Abstract
The ubiquitin-directed AAA-ATPase VCP/p97 facilitates degradation of damaged or misfolded proteins in diverse cellular stress response pathways. Resolving the complexity of its interactions with partner and substrate proteins and understanding its links to stress signaling is therefore a major challenge. Here, we used affinity-purification SWATH mass spectrometry (AP-SWATH) to identify proteins that specifically interact with the substrate-trapping mutant, p97-E578Q. AP-SWATH identified differential interactions over a large detection range from abundant p97 cofactors to pathway-specific partners and individual ligases such as RNF185 and MUL1 that were trapped in p97-E578Q complexes. In addition, we identified various substrate proteins and candidates including the PP1 regulator CReP/PPP1R15B that dephosphorylates eIF2α and thus counteracts attenuation of translation by stress-kinases. We provide evidence that p97 with its Ufd1-Npl4 adapter ensures rapid constitutive turnover and balanced levels of CReP in unperturbed cells. Moreover, we show that p97-mediated degradation, together with a reduction in CReP synthesis, is essential for timely stress-induced reduction of CReP levels and, consequently, for robust eIF2α phosphorylation to enforce the stress response. Thus, our results demonstrate that p97 not only facilitates bulk degradation of misfolded proteins upon stress, but also directly modulates the integrated stress response at the level of signaling.
Keywords: Ubiquitin, Protein Degradation*, Stress response, SWATH-MS, Chaperone*
Key elements of the response to cellular stresses that threaten protein homeostasis include the induction of protein folding factors such as chaperones, the efficient clearance of terminally misfolded proteins and the attenuation of protein synthesis (1). Inhibition of translation is achieved by various stress kinases including the endoplasmic reticulum (ER) 1 stress sensor PERK (2–4). As elements of the integrated stress response (ISR) these kinases converge on phosphorylating the eukaryotic initiation factor 2 alpha (eIF2α) at serine-51 to block global translation at the ribosome (5, 6). The phosphorylation of eIF2α is antagonized by protein phosphatase-1 (PP1) complexes with one of two alternative regulatory subunits. Gadd34/PPP1R15A is induced after stress, and shuts down the stress response to resume protein synthesis (7, 8). In contrast, CReP/PPP1R15B is constitutively expressed and balances eIF2α phosphorylation in unperturbed cells (9).
The clearance of misfolded proteins, on the other hand, is mediated by the ubiquitin-proteasome system (UPS) and autophagy. The AAA+-type ATPase Valosin-containing protein (VCP)/p97 (also called Cdc48 or Ter94) is a critical bottleneck in these processes (10, 11). It helps mobilize and unfold large cohorts of ubiquitin-modified substrates to facilitate their degradation in the proteasome in diverse pathways and compartments ranging from ER-associated degradation (ERAD), ribosomal quality control, mitochondrial stress response to macroautophagy and degradation of chromatin-associated proteins (11–14). Consistent with this, missense mutations in p97 are associated with multisystem proteinopathy-1 in humans (also called IBMPFD/ALS) comprising an inclusion body myopathy, Paget's disease of bone, fronto-temporal dementia and amyotrophic lateral sclerosis (15, 16). Moreover, p97 inhibitors are being explored as cancer drugs based on the rationale that they induce a proteostasis crisis and cell death in cancer cells with extra burden of misfolded proteins, and first inhibitors are moving into clinical trials (17, 18).
p97 has two AAA+ domains, D1 and D2, that form two stacked hexameric rings around a central channel (13). ATP hydrolysis in D2 is most critical for unfolding (19–21). Consistent with this, a variant harboring the E578Q mutation in the Walker B motif of D2 traps substrate proteins (13, 20). A host of accessory proteins cooperate with p97. They include more than thirty cofactor proteins that directly interact with the regulatory N-domain or the C-terminal tail of p97 through dedicated interaction domains, and assist p97 as ubiquitin adapters, targeting factors or regulators (16, 22). However, it is often unclear if and how binding of accessory factors in diverse pathways is linked to substrate unfolding. A second challenge is the identification of critical substrate proteins. Although p97 targets large cohorts of misfolded proteins in established stress pathways, only few regulatory degradation substrates are known so far, and it is unclear if and how p97 governs stress signaling by directly targeting stress regulators.
To tackle the complexity of p97 interactions, we compared p97 wild-type (wt) complexes with complexes containing the p97 substrate-trapping mutant p97-E578Q using affinity purification followed by SWATH mass spectrometry (AP-SWATH). AP-SWATH identified and quantified differential interactions over a large range of interactor abundance. This revealed subsets of cofactors, accessory factors and ubiquitin ligases that were specifically enriched with p97-E578Q indicating that their binding is linked to substrate unfolding. In addition, we isolated a set of p97 substrate candidates. Among them, we identified the PP1 regulator CReP/PPP1R15B as a direct target of the p97-Ufd1-Npl4 cofactor complex. We provide evidence that p97 balances CReP levels in unperturbed cells. Moreover, we demonstrate that p97-mediated degradation of CReP is an important element for the timely reduction of CReP levels after severe stresses and thus for ensuring robust eIF2α phosphorylation to enforce the ISR.
EXPERIMENTAL PROCEDURES
Plasmids
pcDNA5-p97-WT/E578Q-myc-strep/FRT/TO were described previously (23). Expression constructs coding for RNF185 and MUL1 with C-terminal GFP tag were generated by gateway cloning (Invitrogen, Carlsbad, CA) using human ORFeome library vectors (hORFeome V5.1, Open Biosystems; internal IDs: 10236 (RNF185), 6868 (MUL1)). LR recombination was performed with the pcDNA5/FRT/TO/GFP/GW destination vector obtained from pcDNA5/FRT/TO/cSH/GW (24) by substitution of the strep-HA with GFP coding sequence via Gibson Assembly cloning. Full length UBE4B cDNA was amplified from an IMAGE clone (ID 7939541) and cloned into pEGFP-C1 using XhoI and BamHI. Flag-tagged PPP1R15B-mCherry (FLAG_hPPP1R15B_4–713_mCherry_UK1298) was a gift from David Ron (Addgene plasmid # 80707) (25).
Cell Culture and Transfections
HEK293 and HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS) in the presence of penicillin/streptomycin. Stable inducible HEK293 cell lines expressing p97-WT-myc-strep or p97-EQ-myc-strep were generated with the Flp-In T-Rex system (Invitrogen) according to the manufacturer's protocol. Stable cell lines were maintained in culture medium as described above, supplemented with 15 μg/ml blasticidin S and 100 μg/ml hygromycin B. Expression was induced with 1 μg/ml doxycycline. HEK293 and HeLa cells were transiently transfected with plasmids using JetPRIME (Polyplus, Illkirch-Graffenstaden, France) or Lipofectamine2000 reagents (Invitrogen), respectively, 1 day after seeding. Medium was changed 4 h after transfection. Cells were analyzed after 24 h.
RNAi
The siRNA oligomers targeting Ufd1 (#1: GUGGCCACCUACUCCAAAUTT; #2: CUACAAAGAACCCGAAAGATT), p47 (AGCCAGCUCUUCCAUCUUATT), β-TrCP 1/2 (GUGGAAUUUGUGGAACAUCTT), CReP (AAGGGAUGGAUGCAGGUUCCATT) and a nontargeting control oligomer (UUCUCCGAACGUGUCACGUTT) were purchased from Microsynth (Balgach, Switzerland) and were characterized previously (9, 23, 26, 27). Reverse transfection into HEK293 cells was performed with 10 nm final concentration using Lipofectamine RNAiMax (Invitrogen). Cells were analyzed after 48 h or as indicated.
Pharmacological and Stress Treatments
For cycloheximide chase experiments, HEK293 cells were treated with 50 μg/ml CHX (Sigma, St. Louis, MO) for indicated times. p97 and proteasome inhibitors were added for 1 h (or as indicated) using NMS-873 (5 μm; Sigma) and CB-5083 (5 μm; Selleckchem, Houston, TX) or MG132 (20 μm; Merck Millipore, Darmstadt, Germany). For pharmacological stress induction, cells were exposed to 0.5 mm sodium arsenite, 50 μg/ml tunicamycin (both Sigma) or 10 μm thapsigargin (Diagonal, Münster, Germanyl) for the indicated period. UV irradiation was performed by removing medium from adherent cells and covering them in 37 °C PBS. Cells were exposed to 300 J/m2 UV-C (254 nm) and PBS was replaced by the previously removed media (for simultaneous inhibitor treatment drugs were added directly after UV exposure). Cells were analyzed at indicated time points after irradiation.
Immunoprecipitation and Immunoblotting
Cells were harvested 24 h after transient transfection and/or 20 h after induction with doxycycline in IP buffer (150 mm KCl, 5 mm MgCl2, 50 mm Tris-HCl pH 7.4, 1% Triton X-100, 5% glycerol, 2 mm β-mercaptoethanol supplemented with cOmplete EDTA-free protease inhibitors and PhosSTOP (Roche, Basel, Switzerland)) and incubated on ice for 20 min. To stabilize ubiquitination of CReP, the IP buffer was additionally supplemented with 10 mm N-ethylmaleimide (Sigma). Lysates were cleared by centrifugation (15 min, 17,000 × g, 4 °C) and protein concentration was determined in a BCA assay (Interchim, Montlucon, France). For degradation assays, 25 μg protein/sample were directly resolved by SDS-PAGE and analyzed by Western blotting. Immunoprecipitation and pulldown experiments were performed using Strep-Tactin Sepharose (IBA BioTAGnology, Göttingen, Germany), anti-GFP nanobodies or specific antibodies and Protein G Sepharose (GE Healthcare, Chicago, IL). Affinity purifications were performed for 2 h at 4 °C rotating (strep-/GFP pulldown assays) or 2 h incubation with the indicated primary antibodies on ice and additionally with Protein G Sepharose for 1 h rotating at 4 °C to capture antigen-antibody complexes. Beads were washed five times in IP buffer and eluted by boiling in 2 × SDS sample buffer. Samples were resolved by SDS-PAGE and transferred to nitrocellulose membranes (Amersham Biosciences, GE Healthcare). Immunoblot analysis was performed with the indicated antibodies and visualized by chemiluminescence. Quantification of Western Blots was performed by capturing antibody signal either with a CCD camera (Amersham Biosciences Imager 600 (GE Healthcare)) and/or with an Odyssey CLx fluorescence reader (LI-COR, Lincoln, NE) whereas image analysis was performed with the TotalLab Quant software.
Fluorescence Microscopy
HeLa or HEK293 cells were fixed in 4% paraformaldehyde, permeabilized with 0.1% Triton X-100 in PBS, blocked by 3% bovine serum albumin in PBS containing 0.1% Triton X-100 and 0.1% saponin. Confocal laser scanning microscopy was performed on a TCS SP5 AOBS system equipped with standard PMT detectors as well as sensitive HyD detectors (Leica Microsystems, Wetzlar, Germany). Images were acquired using an HCX PL APO 63×/1.4NA oil-immersion objective. Lasers used for excitation were DPSS 561 nm (Alexa Fluor® 568, mCherry), Ar 488 nm (GFP) and Diode 405 nm (DAPI). Acquisition and hardware was controlled by LAS AF software (Leica Microsystems).
Antibodies
Antibodies used for Western blot analysis against p97 (HME8), UBXD1 (E43), Ufd1 (HME14), Npl4 (HME18), p47 (HME22) and p37 (20880) were described previously (23, 28, 29). Anti-UBXD4 (25272) antibody was raised in rabbits against purified recombinant GST-tagged proteins (BioGenes, Berlin, Germany). Anti-UBXD7 (S409D) antibody was generated and obtained from the MRCprotein phosphorylation unit, Dundee (G. Alexandru). Anti-PLAA (Y102) was purchased from Abcam (Cambridge, UK). Mouse anti-ubiquitin (clone FK2, 04–263) and rabbit anti-Histone H3 (06–755) were purchased from Millipore. Anti-PPP1R15B (CReP) was from Proteintech (Rosemont, IL) (14634–1-AP). Rabbit monoclonal anti-phosphoS51-eIF2α (#3398) and anti-phosphoS317Chk1 (#2344) antibodies were purchased from Cell Signaling (Danvers, MA). Mouse monoclonal anti-eIF2α (d-3, sc-133132) and anti-HSC 70 (B-6, sc-7298) antibodies were from Santa Cruz Biotechnology (Dallas, TX). Monoclonal anti-Flag M2 (F3165), anti-myc (M4439), anti-α-tubulin (T5168) and anti-GAPDH (G8795) were from Sigma-Aldrich. Anti-GFP was from Roche. HRP-coupled secondary antibodies were purchased from Bio-Rad (Hercules, CA) and IRDye®-conjugated secondary antibodies from LI-COR. Antibodies used for immunofluorescence stainings were anti-Tom20 (Santa Cruz, sc-11415) and anti-Calnexin (Enzo Life Sciences, ADI-SPA-860-F), and secondary antibodies conjugated to Alexa Fluor® 568 fluorophore (Invitrogen).
Statistical Analysis of Western Blots
All experiments were performed in triplicate or as indicated, with error bars denoting s.e.m. Statistical analyses were carried out with the unpaired two-sided t test. p < 0.05 (*) was considered statistically significant.
Size Exclusion Chromatography
Size exclusion chromatography was performed on a Superose 6 10/300 GL column at 20 °C in 50 mm HEPES pH 7.5, 150 mm NaCl, 5 mm MgCl2, 0.5% NP40, 1 mm DTT, 25 mm β-Glycerophosphate, 10 μm leupeptin/pepstatin. 3.5 mg cleared cell lysates from stable HEK293 p97-WT/EQ cell lines (24 h after induction with doxycycline) were separated into 13 fractions and analyzed by Western blotting.
AP-SWATH Analysis
Cells were lysed on ice in HNN lysis buffer (0.5% IGEPAL ca-630, 50 mm HEPES, pH 7.5, 150 mm NaCl, 50 mm NaF, 200 μm NaVO3, 0.5 mm PMSF, 1.2 μm Avidin, and protease inhibitor mixture (Sigma)) and centrifuged at 16,100 × g for 15 min at 4 °C. Single step affinity purification from the supernatants via the streptavidin binding peptide tag, TCA precipitation, trypsin digestion, and solid phase extraction of peptides were performed as previously described (30). Samples from affinity purifications of p97-wild type or p97-E578Q, in addition to GFP control purifications were analyzed both, in data dependent acquisition (DDA) mode for spectral library building, and SWATH-MS mode for quantitative analysis. MS data in DDA and SWATH-MS modes were acquired using a Sciex 5600 TripleTOF mass spectrometer interfaced to an Eksigent NanoLC Ultra using acquisition parameters as previously described (30) with the exception that for SWATH analysis 64 variable width precursor isolation windows were used (31). Spectral library building from DDA data was performed using the human canonical UniProt/SwissProt database (March 2013) supplemented with common contaminants and iRT peptide sequences containing 20,318 target and 20,283 decoy entries as described (32) except 3 search engines were used (Tandem - version JACKHAMMER 2013.06.15, with and without k-score plugin, and Comet 2014.02 rev. 2, post-processing and spectral library building with Trans Proteomic Pipeline v4.7.0) (33–35), semitryptic digestion was specified and 2 missed cleavages allowed, oxidation of methionine was set as a variable modification, carbamidomethylation was set as a fixed modification, mass tolerance was set to 50 ppm (precursor) and 100 ppm (fragment), the protein false discovery was set to 5% estimated using Mayu v1.08 (iProphet probability threshold > 0.554009, 0.28% PSM FDR, and 0.6% peptide FDR), the top 5 most intense transitions per precursor including neutrals losses were selected, and decoys for OpenSWATH analysis were generated using the “shuffle” method. Peak lists for database searching were generated using the qtofpeakpicker tool in Proteowizard v 3.0.11026. Only peptides with a unique mapping in the database (proteotypic) were used for analysis. To improve coverage of the spectral library we added DDA data from affinity purifications of 23 p97 cofactors (NSFL1C, UFD1L, NPLOC4, YOD1, PLAA, UBXN7, ASPSCR1, FAF1, UBXN1, UBXN2B, SPRTN, NGLY1, UBXN10, VCPIP1, ZFAND2B, UBXN11, OTUD7B, RNF31, ATXN3, UBXN2A, UBXN6, FAF2, and SVIP).
The SWATH-MS data was analyzed using OpenSWATH (v2.1) essentially as described (30, 36). FDR was controlled in the experiment-wide context at 1% at the peptide query level and in the global context at 1% at the peptide query and protein levels (37). Peak groups were further aligned using the TRIC algorithm with the LocalMST and lowess options with a target FDR of 1%. Protein abundances were computed using the 'best flyer peptide' approach by summing the top 5 most intense fragments from the top 3 most intense peptides using aLFQ (38, 39). After log2 tranformation and median normalization, we prefiltered the data by first removing any protein that was not identified in at least 2 out of 3 replicates in either p97-WT or p97-EQ. We removed contaminant proteins by then computing the median log2 fold change and t-tests (equal variance assumed) between either p97-WT or p97-EQ and GFP controls where log2 fold change > 2 and p value < 0.05 was considered a significant interaction. We further insisted proteins whose median log2 abundance in either p97-WT or p97-EQ must be display at least a 2-fold increase over the maximum abundance from a set of 9 GFP controls purifications from a previous study (30). Median log2 fold change and t-tests were also computed between p97-EQ and p97-WT where log2 fold change > 2 or < 2 and p value < 0.05 was considered a significant change in the interaction of a protein with p97 between the wild-type and mutant conditions. Missing protein abundance values were imputed using the minimum abundance from the quantitative matrix for calculating statistics.
Experimental Design and Statistical Rationale for AP-SWATH Experiments
Samples from affinity purifications of 3 biological replicates of p97 wild-type and 3 biological replicates of p97-E578Q, in addition to 2 biological replicates of GFP control purifications (one GFP replicate was discarded because of experimental error) were analyzed both in data dependent acquisition (DDA) mode and SWATH-MS mode. To improve power in contaminant filtering we supplemented the analysis with data from 9 additional independent biological replicate GFP control purifications. As we expected that the effect sizes, both from a contaminant filtering perspective, and from a wild-type versus mutant comparison perspective, should be large, we reasoned that this level of replication would be sufficient to detect significant changes. Multiple hits were validated by orthogonal biochemical methods as described.
RESULTS
SWATH-MS Analysis of VCP/p97 Wild-type and a Substrate-trapping Mutant
Previous MS-based measurements identified a number of factors that interact with p97 or its cofactors, but did not specifically link this analysis to ATP hydrolysis and thus biochemical activity of p97 (41, 42). The strategy applied in the present study was to use the substrate-trapping mutant p97-E578Q (p97-EQ) as bait protein and to quantitatively compare its interactors with those of wild-type p97 (p97-WT). With this approach, we aimed to specifically “freeze” p97-mediated unfolding reactions and entangle substrates as well as associated cofactors and regulators, whose identification could reveal previously unidentified links of p97 to relevant signaling pathways. We combined this approach with SWATH-MS analysis (43), a data independent acquisition method that deterministically records fragment ion spectra for all detectable peptide precursors which are later analyzed in a targeted fashion with reference to spectral libraries where the mass spectrometric and chromatographic coordinates for peptides of interest have been established. SWATH-MS combines the advantages of data dependent acquisition (high throughput) with those of selected reaction monitoring (high reproducibility, consistency, and dynamic range) and we have previously established the advantages of this method for quantitative analysis of affinity purification samples (30). We used previously described stable cell lines that express strep-tagged p97-WT or p97-EQ upon doxycycline induction at near endogenous levels, which integrate into endogenous p97 hexamers (see Fig. 1A for scheme) (23).
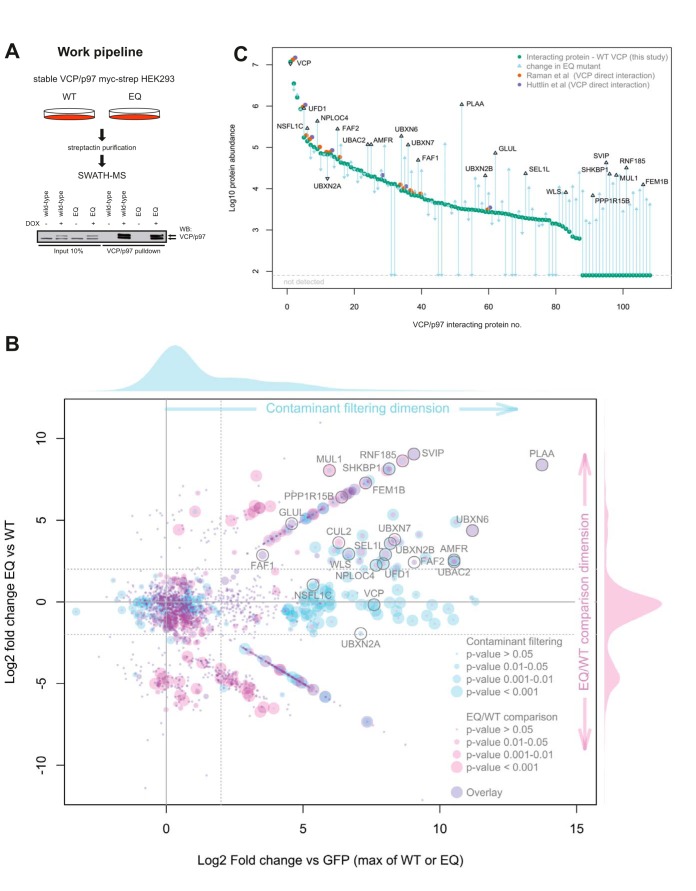
Fig. 1.
Comparative SWATH-MS of VCP/p97 wild-type and the substrate-trapping mutant, VCP/p97-E578Q, identifies differential binding of subsets of cofactors, ubiquitin ligases and potential substrates. A, Work flow. Isogenic stable HEK293 cells were induced to express strep-tagged VCP/p97 wild-type (p97-WT) or VCP/p97-E578Q (p97-EQ). p97 was affinity-purified from detergent extracts and associated proteins analyzed by SWATH-MS. Western blot analysis of overexpressed and endogenous p97 (indicated by arrows). B, Scatterplot of interactors as identified by SWATH analysis. The Y-axis represents the fold change ratio of VCP/p97-EQ versus VCP/p97-WT. The X-axis represents the fold change of VCP/p97-WT or VCP/p97-EQ versus GFP-pulldown controls. p values for contaminant filtering and EQ/WT comparison visualized as indicated. Individual candidates mentioned in the text are annotated. See supplemental Table S1 for full list. The proteins falling along the diagonal lines in either the upper right or lower right quadrant are those which have only been identified and quantified in either the VCP/p97-EQ, or VCP/p97-WT purifications, respectively. C, Plot representing the 108 high confidence hits arranged according to abundance in VCP/p97-WT and changes in VCP/p97-EQ are indicated by arrows. Note the high dynamic range over >4 log10. Individual candidates mentioned in the text are annotated. Observations of a given protein interaction in previous studies are annotated as indicated.
To generate a spectral library for peptides from the proteins of interest, we first analyzed affinity purifications of the p97-WT, p97-EQ, and GFP control purifications by data dependent acquisition (DDA) mass spectrometry. To increase the coverage of relevant proteins in the spectral library we supplemented the DDA analysis with purifications of 23 cofactors from the p97 system. Database searching and spectral library generation (32) was performed resulting in a spectral library containing coordinates for 3,839 proteins represented by 36,540 uniquely mapping peptides. SWATH-MS data was acquired for p97-WT, p97-EQ, and GFP control purifications and quantitative data was extracted by targeted analysis based on the spectral library resulting in a quantitative data matrix containing 1,937 proteins (supplemental Table S1). To filter out contaminant proteins we made a direct comparison of p97-WT or p97-EQ with GFP control purifications that resulted in the detection of 108 high-confidence interactions. To determine which of these interacting proteins might be p97 substrates or otherwise modulated by the EQ mutation we then compared quantitatively proteins co-purified in the p97-EQ or p97-WT conditions. Of the high confidence interacting proteins, 43 displayed an increase in binding to p97-EQ (p value < 0.05) by at least 2 log2, whereas 8 were reduced by at least 2 log2 compared with p97-WT (Fig. 1B). As expected, among the proteins with the strongest increase was PLAA, a cofactor known to specifically bind p97-EQ both in cell lysates and with purified proteins (23, 44). Moreover, we detected other established and previously biochemically validated p97 cofactors such as SVIP, UBXN6/UBXD1, UBXN7, FAF1, and UBXN2B/p37 (22). In addition, factors involved in p97-regulated ERAD such as AMFR/gp78, FAF2/UBXD8, UFD1L/Ufd1, NPLOC4/Npl4, SEL1L and UBAC2, were consistently increased with p97-EQ, suggesting that they were trapped in the segregation processes along with ubiquitinated substrates. Not all known established cofactors were increased in p97-EQ suggesting functional differences in how they cooperate with p97. Even related cofactors such as the SEP domain-containing proteins UBXN2B/p37 (∼7-fold increase), UBXN2A/UBXD4 (∼3.7-fold decrease) and NSFL1C/p47 (no change) showed largely divergent binding preference.
Of note, the AP-SWATH analysis allowed a high dynamic range of detection covering 4 orders of magnitude from the bait protein p97 and known abundant cofactors such as Ufd1 to low abundant proteins lacking dedicated p97-binding domains (Fig. 1C). Thus, our analysis detected many additional interactors not identified in p97 pulldowns of other studies (41, 45). The comparison between our results and selected previous studies (41, 46, 47) is shown in supplemental Fig. S1 (data in supplemental Table S2). Among the low abundance interactors were certain ubiquitin ligases or ligase components such as RNF185, MUL1, or the CUL2 adapter FEM1B that were specifically trapped with the mutant with log2-fold increases ranging from 2 to 9 indicating that they cooperate intimately with p97.
Validation of Differential Cofactor Binding and Ubiquitin Ligase Interaction
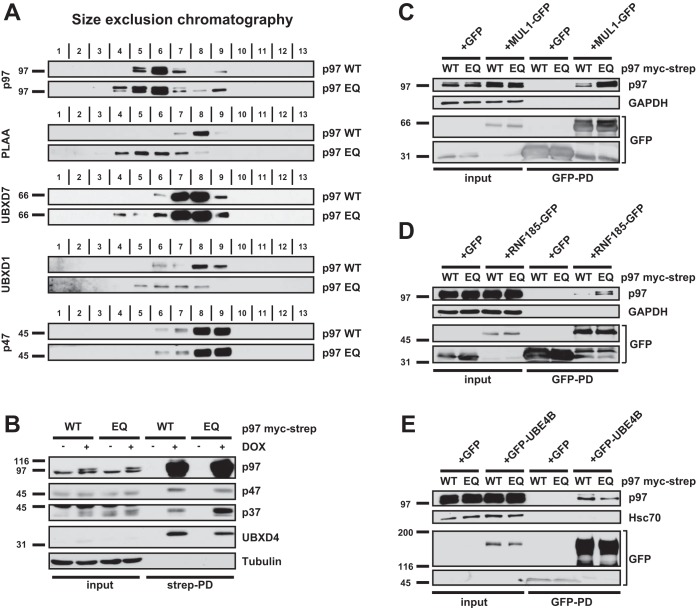
We first validated the AP-SWATH results using gel filtration of lysate from p97-WT and p97-EQ expressing HEK293 cells for known p97 cofactors with available antibodies. As expected, p97-EQ was partially shifted to higher molecular weight fractions compared with p97-WT indicating that it was engaged more tightly in larger complexes (Fig. 2A). Importantly, PLAA and UBXD1 quantitatively shifted to high molecular weight fractions and UBXD7 at least in part co-migrated with p97 fractions. p47 was not bound to p97 under the stringent conditions of gel filtration, as observed earlier (48). We therefore tested the interaction of p47 and the other SEP domain proteins by co-immunoprecipitation of p97-WT and p97-EQ followed by Western blotting. This confirmed that p47 binding was unchanged, p37 was increased and UBXD4 was decreased in p97-EQ pulldowns compared with p97-WT (Fig. 2B). These results validate the results of the AP-SWATH analysis and confirm an intriguing heterogeneity among cofactors with respect to binding in substrate-trapping complexes.
Fig. 2.
Validation of differential binding to cofactors and additional interactors. A, Gel filtration of lysate from cells expressing p97-WT or p97-EQ. Fraction numbers and antibodies used for Western blotting are indicated. Note the shift of p97, PLAA, UBXD1 and UBXD7 in p97-EQ lysate toward the high molecular weight fraction, whereas p47 remain unbound under the stringent conditions. B, Affinity purification of p97-WT or p97-EQ and detection of associated SEP-domain proteins p47, p37 and UBXD4. p97 variants were isolated using streptactin beads and associated proteins analyzed by Western blotting as indicated. C, MUL1-GFP, (D) RNF185-GFP, and (E) GFP-UBE4B were transiently expressed in p97-WT or p97-EQ cells, immunoprecipitated and associated p97 analyzed by Western blotting. Note that MUL1 and RNF185, but not UBE4B, showed increased binding of p97-EQ thus confirming SWATH data.
We next tested low abundance interactors with highly increased binding to p97-EQ according to the AP-SWATH analysis. This included the ubiquitin ligases that have not been linked to p97 such as the mitochondrial MUL1 and RNF185 (both >8 log2 fold increases). MUL1 and RNF185 have both been associated with regulation of mitophagy, whereas RNF185 has also been linked to ERAD (49, 50, 51). Overexpression in p97-WT or p97-EQ expressing cells followed by co-immunoprecipitation of the candidates confirmed increased binding to p97-EQ in both cases (Fig. 2C and 2D). This was also shown in HeLa cells, where correct subcellular localization of overexpressed MUL1 and RNF185 to mitochondria and ER, respectively (50, 51), was confirmed by microscopy (supplemental Fig. S2A–S2D). In contrast, the ubiquitin ligase UBE4B was detected but did not show significantly differential binding in the AP-SWATH analysis and was not increased by the mutation in co-immunoprecipitations (Fig. 2E). Again, expected nuclear localization of the overexpressed UBE4B was confirmed by microscopy (supplemental Fig. S2E).
p97 Targets the Ubiquitinated Protein Phosphatase-1 Regulator CReP/PPP1R15B
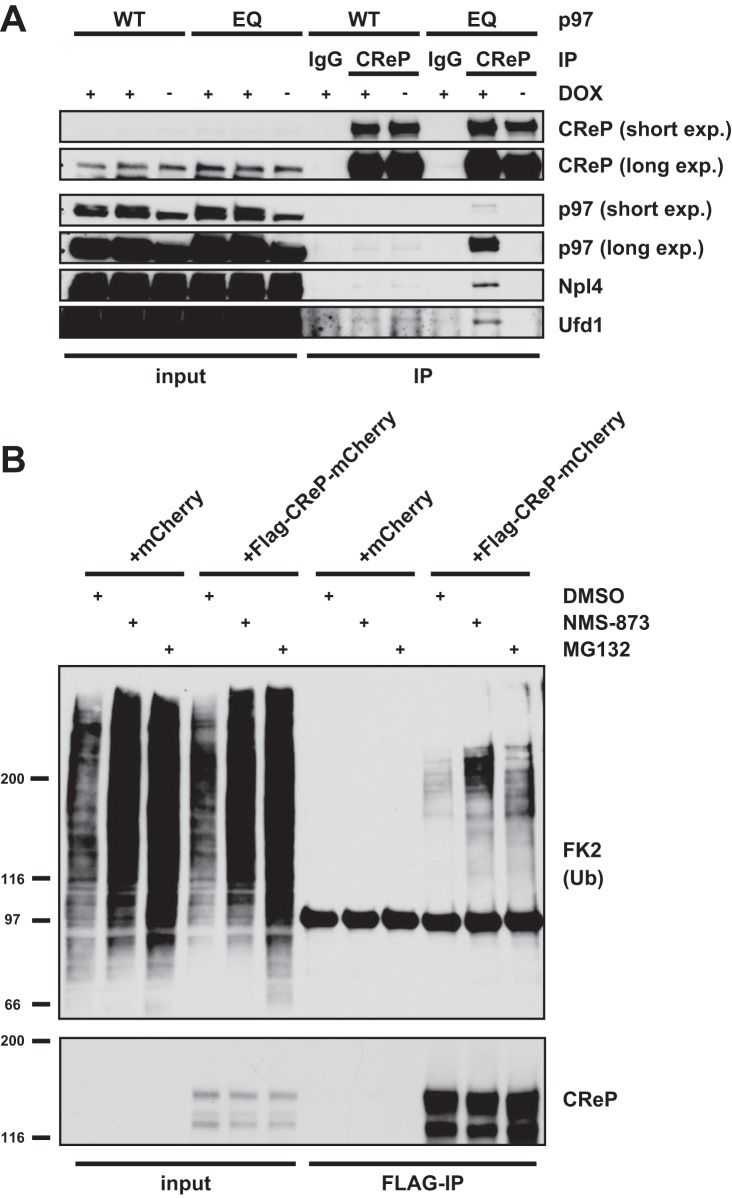
Among the low abundance interactors, a number of proteins were identified that were not known to be p97 cofactors or ubiquitin regulators, and could therefore constitute novel substrates (Fig. 1B and 1C and supplemental Table S1). Of note, glutamine synthetase (annotated as GLUL), which was identified to be increased by ∼30-fold in our screen, was only recently described as a target of p97-mediated degradation (52). Other strong candidates included membrane trafficking or signaling regulators such as SHKBP1 (SH3KBP1 binding protein 1), WLS (wntless Wnt ligand secretion mediator), EPSTI1 (Epithelial-stromal interaction protein 1), TRAFD1 (TRAF-type zinc finger domain containing protein 1) as well as the unconventional myosin MYO5C. We focused on CReP/PPP1R15B, which was exclusively detected in p97-EQ pulldowns (Fig. 1B and 1C), because it is a modulator of the integrated stress response and has not been linked to p97. Notably, in the AP-SWATH analysis, only a single proteotypic peptide was detected which, however, displayed a consistent and confident signal in p97-EQ isolations (median protein level q-value = 2.45E-04) but was not detected in p97-WT (supplemental Fig. S3). Further, according to PeptideAtlas (53), the single peptide we detected is ranked as the highest for predicted observability and among the top few peptides in empirical observations in mass spectrometry experiments where CReP was detected. This is consistent with the view that CReP is very low abundant in our p97-EQ affinity purification sample and can only be observed by the best MS responding peptide. To validate this result, we first immunoprecipitated endogenous CReP from cell lysate expressing p97 variants and confirmed binding to p97-EQ, but not p97-WT (Fig. 3A). Efficient extraction of endogenous CReP in our lysate was confirmed (supplemental Fig. S4A). The CReP antibody detected a second faster-migrating band that was confirmed to be nonspecific using siRNA-mediated depletion of CReP (supplemental Fig. S4B).
Fig. 3.
CReP/PPP1R15B is a direct target of the p97-Ufd1-Npl4 complex. A, Coimmunoprecipitation of endogenous CReP from lysate of p97-WT or p97-EQ cells. Cells were induced to express the p97 variants or left untreated (± DOX). Isolates were analyzed by Western blotting with indicated antibodies. Note the association of p97, Ufd1 and Npl4 in the presence of p97-EQ. B, CReP accumulates in ubiquitinated form upon inhibition of p97 or the proteasome. Flag-tagged CReP-mCherry or mCherry alone was transiently expressed, and cells were treated with the p97 inhibitor NMS-873 (5 μm), the proteasome inhibitor MG132 (20 μm), or vehicle alone (DMSO), for 1 h before lysis. CReP was immunoprecipitated and probed with ubiquitin antibodies by Western blotting.
The Ufd1-Npl4 heterodimer is the major p97 adapter for polyubiquitinated substrate proteins that are destined for degradation in the proteasome (16, 22). We therefore asked whether it is associated also with CReP from the p97-EQ lysates. Indeed, Western blot analysis revealed association with Ufd1 and Npl4 (Fig. 3A). Consistent with that, we also detected polyubiquitination in immunoprecipitations of overexpressed CReP (Fig. 3B). Expected localization of the overexpressed CReP was confirmed by fluorescence microscopy (supplemental Fig. S4C). Importantly, the ubiquitination signal increased after inhibition of p97 or the proteasome (Fig. 3B), suggesting that ubiquitinated CReP is turned over by the proteasome with the help of p97 and, thus, that CReP is a substrate of p97-Ufd1-Npl4.
The p97-Ufd1-Npl4 Complex is Required for Constitutive Degradation of CReP
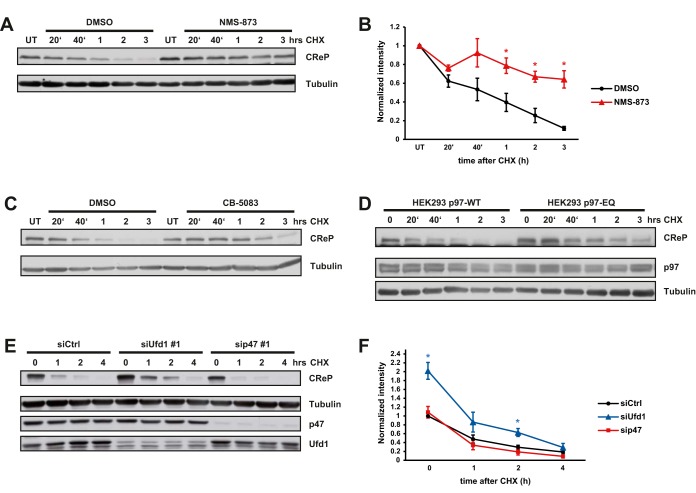
To further explore this, we first analyzed the role of p97 in constitutive degradation of CReP. A cycloheximide (CHX) chase in control cells confirmed its high turnover rate (9), as CReP levels dropped to undetectable levels within 3 h (Fig. 4A and 4B). Crucially, pharmacological inactivation of p97 with two unrelated inhibitors, NMS-873 or CB-5083 (17, 18), stabilized CReP under these conditions (Fig. 4A–4C). Likewise, induction of the dominant-negative p97-EQ trapping mutant in our stable cell line again significantly delayed degradation of CReP compared with p97-WT induction (Fig. 4D). To clarify whether the Ufd1-Npl4 adapter was required, we depleted Ufd1, or the alternative p47 cofactor as control with established siRNAs. Depletion of Ufd1, but not of p47, over 2 days already led to an increase of CReP levels compared with control depletion (Fig. 4E and 4F). Moreover, it delayed degradation in the CHX chase, whereas depletion of p47 had no effect (Fig. 4E and 4F). This was confirmed with an independent Ufd1 siRNA (supplemental Fig. S5). These data suggest that the Ufd1-Npl4 adapter assists p97 in the constitutive degradation of CReP.
Fig. 4.
p97 and Ufd1 are essential for constitutive degradation of CReP. A, Cycloheximide (CHX) chase experiments in the presence of the p97 inhibitor NMS-873 (5 μm) or DMSO alone. Cells were lysed at indicated time points after CHX addition and lysates subjected to Western blotting. Tubulin was probed as loading control. UT = untreated. B, Quantification of chemiluminescence in three independent experiments. CReP Western blotting signals were normalized to loading control and values displayed relative to those in untreated (UT) cells. Error bars, s.e.m., (*) p value < 0.05. C, As in (A), but with the unrelated competitive p97 inhibitor CB-5083 (5 μm). D, CHX chase experiments as in (A) were conducted in cells expressing p97-WT or the dominant-negative p97-EQ. Cells at time point 0 were mock (DMSO) treated. E, CHX chase experiments were perfomed in cells treated with control siRNA, or siRNA directed against Ufd1 or p47 for 48 h. Depletion efficiency of Ufd1 and p47 was monitored with specific antibodies as indicated. Cells at time point 0 were mock (DMSO) treated. F, Quantification of chemiluminescence in four independent experiments. CReP Western blotting signals were normalized to loading control and siRNA control at time point 0. Error bars, s.e.m., (*) p value < 0.05.
p97-Ufd1-Npl4 Facilitated Degradation is Essential for the Drop of CReP Levels and a Robust Increase in eIF2α Phosphorylation after UV and Arsenite Treatment
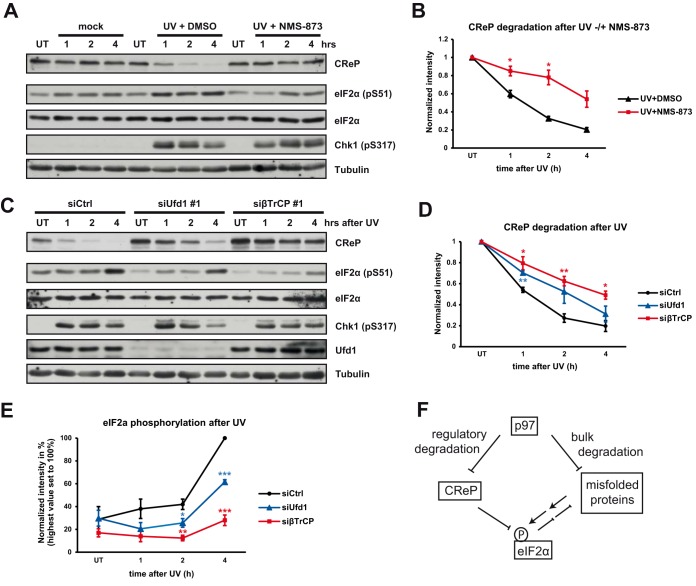
We next sought to clarify the relevance of p97-Ufd1-Npl4-mediated degradation of CReP in the integrated stress response. Under nonstress conditions, the PP1-CReP complex constitutively dephosphorylates eIF2α to antagonize stress kinases and maintain protein synthesis (9). Although CReP levels remain largely stable upon ER stress (9), CReP is quantitatively degraded upon exposure to UV (54). We confirmed stable levels upon thapsigargin and tunicamycin treatments, as well as CReP degradation upon UV exposure (Fig. 5A, 5B and supplemental Fig. S6A). In addition, we observed rapid CReP degradation also upon arsenite treatment (supplemental Fig. S6A). UV exposure did not appear to accelerate the CReP degradation rate when compared with addition of CHX, and the two treatments were not additive (supplemental Fig. S6B), suggesting that the decline of CReP levels under stress is a result of both degradation and inhibition of CReP synthesis. Of note, the degradation of CReP after UV exposure was largely inhibited by NMS-873, and NMS-873 affected degradation also after arsenite treatment (Fig. 5A, 5B and supplemental Fig. S6C), demonstrating that p97 is essential for the stress-induced quantitative removal of CReP. In both conditions, stress-induced phosphorylation of eIF2α was attenuated when p97 was inhibited by NMS-873 (Fig. 5A, and supplemental Fig. S6C), correlating with the failure to degrade CReP and, thus, with persistent PP1-CReP activity that antagonizes eIF2α phosphorylation.
Fig. 5.
The p97-Ufd1-Npl4 complex is required for the stress-induced drop in CReP levels and the robust phosphorylation of eIF2α after UV irradiation. A, Quantitative removal of CReP after UV-induced stress. Cells were either mock-treated, or UV-irradiated in the presence of p97 inhibitor NMS-873 (5 μm) or vehicle alone (DMSO). Cells were lysed at indicated time points after irradiation, and lysates probed with indicated antibodies. Chk1 phosphorylation confirms UV-induced damage. Note the rapid decline of CReP after irradiation that was blocked by NMS-873, whereas DNA damage signaling was unaffected according to Chk1 phosphorylation. UT = untreated. B, Quantification of (A). CReP Western blotting signals were normalized to loading control and values were displayed relative to those in untreated cells. n = 3 independent experiments, error bars, s.e.m., (*) p value < 0.05. C, Ufd1 and β-TrCP depletion delays stress-induced degradation of CReP and prevents robust phosphorylation of eIF2α. Experiments as in (A) after treatment of cells with control, Ufd1 or β-TrCP siRNAs for 48 h. Note the reduced phosphorylation of eIF2α. UT = untreated (not irradiated). D, Quantification of stress-induced CReP degradation in experiments as in (C). CReP Western blotting signals were normalized to loading control and values were displayed relative to those in untreated cells. n = 3 independent experiments, error bars, s.e.m., (*) p value < 0.05, (**) p value < 0.01. E, Quantification of phospho-eIF2α signals in experiments as in (C). Values were normalized to loading control and displayed relative to the maximum stimulation in control cells. n = 4 independent experiments, error bars, s.e.m., (*) p value < 0.05, (**) p value < 0.01, (***) p value < 0.001. F, Model for how p97 affects eIF2α phosphorylation and protects cells from protein stress. p97 maintains protein homeostasis by bulk degradation of misfolded proteins in a variety of pathways and compartments including the ER. In addition, p97 facilitates CReP degradation. This is required to balance CReP levels in unperturbed cells and to ensure its quantitative removal after certain stresses in order to enforce stress-kinase mediated eIF2α phosphorylation.
We next monitored the effect of Ufd1 depletion on stress-induced CReP degradation. Ufd1 depletion delayed CReP degradation after UV irradation (Fig. 5C and 5D) showing that Ufd1 not only assists p97 in the constitutive degradation, but also in the stress-induced degradation of CReP. This mirrored the effect of depleting the ubiquitin ligase component β-TrCP (Fig. 5C), which has previously been shown to trigger CReP degradation (54–57). Of note, the stress-induced induction of eIF2α phosphorylation was attenuated in Ufd1 and β-TrCP-depleted cells (Fig. 5C and 5E), again correlating with the delayed degradation of CReP. Thus, the role of the p97-Ufd1-Npl4 complex in facilitating CReP degradation is critical for the timely decline of CReP levels after cellular stress to ensure robust eIF2α phosphorylation and thereby to enforce the integrated stress response that attenuates protein synthesis.
DISCUSSION
VCP/p97 has fundamental and far-reaching roles in maintaining protein homeostasis particularly under cellular stress conditions. In this study, we combined AP-SWATH with the use of a p97 substrate-trapping mutant to further explore the complexity of the p97 system and how it achieves this task. Biochemical validation of the acquired AP-SWATH data demonstrated the high accuracy of the results reflected in differential binding of known cofactors and additional factors over 4 orders of magnitude of dynamic range demonstrating the suitability and reliability of AP-SWATH for comparison of interactomes from different conditions. The increased dynamic range of quantification, in combination with the use of a substrate-trapping mutant, has allowed us to substantially expand on the set of AP-MS derived p97 protein interactions with respect to either a recent large-scale protein interaction screen (47), or a study focused on p97 (41).
We found that many but not all known p97 cofactors are trapped by the mutant pointing to differences in how cofactors cooperate with p97 despite similar interaction domains. In addition, a number of other factors including E3 ubiquitin ligases were specifically trapped by p97-EQ. This likely reflects the formation of substrate-induced, transient p97-cofactor complexes that only disassemble when the substrate is appropriately unfolded or segregated by p97. Consistent with this notion, we find many proteins involved in ERAD that were trapped ranging from the relevant ligases on the cytosolic side up to SEL1L on the luminal side of the ER. This suggests that p97-EQ stabilizes transient protein interactions in ERAD by “freezing” the substrate in the process of retro-translocation. Likewise, we find that p97-EQ strongly traps ubiquitin ligases such as MUL1 or RNF185, which have both been associated with mitochondrial quality control (49, 50). This points to intriguing mechanisms that link ubiquitin-modification with unfolding also in this pathway.
Although the p97 substrate proteome is highly complex with thousands of heterogeneous substrates in different pathways, the AP-SWATH approach with its high dynamic range also identified individual substrate proteins. Among them, glutamine synthetase was only recently shown to be a direct target of p97 (52). Other highly scoring and low abundant candidates still need to be confirmed, such as the modulator of EGF receptor transport, SHKBP1, a transport factor of Wnt proteins, WLS, the unconventional MYO5C or the NFkB regulators TRAFD1 or EPSTI1. They represent novel and promising directions for understanding p97 function in signaling and membrane transport. Of note, our approach led us to discover the eIF2α-specific PP1 regulator CReP as a substrate of p97. The evidence for CReP being a direct substrate is compelling as we show interaction of CReP with p97 and the Ufd1-Npl4 adapter, p97-Ufd1-Npl4 dependent degradation of CReP, and the accumulation of ubiquitinated CReP in the absence of p97 activity. Importantly, this finding reveals a direct involvement of p97 in stress signaling as an unanticipated aspect of p97 physiology. Phosphorylation of eIF2α is fine-tuned by stress kinases and the antagonizing PP1-CReP complex. CReP has a high constitutive turnover to facilitate responsiveness to regulation. We demonstrate that p97 ensures the high turnover and thus balances CReP levels in unperturbed cells. Moreover, we show here that stress-induced quantitative degradation of CReP requires the p97-Ufd1-Npl4 complex to allow robust phosphorylation of eIF2α in stressed cells, demonstrating that p97 enforces the integrated stress response on the level of signaling.
Intriguingly, CReP degradation is triggered by the SCF-β-TrCP ubiquitin ligase complex (54–57) as is degradation of two other p97-substrates, IkBα and CDC25A (58, 59). This reveals how p97 function is intertwined with stress signaling. DNA-damage induced degradation of CDC25A halts cell cycle progression, whereas CReP degradation is part of the integrated stress response that governs global protein synthesis through regulation of eIF2α phosphorylation. p97 has therefore an unanticipated dual role in maintaining cellular homeostasis (see model Fig. 5F). On one hand, it ensures clearance of damaged and misfolded proteins in a variety of pathways and compartments including the ER. This prevents activation of stress kinases such as PERK that phosphorylate eIF2α and attenuate translation. On the other hand, p97 ensures constitutive turnover of the eIF2α phosphatase component CReP. This is important to balance CReP activity in unperturbed cells, but also to ensure the timely drop in CReP levels under certain stresses to enforce phosphorylation of eIF2α. The quantitative removal of cellular CReP appears to be the result of both the efficient turnover by p97 and a block of CReP synthesis. Inhibitors of eIF2α dephosphorylation have been shown to protect against protein stress (60, 61). Conversely, p97 inhibitors aimed at triggering a proteostasis crisis and cell death in cancer cells (17, 62) will therefore function by perturbing both branches, bulk degradation of damaged proteins and protective signaling of the integrated stress response.
DATA AVAILABILITY
The mass spectrometry proteomics data has been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE (40) partner repository with the data set identifier PXD007876.
Supplementary Material
Acknowledgments
We thank Johannes van den Boom for critical reading and Chrisovalantis Papadopoulos for technical advice.
Footnotes
* The work was funded by DFG grant Me1626/3 to HM. BCC was supported by a Swiss National Science Foundation Ambizione grant (PZ00P3_161435). The group of RA was supported by ERC grants Proteomics v3.0 (AdG-233226 Proteomics v.3.0) and AdG-670821 Proteomics 4D), and the Swiss National Science Foundation (SNSF) grant number: 31003A_166435.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- ER
- endoplasmic reticulum
- AAA
- ATPases associated with diverse cellular activities
- ALS
- amyotrophic lateral sclerosis
- AMFR
- Autocrine motility factor receptor
- AP
- affinity purification
- As
- sodium arsenite
- ASPSCR1
- Alveolar soft part sarcoma chromosomal region candidate gene 1 protein
- ATXN3
- Ataxin-3
- BCA
- bicinchoninic acid
- β-TrCP
- beta-transducin repeat-containing protein
- cdc
- cell division cycle
- Chk1
- checkpoint kinase-1
- CHX
- cycloheximide
- CReP
- constitutive repressor of eIF2α phosphorylation
- CUL
- cullin
- DDA
- data dependent acquisition
- DOX
- doxycycline
- E3
- ubiquitin ligase
- EGF
- epidermal growth factor
- eIF2α
- eukaryotic translation initiation factor 2A
- EPSTI1
- Epithelial-stromal interaction protein 1
- ERAD
- ER-associated degradation
- EQ
- ATPase-deficient p97-E578Q mutant
- exp
- exposure
- FAF
- Fas-associated factor
- FDR
- False discovery rate
- FEM1B
- Fem-1 Homolog B
- GADD34
- Growth arrest and DNA damage-inducible protein
- GFP
- green fluorescent protein
- GLUL
- glutamine synthetase
- HEK
- human embryonic kidney
- HRP
- horseradish peroxidase
- IBMPFD
- inclusion body myopathy with early-onset Paget disease and frontotemporal dementia
- IkBa
- NF-kappa-B inhibitor alpha
- IP
- immunoprecipitation
- iRD
- indexed retention time
- ISR
- integrated stress response
- log
- logarithm
- MUL1
- Mitochondrial ubiquitin ligase activator of NFKB 1
- MYO5C
- Unconventional myosin-Vc
- NFkB
- nuclear factor NF-kappa-B
- NGLY1
- N-glycanase 1
- Npl4
- Nuclear protein localization protein 4 homolog
- NSFL1C
- N-ethylmaleimide-sensitive factor (NSF)L1 cofactor
- OTU
- ovarian tumor DUB domain
- PD
- pulldown
- PERK
- PKR-like ER kinase
- PKR
- protein kinase repressor
- PLAA
- phospholipase A-2-activating protein
- PP1
- Protein phosphatase-1
- ppm
- parts per million
- PPP1R
- Protein phosphatase 1 regulatory subunit
- psm
- peptide spectrum match
- RNF31
- RING finger protein 31
- RNF185
- RING finger protein 185
- SCF
- Skp1–Cullin–F-box
- SEL1L
- Protein sel-1 homolog 1
- s.e.m.
- standard error of the mean
- SEP
- Shp1-eyc-p47 domain
- SH3KBP1
- SH3 Domain Containing Kinase Binding Protein 1
- SHKBP1
- SH3KBP1-binding protein 1
- siRNA
- small interfering RNA
- SPRTN
- SprT-like domain-containing protein Spartan
- strep
- streptavidin
- SVIP
- small VCP/p97-interacting protein
- SWATH
- Sequential Window Acquisition of all Theoretical Mass Spectra
- Tg
- thapsigargin
- Tm
- tunicamycin
- TRAFD1
- TRAF-type zinc finger domain containing protein 1
- TRIC
- TRansfer of Identification Confidence
- Ub
- ubiquitin
- UBAC2
- Ubiquitin-associated domain-containing protein 2
- UBX
- ubiquitin domain-X
- UBE
- ubiquitin-conjugating enzyme
- UBXD
- UBX domain-containing protein
- Ufd
- Ubiquitin fusion degradation protein
- UPS
- ubiquitin-proteasome system
- UT
- untreated
- VCP
- Valosin-containing protein
- VCPIP1
- Valosin-containing protein p97/p47 complex-interacting protein 1
- WLS
- wntless Wnt ligand secretion mediator
- WT
- wild-type
- YOD1
- yeast OTU1 deubiquitinating enzyme 1 homolog
- ZFAND2B
- AN1-type zinc finger protein 2B.
REFERENCES
- 1. Harding H. P., Calfon M., Urano F., Novoa I., and Ron D. (2002) Transcriptional and translational control in the Mammalian unfolded protein response. Annu. Rev. Cell Dev. Biol. 18, 575–599 [DOI] [PubMed] [Google Scholar]
- 2. Harding H. P., Zhang Y., Bertolotti A., Zeng H., and Ron D. (2000) Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell 5, 897–904 [DOI] [PubMed] [Google Scholar]
- 3. Scheuner D., Song B., McEwen E., Liu C., Laybutt R., Gillespie P., Saunders T., Bonner-Weir S., and Kaufman R. J. (2001) Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell 7, 1165–1176 [DOI] [PubMed] [Google Scholar]
- 4. Harding H. P., Zhang Y., Zeng H., Novoa I., Lu P. D., Calfon M., Sadri N., Yun C., Popko B., Paules R., Stojdl D. F., Bell J. C., Hettmann T., Leiden J. M., and Ron D. (2003) An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11, 619–633 [DOI] [PubMed] [Google Scholar]
- 5. Pavitt G. D., and Ron D. (2012) New insights into translational regulation in the endoplasmic reticulum unfolded protein response. Cold Spring Harb. Perspect. Biol. 4, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Holcik M., and Sonenberg N. (2005) Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 6, 318–327 [DOI] [PubMed] [Google Scholar]
- 7. Connor J. H., Weiser D. C., Li S., Hallenbeck J. M., and Shenolikar S. (2001) Growth arrest and DNA damage-inducible protein GADD34 assembles a novel signaling complex containing protein phosphatase 1 and inhibitor 1. Mol. Cell. Biol. 21, 6841–6850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Novoa I., Zhang Y., Zeng H., Jungreis R., Harding H. P., and Ron D. (2003) Stress-induced gene expression requires programmed recovery from translational repression. EMBO J. 22, 1180–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jousse C., Oyadomari S., Novoa I., Lu P., Zhang Y., Harding H. P., and Ron D. (2003) Inhibition of a constitutive translation initiation factor 2alpha phosphatase, CReP, promotes survival of stressed cells. J. Cell Biol. 163, 767–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stolz A., Hilt W., Buchberger A., and Wolf D. H. (2011) Cdc48: a power machine in protein degradation. Trends Biochem. Sci. 36, 515–523 [DOI] [PubMed] [Google Scholar]
- 11. Meyer H., Bug M., and Bremer S. (2012) Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat. Cell Biol. 14, 117–123 [DOI] [PubMed] [Google Scholar]
- 12. Vembar S. S., and Brodsky J. L. (2008) One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xia D., Tang W. K., and Ye Y. (2016) Structure and function of the AAA+ ATPase p97/Cdc48p. Gene 583, 64–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Franz A., Ackermann L., and Hoppe T. (2016) Ring of change: CDC48/p97 drives protein dynamics at chromatin. Front. Gen. 7, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kimonis V. E., Fulchiero E., Vesa J., and Watts G. (2008) VCP disease associated with myopathy, Paget disease of bone and frontotemporal dementia: review of a unique disorder. Biochim. Biophys. Acta 1782, 744–748 [DOI] [PubMed] [Google Scholar]
- 16. Meyer H., and Weihl C. C. (2014) The VCP/p97 system at a glance: connecting cellular function to disease pathogenesis. J. Cell Sci. 127, 3877–3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Anderson D. J., Le Moigne R., Djakovic S., Kumar B., Rice J., Wong S., Wang J., Yao B., Valle E., Kiss von Soly S., Madriaga A., Soriano F., Menon M. K., Wu Z. Y., Kampmann M., Chen Y., Weissman J. S., Aftab B. T., Yakes F. M., Shawver L., Zhou H. J., Wustrow D., and Rolfe M. (2015) Targeting the AAA ATPase p97 as an Approach to Treat Cancer through Disruption of Protein Homeostasis. Cancer Cell 28, 653–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Magnaghi P., D'Alessio R., Valsasina B., Avanzi N., Rizzi S., Asa D., Gasparri F., Cozzi L., Cucchi U., Orrenius C., Polucci P., Ballinari D., Perrera C., Leone A., Cervi G., Casale E., Xiao Y., Wong C., Anderson D. J., Galvani A., Donati D., O'Brien T., Jackson P. K., and Isacchi A. (2013) Covalent and allosteric inhibitors of the ATPase VCP/p97 induce cancer cell death. Nat. Chem. Biol. 9, 548–556 [DOI] [PubMed] [Google Scholar]
- 19. Bodnar N. O., and Rapoport T. A. (2017) Molecular mechanism of substrate processing by the Cdc48 ATPase complex. Cell 169, 722–735 e729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ye Y., Meyer H. H., and Rapoport T. A. (2003) Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J. Cell Biol. 162, 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Blythe E. E., Olson K. C., Chau V., and Deshaies R. J. (2017) Ubiquitin- and ATP-dependent unfoldase activity of P97/VCP*NPLOC4*UFD1L is enhanced by a mutation that causes multisystem proteinopathy. Proc. Natl. Acad. Sci. U.S.A. 114, E4380–E4388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Buchberger A., Schindelin H., and Hanzelmann P. (2015) Control of p97 function by cofactor binding. FEBS Lett. 589, 2578–2589 [DOI] [PubMed] [Google Scholar]
- 23. Ritz D., Vuk M., Kirchner P., Bug M., Schütz S., Hayer A., Bremer S., Lusk C., Baloh R. H., Lee H., Glatter T., Gstaiger M., Aebersold R., Weihl C. C., and Meyer H. (2011) Endolysosomal sorting of ubiquitinated caveolin-1 is regulated by VCP/p97 and UBXD1 and impaired by VCP disease mutations. Nat. Cell Biol. 13, 1116–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Varjosalo M., Sacco R., Stukalov A., van Drogen A., Planyavsky M., Hauri S., Aebersold R., Bennett K. L., Colinge J., Gstaiger M., and Superti-Furga G. (2013) Interlaboratory reproducibility of large-scale human protein-complex analysis by standardized AP-MS. Nat. Methods 10, 307–314 [DOI] [PubMed] [Google Scholar]
- 25. Chen R., Rato C., Yan Y., Crespillo-Casado A., Clarke H. J., Harding H. P., Marciniak S. J., Read R. J., and Ron D. (2015) G-actin provides substrate-specificity to eukaryotic initiation factor 2alpha holophosphatases. eLife 4, 1–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dobrynin G., Popp O., Romer T., Bremer S., Schmitz M. H., Gerlich D. W., and Meyer H. (2011) Cdc48/p97-Ufd1-Npl4 antagonizes Aurora B during chromosome segregation in HeLa cells. J. Cell Sci. 124, 1571–1580 [DOI] [PubMed] [Google Scholar]
- 27. Busino L., Donzelli M., Chiesa M., Guardavaccaro D., Ganoth D., Dorrello N. V., Hershko A., Pagano M., and Draetta G. F. (2003) Degradation of Cdc25A by beta-TrCP during S phase and in response to DNA damage. Nature 426, 87–91 [DOI] [PubMed] [Google Scholar]
- 28. Meyer H. H., Shorter J. G., Seemann J., Pappin D., and Warren G. (2000) A complex of mammalian Ufd1 and Npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. EMBO J. 19, 2181–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kress E., Schwager F., Holtackers R., Seiler J., Prodon F., Zanin E., Eiteneuer A., Toya M., Sugimoto A., Meyer H., Meraldi P., and Gotta M. (2013) The UBXN-2/p37/p47 adaptors of CDC-48/p97 regulate mitosis by limiting the centrosomal recruitment of Aurora A. J. Cell Biol. 201, 559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Collins B. C., Gillet L. C., Rosenberger G., Rost H. L., Vichalkovski A., Gstaiger M., and Aebersold R. (2013) Quantifying protein interaction dynamics by SWATH mass spectrometry: application to the 14-3-3 system. Nat. Methods 10, 1246–1253 [DOI] [PubMed] [Google Scholar]
- 31. Collins B. C., Hunter C. L., Liu Y., Schilling B., Rosenberger G., Bader S. L., Chan D. W., Gibson B. W., Gingras A. C., Held J. M., Hirayama-Kurogi M., Hou G., Krisp C., Larsen B., Lin L., Liu S., Molloy M. P., Moritz R. L., Ohtsuki S., Schlapbach R., Selevsek N., Thomas S. N., Tzeng S. C., Zhang H., and Aebersold R. (2017) Multi-laboratory assessment of reproducibility, qualitative and quantitative performance of SWATH-mass spectrometry. Nat. Commun. 8, 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schubert O. T., Gillet L. C., Collins B. C., Navarro P., Rosenberger G., Wolski W. E., Lam H., Amodei D., Mallick P., MacLean B., and Aebersold R. (2015) Building high-quality assay libraries for targeted analysis of SWATH MS data. Nat. Protocols 10, 426–441 [DOI] [PubMed] [Google Scholar]
- 33. Craig R., Cortens J. P., and Beavis R. C. (2004) Open source system for analyzing, validating, and storing protein identification data. J. Proteome Res. 3, 1234–1242 [DOI] [PubMed] [Google Scholar]
- 34. MacLean B., Eng J. K., Beavis R. C., and McIntosh M. (2006) General framework for developing and evaluating database scoring algorithms using the TANDEM search engine. Bioinformatics 22, 2830–2832 [DOI] [PubMed] [Google Scholar]
- 35. Eng J. K., Jahan T. A., and Hoopmann M. R. (2013) Comet: an open-source MS/MS sequence database search tool. Proteomics 13, 22–24 [DOI] [PubMed] [Google Scholar]
- 36. Rost H. L., Rosenberger G., Navarro P., Gillet L., Miladinovic S. M., Schubert O. T., Wolski W., Collins B. C., Malmstrom J., Malmstrom L., and Aebersold R. (2014) OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nat. Biotechnol. 32, 219–223 [DOI] [PubMed] [Google Scholar]
- 37. Rosenberger G., Bludau I., Schmitt U., Heusel M., Hunter C. L., Liu Y., MacCoss M. J., MacLean B. X., Nesvizhskii A. I., Pedrioli P. G. A., Reiter L., Rost H. L., Tate S., Ting Y. S., Collins B. C., and Aebersold R. (2017) Statistical control of peptide and protein error rates in large-scale targeted data-independent acquisition analyses. Nat. Methods 14, 921–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schubert O. T., Ludwig C., Kogadeeva M., Zimmermann M., Rosenberger G., Gengenbacher M., Gillet L. C., Collins B. C., Rost H. L., Kaufmann S. H., Sauer U., and Aebersold R. (2015) Absolute Proteome Composition and Dynamics during Dormancy and Resuscitation of Mycobacterium tuberculosis. Cell Host Microbe 18, 96–108 [DOI] [PubMed] [Google Scholar]
- 39. Rosenberger G., Ludwig C., Rost H. L., Aebersold R., and Malmstrom L. (2014) aLFQ: an R-package for estimating absolute protein quantities from label-free LC-MS/MS proteomics data. Bioinformatics 30, 2511–2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vizcaino J. A., Cote R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Perez-Riverol Y., Reisinger F., Rios D., Wang R., and Hermjakob H. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–D1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Raman M., Sergeev M., Garnaas M., Lydeard J. R., Huttlin E. L., Goessling W., Shah J. V., and Harper J. W. (2015) Systematic proteomics of the VCP-UBXD adaptor network identifies a role for UBXN10 in regulating ciliogenesis. Nat. Cell Biol. 17, 1356–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Alexandru G., Graumann J., Smith G. T., Kolawa N. J., Fang R., and Deshaies R. J. (2008) UBXD7 Binds Multiple Ubiquitin Ligases And Implicates p97 in HIF1alpha Turnover. Cell 134, 804–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gillet L. C., Navarro P., Tate S., Rost H., Selevsek N., Reiter L., Bonner R., and Aebersold R. (2012) Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics 11, O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Papadopoulos C., Kirchner P., Bug M., Grum D., Koerver L., Schulze N., Poehler R., Dressler A., Fengler S., Arhzaouy K., Lux V., Ehrmann M., Weihl C. C., and Meyer H. (2017) VCP/p97 cooperates with YOD1, UBXD1 and PLAA to drive clearance of ruptured lysosomes by autophagy. EMBO J. 36, 135–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Huttlin E. L., Bruckner R. J., Paulo J. A., Cannon J. R., Ting L., Baltier K., Colby G., Gebreab F., Gygi M. P., Parzen H., Szpyt J., Tam S., Zarraga G., Pontano-Vaites L., Swarup S., White A. E., Schweppe D. K., Rad R., Erickson B. K., Obar R. A., Guruharsha K. G., Li K., Artavanis-Tsakonas S., Gygi S. P., and Harper J. W. (2017) Architecture of the human interactome defines protein communities and disease networks. Nature 545, 505–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Christianson J. C., Olzmann J. A., Shaler T. A., Sowa M. E., Bennett E. J., Richter C. M., Tyler R. E., Greenblatt E. J., Harper J. W., and Kopito R. R. (2012) Defining human ERAD networks through an integrative mapping strategy. Nat. Cell Biol. 14, 93–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Huttlin E. L., Ting L., Bruckner R. J., Gebreab F., Gygi M. P., Szpyt J., Tam S., Zarraga G., Colby G., Baltier K., Dong R., Guarani V., Vaites L. P., Ordureau A., Rad R., Erickson B. K., Wuhr M., Chick J., Zhai B., Kolippakkam D., Mintseris J., Obar R. A., Harris T., Artavanis-Tsakonas S., Sowa M. E., De Camilli P., Paulo J. A., Harper J. W., and Gygi S. P. (2015) The BioPlex network: a systematic exploration of the human interactome. Cell 162, 425–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xue L., Blythe E. E., Freiberger E. C., Mamrosh J. L., Hebert A. S., Reitsma J. M., Hess S., Coon J. J., and Deshaies R. J. (2016) Valosin-containing protein (VCP)-adaptor interactions are exceptionally dynamic and subject to differential modulation by a VCP Inhibitor. Mol. Cell. Proteomics 15, 2970–2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tang F., Wang B., Li N., Wu Y., Jia J., Suo T., Chen Q., Liu Y. J., and Tang J. (2011) RNF185, a novel mitochondrial ubiquitin E3 ligase, regulates autophagy through interaction with BNIP1. PLoS ONE 6, e24367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yun J., Puri R., Yang H., Lizzio M. A., Wu C., Sheng Z. H., and Guo M. (2014) MUL1 acts in parallel to the PINK1/parkin pathway in regulating mitofusin and compensates for loss of PINK1/parkin. eLife 3, e01958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. El Khouri E., Le Pavec G., Toledano M. B., and Delaunay-Moisan A. (2013) RNF185 is a novel E3 ligase of endoplasmic reticulum-associated degradation (ERAD) that targets cystic fibrosis transmembrane conductance regulator (CFTR). J. Biol. Chem. 288, 31177–31191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nguyen T. V., Li J., Lu C. J., Mamrosh J. L., Lu G., Cathers B. E., and Deshaies R. J. (2017) p97/VCP promotes degradation of CRBN substrate glutamine synthetase and neosubstrates. Proc. Natl. Acad. Sci. U.S.A. 114, 3565–3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Farrah T., Deutsch E. W., Hoopmann M. R., Hallows J. L., Sun Z., Huang C. Y., and Moritz R. L. (2013) The state of the human proteome in 2012 as viewed through PeptideAtlas. J. Proteome Res. 12, 162–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Loveless T. B., Topacio B. R., Vashisht A. A., Galaang S., Ulrich K. M., Young B. D., Wohlschlegel J. A., and Toczyski D. P. (2015) DNA damage regulates translation through beta-TRCP targeting of CReP. PLoS Gen. 11, e1005292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Coyaud E., Mis M., Laurent E. M., Dunham W. H., Couzens A. L., Robitaille M., Gingras A. C., Angers S., and Raught B. (2015) BioID-based identification of Skp Cullin F-box (SCF)beta-TrCP1/2 E3 ligase substrates. Mol. Cell. Proteomics 14, 1781–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Low T. Y., Peng M., Magliozzi R., Mohammed S., Guardavaccaro D., and Heck A. J. (2014) A systems-wide screen identifies substrates of the SCFbetaTrCP ubiquitin ligase. Sci. Signaling 7, rs8. [DOI] [PubMed] [Google Scholar]
- 57. Kim T. Y., Siesser P. F., Rossman K. L., Goldfarb D., Mackinnon K., Yan F., Yi X., MacCoss M. J., Moon R. T., Der C. J., and Major M. B. (2015) Substrate trapping proteomics reveals targets of the betaTrCP2/FBXW11 ubiquitin ligase. Mol. Cell. Biol. 35, 167–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li J. M., Wu H., Zhang W., Blackburn M. R., and Jin J. (2014) The p97-UFD1L-NPL4 protein complex mediates cytokine-induced IkappaBalpha proteolysis. Mol. Cell. Biol. 34, 335–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Riemer A., Dobrynin G., Dressler A., Bremer S., Soni A., Iliakis G., and Meyer H. (2014) The p97-Ufd1-Npl4 ATPase complex ensures robustness of the G 2/M checkpoint by facilitating CDC25A degradation. Cell Cycle 13, 919–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Das I., Krzyzosiak A., Schneider K., Wrabetz L., D'Antonio M., Barry N., Sigurdardottir A., and Bertolotti A. (2015) Preventing proteostasis diseases by selective inhibition of a phosphatase regulatory subunit. Science 348, 239–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Boyce M., Bryant K. F., Jousse C., Long K., Harding H. P., Scheuner D., Kaufman R. J., Ma D., Coen D. M., Ron D., and Yuan J. (2005) A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science 307, 935–939 [DOI] [PubMed] [Google Scholar]
- 62. Deshaies R. J. (2014) Proteotoxic crisis, the ubiquitin-proteasome system, and cancer therapy. BMC Biol. 12, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data has been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE (40) partner repository with the data set identifier PXD007876.