Abstract
Germline SDHA mutations are reported in a minority of pheochromocytoma/paraganglioma (PPGL) cases but are associated with an increased risk of malignancy, leading some to advocate cascade genetic testing and surveillance screening of “at-risk” first-degree relatives. However, such approaches rely on accurate estimates of variant pathogenicity and disease penetrance, which may have been subject to ascertainment and reporting biases, although the recent provision of large population-based DNA sequence data sets may provide a potentially unbiased resource to aid variant interpretation. Thus, the aim of the current study was to evaluate the pathogenicity and penetrance of SDHA variants reported in literature-based PPGL cases by comparing their frequency to those occurring in the Genome Aggregation Database (GnomAD) data set, which provides high-quality DNA sequence data on 138,632 individuals. In total, 39 different missense or loss-of-function (LOF) SDHA variants were identified in 95 PPGL index cases. Notably, many of the PPGL-associated SDHA alleles were observed at an unexpectedly high frequency in the GnomAD cohort, with ~1% and ~0.1% of the background population harboring a rare missense or LOF variant, respectively. Although the pathogenicity of several SDHA alleles was supported by significant enrichment in PPGL cases relative to GnomAD controls, calculations of disease penetrance based on allele frequencies in the respective cohorts resulted in much lower estimates than previously reported, ranging from 0.1% to 4.9%. Thus, although this study provides support for the etiological role of SDHA in PPGL formation, it suggests that most variant carriers will not manifest PPGLs and are unlikely to benefit from periodic surveillance screening.
Keywords: GnomAD, mutation, paraganglioma, penetrance, pheochromocytoma, SDHA
Germline SDHA variants are enriched in patients with pheochromocytoma/paraganglioma but are associated with very low disease penetrance such that most variant carriers will not manifest disease.
Pheochromocytomas and paragangliomas (PPGLs) are highly heritable tumors but display marked genetic heterogeneity such that ~35% of cases harbor germline mutations in one of ≥15 genes [1–4]. Consequently, genetic testing is recommended in all affected patients with PPGLs irrespective of a relevant family history and increasingly relies on comprehensive disease-targeted gene panels employing next-generation sequencing [1]. Indeed, the identification of a germline mutation in one of the PPGL susceptibility genes may not only have important clinical implications for the patient but may also facilitate cascade testing and periodic surveillance of “at-risk” first-degree relatives, although the appropriate implementation of such screening programs relies on accurate estimates of variant pathogenicity and disease penetrance [1].
Germline mutations in components of the succinate dehydrogenase complex are responsible for a significant proportion of nonsyndromic PPGL cases [5–7]. In particular, mutations in SDHB and SDHD have been established in large numbers of PPGL cases, providing unequivocal evidence of pathogenicity. Furthermore, the evaluation of cohorts of SDHB and SDHD variant carriers has facilitated reliable estimates of disease penetrance (i.e., ~20% and ~45% by age 60 years for SDHB and SDHD, respectively), thereby supporting the likely effectiveness of periodic surveillance of “at-risk” individuals [5, 8, 9].
In comparison with SDHB and SDHD, mutations in the SDHA subunit have only relatively recently been described in patients with PPGLs and are reported to occur at a markedly lower frequency [10–13]. Furthermore, the absence of a relevant family history in most cases indicates a reduced disease penetrance [3, 7, 12, 14]. However, patients with PPGLs harboring SDHA mutations are also reported to have an increased risk of malignancy, leading several experts to advocate cascade genetic testing in first-degree relatives to facilitate downstream surveillance screening [15, 16]. Such an approach appears to be supported by the recent reporting of large PPGL series in which 3% to 7% of PPGL cases harbored SDHA variants, with estimates of disease penetrance in variant carriers ranging from 10% to 30% [16, 17].
Large-scale population-level DNA sequence data sets provide an invaluable resource to investigate the potential causality of germline variants in hereditary monogenic disease [18, 19]. In particular, quantifying the spectrum and frequency of rare-coding variants in the background population provides a potentially unbiased approach to help establish variant pathogenicity and penetrance, and it avoids many of the ascertainment and reporting biases that may have hampered earlier genetic studies. Indeed, these approaches have enabled the reevaluation of genetic variants associated with several monogenic disease phenotypes, including cardiomyopathy, prion disease, and, more recently, hereditary endocrine disorders [19–22].
Thus, in the current study, we aimed to use the Genome Aggregation Database (GnomAD), which provides high-quality genetic data on 138,632 individuals, to evaluate the pathogenicity and penetrance of germline SDHA variants reported in prior studies of PPGLs.
1. Materials and Methods
A. Ascertainment of Cases
PubMed was used to identify PPGL cases reported in the literature in association with heterozygous germline SDHA variants (up to February 2018). Only apparently unrelated index cases were included in the analysis. Relevant demographic and clinical information was recorded, as was the presence or absence of a relevant family history [i.e., PPGL/gastrointestinal stromal tumor (GIST) or other relevant cancer]. SDHA variants were recorded according to the canonical transcript ENST00000264932. Variants reported as benign or likely benign were recorded but excluded from the overall analysis, whereas variants allocated as “variants of uncertain significance” were included in the analysis but identified as such throughout. The overall frequency of germline SDHA variants in PPGL cases was estimated from cohorts reporting germline SDHA sequencing results for ≥50 individuals.
B. Database Analysis
The GnomAD database provides high-quality variant calls on 138,632 individuals comprising 123,136 exomes and 15,496 genomes. Details of the data set, including sequencing, filtering, and calibration methods, are available at http://gnomad.broadinstitute.org [18]. All SDHA missense and loss-of-function (LOF) variants in the GnomAD cohort were identified (accessed July 2017 to January 2018). LOF variants comprised all single-nucleotide variants predicted to result in nonsense amino acid changes or disruption to canonical donor or acceptor splice sites, as well as small insertions and/or deletions (indels) predicting a frameshift in the encoded protein. Rare SDHA variants were defined as having an allele frequency (AF) <0.05% (i.e., affecting ≤1 in 1000 of the population). Each of the GnomAD missense SDHA variants (n = 357) was evaluated using the computational tools SIFT (http://sift.jcvi.org), Provean (http://provean.jcvi.org), and PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2). The frequency of the literature-based PPGL SDHA variants, together with those reported as pathogenic/likely pathogenic in the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/), was ascertained in the GnomAD data set. Finally, manual visualization of the variant sequences alongside a multiple-sequence alignment file was used to minimize the possibility that SDHA variants represented false-positive artifacts from known pseudogenes (SDHAP1, SDHAP2, SDHAP3). Notably, all rare (AF < 0.05%) missense and LOF SDHA variants observed in the GnomAD database occurred only in the heterozygous state. Therefore, the number of times each variant was observed was equal to the number of individuals in GnomAD carrying the variant.
C. Comparison of SDHA Variant Frequency in PPGL and GnomAD Cohorts and Estimates of Disease Penetrance
Odds ratios (together with 95% CIs) comparing the frequency of SDHA variants between PPGL and GnomAD cohorts were calculated at http://www.hutchon.net/confidor.htm. Estimates of penetrance were based on the Bayes theorem employing the following validated model [23]:
in which D = disease, G = genotype (i.e., variant under study), and = absence of disease. Thus, in this model, the penetrance or lifetime risk of disease given a specific genotype [P(D|G)] is equal to the genotype frequency in cases [P(G|D)] multiplied by the baseline lifetime risk of disease [P(D)], divided by overall genotype frequency in the cohort, which is the sum of the joint probabilities of the disease genotype frequency in cases [P(G|D)P(D)] and controls [23, 24]. However, we modified the approach such that we used allele frequency in place of genotype frequency [20]. CIs were established based on previously reported methods [24], although we adopted a conservative approach using 95% binomial exact CIs for case and control allele frequencies (calculated from the Clopper-Pearson exact method). Thus, the lower bounds for each penetrance CI were derived by using the lower bound for case and upper bound for control allele frequencies, whereas the upper bounds for each penetrance CI were derived from the upper bound for case and lower bound for control allele frequencies [20, 24]. Use of the 95% CI for both case and control allele frequencies ensures the coverage of the CIs for penetrance will be in excess of 95%. Penetrance estimates were established for individual and combined PPGL cohorts using the complete GnomAD data set as well as subpopulations based on the GnomAD and Exome Aggregation Consortium cohorts.
2. Results
A. PPGL-Associated SDHA Mutations
A total of 95 PPGL index cases with rare heterozygous SDHA variants were identified with equal sex distribution and mean age of 40 years (range, 15 to 81 years) (Table 1, Supplemental Table 1). Of these, ~50% presented with head and neck paraganglioma, whereas the remainder manifested either pheochromocytoma or extra-adrenal paraganglioma. Notably, only two index cases had a positive family history of either PPGL or GIST, and in each case, this represented a single affected relative. A small number of additional index cases reported a possible relevant family history, reflecting a single first-degree relative affected with a potentially relevant cancer (e.g., renal cell carcinoma), although in several cases, the SDHA carrier status of the affected relative was unknown and in one case was confirmed to be negative.
Table 1.
Summary of PPGL Index Cases Associated With SDHA Variants Reported in Literature
| Characteristic | Value |
|---|---|
| Index cases, No. | 95 |
| Age, mean (range), y | 40.0 (15–81) |
| Female/male, No. | 48/47 |
| Primary tumor site, No. (%)a | |
| Head and neck PGL | 47 (49) |
| Other PGLb | 30 (31) |
| Pheochromocytoma | 19 (20) |
| Family history, No. | |
| Positive for PPGL/GISTc | 2 |
| Positive for other relevant tumor typesc,d | 7 |
| SDHA variant type, No. (%) | |
| LOFe | 59 (62) |
| Missense | 36 (38) |
| Unique SDHA variants, No. | 39 |
Abbreviation: PGL, paraganglioma.
One patient had both a mediastinal PGL and carotid body tumor, resulting in n = 96 for primary tumor site.
Refers to all extra-adrenal paragangliomas excluding those affecting the head and neck.
In each index case with an apparent positive family history, a single affected family member was reported.
Other relevant tumors include pituitary adenoma and renal cell carcinoma. However, in some instances, the SDHA carrier status of the family member was unknown and in at least one case was known to be negative.
LOF variants include single-nucleotide variants resulting in nonsense amino acid change or canonical splice site disruption, as well as insertions and/or deletions (indels) resulting in a frameshift and premature truncation of the encoded protein.
Among the 95 index cases, 39 different germline heterozygous SDHA variants were observed. No patients were observed to harbor homozygous or compound heterozygous SDHA mutations. Overall, 62% of individuals harbored LOF SDHA variants (i.e., nonsense, splice site, or frameshift), with the remainder expressing missense variants. Notably, >40% of all index cases harbored the p.Arg31Ter nonsense mutation, although >50% of these cases were reported from a single series [17]. Several additional recurrent SDHA mutations were observed, including both LOF and missense variants, although most individual SDHA variants [28/39 (72%)] were observed in only single PPGL cases (Supplemental Fig. 1).
To establish the frequency of germline SDHA variants in patients with PPGL, we identified cohorts of >50 PPGL index cases in which complete SDHA sequencing was reported (Supplemental Table 2). Six studies were included, representing 1959 PPGL cases [16, 17, 25–28]. In this combined cohort, 3.6% of all patients with PPGLs harbored a heterozygous SDHA variant, although the frequency varied markedly between series (range, 0% to 7.6%). When evaluated by tumor site, the highest frequency of SDHA variants was observed in individuals with head and neck paraganglioma (overall, 6.3%; range, 0% to 12.1%), whereas the lowest frequency was observed in patients with adrenal pheochromocytoma (overall, 0.9%; range, 0% to 2.1%) (Supplemental Table 2).
B. SDHA Variants in the GnomAD Cohort
A high cumulative frequency of SDHA rare coding-region variation was observed in the GnomAD population, with ~1% of individuals in the cohort harboring a rare heterozygous SDHA missense variant (i.e., AF <0.05%), whereas strikingly, ~1 in every 1000 individuals carried a heterozygous LOF SDHA allele. Notably, when all GnomAD missense SDHA variants were evaluated using SIFT, Polyphen2, and Provean computational tools (n = 357), >75% were predicted to be potentially damaging (i.e., by one or more programs), with only ~22% predicted benign by each of SIFT, Polyphen-2, and Provean.
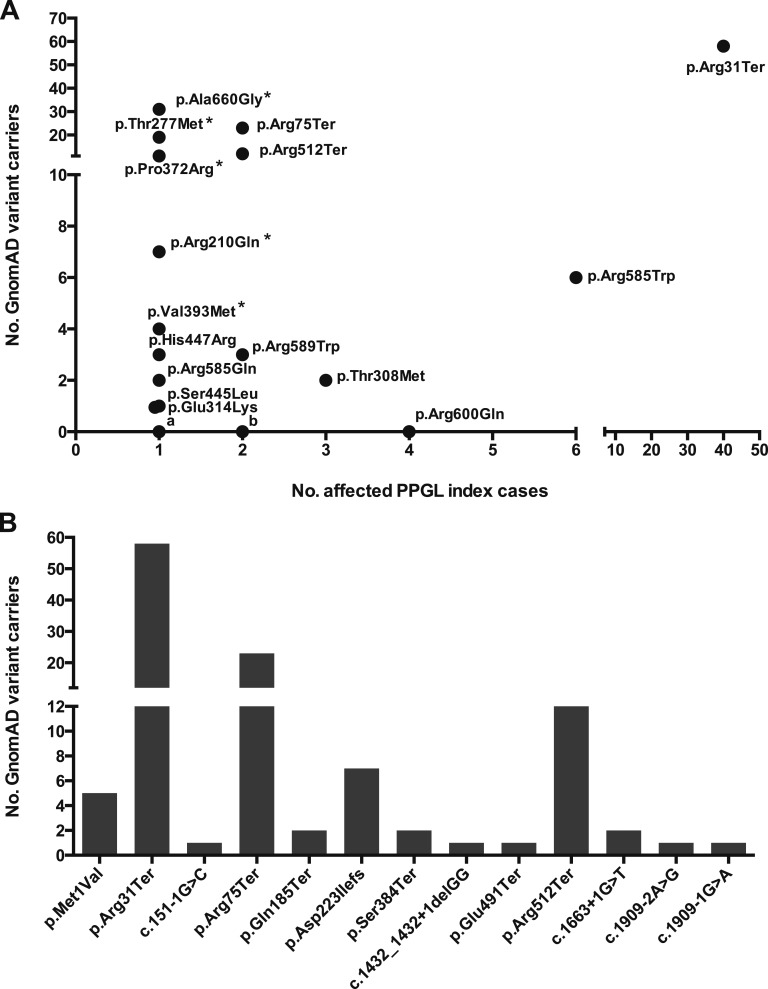
Next, the number of expected individuals at risk for PPGLs due to SDHA mutations in the GnomAD cohort was established. Thus, using the upper and lower bounds of PPGL disease incidence (i.e., 2 to 5/1,000,000/y) and SDHA mutation frequency in PPGL cases (i.e., 1% to 7%), a maximum of ~4 cases (range, 0.4 to 3.8) were predicted (Supplemental Table 3). However, when the GnomAD data set was examined for individuals harboring PPGL-associated SDHA variants, the number observed was several orders of magnitude higher than predicted, with ~1 in every 750 of the GnomAD cohort carrying a potentially pathogenic variant. In total, 15 of 39 (40%) of the different PPGL-associated SDHA alleles were observed in individuals in the GnomAD cohort, of which the p.Arg31Ter occurred at the highest frequency (Fig. 1A). A similar high number of GnomAD individuals harboring LOF variants reported as pathogenic in the ClinVar database were also observed (Fig. 1B). Taken together, the marked excess of individuals with deleterious SDHA alleles in the GnomAD cohort indicated either widespread variant misclassification or low disease penetrance.
Figure 1.
Frequency of PPGL-associated SDHA variants in the GnomAD cohort. (A) Of the 39 individual SDHA variants reported in the literature in association with PPGL index cases, 15 (~40%) were observed in the GnomAD database. Strikingly, the p.Arg31Ter variant was observed at the highest frequency in both the PPGL and GnomAD cohorts. Several additional SDHA variants were observed recurrently in both disease and control cohorts (e.g., Arg585Trp, pThr308Met, p.Arg75Ter, p.Arg512Ter, p.Arg589Trp). In total, 183 individuals in the GnomAD database harbored one of the literature-associated PPGL-associated SDHA variants, representing ~1 in 750 of the GnomAD population. Excluding SDHA alleles reported as “variants of uncertain significance” in their original report (marked with an “*”), 111 GnomAD individuals harbored likely causative SDHA variants (i.e., ~1 in 1250). In contrast, 24 of the PPGL-associated SDHA variants were not observed in the GnomAD database. Of these, 19 variants were observed in single PPGL index cases (denoted “a”), 4 variants were observed twice (denoted “b”), and the p.Arg600Gln variant was observed in 4 unrelated PPGL cases. (B) All pathogenic or likely pathogenic LOF SDHA variants reported in the ClinVar database in association with PPGL/Hereditary Cancer Predisposition were identified and their frequency in the GnomAD cohort evaluated. In total, 33 putative LOF alleles were identified, of which 13 were observed in the GnomAD database (shown above). Overall, 116 individuals in the GnomAD cohort harbored one of the LOF SDHA alleles, equating to ~1 in 1200 of the cohort. All PPGL-associated variants were observed in the heterozygous state and are described relative to the canonical transcript ENST00000264932.
C. Pathogenicity and Penetrance of SDHA Variants
Having observed the high background frequency of individuals harboring potentially pathogenic SDHA variants in the GnomAD population, we next evaluated whether there was a significant excess of SDHA variants in the literature-based PPGL disease cohort relative to the GnomAD controls. We initially focused these studies on p.Arg31Ter as this PPGL-associated variant was observed at the highest frequency in both disease and control cohorts (Fig. 1A). Thus, using the combined cohort of 1959 PPGL cases (Supplemental Table 2), a significant excess of the p.Arg31Ter variant was observed in the disease population relative to the GnomAD controls (Supplemental Table 4), whereas a similar excess was demonstrated in individual PPGL cohorts compared with both “Global” and “European” GnomAD populations, although the extent of enrichment varied between series (Supplemental Table 4). Similarly, a marked excess of LOF SDHA variants was observed in the PPGL cohorts when evaluated cumulatively, despite the high number of individuals harboring LOF alleles in the GnomAD population (Supplemental Table 4). Thus, together, these studies provide additional strong support for an etiological role for SDHA in PPGL formation.
Finally, we established variant-level estimates of disease penetrance using the allele frequencies of the respective SDHA variants observed in disease (i.e., literature-based cohort) and control populations (i.e., GnomAD-based cohorts) (Supplemental Tables 5 and 6). Strikingly, the penetrance estimates were very low, with the majority ranging from 0.1% to 2%, although these varied according to the PPGL and control cohorts used (Table 2). For example, the highest estimates of penetrance were observed for the nonsense p.Arg31Ter variant in the PPGL cohort reported by van der Tuin et al. [17] (range, 1.7% to 4.9%) (Table 2), although these estimates remain considerably lower than those reported in the literature.
Table 2.
Penetrance Estimates for Recurrent PPGL-Associated SDHA Variants
| SDHA Variant Penetrance (%) | Cumulative LOF SNV Penetrance (%) a | ||||
|---|---|---|---|---|---|
| Arg31Ter | Arg75Ter | Arg512Ter | Arg585Trp b | ||
| Combined PPGL cohortc (n = 1959) | |||||
| vs GnomAD global | 0.90 (0.47–1.69) | 0.15 (0.01–0.86) | 0.15 (0.00–1.59) | 0.58 (0.03–5.47) | 0.64 (0.39–1.01) |
| vs GnomAD Europeand | 0.46 (0.24–0.88) | 0.09 (0.01–0.58) | 0.08 (0.00–0.92) | 0.40 (0.02–5.07) | 0.39 (0.23–0.63) |
| vs GnomAD Genomese | 0.59 (0.21–1.73) | 0.20 (0.01–5.56) | 0.05 (0.00–0.10) | 0.39 (0.00–36.0) | 0.47 (0.22–1.05) |
| vs ExAC non-TCGAf | 1.34 (0.55–3.34) | 0.07 (0.01–0.43) | 0.11 (0.00–1.68) | 0.45 (0.02–7.33) | 0.60 (0.33–1.06) |
| Bausch et al. (16) PPGL cohort (n = 972) | |||||
| vs GnomAD global | 0.31 (0.08–0.93) | 0.30 (0.02–1.72) | — | — | 0.34 (0.15–0.70) |
| vs GnomAD European | 0.16 (0.03–0.48) | 0.19 (0.01–1.17) | — | — | 0.20 (0.09–0.44) |
| vs GnomAD Genomes | 0.20 (0.03–0.96) | 0.40 (0.01–10.6) | — | — | 0.25 (0.08–0.72) |
| vs ExAC non-TCGA | 0.45 (0.09–1.86) | 0.14 (0.01–0.85) | — | — | 0.31 (0.12–0.73) |
| van der Tuin et al. (17) PPGL cohort (n = 393) | |||||
| vs GnomAD global | 3.38 (1.69–6.41) | — | 0.72 (0.01–7.43) | 2.85 (0.16–22.3) | 1.77 (0.99–3.05) |
| vs GnomAD European | 1.74 (0.86–3.43) | — | 0.40 (0.01–4.44) | 1.97 (0.09–21.0) | 1.08 (0.58–1.92) |
| vs GnomAD Genomes | 2.21 (0.78–6.58) | — | 0.25 (0.00–4.78) | 1.93 (0.04–73.9) | 1.33 (0.56–3.15) |
| vs ExAC non-TCGA | 4.9 (1.96–12.1) | — | 0.56 (0.01–7.86) | 2.20 (0.09–28.2) | 1.66 (0.83–3.18) |
| vs van der Tuin et al. control cohortg | 1.91 (0.57–7.34) | — | — | — | — |
Penetrance estimates are expressed as percent (95% CI). The methods for calculating penetrance estimates together with the respective CIs are described in the Materials and Methods. Baseline lifetime risk of PPGL was estimated to be 0.025% (i.e., 1/4000) based on a midrange incidence estimate of 3 to 3.5/1,000,000 and a ~80-year window of disease susceptibility. Additional details are provided in the footnotes to Supplemental Table 3. The absence of a penetrance estimate (marked —) indicates the absence of the SDHA variant in the respective case and/or control cohort or insufficient information to establish control allele frequencies.
The LOF single-nucleotide variant (SNV) penetrance estimate accounts for the cumulative frequencies of all nonsense and canonical splice site SDHA variants in the respective disease and control cohorts.
The CIs associated with the penetrance estimates for the Arg585Trp variant are noted to be very wide. Notably, this variant was associated with very low variant allele counts (i.e., one or two) in disease and/or control subpopulations, giving rise to large 95% binomial exact CIs for the respective case and control allele frequencies.
Combined cohort as described in Supplemental Tables 2 and 5.
GnomAD European cohort selected to represent most suitable comparator group as each of the combined and individual PPGL cohorts included individuals of predominantly European origin.
GnomAD Genomes cohort was used to reduce the potential for any confounding from the inclusion of samples for The Cancer Genome Atlas (TCGA). Although GnomAD contains 7208 samples from the TCGA database, none are represented by the 15,496 individuals in whom whole-genome sequencing was undertaken (personal correspondence from GnomAD curators).
Exome Aggregation Consortium (ExAC) non-TCGA cohort provides an alternative comparator group in which all TCGA samples (n = 7601) have been removed from the ExAC population, leaving a remaining cohort of 53,105. This was used to establish population allele frequencies with reduced susceptibility to confounding from the inclusion germline samples from individuals with cancer.
The series reported by van der Tuin et al. (17) reported an “in-house” whole-exome control population in which the frequency of the Arg31Ter variant was established. However, no data were provided on other LOF alleles to allow additional allele frequencies to be established.
3. Discussion
The successful implementation of clinical genetic testing requires accurate estimates of variant pathogenicity and disease penetrance to provide appropriate management of the patient and wider family. In this study, we combined genetic data from large disease and population-based control cohorts to provide additional insight into the role of the SDHA gene in PPGL formation. Most notably, these studies provide support for the role of SDHA in PPGL tumorigenesis while simultaneously demonstrating that variants in SDHA are likely associated with a much lower disease penetrance than those reported for other components of the succinate dehydrogenase complex (i.e., SDHB/SDHD). Thus, SDHA appears to act as a low-penetrance risk allele for PPGL formation.
Several important features emerged during this study. First, the observation that most PPGL index cases reported no positive family history of PPGLs/GISTs is supportive of the low disease penetrance, although it is noteworthy that several individuals reported family members with other tumor types, including renal cell carcinoma, in which SDHA has been implicated [7]. Thus, future studies should aim to define the full range of tumor phenotypes associated with SDHA mutation, which likely extend beyond PPGLs and GISTs. Another striking feature of the current study was the high frequency of the p.Arg31Ter variant, which was observed in a disproportionate number of PPGL cases relative to other LOF alleles. Although this may partly reflect the makeup of the specific populations under study (i.e., a high number of p.Arg31Ter cases from the Netherlands), it suggests there may be variant-specific factors that increase tumor risk (e.g., cis-acting genetic elements not captured in the current study or influences of the truncating variant on expression of the wild-type allele).
The current analysis only allowed penetrance estimates for SDHA variants observed in both disease and control cohorts, with the most reliable estimates (i.e., narrow CIs) obtained for those observed multiple times. Thus, although 15 of 39 unique PPGL-associated SDHA variants occurred in the GnomAD population, the remaining 24 variants were not observed. However, ~80% (19/24) of these variants were observed in single patients with PPGLs, and the appropriate interpretation of such variants remains challenging. For example, ~50% of the different nonsynonymous SDHA variants observed in the GnomAD population occurred in single individuals, and consequently, it may not be possible to distinguish disease-causing mutations from those very rare “background” coding variants identified incidentally. In this regard, missense variants present a particular challenge, and it is notable that a high number of such variants were observed in PPGL index cases, frequently affecting single individuals. Thus, the high background frequency of rare missense SDHA variants observed in GnomAD indicates that some of the PPGL-associated missense variants may have been susceptible to misclassification. Furthermore, we demonstrate the limited specificity of the computational tools frequently used in support of variant pathogenicity, with most GnomAD missense SDHA variants predicted to be deleterious by at least one of the prediction programs. Thus, any high-volume genetic testing for SDHA should anticipate the identification of rare missense SDHA variants, including those not previously observed in control cohorts, which will remain problematic for interpretation.
There are several potential limitations to the current study. The estimates of penetrance rely on accurate variant allele frequencies in both disease and control cohorts. In the current study, we ascertained the SDHA allele frequency in PPGL cases using both individual and combined PPGL cohorts. However, several of the larger cohorts excluded individuals with mutations in more common PPGL-associated genes [16, 17], which in turn will overstate the frequency of SDHA variants in unselected PPGL cases (i.e., the true denominator will be underrepresented). Furthermore, it is also possible that these cohorts included PPGL cases in which a genetic diagnosis was considered more likely. Thus, each of these potential limitations will likely overestimate the SDHA mutation frequency in cases, and as a consequence, our low estimates of disease penetrance may in fact be overstated. In the future, accurate estimates of SDHA mutation frequency in PPGL cases will require the systematic sequencing of large unselected PPGL cohorts. Similarly, for accurate estimates of penetrance, it is necessary that disease and control cohorts are closely matched in terms of population stratification. To address this potential issue, we established allele frequencies not only for the complete GnomAD population but also for additional control cohorts selected to act as suitable comparators (e.g., “GnomAD European” population). Likewise, to ensure our results were not confounded by the unintentional enrichment for relevant disease phenotypes within the control cohort, we evaluated subpopulations in which individuals with known cancers were excluded, and reassuringly, the penetrance estimates based on these groups did not differ markedly from the larger cohorts.
In summary, these studies support a clear etiological role for SDHA in PPGL tumorigenesis while simultaneously indicating that most pathogenic SDHA alleles are associated with very low disease penetrance. These studies suggest that undertaking predictive testing in first-degree relatives with subsequent clinical, biochemical, and radiological surveillance in variant carriers is unlikely to provide an effective strategy for PPGL detection. Thus, based on these population-level genetic data, testing of asymptomatic first-degree relatives is not currently recommended. However, it is likely that future large-scale sequencing projects, coupled with detailed phenotype data, will more accurately elucidate the risks in variant carriers. Furthermore, identifying the potential genetic and/or environmental factors that influence disease expression should be a priority for future studies.
Supplementary Material
Acknowledgments
Financial Support: P.J.N. holds a Scottish Senior Clinical Fellowship funded by the Chief Scientist Office/NHS Research Scotland and the University of Dundee (SCAF/15/01), Scotland. P.M. received a vacation scholarship from the Dundee Clinical Academic Tract Scheme.
Disclosure Summary: The authors have nothing to disclose.
Glossary
Abbreviations:
- AF
allele frequency
- GIST
gastrointestinal stromal tumor
- GnomAD
Genome Aggregation Database
- LOF
loss of function
- PPGL
pheochromocytoma/paraganglioma
References and Notes
- 1. Toledo RA, Burnichon N, Cascon A, Benn DE, Bayley JP, Welander J, Tops CM, Firth H, Dwight T, Ercolino T, Mannelli M, Opocher G, Clifton-Bligh R, Gimm O, Maher ER, Robledo M, Gimenez-Roqueplo AP, Dahia PL; NGS in PPGL (NGSnPPGL) Study Group . Consensus statement on next-generation-sequencing-based diagnostic testing of hereditary phaeochromocytomas and paragangliomas. Nat Rev Endocrinol. 2016;13(4):233–247. [DOI] [PubMed] [Google Scholar]
- 2. Favier J, Amar L, Gimenez-Roqueplo AP. Paraganglioma and phaeochromocytoma: from genetics to personalized medicine. Nat Rev Endocrinol. 2014;11(2):101–111. [DOI] [PubMed] [Google Scholar]
- 3. Crona J, Taïeb D, Pacak K. New perspectives on pheochromocytoma and paraganglioma: toward a molecular classification. Endocr Rev. 2017;38(6):489–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fishbein L, Leshchiner I, Walter V, Danilova L, Robertson AG, Johnson AR, Lichtenberg TM, Murray BA, Ghayee HK, Else T, Ling S, Jefferys SR, de Cubas AA, Wenz B, Korpershoek E, Amelio AL, Makowski L, Rathmell WK, Gimenez-Roqueplo AP, Giordano TJ, Asa SL, Tischler AS, Pacak K, Nathanson KL, Wilkerson MD; Cancer Genome Atlas Research Network . Comprehensive molecular characterization of pheochromocytoma and paraganglioma. Cancer Cell. 2017;31(2):181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Andrews KA, Ascher DB, Pires DEV, Barnes DR, Vialard L, Casey RT, Bradshaw N, Adlard J, Aylwin S, Brennan P, Brewer C, Cole T, Cook JA, Davidson R, Donaldson A, Fryer A, Greenhalgh L, Hodgson SV, Irving R, Lalloo F, McConachie M, McConnell VPM, Morrison PJ, Murday V, Park SM, Simpson HL, Snape K, Stewart S, Tomkins SE, Wallis Y, Izatt L, Goudie D, Lindsay RS, Perry CG, Woodward ER, Antoniou AC, Maher ER. Tumour risks and genotype-phenotype correlations associated with germline variants in succinate dehydrogenase subunit genes SDHB, SDHC and SDHD. J Med Genet. 2018;55(6):384–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Evenepoel L, Papathomas TG, Krol N, Korpershoek E, de Krijger RR, Persu A, Dinjens WN. Toward an improved definition of the genetic and tumor spectrum associated with SDH germ-line mutations. Genet Med. 2014;17(8):610–620. [DOI] [PubMed] [Google Scholar]
- 7. Benn DE, Robinson BG, Clifton-Bligh RJ. 15 Years of paraganglioma: clinical manifestations of paraganglioma syndromes types 1–5. Endocr Relat Cancer. 2015;22(4):T91–T103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rijken JA, Niemeijer ND, Jonker MA, Eijkelenkamp K, Jansen JC, van Berkel A, Timmers HJLM, Kunst HPM, Bisschop PHLT, Kerstens MN, Dreijerink KMA, van Dooren MF, van der Horst-Schrivers ANA, Hes FJ, Leemans CR, Corssmit EPM, Hensen EF. The penetrance of paraganglioma and pheochromocytoma in SDHB germline mutation carriers. Clin Genet. 2017;93(1):60–66. [DOI] [PubMed] [Google Scholar]
- 9. Plouin PF, Amar L, Dekkers OM, Fassnacht M, Gimenez-Roqueplo AP, Lenders JW, Lussey-Lepoutre C, Steichen O; Guideline Working Group . European Society of Endocrinology Clinical Practice Guideline for long-term follow-up of patients operated on for a phaeochromocytoma or a paraganglioma. Eur J Endocrinol. 2016;174(5):G1–G10. [DOI] [PubMed] [Google Scholar]
- 10. Welander J, Garvin S, Bohnmark R, Isaksson L, Wiseman RW, Söderkvist P, Gimm O. Germline SDHA mutation detected by next-generation sequencing in a young index patient with large paraganglioma. J Clin Endocrinol Metab. 2013;98(8):E1379–E1380. [DOI] [PubMed] [Google Scholar]
- 11. Dwight T, Mann K, Benn DE, Robinson BG, McKelvie P, Gill AJ, Winship I, Clifton-Bligh RJ. Familial SDHA mutation associated with pituitary adenoma and pheochromocytoma/paraganglioma. J Clin Endocrinol Metab. 2013;98(6):E1103–E1108. [DOI] [PubMed] [Google Scholar]
- 12. Korpershoek E, Favier J, Gaal J, Burnichon N, van Gessel B, Oudijk L, Badoual C, Gadessaud N, Venisse A, Bayley JP, van Dooren MF, de Herder WW, Tissier F, Plouin PF, van Nederveen FH, Dinjens WN, Gimenez-Roqueplo AP, de Krijger RR. SDHA immunohistochemistry detects germline SDHA gene mutations in apparently sporadic paragangliomas and pheochromocytomas. J Clin Endocrinol Metab. 2011;96(9):E1472–E1476. [DOI] [PubMed] [Google Scholar]
- 13. Burnichon N, Brière JJ, Libé R, Vescovo L, Rivière J, Tissier F, Jouanno E, Jeunemaitre X, Bénit P, Tzagoloff A, Rustin P, Bertherat J, Favier J, Gimenez-Roqueplo AP. SDHA is a tumor suppressor gene causing paraganglioma. Hum Mol Genet. 2010;19(15):3011–3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Casey RT, Ascher DB, Rattenberry E, Izatt L, Andrews KA, Simpson HL, Challis B, Park SM, Bulusu VR, Lalloo F, Pires DEV, West H, Clark GR, Smith PS, Whitworth J, Papathomas TG, Taniere P, Savisaar R, Hurst LD, Woodward ER, Maher ER. SDHA related tumorigenesis: a new case series and literature review for variant interpretation and pathogenicity. Mol Genet Genomic Med. 2017;5(3):237–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tufton N, Ghelani R, Srirangalingam U, Kumar AV, Drake WM, Iacovazzo D, Skordilis K, Berney D, Al-Mrayat M, Khoo B, Akker SA. SDHA mutated paragangliomas may be at high risk of metastasis. Endocr Relat Cancer. 2017;24(7):L43–L49. [DOI] [PubMed] [Google Scholar]
- 16. Bausch B, Schiavi F, Ni Y, Welander J, Patocs A, Ngeow J, Wellner U, Malinoc A, Taschin E, Barbon G, Lanza V, Söderkvist P, Stenman A, Larsson C, Svahn F, Chen JL, Marquard J, Fraenkel M, Walter MA, Peczkowska M, Prejbisz A, Jarzab B, Hasse-Lazar K, Petersenn S, Moeller LC, Meyer A, Reisch N, Trupka A, Brase C, Galiano M, Preuss SF, Kwok P, Lendvai N, Berisha G, Makay Ö, Boedeker CC, Weryha G, Racz K, Januszewicz A, Walz MK, Gimm O, Opocher G, Eng C, Neumann HPH; European-American-Asian Pheochromocytoma-Paraganglioma Registry Study Group . Clinical characterization of the pheochromocytoma and paraganglioma susceptibility genes SDHA, TMEM127, MAX, and SDHAF2 for gene-informed prevention. JAMA Oncol. 2017;3(9):1204–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. van der Tuin K, Mensenkamp AR, Tops CMJ, Corssmit EPM, Dinjens WN, van de Horst-Schrivers AN, Jansen JC, de Jong MM, Kunst HPM, Kusters B, Leter EM, Morreau H, van Nesselrooij BMP, Oldenburg RA, Spruijt L, Hes FJ, Timmers HJLM. Clinical aspects of SDHA-related pheochromocytoma and paraganglioma: a nationwide study. J Clin Endocrinol Metab. 2017;103(2):438–445. [DOI] [PubMed] [Google Scholar]
- 18. Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won HH, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG; Exome Aggregation Consortium . Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Newey PJ, Berg JN, Zhou K, Palmer CNA, Thakker RV. Utility of population-level DNA sequence data in the diagnosis of hereditary endocrine disease. J Endocr Soc. 2017;1(12):1507–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Minikel EV, Vallabh SM, Lek M, Estrada K, Samocha KE, Sathirapongsasuti JF, McLean CY, Tung JY, Yu LP, Gambetti P, Blevins J, Zhang S, Cohen Y, Chen W, Yamada M, Hamaguchi T, Sanjo N, Mizusawa H, Nakamura Y, Kitamoto T, Collins SJ, Boyd A, Will RG, Knight R, Ponto C, Zerr I, Kraus TF, Eigenbrod S, Giese A, Calero M, de Pedro-Cuesta J, Haïk S, Laplanche JL, Bouaziz-Amar E, Brandel JP, Capellari S, Parchi P, Poleggi A, Ladogana A, O’Donnell-Luria AH, Karczewski KJ, Marshall JL, Boehnke M, Laakso M, Mohlke KL, Kähler A, Chambert K, McCarroll S, Sullivan PF, Hultman CM, Purcell SM, Sklar P, van der Lee SJ, Rozemuller A, Jansen C, Hofman A, Kraaij R, van Rooij JG, Ikram MA, Uitterlinden AG, van Duijn CM, Daly MJ, MacArthur DG; Exome Aggregation Consortium (ExAC) . Quantifying prion disease penetrance using large population control cohorts. Sci Transl Med. 2016;8(322):322ra9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Walsh R, Thomson KL, Ware JS, Funke BH, Woodley J, McGuire KJ, Mazzarotto F, Blair E, Seller A, Taylor JC, Minikel EV, MacArthur DG, Farrall M, Cook SA, Watkins H; Exome Aggregation Consortium . Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet Med. 2016;19(2):192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Manrai AK, Funke BH, Rehm HL, Olesen MS, Maron BA, Szolovits P, Margulies DM, Loscalzo J, Kohane IS. Genetic misdiagnoses and the potential for health disparities. N Engl J Med. 2016;375(7):655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stessman HAF, Willemsen MH, Fenckova M, Penn O, Hoischen A, Xiong B, Wang T, Hoekzema K, Vives L, Vogel I, Brunner HG, van der Burgt I, Ockeloen CW, Schuurs-Hoeijmakers JH, Klein Wassink-Ruiter JS, Stumpel C, Stevens SJC, Vles HS, Marcelis CM, van Bokhoven H, Cantagrel V, Colleaux L, Nicouleau M, Lyonnet S, Bernier RA, Gerdts J, Coe BP, Romano C, Alberti A, Grillo L, Scuderi C, Nordenskjöld M, Kvarnung M, Guo H, Xia K, Piton A, Gerard B, Genevieve D, Delobel B, Lehalle D, Perrin L, Prieur F, Thevenon J, Gecz J, Shaw M, Pfundt R, Keren B, Jacquette A, Schenck A, Eichler EE, Kleefstra T. Disruption of POGZ is associated with intellectual disability and autism spectrum disorders. Am J Hum Genet. 2016;98(3):541–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rosenfeld JA, Coe BP, Eichler EE, Cuckle H, Shaffer LG. Estimates of penetrance for recurrent pathogenic copy-number variations. Genet Med. 2012;15(6):478–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sbardella E, Cranston T, Isidori AM, Shine B, Pal A, Jafar-Mohammadi B, Sadler G, Mihai R, Grossman AB. Routine genetic screening with a multi-gene panel in patients with pheochromocytomas. Endocrine. 2017;59(1):175–182. [DOI] [PubMed] [Google Scholar]
- 26. Currás-Freixes M, Inglada-Pérez L, Mancikova V, Montero-Conde C, Letón R, Comino-Méndez I, Apellániz-Ruiz M, Sánchez-Barroso L, Aguirre Sánchez-Covisa M, Alcázar V, Aller J, Álvarez-Escolá C, Andía-Melero VM, Azriel-Mira S, Calatayud-Gutiérrez M, Díaz JA, Díez-Hernández A, Lamas-Oliveira C, Marazuela M, Matias-Guiu X, Meoro-Avilés A, Patiño-García A, Pedrinaci S, Riesco-Eizaguirre G, Sábado-Álvarez C, Sáez-Villaverde R, Sainz de Los Terreros A, Sanz Guadarrama Ó, Sastre-Marcos J, Scolá-Yurrita B, Segura-Huerta Á, Serrano-Corredor ML, Villar-Vicente MR, Rodríguez-Antona C, Korpershoek E, Cascón A, Robledo M. Recommendations for somatic and germline genetic testing of single pheochromocytoma and paraganglioma based on findings from a series of 329 patients. J Med Genet. 2015;52(10):647–656. [DOI] [PubMed] [Google Scholar]
- 27. Rattenberry E, Vialard L, Yeung A, Bair H, McKay K, Jafri M, Canham N, Cole TR, Denes J, Hodgson SV, Irving R, Izatt L, Korbonits M, Kumar AV, Lalloo F, Morrison PJ, Woodward ER, Macdonald F, Wallis Y, Maher ER. A comprehensive next generation sequencing-based genetic testing strategy to improve diagnosis of inherited pheochromocytoma and paraganglioma. J Clin Endocrinol Metab. 2013;98(7):E1248–E1256. [DOI] [PubMed] [Google Scholar]
- 28. Piccini V, Rapizzi E, Bacca A, Di Trapani G, Pulli R, Giachè V, Zampetti B, Lucci-Cordisco E, Canu L, Corsini E, Faggiano A, Deiana L, Carrara D, Tantardini V, Mariotti S, Ambrosio MR, Zatelli MC, Parenti G, Colao A, Pratesi C, Bernini G, Ercolino T, Mannelli M. Head and neck paragangliomas: genetic spectrum and clinical variability in 79 consecutive patients. Endocr Relat Cancer. 2012;19(2):149–155. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.