Abstract
Plant regulatory small RNAs (sRNAs), which include most microRNAs (miRNAs) and a subset of small interfering RNAs (siRNAs), such as the phased siRNAs (phasiRNAs), play important roles in regulating gene expression. Although generated from genetically distinct biogenesis pathways, these regulatory sRNAs share the same mechanisms for post-translational gene silencing and translational inhibition. psRNATarget was developed to identify plant sRNA targets by (i) analyzing complementary matching between the sRNA sequence and target mRNA sequence using a predefined scoring schema and (ii) by evaluating target site accessibility. This update enhances its analytical performance by developing a new scoring schema that is capable of discovering miRNA–mRNA interactions at higher ‘recall rates’ without significantly increasing total prediction output. The scoring procedure is customizable for the users to search both canonical and non-canonical targets. This update also enables transmitting and analyzing ‘big’ data empowered by (a) the implementation of multi-threading chunked file uploading, which can be paused and resumed, using HTML5 APIs and (b) the allocation of significantly more computing nodes to its back-end Linux cluster. The updated psRNATarget server has clear, compelling and user-friendly interfaces that enhance user experiences and present data clearly and concisely. The psRNATarget is freely available at http://plantgrn.noble.org/psRNATarget/.
INTRODUCTION
Plant regulatory small RNAs (sRNAs) are produced from double-stranded RNA duplexes or hairpin single-stranded RNA precursors by the endonuclease activities of Dicer-like (DCL) proteins. sRNAs produced from double-stranded duplexes are referred to as small interfering RNAs (siRNAs), while sRNAs produced from the stem-loops of single-stranded precursors are referred to as micro RNAs (miRNAs) (1). Plant regulatory sRNAs control a range of cellular and developmental functions, including plant cellular defense mechanisms against RNA viruses, transcriptional gene silencing by guiding heterochromatin formation at homologous loci, and sRNA-mediated DNA methylation (1). Among the many roles of plant sRNAs, post-transcriptional gene silencing and translational inhibition guided by miRNA and phased small interfering RNA (phasiRNA) are the two most widely studied mechanisms, in which miRNA and phasiRNA share the same targeting mechanism (2). As the majority of plant regulatory sRNAs are miRNAs, we hereafter use the miRNA target prediction to represent the plant regulatory sRNA target analysis.
Plant miRNAs, of which the lengths range from 20 to 22 nucleotides, regulate gene expression post-transcriptionally or translationally through binding with the Argonaute (AGO) protein and targeting Messenger RNAs (mRNAs) (3,4). Most of the plant miRNA targets identified to date have extensive complementarity to their cognate miRNA mature sequences, yet perfect matches between them were not often found (1,4,5).
Dozens of miRNA-target complementary match patterns have been either validated or further reported since the early study of plant miRNAs (6). Schwab et al. reported that up to four mismatches may be allowed without reducing the efficiency of miRNA–mRNA pairing (7). The authors also extended the mismatch-sensitive seed region (i.e., the critical miRNA-target pairing region) to include the positions 2–12, excluding the cleavage site at the nucleotides 10 and 11. Additionally, it has been reported that G:U mismatches behave similarly to other mismatches (7). Mallory et al. reported a more complex miRNA-target transcript matching pattern based on statistical analyses of 51 confirmed conserved miRNA targets. They observed that non-paired nucleotides were most common at the ends of the duplexes (the positions 1, 2, 20 and 21) as well as at the positions 14 and 15, while non-paired nucleotides were the rarest along the positions 3–10 of the miRNA. No mismatches, G:U wobble pairs, or bulges were present in any miRNA targets bound at the positions 3–4, 7–10 and 9–10 (5). Axtell et al. summarized the match/mismatch patterns between miRNAs and target transcripts, which were experimentally validated prior to 2013 (1). In their analysis, the critical miRNA-target pairing region (seed region) spans the nucleotides 2–13, in which only a single mismatch is permitted. The above match patterns suggest complementarity analysis to be the most effective way to identify potential miRNA target genes (8).
Scoring schemas encoding the match patterns have been widely adopted in the complementarity analysis between plant miRNAs and targets (9,10). Yet, most of these tools utilize a built-in non-customizable scoring schema for target prediction, which poses a challenge to analyze the miRNA–mRNA interactions that have very low complementarity, such as the non-canonical targets described by Brousse et al. (11).
We developed the psRNATarget web server to enable the identification of target genes of the plant miRNAs (12). The psRNATarget uses a mismatch-sensitive ‘seed’ region; specifically, the region spanning the nucleotides 2–7 originally defined in the vertebrate miRNA studies (13). Importantly, the complementary base pairing in this seed region is weighted so that any mismatch is more heavily penalized. psRNATarget also estimates mRNA target accessibility, which is the energy required to open the secondary structure of mRNA around the target region for target site exposure (12). The original psRNATarget web server has been working perfectly to find most of validated miRNA–mRNA interactions; yet, it fails to report those recently published miRNA targets either with very low complementary matches or high target accessibility values in our benchmark dataset (Supplementary Table S1). In addition, as a web-based analysis tool, the original psRNATarget suffers from an important limitation: the ability to reliably upload large datasets, such as the large amounts of sequencing data produced from the next generation sequencing (NGS) platforms, through web browsers. Here, we report the release of a new high-performance psRNATarget web server that is capable of predicting sRNA targets at higher ‘recall rate,’ and with enhanced capability for ‘big’ data uploading and analysis.
METHODS
Implementation and infrastructure
The psRNATarget analysis server consists of a back-end pipeline, which was developed in Java and deployed on a high performance Linux cluster, and front-end web interfaces. We also developed an enhanced job queue management system to manage user-submitted analysis requests and to send back session IDs, which can be used to track job progress and retrieve final results. Submitted jobs are assigned to one of four individual job queues according to the size of the data, preventing large jobs from jamming the back-end analysis pipeline.
The back-end pipeline searches potential target candidates for given miRNAs based on the user customizable complementary matching scoring schema. First, the pipeline employs ssearch36, a component of the FASTA package (14) for sequence alignment between miRNA and candidate targets. The ssearch36 features a SSE2 accelerated Smith-Waterman implementation with much better performance for short sequence alignment compared with the NCBI BLAST (15). This is particularly important for analyzing short mature miRNA sequences. Next, the energy required to unfold secondary structure around the target site, defined as target site accessibility, is calculated using the RNAup program in the Vienna Package (16), as described in the first release of psRNATarget (12). This step is optional in the new scoring schema, however.
The psRNATarget webserver was developed in Flask, a Python web framework. Popular JavaScript and CSS libraries, jQuery and Bootstrap were used to generate user friendly, interactive HTML5 web interfaces. The new psRNATarget release implements a stable, large data upload page, in which up to hundreds of gigabytes of data can be uploaded by multiple uploading threads simultaneously using the HTML5 file API. The maximum file size permitted to be uploaded for analysis is only subject to the analytic capability of the back-end pipeline.
Inputs and outputs
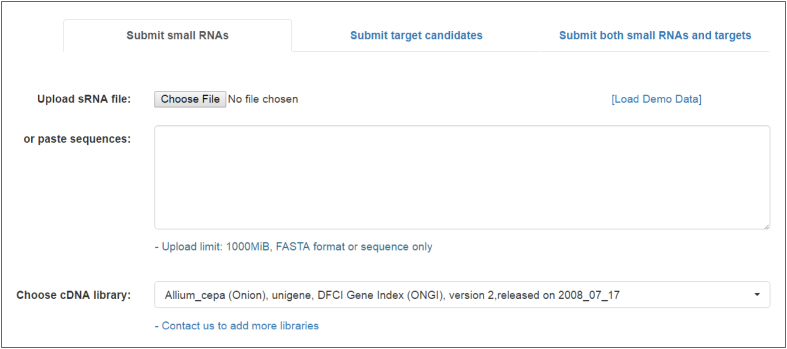
The psRNATarget homepage includes three functional tabs that allow users to upload and search their miRNAs against the preloaded target transcript libraries, to upload and search candidate target transcripts against published miRNA sequences downloaded from the miRBase (17), or to upload both miRNA and target sequences and search for potential miRNA–mRNA interactions between them (Figure 1). The latest release of psRNATarget has significantly more preloaded target libraries than the previous version did. As one example, it includes all transcript libraries from the JGI Phytozome Release 12 (18).
Figure 1.
A screenshot showing the three functional tabs of the psRNATarget, allowing users to (i) upload and search miRNAs against the pre-loaded target transcript libraries, (ii) upload and search target candidates against published miRNA sequences downloaded from miRBase or (iii) upload both miRNA and target sequences and search for potential miRNA-target pairs.
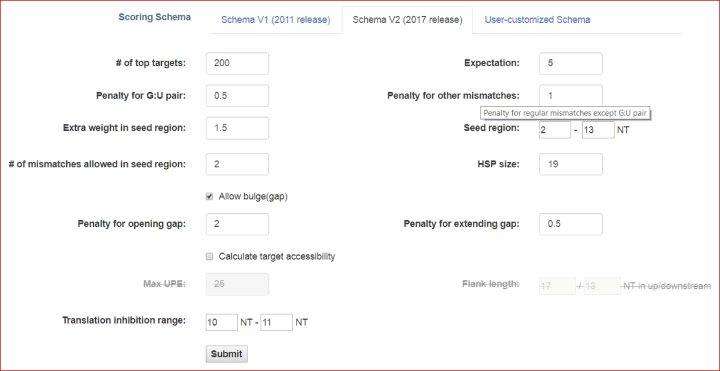
The new scoring schema, V2 is set as the default scoring schema of the updated psRNATarget sever; however, users can choose the scoring schema defined in the previous version (i.e. the V1 schema) if desired. In the new release, the scoring rules are fully customizable. This provides additional flexibilities to meet the special requirements of the end-users. Users may further adjust individual parameters, such as disabling target accessibility estimation to accelerate the analysis, treating G:U pairs as other mismatches, restricting the maximum number of mismatches in the seed region, or minimizing the gap-extending penalty to allow long bulges on the miRNA or target sequence. To assist users in customizing their search parameters, we created popup tips for all customizable options, which can be accessed by leaving the mouse cursor on the label of the individual option for more than one second (Figure 2).
Figure 2.
A screenshot of the psRNATarget web interface for choosing complementary matching scoring schema and customizing both required and optional parameters. A context help prompt will appear when the user leaves the mouse cursor on the text labels of any input field for more than one second.
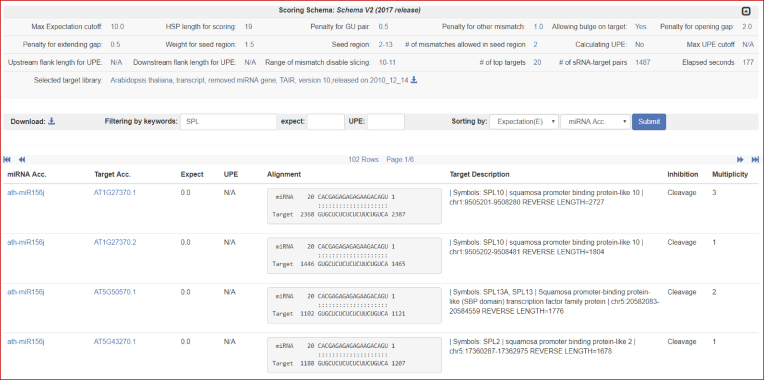
The analysis output page varies depending on the number of predicted miRNA–mRNA interactions in the new release. For output comprising of less than 100,000 miRNA-target in-silico interactions, the updated release of psRNATarget provides a paginated HTML table to display the analysis result. Users can also use the integrated search and sort functions to further filter the predicted miRNA–mRNA interactions (Figure 3). However, to improve the server response time and user experience, only a batch result download link is made available for the output comprising of more than 100,000 miRNA-target in-silico interactions.
Figure 3.
A screenshot of the psRNATarget output page. Users can use the integrated search and sort functions to further filter the predicted miRNA–mRNA interactions.
RESULTS: WHAT’S NEW IN THE LATEST RELEASE?
The new psRNATarget introduces significantly improved analytical capabilities, which can be categorized into two main functionalities: an improved complementary matching scoring schema and the ability to upload and handle much larger datasets compared to the previous release. The updated psRNATarget server also has clear, compelling and user-friendly interfaces that enhance user experiences and present data clearly and concisely.
The new scoring schema is capable of discovering miRNA–mRNA interactions at higher recall rates
The first version of psRNATarget searches target candidates using the scoring schema proposed by Zhang (9). The new release has an improved scoring schema V2 to cover more validated miRNA–mRNA interactions without significantly increasing the final prediction output.
The differences between the scoring schema V1 and V2 are summarized as following. Firstly, target accessibility analysis is disabled by default to include those validated miRNA–mRNA interactions with target accessibility values beyond the default cutoff defined in the scoring schema V1 (Supplementary Table S2). Secondly, in the scoring schema V2, the seed region has been extended to 2–13 bp and only two mismatches (excluding G:U pair) in this region are allowed based on the published plant miRNA target recognition patterns (1). The more stringent seed region scoring rule will be helpful to control the size of total prediction output when the target accessibility is disabled. Thirdly, to accommodate the miRNA–mRNA interactions with long gap, such as the non-canonical target reported recently (11), the penalty for opening gap has been increased to 2, but the penalty for extending gap is decreased to 0.5. Lastly, the maximum expectation in scoring schema V2 can be set to 10.0 instead of 5.0 in scoring schema V1 in case users desire to search those targets with poor complementary matching with miRNA or long gap in alignment.
We created an Arabidopsis benchmark dataset (Supplementary Table S1), comprising of 147 validated miRNA–mRNA interactions, after systematical analyzing literatures and curating published databases. In this benchmark dataset, only the targets with strong evidence from either 5′-RACE experiments or degradome analysis were included as experimentally validated miRNA–mRNA interactions. Since only a limited number of experimentally validated miRNA–mRNA interactions are available to date, computing the commonly adopted performance measurements, such as false positive rate, ROC curve, are not applicable. This is because the pure positive dataset (the validated target genes) is too small and incomplete to generate a reliable estimation value and, more critically, the pure negative dataset is absent. Therefore, ‘recall rate,’ defined as ratio of the predicted also validated miRNA–mRNA interactions over all the validated interactions in our benchmark dataset, might be a reasonable measurement for us to demonstrate the performance of psRNATarget. Meanwhile, the total predictions should also be considered a measurement if the ‘recall rates’ are close because fewer predictions in the output would indicate higher ‘precision.’
Using the improved scoring schema V2, we were able to identify 143 of 147 total validated miRNA–mRNA interactions in the Arabidopsis benchmark dataset with the default cutoff (i.e. expectation is less than or equal to 5.0) making its ‘recall rate’ 6% higher than the original scoring schema V1 in the 2011 release with the same cutoff (Table 1). The scoring schema V2 not only correctly predicted all miRNA–mRNA interactions, which had been predicted by the schema V1, but also retrieved nine more validated miRNA–mRNA interactions (Supplementary Table S2). Only four miRNA–mRNA interactions failed to be reported by the new scoring schema; two of them, including the recently reported non-canonical target, can actually be detected by changing the cutoff value or increasing the maximum number of targets allowed for each miRNA, while the 2011 release with old V1 scoring schema failed to report these targets even after changing these parameters (Supplementary Table S2).
Table 1. The performance comparison between release 2017 and release 2011 using default scoring schema and 147 validated miRNA–mRNA interactions in our Arabidopsis benchmark dataset.
| Recalled interactions | Recall rate (%) | Total predictions in the output | |
|---|---|---|---|
| 2011 Release, Schema V1 | 134 | 91.1 | 9,204 |
| 2017 Release, Schema V2 | 143 | 97.3 | 9,654 |
*The comparison was performed between 65 unique miRNAs/ta-siRNAs in the benchmark dataset (Supplementary Table S1) and the Arabidopsis TAIR10 transcripts. Maximum expectation was set to 5.0 and the maximum number of allowed top targets for each miRNA was set to 200 for both scoring schemas.
To further demonstrate the performance of psRNATarget in analyzing miRNA–mRNA interactions in non-Arabidopsis species, we analyzed another benchmark dataset, referred to as rice miRNA–mRNA interaction data, which was collected from the supplemental data (the additional file 8) of the paper published by Srivastava et al. (8). The new psRNATarget was able to recall much more validated miRNA–mRNA interactions without significantly increasing the total predictions when searching the same rice miRNAs in the rice miRNA–mRNA interaction data against the same rice transcript library, JGI rice Phytozome 12 genome annotation. Specifically, the recall rate was estimated at 82.7% by the psRNATarget 2017 release compared with the 62.5% recall rate by the psRNATarget 2011 release (Table 2), validating the new psRNATarget's improved performance in analyzing both Arabidopsis and non-Arabidopsis miRNA–mRNA interactions.
Table 2. The performance comparison between release 2011 and release 2017 using default scoring schema and 52 validated miRNA–mRNA interactions in rice benchmark dataset.
| Recalled interactions | Recall rate (%) | Total predictions in the output | |
|---|---|---|---|
| 2011 Release, Schema V1 | 33 | 63.5 | 3,286 |
| 2017 Release, Schema V2 | 43 | 82.7 | 4,162 |
*The comparison was performed between 26 unique miRNAs in the rice benchmark dataset (Srivastava et al, 2014, Additional file 8) (8) and the rice JGI Phytozome 12 transcripts. Maximum expectation was set to 5.0 and the maximum number of allowed top targets for each miRNA was set to 200 for both scoring schemas.
Using our Arabidopsis benchmark dataset, prediction performances were also compared between the new psRNATarget and the standalone pipeline, TargetFinder (19). Both applications delivered similar ‘recall rate’ and total predictions when we chose to balance the ‘recall rate’ and precision through adjusting score cutoff (Supplementary Table S3). However, if we chose to maximize ‘recall rate’ to cover more validated interactions, the new psRNATarget will recall slightly more validated interactions and report much less total predictions, which in turn indicate much higher precision (Supplementary Table S3). We further compared performances between new psRNATarget and the TAPIR (20) using its fast mode since the hybrid mode stringently restricts the size of data that can be analyzed each time due to slow computational speed of the algorithm. We noticed that ‘recall rate’ of TAPIR is significantly lower than the other two applications even after we adjusted the cutoff thresholds to maximize the ‘recall rate.’ However, TAPIR generated much less predictions than the psRNATarget 2017 release and TargetFinder did, indicating higher prediction precision (Supplementary Table S3).
The new release of psRNATarget enables ‘big’ data uploading and analysis
Emerging next generation sequencing (NGS) technologies often produce ‘big’ datasets that challenge the downstream bioinformatics analyses, such as miRNA regulatory target analysis. However, the standard HTTP protocol was not designed to transmit such large data files over the internet browsers. The submissions of the gigabytes level data through web browser often fail due to the interruptions of network connections. The new psRNATarget has been optimized for uploading such ‘big’ datasets. We have implemented a stable and resumable multithreading chunked data uploading module via HTTP. The new data upload page utilizes an HTML5 File API to split each data file into small chunks and transmit these chunks in parallel to increase the upload robustness and upload speed. If a single chunk upload fails, the upload is automatically reattempted until succeed. This function was designed to overcome the HTTP protocol restrictions on data uploading including the size limit, low utilization of bandwidth, and the inability to resume upload if interrupted.
In addition, we also allocated hundreds more CPU cores to the back-end Linux cluster to significantly enhance the computing capacity of the psRNATarget. The Linux cluster now has ∼1200 CPU Cores to accommodate the needs of high performance parallel computing. With these improvements, the new psRNATarget can accept and analyze much larger datasets than the 2011 release.
DISCUSSION
Srivastava et al. evaluated the performance of 11 online or stand-alone computational tools, including the psRNATarget server (8), and concluded that the TargetFinder was most efficient in predicting ‘true-positive’ targets in Arabidopsis miRNA–mRNA interactions. For non-Arabidopsis targets, combining the results of TargetFinder (19) and the psRNATarget 2011 release delivers the highest ‘true positive’ coverage, whereas the intersection of psRNATarget and TAPIR (20) outputs deliver the best ‘precise’ predictions. As described in the Result section, the new psRNATarget performed better in ‘recall rate’ or prediction precision compared to the other two outstanding software applications. Furthermore, TargetFinder is a stand-alone software application that requires installation and data preparation, and TAPIR is a web-based tool that also requires data preparation due to lack of a library preloading function. TAPIR is also restricted by its limited data analysis capacity; users can only submit a small-sized set data for each analysis. Overall, our performance comparison continuously demonstrates that the new psRNATarget is one of the best choices to search, rank and filter plant miRNA target genes.
Among the developed plant miRNA target analysis tools, only a few provide online analysis services (8). Furthermore, the updated psRNATarget is the only one capable of uploading and analyzing ‘big’ data, enabled by two key technologies. First, the newly developed multi-threading chunked file uploading module using the HTML5 file chunking API, integrated with customized JavaScript code, facilitates the upload of ‘big’ datasets without extra security risks, such as those associated with Flash/Java plugins. Theoretically, the size of uploading file in the new psRNATarget is only restricted by the amount of disk space on the host server. Enabling multi-threading chunked file uploading is critical for web-based data analysis services dealing with ‘big’ data, which can be readily generated by NGS technologies nowadays. Second, the updated psRNATarget server is empowered by high performance computing capacity of the back-end Linux cluster, which significantly accelerates its analytic procedure, mainly the time-consuming Smith-Waterman search. Hundreds more computing nodes have been added into our cluster, and more will be added when additional CPU resource becomes available, leading to a significant increase (about three times) in analysis capacity and capability compared to our original psRNATarget web server published in 2011.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation [DBI-0960897 and DBI-1458597 to P.X.Z.]; Noble Research Institute. Funding for open access charge: National Science Foundation [DBI-0960897 and DBI-1458597 to P.X.Z.]; Noble Research Institute
Conflict of interest statement. None declared.
REFERENCES
- 1. Axtell M.J. Classification and comparison of small RNAs from plants. Annu. Rev. Plant Biol. 2013; 64:137–159. [DOI] [PubMed] [Google Scholar]
- 2. Fei Q., Xia R., Meyers B.C.. Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell. 2013; 25:2400–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cuperus J.T., Carbonell A., Fahlgren N., Garcia-Ruiz H., Burke R.T., Takeda A., Sullivan C.M., Gilbert S.D., Montgomery T.A., Carrington J.C.. Unique functionality of 22 nt miRNAs in triggering RDR6-Dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat. Struct. Mol. Biol. 2010; 17:997–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rajagopalan R., Vaucheret H., Trejo J., Bartel D.P.. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006; 20:3407–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mallory A.C., Reinhart B.J., Jones-Rhoades M.W., Tang G., Zamore P.D., Barton M.K., Bartel D.P.. MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. EMBO J. 2004; 23:3356–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004; 116:281–297. [DOI] [PubMed] [Google Scholar]
- 7. Schwab R., Palatnik J.F., Riester M., Schommer C., Schmid M., Weigel D.. Specific effects of microRNAs on the plant transcriptome. Dev. Cell. 2005; 8:517–527. [DOI] [PubMed] [Google Scholar]
- 8. Srivastava P.K., Moturu T.R., Pandey P., Baldwin I.T., Pandey S.P.. A comparison of performance of plant miRNA target prediction tools and the characterization of features for genome-wide target prediction. BMC Genomics. 2014; 15:348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang Y. miRU: an automated plant miRNA target prediction server. Nucleic Acids Res. 2005; 33:W701–W704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009; 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brousse C., Liu Q., Beauclair L., Deremetz A., Axtell M.J., Bouche N.. A non-canonical plant microRNA target site. Nucleic Acids Res. 2014; 42:5270–5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dai X., Zhao P.X.. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 2011; 39:W155–W159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lewis B.P., Burge C.B., Bartel D.P.. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are MicroRNA targets. Cell. 2005; 120:15–20. [DOI] [PubMed] [Google Scholar]
- 14. Pearson W.R., Wood T., Zhang Z., Miller W.. Comparison of DNA sequences with protein sequences. Genomics. 1997; 46:24–36. [DOI] [PubMed] [Google Scholar]
- 15. Dai X., Zhuang Z., Zhao P.X.. Computational analysis of miRNA targets in plants: current status and challenges. Brief Bioinform. 2011; 12:115–121. [DOI] [PubMed] [Google Scholar]
- 16. Muckstein U., Tafer H., Hackermuller J., Bernhart S.H., Stadler P.F., Hofacker I.L.. Thermodynamics of RNA-RNA binding. Bioinformatics. 2006; 22:1177–1182. [DOI] [PubMed] [Google Scholar]
- 17. Kozomara A., Griffiths-Jones S.. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011; 39:D152–D157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N. et al. . Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012; 40:D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fahlgren N., Howell M.D., Kasschau K.D., Chapman E.J., Sullivan C.M., Cumbie J.S., Givan S.A., Law T.F., Grant S.R., Dangl J.L. et al. . High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS One. 2007; 2:e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bonnet E., He Y., Billiau K., Van de Peer Y.. TAPIR, a web server for the prediction of plant microRNA targets, including target mimics. Bioinformatics. 2010; 26:1566–1568. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.