We performed whole genome sequencing on Clostridium difficile isolates from a single-center cohort of children with C. difficile infection (CDI). Putative transmission events were identified only among 11.9% of CDIs. Transmission among community- and healthcare facility–associated CDIs were equally infrequent.
Keywords: Clostridium difficile, transmission, pediatrics, whole genome sequencing, genomics
Abstract
Background
Although pediatric Clostridium difficile infections (CDIs) are increasing, C. difficile transmission patterns among children are poorly understood.
Methods
We performed whole genome sequencing (WGS) on C. difficile isolates collected from children diagnosed with CDI between December 2012 and December 2013 at a single academic medical center. Genome sequences of isolates from CDIs diagnosed ≥8 weeks after study initiation were compared to all study isolate genome sequences. Among patients with isogenic isolates (≤2–3 core genome single nucleotide variants [SNVs] identified by pairwise SNV analyses), common inpatient and/or outpatient healthcare exposures were investigated.
Results
Among 131 CDIs in 107 children, WGS identified 104 genetically distinct isolates. Of 84 incident CDIs occurring ≥8 weeks after study initiation, only 10 (11.9%) were caused by a strain isogenic to another cohort CDI isolate (putative transmission events). Proportions of each CDI class putatively associated with transmission were hospital-onset healthcare facility–associated (HCFA), 2/16 (12.5%); community-onset HCFA, 1/17 (5.9%); indeterminate, 1/11 (9.1%); community-associated (CA), 5/40 (12.5%); and recurrent, 1/21 (4.8%). Transmission events among CA and HCFA CDIs were similarly infrequent (5/40 [12.5%] vs 3/33 [9.1%]; P = .64). Shared healthcare facility exposures were only identified among 7/10 putative transmission events. Potential community transmission (same postal code) was not identified.
Conclusions
WGS identified a highly diverse group of C. difficile isolates among children with CDI, including those with HCFA CDI. Clostridium difficile transmission among symptomatic children was very uncommon. Among putatively transmitted cases, investigation of shared healthcare exposures often did not identify a potential transmission source.
Clostridium difficile infection (CDI) is the most common healthcare-associated infection among US adults [1]. The Centers for Diseases Control and Prevention (CDC) [2] classifies CDI as a public health threat that requires “urgent and aggressive action.” CDI prevention strategies require a thorough understanding of C. difficile transmission patterns. Although isolation of patients with CDI is considered an essential infection prevention measure, a recent investigation using whole genome sequencing (WGS) revealed that only a minority of adult CDIs are transmitted from other symptomatic patients [3]. Thus, identifying other sources of transmission is necessary to develop novel and more effective prevention strategies.
Pediatric CDI is also increasing [4], but C. difficile transmission patterns among children are poorly understood. Although the vast majority of pediatric CDIs are community associated (CA) [5], many children with CA CDI have had recent outpatient healthcare exposures [6, 7], suggesting that outpatient settings may also be sites of transmission. Our primary objective in this study was to assess patterns of C. difficile transmission among children. Specifically, using WGS, we measured the prevalence of genetically related strains among a cohort of children with CDI and we determined whether shared inpatient and outpatient healthcare exposures were associated with transmission.
METHODS
Patients and Setting
Clostridium difficile isolates saved from a previous study [8] were originally collected from patients aged ≥12 months diagnosed with CDI using tcdB polymerase chain reaction (PCR; Cepheid, Sunnyvale, California) at the Ann & Robert H. Lurie Children’s Hospital in Chicago, Illinois, between 9 December 2012 and 8 December 2013. Patients were included if they had diarrhea (ie, diarrhea or unformed stool documented in the medical record irrespective of number of stools per day) or ileus, CDI diagnosis documented by the healthcare provider or hospital infection control personnel, and/or received CDI treatment. CDI cases that represented a second positive test within 2 weeks of the original infection were also excluded unless there was resolution and recurrence of symptoms. CDI cases were classified as hospital-onset (HO) healthcare facility–associated (HCFA), community-onset (CO) HCFA, indeterminate, CA, and recurrent using standard definitions, as previously described [9]. The Lurie Children’s Institutional Review Board waived informed consent.
Whole Genome Sequencing
Study isolates had previously been characterized by restriction endonuclease analysis (REA) [8]. Genomic DNA was extracted from C. difficile isolates using the BiOstic Bacteremia DNA Isolation Kit (MO BIO Laboratories, Carlsbad, California). Paired-end sequencing libraries were prepared using the Nextera XT DNA Library Prep Kit (Illumina, San Diego, California), and WGS was performed using Illumina MiSeq to produce paired-end 300 base pair (bp) reads. De novo genome assembly was performed using SPAdes (v3.9.1; http://cab.spbu.ru/software/spades/) [10]. In silico multilocus sequence typing (MLST) [11] was performed using PubMLST (https://pubmlst.org/cdifficile/), which permitted isolate sequence type (ST) and clade assignment. The 131 isolate sequences included in this study have been deposited in DDBJ/ENA/GenBank under accession numbers listed in the Supplementary Material.
Isolate genetic relatedness was determined by performing pairwise comparisons of single nucleotide variants (SNVs) among strains, as adapted from methods described by Eyre et al [3]. Illumina reads were trimmed and filtered for low-quality bases and adapter sequences using Trimmomatic v0.36 [12]. Reads were then aligned to the chromosomal sequence of the clade-specific reference strain using Stampy (v1.0.29) with an expected substitution rate setting of 0.01. The reference strain used for each isolate alignment was based on the major C. difficile clade assignment as determined by MLST: clade 1-630 (GenBank accession number AM180355.1), clade 2-R20291 (GenBank accession number FN545816.1), clade 4-M68 (GenBank accession number FN668375.1), and clade 5- M120 (GenBank accession number FN665653.1). SNVs relative to the reference were called using the mpileup function of samtools (v0.1.19-44428cd) with the following options: -E (recalculate extended BAQ), -M 0 (cap mapping quality at 0), -Q 25 (skip bases with BAQ less than 25), -q 30 (skip alignments with mapQ less than 30), -m 2 (minimum gapped reads for indel candidates of 2), -D (output per-sample DP in binary call format [BCF]), -S (output per-sample strand bias P-value in BCF), and -g (generate BCF output). SNVs were filtered if they failed to meet 1 or more of the following criteria: minimum SNV quality score of 200, minimum read consensus of 75%, minimum of 5 reads covering the SNV position, maximum of 3 times the median read depth of the total alignment, minimum of 1 read in either direction covering the SNV position, homozygous under the diploid model, and not within a repetitive region as determined by BLAST alignment of fragments of the clade-specific reference strain sequence against itself. For each strain, the clade-specific reference strain sequence was used as the base sequence. Any positions with SNVs that passed the above filters were changed to the SNV base. Any positions with SNVs that did not pass the above filters were changed to a missing base character. Any non-SNV position with coverage of fewer than 5 reads was changed to a missing base character. After filtering, positions with a base in <100% of all genomes in the clade were excluded (ie, included only core genome). To minimize the impact of recombination on SNV counts, ClonalFrameML (v1.0-16-g30da94a) was used to identify regions of potential recombination, which were then masked in the alignment. Isolates were considered isogenic if they differed by ≤2 core genome SNVs (isolates collected <124 days apart) or ≤3 core genome SNVs (isolates collected 124–364 days apart), based on analyses of C. difficile evolutionary rate performed by Eyre et al [3]. Initially validated in adult patients, this definition was also more recently validated in infants [13].
Investigation of Putative Transmission
To identify the frequency of putative transmission events, isolates collected between February 2013 and December 2013 (ie, at least 8 weeks after study initiation) were compared to all isolates collected during the entire 12-month study period. This 8-week period was chosen because transmission events that occurred prior to study initiation that resulted in CDI early in the study could be missed. Current CDI surveillance definitions [9] suggest C. difficile has a maximum incubation period of 4–12 weeks (ie, definitions allow a 4-week incubation period to definitively identify HCFA CDI and 12-week incubation period to definitively identify CA CDI). Only incident CDIs (ie, first-time CDIs or nonrecurrent CDIs diagnosed at least 8 weeks after a previous CDI) were included in the measurement of transmission frequency. Among patients with isogenic isolates, shared inpatient and/or outpatient exposures that occurred between 8 weeks prior to the first CDI and the day of the most recent CDI in each putative transmission event were investigated. Among patients with multiple shared exposures, the most plausible transmission scenario was assigned based on the likelihood of direct patient interaction (ie, receiving care in the same unit on the same day), a shared healthcare worker with direct contact with both patients (ie, receiving care in the same unit or facility on the same day), and/or a shared healthcare environment (ie, receiving care in the same unit or facility on different days). Potential community transmission was identified if patients shared a postal code. The proportion of each CDI epidemiologic classification type associated with putative transmission was determined. Total HCFA CDI was the sum of HO- and CO-HCFA CDI cases. Proportions were compared using χ2 test with Stata/IC statistical software, version 12.1 (StataCorp, College Station, Texas). Two-sided P values < 0.05 were considered statistically significant.
RESULTS
Of 189 CDI cases that met inclusion criteria, 156 (82.5%) stools were available for culture, and C. difficile was isolated from 131 (84.0%) stools from 107 unique patients. Median age (at time of first CDI during the study period if a patient had multiple CDIs) was 8.4 years, and 67 (62.6%) were male. Common comorbidities were malignancy (n = 27, 25.2%), inflammatory bowel disease (n = 23, 21.5%), solid organ transplant (n = 11, 10.3%), and gastrostomy tube dependence (n = 16, 15.0%).
The 131 isolates underwent WGS. In sum, median read coverage was 71× (range, 25×–393×); median contig number (from de novo assembly of the reads) was 173 (range, 65–1183); and median N50 (ie, a statistic that indicates the value at which 50% of the entire assembly is contained in contigs of at least this length) was 114196 bp (range, 5540–519873). After masking recombinant regions, the numbers of core genome variant loci were 26970 among 106 clade 1 strains, 16035 among 13 clade 2 strains, 86 among 8 clade 4 strains, and 344 among 4 clade 5 strains. The Supplementary Material lists the REA group, ST, MLST clade, sequencing, and assembly statistics of each isolate. The predominant REA group/ST was DH/ST-42 (clade 1). Of note, only 1 patient in this study had CDI caused by BI/ST-1 (ie, NAP1/027, clade 2).
Among 131 CDIs in 107 children, WGS identified 104 genetically distinct isolates. Only 8 of these isogenic isolates were identified in more than 1 patient (ie, putative transmission events), and only 2 of these 8 isogenic isolates caused CDI in more than 2 patients (ie, 3 patients each). Thus, large clusters of CDI caused by a single clone were not identified. Interestingly, in one putative transmission event, isogenic strains of REA group M, a nontoxigenic strain, were isolated. The lack of tcdB in each strain was confirmed by WGS, indicating that this transmitted strain was not responsible for CDI symptoms. WGS confirmed that all other transmitted strains were toxigenic. Among the remaining 96 strains not putatively associated with transmission, 7 were confirmed by WGS to be nontoxigenic.
Of 131 CDIs that occurred during the 12-month study period, 26 (19.8%) occurred in the first 8 weeks and 105 (80.2%) occurred after 8 weeks from study initiation. Of these 105 CDIs, 84 (80.0%) were incident CDIs. In total, among these 84 incident CDIs, only 10 (11.9%) putative transmission events were identified based on pairwise SNV comparisons between these 84 CDI isolates and all 131 CDI isolates collected during the study period. Proportions of each CDI epidemiologic classification type putatively associated with transmission were HO-HCFA, 2/16 (12.5%); CO-HCFA, 1/17 (5.9%); indeterminate, 1/11 (9.1%); and CA, 5/40 (12.5%). In addition, 1/21 (4.8%) patient with recurrent CDI acquired a new strain that was isogenic to a strain isolated from another patient in this cohort. Putative transmission events among CA CDI and HCFA CDI were similarly infrequent (5/40 [12.5%] vs 3/33 [9.1%]; P = .64).
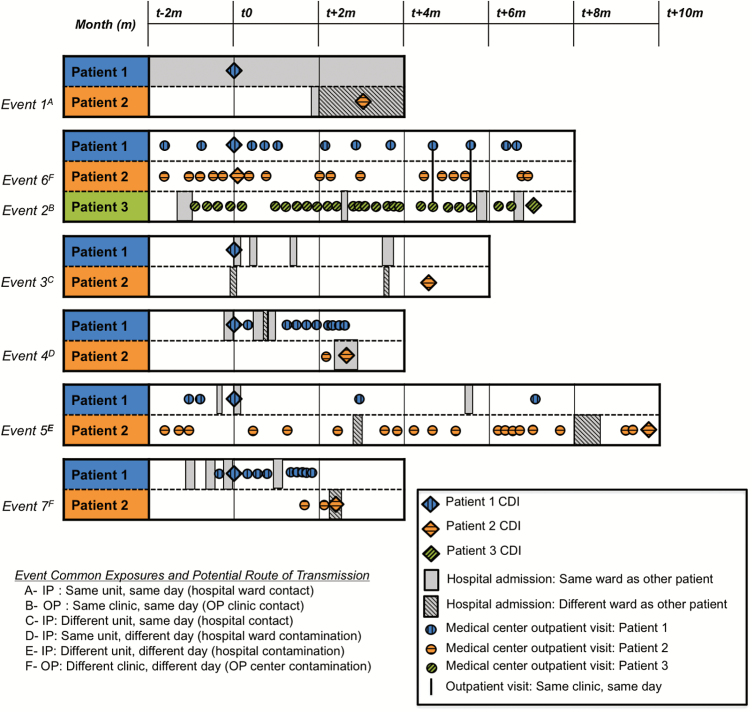
Figure 1 illustrates CDI and healthcare exposure chronology among patients involved in 7 putative transmission events; no epidemiologic link was identified in 3/10 events. Median time between CDI diagnosis of patients involved in these putative transmission events was 84 days (range, 1–300 days). Table 1 lists the most plausible epidemiologic link between patients based on shared inpatient and outpatient exposures.
Figure 1.
Chronology of Clostridium difficile infections and associated healthcare exposures among patients involved in the 7 putative transmission events where an epidemiologic link was identified. Event numbers are referred to in Table 1. Abbreviations: CDI, Clostridium difficile infection; IP, inpatient; OP, outpatient.
Table 1.
Shared Inpatient, Outpatient, and Community Exposures Among 10 Cases of Clostridium difficile Transmission
| Event Numbera | Potential Epidemiologic Link | n (%) | Clostridium difficile Restriction Endonuclease Analysis Group/Sequence Typeb (days between C. difficile infections) |
|---|---|---|---|
| 1 | IP: same unit, same day (hospital ward contact) | 1 (10) | DH/ST-42 (96d) |
| 2 | OP: same clinic, same day (outpatient clinic contact) | 1 (10) | G/ST-8 (187d) |
| 3 | IP: different unit, same day (hospital contact) | 1 (10) | N/ST-10 (148d) |
| 4 | IP: same unit, different dayc (hospital ward contamination) | 1 (10) | AL/ST-58 (72d) |
| 5 | IP: different unit, different dayd (hospital contamination) | 1 (10) | M/ST-15e (300d) |
| OP: same clinic, different day (outpatient clinic contamination) | 0 | … | |
| OP: different clinic, same day (outpatient center contact) | 0 | … | |
| 6–7 | OP: different clinic, different dayf (outpatient center contamination) | 2 (20) | G/ST-8 (1d), DH/ST-42 (65d) |
| Community (same postal code) | 0 | ||
| 8–10 | None identified | 3 (30) | N/ST-10 (21d), AH/ST-67 (1d), Y/ST-2 (242d) |
Abbreviations: IP, inpatient; OP, outpatient.
aEvent numbers are referred to in Figure 1.
bBy multilocus sequence typing (https://pubmlst.org/cdifficile).
cThirty-eight days between hospital ward encounters.
dSixty-nine days between hospital encounters.
eNontoxigenic C. difficile strain.
fOne day between outpatient center encounters for both events.
DISCUSSION
In our large, single-center cohort of children with CDI, using WGS, we identified a highly diverse group of C. difficile isolates, including among children with HCFA CDI. Thus, inferred C. difficile direct/indirect transmission among symptomatic children with CDI was very uncommon. Epidemiologic investigation frequently failed to identify a potential transmission source; patients in 3/10 of the putative transmission events had no previous Lurie Children’s inpatient or outpatient exposures prior to CDI diagnosis. While community exposures would be suspected in these cases, these patients had different postal codes and even resided in different counties. Furthermore, while those in 2/10 transmission events shared outpatient exposures, these outpatient visits were in different specialty clinics on different days, albeit in the same healthcare facility and 1 day apart in each case. In one case, patients were seen in the oncology outpatient clinic on the same day, suggesting possible transmission in this setting. This finding, along with other studies that have documented frequent outpatient healthcare visits among children [6, 7] and adults [14] with CA CDI, reinforces the need for good standard infection prevention and control practices in outpatient settings. This is particularly important for a pediatric oncology population with frequent C. difficile colonization and prolonged shedding after CDI [15].
Our data suggest that possible direct/indirect transmission among children with CDI, occurring in approximately 12% of our CDI cases, is even less frequent than the low transmission frequency reported among adults. Relatively infrequent transmission may be related to differences in CDI molecular epidemiology and strain-specific differences in transmissibility (described in more detail below); our hospital policy of early implementation of contact isolation for patients with diarrhea prior to and irrespective of results from stool testing; and/or good compliance with standard infection prevention and control practices. A whole-genome investigation of adult CDI in the United Kingdom identified genetic relatedness among strains that caused 35% [3] and 19% [16] of CDIs, respectively. Similar to our study, these were not designed to identify other sources of transmission. However, both studies hypothesized that patients colonized with C. difficile may contribute to transmission in the healthcare facility. Mawer et al [16] demonstrated that transmission from patients whose stool was negative for C. difficile toxin occurred 3 times less frequently than from patients whose stool was fecal toxin positive, but they did not determine whether a negative fecal toxin result represented a false-negative test (possibly in the setting of low colony counts, less toxin, and less frequent transmission) or a patient with C. difficile colonization and an alternate diarrheal etiology. Kong et al reported a slightly higher rate of transmission (40% by WGS) in the midst of a BI/NAP1/027 outbreak in their Canadian hospital, but putative transmission among asymptomatic patients was infrequent (3%) [17]. It is important to note that similar to these aforementioned studies, we used bioinformatics approaches to minimize the impact of genetic recombination on SNV identification. We also sought to use clade-specific reference strains for read alignments in order to reduce misalignments and maximize core genome representation.
Of note, only 1 of our study patients had CDI caused by BI/NAP1/027/ST-1, as previously reported [8]. Although this may limit generalizability of our findings to the aforementioned adult studies, which had a higher proportion of BI/NAP1/027/ST-1, our molecular epidemiology is closely aligned to recent trends in CDI epidemiology among US adults. Recent CDC Emerging Infections Program CDI surveillance data [18, 19] from 10 US states suggest that BI/NAP1/027/ST-1 prevalence declined between 2012 and 2014. Furthermore, similar to our cohort, ribotype 106 (also identified as REA group DH) has emerged as the predominant strain that causes CDI among US adults [18, 19].
We coincidentally identified direct/indirect C. difficile transmission among colonized patients. In 2 patients, an isogenic strain of REA group M was identified. This nontoxigenic strain colonized each patient; by chance, this strain was selected from stool culture from each patient. Thus, the toxigenic strain that caused CDI in those patients was not identified, although diarrhea related to CDI may have contributed to transmission of the colonizing nontoxigenic strain. Frequent C. difficile colonization in young and hospitalized children is well described [20], and the contribution of community and healthcare facility transmission of C. difficile among asymptomatic carriers warrants further investigation. Asymptomatic carriers may serve as intermediates in the transmission chain among children with CDI, particularly those transmission events with long time intervals between CDIs and/or those who lack identified shared exposures. While some studies of adults have limited the maximum infectious period at 8 weeks [3, 21], our group [8] and others [15] have demonstrated prolonged periods of carriage following CDI in pediatric patients, suggesting that an infectious period may last well beyond 8 weeks. Nonetheless, given the vast diversity of C. difficile isolates in this study, these putative transmission events do not commonly result in sustained transmission of a single clone to multiple patients.
It is also possible that healthcare facility transmission events are uncommon, particularly in the absence of an outbreak or high endemic rate of CDI. Rather, symptomatic patients may become colonized in the community and develop CDI with their colonizing strains after hospitalization when exposed to antibiotics or other medications that increase CDI risk. In adults, C. difficile colonization present at the time of hospital admission is a significant risk factor for developing CDI after admission [22]. These potential scenarios (ie, asymptomatic carrier transmission vs CDI caused by strains that are present upon admission) are very important to discern, because if pediatric CDIs are commonly caused by strains that patients are carrying at the time of admission, then misattribution of healthcare acquisition of C. difficile results in erroneous measurement of HO-HCFA CDI.
This study has several limitations. Harboring of isogenic strains does not necessarily prove transmission. Exposure to a similar community or healthcare facility reservoir could also result in identification of isogenic strains. Not every child with CDI during the study period was included because either stool was not saved or stool culture was negative for C. difficile. CDI was diagnosed using tcdB PCR, which has less diagnostic predictive value than toxin enzyme immunoassay [20, 23]. Although our clinical microbiology laboratory restricts testing only to unformed stools (irrespective of number of stools per day), we cannot rule out the possibility that some patients had asymptomatic C. difficile carriage along with an alternate diarrheal etiology. Only 1 colony was selected from each stool culture, similar to prior studies (as well as standard clinical microbiology practice) [3, 16]. However, coinfection with multiple strains, which is uncommon (<10%), may have obscured some cases of transmission [3]. This study began 6 months after a new hospital was opened. Because healthcare facility environmental contamination may increase over time, performance in a new healthcare facility may limit external validity. Although a strength of this study was its investigation of healthcare exposures at the main hospital and 14 affiliated outpatient centers, we were unable to account for outpatient exposures outside of the Lurie Children’s network. Because this is a retrospective study, environmental samples from inpatient and outpatient sites were not available to assess for the presence of putatively transmitted C. difficile isolates.
In summary, WGS identified a highly diverse group of C. difficile isolates among children with CDI, including those with HCFA CDI. Clostridium difficile transmission among symptomatic children was very uncommon; among putatively transmitted cases, investigation of shared healthcare exposures often did not identify a potential source of transmission. Additional investigation is required to delineate the role of common environmental sources and asymptomatic carriers in C. difficile transmission among children, as well as to determine the frequency of HCFA CDI being caused by strains that pediatric patients are carrying at the time of admission.
Supplementary Material
Notes
Acknowledgments. We acknowledge Katherine Murphy and the NUSeq Core at Northwestern University Feinberg School of Medicine for their assistance with performance of whole genome sequencing.
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health (NIH).
Financial support. This work was supported by grants from the Thrasher Research Fund (Early Career Award 11854 to L. K. K.), the National Institute of Allergy and Infectious Diseases at the National Institutes of Health (K23 AI123525 to L. K. K.), and the American Cancer Society (MRSG-13-220-01 to E. A. O.). Research reported in this publication was supported, in part, by the NIH’s National Center for Advancing Translational Sciences (grant UL1TR001422).
Potential conflicts of interest. L. K. K. is a scientific advisor for Actelion and has received research supplies from Alere. L. K. K. and S. J. P. have received research grants from Merck and Cubist. D. N. G. holds patents for the prevention of Clostridium difficile infection; is a consultant for Sanofi Pasteur, DaVolterra, MGB, and Pfizer; and is an advisory board member of Merck, Rebiotix, Summit, and Actelion. E. A. O is a scientific advisor for Gladius Pharmaceuticals. All remaining authors: No reported conflicts of Interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Magill SS, Edwards JR, Bamberg W, et al. ; Emerging Infections Program Healthcare-Associated Infections and Antimicrobial Use Prevalence Survey Team Multistate point-prevalence survey of health care-associated infections. N Engl J Med 2014; 370:1198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States, 2013 Available at: http://www.cdc.gov/drugresistance/pdf/ar-threats-2013–508.pdf. Accessed 11 December 2017.
- 3. Eyre DW, Cule ML, Wilson DJ, et al. Diverse sources of C. difficile infection identified on whole-genome sequencing. N Engl J Med 2013; 369:1195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tamma PD, Sandora TJ. Clostridium difficile infection in children: current state and unanswered questions. J Pediatric Infect Dis Soc 2012; 1:230–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wendt JM, Cohen JA, Mu Y, et al. Clostridium difficile infection among children across diverse US geographic locations. Pediatrics 2014; 133:651–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Crews JD, Anderson LR, Waller DK, Swartz MD, DuPont HL, Starke JR. Risk factors for community-associated Clostridium difficile-associated diarrhea in children. Pediatr Infect Dis J 2015; 34:919–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kociolek LK, Patel SJ, Zheng X, Todd KM, Shulman ST, Gerding DN. Clinical and microbiologic assessment of cases of pediatric community-associated Clostridium difficile infection reveals opportunities for improved testing decisions. Pediatr Infect Dis J 2016; 35:157–61. [DOI] [PubMed] [Google Scholar]
- 8. Kociolek LK, Patel SJ, Shulman ST, Gerding DN. Molecular epidemiology of Clostridium difficile infections in children: a retrospective cohort study. Infect Control Hosp Epidemiol 2015; 36:445–51. [DOI] [PubMed] [Google Scholar]
- 9. McDonald LC, Coignard B, Dubberke E, Song X, Horan T, Kutty PK; Ad Hoc Clostridium difficile Surveillance Working Group Recommendations for surveillance of Clostridium difficile-associated disease. Infect Control Hosp Epidemiol 2007; 28:140–5. [DOI] [PubMed] [Google Scholar]
- 10. Bankevich A, Nurk S, Antipov D, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012; 19:455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jolley KA, Maiden MC. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 2010; 11:595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stoesser N, Eyre DW, Quan TP, et al. ; Modernising Medical Microbiology Informatics Group Epidemiology of Clostridium difficile in infants in Oxfordshire, UK: risk factors for colonization and carriage, and genetic overlap with regional C. difficile infection strains. PLoS One 2017; 12:e0182307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chitnis AS, Holzbauer SM, Belflower RM, et al. Epidemiology of community-associated Clostridium difficile infection, 2009 through 2011. JAMA Intern Med 2013; 173:1359–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dominguez SR, Dolan SA, West K, et al. High colonization rate and prolonged shedding of Clostridium difficile in pediatric oncology patients. Clin Infect Dis 2014; 59:401–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mawer DPC, Eyre DW, Griffiths D, et al. Contribution to Clostridium difficile transmission of symptomatic patients with toxigenic strains who are fecal toxin negative. Clin Infect Dis 2017; 64:1163–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kong LY, Eyre D, Corbeil J, et al. Clostridium difficile: investigating transmission patterns between symptomatic and asymptomatic patients using whole genome sequencing [Oral Abstract 77, IDWeek, October 4–8, 2017, San Diego, CA]. Open Forum Infect Dis 2017; 4(Suppl 1): S1. [Google Scholar]
- 18. Paulick A, Karlsson M, Albreght V, et al. The Role of Ribotype 106 as a Cause of Clostridium difficile Infection in the United States, 2012–2014 [abstract PIII-7]. In: Program and Abstract Book of the 13th Biennial Congress of the Anaerobe Society of the Americas (Nashville). Los Angeles, CA: Anaerobe Society of the Americas, 2016:197. [Google Scholar]
- 19. Karlsson M, Paulick A, Albreght V, Granade M, Guh A, Rasheed JK. Molecular epidemiology of Clostridium difficile isolated in the United States, 2014 [abstract PIII-4]. In: Program and Abstract Book of the 13th Biennial Congress of the Anaerobe Society of the Americas (Nashville). Los Angeles, CA: Anaerobe Society of the Americas, 2016:194. [Google Scholar]
- 20. Kociolek LK. Strategies for optimizing the diagnostic predictive value of Clostridium difficile molecular diagnostics. J Clin Microbiol 2017; 55:1244–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kumar N, Miyajima F, He M, et al. Genome-based infection tracking reveals dynamics of Clostridium difficile transmission and disease recurrence. Clin Infect Dis 2016; 62:746–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tschudin-Sutter S, Carroll KC, Tamma PD, et al. Impact of toxigenic Clostridium difficile colonization on the risk of subsequent C. difficile infection in intensive care unit patients. Infect Control Hosp Epidemiol 2015; 36:1324–9. [DOI] [PubMed] [Google Scholar]
- 23. Polage CR, Gyorke CE, Kennedy MA, et al. Overdiagnosis of Clostridium difficile infection in the molecular test era. JAMA Intern Med 2015; 175:1792–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.