Abstract
Background:
Musashi1 (MSI1) is a characteristic stem cell marker that regulates the balance between cell self-renewal and differentiation. Evidence has identified MSI1 as a pivotal oncogenic regulator in diverse malignancies. However, little evidence uncovers the role of genetic variations of MSI1 gene in cancer etiology.
Objective:
The aim of this study was to investigate the association between genetic variants in the MSI1 gene and lung cancer risk.
Methods:
Based on a two-stage retrospective study with a total of 1559 patients with lung cancer and 1667 healthy controls, we evaluated the relevance between three putative functional SNPs in the MSI1 promoter (i.e., -2696T>C[rs7959801], -2297T>C[rs3742038] and -1081C>T[rs34570155]) and lung cancer risk.
Results:
We found that the SNP rs7959801T>C was significantly associated with lung cancer susceptibility. Compared to those with rs7959801TT wild-genotype, individuals with CT/CC variant genotypes exerted consistently beneficial roles in lung cancer risk in the discovery set (adjusted odd ratios [OR] = 0.67; 95% confidence interval [CI] = 0.57-0.80), and in the validation set (OR=0.69; 95%CI=0.54-0.88). Functional assays indicated that the allele transformation from T to C in rs7959801 of MSI1 gene arrestingly decreased its transcription activity in vitro. Furthermore, the expression levels of MSI1 were significantly lower in the patients with CT/CC variants than in those who were with TT genotype.
Conclusion:
Our findings suggested that the rs7959801T>C polymorphism in the MSI1 promoter conferred a decreased risk to lung cancer by reducing the expression of MSI1 and it may be a promising indicator for lung cancer predisposition.
Keywords: Lung cancer, Musashi1, Promoter, Genetic variant, Susceptibility, Case-control study
1. Introduction
Lung cancer is the most common cancer and ranks the leading cause of cancer-related mortality worldwide [1]. In the past decades, the prevalence of lung cancer is being in rapid rise in China [2, 3]. Although tremendous genetic variants such as Single Nucleotide Polymorphism (SNP) have been reported to be associated with lung cancer risk and progression based on a serial of Genome-Wide Association Studies (GWASs) [4-6], the molecular mechanisms of how these genetic loci participate in cancer initiation and development are still largely unclear. Recent studies indicate that cancer heterogeneity is a result of the hierarchical organization of tumor cells by a portion of cells with stem/progenitor cell features termed as Cancer Stem Cells (CSCs) [7]. Meaningfully, CSCs have been identified to play a critical role in tumor initiation, maintenance and metastasis [8-12], as well as in relapse and drug resistance [13, 14]. Furthermore, evidence reports that genetic variants in key genes of CSCs could influence their biological functions and thus be associated with cancer susceptibility [15]. Above evidence might provide a novel approach to seek especial and functional biomarkers for cancer predisposition.
Musashi RNA binding protein 1(Musashi1, also known as MSI1, OMIM#603328) is deemed to an important cell surface marker of CSCs that functions as a regulator for maintenance of the stem-cell state, differentiation, and tumorigenesis by repressing translation of its downstream targets [16, 17]. Growing evidence reveals that overexpression of MSI1 results in cell proliferation and apoptosis via the translational inhibition of Numb, p21WAF and Dickkopf3, which are well-known factors in cancer stem cell biology and cell cycle regulation [17-19]. In addition, aberrant expression of MSI1 has been reported to be associated with many malignancies including hepatoma [20], gliomas [21], colorectal adenoma [22] and lung cancer [23]. All these findings suggest that MSI1 may act as an oncogene.
Human MSI1 gene locates at chromosome 12q24, contains 15 exons, and encodes a 362-amino acids protein [24]. The correlations between genetic variations in MSI1 and cancer risk have not yet been established comprehensively. Recently, one study indicated that a SNP in the 3’-UTR of MSI1 gene was associated with an increased risk of lung cancer in Chinese population [25]. However, no evidence was presented to identify association of polymorphisms in other functional region of MSI1 gene to human diseases. Because of the fact that the promoter plays an important role in gene’s genomic stability, transcriptional efficiency and eventual protein expression; genetic variants in the promoter region may have influence on transcriptional modulation of the gene and thus contribute to disease susceptibility. Herein, we hypothesized that polymorphisms in the promoter of MSI1 gene were associated with lung cancer risk though affecting gene’s expression.
In the current study, we genotyped three potentially functional SNPs in the MSI1 promoter (i.e., -2696T>C[rs7959 801], -2297T>C[rs3742038] and -1081C>T [rs34570155]), and assessed their associations with lung cancer risk based on a two-stage retrospective study in southern and eastern Chinese populations. For the promising causal SNPs, we further performed a serial of experiments to decode their molecular functions on lung tumorigenesis.
2. Materials and Methods
2.1. Study Subjects
The studied populations conducted in southern and eastern China have been described previously [26-28]. In brief, 1056 primary lung cancer cases and 1056 cancer-free controls recruited from Guangzhou city were used as discovery set; 503 patients with lung cancer and 623 healthy controls from Suzhou city were served as validation set. All participants are generally unrelated Han Chinese and none had blood transfusions in the last 6 months. Each subject was asked to provide data on individual’s demographic and clinical characteristics, and to donate 5 ml blood after obtaining their written informed consents. The detailed information of subjects’ recruitment and definitions of variables was presented previously [26-28]. The study was approved by the Institutional Review Boards of Guangzhou Medical University (Ethics Committee of Guangzhou Medical University: GZMC2007-07-0676) and Soochow University (Ethics Committee of Soochow University: SZUM2008031233).
2.2. SNP Selection and Genotyping
In current study, those SNPs that were located in the promoter region of MSI1 gene [24] and had Minor Allele Frequency (MAF) >5% in Chinese population were screened based on the dbSNP database (http://www.ncbi.nlm.nih.gov/). Finally, three common SNPs in the MSI1 promoter (i.e., -2696T>C[rs7959801],-2297T>C[rs3742038] and -1081C>T [rs34570155]) were selected.
The genomic DNA from subjects’peripheral blood was extracted with routine method. The genotypes of the studied SNPs were measured using the TaqMan allelic discrimination assay performed in the ABI PRISM 7500 Sequence Detection Systems (Applied Biosystems, Foster City, CA, USA). The primers and probes for the SNPs were purchased from Life Technologies company (Applied Biosystems).
Approximately 10% samples for each SNP were randomly selected to re-genotyping, and the results were in 100% concordance.
2.3. Construction of Reporter Plasmids
Two luciferase reporter plasmids containing rs7959801T or C allele were further constructed to evaluate the effect of this SNP on transcription activity of the MSI1 (Fig. 1A). The expatiation of the assay procedures was described elsewhere [29]. In short, a fragment of a total of 2950bp for MSI1 core promoter (from -2914 to +36 nucleotides relative to the transcription start site) with rs7959801T allele was amplified from subjects with rs7959801TT genotype, using the forward primer 5′-CGGGGTACCTTTACTATGAGGATCAT GAGTTTAC-3′and reverse primer 5′-ACGCGTCGACCC GCTCGAGGGAGGCGAGGCCGGGCTGGG-3′, including KpnI and XhoI enzymes restriction sites. The amplified product was cleaved with the KpnI and XhoI enzymes (New England, BioLabs, Ipswich, MA) and then ligated to pGL3 basic vector (Promega, Madison, WI) by T4 DNA ligase (New England BioLabs). The construct with C allele was obtained from wild-type construct by site-directed mutagenesis using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). All constructs were sequenced to confirm the allele, the orientation and integrity of each insert.
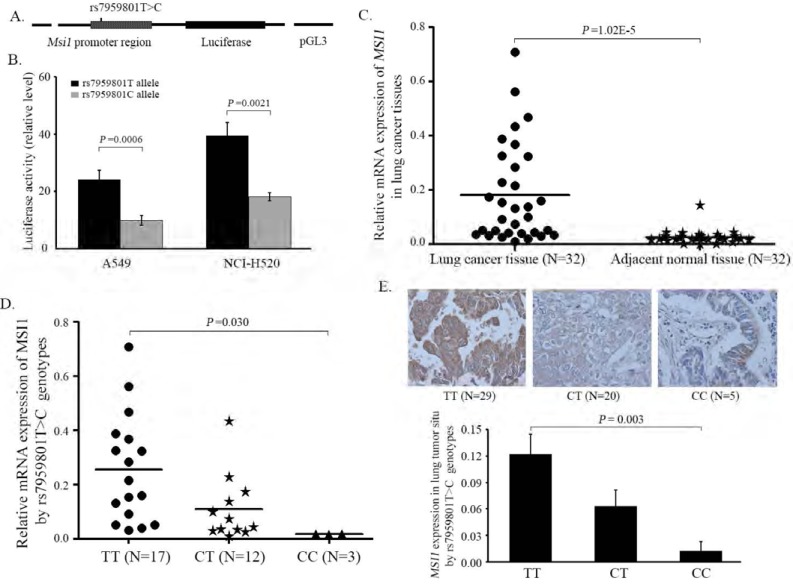
Fig. (1).
The effect of rs7959801T>C genotypes on MSI1 biological phenotypes. (A) Schematic diagram of the reporter constructs containing 2950 bp promoter of MSI1 with rs7959801T or C allele. (B) Luciferase activity of the MSI1 promoter constructs in two lung cancer cell lines (A549 and NCI-H520). The luciferase activity of each construct was normalised against the internal control of renilla luciferase. Columns, mean from three independent experiments; bars, standard deviation; Student’s t-test was used to compare the expression levels of different constructs. (C) mRNA expression of MSI1 in lung cancer tissues and adjacent normal lung tissues. (D) MSI1 mRNA expression in lung cancer tissues with rs7959801T>C genotypes. (E) Protein expressions in lung cancer tissues in situ using the Image Pro Plus software to score the MSI1 expression (magnification ×200). The comparisions of MSI1 expression in different genotypes were measured with one-way ANOVA test.
2.4. Luciferase Assays
The luciferase assays were executed as described previously [26, 30]. In brief, two human lung cancer cell lines, A549 and NCI-520 were cultured into 24-well plates and then transfected with 1.5μg reporter plasmids (T or C allele) and 10 ng pRL-TK plasmids (Promega, Madison,WI) using lipofectamine 2000 Transfection Reagent (Invitrogen, Carlsbad, CA). The activity of each reporter with firefly luciferase and the internal standard with Renilla luciferase was quantified with a Dual-Luciferase Reporter Assay System (Promega).
2.5. Quantitative Real-time PCR
Total RNA was dissociated from thirty-two lung cancer tissues and their peripheral normal tissues that were collected during the surgical excision from the first hospital, second hospital and the tumor hospital affiliated to Guangzhou Medical University according to routine method [28, 31]. All tumor samples were histologically confirmed and genotypes of these samples were detected by sequencing. 2 µg total RNA was then reversely transcribed to complementary DNA using random primers and Superscript II (Invitrogen). The mRNA expression of MSI1 was quantified with the primers (forward: 5′-GAGGGTTCGG GTTTGTCACG-3′ and reverse: 5′-GGCGACATCACCTCCTTTGG-3′) in ABI Prism 7500 sequence detection system based on the SYBR-Green method and β-actin was used as an internal reference gene using the primers: 5′-GGCGGCACCACCAT GTACCCT-3′ and 5′-AGGGGCCGGACTCGTCATACT-3′. Method of 2-∆ct was used to calculate the relative level of MSI1.
2.6. Immunohistochemistry
Immunohistochemistry was performed following standard procedures [31]. In short, sections were dewaxed, and rehydrated by sequential immersion in xylene, graded ethanol and water. The sections were then boiled in 10 mmol/L citrate buffer (pH 6.0) in a microwave oven for antigen retrieval. The slides were treated with 3% hydrogen peroxide in methanol to quench endogenous peroxidase activity, followed by incubation with 1% fish skin gelatin to block the nonspecific staining. Sections were then incubated overnight at 4°C with primary antibody of MSI1 (Abcam, Cambridge, USA; 1:300 dilution). After washing with Phosphate-Buffered Saline (PBS), the sections was treated with biotinylated goat anti-rabbit IgG for 30 min at room temperature, rinsed with PBS and followed by an avidinbiotin-peroxidase conjugate for 30 min. Reaction products were observed by incubation with diaminobenzidine. Negative controls were treated identically but with the primary antibodies omitted. The immunohistochemical reactions were then visualized under high-power magnification (×200) using an Olympus microscope. Meanwhile, all fifty-four tissue samples were genotyped. Staining intensity score of the MSI1 expression was calculated using the Image Pro Plus software, and percentage of positive cells examined was scored as 0 (negative), 1 (<10%), 2 (11-50%), 3 (51-80%) and 4 (>80%). The intensity scores and fraction of positive cell were multiplied to represent the relative level of MSI1 expression.
2.7. Statistical Analysis
The chi-square test was used to test the distributions of selected characteristics and genotypes between cases and controls. Logistic regression models with or without adjusting for surrounding factors were used to estimate the association between each SNP and lung cancer risk. Breslow-Day test was used to test the homogeneity of the results between stratum-ORs. A multiplicative interaction model was applied to evaluate the possible gene-environment interactions [28]. The statistical power was calculated using the PS Software. The False Positive Report Probability (FPRP) test was applied to detect the false-positive association findings [32]. The Student’s t test was used to assess the transcriptional levels between different luciferase constructs. The one-way analysis of variance (ANOVA) test and the linear regression model, which has been adjusted for surrounding factors, were used to evaluate the association between MSI1 genotypes and MSI1 expression. All tests were two-sided and operated using the SAS software (version 9.3; SAS Institute), and P <0.05 was defined as statistically significant.
3. Results
3.1. Associations Between MSI1 SNPs and Risk of Lung Cancer
The information of demographic characteristics in the studied populations were described previously [28, 31, 33] and presented in Table S1 (129.1KB, pdf) . In the discovery set, we only observed a significant correlation between the SNP rs7959801T>C and lung cancer risk. As shown in Table 1, compared to those with rs7959801TT wild-genotype, the carriers with CT heterozygous genotype had a decreased risk of lung cancer (odds ratio [OR] =0.64, 95% confidence interval [CI] =0.53-0.76; P =1.02E-6).
Table 1.
The genotype frequencies of studied SNPs in Musashi-1 promoter and their associations with lung cancer risk.
| Genotypes/Alleles | Discovery Set (Southern Chinese) | - | Validation Set (Eastern Chinese) | - | Merged Set | - | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - |
Case
(n=1056) n(%) |
Control a
(n=1056) n(%) |
Adjusted
OR (95% CI)b |
P value |
Case
(n=503) n(%) |
Control a
(n=623) n(%) |
Adjusted
OR (95% CI)b |
P value |
Case
(n=1559) n(%) |
Control
(n=1679) n(%) |
Adjusted
OR (95% CI)b |
P value |
| rs7959801T>C | - | - | - | - | - | - | - | - | - | - | - | - |
| TT | 563(53.3) | 460(43.6) | 1.00 (ref.) | - | 278(55.3) | 287(46.1) | 1.00 (ref.) | - | 841(53.9) | 747(44.5) | 1.00(ref.) | - |
| CT | 385(36.5) | 492(46.6) | 0.64(0.53-0.76) | 1.02E-6 | 185(36.8) | 270(43.3) | 0.71(0.55-0.91) | 0.007 | 570(36.6) | 762(45.4) | 0.66(0.57-0.77) | 4.23E-8 |
| CC | 108(10.2) | 104(9.8) | 0.85(0.63-1.15) | 0.278 | 40(7.9) | 66(10.6) | 0.63(0.41-0.97) | 0.031 | 148(9.5) | 170(10.1) | 0.78(0.61-0.99) | 0.048 |
| Group test P value | - | - | 7.89E-6 | - | - | - | 0.008 | - | - | - | 2.56E-7 | - |
| Additive model | - | - | - | - | - | - | - | - | - | - | - | - |
| CC vs. CT vs. TT |
- | - | 0.81(0.71-0.92) | 0.001 | - | - | 0.76(0.63-0.91) | 0.003 | - | - | 0.79(0.71-0.88) | 1.43E-5 |
| Dominant model | - | - | - | - | - | - | - | - | - | - | - | - |
| TT | 563(53.3) | 460(43.6) | 1.00 (ref.) | - | 278(55.3) | 287(46.1) | 1.00 (ref.) | - | 841(53.9) | 747(44.5) | 1.00 (ref.) | - |
| CT+CC | 493(46.7) | 596(56.4) | 0.67(0.57-0.80) | 6.25E-6 | 225(44.7) | 336(53.9) | 0.69(0.54-0.88) | 0.002 | 718(46.1) | 932(55.5) | 0.68(0.59-0.79) | 1.18E-7 |
| - | - | - | - | - | - | - | - | - | - | - | - | - |
| rs3742038T>C | - | - | - | - | - | - | - | - | - | - | - | - |
| TT | 960(90.9) | 935(88.5) | 1.00 (ref.) | - | 440(87.5) | 547(87.8) | 1.00 (ref.) | - | 1400(89.8) | 1482(88.3) | 1.00(ref.) | - |
| CT | 87(8.2) | 114(10.8) | 0.74(0.55-1.01) | 0.052 | 57(11.3) | 73(11.7) | 0.94(0.64-1.36) | 0.736 | 144(9.2) | 187(11.1) | 0.82(0.65-1.03) | 0.081 |
| CC | 9(0.9) | 7(0.7) | 1.25(0.46-3.38) | 0.657 | 6(1.2) | 3(0.5) | 2.17(0.53-8.83) | 0.203 | 15(1.0) | 10(0.6) | 1.55(0.69-3.47) | 0.259 |
| Group test P value | - | - | 0.122 | - | - | - | 0.406 | - | - | - | 0.107 | - |
| Additive model | - | - | - | - | - | - | - | - | - | - | - | - |
| CC vs. CT vs. TT |
- | - | 0.83(0.64-1.07) | 0.141 | - | - | 1.04(0.75-1.44) | 0.824 | - | - | 0.90(0.74-1.11) | 0.338 |
| Dominant model | - | - | - | - | - | - | - | - | - | - | - | - |
| TT | 960(90.9) | 935(88.5) | 1.00 (ref.) | - | 440(87.5) | 547(87.8) | 1.00 (ref.) | - | 1400(89.8) | 1482(88.3) | 1.00 (ref.) | - |
| CT+CC | 96(9.1) | 121(11.5) | 0.77(0.58-1.03) | 0.076 | 63(12.5) | 76(12.2) | 0.99(0.69-1.42) | 0.951 | 159(10.2) | 197(11.7) | 0.85(0.68-1.07) | 0.163 |
| - | - | - | - | - | - | - | - | - | - | - | - | - |
| rs34570155C>T | - | - | - | - | - | - | - | - | - | - | - | - |
| CC | 510(48.3) | 478(45.3) | 1.00 (ref.) | - | 233(46.3) | 293(47.0) | 1.00 (ref.) | - | 743(47.6) | 771(45.9) | 1.00(ref.) | - |
| TC | 404(38.3) | 446(42.2) | 0.85(0.71-1.02) | 0.086 | 196(39.0) | 256(41.1) | 0.98(0.76-1.28) | 0.928 | 600(38.5) | 702(41.8) | 0.89(0.77-1.04) | 0.113 |
| TT | 142(13.4) | 132(12.5) | 1.02(0.79-1.34) | 0.952 | 74(14.7) | 74(11.9) | 1.29(0.89-1.87) | 0.347 | 216(13.9) | 206(12.3) | 1.11(0.89-1.38) | 0.442 |
| Group test P value | - | - | 0.176 | - | - | - | 0.366 | - | - | - | 0.116 | - |
| Additive model | - | - | - | - | - | - | - | - | - | - | - | - |
| TT vs. TC vs. CC |
- | - | 0.97(0.85-1.09) | 0.571 | - | - | 1.09(0.92-1.30) | 0.306 | - | - | 1.01(0.91-1.11) | 0.950 |
| Dominant model | - | - | - | - | - | - | - | - | - | - | - | - |
| CC | 510(48.3) | 478(45.3) | 1.00 (ref.) | - | 233(46.3) | 293(47.0) | 1.00 (ref.) | - | 1182(75.8) | 1343(80.0) | 1.00 (ref.) | - |
| TC+TT | 546(51.7) | 578(54.7) | 0.89(0.75-1.06) | 0.188 | 270(53.7) | 330(53.0) | 1.06(0.83-1.34) | 0.655 | 377(24.2) | 336(20.0) | 0.94(0.82-1.08) | 0.321 |
Bold type: statistically significant, P < 0.05.
a The observed genotype frequencies among the control subjects were in agreement with the Hardy-Weinberg equilibrium (p2+2pq+q2 = 1) in the control subjects of both sets (P >0.05 for all).
bAdjusted in a logistic regression model that included age, sex, smoking status, alcohol use and family history of cancer.
The results were further confirmed in the validation set as the individuals with CT variant genotype exerted a 28% decreased risk of lung cancer when compared to the subjects with TT common genotype (OR=0.71, 95%CI=0.55-0.91; P=0.007). The heterogeneity test showed that the above associations in two populations were homogeneous (P =0.783). We then combined the two datasets to increase the study power and found that compared to those with rs7959801TT wild-genotype, the individuals with CT (OR =0.66, 95%CI =0.57-0.77; P =4.23E-8) or CC (OR =0.78, 95%CI =0.61-0.99; P =0.048) variant genotype had a significantly decreased risk for lung cancer, respectively. Also, in the dominant model, the carriers with CT/CC variants had a prominent declining risk of lung cancer compared to the carriers with TT wild-genotype (OR =0.68, 95%CI =0.59-0.79; P =1.18E-7). However, for other SNPs rs3742038T>C and rs34570155C>T, no notable relevance with risk of lung cancer was observed in both two datasets.
3.2. Stratification Analysis of Relationships Between MSI1 rs7959801T>C Genotypes and Lung Cancer Risk
Among all the assumed genetic models, the hereditary effect of rs7959801T>C on lung cancer risk was best in the dominant model as it showed the smallest Akaike’s Information Criterion (AIC) value. Hence, we further performed the stratified analysis for relationships between rs7959801T>C and lung cancer risk using dominant model. As shown in Table 2, although the quantitative effects of rs7959801C variants on cancer risk presented by stratum-ORs were in some difference, the heterogeneity test indicated that no significant deviations occurred in these subgroups (P>0.05 for all). Moreover, no remarkable interaction was found for this SNP with surrounding factors on lung cancer risk (P>0.05 for all).
Table 2.
Stratification analysis of the Musashi-1 rs7959801T>C genotypes by selected variables in lung cancer patients and controls.
| - | Cases (n = 1559) | - | Controls (n = 1679) |
Adjusted OR
(95% CI) a |
Phomob | Pinterc | ||
|---|---|---|---|---|---|---|---|---|
|
CT+CC
n (%) |
TT
n (%) |
- |
CT+CC
n (%) |
TT
n (%) |
CT+CC vs. TT | - | - | |
| Age (years) | - | - | - | - | - | - | 0.099 | 0.271 |
| ≤ 60 | 367(45.4) | 442(54.6) | - | 505(57.6) | 372(42.4) | 0.61(0.51-0.75) | - | - |
| > 60 | 351(46.8) | 399(53.2) | - | 427(53.2) | 375(46.8) | 0.75(0.62-0.92) | - | - |
| Sex | - | - | - | - | - | - | 0.909 | 0.945 |
| Male | 505(46.3) | 586(53.7) | - | 662(55.9) | 523(44.1) | 0.68(0.58-0.80) | - | - |
| Female | 213(45.5) | 255(54.5) | - | 270(54.7) | 224(45.3) | 0.69(0.53-0.89) | - | - |
| Smoking status | - | - | - | - | - | - | 0.439 | 0.374 |
| Ever | 373(45.3) | 451(54.7) | - | 429(56.1) | 336(43.9) | 0.65(0.53-0.79) | - | - |
| Never | 345(46.9) | 390(53.1) | - | 503(55.0) | 411(45.0) | 0.72(0.59-0.88) | - | - |
| Drinking status | - | - | - | - | - | - | 0.250 | 0.268 |
| Ever | 123(42.0) | 170(58.0) | - | 190(55.6) | 152(44.4) | 0.58(0.42-0.80) | - | - |
| Never | 595(47.0) | 671(53.0) | - | 742(55.5) | 595(44.5) | 0.71(0.61-0.83) | - | - |
| Family history of cancer | - | - | - | - | - | - | 0.945 | 0.882 |
| Yes | 54(41.9) | 75(58.1) | - | 76(51.7) | 71(48.3) | 0.64(0.39-1.05) | - | - |
| No | 664(46.4) | 766(53.6) | - | 856(55.9) | 676(44.1) | 0.69(0.59-0.79) | - | - |
| Family history of lung cancer | - | - | - | - | - | - | 0.430 | 0.677 |
| Yes | 20(38.5) | 32(61.5) | - | 24(55.8) | 19(44.2) | 0.36(0.14-0.96) | - | - |
| No | 698(46.3) | 809(53.7) | - | 908(55.5) | 728(44.5) | 0.69(0.60-0.79) | - | - |
| Histological types | - | - | - | 932(55.5) | 747(44.5) | - | 0.324 | - |
| Adenocarcinoma | 284(46.2) | 331(53.8) | 0.68(0.57-0.82) | - | - | |||
| Squamous cell carcinoma | 240(45.5) | 287(54.5) | 0.67(0.55-0.82) | - | - | |||
| Large cell carcinoma | 34(51.5) | 32(48.5) | 0.84(0.52-1.38) | - | - | |||
| Small cell lung cancer | 85(44.0) | 108(56.0) | 0.62(0.46-0.84) | - | - | |||
| Other carcinomasc | 75(47.5) | 83(52.5) | 0.72(0.52-1.00) | - | - | |||
| Stages | - | - | - | 0.459 | - | |||
| I | 97(48.5) | 103(51.5) | 932(55.5) | 747(44.5) | 0.75(0.56-1.01) | - | - | |
| II | 58(39.5) | 89(60.5) | 0.51(0.36-0.73) | - | - | |||
| III | 231(47.1) | 259(52.9) | 0.71(0.58-0.87) | - | - | |||
| IV | 332(46.0) | 390(54.0) | 0.68(0.57-0.81) | - | - | |||
Bold type: statistically significant, P < 0.05.
a ORs were adjusted for age, sex, and smoking status, and alcohol use, family history of cancer in a logistic regression model.
b P value of homogeneity test between strata for the related ORs of rs7959801T>C (rs7959801 CT+CC vs. TT genotypes).
c P value of test for the multiplicative interaction between rs7959801T>C genotypes and selected variables on cancer risk in logistic regression models.
3.3. Effects of the rs7959801T>C Genotypes on MSI1 Transcriptional Activity
Two luciferase reporter constructs comprised of rs7959801T or C allele were generated and assayed to evaluate the influence of this SNP on MSI1 transcriptional activity. As shown in Fig. (1B), the transcriptional level of the reporter gene integrated the MSI1 promoter with rs7959801C allele was markedly lower than the construct with T allele both in A549 cell and NCI-520 cell (P value equals to 0.0006 and 0.0021, respectively).
3.4. Correlation Between rs7959801T>C Genotypes and MSI1 Expression
As shown in Fig. (1C), we observed that the expression of MSI1 was significantly higher in lung cancer tissues than in their adjacent normal tissues (paired t test: P =1.02E-5). We further found that the MSI1 mRNA levels were in negative correlation with the number of rs7959801C variant allele (ANOVA test: P =0.030; lineal regression: P = 0.009; Fig. 1D). The protein expression of MSI1 in fifty-four lung cancer tissues in situ confirmed the above findings as the Fig. (1E) indicated that the tissues with rs7959801C variant genotype (CT or CC) exerted a strikingly decreased MSI1 expression compared to those with TT wild-genotype (ANOVA test: P =0.003; lineal regression: P =0.0008).
4. Discussion
In this study, we assessed the correlation between genetic variations in MSI1 promoter and the risk of lung cancer in Chinese populations and found that the SNP rs7959801T>C exerted a rewarding effect on developing lung cancer. Mechanical experiments revealed that the substitute from rs7959801T to C could remarkably decrease the expression of the MSI1 gene both in mRNA and protein level. To the best of our knowledge, this is the first study to show the relationship between functional SNP in the promoter of MSI1 gene and lung cancer susceptibility.
Evidence has been considered CSC markers as an important component in the maintenance of self-renewal and resistance to apoptosis pathway activation in these cells [34]. Several studies indicate that CSCs are rich in tumorspheres and give rise to tumors [35]. Notably, recent reports have demonstrated the existence of CSC in lung cancer [36]. As a pivotal molecule for CSC, MSI1 has been identified to play a critical role in tumorigenesis and tumor progression [37, 38]. Abnormal over-expression of MSI1 occurred in different types of human malignancies including lung cancer [20, 21, 23]. For example, one study indicated that the expression of
MSI1 was enriched in lung cancer cells and tissue specimens. Silencing of MSI1 gene by shRNA lentivirus-mediated could reduce spheroid colony proliferation and inhibited cell growth via the reduction of nuclear localization of β-catenin and inhibition of the processing of intracellular Notch [39]. In addition, another study showed a close link of increased MSI1 expression to the development of lung cancer [40]. In the current study, we consistently found that the expression of MSI1 was higher in lung cancer tissues than in their adjacent normal tissues. All above results suggested that the MSI1 might act as oncogene in lung tumorigenesis.
Emerging studies have identified that genetic variants in the potentially functional area of genes could influence gene’s structure, function or expression and thus cause unexpected consequence [31, 41]. And also, evidence reported that several SNPs in MSI1 was associated with various human diseases [42]. However, up to now, few studies have interpreted the potential effects of genetic variants in MSI1 gene on cancer susceptibility. There was only one study that reported a SNP in the 3’-UTR of MSI1 associated with an increased risk of lung cancer [25]. Nevertheless, no further evidence shows how this SNP contributes to lung cancer development. Herein, based on the candidate gene approach that is economical and has rather high statistical power than GWAS and Quantitative Trait Locus (QTL) approaches, we tested the association between three putative functional SNPs in the promoter of MSI1 gene and lung cancer risk in discovery set and then validated in other independent population. We found that the SNP rs7959801T>C was significantly associated with a decreased risk of lung cancer. The alteration from rs7959801T to C could decreased the expression of MSI1, which might decrease the number or undermine the biological function of lung CSCs and thus exert a decreased risk for lung cancer. All these findings indicated that the SNP rs7959801T>C of MSI1 may be a useful biomarker for lung cancer susceptibility in Chinese.
Several deficiencies and limitations should be concerned in the present study. Because of that there were two hospital-based retrospective studies, restricted Chinese Han populations; some selection biases were unavoidable in this study. In fact, the genotype frequencies of all selected SNPs among controls fitting the Hardy-Weinberg equilibrium law indicate the randomness of subjects selection. Also, we have obtained concordant results of the association between rs7959801T>C and lung cancer risk both in two populations accompanied by a 99.9% statistically study power. In addition, it yielded a value of 0.000 with a 0.001 prior probability lower than the preset FPRP-level criterion of 0.20, suggesting that this finding is noteworthy. Furthermore, the association between the SNP rs7959801T>C and risk of lung cancer had been supported by a serial of functional assays, further proving that our results were not achieved by chance. Even so, to further better reveal the effect of rs7959801 T>C in the promoter of MSI1 gene on lung carcinogenesis, in vivo experiment such as xenograft models should be performed.
Conclusion
In summary, our findings provide the first evidence that the SNP rs7959801T>C in the promoter of MSI1 gene confers a rewarding role in lung cancer risk by reducing MSI1 expression, which may be a functional biomarker for the risk of lung cancer. Further studies with larger population-based studies in different ethnic groups and in vivo animal experiments are needed to validate our findings.
ACKNOWLEDGEMENTS
This study was supported by the National Natural Science Foundation of China (No. 81473040, 81673267, 81273149, 81402753, 81672303, 81602289) and Yangcheng Scholar Grants (1201541589). We thank Dr. Zhanhong Xie and Ms. Wanmin Zeng for their assistance in recruiting the subjects.
List of Abbreviations
- CSCs
Cancer Stem Cells
- GWASs
Genome-Wide Association Studies
- Musashi1
Musashi RNA binding protein 1
- OR
Odd Ratio
- SNPs
Single Nucleotide Polymorphisms
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publisher’s website along with the published article.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The study was approved by the Institutional Review Boards of Guangzhou Medical University (Ethics Committee of Guangzhou Medical University: GZMC2007-07-0676) and Soochow University (Ethics Committee of Soochow University: SZUM2008031233).
HUMAN AND ANIMAL RIGHTS
No animals were used in this study. All human research procedures followed were in accordance with the Helsinki Declaration of 1975, as revised in 2008.
CONSENT FOR PUBLICATION
Written informed consent was obtained from each individual. After given a written informed consent, each participant was scheduled for an interview with a structured questionnaire to provide data on smoking status, alcohol use and other factors including family history of cancer.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Zheng R., Zeng H., Zhang S., Chen T., Chen W. National estimates of cancer prevalence in China, 2011. Cancer Lett. 2015;370(1):33–38. doi: 10.1016/j.canlet.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Chen W., Zheng R., Zeng H., Zhang S. Epidemiology of lung cancer in China. Thorac. Cancer. 2015;6(2):209–215. doi: 10.1111/1759-7714.12169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hung R.J., McKay J.D., Gaborieau V., Boffetta P., Hashibe M., Zaridze D., Mukeria A., Szeszenia-Dabrowska N., Lissowska J., Rudnai P., Fabianova E., Mates D., Bencko V., Foretova L., Janout V., Chen C., Goodman G., Field J.K., Liloglou T., Xinarianos G., Cassidy A., McLaughlin J., Liu G., Narod S., Krokan H.E., Skorpen F., Elvestad M.B., Hveem K., Vatten L., Linseisen J., Clavel-Chapelon F., Vineis P., Bueno-de-Mesquita H.B., Lund E., Martinez C., Bingham S., Rasmuson T., Hainaut P., Riboli E., Ahrens W., Benhamou S., Lagiou P., Trichopoulos D., Holcatova I., Merletti F., Kjaerheim K., Agudo A., Macfarlane G., Talamini R., Simonato L., Lowry R., Conway D.I., Znaor A., Healy C., Zelenika D., Boland A., Delepine M., Foglio M., Lechner D., Matsuda F., Blanche H., Gut I., Heath S., Lathrop M., Brennan P. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452(7187):633–637. doi: 10.1038/nature06885. https:// www.nature.com/articles/nature06885 [DOI] [PubMed] [Google Scholar]
- 5.Dong J., Hu Z., Wu C., Guo H., Zhou B., Lv J., Lu D., Chen K., Shi Y., Chu M., Wang C., Zhang R., Dai J., Jiang Y., Cao S., Qin Z., Yu D., Ma H., Jin G., Gong J., Sun C., Zhao X., Yin Z., Yang L., Li Z., Deng Q., Wang J., Wu W., Zheng H., Zhou G., Chen H., Guan P., Peng Z., Chen Y., Shu Y., Xu L., Liu X., Liu L., Xu P., Han B., Bai C., Zhao Y., Zhang H., Yan Y., Amos C.I., Chen F., Tan W., Jin L., Wu T., Lin D., Shen H. Association analyses identify multiple new lung cancer susceptibility loci and their interactions with smoking in the Chinese population. Nat. Genet. 2012;44(8):895–899. doi: 10.1038/ng.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu Z., Wu C., Shi Y., Guo H., Zhao X., Yin Z., Yang L., Dai J., Hu L., Tan W., Li Z., Deng Q., Wang J., Wu W., Jin G., Jiang Y., Yu D., Zhou G., Chen H., Guan P., Chen Y., Shu Y., Xu L., Liu X., Liu L., Xu P., Han B., Bai C., Zhao Y., Zhang H., Yan Y., Ma H., Chen J., Chu M., Lu F., Zhang Z., Chen F., Wang X., Jin L., Lu J., Zhou B., Lu D., Wu T., Lin D., Shen H. A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12.2 in Han Chinese. Nat. Genet. 2011;43(8):792–796. doi: 10.1038/ng.875. [DOI] [PubMed] [Google Scholar]
- 7.Easwaran H., Tsai H.C., Baylin S.B. Cancer epigenetics: Tumor heterogeneity, plasticity of stem-like states, and drug resistance. Mol. Cell. 2014;54(5):716–727. doi: 10.1016/j.molcel.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dick J.E. Stem cell concepts renew cancer research. Blood. 2008;112(13):4793–4807. doi: 10.1182/blood-2008-08-077941. [DOI] [PubMed] [Google Scholar]
- 9.Visvader J.E., Lindeman G.J. Cancer stem cells in solid tumours: Accumulating evidence and unresolved questions. Nat. Rev. Cancer. 2008;8(10):755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 10.Pardal R., Clarke M.F., Morrison S.J. Applying the principles of stem-cell biology to cancer. Nat. Rev. Cancer. 2003;3(12):895–902. doi: 10.1038/nrc1232. [DOI] [PubMed] [Google Scholar]
- 11.Al-Hajj M., Clarke M.F. Self-renewal and solid tumor stem cells. Oncogene. 2004;23(43):7274–7282. doi: 10.1038/sj.onc.1207947. [DOI] [PubMed] [Google Scholar]
- 12.Rosen J.M., Jordan C.T. The increasing complexity of the cancer stem cell paradigm. Science. 2009;324(5935):1670–1673. doi: 10.1126/science.1171837. http://science.sciencemag.org/content/324/5935/1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song L.L., Miele L. Cancer stem cells-an old idea that’s new again: Implications for the diagnosis and treatment of breast cancer. Expert Opin. Biol. Ther. 2007;7(4):431–438. doi: 10.1517/14712598.7.4.431. [DOI] [PubMed] [Google Scholar]
- 14.Zhou B.B., Zhang H., Damelin M., Geles K.G., Grindley J.C., Dirks P.B. Tumour-initiating cells: Challenges and opportunities for anticancer drug discovery. Nat. Rev. Drug Discov. 2009;8(10):806–823. doi: 10.1038/nrd2137. [DOI] [PubMed] [Google Scholar]
- 15.Chattopadhyay I., Singh A., Phukan R., Purkayastha J., Kataki A., Mahanta J., Saxena S., Kapur S. Genome-wide analysis of chromosomal alterations in patients with esophageal squamous cell carcinoma exposed to tobacco and betel quid from high-risk area in India. Mutat. Res. 2010;696(2):130–138. doi: 10.1016/j.mrgentox.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 16.Okano H., Imai T., Okabe M. Musashi: A translational regulator of cell fate. J. Cell Sci. 2002;115(Pt 7):1355–1359. doi: 10.1242/jcs.115.7.1355. [DOI] [PubMed] [Google Scholar]
- 17.Imai T., Tokunaga A., Yoshida T., Hashimoto M., Mikoshiba K., Weinmaster G., Nakafuku M., Okano H. The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol. Cell. Biol. 2001;21(12):3888–3900. doi: 10.1128/MCB.21.12.3888-3900.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Battelli C., Nikopoulos G.N., Mitchell J.G., Verdi J.M. The RNA-binding protein Musashi-1 regulates neural development through the translational repression of p21WAF-1. Mol. Cell. Neurosci. 2006;31(1):85–96. doi: 10.1016/j.mcn.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 19.Wang X.Y., Yin Y., Yuan H., Sakamaki T., Okano H., Glazer R.I. Musashi1 modulates mammary progenitor cell expansion through proliferin-mediated activation of the Wnt and Notch pathways. Mol. Cell. Biol. 2008;28(11):3589–3599. doi: 10.1128/MCB.00040-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shu H.J., Saito T., Watanabe H., Ito J.I., Takeda H., Okano H., Kawata S. Expression of the Musashi1 gene encoding the RNA-binding protein in human hepatoma cell lines. Biochem. Biophys. Res. Commun. 2002;293(1):150–154. doi: 10.1016/S0006-291X(02)00175-4. [DOI] [PubMed] [Google Scholar]
- 21.Toda M., Iizuka Y., Yu W., Imai T., Ikeda E., Yoshida K., Kawase T., Kawakami Y., Okano H., Uyemura K. Expression of the neural RNA-binding protein Musashi1 in human gliomas. Glia. 2001;34(1):1–7. doi: 10.1002/glia.1034. [DOI] [PubMed] [Google Scholar]
- 22.Schulenburg A., Cech P., Herbacek I., Marian B., Wrba F., Valent P., Ulrich-Pur H. CD44-positive colorectal adenoma cells express the potential stem cell markers musashi antigen (msi1) and ephrin B2 receptor (EphB2). J. Pathol. 2007;213(2):152–160. doi: 10.1002/path.2220. [DOI] [PubMed] [Google Scholar]
- 23.Moreira A.L., Gonen M., Rekhtman N., Downey R.J. Progenitor stem cell marker expression by pulmonary carcinomas. Mod. Pathol. 2010;23(6):889–895. doi: 10.1038/modpathol.2010.68. [DOI] [PubMed] [Google Scholar]
- 24.Feng G.J., Ming Z.W., Xiang G., Ning L., Shou L.J. Cloning and functional analysis of the promoter region of an intestinal stem cell specific expressed gene, Musashi-1. Parenter. Enteral Nutr. 2006;13(5):257–260. [Google Scholar]
- 25.Wang X., Hu J.F., Tan Y., Cui J., Wang G., Mrsny R.J., Li W. Cancer stem cell marker Musashi-1 rs2522137 genotype is associated with an increased risk of lung cancer. PLoS One. 2014;9(5):e95915. doi: 10.1371/journal.pone.0095915. http://journals.plos.org/plosone/article?id =10.1371/journal.pone.0095915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu B., Yang L., Huang B., Cheng M., Wang H., Li Y., Huang D., Zheng J., Li Q., Zhang X., Ji W., Zhou Y., Lu J. A functional copy-number variation in MAPKAPK2 predicts risk and prognosis of lung cancer. Am. J. Hum. Genet. 2012;91(2):384–390. doi: 10.1016/j.ajhg.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang L., Liu B., Huang B., Deng J., Li H., Yu B., Qiu F., Cheng M., Wang H., Yang R., Yang X., Zhou Y., Lu J. A functional copy number variation in the WWOX gene is associated with lung cancer risk in Chinese. Hum. Mol. Genet. 2013;22(9):1886–1894. doi: 10.1093/hmg/ddt019. [DOI] [PubMed] [Google Scholar]
- 28.Lu J., Yang L., Zhao H., Liu B., Li Y., Wu H., Li Q., Zeng B., Wang Y., Ji W., Zhou Y. The polymorphism and haplotypes of PIN1 gene are associated with the risk of lung cancer in Southern and Eastern Chinese populations. Hum. Mutat. 2011;32(11):1299–1308. doi: 10.1002/humu.21574. [DOI] [PubMed] [Google Scholar]
- 29.Qiu F., Yang L., Fang W., Li Y., Yang R., Yang X., Deng J., Huang B., Xie C., Zhou Y., Lu J. A functional polymorphism in the promoter of ERK5 gene interacts with tobacco smoking to increase the risk of lung cancer in Chinese populations. Mutagenesis. 2013;28(5):561–567. doi: 10.1093/mutage/get033. [DOI] [PubMed] [Google Scholar]
- 30.Liu B., Chen D., Yang L., Li Y., Ling X., Liu L., Ji W., Wei Y., Wang J., Wei Q., Wang L., Lu J. A functional variant (-1304T>G) in the MKK4 promoter contributes to a decreased risk of lung cancer by increasing the promoter activity. Carcinogenesis. 2010;31(8):1405–1411. doi: 10.1093/carcin/bgq126. [DOI] [PubMed] [Google Scholar]
- 31.Cheng M., Yang L., Yang R., Yang X., Deng J., Yu B., Huang D., Zhang S., Wang H., Qiu F., Zhou Y., Lu J. A microRNA-135a/b binding polymorphism in CD133 confers decreased risk and favorable prognosis of lung cancer in Chinese by reducing CD133 expression. Carcinogenesis. 2013;34(10):2292–2299. doi: 10.1093/carcin/bgt181. [DOI] [PubMed] [Google Scholar]
- 32.Wacholder S., Chanock S., Garcia-Closas M., El Ghormli L., Rothman N. Assessing the probability that a positive report is false: An approach for molecular epidemiology studies. J. Natl. Cancer Inst. 2004;96(6):434–442. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang L., Li Y., Cheng M., Huang D., Zheng J., Liu B., Ling X., Li Q., Zhang X., Ji W., Zhou Y., Lu J. A functional polymorphism at microRNA-629-binding site in the 3′-untranslated region of NBS1 gene confers an increased risk of lung cancer in Southern and Eastern Chinese population. Carcinogenesis. 2012;33(2):338–347. doi: 10.1093/carcin/bgr272. [DOI] [PubMed] [Google Scholar]
- 34.Keysar S.B., Jimeno A. More than markers: Biological significance of cancer stem cell-defining molecules. Mol. Cancer Ther. 2010;9(9):2450–2457. doi: 10.1158/1535-7163.MCT-10-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamashita T., Honda M., Nakamoto Y., Baba M., Nio K., Hara Y., Zeng S.S., Hayashi T., Kondo M., Takatori H., Yamashita T., Mizukoshi E., Ikeda H., Zen Y., Takamura H., Wang X.W., Kaneko S. Discrete nature of EpCAM+ and CD90+ cancer stem cells in human hepatocellular carcinoma. Hepatology. 2013;57(4):1484–1497. doi: 10.1002/hep.26168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeh C.T., Wu A.T., Chang P.M., Chen K.Y., Yang C.N., Yang S.C., Ho C.C., Chen C.C., Kuo Y.L., Lee P.Y., Liu Y.W., Yen C.C., Hsiao M., Lu P.J., Lai J.M., Wang L.S., Wu C.H., Chiou J.F., Yang P.C., Huang C.Y. Trifluoperazine, an antipsychotic agent, inhibits cancer stem cell growth and overcomes drug resistance of lung cancer. Am. J. Respir. Crit. Care Med. 2012;186(11):1180–1188. doi: 10.1164/rccm.201207-1180OC. [DOI] [PubMed] [Google Scholar]
- 37.Sureban S.M., May R., George R.J., Dieckgraefe B.K., McLeod H.L., Ramalingam S., Bishnupuri K.S., Natarajan G., Anant S., Houchen C.W. Knockdown of RNA binding protein musashi-1 leads to tumor regression in vivo. Gastroenterology. 2008;134(5):1448–1458. doi: 10.1053/j.gastro.2008.02.057. [DOI] [PubMed] [Google Scholar]
- 38.Nishimoto Y., Okano H. New insight into cancer therapeutics: Induction of differentiation by regulating the Musashi/Numb/Notch pathway. Cell Res. 2010;20(10):1083–1085. doi: 10.1038/cr.2010.122. [DOI] [PubMed] [Google Scholar]
- 39.Wang X.Y., Yu H., Linnoila R.I., Li L., Li D., Mo B., Okano H., Penalva L.O., Glazer R.I. Musashi1 as a potential therapeutic target and diagnostic marker for lung cancer. Oncotarget. 2013;4(5):739–750. doi: 10.18632/oncotarget.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanai R., Eguchi K., Takahashi M., Goldman S., Okano H., Kawase T., Yazaki T. Enhanced therapeutic efficacy of oncolytic herpes vector G207 against human non-small cell lung cancer--expression of an RNA-binding protein, Musashi1, as a marker for the tailored gene therapy. J. Gene Med. 2006;8(11):1329–1340. doi: 10.1002/jgm.965. [DOI] [PubMed] [Google Scholar]
- 41.Qiu F., Yang L., Lu X., Chen J., Wu D., Wei Y., Nong Q., Zhang L., Fang W., Chen X., Ling X., Yang B., Zhang X., Zhou Y., Lu J. The MKK7 p.Glu116Lys rare variant serves as a predictor for lung cancer risk and prognosis in Chinese. PLoS Genet. 2016;12(3):e1005955. doi: 10.1371/journal.pgen.1005955. http://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1005955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.den Hoed M., Eijgelsheim M., Esko T., Brundel B.J., Peal D.S., Evans D.M., Nolte I.M., Segre A.V., Holm H., Handsaker R.E., Westra H.J., Johnson T., Isaacs A., Yang J., Lundby A., Zhao J.H., Kim Y.J., Go M.J., Almgren P., Bochud M., Boucher G., Cornelis M.C., Gudbjartsson D., Hadley D., van der Harst P., Hayward C., den Heijer M., Igl W., Jackson A.U., Kutalik Z., Luan J., Kemp J.P., Kristiansson K., Ladenvall C., Lorentzon M., Montasser M.E., Njajou O.T., O’Reilly P.F., Padmanabhan S., St. Pourcain B., Rankinen T., Salo P., Tanaka T., Timpson N.J., Vitart V., Waite L., Wheeler W., Zhang W., Draisma H.H., Feitosa M.F., Kerr K.F., Lind P.A., Mihailov E., Onland-Moret N.C., Song C., Weedon M.N., Xie W., Yengo L., Absher D., Albert C.M., Alonso A., Arking D.E., de Bakker P.I., Balkau B., Barlassina C., Benaglio P., Bis J.C., Bouatia-Naji N., Brage S., Chanock S.J., Chines P.S., Chung M., Darbar D., Dina C., Dorr M., Elliott P., Felix S.B., Fischer K., Fuchsberger C., de Geus E.J., Goyette P., Gudnason V., Harris T.B., Hartikainen A.L., Havulinna A.S., Heckbert S.R., Hicks A.A., Hofman A., Holewijn S., Hoogstra-Berends F., Hottenga J.J., Jensen M.K., Johansson A., Junttila J., Kaab S., Kanon B., Ketkar S., Khaw K.T., Knowles J.W., Kooner A.S., Kors J.A., Kumari M., Milani L., Laiho P., Lakatta E.G., Langenberg C., Leusink M., Liu Y., Luben R.N., Lunetta K.L., Lynch S.N., Markus M.R., Marques-Vidal P., Mateo Leach I., McArdle W.L., McCarroll S.A., Medland S.E., Miller K.A., Montgomery G.W., Morrison A.C., Muller-Nurasyid M., Navarro P., Nelis M., O’Connell J.R., O’Donnell C.J., Ong K.K., Newman A.B., Peters A., Polasek O., Pouta A., Pramstaller P.P., Psaty B.M., Rao D.C., Ring S.M., Rossin E.J., Rudan D., Sanna S., Scott R.A., Sehmi J.S., Sharp S., Shin J.T., Singleton A.B., Smith A.V., Soranzo N., Spector T.D., Stewart C., Stringham H.M., Tarasov K.V., Uitterlinden A.G., Vandenput L., Hwang S.J., Whitfield J.B., Wijmenga C., Wild S.H., Willemsen G., Wilson J.F., Witteman J.C., Wong A., Wong Q., Jamshidi Y., Zitting P., Boer J.M., Boomsma D.I., Borecki I.B., van Duijn C.M., Ekelund U., Forouhi N.G., Froguel P., Hingorani A., Ingelsson E., Kivimaki M., Kronmal R.A., Kuh D., Lind L., Martin N.G., Oostra B.A., Pedersen N.L., Quertermous T., Rotter J.I., van der Schouw Y.T., Verschuren W.M., Walker M., Albanes D., Arnar D.O., Assimes T.L., Bandinelli S., Boehnke M., de Boer R.A., Bouchard C., Caulfield W.L., Chambers J.C., Curhan G., Cusi D., Eriksson J., Ferrucci L., van Gilst W.H., Glorioso N., de Graaf J., Groop L., Gyllensten U., Hsueh W.C., Hu F.B., Huikuri H.V., Hunter D.J., Iribarren C., Isomaa B., Jarvelin M.R., Jula A., Kahonen M., Kiemeney L.A., van der Klauw M.M., Kooner J.S., Kraft P., Iacoviello L., Lehtimaki T., Lokki M.L., Mitchell B.D., Navis G., Nieminen M.S., Ohlsson C., Poulter N.R., Qi L., Raitakari O.T., Rimm E.B., Rioux J.D., Rizzi F., Rudan I., Salomaa V., Sever P.S., Shields D.C., Shuldiner A.R., Sinisalo J., Stanton A.V., Stolk R.P., Strachan D.P., Tardif J.C., Thorsteinsdottir U., Tuomilehto J., van Veldhuisen D.J., Virtamo J., Viikari J., Vollenweider P., Waeber G., Widen E., Cho Y.S., Olsen J.V., Visscher P.M., Willer C., Franke L., Global B.C., Consortium C.A., Erdmann J., Thompson J.R., Consortium P.G., Pfeufer A., Consortium Q.G., Sotoodehnia N., Consortium Q.-I., Newton-Cheh C., Consortium C.-A., Ellinor P.T., Stricker B.H., Metspalu A., Perola M., Beckmann J.S., Smith G.D., Stefansson K., Wareham N.J., Munroe P.B., Sibon O.C., Milan D.J., Snieder H., Samani N.J., Loos R.J. Identification of heart rate-associated loci and their effects on cardiac conduction and rhythm disorders. Nat. Genet. 2013;45(6):621–631. doi: 10.1038/ng.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material is available on the publisher’s website along with the published article.