Sci Technol 3:37–58. doi: 10.1146/annurev-food-022811-101255. [DOI] [PubMed] [Google Scholar]
- 33.Whidbey C, Harrell MI, Burnside K, Ngo L, Becraft AK, Iyer LM, Aravind L, Hitti J, Adams Waldorf KM, Rajagopal L. 2013. A hemolytic pigment of group B Streptococcus allows bacterial penetration of human placenta. J Exp Med 210:1265–1281. doi: 10.1084/jem.20122753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosa-Fraile M, Dramsi S, Spellerberg B. 2014. Group B streptococcal haemolysin and pigment, a tale of twins. FEMS Microbiol Rev 38:932–946. doi: 10.1111/1574-6976.12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pritzlaff CA, Chang JC, Kuo SP, Tamura GS, Rubens CE, Nizet V. 2001. Genetic basis for the beta-haemolytic/cytolytic activity of group B Streptococcus. Mol Microbiol 39:236–247. doi: 10.1046/j.1365-2958.2001.02211.x. [DOI] [PubMed] [Google Scholar]
- 36.Barnett TC, Cole JN, Rivera-Hernandez T, Henningham A, Paton JC, Nizet V, Walker MJ. 2015. Streptococcal toxins: role in pathogenesis and disease. Cell Microbiol 17:1721–1741. doi: 10.1111/cmi.12531. [DOI] [PubMed] [Google Scholar]
- 37.Liu GY, Doran KS, Lawrence T, Turkson N, Puliti M, Tissi L, Nizet V. 2004. Sword and shield: linked group B streptococcal beta-hemolysin/cytolysin and carotenoid pigment function to subvert host phagocyte defense. Proc Natl Acad Sci U S A 101:14491–14496. doi: 10.1073/pnas.0406143101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McDonnell G, Russell AD. 1999. Antiseptics and disinfectants: activity, action, and resistance. Clin Microbiol Rev 12:147–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bacon JA, Ulrich RG, Davis JP, Thomas EM, Johnson SS, Conder GA, Sangster NC, Rothwell JT, McCracken RO, Lee BH, Clothier MF, Geary TG, Thompson DP. 1998. Comparative in vitro effects of closantel and selected beta-ketoamide anthelmintics on a gastrointestinal nematode and vertebrate liver cells. J Vet Pharmacol Ther 21:190–198. doi: 10.1046/j.1365-2885.1998.00139.x. [DOI] [PubMed] [Google Scholar]
- 40.Fuller JR, Vitko NP, Perkowski EF, Scott E, Khatri D, Spontak JS, Thurlow LR, Richardson AR. 2011. Identification of a lactate-quinone oxidoreductase in Staphylococcus aureus that is essential for virulence. Front Cell Infect Microbiol 1:19. doi: 10.3389/fcimb.2011.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Colussi T, Parsonage D, Boles W, Matsuoka T, Mallett TC, Karplus PA, Claiborne A. 2008. Structure of alpha-glycerophosphate oxidase from Streptococcus sp.: a template for the mitochondrial alpha-glycerophosphate dehydrogenase. Biochemistry 47:965–977. doi: 10.1021/bi701685u. [DOI] [PubMed] [Google Scholar]
- 42.Kerscher S, Dröse S, Zickermann V, Brandt U. 2008. The three families of respiratory NADH dehydrogenases. Results Probl Cell Differ 45:185–222. doi: 10.1007/400_2007_028. [DOI] [PubMed] [Google Scholar]
- 43.Björklöf K, Zickermann V, Finel M. 2000. Purification of the 45 kDa, membrane bound NADH dehydrogenase of Escherichia coli (NDH-2) and analysis of its interaction with ubiquinone analogues. FEBS Lett 467:105–110. doi: 10.1016/S0014-5793(00)01130-3. [DOI] [PubMed] [Google Scholar]
- 44.Yano T, Li LS, Weinstein E, Teh JS, Rubin H. 2006. Steady-state kinetics and inhibitory action of antitubercular phenothiazines on Mycobacterium tuberculosis type-II NADH-menaquinone oxidoreductase (NDH-2). J Biol Chem 281:11456–11463. doi: 10.1074/jbc.M508844200. [DOI] [PubMed] [Google Scholar]
- 45.Liu J, Krulwich TA, Hicks DB. 2008. Purification of two putative type II NADH dehydrogenases with different substrate specificities from alkaliphilic Bacillus pseudofirmus OF4. Biochim Biophys Acta 1777:453–461. doi: 10.1016/j.bbabio.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bergsma J, Van Dongen MB, Konings WN. 1982. Purification and characterization of NADH dehydrogenase from Bacillus subtilis. Eur J Biochem 128:151–157. doi: 10.1111/j.1432-1033.1982.tb06945.x. [DOI] [PubMed] [Google Scholar]
- 47.Rezaïki L, Lamberet G, Derré A, Gruss A, Gaudu P. 2008. Lactococcus lactis produces short-chain quinones that cross-feed group B Streptococcus to activate respiration growth. Mol Microbiol 67:947–957. doi: 10.1111/j.1365-2958.2007.06083.x. [DOI] [PubMed] [Google Scholar]
- 48.Fernandez A, Lechardeur D, Derré-Bobillot A, Couvé E, Gaudu P, Gruss A. 2010. Two coregulated efflux transporters modulate intracellular heme and protoporphyrin IX availability in Streptococcus agalactiae. PLoS Pathog 6:e1000860. doi: 10.1371/journal.ppat.1000860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shabayek S, Spellerberg B. 2017. Acid stress response mechanisms of group B streptococci. Front Cell Infect Microbiol 7:395. doi: 10.3389/fcimb.2017.00395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Derré-Bobillot A, Cortes-Perez NG, Yamamoto Y, Kharrat P, Couvé E, Da Cunha V, Decker P, Boissier MC, Escartin F, Cesselin B, Langella P, Bermúdez-Humarán LG, Gaudu P. 2013. Nuclease A (Gbs0661), an extracellular nuclease of Streptococcus agalactiae, attacks the neutrophil extracellular traps and is needed for full virulence. Mol Microbiol 89:518–531. doi: 10.1111/mmi.12295. [DOI] [PubMed] [Google Scholar]
- 51.Shearer MJ, Newman P. 2014. Recent trends in the metabolism and cell biology of vitamin K with special reference to vitamin K cycling and MK-4 biosynthesis. J Lipid Res 55:345–362. doi: 10.1194/jlr.R045559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brinster S, Lamberet G, Staels B, Trieu-Cuot P, Gruss A, Poyart C. 2009. Type II fatty acid synthesis is not a suitable antibiotic target for Gram-positive pathogens. Nature 458:83–86. doi: 10.1038/nature07772. [DOI] [PubMed] [Google Scholar]
- 53.Ratliff BB, Abdulmahdi W, Pawar R, Wolin MS. 2016. Oxidant mechanisms in renal injury and disease. Antioxid Redox Signal 25:119–146. doi: 10.1089/ars.2016.6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Horsburgh MJ, Clements MO, Crossley H, Ingham E, Foster SJ. 2001. PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect Immun 69:3744–3754. doi: 10.1128/IAI.69.6.3744-3754.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Henningham A, Döhrmann S, Nizet V, Cole JN. 2015. Mechanisms of group A Streptococcus resistance to reactive oxygen species. FEMS Microbiol Rev 39:488–508. doi: 10.1093/femsre/fuu009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.American Society of Health-System Pharmacists, Inc 2009. Zafirlukast. American Society of Health-System Pharmacists, Inc, Bethesda, MD: https://medlineplus.gov/druginfo/meds/a697007.html. Accessed 24 October 2017 [Google Scholar]
- 57.Dekhuijzen PN, Koopmans PP. 2002. Pharmacokinetic profile of zafirlukast. Clin Pharmacokinet 41:105–114. doi: 10.2165/00003088-200241020-00003. [DOI] [PubMed] [Google Scholar]
- 58.Pinault L, Han JS, Kang CM, Franco J, Ronning DR. 2013. Zafirlukast inhibits complexation of Lsr2 with DNA and growth of Mycobacterium tuberculosis. Antimicrob Agents Chemother 57:2134–2140. doi: 10.1128/AAC.02407-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gerits E, Van der Massen I, Vandamme K, De Cremer K, De Brucker K, Thevissen K, Cammue BPA, Beullens S, Fauvart M, Verstraeten N, Michiels J, Roberts M. 2017. In vitro activity of the antiasthmatic drug zafirlukast against the oral pathogens Porphyromonas gingivalis and Streptococcus mutans. FEMS Microbiol Lett 364. doi: 10.1093/femsle/fnx005. [DOI] [PubMed] [Google Scholar]
- 60.Calhoun WJ. 1998. Summary of clinical trials with zafirlukast. Am J Respir Crit Care Med 157:S238–S246. doi: 10.1164/ajrccm.157.6.mar6. [DOI] [PubMed] [Google Scholar]
- 61.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gaillot O, Poyart C, Berche P, Trieu-Cuot P. 1997. Molecular characterization and expression analysis of the superoxide dismutase gene from Streptococcus agalactiae. Gene 204:213–218. doi: 10.1016/S0378-1119(97)00548-9. [DOI] [PubMed] [Google Scholar]
- 63.Biswas I, Gruss A, Ehrlich SD, Maguin E. 1993. High-efficiency gene inactivation and replacement system for Gram-positive bacteria. J Bacteriol 175:3628–3635. doi: 10.1128/jb.175.11.3628-3635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Poyart C, Trieu-Cuot P. 1997. A broad-host-range mobilizable shuttle vector for the construction of transcriptional fusions to beta-galactosidase in Gram-positive bacteria. FEMS Microbiol Lett 156:193–198. doi: 10.1111/j.1574-6968.1997.tb12726.x. [DOI] [PubMed] [Google Scholar]
- 65.Nizet V, Gibson RL, Chi EY, Framson PE, Hulse M, Rubens CE. 1996. Group B streptococcal beta-hemolysin expression is associated with injury of lung epithelial cells. Infect Immun 64:3818–3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cretenet M, Le Gall G, Wegmann U, Even S, Shearman C, Stentz R, Jeanson S. 2014. Early adaptation to oxygen is key to the industrially important traits of Lactococcus lactis ssp. cremoris during milk fermentation. BMC Genomics 15:1054. doi: 10.1186/1471-2164-15-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S. agalactiae amino acid sequence shows high identity to C. thermarum and S. aureus NDH-2s. Identical residues among the three sequences are shaded in red. The figure was made using ESPript (67). Download FIG S1, TIF file, 1.4 MB (1.4MB, tif) .
Copyright © 2018 Lencina et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The Δndh growth phenotype is complemented by a plasmid-carried copy of ndh. (A and B) Final growth (A) and pH (B) at 8 h of culture under respiration permissive (white bars) or nonpermissive (black bars) conditions for WT and Δndh strains carrying the empty vector (p) or ndh-expressing vector (p-ndh). Data are plotted as the average ± SD from three independent experiments. *, P = 0.0001 via two-way ANOVA with Dunnett’s posttest compared to wild-type nonpermissive. Download FIG S2, TIF file, 0.2 MB (211.6KB, tif) .
Copyright © 2018 Lencina et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The Δndh, Δnox2, and ΔcydA mutant strains are not attenuated for virulence in blood, liver, or brain. Mice were infected with 2 × 106 CFU of S. agalactiae, and bacteria were enumerated at 24, 48, and 72 h postinfection in blood (A), liver (B), and brain (C). Bacterial fitness of mutants is shown as a ratio of the log10 number (CFU/gram) of mutant to WT bacteria. *, P < 0.05 for the mutant/WT ratio by Kruskal-Wallis test followed by Dunn’s multiple comparisons. # denotes a median bacterial load (CFU/milliliter) of zero for the group. Download FIG S3, TIF file, 0.3 MB (269.1KB, tif) .
Copyright © 2018 Lencina et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.