Abstract
Summary
Mosdepth is a new command-line tool for rapidly calculating genome-wide sequencing coverage. It measures depth from BAM or CRAM files at either each nucleotide position in a genome or for sets of genomic regions. Genomic regions may be specified as either a BED file to evaluate coverage across capture regions, or as a fixed-size window as required for copy-number calling. Mosdepth uses a simple algorithm that is computationally efficient and enables it to quickly produce coverage summaries. We demonstrate that mosdepth is faster than existing tools and provides flexibility in the types of coverage profiles produced.
Availability and implementation
mosdepth is available from https://github.com/brentp/mosdepth under the MIT license.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Measuring the depth of sequencing coverage is critical for genomic analyses such as calling copy-number variation (CNV), e.g. by cn.mops (Klambauer et al., 2012), quality control (Pedersen et al., 2017), and determining which genomic regions have too low, or too high (Li, 2014) coverage for reliable variant calling. Given the scope of applications for coverage profiles, there are several existing tools that calculate genome-wide coverage. Samtools depth (Li et al., 2009) outputs per-base coverage; BEDTools genomecov (Quinlan and Hall, 2010; Quinlan, 2014) can output per-region or per-base coverage; Sambamba (Tarasov et al., 2015) also provides per-base and per-window depth calculations. The need for efficient coverage calculation increases with the number and depth of whole genome sequences, and existing methods require roughly an hour or more of computation for a typical human genome with 30× coverage. Here, we introduce mosdepth and show that it is faster than existing methods and has additional utility.
2 Materials and methods
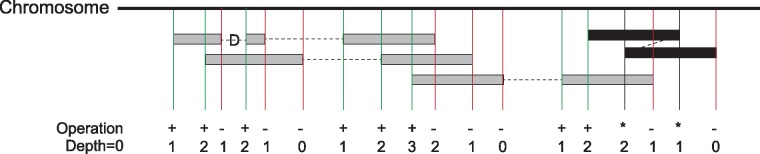
Mosdepth uses HTSLib (http://www.htslib.org/) via the nim programming language (https://nim-lang.org); it expects the input BAM or CRAM file to be sorted by position. In contrast to samtools, which uses a ‘pileup’ engine that tracks each nucleotide in every read, mosdepth only tracks chunks of read alignments. Only the start and end position of each chunk of an alignment (each alignment may have multiple chunks if it is split by a deletion or other event) are tracked in an array (of 32 bit integers) whose size is the length of the chromosome. For each chunk of an alignment to the reference genome, mosdepth increments the start and decrements the end for the the value at the index in the array corresponding to that chromosomal position (Fig. 1). It avoids double-counting coverage when the ends of a paired-end sequencing fragment have overlapping alignments (Fig. 1, black alignment). Once the coverage array has tracked all alignment starts and ends in a BAM or CRAM file, the depth at a particular position is calculated as the cumulative sum of all array positions preceding it (a similar algorithm is used in BEDTools which track starts and ends separately).
Fig. 1.
Mosdepth coverage calculation algorithm. An array the size of the current chromosome is allocated. As each alignment is read from a position-sorted BAM or CRAM file, the value at each start is incremented and the value at each stop is decremented. As illustrated by the alignment with a deletion (D) CIGAR operation, each alignment may have multiple starts and ends. If the leftmost read (the one seen first) of a paired-end alignment has an end that overlaps the position of its mate (which is given as a field in the BAM record) then it is stored in a hash-table until its mate is seen. At that time, the overlap between the mates is calculated, the regions of overlap are decremented and the item is removed from the hash. This prevents double counting coverage from two ends of the same paired-end DNA fragment (black alignment, ‘*’ operation means no coverage increment or decrement is made). Once all reads for a chromosome are consumed, the per-base coverage is simply the cumulative sum of the preceding positions
The coverage along a chromosome is calculated in place by replacing the composite start and end counts with the cumulative sum up to each element in the array. Once complete, the coverage of a region is simply the mean of the elements in the array spanning from start to end. This makes it possible to calculate coverage extremely quickly, even for millions of small regions. This setup is also amenable to rapid calculation of a genome’s coverage distribution: that is, the number of bases covered by a given number of reads across the genome or in the given regions. The distribution calculation requires an extra iteration through the array that counts the occurrence of each coverage value. The mosdepth method does require more memory–for the 249 megabase chromosome 1 in the human genome, it will require about 1GB of memory, however, that number is not dependent on the depth of coverage or number of alignments. Despite its flexibility, mosdepth is easy to use and understand (see Supplementary Material for example uses).
3 Results
We compared the time and memory requirements of mosdepth (v0.1.6) to samtools (v1.5), BEDTools (v2.26.0) and sambamba (v0.6.6) on a BAM with about 30× coverage from the Simons Genome Diversity Panel (Mallick et al., 2016) (Supplementary Material). With a single CPU, mosdepth is faster than existing tools, and can be even faster with multiple decompression threads (Table 1). Results for CRAM and for other options such as window-based depth calculations are shown in Supplementary Table S1. At four threads, there is no additional benefit to adding more decompression threads as shown in Supplementary Figure S1.
Table 1.
Comparison of depth tools for time and memory use on a 30× BAM
| Tool | Threads | Relative time | Time (hh:mm:ss) | Memory (MiB) |
|---|---|---|---|---|
| Mosdepth | 1 | 1 | 25:23 | 1196 |
| Mosdepth | 3 | 0.57 | 14:27 | 1196 |
| Samtools | 1 | 1.98 | 50:12 | 27 |
| Sambamba | 1 | 5.71 | 2:24:53 | 166 |
| BEDtools | 1 | 5.31 | 2:14:44 | 1908 |
Note: Mosdepth and BEDTools use much more memory, but mosdepth is nearly twice as fast as the next fastest tool, samtools. The threads column reflects the number of threads for BAM/CRAM decompression.
To evaluate consistency between the tools, we compared the output to samtools depth. Mosdepth cannot include or exclude individual bases because of low base-quality (BQ) as can samtools depth. In contrast, samtools depth cannot avoid double-counting overlapping regions unless the BQ cutoff is set to a value > 0. Therefore, we compared mosdepth without mate overlap correction to samtools depth with a BQ cutoff of 0 for chromosome 22 of the dataset used for Table 1. With this comparison set up to evaluate differences, we found no discrepancies in reported depth among the tools for the entire chromosome.
4 Discussion
Mosdepth is a quick, convenient tool for genome-wide depth calculation. The optional coverage distribution is useful for quality control and the depth output is applicable without further processing as input to many CNV detection tools. While the method it employs requires greater memory use, it makes the implementation simple and fast, enables a straightforward coverage distribution calculation, and expedites the depth calculations for even millions of regions. Mosdepth is useful for exome, whole-genome, and targeted sequencing projects. It is available from source-code, as a binary, and from bioconda (https://bioconda.github.io/).
Funding
This research was supported by awards to A.R.Q. from the US National Human Genome Research Institute (NIH R01HG006693 and NIH R01HG009141), the US National Institute of General Medical Sciences (NIH R01GM124355) and the US National Cancer Institute (NIH U24CA209999).
Conflict of Interest: none declared.
Supplementary Material
References
- Klambauer G. et al. (2012) cn.MOPS: mixture of poissons for discovering copy number variations in next-generation sequencing data with a low false discovery rate. Nucleic Acids Res., 40, e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. (2014) Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics, 30, 2843–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. (2009) The sequence alignment/map format and samtools. Bioinformatics, 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick S. et al. (2016) The simons genome diversity project: 300 genomes from 142 diverse populations. Nature, 538, 201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen B.S. et al. (2017) Indexcov: fast coverage quality control for whole-genome sequencing. Gigascience, 11, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A.R. (2014) Bedtools: the swiss-army tool for genome feature analysis. Curr. Protoc. Bioinformatics, 47. doi:10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. (2010) Bedtools: a flexible suite of utilities for comparing genomic features. Bioinformatics, 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasov A. et al. (2015) Sambamba: fast processing of ngs alignment formats. Bioinformatics, 31, 2032–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.