Abstract
The Endoplasmic reticulum aminopeptidase I (ERAP1) trims peptides to their optimal size for binding to Major Histocompatibility Complex class I proteins. The natural polymorphism of this enzyme is associated with ankylosing spondylitis (AS) in epistasis with the major risk factor for this disease, HLA-B*27, suggesting a direct relationship between AS and HLA-B*27-bound peptides. Three polymorphisms that affect peptide trimming protect from AS: K528R, D575N/R725Q, and Q730E. We characterized and ranked the effects of each mutation, and their various combinations, by quantitative comparisons of the HLA-B*27 peptidomes from cells expressing distinct ERAP1 variants. Five features were examined: peptide length, N-terminal flanking residues, N-terminal residues of the natural ligands, internal sequences and affinity for B*27:05. Polymorphism at residue 528 showed the largest influence, affecting all five features regardless of peptide length. D575N/R725Q showed a much smaller effect. Yet, when co-occurring with K528R, it further decreased ERAP1 activity. Polymorphism at residue 730 showed a significant influence on peptide length, because of distinct effects on trimming of nonamers compared with longer peptides. Accordingly, multiple features were affected by the Q730E mutation in a length-dependent way. The alterations induced in the B*27:05 peptidome by natural ERAP1 variants with different K528R/Q730E combinations reflected separate and additive effects of both mutations. Thus, the influence of ERAP1 on HLA-B*27 is very diverse at the population level, because of the multiplicity and complexity of ERAP1 variants, and to the distinct effects of their co-occurring polymorphisms, leading to significant modulation of disease risk among HLA-B*27-positive individuals.
Keywords: Immunology*, Peptides*, Inflammation, Enzymes*, Peptidomics, ankylosing spondylitis, antigen processing, Erap1, HLA-B27
ERAP1 is a polymorphic aminopeptidase of the endoplasmic reticulum expressed in all individuals, whose best characterized function is to trim peptides to their proper length for Major Histocompatibility Complex Class I (MHC-I) 1 molecules (1, 2). The absence of this enzyme results in substantially altered MHC-I peptidomes characterized by longer and lower affinity peptides, and reduced MHC-I expression (3–8). Major features of ERAP1 include its capacity to cleave virtually all peptide bonds except those involving Pro, a preference for hydrophobic N-terminal (P1) residues, low efficiency toward acidic and basic ones (9–12), and, uniquely among aminopeptidases, a limitation of its activity determined by substrate length, a feature known as the molecular ruler mechanism. This consists in that the enzyme efficiently trims 10-mers and longer peptides, but its activity toward 9-mers is significantly reduced, becoming virtually inactive for shorter peptides (13). Natural ERAP1 variants are complex allotypes characterized by multiple amino acid changes arising by single nonsynonymous single nucleotide polymorphisms (SNPs), some of which affect the enzymatic activity. There are 10 major ERAP1 variants in human populations, usually referred to as ERAP1 haplotypes (Hap), or combinations of SNPs, designated as Hap1 to Hap10 (14).
The association of ERAP1 polymorphism with ankylosing spondylitis (AS) in epistasis with the major risk factor for this disease, HLA-B*27, was revealed by genome-wide association studies (15, 16), and was followed by similar findings in other MHC-I associated diseases, such as psoriasis (17) and Behçet's disease (18). The association with AS follows a two-mutation model consistent with a primary protective effect of the K528R change and a secondary effect of D575N/R725Q, the latter tagged by the rs10050860 SNP, at codon 575. HLA-B*27-positive individuals homozygous for both polymorphisms are about 25% less likely to develop AS that individuals homozygous for the corresponding risk alleles (16). A recent study suggests that the association of rs10050860 may be accounted for by strong linkage disequilibrium with a splice-altering variant, rs7063, influencing protein levels (19).
Multiple studies that analyzed the association of natural ERAP1 haplotypes with AS, reviewed in (14), showed that one or more of the Hap1-Hap3 haplotypes, all including the risk polymorphisms at residues 528, 575, and 725, were associated with increased risk of AS, whereas Hap10, including the protective polymorphisms at all three positions, was protective. In early genetic studies the Q730E polymorphism showed one of the highest associations with AS, Q730 being the risk variant (15, 20). These studies did not specifically address the linkage disequilibrium among missense polymorphisms within the ERAP1 gene. A more recent fine mapping study (21) restricted the AS susceptibility region of ERAP1 to a relatively short span of DNA, including both the missense SNPs rs30187 (K528R) and rs10050860 (D575N), but not rs27044 (Q730E), raising the possibility that the latter might not be directly involved in AS.
The epistatic association of ERAP1 with AS and other inflammatory diseases (22) suggests a pathogenetic role of peptide processing, emphasizing the need to characterize the effects of ERAP1 polymorphism on MHC-I bound peptidomes. The issue is not simple because of the distinct influence of individual polymorphisms on the enzymatic activity of ERAP1 and the likely interplay of effects of these changes in the natural ERAP1 haplotypes. This is suggested by observations that the activity of a given ERAP1 variant is the result of its combination of polymorphic residues (23, 24). These studies pointed to the necessity of directly defining the effect of natural ERAP1 haplotypes on the HLA-B*27 peptidome in live cells with distinct ERAP1 backgrounds.
Two previous studies from our laboratory addressed this issue (25, 26), demonstrating the significant influence of disease-associated ERAP1 variants on the HLA-B*27 peptidome. They also showed a dominant effect of the K528R change over D575N/R725Q, in agreement with in vitro studies showing that polymorphism at residue 528 is a major determinant of ERAP1 activity (27–29). Yet, our earlier studies used a relatively low-resolution peptidomic approach, which involved the direct identification of a reduced number of peptides, limiting statistical assessments and the detection of effects on the peptidome to the very major ones. Moreover, in the first of those studies (25), the presence of ERAP2 was not fully considered. This enzyme trims basic and few other P1 residues (10, 22), is also involved in the processing of MHC-I ligands (10, 30–32), is not expressed in at least 25% of individuals (33) and has a significant influence on the HLA-B*27 peptidome (34, 35).
Thus, in the present study we undertook a systematic and comprehensive analysis of the effects of AS-associated ERAP1 polymorphism on the HLA-B*27 peptidome. We compared the B*27:05 peptidomes from ERAP2-positive cells expressing distinct ERAP1 haplotypes by means of a homogeneous and high-resolution immunopeptidomic approach. Through the identification of several thousands of peptides from each cell line obtained in uniform conditions, we were able to characterize ERAP1-dependent changes affecting multiple features of the peptidome with high reliability and statistical strength. By selecting cell lines with ERAP1 variants showing different combinations of polymorphic residues, we could assign distinct effects to individual disease-associated changes, rank their relative contribution to shaping the HLA-B*27 peptidome, and define the interactive effects among co-occurring polymorphisms in the natural ERAP1 haplotypes.
MATERIALS AND METHODS
Cell Lines, and Western Blots
Four HLA-B*27:05-positive lymphoblastoid cell lines (LCL) were used in this study: LCL 6370 (HLA-A*02, *68; B*27:05, *44; C*02, *07), LG2 (HLA-A*02, B*27:05, C*01), P50 (HLA-A*23, *24; B*27:05, *14; C*02, *08) and C1R05 (B*27:05, *35:03low, C*04) (Table I). The latter is a transfectant line derived from HMy2.C1R cells, which are defective in their endogenous HLA-A, B genes (36). The four cell lines are ERAP2-positive and express similar levels of this enzyme (26, 35). They are homozygous at the ERAP1 locus and express distinct ERAP1 allotypes (26, 34). The cells were cultured in RPMI 1640 medium with 10% fetal bovine serum (Biowest, Nuaillé, France), 25 mm HEPES buffer, 2 mm l-glutamine, penicillin and streptomycin. ME1 (IgG1), an anti-HLA-B*07/B*27/B*22 monoclonal antibody (mAb) that recognizes HC/β2m/peptide complexes (37), was used for immunopurification of HLA-B*27. Among the non-B*27 MHC-I molecules expressed in the cell lines used in this study, ME1 is known to have a weak cross-reaction only with HLA-B*14 (38, 39). Besides this antibody, W6/32 (IgG2a), which recognizes a monomorphic determinant of HLA-A,B,C molecules (40) was used in flow cytometry.
Table I. ERAP1 and ERAP2 phenotypes of the cell lines used in this studya.
| Comp. | ERAP1 | 12 | 56 | 127 | 276 | 346 | 349 | 528 | 575 | 725 | 730 | ERAP2 | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6370 | Hap2 | T | E | R | I | G | M | K | D | R | Q | + | (34) |
| LG2 | Hap3 | T | E | R | I | G | M | K | D | R | E | + | (26) |
| LG2 | Hap3 | T | E | R | I | G | M | K | D | R | E | + | |
| C1R05 | Hap8 | T | E | P | M | G | M | R | D | R | E | + | (25) |
| 6370 | Hap2 | T | E | R | I | G | M | K | D | R | Q | + | |
| C1R05 | Hap8 | T | E | P | M | G | M | R | D | R | E | + | |
| C1R05 | Hap8 | T | E | P | M | G | M | R | D | R | E | + | |
| P50 | Hap10 | T | E | P | I | G | V | R | N | Q | E | + | |
| 6370 | Hap2 | T | E | R | I | G | M | K | D | R | Q | + | |
| P50 | Hap10 | T | E | P | I | G | V | R | N | Q | E | + | (26) |
| LG2 | Hap3 | T | E | R | I | G | M | K | D | R | E | + | |
| P50 | Hap10 | T | E | P | I | G | V | R | N | Q | E | + |
a Haplotypes and amino acid residues associated with AS risk are in boldface and underlined.
Western blotting was done and quantified as previously described (25), using the mAbs 6H9, 3F5 (both from RD systems, Minneapolis, MN), and GTU88 (Sigma) to reveal ERAP1, ERAP2 and γ-tubulin, respectively.
Flow Cytometry
About 2 × 105 cells where washed twice with 200 μl of PBS, centrifuged at 1500 RPM for 2 min. and incubated with 50 μl of ME1 or W6/32 for 20 min at 4 °C (10 and 20 μg/ml respectively). The cells were washed twice with 200 μl of PBS, incubated with FITC-conjugated anti-mouse IgG secondary antibody (eBioscience, San Diego, CA) at 1 μg/ml for 20 min. at 4 °C, and washed twice with PBS. The detection was made using a FACSCalibur instrument (BD Bioscinece, San José, CA) with CellQuest Pro version. All data were analyzed using the FlowJo software, version 7.5 (Tree Star, Inc, Ashland, OR).
Isolation of HLA-A*B*27:05-bound Peptidomes
About 1 × 109 cells were lysed at 4 °C in 150 mm NaCl, 20 mm Tris-HCl, pH 7.5, 1% Igepal CA-630 (Sigma-Aldrich) and a mixture of protease inhibitors (Roche, Mannheim, Germany). After centrifugation, the supernatant was passed through a column containing the ME1 mAb bound to CNBr-activated Sepharose 4B (GE Healthcare, Buckinghamshire, UK) and washed with 20 column volumes each of 20 mm Tris-HCl, pH 8.0 containing: 1) 150 mm NaCl, 2) 400 mm NaCl, 3) 150 mm NaCl, and 4) 40 column volumes of buffer without NaCl. Peptides were eluted with 1% TFA (Sigma-Aldrich) at room temperature, filtered through Vivaspin 2, cutoff 5000 Da (Sartorius Stedim Biotech, Gottingen, Germany), concentrated in a SpeedVac and subjected to reversed phase purification using OMIX tips (Varian Inc. Palo Alto, CA) by elution with 50% acetonitrile, 0.1% TFA in water. The eluted peptides were dried and stored at −20 °C. For each LCL, three preparations were independently obtained from the same cell amounts.
Mass Spectrometry
The recovered peptides were analyzed in a Q-Exactive-Plus mass spectrometer fitted with Ultimate 3000 RSLC nanocapillary UHPLC (Thermo Fisher Scientific, Waltham, MA). The peptides were resolved on a capillary column (75 μm ID and ≈20 cm long) pressure packed as in (41) with C18 reversed-phase 3.5 μm beads (Reprosil-C18-Aqua, Dr. Maisch GmbH, Ammerbuch-Entringen, Germany), using a 7–28% acetonitrile gradient with 0.1% formic acid during 180 min followed by 28–95% during 15 min. The flow rate was 0.15 μl/min. The dynamic exclusion was set to 20 s and the automatic gain control value for the full MS was set to 3 × 106. The selected masses were fragmented from the survey scan of mass-to-charge ratio (m/z) 300–1800 AMU at resolution 70,000. Data was acquired using a data-dependent “top-10” method, fragmenting the peptides by higher-energy collisional dissociation. MS/MS spectra were acquired starting at m/z 200 with a resolution of 17,500. The target value of the MS/MS was set to 1 × 105 and the isolation window to 1.8 m/z. The maximum injection time was set to 100 ms and normalized collision energy to 25 eV. No fragmentation was performed for peptides with unassigned precursor ion charge states or charge states of four and above. The peptide match option was set to Preferred. Fragmented masses were dynamically excluded from further selection for fragmentation for 20 s.
Experimental Design and Statistical Rationale
HLA-B*27-bound peptides were isolated from three independent preparations of each of the four LCL used in this study, to provide for biological replicates. About 1 × 109 cells were used for each preparation. The peptides were fragmented by mass spectrometry, as detailed above, and their sequences were assigned, from the MS/MS spectra, using the MaxQuant software (version 1.5.0.25) (42) with the Andromeda search engine (43) and the human UniProt/Swiss-Prot database (release 2015_07, 20197 entries) under the following parameters: precursor ion mass and fragment mass tolerance 20 ppm, false discovery rate (FDR) 0.05 for peptide-spectrum matching. N-terminal acetylation, Gln-to-pyroglutamic cyclation and Met oxidation were included as variable modifications. No fixed modifications were included. Identifications derived from the reverse database and known contaminants were eliminated. Neither the specificity of proteases used to generate peptides, nor the missed/nonspecific cleavages permitted applied to our analysis, as it concerns peptides endogenously generated in live cells. Peptides in the length range of 8 to 14 amino acid residues, corresponding to most MHC-I ligands, with Arg or Gln at peptide position (P) 2, which is the major anchor motif of HLA-B*27, were assigned as B*27:05 ligands. This selection removes most of the contaminant peptides unrelated to MHC-I and to non-B*27 MHC-I ligands.
To assess the effect of the ERAP1 background in each LCL on the B*27:05 peptidome we carried out quantitative comparisons of the shared ligands between cell line pairs. Six such comparisons were carried out, each one addressing the effects of ERAP1 polymorphisms (Table I). Prior to these comparisons, the ion peak intensity of each peptide in each cell line was normalized to the total intensity of the ion peaks corresponding to all the identified peptides in that cell line after filtering for length and P2 residues as described above, so that the intensity of irrelevant signals was not considered. This normalization also corrects for differences in peptide yields arising, for instance, from distinct HLA-B*27 expression among cell lines. The normalization was done in each individual experiment and the normalized ion peak intensities assigned to the peptides from each cell line were the mean values from the three experiments. The ratio between the normalized intensities of a given peptide in the two cell lines (IR) was taken as an indication of the relative amounts of that peptide.
The shared peptides between any given cell line pair were classified as follows. Those that were more abundant in one cell line, relative to the other (IR>1.0), were subdivided in those with IR>1.0 to 1.5 (hereafter abbreviated as IR>1.0–1.5) and those with IR>1.5. The former subset included the peptides with similar or slightly increased levels in one cell line relative to the other. The IR>1.5 subset included those peptides showing higher expression differences in each cell line compared with the other. This classification is based on our previous studies showing that ERAP1/ERAP2-dependent differences in MHC-I peptidomes are best revealed among peptides in the IR>1.5 subset and are usually attenuated or absent altogether in the IR>1.0–1.5 group. This is expected from the fact that quantitative effects of ERAP1 polymorphism on MHC-I ligands should be mainly observed among peptides showing larger differences in relative amounts between the cell lines compared. The relationship of this classification to the statistical significance of the differences in peptide amounts was established by means of Volcano plots for each pairwise comparison. In these analyses the Student's t test was used to assess the statistical significance of the expression differences of individual peptides in the two cell lines compared.
Recombinant ERAP1 Proteins and Hydrolytic Assay
The purification of recombinant ERAP1 proteins and the hydrolysis assay with the fluorogenic substrate Leu-7-amido-4-methyl-coumarine (l-AMC) were carried out exactly as previously described (23).
Classification of Amino Acid Residues According to ERAP1 Susceptibility
Amino acid residues were classified according to their susceptibility to ERAP1 (12), as ERAP1-susceptible (A, C, L, M, Y), intermediate (F, G, H, I, N, Q, S, T), or resistant (D, E, K, P, R, V, W), as previously described (25).
Binding Affinity
Theoretical binding affinities of B*27:05 ligands were calculated using the NetMHCcons 1.1 Server (http://www.cbs.dtu.dk/services/NetMHCcons/), which integrates three different algorithms, as described elsewhere (44).
Statistical Analyses
Differences in peptide length and residue frequencies were assessed by the χ2 test with Bonferroni correction, when applicable. Differences in binding affinity were analyzed by the Mann-Whitney U test. The Student's t test was used to assess the statistical significance of the expression differences of individual peptides among cell lines. p < 0.05 was considered as statistically significant in all cases.
RESULTS
ERAP1, ERAP2, and HLA-B*27 Expression in HLA-B*27:05 Positive Cell Lines
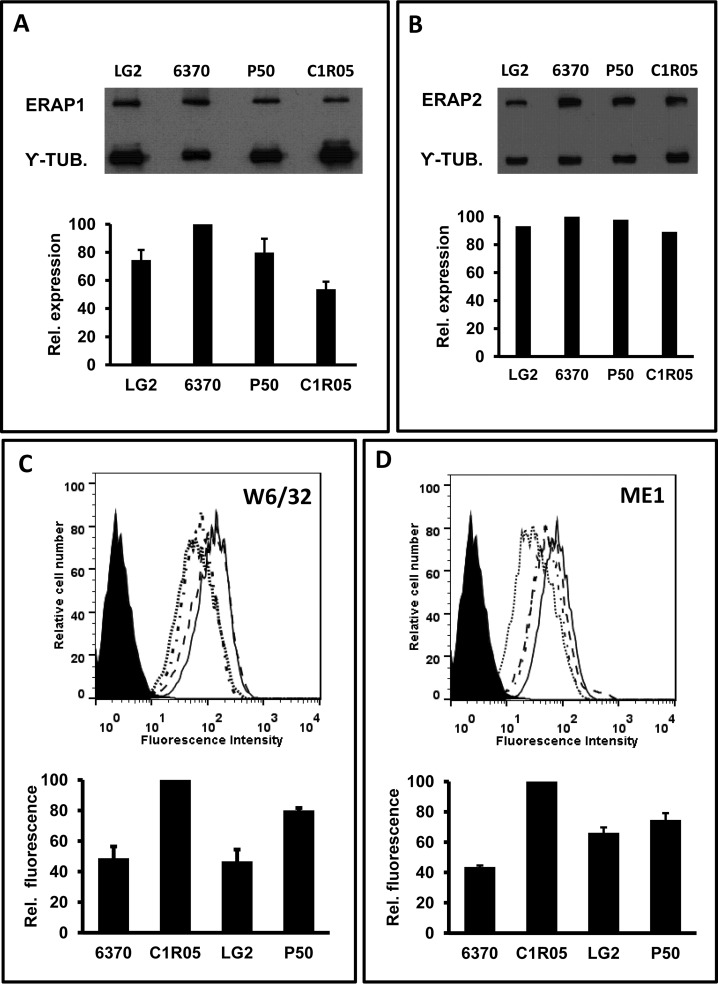
Although the expression of ERAP1 and ERAP2 in the four cell lines used in this study were separately assessed in previous reports (26, 35), their relative protein levels were jointly determined by Western blotting in the context of the present study (Fig. 1A–1B). For ERAP1 they were the following: LCL 6370 (100), LG2 (75 ± 7), P50 (80 ± 10), C1R05 (54 ± 5). ERAP2 expression was similar in the four cell lines, as previously reported.
Fig. 1.
ERAP1, ERAP2 and MHC-I expression in LCL. A, Expression of ERAP1 proteins in the indicated cell lines. A representative experiment (top) and the mean ± standard deviation of 3 independent analyses (bottom) are shown. The results are expressed as the amounts of ERAP1 relative to the cell line with the highest expression of the enzyme. B, ERAP2 protein expression in the same cell lines; conventions are as in panel A, except that a single experiment (top) and its quantization (bottom) is shown, because the similar expression of ERAP2 in these four cell lines was previously reported (26, 35). C, Flow cytometry analysis of the surface expression of MHC-I antigens in LCL 6370 (. . . .), C1R05 (_____), LG2 (.-.-.-), and P50 (—), as assessed with the W6/32 mAb. A representative measurement (top) and the mean ± standard deviation of triplicate samples (bottom) is shown, with mean fluorescent values made relative to the cell line with the highest MHC-I expression. D, Flow cytometry analysis of the surface expression of HLA-B*27 as assessed with the ME1 mAb. Conventions are as in panel C. Note that P50 expresses HLA-B*14, which shows some reactivity with ME1.
Total MHC-I expression was highest in the C1R05 transfectant, followed by P50, and was lowest and similar for LCL 6370 and LG2 (Fig. 1C). Accordingly, HLA-B*27 expression was highest for C1R05, followed by LG2 and P50. LG2 is homozygous for HLA-B*27 and its expression was roughly double compared with the heterozygous LCL 6370. Although P50 is also heterozygous for HLA-B*27, its relatively high staining with ME1 is explained both by the higher overall expression of MHC-I molecules (Fig. 1C) and by the moderate cross-reaction of ME1 with HLA-B*14 (38, 39), which is also expressed in this cell line.
Identification of HLA-B*27:05 Ligands Presented in Various ERAP1 Contexts
The B*27:05 peptidomes from LCL 6370, LG2, C1R05 and P50, isolated from three independent preparations for each cell line, were analyzed by MS. A total of 9162 peptides with 8 to 14 amino acid residues and Arg2 or Gln2 were assigned as B*27:05 ligands in one or more of the four LCL, of which 6894 (LCL 6370), 6094 (LG2), 7205 (C1R05), and 6620 (P50) were detected in the individual cell lines (supplemental Table S1). The molecular weight and length distributions of the identified B*27:05 ligands were very similar among cell lines, although a small increase of 9-mers, and a corresponding decrease of longer peptides, was observed in LG2 (supplemental Fig. S1).
Quantitative Effects of ERAP1 Polymorphism on the B*27:05 Peptidome
In the following analyses we examined the effects of distinct ERAP1 polymorphisms on the amounts of B*27:05 ligands. To this end, we compared the B*27:05 peptidomes from ERAP2-positive cell line pairs differing in their ERAP1 variants. Six such comparisons were carried out (Table I) using the experimental strategy described above (see Methods: Experimental Design and Statistical rationale). We focused on the shared peptides between any given cell line pair. The intensity ratio of a given peptide in the two cell lines, IR, was taken as an indication of the relative amounts of that peptide in both cell lines (supplemental Table S1).
The peptides that were more abundant in one cell line, relative to the other (IR>1.0), were subdivided in those with IR>1.0–1.5 and those with IR>1.5 (supplemental Table S2). The relationship of this classification to the statistical significance of the differences in peptide amounts is shown in supplemental Fig. S2. Peptides found only in one of the two cell lines were excluded from these analyses and were separately analyzed.
In all comparisons five features were analyzed: peptide length, N-terminal flanking residues (P-1), which reflect influence on epitope generation, P1 residues, which reflect influence on epitope destruction, residue frequencies at positions downstream the N terminus, which reflect additional peptide alterations, and theoretical binding affinity, which reflects global peptidome optimization. We have previously demonstrated that ERAP1/2-independent effects between LCL have no detectable effect on the HLA-B*27 peptidome in our experimental setup (25, 34, 35). A summary of the statistically significant differences observed in the peptide features analyzed in all pairwise comparisons, which are described in detail below, is given in Table II.
Table II. Summary of statistically significant differences in pairwise comparisons among LCLa.
| IR>1 |
Lengthb |
P(-1) residues with higher ERAP1 susceptibilityc |
P1 residues with higher ERAP1 susceptibilityc |
Increased affinity |
||||
|---|---|---|---|---|---|---|---|---|
| Comparison | Total | Total | Total | 9-Mers | ≥10-Mers | Total | 9-Mers | ≥10-Mers |
| 6370/LG2 | 1.4E-33 | NS | 2.2E-03 | 3.2E-05 | 2.8E-04 | 1.6E-02 | <1.0E-04 | 1.9E-02 |
| LG2/C1R05 | 1.6E-25 | 7.1E-03 | 3.2E-12 | 1.5E-16 | 2.5E-05 | <1.0E-04 | ||
| 6370/C1R05 | 1.1E-08 | 0.03 | 1.3E-19 | 3.9E-12 | 9.3E-13 | <1.0E-04 | ||
| 6370/P50 | 2.5E-31 | 1.4E-08 | 5.5E-18 | 4.7E-13 | 1.7E-13 | <1.0E-04 | ||
| LG2/P50 | 7.1E-69 | 3.5E-07 | 1.4E-10 | 4.2E-20 | 3.7E-03 | <1.0E-04 | ||
| C1R05/P50 | NS | NS | 8.5E-08 | 7.3E-04 | 4.6E-05 | <1.0E-04 | ||
a The data correspond to the IR>1.0 subsets of B*27:05 ligands in all comparisons. Statistical values in boldface or underlined are increased in the equally coded cell lines.
b Values correspond to increased percentage of 9-mers.
c For the purpose of this table, to simplify the representation, residues were subdivided in only two groups, with the residues of intermediate susceptibility to ERAP1 (F, G, H, I, N, Q, S, T) being joined to those of either low (D, E, K, P, R, V, W) or high susceptibility (A, C, L, M, Y) depending on the tendencies observed in Figs. 2–8. The statistical differences refer to the joint frequency of residues in each category.
LCL 6370/LG2 (Hap2/Hap3)
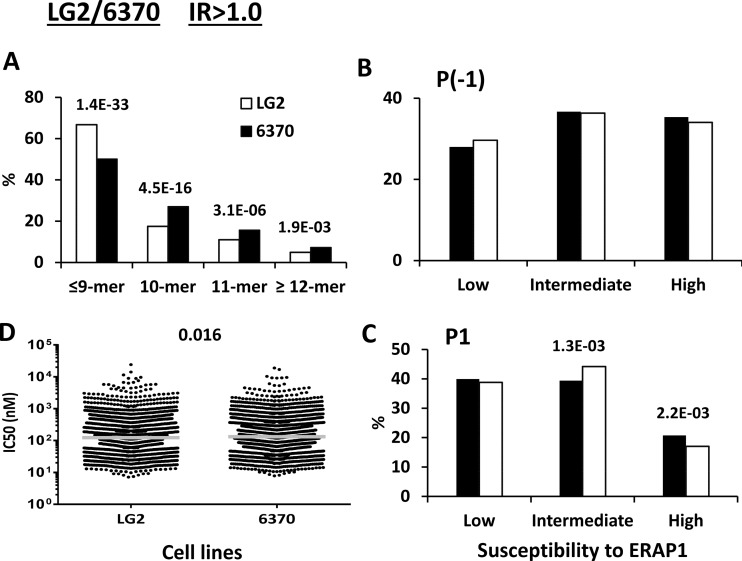
This comparison allowed us to assess the effect of the Q730E change in ERAP1 (Table I). The peptides with IR>1.0 showed a significantly higher percentage of peptides ≤9-mers (8-mers were <1% in all cases) in LG2 (E730), and a corresponding decrease of longer peptides, relative to LCL 6370 (Q730) (Fig. 2A). This difference was larger between the IR>1.5 subsets, and virtually nonexistent between the IR>1.0–15 subsets of both cell lines (supplemental Fig. S3A). No differences were observed in the P(-1) frequencies, indicating that the Q730E change has little influence on epitope generation as a function of the susceptibility of P(-1) residues to ERAP1 (Fig. 2B, supplemental Fig. S3B). In contrast, a decrease of P1 residues with high susceptibility to this enzyme was found in the IR>1.5, but not in the IR>1.0–1.5 subset of LG2 (supplemental Fig. S3C). The same effect was observed among the whole set of predominant peptides (IR>1.0) from this cell line, which included both subsets (Fig 2C). In this and all subsequent comparisons the differences observed between the IR>1.0 peptide sets reflect mainly those in the IR>1.5 subsets but are often somewhat attenuated because of the lack of differences between the IR>1.0–1.5 subsets. Thus, E730 in LG2 confers higher trimming activity, which is reflected both in higher amounts of shorter peptides and increased destruction of B*27:05 ligands with ERAP1-susceptible P1 residues. These alterations only marginally affected the global affinity of the B*27:05 peptidome (Fig. 2D, supplemental Fig. S3D).
Fig. 2.
Comparison of the shared peptides between LCL LG2 and 6370: IR>1.0 subsets. A total of 2444 (LG2: white bars) and 2869 (6370: black bars) peptides were included in the IR>1.0 subset from each cell line. A, Peptide length distribution, B, Percentage of P(-1) residues with low, intermediate and high susceptibility to ERAP1, C, Percentage of P1 residues with low, intermediate and high susceptibility to ERAP1, D, theoretical affinity of the peptides for B*27:05, as estimated by their IC50 values. When the differences reached statistical significance, this was indicated by their p values.
LG2/C1R05 (Hap3/Hap8)
This comparison allowed us to assess the effect of the R127P/I276M/K528R changes in ERAP1, in the E730 context (Table I). Previous in vitro analyses failed to show any effect of the R127P change on substrate trimming (27, 34) and there is also no evidence that the non-AS-associated I276M mutation has any functional effect. Thus, the major influence of these mismatches is probably conferred by the K528R change.
A significantly higher percentage of peptides ≤9-mers, to the expense of longer peptides, was observed in LG2 (K528), relative to C1R05 (R528) cells, both in the IR>1.0 and IR>1.5 subsets, but not in the IR>1.0–1.5 subsets, reflecting increased trimming by the K528 variant (Fig. 3A and supplemental Fig. S4A). This effect was comparable to that conferred by the Q730E change. However, the risk alleles in both positions (K528 and Q730) showed opposite effects.
Fig. 3.
Comparison of the shared peptides between LCL LG2 and C1R05: IR>1. 0 subsets. A total of 2215 (LG2: black bars) and 2801 (C1R05: white bars) peptides were included in the IR>1.0 subset from each cell line. A, Peptide length distribution, B, Percentage of P(-1) residues with low, intermediate and high susceptibility to ERAP1, C, Percentage of P1 residues with low, intermediate and high susceptibility to ERAP1, D, theoretical affinity of the peptides for B*27:05, as estimated by their IC50 values. When the differences reached statistical significance, this was indicated by their p values.
The higher activity of the K528 variant in LG2 was also reflected in a lower frequency of P(-1) and P1 residues with high sensitivity to ERAP1 trimming and a corresponding increase of ERAP1-resistant residues (Fig 3B–3C and supplemental Fig. S4B–S4C). These effects were clearly higher than those observed across the Q730E change (Fig. 2B–2C), indicating that the K528R and Q730E alter the generation/destruction balance of B*27:05 ligands in very distinct ways. The high activity conferred by K528 resulted in a significantly higher affinity of the peptides in the IR>1.0 and IR>1.5 subsets from LG2, relative to C1R05 (Fig. 3D and supplemental Fig. S4D).
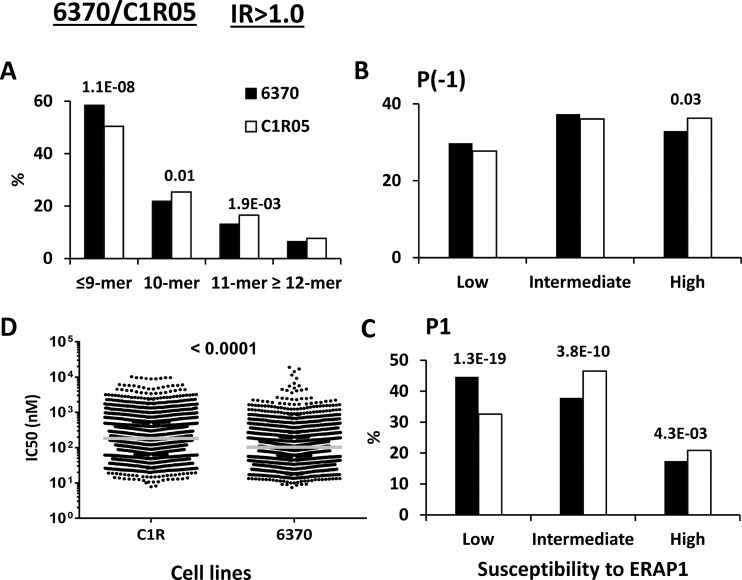
LCL 6370/C1R05 (Hap2/Hap8)
This comparison allowed us to assess the added effect of the Q730E change on the R127P/I276M/K528R mismatch (Table I). As expected, a higher percentage of peptides ≤9-mers, to the expense of longer peptides, was observed in LCL 6370 (K528/Q730), relative to C1R05 (R528/E730), both in the IR>1.0 and IR>1.5 subsets, consistent with the presence of K528 in the ERAP1 variant of the former LCL (Fig. 4A and supplemental Fig. S5A). However, these differences were smaller than those in LG2/C1R05 (KE/RE), strongly suggesting that Q730 in LCL 6370 diminish the length effects of K528.
Fig. 4.
Comparison of the shared peptides between LCL 6370 and C1R05: IR>1. 0 subsets. A total of 2246 (6370: black bars) and 3070 (C1R05: white bars) peptides were included in the IR>1.0 subset from each cell line. A, Peptide length distribution, B, Percentage of P(-1) residues with low, intermediate and high susceptibility to ERAP1, C, Percentage of P1 residues with low, intermediate and high susceptibility to ERAP1, D, theoretical affinity of the peptides for B*27:05, as estimated by their IC50 values. When the differences reached statistical significance, this was indicated by their p values.
The effects on P(-1) residues were somewhat lower, and those on P1 higher (Fig. 4B–4C and supplemental Fig. S5B–S5C), than in LG2/C1R05 (Fig. 3B and supplemental Fig. S4B). This is consistent with a length-dependent effect of the Q730E change on trimming.
The global affinity of the predominant peptides was higher in LCL 6370 than in C1R05 (Fig. 4D and supplemental Fig. S5D), to an extent like that in LG2/C1R05. This is consistent with the small effects of Q730E on the affinity of the B*27:05 peptidome (Fig. 2D).
The increased expression of ERAP1 in LG2 and 6370 cells, compared with C1R05, is frequently observed among ERAP1 variants with K528, relative to those with R528 (20, 45), which enhances the effect of this mutation.
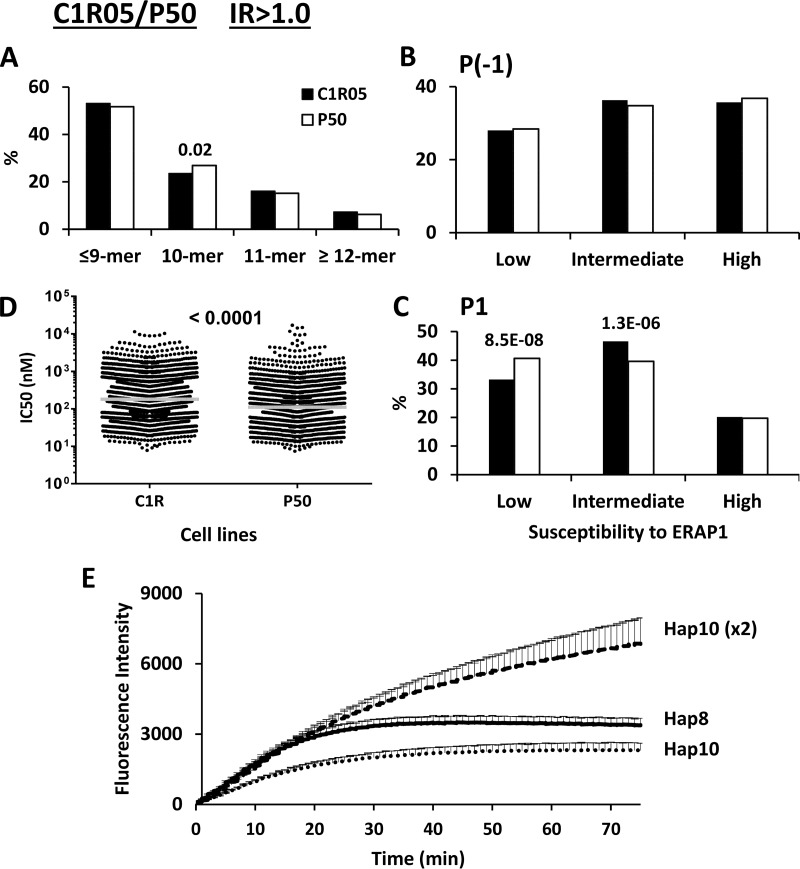
C1R05/P50 (Hap8/Hap10)
This comparison intended to address the joint effect of the M276I/M349V/D57N/R725Q changes in ERAP1, upon concordance at residues R528 and E730 (Table I). Peptide length distribution and P(-1) usage were very similar among the predominant B*27:05 ligands from both cell lines (Fig. 5A–5B and supplemental Fig. S6A–S6B). In contrast, an increase of ERAP1-resistant P1 residues, with a concomitant decrease of those with intermediate susceptibility, as well as increased affinity, was observed in P50, relative to C1R05, in the IR>1.0 and IR>1.5 subsets, but not in the IR>1.0–1.5 subsets (Fig. 5C–5D and supplemental Fig. S6C–S6D).
Fig. 5.
Comparison of the shared peptides between LCL C1R05 and P50: IR>1.0 subsets. A total of 3098 (C1R05: black bars) and 2262 (P50: white bars) peptides were included in the IR>1.0 subset from each cell line. A, Peptide length distribution, B, Percentage of P(-1) residues with low, intermediate and high susceptibility to ERAP1, C, Percentage of P1 residues with low, intermediate and high susceptibility to ERAP1, D, theoretical affinity of the peptides for B*27:05, as estimated by their IC50 values, E, Hydrolytic activity of recombinant variants of ERAP1 (Hap8 and Hap10) toward l-AMC at the same or at a double (×2) E:S ratio. When the differences in panels A–D reached statistical significance, this was indicated by their p values.
To address the possibility that these effects were because of the 1.5 to 2-fold higher ERAP1 expression in P50 cells, relative to C1R05, recombinant Hap8 and Hap10 variants of ERAP1 were tested for their hydrolytic activity toward l-AMC. At the same enzyme-to-substrate (E:S) ratio Hap8 showed slightly higher trimming, compared with Hap10, as previously reported (46). However, at a double E:S ratio Hap10 showed higher hydrolytic activity than Hap8 (Fig 5E). Thus, the differences observed between the B*27:05 peptidomes from C1R05 and P50 are probably because of differences in ERAP1 expression, rather than to polymorphism between the two enzyme variants.
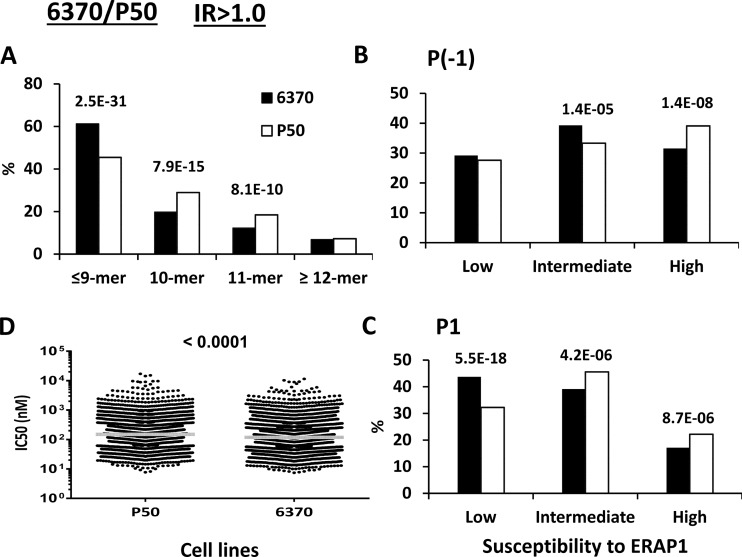
LCL 6370/P50 (Hap2/Hap10)
This comparison addressed the effect of a major AS-associated ERAP1 haplotype, Hap2, relative to the AS-protective haplotype, Hap10 (Table I). It allowed us to assess the effect of the M349V/D575N/R725Q changes when they occur along R127P/K528R/Q730E, whose effects were examined in the 6370/C1R05 comparison. A comparative analysis of B*27:05 ligands from LCL 6370 and P50, involving much less peptides, was previously reported (35).
The higher activity of ERAP1 in LCL 6370, relative to P50, was obvious from its effects, observed in both the IR>1.0 and IR>1.0–1.5 subsets, on increasing the percentage of peptides ≤9-mers (Fig. 6A and supplemental Fig. S7A), and in the lower percentages of B*27:05 ligands with ERAP1-sensitive P(-1) or P1 residues (Fig. 6B–6C and supplemental Fig. S7B–S7C), resulting in a B*27:05 peptidome of higher affinity (Fig. 6D and supplemental Fig. S7D).
Fig. 6.
Comparison of the shared peptides between LCL 6370 and P50: IR>1.0 subsets. A total of 2783 (6370: black bars) and 2756 (P50: white bars) peptides were included in the IR>1.0 subset from each cell line. A, Peptide length distribution, B, Percentage of P(-1) residues with low, intermediate and high susceptibility to ERAP1, C, Percentage of P1 residues with low, intermediate and high susceptibility to ERAP1, D, theoretical affinity of the peptides for B*27:05, as estimated by their IC50 values. When the differences reached statistical significance, this was indicated by their p values.
The differences in length and P(-1) usage were larger than those observed in 6370/C1R05 (Fig. 4 and supplemental Fig. S5), indicating that the additional M349V/D575N/R725Q changes in P50 further diminish ERAP1 activity.
LG2/P50 (Hap3/Hap10)
Like the previous one, this comparison addressed the effect of another major AS-associated ERAP1 haplotype, Hap3, relative to Hap10 (Table I). It allowed us to assess the effect of the M349V/D575N/R725Q changes when they occur along R127P/K528R, whose effects were examined in the LG2/C1R05 comparison (Fig. 3 and supplemental Fig. S4). The major difference between the 6370/P50 and LG2/P50 comparisons is the concordance of E730 in the latter cell line pair.
The differences between the B*27:05 ligands from LG2 and P50 in the IR>1.0 an IR>1.5 subsets were larger in peptide length and in the trimming of P(-1) residues (Fig. 7A–7B and supplemental Fig. S8A–S8B) than those in LG2/C1R05 (Figs. 3 and supplemental Fig. S4). Thus, again, the M349V/D575N/R725Q changes add to the dominant effect of K528R in decreasing the enzymatic activity of ERAP1. Yet, the alterations in P1 frequencies and affinity were similar or slightly smaller in LG2/P50 (Fig. 7C–7D and supplemental Fig. S8C–S8D) compared with LG2/C1R05.
Fig. 7.
Comparison of the shared peptides between LCL LG2 and P50: IR>1.0 subsets. A total of 2519 (LG2: black bars) and 2531 (P50: white bars) peptides were included in the IR>1.0 subset from each cell line. A, Peptide length distribution, B, Percentage of P(-1) residues with low, intermediate and high susceptibility to ERAP1, C, Percentage of P1 residues with low, intermediate and high susceptibility to ERAP1, D, theoretical affinity of the peptides for B*27:05, as estimated by their IC50 values. When the differences reached statistical significance, this was indicated by their p values.
Residue 730 Modulates the Effects of ERAP1 on the HLA-B*27 Peptidome as a Function of Peptide Length
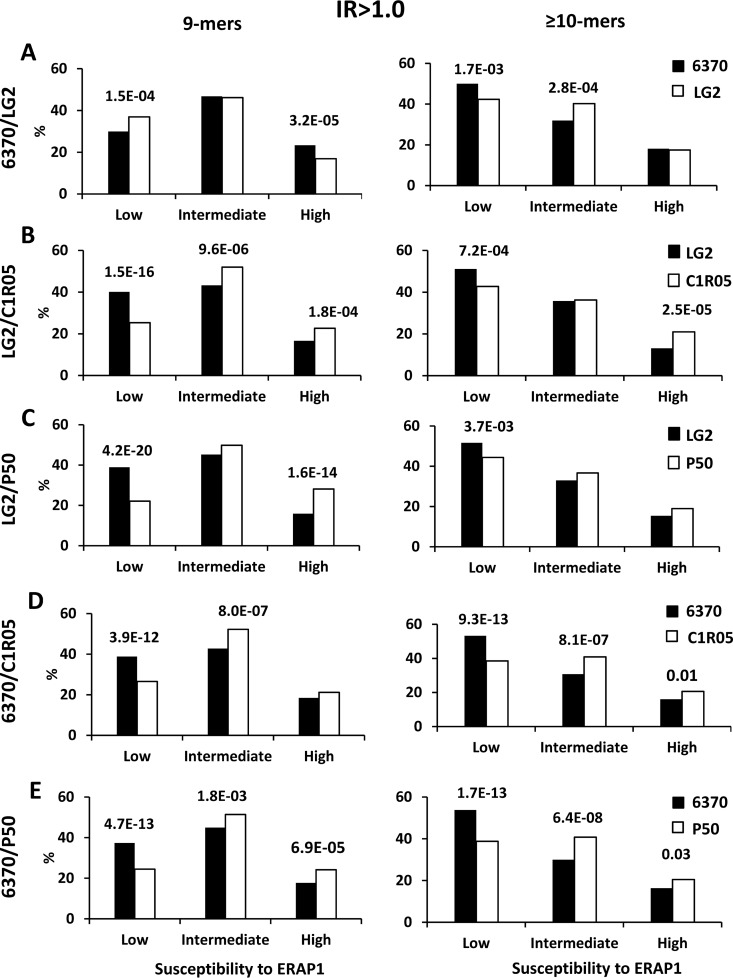
A previous study using poly-Gly peptide analogs and in vitro digestions showed that the Q730E polymorphism affects substrate length selection by increasing ERAP1 preference for shorter substrates (47). To establish the significance of this effect in live cells we examined the differences in P1 residue frequencies and affinity between the B*27:05 ligands in the IR>1.0 subsets from each cell line, separately considering the 9-mers and the peptides ≥10-mers in each pairwise comparison (Fig. 8). In LCL 6370/LG2 (Fig 8A), the percentage of 9-mers with ERAP1-resistant residues was higher in LG2 (E730), because of increased destruction of 9-mers with ERAP1-susceptible residues, indicating higher activity of the E730 variant for these peptides. The opposite situation was found for peptides ≥10-mers, where the frequency of ERAP1-resistant residues was higher in LCL 6370 (Q730). This result confirms that the Q730E change in ERAP1 leads to higher activity toward 9-mers and lower trimming of longer peptides, demonstrating the significant influence of this polymorphism on the length of HLA-B*27 ligands in live cells.
Fig. 8.
Length-dependent effects of ERAP1 polymorphism on P1 residue usage. Percentage of P1 residues of 9-mers (left panels) and ≥10-mers (right panels) from the IR>1.0 subsets with low, intermediate and high susceptibility to ERAP1. A, A total of 1431 (from LCL 6370: Black bars) and 1616 (from LG2: White bars) 9-mers, and 1431 and 813 peptides ≥10-mers, respectively, were compared. B, A total of 1443 (LG2: Black bars) and 1417 (C1R05: White bars) 9-mers, and 761 and 1380 peptides ≥10-mers, respectively, were included. C, A total of 1720 (LG2: Black bars) and 1117 (P50: White bars) 9-mers, and 786 and 1413 peptides ≥10-mers, respectively, were included. D, A total of 1432 (LCL 6370: Black bars) and 1543 (C1R05: White bars) 9-mers, and 1024 and 1522 peptides ≥10-mers, respectively, were included. E, A total of 1689 (LCL 6370: Black bars) and 1251 (P50: White bars) 9-mers, and 1079 and 1504 peptides ≥10-mers, respectively, were included. When the differences reached statistical significance, this was indicated by their p values.
In LG2/C1R05 (Fig. 8B) and LG2/P50 (Fig. 8C), all concordant in E730, the frequency of ERAP1-resistant P1 residues was much higher among 9-mers than among longer peptides. This can be explained by the concurrence of two effects adding to each other in both comparisons. The first one is that the lower activity of ERAP1 because of the K528R and other polymorphisms in C1R05 and P50 becomes more critical for 9-mers, because they are close to the lowest substrate length suitable for this enzyme. The second effect is that E730 in ERAP1 confers higher trimming efficiency for 9-mers, relative to longer peptides.
The influence of the Q730E polymorphism on the effects of K528R became apparent in the LCL 6370/C1R05 and 6370/P50 comparisons (Fig. 8D–8E). In both cases the frequencies of ERAP1-resistant P1 residues in 9-mers and peptides ≥10-mers were similarly increased in LCL 6370 (K528/Q730), relative to C1R05 or P50 (both with R528/E730). This can be explained by the fact that K528 confers higher trimming than R528, regardless of peptide size. However, Q730 confers lower trimming than E730 for 9-mers and higher trimming of longer ones, so that the activity differences between K528/Q730 and R528/E730 are larger for peptides ≥10-mers, resulting in similar effects on P1 residue frequencies regardless of peptide length.
These length-dependent effects of Q730E resulted in a distinct influence on affinity when 9-mers or peptides ≥10-mers were separately considered. The effect was most dramatic upon concordance at other positions, as in the 6370/LG2 comparison (supplemental Fig. S9): the affinity of 9-mers was higher for LG2 (E730) and that of peptides ≥10-mers was higher for LCL 6370 (Q730).
Influence of ERAP1 Polymorphism on the Sequence of B*27:05 Ligands Downstream the N Terminus
The residue frequencies of B*27:05 9-mers in the IR>1.0 were analyzed in all pairwise comparisons. Statistically significant differences were found at most peptide positions with a pattern that was different in each cell line pair (Table III). These results indicate that ERAP1 polymorphism shapes the HLA-B*27 peptidome not only by differential trimming of P(-1) and P1 residues, but also by favoring or disfavoring peptides with distinct internal sequences.
Table III. Statistically significant differences in residue usage at all peptide positions among shared nonamers in the IR>1.0 subsets in the indicated LCL comparisonsa.
| LCL | P1 | P2 | P3 | P4 | P5 | P6 | P7 | P8 | P9 |
|---|---|---|---|---|---|---|---|---|---|
| 6370/LG2 | A | F | G,L | K | |||||
| LG2/C1R05 | A,E,N,S,K,R | D,F,M | P | P,L | L | V | |||
| 6370/C1R05 | E,N,S,K,R | D,E | P,K | D,E,G,I | R | K,L,V,M | |||
| C1R05/P50 | R,F,L | D | P | I,D,E,W | A | K | |||
| 6370/P50 | A,S,F,K,L,R | A | V | K | |||||
| LG2/P50 | A,S,F,K,R | A,F | K,N,L | K | K | K |
a Residues underlined or not are those that are increased in the correspondingly coded LCL.
Peptides Selectively Found in Individual Cell Lines
Peptides selectively found in only one cell line in pairwise comparisons ranged about 13% to 30% percent of the total peptides in that cell line (supplemental Table S2). These peptides showed differences both in length and frequency of P1 residues with distinct susceptibility to ERAP1 that were generally very similar as those observed when the shared peptides predominant in the same cells lines (IR>1.0) were compared (supplemental Fig. S10).
It is important to note that in our MS analyses failure to detect a peptide in a given pool does not mean that the peptide is absent. Thus, these subsets encompass: (1) truly cell-specific ligands, including peptides specifically generated in one particular ERAP1 context, (2) occasional contaminating ligands of non-B*27 MHC-I molecules not shared between the cell lines compared, (3) peptides that are expressed in very low amounts or were fortuitously undetected in one of the cell lines. Except for C1R05, from which many more peptides were identified, the percentage of peptides found only in one of the four cell lines compared was small: 2.3% (LG2), and 4.7% (LCL 6370 and P50). These numbers might reflect somewhat better the maximal percentages of epitopes specifically generated in a given ERAP1context. Yet, for the reasons explained above, the actual number of ERAP1 haplotype-specific peptides is probably much lower.
DISCUSSION
This study provides a comprehensive characterization of the effects of ERAP1 polymorphism on the B*27:05 peptidome in live cells, effects which are presumably at the basis of the epistatic association of both molecules with AS. The main features defining the association of ERAP1 with this disease can be summarized as follows: (1) one or more of the Hap1-Hap3 haplotypes predispose to AS whereas Hap10 is protective (14), (2) the K528R polymorphism is a major factor influencing the association, with K528 being the risk variant (16), (3) a secondary factor influencing AS risk is the presence of D575/R725 (16), but this may actually be because of a linked polymorphism influencing protein levels (19), (4) on the basis of fine-mapping studies, it seems plausible that the Q730E polymorphism might not be directly associated with AS (21). Thus, it is important to address the correspondence between the genetic contribution of ERAP1 polymorphisms to AS and their biochemical effects because such correspondence will highlight the changes in the peptidome that may be more relevant to the pathological role of HLA-B*27.
Previous studies both in vitro (16, 27–29) and on live cells (26) have established a major effect of the K528R polymorphism on ERAP1 activity and on the HLA-B*27 peptidome, with the protective R528 variant leading to decreased activity. Although the D575N/R725Q changes lead to lower activity in vitro (24), a previous study in live cells failed to detect any significant effect (26). Concerning Q730E, in vitro studies with poly-Gly peptides revealed that the Q730 variant showed higher activity than E730 for 10-mers and longer substrates, and lower activity for 9-mers and shorter peptides. This polymorphism influenced the surface expression of free HLA-B*27 heavy chains on monocytes in one study (48), but its effects on MHC-I-bound peptidomes in live cells has not been previously explored.
We addressed two main issues: (1) to distinguish and rank the effects of each of the Q730E, K528R, and D575N/R725Q polymorphisms on the B*27:05 peptidome, and (2) to characterize their joint effects in the risk (Hap2, Hap3) and protective (Hap10) haplotypes.
Our study was mostly focused on the quantitative differences among shared HLA-B*27 ligands expressed in distinct ERAP1 contexts for two reasons. The first one is the logical assumption that the same rules determining quantitative differences in the amounts of certain ligands as a function of the ERAP1 polymorphism should apply, in the extreme, to epitopes that are presented in one context but not in other. The second reason is that in our analyses, failure to detect a peptide in a given pool does not necessarily mean that the peptide is absent. Therefore, we cannot determine how many peptides are specifically generated or destroyed in a given context. Our study was not designed for that purpose, but to address the effects of ERAP1 variants on the amount of peptides with given features, resulting from changes in their generation/destruction balance.
The ERAP1 polymorphisms analyzed influenced the B*27:05 peptidome at five levels: peptide length, frequency of P(-1), P1, and internal residues, and peptide affinity. In general, the effects on P(-1) residues were smaller than on P1 suggesting that, for N-terminal residues with equal susceptibility to trimming, ERAP1 polymorphism has more influence on modulating epitope destruction than epitope generation. Nonamers are generally less efficiently trimmed than longer peptides, because they are close to the lowest substrate length handled by this enzyme (13). Thus, activity differences among ERAP1 variants are likely to affect more drastically the relative trimming of distinct N-terminal residues of natural B*27:05 ligands, particularly 9-mers, than those of their precursors. The altered frequencies of internal residues may reflect a direct influence of ERAP1 polymorphism on substrate handling because binding and trimming by ERAP1is influenced by the internal sequence of the substrate (49), be a compensatory effect for the changes in binding affinity to B*27:05 induced by P1 alterations, or both.
Key to understanding the effects described in this study was that the Q730E polymorphism alone induced distinct alterations depending on peptide length, with the E730 variant leading to a substantially increased abundance of 9-mers. This observation is fully consistent with the activity pattern of the Q730 and E730 variants defined in vitro (47) and, for the first time to our knowledge, demonstrates that this pattern has general significance and large effects in a natural peptidome processed in live cells. This mutation had moderate effects on P1 residue usage and affinity on the whole of peptidome, but this was because of the fact that such effects were opposite for 9-mers and peptides ≥10-mers, again reflecting the distinct length-dependent activity of these variants.
The effects of K528R, although comparable in magnitude to those of E730Q on peptide length, were substantially larger on all other features examined, reflecting the higher activity of the AS-associated K528 variant. It also revealed similar trends for peptides with different lengths, again in agreement with studies using poly-Gly analogs (47). In our previous study (26), carried out at lower resolution and with much fewer peptides identified, the effects of K528R on P1 residue usage were detected, but those on P(-1) and peptide length went unnoticed.
The molecular bases for the distinct effects of the Q730E and K528R changes have been analyzed in detail by Stratikos and colleagues (47, 50). Residue 528 is in an interdomain region of ERAP1 and has a major effect on the dynamics of the conformational transitions driving the acquisition of enzymatic activity. In contrast, residue 730 is in the substrate-binding site, has less influence on the conformational dynamics of the molecule and a more prominent effect on substrate binding.
Concerning D575N/R725Q, its effects could be better assessed by comparing the results in LG2/P50, where these changes co-occurred with K528R, and LG2/C1R05, with the K528R mismatch and concordance at residues 575 and 725 (Table I). These comparisons demonstrated that D575N/R725Q negatively influenced the enzymatic activity of ERAP1, adding to the substantially higher effect of K528R.
The length-dependent effects of the Q730E polymorphism on the B*27:05 peptidome imply that the influence of all other changes analyzed in the pairwise comparisons must take into account the concordance or discordance at residue 730. ERAP1 activity is E>Q730 for peptides ≤9-mers and E<Q730 for longer peptides, but K>R528 independently of peptide length. Thus, the following activity ranking of ERAP1 variants can be established: KE>KQ>RE>RQ for peptides ≤9-mers, and KQ>KE>RQ>RE for longer peptides. In LG2/C1R05 and LG2/P50 (KE versus RE in both cases), the effect of E730 on favoring trimming of 9-mers more than longer peptides implies that the higher activity of K528 in LG2 will be further increased by E730 for 9-mers more than for peptides ≥10-mers, as was actually observed in the much higher effects on P1 residue usage for 9-mers in both comparisons. Discordance at residue 730 in 6370/C1R and 6370/P50 (KQ versus RE in both) implies that Q730E will increase the differences in enzymatic activity and trimming for peptides ≥10-mers (KQ≫RE) but will attenuate this effect on 9-mers and shorter peptides (KQ>RE). As a result, the differences in P1 residue usage were similar for 9-mers and longer peptides in both comparisons. This contrasting effect was also reflected on peptide length: the increase of 9-mers was substantially higher in LG2/P50 and LG2/C1R than in 6370/P50 and 6370/C1R05, respectively (Table II).
The combined influence of K528R and Q730E on the B*27:05 peptidome is consistent with enzymological studies suggesting that the effects of both mutations are independent and additive (47). Our results reflect the full dimension of these combined effects on a highly complex MHC-I-bound peptidome expressed on live cells.
There is a general concordance between the biochemical and genetic data concerning the role of functional ERAP1 polymorphism. This applies to the major role of K528R on the genetic risk of ERAP1 in AS, on enzymatic activity and on the B*27:05 peptidome. The secondary role of D575N/R725Q both on AS risk and on the B*27:05 peptidome adds to this concordance, although in this case the genetic influence of this polymorphism, as tagged by the rs10050860 SNP (D575N), may be determined by another polymorphism in tight linkage disequilibrium, in particular the splice-altering SNP rs7063 (19).
The relationship between the role of Q730E in AS and the biochemical effects of this polymorphism is more difficult to assess. Its substantial influence on the B*27:05 peptidome, suggests that the Q730E change may directly influence AS susceptibility. Yet, unlike other polymorphisms in which the protective alleles are “loss-of function” variants leading to decreased enzymatic activity, the protective E730 allotype shows higher activity than the risk counterpart Q730 for 9-mers, which account for most of the HLA-B*27 peptidome. Indeed, the patterns of peptide length distribution and P1 residue usage among 9-mers were very similar for the risk variant K528 and the protective E730. This functional paradox might be resolved if the AS protective effect of Q730E suggested by earlier studies (15, 20) would be because of linkage disequilibrium with the neighbor AS-associated region defined by more recent fine-mapping (21), which does not include rs27044 (Q730E). Studies on haplotype associations, reviewed in (14), established the protective role of Hap10 (with E730) and suggested that Hap1-Hap3 predispose to AS. These haplotypes are discordant at residue 730: Q730 in Hap1 and Hap2, E730 in Hap3. To our knowledge, no genetic studies involving the association of whole ERAP1 haplotypes with AS have specifically addressed whether Hap2 and Hap3, discordant only at Q730E, differ in the strength of their association with AS.
In conclusion, the epistatic association of ERAP1 with AS can be rationalized based on a complex and significant influence of the enzymatic polymorphism on the HLA-B*27 peptidome. This influence affects multiple peptide features, altering the generation/destruction balance of a large fraction of the peptidome and its affinity. Distinct polymorphic residues act differently on each of these features and their contribution seems to be, to a large extent, additive. Thus, the differential effects of AS-predisposing and AS-protective haplotypes on the B*27:05 peptidome is the combined result of the distinct influence, both quantitative and qualitative, of the polymorphisms involved. The magnitude of this changes can profoundly affect antigen presentation and, presumably, other pathogenetic features of HLA-B*27, such as folding and cell surface stability, providing a peptide-mediated basis for the joint association of ERAP1 and HLA-B*27 with AS.
In at least two other MHC-I associated inflammatory disorders, psoriasis and Behçet′s disease, ERAP1 is associated with disease in epistasis with the risk MHC-I alleles, C*06:02 and B*51:01, respectively (17, 18). Like in AS, The risk ERAP1 haplotypes in psoriasis seem to be Hap1–3, with Hap2 being an important risk factor at least among East Asians, and Hap10 being protective (14). The features of the C*06:02 peptidome were recently determined (51). Similarly to HLA-B*27, Arg2 was a major motif, whereas aromatic residues were predominant at P1. To our knowledge the influence of ERAP1 on the C*06:02 peptidome has not yet been experimentally addressed. Yet, compared with HLA-B*27, the distinct susceptibility of the preferred P1 residues to ERAP1 may imply large differences in the over-trimming and destruction of C*06:02 ligands, depending on these residues and the enzymatic activity of ERAP1 variants. In contrast to AS and psoriasis, the low activity Hap10 variant of ERAP1 is the risk factor for Behçet's disease (52). The peptidome of B*51, an allotype strongly associated with this disease, consists mainly of two subpeptidomes, with Pro2 and Ala2 respectively, which differ in their susceptibility to ERAP1 and in their binding affinity (46). The main effect of Hap10 on this peptidome is to upset the relative amounts of both subpeptidomes, resulting in a peptidome of globally lower affinity, which is predicted to alter NK recognition of HLA-B*51 (53). Thus, although detailed studies, such as those reported here for HLA-B*27, have not yet been performed on other disease-associated MHC-I allotypes, ERAP1 polymorphism can significantly alter the nature of their peptidomes, changing in this way their immunological features and providing a sound basis for the epistatic association of ERAP1 and MHC-I with distinct inflammatory diseases.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (http://www.ebi.ac.uk/pride) with the dataset identifier PXD008500.
Supplementary Material
Footnotes
* This work is supported by grants SAF2014/51931-R (Plan Nacional de I+D+i) to JALC, Israel Science Foundation grant 1435/16 to AA, and an institutional grant of the Fundación Ramón Areces to the Centro de Biología Molecular Severo Ochoa.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- MHC-I
- Major Histocompatibility Complex class I
- AS
- ankylosing spondylitis
- E:S
- enzyme/substrate
- IR
- intensity ratio
- L-AMC
- Leu-7-amido-4-methyl-coumarine
- mAb
- monoclonal antibody
- P(-1)
- N-terminal flanking residue
- P1
- N-terminal residue
- SNP
- single nucleotide polymorphism.
REFERENCES
- 1. Saric T., Chang S. C., Hattori A., York I. A., Markant S., Rock K. L., Tsujimoto M., and Goldberg A. L. (2002) An IFN-gamma-induced aminopeptidase in the ER, ERAP1, trims precursors to MHC class I-presented peptides. Nat. Immunol. 3, 1169–1176 [DOI] [PubMed] [Google Scholar]
- 2. Serwold T., Gonzalez F., Kim J., Jacob R., and Shastri N. (2002) ERAAP customizes peptides for MHC class I molecules in the endoplasmic reticulum. Nature 419, 480–483 [DOI] [PubMed] [Google Scholar]
- 3. Hammer G. E., Gonzalez F., Champsaur M., Cado D., and Shastri N. (2006) The aminopeptidase ERAAP shapes the peptide repertoire displayed by major histocompatibility complex class I molecules. Nat. Immunol. 7, 103–112 [DOI] [PubMed] [Google Scholar]
- 4. Hammer G. E., Gonzalez F., James E., Nolla H., and Shastri N. (2007) In the absence of aminopeptidase ERAAP, MHC class I molecules present many unstable and highly immunogenic peptides. Nat. Immunol. 8, 101–108 [DOI] [PubMed] [Google Scholar]
- 5. Yan J., Parekh V. V., Mendez-Fernandez Y., Olivares-Villagomez D., Dragovic S., Hill T., Roopenian D. C., Joyce S., and Van Kaer L. (2006) In vivo role of ER-associated peptidase activity in tailoring peptides for presentation by MHC class Ia and class Ib molecules. J. Exp. Med. 203, 647–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blanchard N., Kanaseki T., Escobar H., Delebecque F., Nagarajan N. A., Reyes-Vargas E., Crockett D. K., Raulet D. H., Delgado J. C., and Shastri N. (2010) Endoplasmic reticulum aminopeptidase associated with antigen processing defines the composition and structure of MHC class I peptide repertoire in normal and virus-infected cells. J. Immunol. 184, 3033–3042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen L., Fischer R., Peng Y., Reeves E., McHugh K., Ternette N., Hanke T., Dong T., Elliott T., Shastri N., Kollnberger S., James E., Kessler B., and Bowness P. (2014) Critical Role of Endoplasmic Reticulum Aminopeptidase 1 in Determining the Length and Sequence of Peptides Bound and Presented by HLA-B27. Arthritis Rheumatol. 66, 284–294 [DOI] [PubMed] [Google Scholar]
- 8. Barnea E., Melamed K. D., Haimovich Y., Satumtira N., Dorris M. L., Nguyen M. T., Hammer R. E., Tran T. M., Colbert R. A., Taurog J. D., and Admon A. (2017) The HLA-B27 peptidome in vivo in spondyloarthritis-susceptible HLA-B27 transgenic rats and the effect of ERAP1 deletion. Mol. Cell Proteomics 16, 642–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Serwold T., Gaw S., and Shastri N. (2001) ER aminopeptidases generate a unique pool of peptides for MHC class I molecules. Nat. Immunol. 2, 644–651 [DOI] [PubMed] [Google Scholar]
- 10. Saveanu L., Carroll O., Lindo V., Del Val M., Lopez D., Lepelletier Y., Greer F., Schomburg L., Fruci D., Niedermann G., and Van Endert P. M. (2005) Concerted peptide trimming by human ERAP1 and ERAP2 aminopeptidase complexes in the endoplasmic reticulum. Nat. Immunol. 6, 689–697 [DOI] [PubMed] [Google Scholar]
- 11. Goto Y., Tanji H., Hattori A., and Tsujimoto M. (2008) Glutamine-181 is crucial in the enzymatic activity and substrate specificity of human endoplasmic-reticulum aminopeptidase-1. Biochem. J. 416, 109–116 [DOI] [PubMed] [Google Scholar]
- 12. Hearn A., York I. A., and Rock K. L. (2009) The specificity of trimming of MHC class I-presented peptides in the endoplasmic reticulum. J. Immunol. 183, 5526–5536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chang S. C., Momburg F., Bhutani N., and Goldberg A. L. (2005) The ER aminopeptidase, ERAP1, trims precursors to lengths of MHC class I peptides by a “molecular ruler” mechanism. Proc. Natl. Acad. Sci. U.S.A. 102, 17107–17112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ombrello M. J., Kastner D. L., and Remmers E. F. (2015) Endoplasmic reticulum-associated amino-peptidase 1 and rheumatic disease: genetics. Curr. Opin. Rheumatol. 27, 349–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. WTCC Consortium. (2007) Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat. Genet. 39, 1329–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. The TASK and WTCCC2 Consortia. (2011) Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat. Genet. 43, 761–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Strange A., Capon F., Spencer C. C., Knight J., Weale M. E., Allen M. H., Barton A., Band G., Bellenguez C., Bergboer J. G., Blackwell J. M., Bramon E., Bumpstead S. J., Casas J. P., Cork M. J., Corvin A., Deloukas P., Dilthey A., Duncanson A., Edkins S., Estivill X., Fitzgerald O., Freeman C., Giardina E., Gray E., Hofer A., Huffmeier U., Hunt S. E., Irvine A. D., Jankowski J., Kirby B., Langford C., Lascorz J., Leman J., Leslie S., Mallbris L., Markus H. S., Mathew C. G., McLean W. H., McManus R., Mossner R., Moutsianas L., Naluai A. T., Nestle F. O., Novelli G., Onoufriadis A., Palmer C. N., Perricone C., Pirinen M., Plomin R., Potter S. C., Pujol R. M., Rautanen A., Riveira-Munoz E., Ryan A. W., Salmhofer W., Samuelsson L., Sawcer S. J., Schalkwijk J., Smith C. H., Stahle M., Su Z., Tazi-Ahnini R., Traupe H., Viswanathan A. C., Warren R. B., Weger W., Wolk K., Wood N., Worthington J., Young H. S., Zeeuwen P. L., Hayday A., Burden A. D., Griffiths C. E., Kere J., Reis A., McVean G., Evans D. M., Brown M. A., Barker J. N., Peltonen L., Donnelly P., and Trembath R. C. (2010) A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat. Genet. 42, 985–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kirino Y., Bertsias G., Ishigatsubo Y., Mizuki N., Tugal-Tutkun I., Seyahi E., Ozyazgan Y., Sacli F. S., Erer B., Inoko H., Emrence Z., Cakar A., Abaci N., Ustek D., Satorius C., Ueda A., Takeno M., Kim Y., Wood G. M., Ombrello M. J., Meguro A., Gul A., Remmers E. F., and Kastner D. L. (2013) Genome-wide association analysis identifies new susceptibility loci for Behcet's disease and epistasis between HLA-B*51 and ERAP1. Nat. Genet. 45, 202–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hanson A. L., Cuddihy T., Haynes K., Loo D., Morton C. J., Oppermann U., Leo P., Thomas G. P., Le Cao K. A., Kenna T. J., and Brown M. A. (2018) Genetic variants in ERAP1 and ERAP2 associated with immune-mediated diseases influence protein expression and isoform profile. Arthritis Rheumatol. 70, 255–265 [DOI] [PubMed] [Google Scholar]
- 20. Harvey D., Pointon J. J., Evans D. M., Karaderi T., Farrar C., Appleton L. H., Sturrock R. D., Stone M. A., Oppermann U., Brown M. A., and Wordsworth B. P. (2009) Investigating the genetic association between ERAP1 and ankylosing spondylitis. Hum. Mol. Genet. 18, 4204–4212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. van de Bunt M., Cortes A., IGAS Consortium, Brown M. A., Morris A. P., and McCarthy M. I. (2015) Evaluating the Performance of Fine-Mapping Strategies at Common Variant GWAS Loci. PLoS. Genet. 11, e1005535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lopez de Castro J. A., Alvarez-Navarro C., Brito A., Guasp P., Martin-Esteban A., and Sanz-Bravo A. (2016) Molecular and pathogenic effects of endoplasmic reticulum aminopeptidases ERAP1 and ERAP2 in MHC-I-associated inflammatory disorders: Towards a unifying view. Mol. Immunol. 77, 193–204 [DOI] [PubMed] [Google Scholar]
- 23. Martin-Esteban A., Gomez-Molina P., Sanz-Bravo A., and Lopez de Castro J. A. (2014) Combined Effects of Ankylosing Spondylitis-associated ERAP1 Polymorphisms Outside the Catalytic and Peptide-binding Sites on the Processing of Natural HLA-B27 Ligands. J. Biol. Chem. 289, 3978–3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Reeves E., Edwards C. J., Elliott T., and James E. (2013) Naturally Occurring ERAP1 Haplotypes Encode Functionally Distinct Alleles with Fine Substrate Specificity. J. Immunol. 191, 35–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Garcia-Medel N., Sanz-Bravo A., Van Nguyen D., Galocha B., Gomez-Molina P., Martin-Esteban A., Alvarez-Navarro C., and López de Castro J. A. (2012) Functional Interaction of the Ankylosing Spondylitis-associated Endoplasmic Reticulum Aminopeptidase 1 Polymorphism and HLA-B27 in Vivo. Mol. Cell. Proteomics. 11, 1416–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sanz-Bravo A., Campos J., Mazariegos M. S., and Lopez de Castro J. A. (2015) Dominant Role of the ERAP1 Polymorphism R528K in Shaping the HLA-B27 Peptidome Through Differential Processing Determined by Multiple Peptide Residues. Arthritis Rheumatol. 67, 692–701 [DOI] [PubMed] [Google Scholar]
- 27. Goto Y., Hattori A., Ishii Y., and Tsujimoto M. (2006) Reduced activity of the hypertension-associated Lys528Arg mutant of human adipocyte-derived leucine aminopeptidase (A-LAP)/ER-aminopeptidase-1. FEBS Lett. 580, 1833–1838 [DOI] [PubMed] [Google Scholar]
- 28. Evnouchidou I., Kamal R. P., Seregin S. S., Goto Y., Tsujimoto M., Hattori A., Voulgari P. V., Drosos A. A., Amalfitano A., York I. A., and Stratikos E. (2011) Cutting Edge: Coding single nucleotide polymorphisms of endoplasmic reticulum aminopeptidase 1 can affect antigenic peptide generation in vitro by influencing basic enzymatic properties of the enzyme. J. Immunol. 186, 1909–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kochan G., Krojer T., Harvey D., Fischer R., Chen L., Vollmar M., von Delft F., Kavanagh K. L., Brown M. A., Bowness P., Wordsworth P., Kessler B. M., and Oppermann U. (2011) Crystal structures of the endoplasmic reticulum aminopeptidase-1 (ERAP1) reveal the molecular basis for N-terminal peptide trimming. Proc. Natl. Acad. Sci. U.S.A. 108, 7745–7750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lorente E., Barriga A., Johnstone C., Mir C., Jimenez M., and Lopez D. (2013) Concerted in vitro trimming of viral HLA-B27-restricted ligands by human ERAP1 and ERAP2 aminopeptidases. PLoS. ONE. 8, e79596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Evnouchidou I., Weimershaus M., Saveanu L., and van Endert P. (2014) ERAP1-ERAP2 Dimerization Increases Peptide-Trimming Efficiency. J. Immunol. 193, 901–908 [DOI] [PubMed] [Google Scholar]
- 32. Chen H., Li L., Weimershaus M., Evnouchidou I., van Endert P., and Bouvier M. (2016) ERAP1-ERAP2 dimers trim MHC I-bound precursor peptides; implications for understanding peptide editing. Sci. Rep. 6, 28902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Andres A. M., Dennis M. Y., Kretzschmar W. W., Cannons J. L., Lee-Lin S. Q., Hurle B., Schwartzberg P. L., Williamson S. H., Bustamante C. D., Nielsen R., Clark A. G., and Green E. D. (2010) Balancing selection maintains a form of ERAP2 that undergoes nonsense-mediated decay and affects antigen presentation. PLoS. Genet. 6, e1001157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Martin-Esteban A., Guasp P., Barnea E., Admon A., and Lopez de Castro J. A. (2016) Functional Interaction of the Ankylosing Spondylitis Associated Endoplasmic Reticulum Aminopeptidase 2 with the HLA-B*27 Peptidome in Human Cells. Arthritis Rheumatol. 68, 2466–2475 [DOI] [PubMed] [Google Scholar]
- 35. Martin-Esteban A., Sanz-Bravo A., Guasp P., Barnea E., Admon A., and Lopez de Castro J. A. (2017) Separate effects of the ankylosing spondylitis associated ERAP1 and ERAP2 aminopeptidases determine the influence of their combined phenotype on the HLA-B*27 peptidome. J. Autoimmun. 79, 28–38 [DOI] [PubMed] [Google Scholar]
- 36. Zemmour J., Little A. M., Schendel D. J., and Parham P. (1992) The HLA-A,B “negative” mutant cell line C1R expresses a novel HLA-B35 allele, which also has a point mutation in the translation initiation codon. J. Immunol. 148, 1941–1948 [PubMed] [Google Scholar]
- 37. Ellis S. A., Taylor C., and McMichael A. (1982) Recognition of HLA-B27 and related antigens by a monoclonal antibody. Hum. Immunol. 5, 49–59 [DOI] [PubMed] [Google Scholar]
- 38. Parham P., Antonelli P., Herzenberg L. A., Kipps T. J., Fuller A., and Ward F. E. (1986) Further studies on the epitopes of HLA-B7 defined by murine monoclonal antibodies. Hum. Immunol. 15, 44–67 [DOI] [PubMed] [Google Scholar]
- 39. Kapasi K., and Inman R. D. (1994) ME1 epitope of HLA-B27 confers class I-mediated modulation of gram- negative bacterial invasion. J. Immunol. 153, 833–840 [PubMed] [Google Scholar]
- 40. Barnstable C. J., Bodmer W. F., Brown G., Galfre G., Milstein C., Williams A. F., and Ziegler A. (1978) Production of monoclonal antibodies to group A erythrocytes, HLA and other human cell surface antigens. New tools for genetic analysis. Cell 14, 9–20 [DOI] [PubMed] [Google Scholar]
- 41. Ishihama Y., Rappsilber J., Andersen J. S., and Mann M. (2002) Microcolumns with self-assembled particle frits for proteomics. J. Chromatogr. A 979, 233–239 [DOI] [PubMed] [Google Scholar]
- 42. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 43. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome. Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 44. Karosiene E., Lundegaard C., Lund O., and Nielsen M. (2012) NetMHCcons: a consensus method for the major histocompatibility complex class I predictions. Immunogenetics 64, 177–186 [DOI] [PubMed] [Google Scholar]
- 45. Costantino F., Talpin A., Evnouchidou I., Kadi A., Leboime A., Said-Nahal R., Bonilla N., Letourneur F., Leturcq T., Ka Z., van Endert P., Garchon H. J., Chiocchia G., and Breban M. (2015) ERAP1 gene expression is influenced by non-synonymous polymorphisms associated with predisposition to spondyloarthritis. Arthritis Rheumatol. 67, 1525–1534 [DOI] [PubMed] [Google Scholar]
- 46. Guasp P., Alvarez-Navarro C., Gomez-Molina P., Martin-Esteban A., Marcilla M., Barnea E., Admon A., and López de Castro J. A. (2016) The peptidome of the Behcet's disease-associated HLA-B*51:01 includes two sub-peptidomes differentially shaped by ERAP1. Arthritis Rheumatol. 68, 505–515 [DOI] [PubMed] [Google Scholar]
- 47. Stamogiannos A., Koumantou D., Papakyriakou A., and Stratikos E. (2015) Effects of polymorphic variation on the mechanism of Endoplasmic Reticulum Aminopeptidase 1. Mol. Immunol. 67, 426–435 [DOI] [PubMed] [Google Scholar]
- 48. Haroon N., Tsui F. W., Uchanska-Ziegler B., Ziegler A., and Inman R. D. (2012) Endoplasmic reticulum aminopeptidase 1 (ERAP1) exhibits functionally significant interaction with HLA-B27 and relates to subtype specificity in ankylosing spondylitis. Ann. Rheum. Dis. 71, 589–595 [DOI] [PubMed] [Google Scholar]
- 49. Evnouchidou I., Momburg F., Papakyriakou A., Chroni A., Leondiadis L., Chang S. C., Goldberg A. L., and Stratikos E. (2008) The internal sequence of the peptide-substrate determines its N-terminus trimming by ERAP1. PLoS. ONE 3, e3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Papakyriakou A., and Stratikos E. (2017) The Role of Conformational Dynamics in Antigen Trimming by Intracellular Aminopeptidases. Front. Immunol. 8, doi: 10.3389/fimmu.2017.00946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mobbs J. I., Illing P. T., Dudek N. L., Brooks A. G., Baker D. G., Purcell A. W., Rossjohn J., and Vivian J. P. (2017) The molecular basis for peptide repertoire selection in the human leucocyte antigen (HLA) C*06:02 molecule. J. Biol. Chem. 292, 17203–17215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Takeuchi M., Ombrello M. J., Kirino Y., Erer B., Tugal-Tutkun I., Seyahi E., Ozyazgan Y., Watts N. R., Gul A., Kastner D. L., and Remmers E. F. (2017) A single endoplasmic reticulum aminopeptidase-1 protein allotype is a strong risk factor for Behcet's disease in HLA-B*51 carriers. Ann. Rheum. Dis. 75, 2208–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Guasp P., Barnea E., Gonzalez-Escribano M. F., Jimenez-Reinoso A., Regueiro J. R., Admon A., and Lopez de Castro J. A. (2017) The Behcet's disease-associated variant of the aminopeptidase ERAP1 shapes a low affinity HLA-B*51 peptidome by differential subpeptidome processing. J. Biol. Chem. 292, 9680–9689 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (http://www.ebi.ac.uk/pride) with the dataset identifier PXD008500.