Abstract
CRISPR‐Cas systems constitute an adaptive immune system that provides acquired resistance against phages and plasmids in prokaryotes. Upon invasion of foreign nucleic acids, some cells integrate short fragments of foreign DNA as spacers into the CRISPR locus to memorize the invaders and acquire resistance in the subsequent round of infection. This immunization step called adaptation is the least understood part of the CRISPR‐Cas immunity. We have focused here on the adaptation stage of Streptococcus thermophilus DGCC7710 type I‐E CRISPR4‐Cas (St4) system. Cas1 and Cas2 proteins conserved in nearly all CRISPR‐Cas systems are required for spacer acquisition. The St4 CRISPR‐Cas system is unique because the Cas2 protein is fused to an additional DnaQ exonuclease domain. Here, we demonstrate that St4 Cas1 and Cas2‐DnaQ form a multimeric complex, which is capable of integrating DNA duplexes with 3′‐overhangs (protospacers) in vitro. We further show that the DnaQ domain of Cas2 functions as a 3′–5′‐exonuclease that processes 3′‐overhangs of the protospacer to promote integration.
Keywords: adaptation, Cas1, Cas2, protospacer, Streptococcus thermophilus
Subject Categories: Chromatin, Epigenetics, Genomics & Functional Genomics; Microbiology, Virology & Host Pathogen Interaction
Introduction
In bacteria and archaea, clustered regularly interspaced short palindromic repeats (CRISPR) together with CRISPR‐associated (Cas) genes constitute an adaptive immune system that provides an acquired resistance against viruses and plasmids 1. Upon invasion of foreign nucleic acids, CRISPR‐Cas system hijacks short fragments of invasive DNA, called protospacers, and integrates them as new spacers into the CRISPR array to memorize the invaders and immunize the host against an invasive agent 2, 3. Spacers acquired in the CRISPR array during this adaptation stage are subsequently used as templates to generate small interfering CRISPR RNA (crRNA) molecules. Together with Cas proteins, crRNAs assemble into effector complexes that provide immunity by degrading viral DNA in the next round of infection 4, 5, 6, 7. Cas1 and Cas2 proteins which are universal to almost all CRISPR‐Cas systems 8, 9 are involved in the spacer acquisition. Cas1 and Cas2 genes in the CRISPR‐Cas locus typically constitute a single operon except of stand‐alone Cas1 progenitors, called Casposons 10. Acquisition of new spacers by the type I CRISPR‐Cas system occurs by two different modes: naïve and primed 11, 12, 13. During the naïve adaptation, bacteria acquire spacers from a foreign DNA source de novo. In contrast, the primed adaptation relies on a pre‐existing (priming) spacer that enables a biased and enhanced uptake of new spacers. Despite the differences, both modes critically depend on Cas1 and Cas2 proteins.
In Escherichia coli (Ec) CRISPR‐Cas system, Cas1 and Cas2 are the only Cas proteins required for the naïve spacer acquisition in vivo 13. In vitro studies revealed that Cas1 and Cas2 form a stable Cas14:Cas22 complex 14. This complex binds a 33 bp oligodeoxynucleotide comprised of 23 bp duplex region flanked by 5 nt protruding ends at 3′ terminus and integrates it into the CRISPR array in the supercoiled plasmid DNA in vitro 15, 16. The integration reaction catalyzed by the Cas1:Cas2 complex proceeds through a direct nucleophilic attack of the 3′‐OH of the protospacer on the phosphodiester bond in the target DNA. In that respect, the mechanism of spacer integration into the CRISPR array is reminiscent of retroviral integrases and transposases 15, 16, 17, 18, 19. Moreover, Ec Cas1:Cas2 complex of the type I‐E CRISPR‐Cas system is capable of spacer integration not only in the supercoiled plasmid but also into the CRISPR array of linear DNA. However, in the latter case, it requires an auxiliary integration host factor (IHF) protein, which binds the leader sequence in the vicinity of the CRISPR array and creates a local DNA curvature to facilitate the Cas1:Cas2 recognition of the leader‐repeat junction and promote integration 18. In contrast to the type I‐E system, Cas1:Cas2 complex of the type II CRISPR‐Cas system is capable of integrating protospacers into linear substrates in vitro in the absence of either IHF or other auxiliary proteins 19, 20, 21.
Although the spacer integration step is relatively well understood, the spacer capture step during which protospacers are generated and processed for integration remains unclear. It has been reported that in the case of naïve adaptation in E. coli, new spacer generation requires the RecBCD recombination complex 22, while in the primed adaptation pathway, auxiliary components like Cascade (CRISPR‐associated complex for antiviral defense) and Cas3 nuclease are necessary alongside Cas1 and Cas2 11, 12, 23. Moreover, in some CRISPR‐Cas systems additional domains such as Cas4 or DnaQ 24, 25, 26 fused to Cas1 or Cas2 are presumably involved in the naïve adaptation; however, their role remains to be established.
In this study, we have focused on the spacer integration step in the Streptococcus thermophilus DGCC7710 type I‐E CRISPR4‐Cas (St4) system 27. Unlike a single‐domain Cas2 in an Ec type I‐E CRISPR‐Cas system, the St4 Cas2 protein is fused to the DnaQ domain, homologous to the ε subunit of the Ec replicative DNA polymerase III. The ε subunit is a 3′–5′ exonuclease, which is responsible for proofreading activity of the DNA polymerase 28, 29. Although the Cas2‐DnaQ fusion has been identified in several type I‐E CRISPR‐Cas systems 25, the exact role of the DnaQ domain in the spacer adaptation remains unknown.
Here, we show that Cas1 and Cas2‐DnaQ proteins of S. thermophilus CRISPR4‐Cas system form a stable complex. The complex is capable of integrating oligonucleotide duplexes with 3′ protruding ends into the supercoiled plasmid; however, the integration efficiency depends on the length of the 3′‐termini flanking the duplex region. We further demonstrate that the DnaQ domain of the Cas2 protein possesses the 3′–5′ exonuclease activity, which is required for processing of the 3′‐ends of spacer precursors to produce optimal substrates for integration.
Results and Discussion
St4 Cas1:Cas2‐DnaQ complex
Cas1 and Cas2 proteins of Ec type I CRISPR‐Cas system form a multimeric Cas14:Cas22 complex 14 and are the only Cas proteins absolutely required for spacer acquisition in vivo 13 and in vitro 16. To probe whether Cas1 and Cas2‐DnaQ from the orthologous St4 CRISPR‐Cas system (Fig 1A) also form a complex, we first engineered StrepII‐tagged Cas1 and 6xHis‐tagged Cas2‐DnaQ variants. Next, we co‐expressed respective proteins in the recombinant E. coli host and isolated proteins using either Strep‐Tactin or Ni2+‐NTA columns and analyzed proteins by SDS–PAGE. We found that Strep‐Cas1 and Cas2‐DnaQ‐His proteins co‐eluted both from Ni2+‐NTA and Strep‐Tactin columns. Subsequent size‐exclusion chromatography step revealed that both proteins eluted as a single peak (Fig 1B) corresponding to molecular mass of 250 ± 30 kDa, which correlates well with theoretical mass of 220 kDa of the Cas14:Cas2‐DnaQ2 complex.
Figure 1. Characterization of the Cas1 and Cas2‐DnaQ proteins of the Streptococcus thermophilus DGCC7710 CRISPR4‐Cas system.
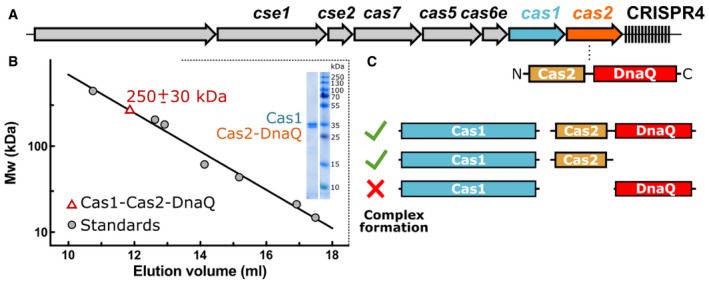
- Schematic organization of the CRISPR4‐Cas locus. The DnaQ domain is fused to the C‐terminus of the Cas2 protein.
- Molecular mass of the St4 Cas1:Cas2‐DnaQ complex obtained by the size‐exclusion chromatography. The estimated molecular mass of the complex is 250 ± 30 kDa, which correlates well with theoretical mass of 220 kDa of the complex. Coomassie blue‐stained sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) of the isolated Cas1:Cas2‐DnaQ complex is shown as an inset.
- Mapping of Cas2 determinants required for the Cas1:Cas2‐DnaQ complex formation. Deletion analysis indicates that Cas1 domain and the N‐Cas2 domain (1–124 aa), but not the DnaQ domain (125–301 aa), are required for complex assembly.
Next, we analyzed whether both domains of Cas2‐DnaQ (N‐Cas2 and C‐DnaQ domains, respectively) are necessary for complex formation. To this end, we engineered Cas2‐DnaQ variants consisting of either N‐Cas2 (1–124 aa) or C‐DnaQ (125–301 aa) domains, co‐expressed individual truncated variants with the full‐length Strep‐Cas1, and subjected to affinity chromatography on the Strep‐Tactin column. The SDS–PAGE analysis revealed that N‐Cas2, but not C‐DnaQ co‐eluted with Strep‐Cas1 (Figs 1C and EV1) indicating that the N‐terminal domain of Cas2 is responsible for the Cas1:Cas2‐DnaQ complex formation.
Figure EV1. Cas1:Cas2‐DnaQ composition analysis.
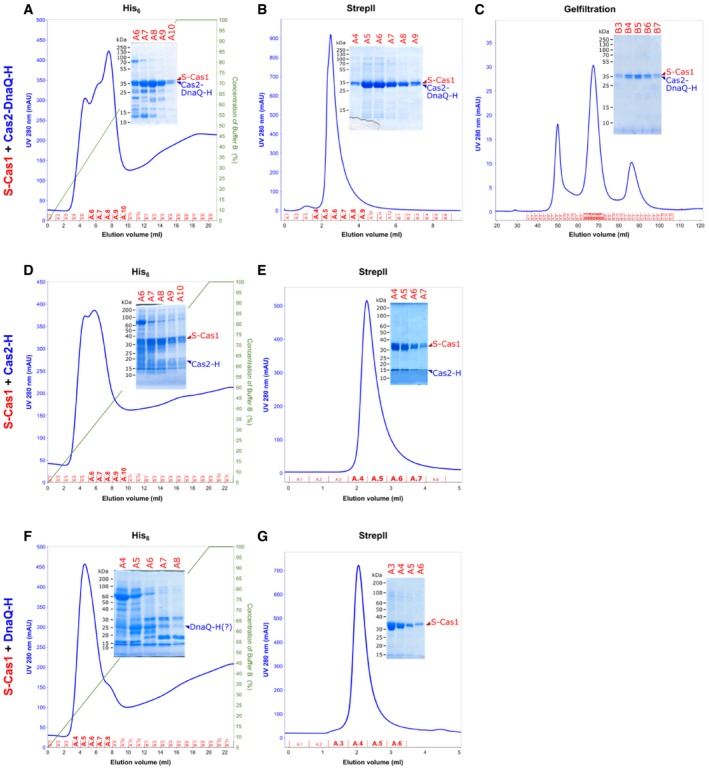
- S‐Cas1 and full‐length Cas2‐DnaQ‐H isolation on His6‐tag affinity column.
- S‐Cas1 and full‐length Cas2‐DnaQ‐H isolation on StrepII affinity column.
- S‐Cas1 and Cas2‐DnaQ‐H proteins elute as a single peak from size‐exclusion column.
- S‐Cas1 and Cas2‐H domain isolation on His6‐tag affinity column.
- S‐Cas1 and Cas2‐H isolation on StrepII affinity column.
- When S‐Cas1 was co‐expressed with DnaQ‐H, only DnaQ‐H was isolated on His6‐tag affinity column.
- When S‐Cas1 was co‐expressed with DnaQ‐H, only S‐Cas1 was isolated on StrepII affinity column.
Spacer integration by the St4 Cas1:Cas2‐DnaQ complex
Escherichia coli Cas1:Cas2 complex is capable of integrating 33 nt oligoduplex into supercoiled plasmid in vitro resulting in plasmid relaxation 16. Subsequent studies revealed that 23 bp DNA oligoduplex containing 5 nt 3′‐overhangs or splayed ends 15, 30 is an optimal substrate for integration. We probed St4 Cas1:Cas2‐DnaQ ability to integrate similar oligoduplexes into a plasmid bearing CRISPR region (Fig 2A and B). The integration activity was undetectable using the 33 bp duplex with blunt ends as the protospacer. However, we found that the St4 Cas1:Cas2‐DnaQ complex integrates oligoduplexes, which have 3′‐overhangs (Fig 2B) with the integration efficiency depending on the overhang length. Further, only Cas1 catalytic activity was necessary for complex to integrate oligoduplexes (Fig 2C).
Figure 2. Spacer integration by the St4 Cas1:Cas2‐DnaQ complex in vitro .
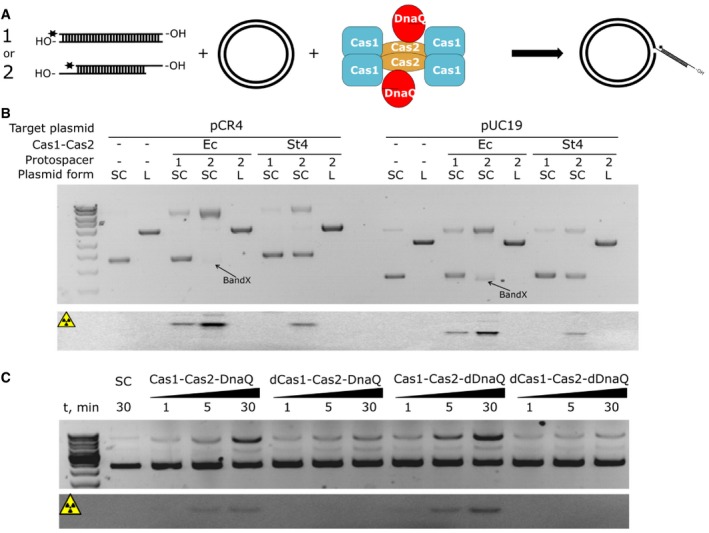
- Schematic representation of the integration assay. Partial integration of oligoduplex into a supercoiled plasmid should result in the plasmid relaxation.
- Cas1:Cas2‐DnaQ integrates the oligoduplex into supercoiled plasmid. To monitor spacer integration, blunt‐ended 33 bp oligoduplex (1) or 23 bp duplex with 5 nt 3′‐overhangs (2) was 5′‐33P labeled and incubated with supercoiled (SC) or linear (L) plasmid containing CRISPR region (pCR4) or control plasmid (pUC19) and Ec Cas1:Cas2 or the St4 Cas1:Cas2‐DnaQ complex. Reaction products were analyzed on the EtBr‐stained agarose gel. Both Ec and St4 Cas1:Cas2 complexes integrate protospacers into plasmids without CRISPR region, but are unable to integrate into linear plasmids. Furthermore, the St4 complex is unable to integrate blunt‐ended protospacers. The product of Ec Cas1:Cas2 disintegration reaction (reverse reaction catalyzed by Ec Cas1:Cas2) is indicated as band X. Lower panel shows reaction products of radioactively labeled protospacer partial integration visualized by phosphorimager. Radioactive bands correspond to relaxed plasmid form and thus integration events.
- Active site of DnaQ domain is dispensable for integration step. Protospacer (2) was incubated with SC pCR4 in the presence of WT Cas1:Cas2‐DnaQ, the Cas1 active site (D227A) mutant (dCas1), the DnaQ active site (D135A) mutant (dDnaQ), or the double mutant. Only when the Cas1 active site was intact, integration reactions were observed. Reaction products were visualized by the EtBr‐stained agarose gel (upper panel) or phosphorimager (bottom panel).
Source data are available online for this figure.
Next, using radioactively labeled oligoduplexes we showed that the supercoiled form of the plasmid was relaxed due to integration events. St4 Cas1:Cas2‐DnaQ complex, similar to Ec Cas1:Cas2, integrated oligoduplex into a plasmid bearing CRISPR region or pUC19 plasmid lacking CRISPR region with similar efficiency. In fact, plasmid supercoiling rather than the CRISPR array was a major requirement for integration events to occur (Fig 2B). However, differently from Ec Cas1:Cas2, the integration by the Sth4 Cas1:Cas2‐DnaQ complex does not result in “band X” (Fig 2B), which is likely the outcome of protospacer disintegration and plasmid re‐ligation resulting in different topoisomers 16. This finding suggests that St4 Cas1:Cas2‐DnaQ complex does not catalyze disintegration of the reaction intermediate under experimental conditions tested. St4 Cas1:Cas2‐DnaQ catalyzed oligoduplex integration reactions in the presence of various divalent metal ions with magnesium ions being optimal cofactors for the integration reaction (Fig EV2A).
Figure EV2. Activity dependence on divalent metal ions and sequence features.
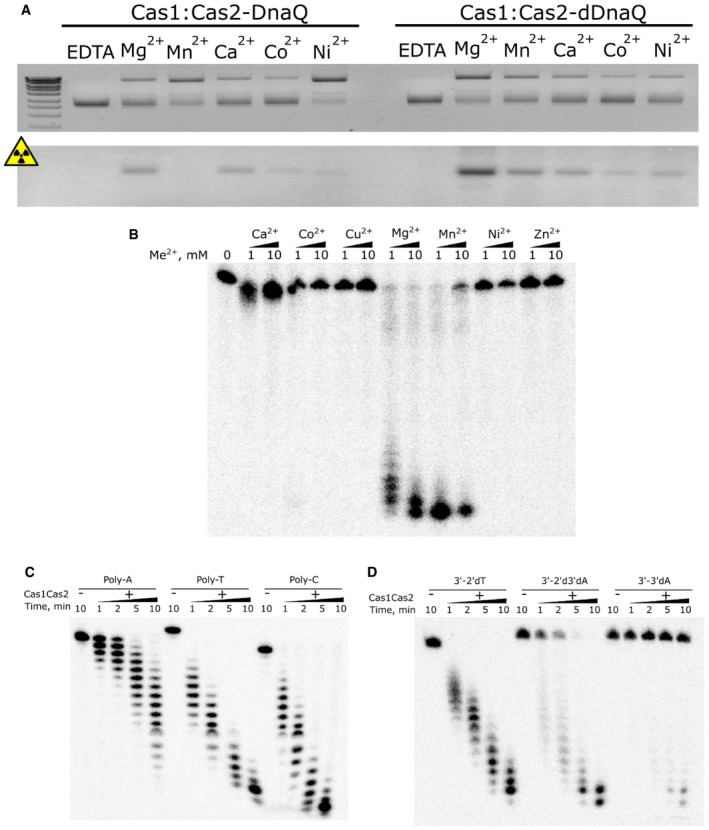
- Integration reactions were performed in the presence of different divalent metal co‐factor using Cas1:Cas2‐DnaQ complex and 23 bp DNA duplex with 5 nt 3′‐overhangs as protospacer. Mg2+, Mn2+, and Ca2+ were determined to be the best cofactors for integration reaction. Mn2+ activates DnaQ domain so strongly that integration becomes undetectable, due to degradation of spacers. Reactions were incubated for 30 min at 42°C.
- Exonuclease reactions were performed in the presence of different divalent metal co‐factor using Cas1:Cas2‐DnaQ complex and 72 nt single‐stranded DNA oligonucleotide. DNA degradation was most efficient with Mg2+ and Mn2+ ions.
- Exonuclease rates of Cas1:Cas2‐DnaQ complex were probed using polyA, polyT, and polyC single‐stranded DNA oligonucleotides.
- Exonuclease rates of Cas1:Cas2‐DnaQ complex were probed using polyA oligonucleotide (2′dT), which was blocked at its 3′‐end with 2′, 3′‐dideoxyadenosine (2′d3′dA) or 3′‐deoxyadenosine (3′dA) using terminal deoxynucleotidyl transferase.
DnaQ domain within the Cas1:Cas2‐DnaQ complex is a 3′–5′ exonuclease
Proteins of DnaQ family exhibit 3′–5′ DNA exonuclease activity 28, 29. To probe whether the DnaQ domain within St4 Cas1:Cas2‐DnaQ complex shows the exonuclease activity, 5′‐end radioactively labeled single‐stranded (ss) and double‐stranded (ds) DNA oligonucleotides were incubated with the complex and reaction products were analyzed by denaturing gel electrophoresis. Both ss and ds oligonucleotides showed a ladder‐like degradation pattern and the size of the degradation products decreased with time consistent with 3′–5′ DNA exonuclease activity. Moreover, D135A mutation at the active site of the DnaQ domain, predicted by multiple sequence alignments (Fig EV3B), abolished DNA degradation (Fig 3A).
Figure EV3. Multiple sequence alignments used in prediction of active site residues of Cas1 and DnaQ domains.
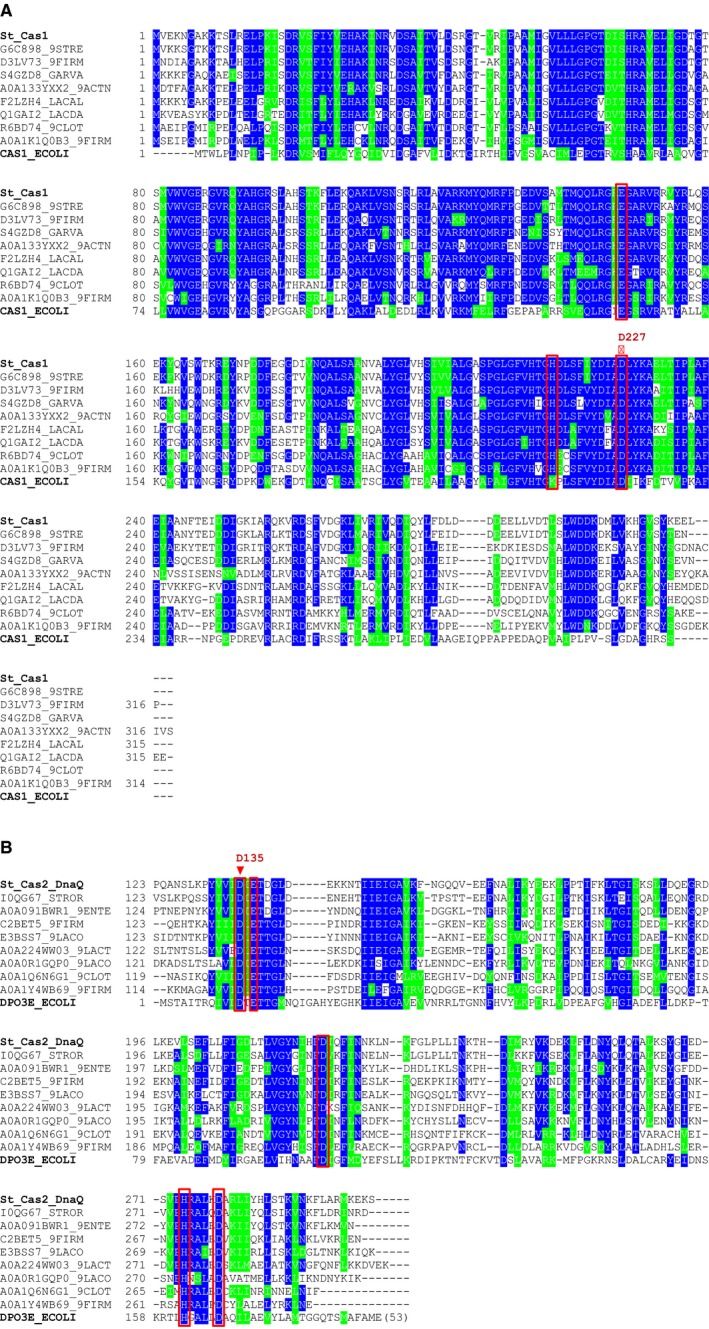
- Alignment of Streptococcus thermophilus Cas1 (St_Cas1) and its homologs including well‐characterized Escherichia coli Cas1 (CAS1_ECOLI). Positions corresponding to the active site residues of E. coli Cas1 are marked with red rectangles. The point mutation in S. thermophilus Cas1 is indicated with the red triangle. St_Cas1 homologs are represented by their Uniprot entry names.
- Alignment of S. thermophilus Cas2‐DnaQ (St_Cas2_DnaQ) and its homologs with E. coli PolIII ε‐subunit (DPO3E_ECOLI). Positions corresponding to the active site residues of ε‐subunit are marked with red rectangles. The amino acid that was mutated in S. thermophilus Cas2‐DnaQ is labeled with the red triangle. Sequences, except for St_Cas2_DnaQ, are represented by their Uniprot entry names.
Figure 3. Nuclease activity of DnaQ domain.
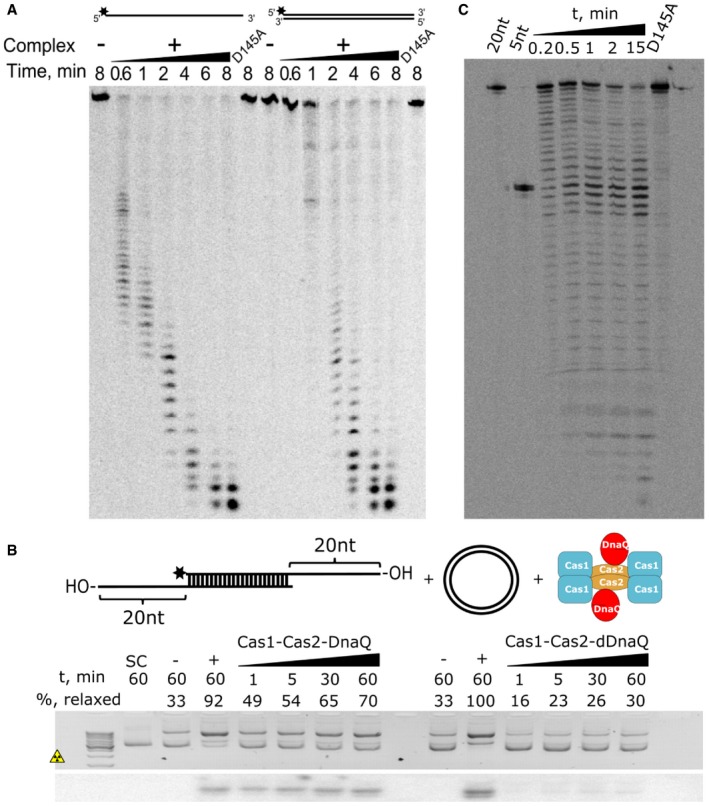
- The St4 Cas1:Cas2‐DnaQ complex shows the 3′‐exonuclease activity on both single‐stranded and double‐stranded DNA substrates. The D135A mutation in the DnaQ active site abolishes exonuclease activity to yield catalytically dead DnaQ (dDnaQ).
- The integration assay using oligoduplexes with 20 nt 3′‐overhangs. Schematic representation of the reaction setup is depicted in the upper panel. Reaction products are visualized by the EtBr‐stained agarose gel (middle panel) or phosphorimager (bottom panel). Reactions without any oligoduplex added were used as negative controls (−), whereas the oligoduplex with 5 nt 3′‐overhangs was used as a positive control (+). The percentage (%) of relaxed plasmid is indicated above each lane.
- DnaQ within Cas1:Cas2‐DnaQ complex trims 3′‐protruding ends of the oligoduplex. Oligoduplex with 20 nt 3′‐overhangs was incubated with WT Cas1:Cas2‐DnaQ or Cas1:Cas2‐dDnaQ prior to integration reactions. Labeled protospacers with 20 nt or 5 nt 3′‐overhangs (20 and 5 nt, respectively) were used as markers.
Source data are available online for this figure.
Next, we aimed to establish requirements for the exonuclease activity of the St4 Cas1:Cas2‐DnaQ complex. Firstly, we screened divalent metal ions as DnaQ cofactors and found that magnesium and manganese ions were the best cofactors for the DnaQ‐dependent oligonucleotide hydrolysis (Figs 3A and EV2B). Secondly, we evaluated DNA sequence requirements for the exonucleolytic activity comparing degradation rates of polyA, polyT, and polyC oligonucleotides. PolyT and polyC oligonucleotides were degraded faster than polyA (Fig EV2C), suggesting that pyrimidines rather than adenine are preferable at the 3′‐end of the DNA. Lastly, we incorporated ddA or 3′‐dA at the 3′‐end of the oligonucleotide and monitored the cleavage rate. In the absence of both 3′‐ and 2′‐OH groups of the ribose, oligonucleotide hydrolysis was markedly reduced, while cleavage was abolished in the presence of the 2′‐OH group (Fig EV2D). Taken together, these data show that DnaQ domain of the St4 Cas1:Cas2‐DnaQ complex exonucleolytically degrades DNA in the 3′−5′ direction. This process is the most efficient when a pyrimidine deoxynucleotide is present at the 3′‐end.
DnaQ domain trims 3′‐ends of protospacers
In the Cas1:Cas2 complex, the Cas1 active site catalyzes 3′‐OH nucleophilic attack of the protospacer on the phosphodiester bond of the target sequence 13, 16. To probe requirements for integration reaction catalyzed by the St4 Cas1:Cas2‐DnaQ complex, we engineered Cas1 (D227A), DnaQ (D135A), or double Cas1 (D227A):Cas2‐DnaQ (D135A) active site mutants guided by multiple sequence alignment (Fig EV3). Mutation in the DnaQ active site had no effect on the integration efficiency suggesting that the DnaQ domain is not necessary for this process. However, we could not detect any integration in the case of Cas1 active site D227A mutant as well as double Cas1 (D227A) and DnaQ (D135A) mutant (Fig 2C), supporting previously published data that Cas1 is responsible for the integrase activity. Interestingly, the St4 Cas1:Cas2‐DnaQ complex having the impaired DnaQ domain of Cas2 integrated duplexes with 5 or 6 nt 3′‐protruding ends with higher efficiency than duplexes with 7 or 8 nt overhangs (Fig EV4B) in contrast to the WT Cas1:Cas2‐DnaQ complex (Fig EV4A).
Figure EV4. Integration efficiencies of protospacers with different 3′‐overhang lengths.
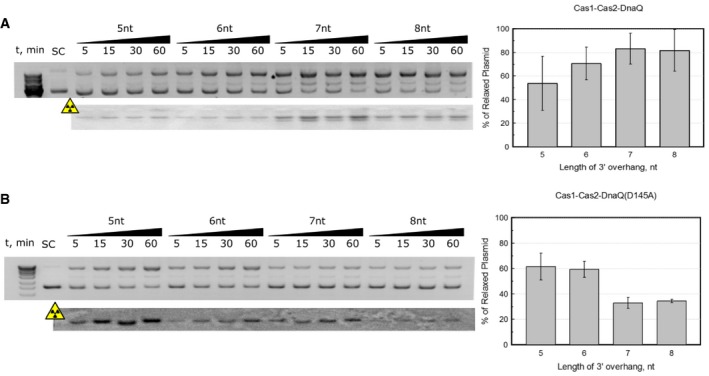
- Integration reactions performed using WT Cas1:Cas2‐DnaQ complex; N = 3; error bars represent standard error of the mean value. This complex integrates duplexes with 7 or 8 nt overhangs better than purported optimal protospacer bearing 5 nt overhangs.
- Integration reactions performed using Cas1:Cas2‐DnaQ (D135A) mutant (inactive DnaQ); N = 3; error bars represent standard error of the mean value. Conversely, DnaQ mutant integrates optimal protospacer noticeably better than those bearing 7 or 8 nt overhangs.
The exonuclease activity of the DnaQ domain (Fig EV2B) and different substrate preferences of WT and DnaQ‐mutant complexes (Fig EV4) suggest that DnaQ domain may be required to process 3′‐overhangs that are too long for efficient integration. Therefore, next we monitored integration efficiencies of the wild‐type complex and the DnaQ‐mutant complex using the 23 bp oligoduplex, which had 20 nt overhangs at both 3′‐ends. We were barely able to detect integration events using the DnaQ active site mutant, whereas the WT Cas1:Cas2‐DnaQ complex efficiently integrated oligoduplexes (Fig 3B). Once we analyzed Cas1:Cas2‐DnaQ exonuclease reaction products of the oligoduplex containing 20 nt overhangs, we found that the WT Cas1:Cas2‐DnaQ complex truncated 20 nt 3′‐overhangs to generate spacers with predominantly 2–8 nt long 3′‐overhangs (Fig 3C). According to the crystal structure of E. coli Cas1:Cas2 15, 30, DNA harboring such 3′ overhangs is the optimal substrate for integration.
To determine whether integrations occurred in the CRISPR array and to establish the length of 3′‐overhangs of the duplexes that were actually integrated, we developed a PCR‐based assay (Fig EV5A and D). We performed PCR using primers binding to the integration oligoduplex and (i) the leader sequence of the CRISPR region or (ii) 2nd spacer of the CRISPR region. Based on the length of PCR products, we concluded that integration events occurred in the vicinity of the first and the second repeats of the CRISPR array (Fig EV5B and E). Next, the PCR products were gel‐purified, cloned into pJET1.2 plasmid, and individual clones were sequenced. Sequencing results revealed that 3′‐overhangs of integrated protospacers ranged from 2 to 7 nt, with 5 nt 3′‐overhang being the most common (Fig EV5C and F). Overall, these data show that the DnaQ domain fused to Cas2 processes the 3′‐ends of the protospacers to produce overhangs optimal for integration. However, there are known instances of nuclease activities of Cas proteins that are not associated with the CRISPR‐Cas function 31, 32, 33. Therefore, further in vivo studies should ascertain whether our findings are applicable for the CRISPR‐Cas adaptation in natural hosts.
Figure EV5. PCR‐based assay for detection of integration into CRISPR locus.
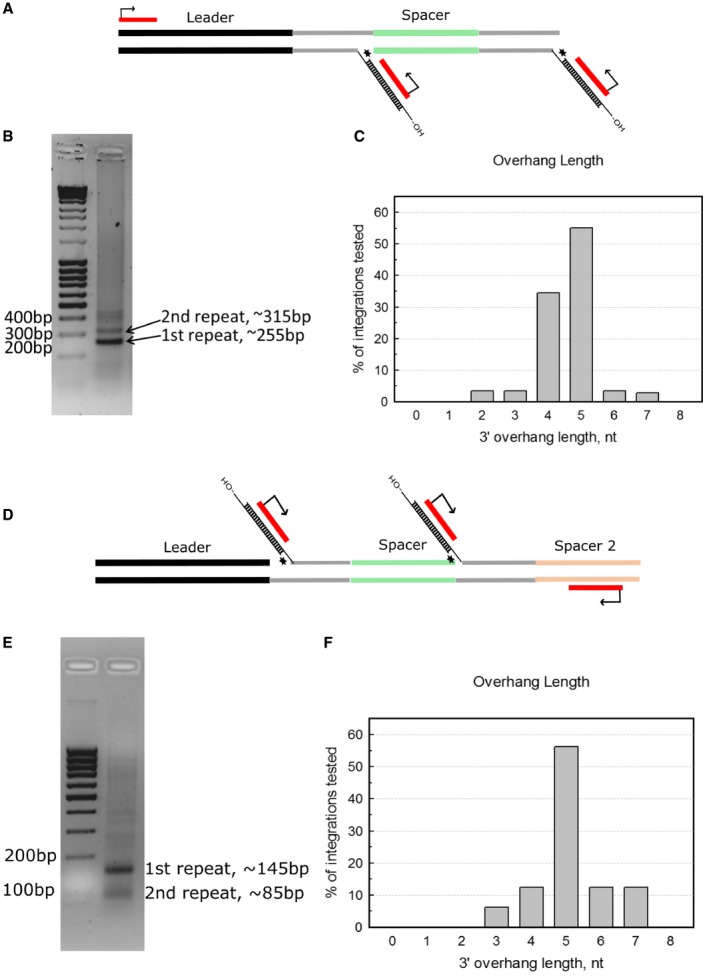
- Integration products were amplified by PCR using primers binding to the leader sequence and protospacers that were integrated to the CRISPR region.
- PCR products are analyzed on 1.5% agarose gel stained with EtBr. 1st repeat and 2nd repeat denoted bands correspond to integrations after the first or second repeats (as drawn in panel A). Lowest band corresponding to integration event in the vicinity of the first repeat was cut out and purified and subsequently cloned into pJET1.2.
- Length distribution of integrated protospacer 3′‐overhangs as determined from Sanger sequencing of separate pJET1.2 clones, n = 29.
- Integration products were amplified by PCR using primers binding to the 2nd spacer and protospacers that were integrated into the CRISPR region.
- PCR products are analyzed on 1.5% agarose gel stained with EtBr. 1st repeat and 2nd repeat denoted bands correspond to integrations after the first or second repeats (as drawn in panel A). Upper band, corresponding to integration event in the vicinity of the first repeat was cut out and purified and subsequently cloned into pJET1.2.
- Length distribution of integrated protospacer 3′‐overhangs as determined from Sanger sequencing of separate pJET1.2 clones, n = 16.
Source data are available online for this figure.
Auxiliary proteins and domains associated with the integration core
Entry of foreign DNA into the cell initiates adaptation stage of CRISPR‐Cas systems, which in type I systems can proceed in two different pathways: (i) naïve, when foreign DNA is novel for CRISPR‐Cas system or (ii) primed, when the CRISPR‐Cas system has already encountered the intruder 2, 3. In the naïve adaptation pathway, protospacers are generated de novo by RecBCD or analogous complexes 22. In the primed adaptation pathway, Cascade recognizes foreign DNA with fully or partially matching protospacer, which primes generation of new protospacers, most likely, by Cas3 degradation 23, 27, 34. Protospacers generated in both of these two pathways are integrated into CRISPR locus by the Cas1:Cas2 core complex (Fig 4).
Figure 4. Schematic representation of the adaptation step in the St4‐like CRISPR‐Cas systems.
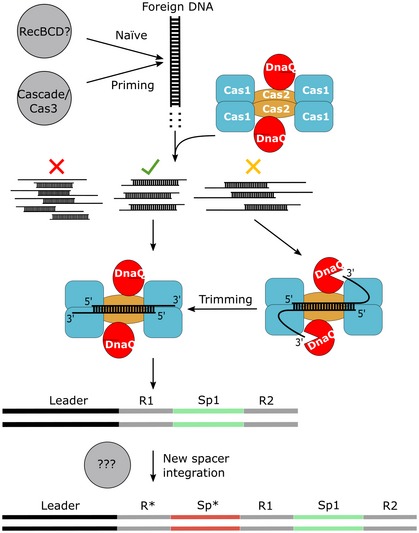
Foreign DNA may be directed to one of the two pathways of CRISPR‐Cas adaptation: (i) naïve, if it is a novel infection and (ii) primed, if it is a repetitive infection. In the naïve pathway, RecBCD or other cellular machinery degrades foreign DNA generating protospacers or its intermediates, while the Cas3/Cascade effector complex presumably generates protospacers in the priming pathway. DNA fragments serving as precursors for protospacers may contain different DNA ends and broadly fall into three categories: (i) optimal protospacers (green check mark), (ii) blunt‐ended or 5′‐overhanged DNA fragments are unproductive protospacer intermediates (red cross), and (iii) DNA fragments with extended 3′‐overhangs are subjected for DnaQ processing to generate optimal spacers (yellow cross). The DnaQ domain trims 3′‐overhangs producing an optimal protospacer, which is bound by the core Cas1:Cas2 complex. In the final stage of the mechanism, the Cas1:Cas2‐DnaQ complex bound to a protospacer uses its integrase activity to insert the protospacer into the CRISPR region at a leader proximal end. Gaps formed during integration reaction are filled in by cellular machinery resulting in the duplicated repeat and a new spacer (*).
Cas1:Cas2 complex is essential, but not always sufficient to achieve spacer integration in vivo. Often, auxiliary proteins or domains are required. For example, in a number of type III CRISPR‐Cas systems Cas1‐reverse transcriptase (Cas1‐RT) fusion enables acquisition of RNA spacers in a reverse transcription‐dependent manner 35. In the type I‐F CRISPR‐Cas systems, Cas2 protein is fused to the Cas3 protein, which is responsible for foreign DNA degradation in type I systems 8, 36. It has been shown that the Cas2‐Cas3 fusion protein forms a complex with Cas1 37 and DNA fragments generated by Cas3 serve as substrates for Cas1:Cas2‐catalyzed integration reaction 23, 34. Furthermore, spacer acquisition in the type I‐F systems via priming pathway occurred to be more than 500‐fold efficient than de novo naïve adaptation 38. It is likely that Cas2‐Cas3 fusion promotes the interaction between interference and integration machineries by placing them in proximity of one another 38, 39, 40.
As determined from crystal structures of Ec Cas1:Cas2 with DNA, an optimal substrate for the integration reaction is the 23 bp DNA oligoduplex containing 5 nt long 3′‐overhangs 15, 30. However, how DNA fragments are generated in both adaptation pathways remains unknown, but we conjecture that those fragments may contain various lengths of duplex region and single‐stranded protrusions; therefore, only a fraction of DNA fragments may meet the requirements for optimal protospacer, which could be integrated by the Cas1:Cas2 core complex. Such restrictions suggest that additional oligonucleotide processing might be required to produce optimal protospacers for integration. In type I‐B system of Haloarcula hispanica, the Cas4 protein may play such a role. It has been shown that Cas4 interacts with Cas1, Cas2, and Csa1 in vitro to form a heterocomplex 41 and is necessary for in vivo spacer acquisition in the CRISPR array of type I‐B system of H. hispanica 24. The Cas4 protein contains a RecB‐like nuclease motif and a [Fe‐S] cluster, which is coordinated by four conserved Cys residues. It possesses a 5′–3′ exonuclease activity 42 and could thus be responsible for the generation of protospacers with 3′‐overhangs from the intermediates with blunt or 5′‐protruding ends. Recently it was shown that Cas4 from type I‐A system from Sulfolobus solfataricus processes protospacers bound by Cas1:Cas2 complexes in a PAM‐dependent manner 43. Authors of that work and several subsequent studies show that Cas4 works as an endonuclease and cleaves 3′‐overhangs at the PAM 43, 44, 45, thus generating overhangs similar to those generated by DnaQ domain.
In contrast to recent findings about Cas4 working as an endonuclease, here we have shown that DnaQ domain fused to Cas2 protein is an exonuclease that degrades DNA from the 3′‐end. Furthermore, the exonuclease activity is prerequisite for efficient integration of DNA substrates with 3′‐protrusions longer than 5 nt. In light of these findings, we propose (Fig 4) that DnaQ domain processes extended 3′‐overhangs of oligonucleotide precursors to produce suitable protospacers for integration.
Materials and Methods
Cloning and mutagenesis of Cas1:Cas2‐DnaQ
Sequences of full‐length Cas1 (WP_024704118.1) and Cas2‐DnaQ (WP_024704117.1) as well as Cas2 (1–124 fragment of full‐length protein) and DnaQ (125–301 fragment of full‐length protein) domains were PCR‐amplified from S. thermophilus DCGG7710 strain and cloned into pET‐SH (modified version of pET‐Duet1 vector (Novagen) where N‐terminal His6‐tag and C‐terminal S‐tag sequences are replaced with StrepII‐tag and His6‐tag, respectively) vector. Cas1 gene was inserted into BamHI and NotI sites, thus fusing N‐terminus of the Cas1 protein with StrepII‐tag, while Cas2‐DnaQ, Cas2, and DnaQ domains were His6‐tagged at C‐terminus by inserted their genes into AatII and XhoI sites of the pET‐SH vector. Residues D227 and D135 of respective active sites of Cas1 and DnaQ were determined by multiple sequence alignments (Fig EV3). Cas1 mutant D227A and Cas2‐DnaQ mutant D135A were obtained by the Quick Change Mutagenesis (QCM) Protocol 46. Sequences of the genes were verified by DNA sequencing.
Cas1:Cas2‐DnaQ complex purification
Expression vector constructs were used to transform E. coli BL21(DE3) strain. Transformed bacteria were grown at 37°C in LB medium until 0.7 OD (600 nm) was reached. Then, medium was cooled to 16°C temperature and proteins were expressed for 17 h by adding 1 mM IPTG. Harvested cells were disrupted by sonication in buffer A (20 mM Tris–HCl (pH 8.0 at 25°C), 500 mM NaCl, 2 mM PMSF), and cell debris was removed by centrifugation. The supernatant containing Cas1 and Cas2‐DnaQ or its domains was loaded onto the Ni2+‐charged HiTrap chelating HP column (GE Healthcare) and eluted with a linear gradient of increasing imidazole in buffer A. The fractions containing Cas1:Cas2‐DnaQ complex were pooled and subsequently loaded onto StrepTrap HP column (GE Healthcare). Proteins were eluted with 2.5 mM of d‐Desthiobiotin in buffer A. Finally, the complex was run through the HiLoad 16/600 Superdex 200 pg column (GE Healthcare) to isolate pure complex.
Analytical gel‐filtration
Gel‐filtration of Cas1:Cas2‐DnaQ complex was carried out at room temperature using Superdex 200 10/300 GL column (GE Healthcare) pre‐equilibrated with a buffer (20 mM Tris–HCl (pH 8.0 at 25°C), 500 mM NaCl). A calibration curve was generated by measuring the elution volumes of a series of standard proteins of known molecular mass (Bio‐Rad). The molecular mass of Cas1:Cas2‐DnaQ complex was calculated by interpolating its elution volume onto the calibration curve.
Protospacer integration assay
Integration reactions were carried out as described in 16. Briefly, 75 nM of Cas1:Cas2‐DnaQ was incubated with ~ 5 nM pUC19‐CR4 or pUC19 plasmid and ~ 500 nM radioactively labeled oligoduplex with various length 3′‐overhangs in an integration buffer (20 mM HEPES‐NaOH, pH 7.5, 25 mM KCl, 10 mM MgCl2, 1 mM DTT, 10% v/v DMSO). Reaction was stopped by the addition of 2% SDS and 10 U of proteinase K (Thermo Fisher Scientific) and incubating for 20 min at 55°C, as per manufacturer's recommendation. Reaction products were analyzed on 1% agarose gel stained with EtBr, which were ran for 1.5 h. Gels were then dried and exposed to phosphorescent screens (FujiFilm). Radioactive bands were visualized on FujiFilm FLA‐5100 scanner.
Nuclease assay
To evaluate the DnaQ nuclease activity, 2 nM of single‐stranded and double‐stranded radioactively labeled oligonucleotides (Table EV1) was used as substrates in the nuclease assay. St4 Cas1:Cas2‐DnaQ nuclease reaction was performed at 37°C for indicated time interval in Nuclease buffer: 10 mM Tris–HCl (pH 8), 10 mM MgCl2, 100 mM NaCl, 10% glycerol, 0.1 mg/ml BSA. Reactions were initiated by addition of complex. The reactions were stopped by addition of 3× stop solution (67.5 mM EDTA, 27% (v/v) glycerol, 0.3% (w/v) SDS).
To analyze the protospacer fate prior to the integration, 25 nM of radioactively labeled protospacer containing 20 nt 3′‐overhangs (Table EV1) was incubated at 42°C with 75 nM of Cas1:Cas2‐DnaQ complex in the integration buffer. Reactions were initiated by the addition of the complex and stopped by adding phenol:chloroform:isoamyl alcohol mixture. Samples were centrifuged and aqueous fraction collected. It was mixed with loading dye containing formamide. Samples were boiled and then cooled to room temperature. Samples were fractionated using denaturing PAGE. Gels were dried and exposed to phosphorescent screens (FujiFilm). Radioactive bands were visualized on FujiFilm FLA‐5100 scanner.
PCR‐based assay for detection of integration into CRISPR locus
Samples from integration reactions were used as PCR templates. PCR was performed using primers binding to the leader sequence of the CRISPR region and the protospacer, which was used for the integration assay. PCR products were analyzed on 1.5% agarose gel stained with EtBr. Bands corresponding to integrations at the first or second repeats were excised from the gel and purified using GeneJET Gel Extraction and DNA Cleanup Micro Kit (Thermo Fisher Scientific). Bands were then cloned into pJET1.2 plasmid (Thermo Fisher Scientific) via blunt ends. Plasmid DNA was isolated from separate clones using GeneJET Plasmid Miniprep Kit (Thermo Fisher Scientific) and sequenced using pJET forward and reverse primers.
Author contributions
GD, TS, GG, and VS designed the experiments. GD, AS, and TS performed the experiments. ČV performed bioinformatics analysis. GD, TS, GG, and VS analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
Conflict of interest
G.G. and V.S are inventors on patent applications related to CRISPR and co‐founders of CasZyme. G.G. is an employee of CasZyme.
Supporting information
Expanded View Figures PDF
Table EV1
Source Data for Expanded View
Review Process File
Source Data for Figure 2
Source Data for Figure 3
Acknowledgements
We thank Algirdas Toleikis and Justas Lavišius for preliminary characterization of the DnaQ domain and the Cas1:Cas2‐DnaQ complex. G.G. acknowledges support from Research Council of Lithuania (grant MIP‐027/2014).
EMBO Reports (2018) 19: e45543
References
- 1. Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315: 1709–1712 [DOI] [PubMed] [Google Scholar]
- 2. Amitai G, Sorek R (2016) CRISPR–Cas adaptation: insights into the mechanism of action. Nat Rev Microbiol 14: 67–76 [DOI] [PubMed] [Google Scholar]
- 3. Sternberg SH, Richter H, Charpentier E, Qimron U (2016) Adaptation in CRISPR‐Cas systems. Mol Cell 61: 797–808 [DOI] [PubMed] [Google Scholar]
- 4. Brouns SJJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJH, Snijders APL, Dickman MJ, Makarova KS, Koonin EV, van der Oost J (2008) Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321: 960–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP (2009) RNA‐guided RNA cleavage by a CRISPR RNA‐Cas protein complex. Cell 139: 945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual‐RNA‐guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gasiunas G, Barrangou R, Horvath P, Siksnys V (2012) Cas9‐crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA 109: E2579–E2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Makarova KS, Haft DH, Barrangou R, Brouns SJJ, Charpentier E, Horvath P, Moineau S, Mojica FJM, Wolf YI, Yakunin AF et al (2011) Evolution and classification of the CRISPR‐Cas systems. Nat Rev Microbiol 9: 467–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJJ, Charpentier E, Haft DH et al (2015) An updated evolutionary classification of CRISPR–Cas systems. Nat Rev Microbiol 13: 722–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Krupovic M, Makarova KS, Forterre P, Prangishvili D, Koonin EV (2014) Casposons: a new superfamily of self‐synthesizing DNA transposons at the origin of prokaryotic CRISPR‐Cas immunity. BMC Biol 12: 36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Datsenko KA, Pougach K, Tikhonov A, Wanner BL, Severinov K, Semenova E (2012) Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat Commun 3: 945 [DOI] [PubMed] [Google Scholar]
- 12. Swarts DC, Mosterd C, van Passel MWJ, Brouns SJJ (2012) CRISPR interference directs strand specific spacer acquisition. PLoS One 7: e35888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yosef I, Goren MG, Qimron U (2012) Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli . Nucleic Acids Res 40: 5569–5576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nuñez JK, Kranzusch PJ, Noeske J, Wright AV, Davies CW, Doudna JA (2014) Cas1–Cas2 complex formation mediates spacer acquisition during CRISPR–Cas adaptive immunity. Nat Struct Mol Biol 21: 528–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nuñez JK, Harrington LB, Kranzusch PJ, Engelman AN, Doudna JA (2015) Foreign DNA capture during CRISPR–Cas adaptive immunity. Nature 527: 535–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nuñez JK, Lee ASY, Engelman A, Doudna JA (2015) Integrase‐mediated spacer acquisition during CRISPR–Cas adaptive immunity. Nature 519: 193–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rollie C, Schneider S, Brinkmann AS, Bolt EL, White MF (2015) Intrinsic sequence specificity of the Cas1 integrase directs new spacer acquisition. Elife 4: e08716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nuñez JK, Bai L, Harrington LB, Hinder TL, Doudna JA (2016) CRISPR immunological memory requires a host factor for specificity. Mol Cell 62: 824–833 [DOI] [PubMed] [Google Scholar]
- 19. Wright AV, Doudna JA (2016) Protecting genome integrity during CRISPR immune adaptation. Nat Struct Mol Biol 23: 876–883 [DOI] [PubMed] [Google Scholar]
- 20. Wright AV, Liu J‐J, Knott GJ, Doxzen KW, Nogales E, Doudna JA (2017) Structures of the CRISPR genome integration complex. Science 679: eaao0679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xiao Y, Ng S, Hyun Nam K, Ke A (2017) How type II CRISPR–Cas establish immunity through Cas1–Cas2‐mediated spacer integration. Nature 550: 137–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Levy A, Goren MG, Yosef I, Auster O, Manor M, Amitai G, Edgar R, Qimron U, Sorek R (2015) CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature 520: 505–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Künne T, Kieper SN, Bannenberg JW, Vogel AIM, Miellet WR, Klein M, Depken M, Suarez‐Diez M, Brouns SJJ (2016) Cas3‐derived target DNA degradation fragments fuel primed CRISPR adaptation. Mol Cell 63: 852–864 [DOI] [PubMed] [Google Scholar]
- 24. Li M, Wang R, Xiang H (2014) Haloarcula hispanica CRISPR authenticates PAM of a target sequence to prime discriminative adaptation. Nucleic Acids Res 42: 7226–7235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Horvath P, Barrangou R (2010) CRISPR/Cas, the immune system of bacteria and archaea. Science 327: 167–170 [DOI] [PubMed] [Google Scholar]
- 26. Plagens A, Tripp V, Daume M, Sharma K, Klingl A, Hrle A, Conti E, Urlaub H, Randau L (2014) In vitro assembly and activity of an archaeal CRISPR‐Cas type I‐A Cascade interference complex. Nucleic Acids Res 42: 5125–5138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sinkunas T, Gasiunas G, Waghmare SPSP, Dickman MJMJ, Barrangou R, Horvath P, Siksnys V (2013) In vitro reconstitution of Cascade‐mediated CRISPR immunity in Streptococcus thermophilus . EMBO J 32: 385–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang Y, Braithwaite DK, Ito J (1997) Evolution of dnaQ, the gene encoding the editing 3′ to 5′ exonuclease subunit of DNA polymerase III holoenzyme in Gram‐negative bacteria. FEBS Lett 400: 94–98 [DOI] [PubMed] [Google Scholar]
- 29. Hamdan S, Carr PD, Brown SE, Ollis DL, Dixon NE (2002) Structural basis for proofreading during replication of the Escherichia coli chromosome. Structure 10: 535–546 [DOI] [PubMed] [Google Scholar]
- 30. Wang J, Li J, Zhao H, Sheng G, Wang M, Yin M, Wang Y (2015) Structural and mechanistic basis of PAM‐dependent spacer acquisition in CRISPR‐Cas systems. Cell 163: 840–853 [DOI] [PubMed] [Google Scholar]
- 31. Beloglazova N, Brown G, Zimmerman MD, Proudfoot M, Makarova KS, Kudritska M, Kochinyan S, Wang S, Chruszcz M, Minor W et al (2008) A novel family of sequence‐specific endoribonucleases associated with the clustered regularly interspaced short palindromic repeats. J Biol Chem 283: 20361–20371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Babu M, Beloglazova N, Flick R, Graham C, Skarina T, Nocek B, Gagarinova A, Pogoutse O, Brown G, Binkowski A et al (2011) A dual function of the CRISPR‐Cas system in bacterial antivirus immunity and DNA repair. Mol Microbiol 79: 484–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nam KH, Ding F, Haitjema C, Huang Q, Delisa MP, Ke A (2012) Double‐stranded endonuclease activity in B. halodurans clustered regularly interspaced short palindromic repeats (CRISPR)‐associated Cas2 protein. J Biol Chem 287: 35943–35952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Semenova E, Savitskaya E, Musharova O, Strotskaya A, Vorontsova D, Datsenko KA, Logacheva MD, Severinov K (2016) Highly efficient primed spacer acquisition from targets destroyed by the Escherichia coli type I‐E CRISPR‐Cas interfering complex. Proc Natl Acad Sci USA 113: 7626–7631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Silas S, Mohr G, Sidote DJ, Markham LM, Sanchez‐Amat A, Bhaya D, Lambowitz AM, Fire AZ (2016) Direct CRISPR spacer acquisition from RNA by a natural reverse transcriptase‐Cas1 fusion protein. Science 351: aad4234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sinkunas T, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V (2011) Cas3 is a single‐stranded DNA nuclease and ATP‐dependent helicase in the CRISPR/Cas immune system. EMBO J 30: 1335–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Richter C, Gristwood T, Clulow JS, Fineran PC (2012) In vivo protein interactions and complex formation in the pectobacterium atrosepticum subtype I‐F CRISPR/Cas system. PLoS One 7: e49549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Staals RHJ, Jackson SA, Biswas A, Brouns SJJ, Brown CM, Fineran PC, Nishihama S, Yoshizuka K, Li X, Kawano T (2016) Interference dominates and amplifies spacer acquisition in a native CRISPR‐Cas system. Nat Commun 23: 127–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Richter C, Dy RL, McKenzie RE, Watson BNJ, Taylor C, Chang JT, McNeil MB, Staals RHJ, Fineran PC (2014) Priming in the Type I‐F CRISPR‐Cas system triggers strand‐independent spacer acquisition, bi‐directionally from the primed protospacer. Nucleic Acids Res 42: 8516–8526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vorontsova D, Datsenko KA, Medvedeva S, Bondy‐Denomy J, Savitskaya EE, Pougach K, Logacheva M, Wiedenheft B, Davidson AR, Severinov K et al (2015) Foreign DNA acquisition by the I‐F CRISPR–Cas system requires all components of the interference machinery. Nucleic Acids Res 43: 10848–10860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Plagens A, Tjaden B, Hagemann A, Randau L, Hensel R (2012) Characterization of the CRISPR/Cas Subtype I‐A system of the hyperthermophilic crenarchaeon Thermoproteus tenax . J Bacteriol 194: 2491–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang J, Kasciukovic T, White MF (2012) The CRISPR associated protein Cas4 Is a 5′ to 3′ DNA exonuclease with an iron‐sulfur cluster. PLoS One 7: e47232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rollie C, Graham S, Rouillon C, White MF (2018) Prespacer processing and specific integration in a Type I‐A CRISPR system. Nucleic Acids Res 46: 1007–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kieper SN, Almendros C, Behler J, McKenzie RE, Nobrega FL, Haagsma AC, Vink JNA, Hess WR, Brouns SJJ (2018) Cas4 facilitates PAM‐compatible spacer selection during CRISPR adaptation. Cell Rep 22: 3377–3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee H, Zhou Y, Taylor DW, Sashital DG (2018) Cas4‐dependent prespacer processing ensures high‐fidelity programming of CRISPR arrays. Mol Cell 70: 48–59.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zheng L, Baumann U, Reymond J‐L (2004) An efficient one‐step site‐directed and site‐saturation mutagenesis protocol. Nucleic Acids Res 32: e115 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Table EV1
Source Data for Expanded View
Review Process File
Source Data for Figure 2
Source Data for Figure 3


