Abstract
Genome-wide association studies have generated over thousands of susceptibility loci for many human complex traits, and yet for most of these associations the true causal variants remain unknown. Tissue/cell type-specific prediction and prioritization of non-coding regulatory variants will facilitate the identification of causal variants and underlying pathogenic mechanisms for particular complex diseases and traits. By leveraging recent large-scale functional genomics/epigenomics data, we develop an intuitive web server, GWAS4D (http://mulinlab.tmu.edu.cn/gwas4d or http://mulinlab.org/gwas4d), that systematically evaluates GWAS signals and identifies context-specific regulatory variants. The updated web server includes six major features: (i) updates the regulatory variant prioritization method with our new algorithm; (ii) incorporates 127 tissue/cell type-specific epigenomes data; (iii) integrates motifs of 1480 transcriptional regulators from 13 public resources; (iv) uniformly processes Hi-C data and generates significant interactions at 5 kb resolution across 60 tissues/cell types; (v) adds comprehensive non-coding variant functional annotations; (vi) equips a highly interactive visualization function for SNP-target interaction. Using a GWAS fine-mapped set for 161 coronary artery disease risk loci, we demonstrate that GWAS4D is able to efficiently prioritize disease-causal regulatory variants.
INTRODUCTION
Since the majority of genome-wide association study (GWAS) risk loci are located in the non-coding genomic region, identifying and interpreting how genetic variants in these loci regulate gene expression and then being able to explain disease susceptibility continues to be a challenge (1–4). An increasing number of studies have shown that associated variants for a particular trait/disease are significantly enriched in certain regulatory signals of the relevant tissues/cell types (5,6). Therefore, integrating GWAS signals with coordinated genomic/epigenomic profiles in specific tissue/cell type provides a promising direction to fine-map the causal regulatory variant (7–10). In addition, connecting regulatory variants to their gene targets under a dynamic cellular environment is experimentally expensive, and computational methods are needed for more accurate predictions (11). However, recent international functional genomic projects, such as ENCODE and the 4D Nucleome project, continuously generate genome-wide chromosome conformation capture data, including Hi-C and ChIA-PET, on widespread tissues/cell types across human organs and development stages, which provides profound opportunities to study the effect of regulatory variants at spatiotemporal levels (12–15).
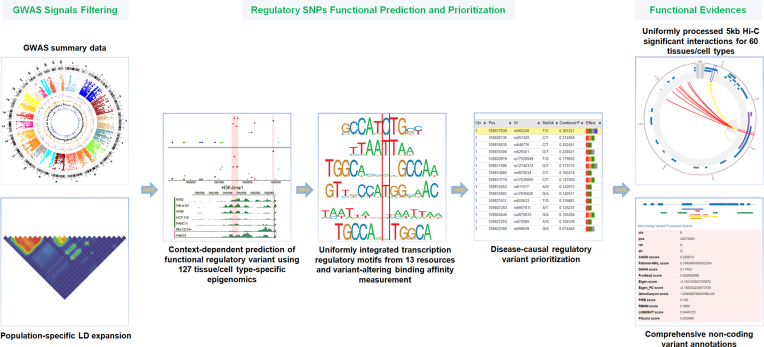
Previously, we developed the web server GWAS3D to detect human regulatory variants by the integrative analysis of genome-wide associations, chromosome interactions and histone modifications (16). This server has been successfully applied for functional fine-mapping and annotation of regulatory variants (17,18). Compared with other widely used tools such as HaploReg (19) and RegulomeDB (20), our web server quantitatively prioritizes all associated SNPs in the linkage disequilibrium (LD) proxy and provides comprehensive functional annotations to interpret variant regulatory effects. During the past few years, several tools and resources have been developed to annotate regulatory variants (21–25). However, online tools that can incorporate tissue/cell-type-specific genomic/epigenomic information toward prioritizing disease-causal regulatory variants are still lacking. In this work, by integrating the latest multidimensional functional genomic resources and our recent regulatory variant prioritization method, cepip (26), we updated our previous web server to systematically analyze GWAS signals and to identify context-specific regulatory variants. The GWAS4D web server is freely available at http://mulinlab.tmu.edu.cn/gwas4d or http://mulinlab.org/gwas4d.
METHODS AND PIPELINE
GWAS4D has been improved substantially in its current form by incorporating uniformly processed genomic/epigenomic data, integrated transcription regulator motif data, and comprehensive functional annotations, as well as our recent prioritization method for regulatory variants. We illustrate the web server data and pipeline in the following sections.
Data collection and processing
Genetics data
Genetic variant and allele information were retrieved from dbSNP150 (27) and 1000 Genomes Project phase 1 release (28). LD was computed by MACH using genotype information from 11 HapMap phases I + II + III subpopulations and four 1000 Genomes Project super populations (AFR, AMR, ASN and EUR) (29,30). GENCODE v27 annotation was used to map genetic variants to gene loci (31).
Tissue/cell-type-specific epigenome data
Consolidated epigenomes from 127 human tissue/cell lines were downloaded from the web portal of the NIH Roadmap Epigenomics project (32), which includes narrow peaks from eight histone modifications ChIP-seq (H3K27ac, H3K27me3, H3K36me3, H3K4me1, H3K4me2, H3K4me3, H3K79me2 and H3K9me3) and DNase-seq. For tissue/cell lines with missing epigenomes, imputed narrow peaks were used.
Tissue/cell-type-specific Hi-C data
Genome-wide in situ or dilution Hi-C raw reads for diverse human tissue/cell lines were collected and downloaded from ENCODE, 4DN, GEO and published literature. To ensure relatively adequate read coverage for chromatin interaction identification at 5 Kb resolution, we required that each biological sample library should contain at least 150M sequenced reads. For the same tissue/cell line with multiple Hi-C libraries from different biological replicates or labs, we only kept the largest sequencing library. Using these criteria, GWAS4D now incorporates 60 Hi-C libraries from 14 human primary tissues and 46 human cell lines (see Supplementary Table S1 for detailed information of the used Hi-C libraries). We uniformly processed these Hi-C data according to the HiC-Pro (33) standard analysis pipeline and considered significant chromatin interaction at 5 Kb resolution using HOMER (34) (see Supplementary Table S1 for the number of significant interactions of each Hi-C library).
Transcriptional regulator motif data
Position frequency matrices of known and inferred transcription regulator motifs were curated and merged from 13 datasets (Supplementary Table S2). Since a regulator may contain redundant or highly similar motifs, it is necessary to reduce the motif volume and select the representative ones. For each transcription regulator, we first used MACRO-APE (35) to calculate the pairwise similarity among collected motifs. We then clustered these motifs using the Calinski-Harabasz index (36) in the R fpc-package by dynamically setting the optimal number of clusters (no more than three clusters). We picked up the one representative motif with the largest information content in each cluster. Finally, GWAS4D compiles 3105 motifs for 1480 transcription regulators, providing an integrated compendium for evaluating variant effects on DNA–protein interactions.
Regulatory variant annotation data
Basic genomic features of genetic variants were retrieved through CADD base-wise annotations (37) and SNVrap (38). Variant allele frequency information was downloaded from gnomAD (39). GWAS4D also integrates multiple functional prediction scores of non-coding variants for all the possible single nucleotide variants from 10 algorithms (Supplementary Table S2). Nonsynonymous mutation deleterious/pathogenic scores were received from the dbNSFP (40). Base-wise evolutionary scores were integrated from GADD and SiPhy (41). In addition, GWAS4D annotates variants using tissue/cell-type-specific regulatory signals, including open chromatin and histone modification data from the Roadmap Epigenomics project, transcription factor binding data from the CistromeDB (42), and CAGE associated transcript signals from the FANTOM CAT (43). Finally, trait/disease-association data were downloaded from GWASdb (44), GWAS Catalog (45), ClinVar (46), COSMIC (47) and GTEx (48) (see Supplementary Table S2 for detailed information of the annotations used).
GWAS statistical fine-mapping data
To validate the usability of the GWAS4D web server, we used a fine-mapped credible set of 161 coronary artery disease (CAD) risk loci from a recent GWAS meta-analysis (49). SNPs that overlapped with protein coding regions and splicing sites were filtered out according to VEP annotation (50). Using the remaining non-coding fine-mapped SNPs, we first evaluated whether the GWAS4D predictions differed when selecting CAD matched and unmatched cell types. We then divided the SNPs into four subsets using PAINTOR posterior probabilities in the whole credible set (confidence >95%, confidence between 50% and 95%, confidence between 10% and 50% and confidence <10%) (51). We investigated whether the GWAS4D prediction scores in the high confidence subset are larger than those in the low confidence subset. Finally, we inspected the ability of GWAS4D to disentangle true disease-causal regulatory variants from a risk locus containing SNPs with similar posterior probabilities of causality.
Context-specific prioritization of regulatory variants from GWAS signals
GWAS4D analyzes and prioritizes human regulatory variants after GWAS statistical mapping. GWAS4D utilizes multiple tissue/cell-type-specific functional evidence sources to fine-map potentially disease-causal regulatory variants. The overall workflow of GWAS4D is shown in Figure 1.
Figure 1.
The workflow of GWAS4D (see the description of prioritization pipeline for details).
SNP filtering and LD expansion
GWAS4D accepts multiple variant description formats including VCF-like, dbSNP ID and coordinate-only. Variants not using the VCF-like format will be automatically lifted to dbSNP150 and assigned respective reference/1st alternative alleles using SNPTracker (52). The web server will discard the variants not mapped onto dbSNP150 or 1000 Genomes Project unless the VCF-like format is used. The GWAS P-value is optimal but could be used to filter out less significant signals. For each leading SNP, GWAS4D can retrieve all linked SNPs in the corresponding LD proxy by a user-defined population and r-squared (r2) cutoff. However, this LD expansion can be disabled when input variants are a statistically fine-mapped GWAS credible set.
Prediction of tissue/cell-type-specific regulatory probability
To predict the regulatory probability of a genetic variant in a particular tissue/cell type, we used our recently developed context-dependent epigenomic weighting method, cepip (26). Given a defined tissue/cell type, GWAS4D utilizes cepip to score each SNP after filtering and LD expansion steps. In general, GWAS4D will report three probabilities: (i) ‘composite P’ shows the likelihood of the variant to be functional in gene regulation by our context-free ensemble method (53); (ii) ‘cell P’ represents the condition-dependent regulatory potential in the current tissue/cell type; (iii) ‘combined P’ is the final regulatory probability that jointly considers both context-free and context-dependent models. For a GWAS trait with unknown causal tissue/cell type, GWAS4D estimates the most relevant tissue/cell type by comparing the normalized mean ‘cell P’ of input GWAS signals across 127 reference tissue/cell types (26).
Prediction of the regulatory effect on TF binding
GWAS4D substantially extends the search pool of transcription regulator motifs and is able to scan as many as 3105 motifs for 1480 regulators. Using the same motif scanning and scoring strategy as in the GWAS3D, GWAS4D evaluates the difference of the transcription regulator binding affinity caused by different alleles from the investigated variant. The statistical significance of the variant effect was measured by a permutated null distribution of binding affinity differences.
Prioritization and annotation of scored SNPs
Given the number (N) of independent regulatory signals in each r2-defined LD proxy of GWAS leading variant, GWAS4D prioritizes all the scored variants and recommends the top N variants according to the ‘combined P’ in the corresponding LD proxy. GWAS4D also investigates whether the ‘cell P’ of the regulatory variant is top ranked across all 127 epigenome datasets to define the likely tissue/cell-type specificity. In addition, GWAS4D reports the top possible transcription regulators altered by the variant effect as well as their relevant motifs. To comprehensively exploit the variant regulatory effect, GWAS4D provides a batch of annotations and functional evidence, including variant genomic features, variant allele frequency, non-coding variant functional prediction scores, conservation scores, chromatin states, transcription factor binding events, expression quantitative trait loci (eQTLs) as well as many trait/disease associations. These annotations were indexed and randomly accessed by Tabix (54).
Linking a regulatory variant to its target loci
After the SNP prioritization step, GWAS4D maps the variant locus to genes using GENCODE annotations and records the chromosome ideogram if the variant locates in the intergenic region. GWAS4D also utilizes significant 5 kb Hi-C interactions of selected tissue/cell types to link the regulatory variant to its target loci. Considering SNP-located 5 kb bin as a virtual 4C viewpoint, the top 20 significant chromatin interactions in the same chromosome were plotted using CHiCP (55). Additionally, GWAS will display the peak signals of active or repressive chromatin marks within the viewpoint and target bins using Roadmap Epigenomics annotation tracks of the selected tissue/cell type.
WEB SERVER DESCRIPTION
Usage and interface
The GWAS4D web server accepts any of the four following GWAS variants input formats: VCF-like, coordinate-only, dbSNP ID and PLINK-like. Since the last three formats do not contain allele information, GWAS4D will supply respective alleles using reference/first alternative alleles in dbSNP 150 and 1000 Genomes Project and will discard variants that cannot map to these two datasets. Therefore, the VCF-like format is the suggested input format. Both plain text and uploaded file of GWAS variants are well supported, and the P-value is an optional input field and is used to filter less significant variants. To search correlated SNPs in the LD proxy of a leading variant, users can define a reference LD dataset and corresponding r2 cutoff on either the 1000 Genomes or HapMap populations. In the case where input variants are statistically fine-mapped GWAS signals, users could skip the LD expansion function by checking ‘No LD Expansion’. By default, GWAS4D assumes that there is only one independent regulatory SNP in each LD proxy of leading variant and outputs the top scored SNP according to its ‘combined P’ value, but users can adjust the number of top prioritized variants in each LD proxy. To perform context-specific prioritization, users can select the epigenomes of a tissue/cell type from 127 reference tissue/cell types or upload narrow peaks of chromatin marks for a particular tissue/cell type. GWAS4D will automatically match an empirically relevant Hi-C dataset upon the selection of the reference tissue/cell type, but users are suggested to manually confirm or upload matched tissue/cell type-specific Hi-C interactions for better interpretation of target regions of regulatory SNP. In addition, GWAS4D allows one to estimate the likely relevant tissue/cell type if the user cannot assign a GWAS trait-associated tissue/cell type. Finally, users are able to customize the list of transcription regulators for motif scanning and to define the significance cutoff of the variant effect on the transcription regulator binding. GWAS4D allows three types of job retrieving methods, including encrypted link, job menu and email notification.
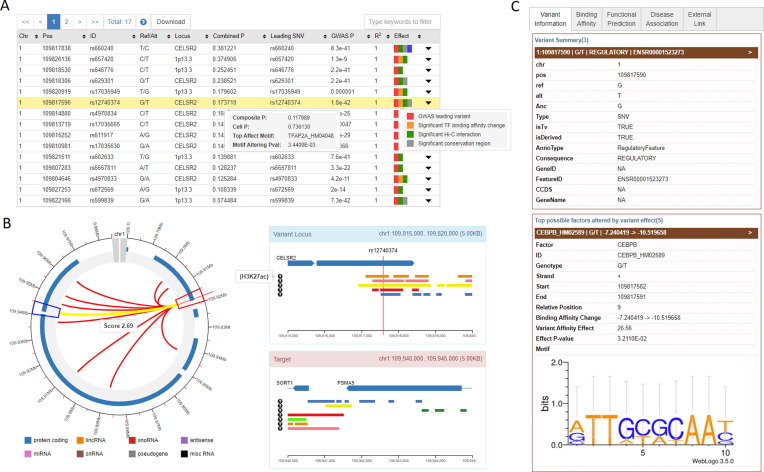
The GWAS4D platform displays the results in a highly interactive and user-friendly interface. Once the submitted job is finished, the URL is redirected to the result page (Figure 2). The left panel of the result page shows a table with the final results of prioritization, in which the regulatory SNPs are ordered according to the ‘combined P’ scores (Figure 2A). This searchable table also summarizes the genomics position of the variant, allele information and the relationship with the leading GWAS SNP. Importantly, to highlight the regulatory effect of each prioritized SNP, color marks were used in the last column of the prioritization table. For example, the SNP with the most significant transcription regulatory binding affinity change will be marked by an orange stamp, and the SNP fetching tissue/cell-type-specific ‘cell P’ under the current condition will be marked by a blue stamp, and the SNP located in the significant chromatin interaction region will be marked by a green stamp. By hovering over each regulatory SNP, users can inspect more information about the predicted regulatory scores. In addition to the detailed description of the SNP regulatory effect, GWAS4D also introduces an interactive circular plot using CHiCP to display the top most significant 5 kb chromatin interactions by assigning virtual 4C viewpoint to the SNP-contained locus. Chromatin marks within the connected 5 kb bins can also be showed when users click on each interaction arc (Figure 2B). The whole prioritization table can be downloaded with a tabular file. The right panel of the results page provides functional evidence and annotations of the regulatory variant in several categories (Figure 2C). The ‘Variant Information’ tab reports the variant's genomic attributes and allele frequency in the world-wide human population; the ‘Functional Evidence’ tab shows information of the SNP-altered transcription regulator binding affinity change, as well as several regulatory signals in the variant locus, such as transcriptional factor ChIP-seq peaks from CistromeDB and CAGE cluster signal from FANTOM CAT; the ‘Functional Prediction’ tab lists base-wise non-coding/coding functional prediction scores and conservation scores from multiple algorithms; the ‘Trait/disease Association’ tab presents trait/disease-associated records from GTEx, GWASdb, ClinVar and COSMIC; the ‘External Link’ tab links the SNP to several commonly used non-coding variant annotation resources including HaploReg (19), RegulomeDB (20), rSNPBase (21) and 3DSNP (23). Users can download all of the functional predictions and annotation information for each prioritized variant by simply clicking the download icon in the variant prioritization table.
Figure 2.
The result pages of the GWAS4D web server. (A) final prioritization table of GWAS4D; (B) virtual 4C circular plot for the top most significant Hi-C interactions between the variant locus and the target regions; (C) functional annotation tabs for prioritized SNPs.
Web server design
The GWAS4D web server was built on a Perl-based web framework, ‘Catalyst’. The annotation information was indexed and retrieved by MySQL and Tabix. The Oracle Grid Engine was used as the job management system for task submission, and JQuery and related JavaScript UI components were used to construct the interactive web pages. GWAS4D now supports no >10 000 significant input variants under the ‘without LD extension’ mode and 2500 significant input variants under the ‘LD extension’ mode. Usually, GWAS4D can finish conventional GWAS jobs in approximately 10 minutes (Supplementary Table S3).
Evaluation
The overall performance of the applied method for regulatory variant prioritization has been benchmarked in our previous publications (26,53). Here, we used a GWAS fine-mapped credible set at 161 CAD risk loci as an example to demonstrate one of the applications and the effectiveness of our GWAS4D web server. We first executed the GWAS4D with the ‘no LD expansion’ mode on 1699 non-coding CAD-associated SNPs using HUVEC, HepG2 and GM12878 epigenomes. We observed that the HUVEC-based ‘combined P’ scores were significantly higher than HepG2, GM12878-based ‘combined P’ scores for these credible SNPs (P-value = 5.2E–5 and 1.2E–10, respectively, Mann–Whitney U test), indicating that selecting relevant tissue/cell types that match GWAS traits/diseases may improve the detection of causal regulatory variants (Supplementary Figure S1A). By partitioning the fine-mapped SNPs into four separate subsets, we also showed that SNPs in the >95% confidence subset obtained larger ‘combined P’ scores than those in the low confidence subsets (P-value = 0.039 by comparing >95% confidence subset with <10% confidence subset, Mann–Whitney U test) (Supplementary Figure S1B). This result suggests that GWAS4D could accurately prioritize disease-causal regulatory variants. In addition, we further exploited whether GWAS4D can distinguish true causal SNPs from a difficult credible set in which highly linked SNPs achieve a similar posterior probability of causality. We selected a credible set containing five SNPs with a posterior probability approximately 0.2 at the SMAD3 locus on chromosome 15. GWAS4D evaluated these five SNPs using the HUVEC cell type and prioritized them according to their ‘combined P’ scores (Supplementary Table S4). The top ranked SNP, rs17293632, did not obtain the highest posterior probability in the previous fine-mapping, but it overlaps with many active regulatory signals (DNA hypersensitive site, H3K27ac, H3K4me1, H3K4me2 and H3K4me3), eQTL and conservative regions. The HUVEC Hi-C data shows that the SNP locus interacts with the SMAD3 alternative promoter region at 5 kb resolution (Supplementary Figure S2A). GWAS4D also identifies that this variant could possibly disrupt the binding of AP-1 transcription factor JunB (Supplementary Figure S2B). Taken together, these results are largely consistent with two recent studies regarding functional validations of rs17293632 at the SMAD3 locus that confers CAD risk (56,57).
Few web servers that can incorporate tissue/cell-type-specific genomic/epigenomic information toward prioritizing and annotating disease-causal regulatory variants. Compared with the available features among several new or commonly used online resources, including HaploReg (19), RegulomeDB (20), rSNPBase (21), 3DSNP (23) and PINES (https://doi.org/10.1101/083642) and FUMA (25), we found that our GWAS4D provides the most comprehensive information and functions in the identification of tissue/cell type-specific regulatory variants (Supplementary Table S5).
CONCLUSION
Recent large-scale functional genomic/epigenomic studies have significantly expanded DNA regulatory codes in diverse human tissue/cell types. Connecting such effects with trait/disease associations to fine-map causal regulatory variants and their target genes is important in the post-GWAS era. By equipping our latest regulatory variant prioritization algorithm, comprehensive and up-to-date functional genomics resources, as well as an interactive user interface, GWAS4D systematically investigates the regulatory effect of GWAS SNPs in a tissue/cell-type-specific manner. Using relevant genomic/epigenomic data that matches GWAS traits/diseases, GWAS4D is able to enumerate and prioritize the likely functional SNPs in the LD proxy of each GWAS signal or re-evaluate the causal probability of regulatory variants based only on statistically fine-mapped GWAS SNPs. Additionally, GWAS4D incorporates our uniformly processed Hi-C chromatin interaction data at 5 kb resolution for 60 human primary tissue/cell lines, which provides a unique compendium to connect the genetic loci of diseases with their regulated targets. We expect GWAS4D to greatly aid researchers in investigating the genetic mechanisms of disease and to create more significant impacts in the era of human non-coding genomics.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31701143]; Talent Excellence Program from Tianjin Medical University; Startup Funding from Tianjin Medical University and the Thousand Youth Talents Plan of Tianjin. Funding for open access charge: National Natural Science Foundation of China [31701143]; Discipline Development Fund from Tianjin Medical University.
Conflict of interest statement. None declared.
REFERENCES
- 1. Lee P.H., Lee C., Li X., Wee B., Dwivedi T., Daly M.. Principles and methods of in-silico prioritization of non-coding regulatory variants. Hum. Genet. 2018; 137:15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li M.J., Yan B., Sham P.C., Wang J.. Exploring the function of genetic variants in the non-coding genomic regions: approaches for identifying human regulatory variants affecting gene expression. Brief. Bioinformatics. 2015; 16:393–412. [DOI] [PubMed] [Google Scholar]
- 3. Edwards S.L., Beesley J., French J.D., Dunning A.M.. Beyond GWASs: illuminating the dark road from association to function. Am. J. Hum. Genet. 2013; 93:779–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boyle E.A., Li Y.I., Pritchard J.K.. An expanded view of complex traits: from polygenic to omnigenic. Cell. 2017; 169:1177–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Trynka G., Sandor C., Han B., Xu H., Stranger B.E., Liu X.S., Raychaudhuri S.. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat. Genet. 2013; 45:124–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Finucane H.K., Bulik-Sullivan B., Gusev A., Trynka G., Reshef Y., Loh P.R., Anttila V., Xu H., Zang C., Farh K. et al. . Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 2015; 47:1228–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Trynka G., Westra H.J., Slowikowski K., Hu X., Xu H., Stranger B.E., Klein R.J., Han B., Raychaudhuri S.. Disentangling the effects of colocalizing genomic annotations to functionally prioritize non-coding variants within complex-trait loci. Am. J. Hum. Genet. 2015; 97:139–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Iotchkova V., Huang J., Morris J.A., Jain D., Barbieri C., Walter K., Min J.L., Chen L., Astle W., Cocca M. et al. . Discovery and refinement of genetic loci associated with cardiometabolic risk using dense imputation maps. Nat. Genet. 2016; 48:1303–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schmidt E.M., Zhang J., Zhou W., Chen J., Mohlke K.L., Chen Y.E., Willer C.J.. GREGOR: evaluating global enrichment of trait-associated variants in epigenomic features using a systematic, data-driven approach. Bioinformatics. 2015; 31:2601–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang C., Zhang S.. Large-scale determination and characterization of cell type-specific regulatory elements in the human genome. J. Mol. Cell Biol. 2017; 9:463–476. [DOI] [PubMed] [Google Scholar]
- 11. Krijger P.H., de Laat W.. Regulation of disease-associated gene expression in the 3D genome. Nat. Rev. Mol. Cell Biol. 2016; 17:771–782. [DOI] [PubMed] [Google Scholar]
- 12. Davis C.A., Hitz B.C., Sloan C.A., Chan E.T., Davidson J.M., Gabdank I., Hilton J.A., Jain K., Baymuradov U.K., Narayanan A.K. et al. . The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 2018; 46:D794–D801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rao S.S., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S. et al. . A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014; 159:1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tang Z., Luo O.J., Li X., Zheng M., Zhu J.J., Szalaj P., Trzaskoma P., Magalska A., Wlodarczyk J., Ruszczycki B. et al. . CTCF-mediated human 3D genome architecture reveals chromatin topology for transcription. Cell. 2015; 163:1611–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dekker J., Belmont A.S., Guttman M., Leshyk V.O., Lis J.T., Lomvardas S., Mirny L.A., O'Shea C.C., Park P.J., Ren B. et al. . The 4D nucleome project. Nature. 2017; 549:219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li M.J., Wang L.Y., Xia Z., Sham P.C., Wang J.. GWAS3D: Detecting human regulatory variants by integrative analysis of genome-wide associations, chromosome interactions and histone modifications. Nucleic Acids Res. 2013; 41:W150–W158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sun C., Molineros J.E., Looger L.L., Zhou X.J., Kim K., Okada Y., Ma J., Qi Y.Y., Kim-Howard X., Motghare P. et al. . High-density genotyping of immune-related loci identifies new SLE risk variants in individuals with Asian ancestry. Nat. Genet. 2016; 48:323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jones G.T., Tromp G., Kuivaniemi H., Gretarsdottir S., Baas A.F., Giusti B., Strauss E., Van’t Hof F.N., Webb T.R., Erdman R. et al. . Meta-analysis of genome-wide association studies for abdominal aortic aneurysm identifies four new disease-specific risk loci. Circ. Res. 2017; 120:341–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ward L.D., Kellis M.. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016; 44:D877–D881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Boyle A.P., Hong E.L., Hariharan M., Cheng Y., Schaub M.A., Kasowski M., Karczewski K.J., Park J., Hitz B.C., Weng S. et al. . Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012; 22:1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guo L., Du Y., Chang S., Zhang K., Wang J.. rSNPBase: a database for curated regulatory SNPs. Nucleic Acids Res. 2014; 42:D1033–D1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guo Y., Conti D.V., Wang K.. Enlight: web-based integration of GWAS results with biological annotations. Bioinformatics. 2015; 31:275–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lu Y., Quan C., Chen H., Bo X., Zhang C.. 3DSNP: a database for linking human noncoding SNPs to their three-dimensional interacting genes. Nucleic Acids Res. 2017; 45:D643–D649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Martin J.S., Xu Z., Reiner A.P., Mohlke K.L., Sullivan P., Ren B., Hu M., Li Y.. HUGIn: Hi-C unifying genomic interrogator. Bioinformatics. 2017; 33:3793–3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Watanabe K., Taskesen E., van Bochoven A., Posthuma D.. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017; 8:1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li M.J., Li M., Liu Z., Yan B., Pan Z., Huang D., Liang Q., Ying D., Xu F., Yao H. et al. . cepip: context-dependent epigenomic weighting for prioritization of regulatory variants and disease-associated genes. Genome Biol. 2017; 18:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K.. dbSNP: the NCBI database of genetic variation. Nucleic acids research. 2001; 29:308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Genomes Project, C. Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A.. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012; 491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li Y., Willer C.J., Ding J., Scheet P., Abecasis G.R.. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genetic Epidemiol. 2010; 34:816–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. International HapMap, C A haplotype map of the human genome. Nature. 2005; 437:1299–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S. et al. . GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012; 22:1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Roadmap Epigenomics, C. Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J. et al. . Integrative analysis of 111 reference human epigenomes. Nature. 2015; 518:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Servant N., Varoquaux N., Lajoie B.R., Viara E., Chen C.J., Vert J.P., Heard E., Dekker J., Barillot E.. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015; 16:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vorontsov I.E., Kulakovskiy I.V., Makeev V.J.. Jaccard index based similarity measure to compare transcription factor binding site models. Algorithms Mol. Biol.: AMB. 2013; 8:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kozak M. “A Dendrite Method for Cluster Analysis” by Calinski and Harabasz: a classical work that is far too often incorrectly cited. Commun. Stat.-Theor. M. 2012; 41:2279–2280. [Google Scholar]
- 37. Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J.. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014; 46:310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li M.J., Wang J.. Current trend of annotating single nucleotide variation in humans–A case study on SNVrap. Methods. 2015; 79–80:32–40. [DOI] [PubMed] [Google Scholar]
- 39. Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B. et al. . Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016; 536:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu X., Wu C., Li C., Boerwinkle E.. dbNSFP v3.0: a one-stop database of functional predictions and annotations for human nonsynonymous and splice-site SNVs. Hum. Mutat. 2016; 37:235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Garber M., Guttman M., Clamp M., Zody M.C., Friedman N., Xie X.. Identifying novel constrained elements by exploiting biased substitution patterns. Bioinformatics. 2009; 25:i54–i62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mei S., Qin Q., Wu Q., Sun H., Zheng R., Zang C., Zhu M., Wu J., Shi X., Taing L. et al. . Cistrome Data Browser: a data portal for ChIP-Seq and chromatin accessibility data in human and mouse. Nucleic Acids Res. 2017; 45:D658–D662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hon C.C., Ramilowski J.A., Harshbarger J., Bertin N., Rackham O.J., Gough J., Denisenko E., Schmeier S., Poulsen T.M., Severin J. et al. . An atlas of human long non-coding RNAs with accurate 5′ ends. Nature. 2017; 543:199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li M.J., Liu Z., Wang P., Wong M.P., Nelson M.R., Kocher J.P., Yeager M., Sham P.C., Chanock S.J., Xia Z. et al. . GWASdb v2: an update database for human genetic variants identified by genome-wide association studies. Nucleic Acids Res. 2016; 44:D869–D876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., Hastings E., Junkins H., McMahon A., Milano A., Morales J. et al. . The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 2017; 45:D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Jang W. et al. . ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018; 46:D1062–D1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Forbes S.A., Beare D., Boutselakis H., Bamford S., Bindal N., Tate J., Cole C.G., Ward S., Dawson E., Ponting L. et al. . COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017; 45:D777–D783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Consortium, G.T., Laboratory, D.A., Coordinating Center -Analysis Working, G., Statistical Methods groups-Analysis Working, G., Enhancing, G.g., Fund, N.I.H.C., Nih/Nci, Nih/Nhgri, Nih/Nimh, Nih/Nida Genetic effects on gene expression across human tissues. Nature. 2017; 550:204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. van der Harst P, Verweij N.. Identification of 64 novel genetic loci provides an expanded view on the genetic architecture of coronary artery disease. Circ. Res. 2018; 122:433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R., Thormann A., Flicek P., Cunningham F.. The ensembl variant effect predictor. Genome Biol. 2016; 17:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kichaev G., Roytman M., Johnson R., Eskin E., Lindstrom S., Kraft P., Pasaniuc B.. Improved methods for multi-trait fine mapping of pleiotropic risk loci. Bioinformatics. 2017; 33:248–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Deng J.E., Sham P.C., Li M.X.. SNPTracker: a swift tool for comprehensive tracking and unifying dbSNP rs IDs and genomic coordinates of massive sequence variants. G3. 2015; 6:205–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li M.J., Pan Z., Liu Z., Wu J., Wang P., Zhu Y., Xu F., Xia Z., Sham P.C., Kocher J.P. et al. . Predicting regulatory variants with composite statistic. Bioinformatics. 2016; 32:2729–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li H. Tabix: fast retrieval of sequence features from generic TAB-delimited files. Bioinformatics. 2011; 27:718–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schofield E.C., Carver T., Achuthan P., Freire-Pritchett P., Spivakov M., Todd J.A., Burren O.S.. CHiCP: a web-based tool for the integrative and interactive visualization of promoter capture Hi-C datasets. Bioinformatics. 2016; 32:2511–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Turner A.W., Martinuk A., Silva A., Lau P., Nikpay M., Eriksson P., Folkersen L., Perisic L., Hedin U., Soubeyrand S. et al. . Functional Analysis of a Novel Genome-Wide association study signal in SMAD3 that confers protection from coronary artery disease. Arterioscle. Thromb. Vasc. Biol. 2016; 36:972–983. [DOI] [PubMed] [Google Scholar]
- 57. Miller C.L., Pjanic M., Wang T., Nguyen T., Cohain A., Lee J.D., Perisic L., Hedin U., Kundu R.K., Majmudar D. et al. . Integrative functional genomics identifies regulatory mechanisms at coronary artery disease loci. Nat. Commun. 2016; 7:12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.