Abstract
Here we present a free interactive web tool to process and visualize proteomics data sets with a single click. GiaPronto can process all proteomics quantification methods, i.e. label-free, SILAC and isobaric labeling, and analyze post-translational modifications (PTMs). The software performs normalization and statistics, assists determination of regulated proteins, biomarkers and Gene Ontology (GO) enrichment, and provides high resolution images and tables for further data analysis. We foresee that GiaPronto will become the most rapid and simple tool for assessing data quality and determining most relevant features of proteomic data sets. GiaPronto is available at giapronto.diskinlab.org.
Keywords: Algorithms, Data standards, Bioinformatics software, SILAC, Label-free quantification, Data Analysis, Data Visualization
Proteomics has become a widely popular discipline for the analysis of biological models and their perturbations. The high sensitivity, accuracy and versatility of mass spectrometry in both identifying and quantifying protein samples and characterizing protein post-translational modifications (PTMs) 1 has evolved proteomics in multiple sub-disciplines. This includes multiple strategies such as peptide centric (bottom-up) and protein centric (top-down) analysis (e.g. for histone analysis (1)); multiple quantification techniques (2), including label-based and label-free; and multiple software tools, including commercial and freely available.
Even though MS based proteomics is versatile, most projects are peptide centric, i.e. bottom-up, because of its relatively simpler workflow, higher sensitivity and higher robustness than the other strategies. The quantification method is not as uniform, as the convenience of one method or the other may vary upon type of sample, number of samples and availability of reagents. For instance, in case of large numbers of samples/replicates labeling techniques are preferred, to mix multiple conditions and reduce the number of LC-MS runs. On the other hand, label-free strategies do not require additional labeling steps and maintain a more linear dynamic range of quantifications (3).
We present GiaPronto, a software developed in Shiny (4) that uses a protein and/or a PTM table as input file to process data and plot a variety of common and less common graphs for the analysis of proteomics data sets. As compared with existing software that provide excellent support for proteomics data analysis, e.g. Perseus (5), GiaPronto is fully automated and provides both quality control and biological interpretation of protein and PTM level data. We foresee GiaPronto will be an appreciated software tool for proteomic data analysis and visualization, mostly because of its simplicity and speed. With this, we aim to address also scientists not specialized in proteomics, who want to incorporate proteomic analysis into their research programs.
EXPERIMENTAL PROCEDURES
Software Design
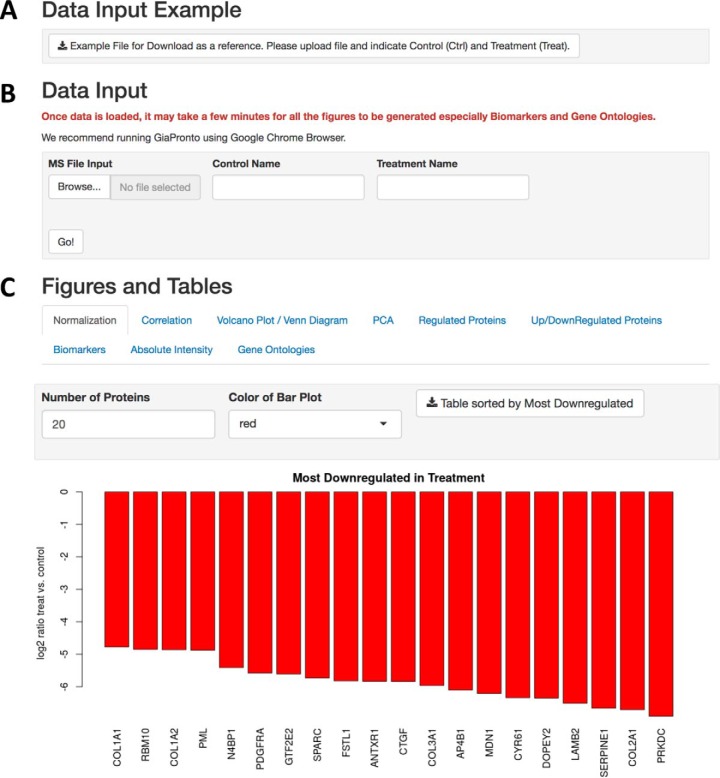
GiaPronto is written in R and the user interface was developed in Shiny. This software is housed on the Amazon Web Server at the Children's Hospital of Philadelphia. The interface provides an example file (Fig. 1A) and was designed using tabs for each customizable figure and legend. The user can indicate the control and treatment conditions for their analysis (Fig. 1B) The figures can be customized based on the p value, number of displayed items and color scheme by the user as indicated (Fig. 1C).
Fig. 1.
Graphical User Interface. A, GiaPronto provides an example file to aid the user in formatting their data for analysis. B, In data input section, the user can specify the control and treatment sample names will be used to generate the graphs, figure legends and Tables in the output. C, The output is partitioned into tabs and each figure is completely interactive where the user can customize the parameters.
Data Analysis
GiaPronto performs normalization if log transformed raw values are normally distributed (common assumption in proteomics analyses). Raw protein/PTM intensities are log2 transformed and normalized by subtracting the average. In case contaminant proteins are specified in the input table, as the MaxQuant output provides, the software will take them into consideration for the normalization, because they contributed to the total protein amount, and then discard them for the actual data analysis. Statistics is assessed using t test.
Software Implementation
The software is optimized to input the raw output of the free and popular proteomics software MaxQuant (6), but any .txt file can be uploaded with very minor edits of the column headers. The software accepts all kinds of quantification methods, including label-free, metabolic labeling (e.g. SILAC) and chemical tags such as isobaric labeling, that are computed from the desired database search software. Moreover, it can perform analysis of both proteome and PTMs. For PTM level analysis, the user must upload a table containing protein (proteingroups.txt) and PTM (e.g. Phospho(STY).txt). The input file requires minimal information; in case of protein data only the UniProt accession number (Protein.ID), gene names (Gene.names) and the quantification values of the different samples is required. Quantification columns begin with iBAQ. followed by the sample name with the replicates indicated by an underscore followed by a number (iBAQ.Samp_1). For SILAC analysis, heavy and light samples must be indicated by iBAQ.H.Samp or iBAQ.L.Samp.
Example Data Set
The data set that will be used throughout this manuscript was obtained from Kulej et al. 2017 (7). This data set was used to assess the accuracy and quality of GiaPronto. To determine changes in protein expression in response to viral infection, human foreskin fibroblast (HFF) cells were infected with HSV-1 strain 17 syn+ and proteins were harvested at 3 and 15 h post infection (hpi). Cells were lysed in cold lysis buffer and digested using Lys-C for three hours. Following digestion, proteins were reduced with 10 mm DTT for one hour at room temperature, alkylated using 20 mm iodoacetamide in dark for 30 min and digested with trypsin (1:50 ratio enzyme to protein) for 12 h at room temperature. For phosphoproteomic analysis, peptides were enriched on titanium dioxide resin and unmodified peptides were washed from the beads. Phosphorylated and unmodified peptides were lyophilized and resuspended in 0.1% trifluroacetic acid in preparation for desalting. Phosphorylated peptides were desalted using R3 resin whereas unmodified peptides were desalted using C18. Samples were dried and resuspended in Buffer A with 0.1% formic acid. Samples were analyzed by Easy-nLC coupled to the Orbitrap Fusion Tribrid Mass Spectrometer (Thermo Scientific, San Jose, CA). Raw files were analyzed using MaxQuant version 1.5.2.8 with a 1% false discovery rate (FDR). The main search peptide tolerance was set to 4.5 ppm and the first search tolerance was set to 20 ppm. More specifically, the spectra were searched against the human and HSV viral proteome using Andromeda. Variable (e.g. methionine oxidation, N-term acetylation, phosphorylation) and fixed (carbamidomethyl cystine) modifications were indicated. Trypsin was designated as the digestive enzyme during database searching and two missed cleavages were permitted. Raw data, annotated spectra, databases and MaxQuant version were uploaded to ProteomeXchange (PXD005467).
RESULTS
Normalization
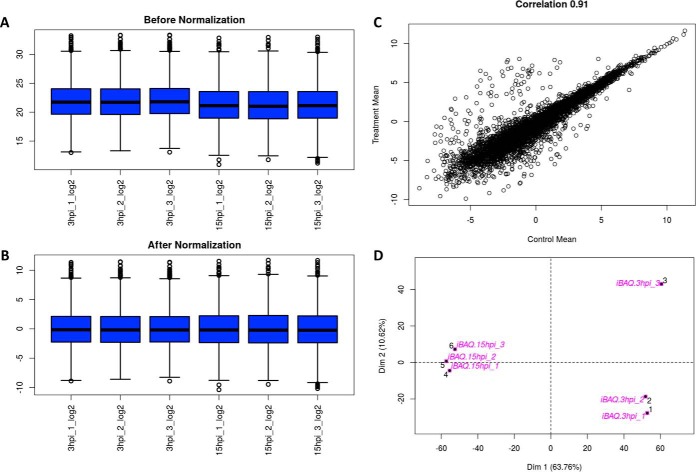
Once the Table is uploaded, GiaPronto automatically performs normalization to adjust MS runs for differences in injection amount and biases in sample preparation. Briefly, raw protein intensities are log2 transformed and normalized by the average of the distribution. Normalization is a common, albeit critical, step of the data processing; if injection biases are not corrected they can cause an inaccurate view of protein regulation and experimental conclusions.
For contaminant proteins, the software includes them for the normalization and then removes them from future analysis. In our example data set, GiaPronto displayed that slightly more material was injected for the 3 hpi samples compared with the 15 hpi (Fig. 2A). After each sample has been normalized by the mean of the distribution, all samples are centered around zero and can be compared with characterize changes in the proteome in response to viral infection (Fig. 2B). For SILAC labeling, each sample is normalized to the sample mean following a log2 transform. Then, the ratio for heavy to light is calculated. On the contrary, for isobaric tag analysis, the software computes first the ratio with the first reporter ion used as reference, and then it performs normalization.
Fig. 2.
Data Quality Assessment. A, GiaPronto generates data distribution figure prior to normalization. B, Then, our software performs normalization by computing the log2 ratio and adjusting to the mean of the distribution so all samples should be centered at zero. C, GiaPronto allows the user to assess correlation between treatment and control so samples that were treated or biologically similar should have a strong correlation. D, Lastly, our software display a principle component analysis plot where replicates of the same sample should cluster together.
For the analysis of PTMs, both the protein table and the table with modified sites must be uploaded. For instance, the MaxQuant output of phosphorylation sites is by default a .txt table named “Phospho(STY).txt”. Where possible, the PTM abundance gets normalized by the respective protein abundance, to accurately determine whether the changes in abundance of a PTM is because of the actual regulation of the modification or to the changes in abundance of the protein. Such “partial normalization” takes place only when the respective protein is quantified in every replicate. In case modified peptides were enriched and analyzed separately from the full proteome, e.g. in case of titanium dioxide (TiO2) enrichment for phosphorylation analysis (8), the protein table should be the LC-MS runs of the full proteome, i.e. the TiO2 flow through of not enriched peptides. Unfortunately, it is rather common that some quantified PTMs do not have the respective protein quantified in the flow-through. In these cases, PTMs that have a respective protein quantified in all runs are normalized, whereas those that do not remain unnormalized. This “partial normalization” is currently the best approach that avoids imputation of missing values, but still guarantees correction for changes in protein abundance where possible. In addition, the user can download tables with all normalized data for additional data analysis.
Quality Control
GiaPronto allows the user to assess the quality of their data. The software displays data distribution figures that can allow the user to determine difference in injection amount, which can allow poor quality runs to be identified. After normalization, we determine the correlation between the data conditions. The correlation value can be specified as pearson, spearman or kendall; if the value is close to 1, this implies that the two conditions are highly similar. The spearman correlation between two variables is equal to the Pearson correlation between the rank values of those two variables. A perfect correlation is when the most abundant protein in control is the most abundant in treatment, and so on for all proteins. The correlation does not necessarily need to be high, but it should be if the two analyzed samples are biologically similar. In our example, there is a strong correlation (R2 = 0.91) between the control (3 hpi) and treatment (15 hpi) with several proteins that have altered abundances in response to viral infection (Fig. 2C).
The Principal Component Analysis (PCA) can be used to determine replicate reproducibility, because conditions that are more similar should cluster together. The graph displays in two dimensions the n-dimensional data set, aiming to simplify the distance between samples into a spatial 2D graph. Replicates of the same sample should cluster close to each other, whereas different condition should be further apart. If replicates do not cluster, it might be appropriate to verify the quality of the LC-MS runs and ensure samples were not mislabeled. In our example data set, proteins extracted and analyzed by MS cluster based on the number of hours post infection. Replicates obtained 15hpi and 3hpi cluster together by hours post infection (Fig. 2D). Overall, this indicates strong reproducibility between biological replicates, which suggests high quality data.
Identification of Relevant Proteins and Biomarkers
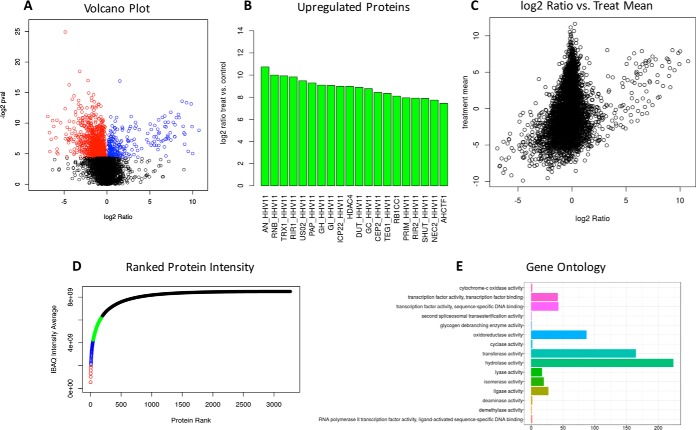
Statistical significance is assessed using the two-tailed t test without multiple testing correction. GiaPronto facilitates the identification of regulated proteins and biomarkers. Proteins with their associated p values were sorted based on the log2 between the two conditions. Proteins are then plotted based on the log2 ratio between proteins obtained 15 hpi and 3 hpi and their p value. This volcano plot allows the user to determine proteins that have a substantial fold change between condition and are statistically significant (Fig. 3A). Our example data shows several proteins that are significantly up-regulated (blue) and downregulated (red) in response to treatment. GiaPronto allows the user to view these proteins as a bar graph and download a table with each protein ID, fold change and p value. Fig. 3B shows an up-regulation of proteins associated with viral infection, which indicates the ability of GiaPronto to identify relevant proteins. Biomarkers are defined as proteins that are up-regulated and abundant in response to viral infection. First, we multiply the log2 ratio by the abundance and rank the resulting proteins. The results are displayed as both a scatterplot which plots proteins based on their log2 ratio and abundance in either treatment or control (Fig. 3C). Next, proteins that are ranked based on these parameters and displayed as bar graphs.
Fig. 3.
Statistical Analysis and Identification of Relevant Proteins. A, Our software uses a t test to determine statistically significant proteins between experimental conditions. A volcano plot is displayed, which allows the user to determine proteins that are both significant and have a substantial fold change between treatment and control. B, GiaPronto displays proteins that are up-regulated and downregulated in response to modulation. C, Lastly, potential biomarkers are determine by examining proteins that are both abundant (intensity of signal) and sample specific (high fold change). The comprehensive list of regulated proteins and biomarkers can be exported in an excel table for additional analysis. D, GiaPronto plots the protein absolute intensity by dividing proteins into quartiles based on their abundance. The top 25% of proteins are displayed in red and the lowest 25% are shown in black. E, Our software performs gene ontologies using clusterProfiler to examine changes in biological process, molecular function and cellular component.
Another helpful approach to visualize the data set from a different perspective is to verify whether a small percentage of proteins represent most of the total protein mass in the sample. To do so, GiaPronto ranks the proteins based on their iBAQ value and it divides them into four quartiles. Each quartile is color coded to simplify the view. In our specific example, the proteins in red represent the top 25% of the protein mass detected by the mass spectrometer. This figure illustrates the differences in protein abundance by showing that a few proteins are so overwhelmingly more abundant (red) than others (black). Overall, this figure can be helpful and allow the user to determine if they need to deplete high abundant proteins prior to analyzing the sample by MS (Fig. 3D). This figure can also give insight into why data sets can vary in the number of protein identifications and abundance, especially if in a specific condition few proteins occupy the most of the total mass.
Gene Ontologies
Gene Ontologies are determined using clusterProfiler and R based package (9). Upregulated protein accession numbers are used to determine which biological processes, molecular functions and cellular compartments are modulated in response to treatment (Fig. 3E). Currently, the software can perform gene ontologies for H. sapiens, M. musculus, C. elegans and D. melanogaster. The output includes two figure representations including one that displays the number of proteins that belong to each group and the other that uses color to represent the p value. These analyses can aid in identifying relevant protein groups that are biologically relevant, which can be informative for validation and follow-up studies.
Downloadable Tables
Throughout the entire result page it is possible to download .xlsx tables for additional analyses not covered in the software or not displayed in the figure representations. For example, the user can export a table of the full analysis in addition to the table normalized data. Next, the user can export a table of all proteins ranked based on the log2 ratio between treatment and control and the p value as calculated by a t test. These tables are available for both relevant proteins and biomarkers. These tables can be further analyzed or imported into other software for further analysis, such as pathway enrichment or network visualization.
In summary, GiaPronto has the capability to analyze all quantification methods and perform both protein and PTM level analysis. In addition, it allows the user to normalize their data, assess the quality and reproducibility between samples, identify relevant proteins/biomarkers and examine gene ontologies.
DISCUSSION
MS is nowadays a widely used technique to identify and quantify proteins and characterize PTMs is many experimental contexts. Because of advances in sensitivity and quantification accuracy, thousands of proteins and PTMs can be identified in a single MS run so extracting biologically relevant protein and PTM changes can be a laborious process that requires training in mass spectrometry and bioinformatics.
Here, we present a free, online, user-friendly software that analyzes proteomics result files (after identification and quantification) in a single click. GiaPronto automatically performs normalization, assesses quality control, identifies relevant/regulated proteins and displays Gene Ontology (GO) enrichment. The software is compatible with any kind of labeling (SILAC and chemical tags) or label-free technique, and allows as input PTM data as well, which are commonly even more challenging to process.
Because our software was built in Shiny with a user-friendly interface, GiaPronto does not require computational experience or expertise and can run on both PC and Macintosh computers that are equipped with an Internet connection. Our output provides customizable, publication quality, figures and many parameters that can be modified by the user. GiaPronto provides a framework that allows rigorous data quality assessment and suggests several ways to represent and interpret MS data. Detailed figure legends are also provided to assist data interpretation for unfamiliar graphical representations. We have created an interface that allows the user to customize color coding, correlation and significant threshold based on the user's preference. Overall, GiaPronto provides a platform for rapid and consistent data analysis that eliminates manual analysis of data that can be an error prone and time-consuming process.
Despite advances in both instrumentation and database searching, there is still an unmet need of making proteomics data analysis as standardized as genomics and accessible for not specialists. Because of this, performing a detailed analysis of these large data sets remains a time-consuming process almost exclusive of proteomics labs. Many institutions are investing in proteomics cores that provide database searching results (protein identifications and quantification). However, it frequently happens that core customers have desire to re-mine data sets and perform further data analysis in addition to the one provided by the core itself. Because GiaPronto performs most of the processing automatically it can assist non-proteomics specialists for data analysis. Our software allows researchers of different fields to incorporate MS into their research programs, which can greatly promote interdisciplinary science.
In summary, GiaPronto is a free and user-friendly software to plot a wide variety of graphs for data analysis of proteomics data sets with a single click. This tool aims to simplify quality control and filtering of proteomics data, for the scientific community not specialized in bioinformatics.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD005467 (https://www.ebi.ac.uk/pride/archive/projects/PXD005467) (7).
Acknowledgments
We thank Samuel Wein for reviewing the coding aspect of this manuscript as well as Zhe Zhang and Yuanchao Zhang for their help with the web server.
Footnotes
* This work was supported by NIH grants R01GM110174 and P01CA196539, and DOD grant W81XWH-113-1-0426. BAG also acknowledges funding from the Leukemia and Lymphoma Society Dr. Robert Arceci Scholar Award. This work was supported by a grant from the W.W. Smith Charitable Trust (SJD). We have declared no conflict of interest.
1 The abbreviations used are:
- PTM
- post-translational modification
- HFF
- human foreskin fibroblast
- GO
- Gene Ontology
- DTT
- dithiothreitol
- FDR
- false discovery rate
- hpi
- hours post-infection
- iBAQ
- intensity-based absolute quantification
- MS
- mass spectrometry
- PCA
- principal component analysis
- PPM
- parts per million
- SILAC
- stable isotope labeling by amino acids in cell culture
- TiO2
- titanium dioxide.
REFERENCES
- 1. Sidoli S., Cheng L., and Jensen O. N. (2012) Proteomics in chromatin biology and epigenetics: Elucidation of post-translational modifications of histone proteins by mass spectrometry. J. Proteomics 75, 3419–3433 [DOI] [PubMed] [Google Scholar]
- 2. Bantscheff M., Lemeer S., Savitski M. M., and Kuster B. (2012) Quantitative mass spectrometry in proteomics: critical review update from 2007 to the present. Anal. Bioanal. Chem. 404, 939–965 [DOI] [PubMed] [Google Scholar]
- 3. Williamson J. C., Edwards A. V., Verano-Braga T., Schwammle V., Kjeldsen F., Jensen O. N., and Larsen M. R. (2016) High-performance hybrid Orbitrap mass spectrometers for quantitative proteome analysis: observations and implications. Proteomics 6, 907–914 [DOI] [PubMed] [Google Scholar]
- 4. Gatto L., and Christoforou A. (2014) Using R and Bioconductor for proteomics data analysis. Biochim. Biophys. Acta 1844, 42–51 [DOI] [PubMed] [Google Scholar]
- 5. Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M. Y., Geiger T., Mann M., and Cox J. (2016) The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 [DOI] [PubMed] [Google Scholar]
- 6. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 7. Kulej K., Avgousti D. C., Sidoli S., Herrmann C., Della Fera A. N., Kim E. T., Garcia B. A., and Weitzman M. D. (2017) Time-resolved Global and Chromatin Proteomics during Herpes Simplex Virus Type 1 (HSV-1) Infection. Mol. Cell. Proteomics 16, S92–S107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thingholm T. E., Jorgensen T. J., Jensen O. N., and Larsen M. R. (2006) Highly selective enrichment of phosphorylated peptides using titanium dioxide. Nat. Protocols 1, 1929–1935 [DOI] [PubMed] [Google Scholar]
- 9. Yu G., Wang L., Han Y., and He Q. (2012) clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD005467 (https://www.ebi.ac.uk/pride/archive/projects/PXD005467) (7).