Abstract
We present a new update to MetaboAnalyst (version 4.0) for comprehensive metabolomic data analysis, interpretation, and integration with other omics data. Since the last major update in 2015, MetaboAnalyst has continued to evolve based on user feedback and technological advancements in the field. For this year's update, four new key features have been added to MetaboAnalyst 4.0, including: (1) real-time R command tracking and display coupled with the release of a companion MetaboAnalystR package; (2) a MS Peaks to Pathways module for prediction of pathway activity from untargeted mass spectral data using the mummichog algorithm; (3) a Biomarker Meta-analysis module for robust biomarker identification through the combination of multiple metabolomic datasets and (4) a Network Explorer module for integrative analysis of metabolomics, metagenomics, and/or transcriptomics data. The user interface of MetaboAnalyst 4.0 has been reengineered to provide a more modern look and feel, as well as to give more space and flexibility to introduce new functions. The underlying knowledgebases (compound libraries, metabolite sets, and metabolic pathways) have also been updated based on the latest data from the Human Metabolome Database (HMDB). A Docker image of MetaboAnalyst is also available to facilitate download and local installation of MetaboAnalyst. MetaboAnalyst 4.0 is freely available at http://metaboanalyst.ca.
INTRODUCTION
MetaboAnalyst is a comprehensive web-based tool suite designed to help users easily perform metabolomic data analysis, visualization, and functional interpretation. It was first introduced in 2009 with a single module for metabolomic data processing and statistical analysis (1). Since then, it has been continuously updated to meet the evolving needs of the metabolomics research community. Version 2.0, which was released in 2012 (2), incorporated three new modules for metabolite set enrichment analysis (MSEA) (3), metabolic pathway analysis (MetPA) (4), as well as advanced two-factor and time-series analyses (MetATT) (5). Version 3.0, which was released in 2015 (6), added the support for biomarker analysis (7), power analysis, and joint pathway analysis supporting both metabolites and genes, coupled with a major upgrade of the underlying web framework.
With each iteration, MetaboAnalyst has grown more popular. To better handle the growing user traffic, MetaboAnalyst has been migrated to a Google cloud server for improved performance and accessibility. According to Google Analytics, over the past 12 months, MetaboAnalyst has processed >1.8 million jobs submitted from ∼60 000 users. Based on our citation analysis for 2017, MetaboAnalyst has been used in at least one-fourth of all metabolomics publications for that year, attesting to its status as one of the preferred tools for metabolomic data analysis. MetaboAnalyst has been used to elucidate key metabolic differences in breast cancer of African-American and Caucasian women (8), to identify highly predictive biomarkers of ketosis in dairy cows (9), to understand alterations in the intestinal metabolome during enteric infections (10), as well as to study many other biological processes and complex diseases (11–14).
However, the field of metabolomics continues to evolve and it is important that MetaboAnalyst also evolves to keep current with the field and its growing user base. For this year's update, MetaboAnalyst has been substantially upgraded to enhance its user interface, to improve reproducibility/transparency, to support batch processing, to provide improved pathway interpretation from untargeted mass spectrometry (MS) data, to support meta-analysis and multi-omics analysis, to expand its underlying knowledgebase, and to support more facile local installations. In particular, the key features of this year's update include:
A companion R package (MetaboAnalystR) and an accompanying R-command history panel to permit more transparent and reproducible analysis;
Significantly expanded libraries of metabolite sets and metabolic pathways to support comprehensive metabolomics data interpretation through functional enrichment analysis;
A new module based on the mummichog (15) algorithm for pathway activity prediction from untargeted metabolomics data;
A new module to support metabolomic biomarker meta-analysis;
A new module to support multi-omics data integration through knowledge-based network analysis and visualization;
Other important updates including a Docker image for facile download and local installation of MetaboAnalyst; as well as providing direct links to several online tools for nuclear magnetic resonance (NMR), gas chromatography–mass spectrometry (GC–MS) and liquid chromatography–mass spectrometry (LC–MS) spectral analysis.
These changes and updates are all contained in MetaboAnalyst 4.0, which is freely available at http://metaboanalyst.ca. For each new module, we have added frequently asked questions (FAQs) and implemented functions for comprehensive analysis report generation. A more detailed description of each of these updates and changes in MetaboAnalyst 4.0 is given below.
Overview of the MetaboAnalyst 4.0 framework
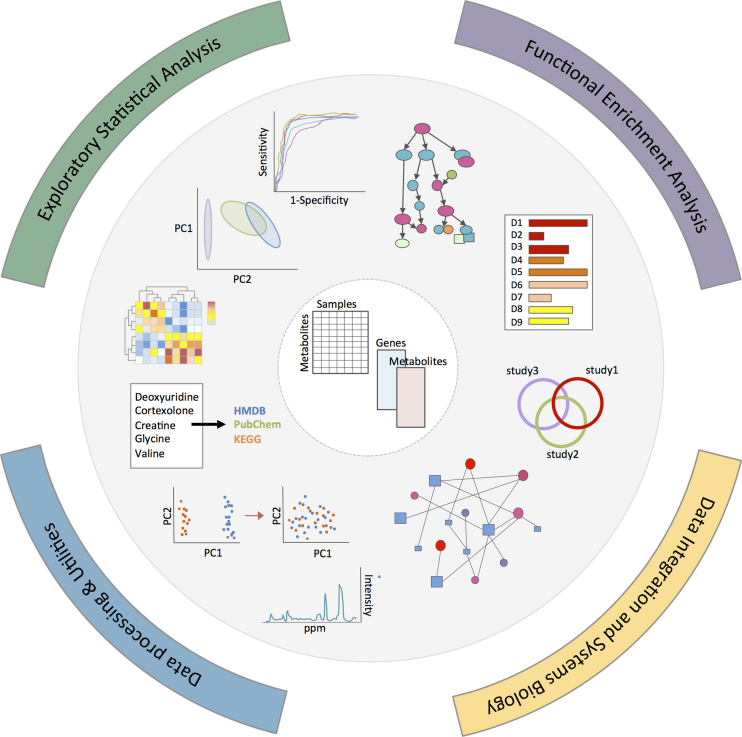
MetaboAnalyst's user interface has been upgraded to provide a more modern ‘look and feel’ while maintains the same easy-to-use modular analytical pipeline. To facilitate navigation, all functions are now organized into 12 modules, which can be arranged into four general categories: (i) exploratory statistical analysis, (ii) functional analysis, (iii) data integration and systems biology and (iv) data processing & utility functions (Figure 1). The exploratory statistical analysis category (general statistics, biomarker analysis, two-factor/time-series analysis and power analysis) can accept data from either targeted or untargeted metabolomics data sets. The functional analysis category has been expanded to include a new module on pathway activity prediction from MS data, in addition to the two existing modules for metabolite set enrichment analysis and pathway analysis of targeted metabolomics data. The data integration and systems biology category now includes three modules (biomarker meta-analysis, joint pathway analysis, and network explorer). Finally, the data processing and other utilities category contains common data processing tools such as compound ID conversion, batch effect correction, as well as links to three easy-to-use, web-based tools including Bayesil (16), GC-AutoFit and XCMS Online (17) for processing and annotation of NMR, GC–MS and LC–MS spectra, respectively.
Figure 1.
Overview of MetaboAnalyst 4.0 framework. The current modules can be organized into four general categories: (i) exploratory statistical analysis, (ii) functional analysis, (iii) data integration & systems biology and (iv) data processing & utility functions.
MetaboAnalystR and improved transparency/reproducibility
Thanks to continuing technological advancements in the field along with very helpful user feedback, many small updates and feature enhancements have taken place over the years to make MetaboAnalyst faster, more intuitive, and more robust. A potential downside associated with this continuous evolution is that it could lead to long-term reproducibility issues due to small changes in the interface or default parameter settings. While this flexibility is one feature that has made MetaboAnalyst so appealing, it has also made it inherently challenging to fully capture all steps required for reproducible analysis in the future. One possible way to alleviate this issue is to host multiple snapshots of the tool created at different time points. However, the maintenance costs associated with such an approach would be prohibitive. Another approach is to improve MetaboAnalyst's transparency throughout the analysis process. Because most of MetaboAnalyst's analytical tools are based on R functions, it would be much more efficient to capture the workflow using R commands embedded with the parameters selected by users. Furthermore, many advanced users of MetaboAnalyst have requested access to its underlying R functions in order to develop more customized data analysis or to perform extensive batch data processing.
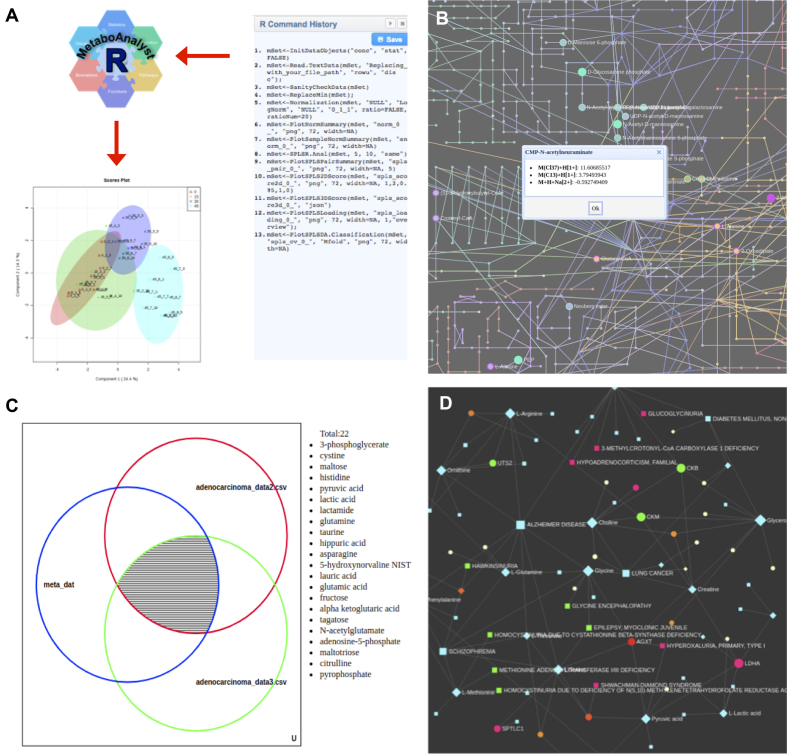
To accommodate these needs (i.e. better support for transparency, flexibility and batch analysis), we have developed the MetaboAnalystR package. This is a companion R-package that permits users to ‘see’ and save the R code that MetaboAnalyst is running in real-time, which can then be used locally to reproduce the analytical workflow. MetaboAnalystR is designed to support more transparent, reproducible yet flexible analysis of metabolomic data within MetaboAnalyst. The R code between MetaboAnalystR and the web server has been extensively modified to ensure that they are fully interchangeable and have identical functionalities across either platform. During each session of data analysis, these R commands are displayed on the right side of each page in the ‘R command history’ sidebar, and each command appears sequentially based on when the command was executed (Figure 2A). MetaboAnalyst also stores the entire R command history as an executable R script that can be downloaded following the completion of each module. This script contains all user-selected parameters and selected tests. We believe that revealing the R code behind MetaboAnalyst improves transparency and allows users to track each step of their analysis in a form (R script) that can be easily shared and reproduced either on the web or locally using the MetaboAnalystR package. Beginner R users will be able to quickly learn the basics of MetaboAnalystR by copying the commands generated via their web-based analysis directly into R and reproduce their analyses; while advanced R users will be able to incorporate MetaboAnalystR package into their analytical workflows or customize the code to suit their needs. We believe that the MetaboAnalystR feature not only captures the workflow for better reproducibility, but also offers greater flexibility for more refined analysis and batch processing.
Figure 2.
Summary of new features introduced in MetaboAnalyst 4.0. (A) An illustration showing the R command history panel and the companion MetaboAnalystR package will allow users to easily reproduce their analyses. In this case, the R command history captures all R commands leading to the generation the PLS-DA 2D score plot, which can then be reproduced in MetaboAnalystR using identical R commands (except the file path parameter for user input). (B) A zoomed-in view of the KEGG metabolic network showing the potential metabolite hits predicted from the mummichog algorithm. Clicking on a highlighted node will display all possible matched adduct forms of the corresponding compound. (C) An interactive Venn diagram showing the results from a biomarker meta-analysis. Clicking on an area will show the corresponding hits. (D) An example of the metabolite-gene-disease interaction network created based on user input.
Obviously, with any major update to a resource like MetaboAnalyst, there is also some concern about the reproducibility (or return-accessibility) of data analyses performed using earlier versions of the server. For instance, due to updates to the underlying metabolite set libraries, the ranks and P-values of the top hits would change for the same input data. To help alleviate this issue, the previous version of MetaboAnalyst (version 3.0) will still be maintained (http://old.metaboanalyst.ca), as long as there is sufficient interest and user traffic.
MetaboAnalyst's knowledgebase update
To address potential issues such as the decline in analysis quality due to a lack of updated annotations (18,19), we have put considerable effort into updating MetaboAnalyst's underlying knowledgebase. The most noteworthy updates are to the compound database used for metabolite name mapping, to the pathway libraries used for metabolic pathway analysis, and to the metabolite sets for functional enrichment analysis. The details of these updates are summarized below.
Compound database
MetaboAnalyst performs in-house mapping of common compound names to a wide variety of database identifiers including KEGG (20), HMDB (21), ChEBI (22), METLIN (23) and PubChem (24) prior to performing any functional analysis. This knowledgebase has been updated with HMDB Version 4.0, including updates of HMDB identifiers and links to other databases. As a result, MetaboAnalyst's compound database has been expanded to include ∼19 000 compounds. They represent the core subset of HMDB compounds (∼114 100) with more detailed annotations relevant for downstream functional analysis.
Metabolite sets and metabolic pathway libraries
MetaboAnalyst's metabolite sets are primarily used by its MSEA module. Many MetaboAnalyst users utilize this module to provide appropriate functional and biological context to their uploaded metabolomic data. Six existing metabolite set libraries, and one new metabolite set library, were updated/created based on HMDB version 4.0. The updated metabolite set libraries include disease sets in blood (344 diseases, increased from 330 diseases), cerebral spinal fluid (166 diseases, increased from 108 diseases), and urine (384 diseases, increased from 290 diseases), as well as location-based metabolite sets (73 organs, biofluids and tissues, increased from 57 organs, biofluids and tissues), pathway-based metabolite sets (147 metabolic pathways, increased from 80 metabolic pathways), single nucleotide polymorphisms (SNPs)-associated metabolite sets (4598 SNPs, increased from 4501 SNPs), and a new metabolite set library on drug-related pathways (461 pathways). It should be noted that these metabolite sets were derived primarily from human-only data. We are currently updating the Pathway Analysis module to support interactive visual analysis of the extensive list of pathways from SMPDB (25).
New module #1: MS peaks to pathways
High-throughput analysis and functional interpretation of untargeted or global MS-based metabolomics data continues to be a major bottleneck in current metabolomics research. Conventional MS-based procedures typically include peak identification, spectral deconvolution, and peak annotation. A number of excellent methods have been developed to deal with the first two tasks (26,27), which typically yield a list of ‘clean’ MS peaks. Peak annotations are then performed manually by searching through a variety of spectral or compound databases. This process can often generate a number of false positives, due to redundancies in masses or the lack of unique MS spectral signatures for many small compounds (28,29). High-resolution MS instruments are increasingly used to reduce these false hits analytically. Computationally, a promising approach is to shift the unit of analysis from individual compounds to individual pathways (or any groups of functionally related compounds which collectively produce more distinctive spectral footprints)—a concept similar to the widely used gene set enrichment analysis or GSEA (30). The mummichog algorithm was an elegant and efficient implementation of this concept that enables direct prediction of pathway activities from high-resolution MS peaks, without performing accurate peak annotation upfront (15). Currently, the algorithm lacks a graphic interface, thus limiting the access to many bench researchers. Due to its popularity and repeated user requests, we added a new module (called ‘MS Peaks to Pathways’) in MetaboAnalyst to support mummichog-based MS peak analysis through user-friendly interface. We re-implemented the mummichog (version 1.0.10) algorithm in R to be consistent with MetaboAnalyst workflow and the aforementioned strategy of reproducibility. The knowledgebase for this module consists of five genome-scale metabolic models obtained from the original Python implementation which have either been manually curated or downloaded from BioCyc, as well as an expanded library of 21 organisms derived from the KEGG metabolic pathways. The inclusion of SMPDB pathways for other model organisms will occur in the next few months (25). While compound identification is generally de-emphasized in mummichog, the post hoc analysis of the matched compounds is critical for downstream validation and interpretation. To address these needs, we implemented a KEGG style global metabolic network to allow users to visualize the overall peak matching patterns as well as to interactively zoom into a particular candidate compound to examine all of its matched isotopic or adduct forms.
To use this module, users must upload a table containing three columns—m/z features, P-values, and statistical scores (e.g. t-scores or fold-change values). If these values have not yet been calculated, users can upload their m/z peak list files or peak tables to MetaboAnalyst's Statistical Analysis module to perform their statistical analysis of choice, then upload these results into the ‘MS Peaks to Pathways’ module. Users need to specify the mass accuracy, the ion mode (positive or negative), and the P-value cutoff to delineate between significantly enriched m/z features and the background universe. Following data upload, users must select an organism (library) from which to perform the untargeted pathway analysis. More detailed explanation of the approach are available in MetaboAnalyst's FAQs, as well as the original publication by Li et al. (15).
The output of the ‘MS Peaks to Pathways’ module consists of a result table containing ranked pathways that are enriched in the user-uploaded data. The table includes the total number of hits, raw P-values (Fisher's exact or hypergeometric test), EASE scores, and P-values modeled on user data using a Gamma distribution. Users can click the ‘View’ link to view the detailed hits for each pathway. A comprehensive table containing the compound matching information for all user-uploaded m/z features is also available for download. Importantly, all of this information (pathways, compounds, and matched peaks) can be intuitively explored within the KEGG global metabolic network (Figure 2B). The page consists of three sections: (i) a top toolbar containing different menus to control various visualization features, (ii) a left-hand panel showing the pathway analysis results and (iii) a central view for interactive visual exploration of the metabolic network. Users can scroll their mouse to zoom in and out of the network view. Clicking on a pathway name on the left panel will highlight all of its compounds within the network. Double-clicking a metabolite node will show all the matching details for the corresponding compound as shown in the dialog (Figure 2B). The current view can be downloaded as either a portable network graphics (PNG) or scalable vector graphics (SVG) file.
New module #2: Biomarker meta-analysis
Biomarker identification continues to be an important area of research in metabolomics (31). However, a major challenge in many metabolomics-based biomarker discovery efforts is the validation of potential metabolic markers (32). Questions have been raised about biomarker consistency and robustness across different metabolomics studies conducted on the same disease. As a result, the importance of external validation to improve statistical power for biomarker validation has been increasingly emphasized (33,34). To address this issue of biomarker validation and reproducibility, there is growing interest among researchers to combine multiple published metabolomics datasets collected under similar conditions. The idea is that this approach would reduce study bias to enable more robust biomarker identification. This practice is often referred to as biomarker ‘meta-analysis’ (35). When executed properly, biomarker meta-analysis can leverage the collective power of multiple independent studies to overcome potential biases and small effect sizes associated with individual datasets. This can significantly improve the precision in identifying true patterns within the data (36–38). However, user-friendly tools dedicated to support biomarker meta-analysis of metabolomic data are currently lacking (39). To address this issue, we have implemented a new module in MetaboAnalyst 4.0 called ‘Biomarker Meta-analysis’. The primary goal of the this module is to provide an easy-to-use interface to support the identification of robust biomarkers through meta-analysis of multiple datasets from independent metabolomics studies. The main steps for using the Biomarker Meta-analysis module are as follows:
Prior to uploading the data, the user should clean all datasets to ensure consistency amongst feature names (compound IDs, spectral bins or peaks) as well as consistency in the class labels (two groups only) across all included studies;
Once the data is cleaned and uploaded, the user can perform standard data processing, normalization, and differential analysis for each individual data set;
Once each individual data set has been processed via step (ii) above, meta-analysis can be performed using one of several statistical options: (a) combining P-values, (b) vote counting or (c) direct merging of data for very similar datasets (38);
After step iii has been completed, the result table containing summary statistics for all significant features is then displayed. Users can then click to view a boxplot summary of any feature across different datasets;
Users can visually explore the meta-analysis results in an interactive Venn diagram to view the shared features among all possible combinations of the datasets. An example is shown in Figure 2C.
New module #3: Network Explorer
Metabolomics is increasingly being used with other omics platforms such as transcriptomics, proteomics, and metagenomics to study complex diseases as well as to gain functional insights into microbial communities. However, integrating multiple omics data and interpreting these results at a systems level has become a significant challenge (40,41). A commonly used strategy is to analyze each set of omics data individually using tools and methods already developed for each field, and then piece together the ‘big picture’ using individual lists of significant features (i.e. metabolites, genes, and proteins). In particular, networks are very intuitive and flexible vehicles to convey our knowledge at a systems level. For instance, known relationships between genes, metabolites, and diseases can be easily represented as knowledge-based networks. By harnessing the power of networks and a priori biological knowledge, these lists of significant features can be co-projected onto the networks to reveal important links between them, as well as their associations with diseases or other interesting phenotypes. Such a comprehensive knowledgebase that connects metabolites with other molecular entities or phenotypes of interest, coupled with the support for interactive network visualization, will be an essential asset to help address current data integration challenges (42). The Network Explorer module has been developed to address this need. The aim of this module is to provide users with an easy-to-use interface that permits the mapping of their metabolites and/or genes (including KEGG orthologs or KOs) onto different types of molecular interaction networks. This network visualization can then be used to gain novel insights or assist users with the development of new hypotheses.
The Network Explorer module complements MetaboAnalyst's Joint Pathway Analysis module by allowing the identification of connections that cross the boundaries of conventional pathways as well as enabling a more global view of the functional changes which may not be obvious when one examines individual pathways. The Network Explorer module currently supports five types of biological networks including the KEGG global metabolic network, a gene-metabolite interaction network, a metabolite-disease interaction network, a metabolite-metabolite interaction network, and a metabolite-gene-disease interaction network. The last four networks are created based on information gathered primarily from HMDB and STITCH databases (43), and are applicable to human studies only.
Users can upload either a list of metabolites, a list of genes, or both. For metabolites, MetaboAnalyst 4.0 currently accepts compound names, HMDB IDs or KEGG compound IDs as metabolite identifiers. For genes, Entrez IDs, ENSEMBL IDs, official gene symbols or KEGG orthologs are currently supported. The uploaded lists of metabolites and genes are then mapped using MetaboAnalyst's internal databases. Following this step, users can select which of the five networks to begin to visually explore their data. On the network visualization page, users can use their mouse or touchpad to zoom in and out, highlight, drag and drop nodes (except the KEGG global metabolic network), or click on a node/edge for further details. Users can also perform functional enrichment analysis and then highlight those metabolites or genes involved in functions of interest on the network. The background color of the network and the color of the nodes/edges can also be customized. An example output from MetaboAnalyst's Network Explorer module is shown in Figure 2D. Each generated network can then be exported as a SVG or PNG image for publication purposes. We believe that the integration of interactive network exploration, functional enrichment analysis, and network topological analysis will provide users with more informative views and richer contextual information to facilitate the generation of testable hypotheses.
Other feature updates
There have been many small updates based on the user suggestions that have accumulated over the past three years. For instance, in the Biomarker Analysis module, many users indicated that they wanted to be able to select features that give information complementary to the biomarkers they already selected. We have therefore added feature similarity information using the cluster membership from k-means analysis to enhance the support for feature selection. Based on additional user feedback, we have also added two variants of the popular partial least square (PLS) methods, including orthogonal PLS and sparse PLS for improved data interpretation and more robust statistical analysis (44,45). For the two-way analysis of variance (ANOVA), we have added support for both type I and type III ANOVA, as well as additional analysis options for different experimental designs. While it is well known that MetaboAnalyst only has limited support for raw spectra profiling, we have attempted to remedy this by adding a ‘Spectral Analysis’ feature to point users to several easy-to-use, web-based tools that are freely available for spectral processing and annotation. It currently contains links to Bayesil (16), GC-AutoFit, and XCMS Online for NMR, GC-MS and LC-MS spectral processing, respectively.
Implementation
MetaboAnalyst 4.0 was implemented based on the PrimeFaces (v6.1) component library (http://primefaces.org/) and R (version 3.4.3). The interactive network visualization was implemented using the sigma.js JavaScript library (http://sigmajs.org). The entire system is hosted on a Google Cloud server with 32GB of RAM and eight virtual CPUs with 2.6 GHz each. The server is currently dealing with 5000∼8000 data analysis jobs submitted from ∼1000 users on a daily basis. For those who wish to use MetaboAnalyst 4.0 locally, we have provided the options to download the MetaboAnalyst as a war file or Docker image. Detailed instructions for download and local installation are provided on the ‘Resources’ page of the web server. The MetaboAnalystR package is available from the GitHub (https://github.com/xia-lab/MetaboAnalystR).
Comparison with other tools
Several web-based as well as several web-enabled tools for metabolomic data analysis have been developed over recent years, including XCMS Online, Workflow4Metabolomics (46), Galaxy-M (47), and Metabox (48). Detailed comparisons between these tools and MetaboAnalyst 4.0, as well as its previous versions are shown in Table 1. Based on this table, it is clear that MetaboAnalyst offers the most comprehensive support for statistical analysis, functional interpretation and integration with other omics data. It is also evident that MetaboAnalyst supports real-time interactive data analysis in way that no other tool currently can. While MetaboAnalyst has been limited in its built-in support for raw spectral processing and annotation, the new ‘Spectral Analysis’ feature help address this shortcoming. Certainly, raw LC-MS spectral processing and analysis has been a major strength of XCMS online, Galaxy-M and Workflow4Metabolomics, and these tools continue to be the ‘go-to’ resources for LC-MS data analysis. Overall, the primary strength of MetaboAnalyst is in its downstream data analysis, just as it is with Metabox. Indeed, the design of Metabox is similar to MetaboAnalyst in that it primarily accepts preprocessed metabolomics data for various statistical computing, functional analysis, and network-based integration. However, as noted in Table 1, no public server is currently available for Metabox and researchers must install it locally in order to use this tool.
Table 1. Comparison of the main features of MetaboAnalyst (versions 1.0 - 4.0) with other web-based or web-enabled tools. Symbols used for feature evaluations with ‘√’ for present, ‘-’ for absent, and ‘+’ for a more quantitative assessment, with more ‘+’ indicating better support.
| Tool name | MetaboAnalyst | XCMS online | Galaxy-M | W4M | Metabox | |||
|---|---|---|---|---|---|---|---|---|
| 4.0 | 3.0 | 2.0 | 1.0 | |||||
| Data processing | ||||||||
| Raw spectra | ++ | + | + | + | +++ | +++ | +++ | - |
| Data filtering | √ | √ | √ | - | √ | √ | - | - |
| Missing-value | √ | √ | √ | √ | - | √ | - | - |
| Normalization | +++ | +++ | ++ | ++ | - | ++ | ++ | ++ |
| Statistical analysis | ||||||||
| Univariate | +++ | +++ | +++ | ++ | + | ++ | ++ | ++ |
| Multivariate | +++ | ++ | ++ | ++ | ++ | + | +++ | + |
| Clustering | +++ | +++ | ++ | ++ | + | - | + | + |
| Classification | ++ | ++ | ++ | ++ | - | - | - | - |
| Power analysis | √ | √ | - | - | - | - | - | √ |
| Biomarker analysis | √ | √ | - | - | - | - | - | - |
| Functional analysis | ||||||||
| Enrichment | +++ | ++ | ++ | - | - | - | - | + |
| Pathway analysis | +++ | ++ | ++ | - | √ | - | - | √ |
| Mummichog | ++ | - | - | - | + | - | - | - |
| Data integration and systems biology | ||||||||
| Joint pathway analysis | √ | √ | - | - | √ | - | - | - |
| Knowledge -based network analysis | √ | - | - | - | - | - | - | √ |
| Correlation-based network analysis | - | - | - | - | - | - | - | √ |
| Biomarker meta-analysis | ++ | - | - | - | + | - | - | - |
• XCMS Online: https://xcmsonline.scripps.edu.
• Galaxy-M: https://github.com/Viant-Metabolomics/Galaxy-M.
• Workflow4Metabolomics (W4M): http://workflow4metabolomics.org/.
• Metabox: http://kwanjeeraw.github.io/metabox/.
CONCLUSIONS
Perhaps the most visible change to MetaboAnalyst is its newly designed web interface, which allows new features to be more easily ‘plugged in’. It also gives more space to permit interactive exploration of large networks as well as to display R command history during data analysis. We believe the latter feature, in combination with the release of the MetaboAnalystR package, will greatly improve reproducibility and transparency during metabolomics data analysis. Many advanced MetaboAnalyst users have felt constrained by the analysis boundaries defined by its web interface, and have asked for a more flexible workflow design and batch processing capabilities. The MetaboAnalystR package addresses these limitations. Users can now create a workflow (R command history) through the web interface, customize the workflow by changing the order of the commands or their parameters, and finally execute the workflow in batch mode using the R package. For those researchers who are already familiar with R programming, it is also possible to directly modify the underlying R code to suit their needs. Another major focus of this MetaboAnalyst's update is the addition of new modules to support further data integration (biomarker meta-analysis and multi-omics analysis), as well as functional analysis for high-resolution untargeted MS data. These additions were made in response to frequent user requests and growing trends seen in metabolomic data analysis practices. Finally, to ensure that the biological interpretation of metabolomic data remains as current and insightful as possible, all of MetaboAnalyst's underlying knowledgebases have been updated. These updates will allow metabolomics researchers to move beyond simply re-iterating common textbook interpretations of metabolism, and give them much more useful insights into complex and relevant biological processes that are ultimately driven by metabolites. Overall, we believe these updates will allow MetaboAnalyst to remain at the cutting edge of computational metabolomics and systems biology, and that it will continue to enable new discoveries and greater insights for a growing number of metabolomics researchers.
FUNDING
McGill University; Natural Sciences and Engineering Research Council of Canada (NSERC); Canadian Institutes of Health Research (CIHR); Genome Canada; Canada Research Chairs Program (CRC). Funding for open access charge: Genome Canada.
Conflict of interest statement. None declared.
REFERENCES
- 1. Xia J., Psychogios N., Young N., Wishart D.S.. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009; 37:W652–W660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Xia J., Mandal R., Sinelnikov I.V., Broadhurst D., Wishart D.S.. MetaboAnalyst 2.0—a comprehensive server for metabolomic data analysis. Nucleic Acids Res. 2012; 40:W127–W133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Xia J., Wishart D.S.. MSEA: a web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res. 2010; 38:W71–W77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xia J., Wishart D.S.. MetPA: a web-based metabolomics tool for pathway analysis and visualization. Bioinformatics. 2010; 26:2342–2344. [DOI] [PubMed] [Google Scholar]
- 5. Xia J., Sinelnikov I.V., Wishart D.S.. MetATT: a web-based metabolomics tool for analyzing time-series and two-factor datasets. Bioinformatics. 2011; 27:2455–2456. [DOI] [PubMed] [Google Scholar]
- 6. Xia J., Sinelnikov I.V., Han B., Wishart D.S.. MetaboAnalyst 3.0—making metabolomics more meaningful. Nucleic Acids Res. 2015; 43:W251–W257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xia J., Broadhurst D.I., Wilson M., Wishart D.S.. Translational biomarker discovery in clinical metabolomics: an introductory tutorial. Metabolomics. 2013; 9:280–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tayyari F., Gowda G.N., Olopade O.F., Berg R., Yang H.H., Lee M.P., Ngwa W.F., Mittal S.K., Raftery D., Mohammed S.I.. Metabolic profiles of triple-negative and luminal A breast cancer subtypes in African-American identify key metabolic differences. Oncotarget. 2018; 9:11677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang G., Dervishi E., Dunn S.M., Mandal R., Liu P., Han B., Wishart D.S., Ametaj B.N.. Metabotyping reveals distinct metabolic alterations in ketotic cows and identifies early predictive serum biomarkers for the risk of disease. Metabolomics. 2017; 13:43. [Google Scholar]
- 10. Reynolds L.A., Redpath S.A., Yurist-Doutsch S., Gill N., Brown E.M., van der Heijden J., Brosschot T.P., Han J., Marshall N.C., Woodward S.E.. Enteric helminths promote Salmonella coinfection by altering the intestinal metabolome. J. Infect. Dis. 2017; 215:1245–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bahado-Singh R.O., Akolekar R., Mandal R., Dong E., Xia J., Kruger M., Wishart D.S., Nicolaides K.. Metabolomic analysis for first-trimester Down syndrome prediction. Am. J. Obstet. Gynecol. 2013; 208:371–378. [DOI] [PubMed] [Google Scholar]
- 12. Cox A.G., Hwang K.L., Brown K.K., Evason K.J., Beltz S., Tsomides A., O’Connor K., Galli G.G., Yimlamai D., Chhangawala S.. Yap reprograms glutamine metabolism to increase nucleotide biosynthesis and enable liver growth. Nat. Cell Biol. 2016; 18:886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Arts R.J., Novakovic B., ter Horst R., Carvalho A., Bekkering S., Lachmandas E., Rodrigues F., Silvestre R., Cheng S.-C., Wang S.-Y.. Glutaminolysis and fumarate accumulation integrate immunometabolic and epigenetic programs in trained immunity. Cell Metab. 2016; 24:807–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Paglia G., Miedico O., Cristofano A., Vitale M., Angiolillo A., Chiaravalle A.E., Corso G., Di Costanzo A.. Distinctive pattern of serum elements during the progression of Alzheimer's disease. Scientific Rep. 2016; 6:22769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li S., Park Y., Duraisingham S., Strobel F.H., Khan N., Soltow Q.A., Jones D.P., Pulendran B.. Predicting network activity from high throughput metabolomics. PLoS Comput. Biol. 2013; 9:e1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ravanbakhsh S., Liu P., Bjordahl T.C., Mandal R., Grant J.R., Wilson M., Eisner R., Sinelnikov I., Hu X., Luchinat C.. Accurate, fully-automated NMR spectral profiling for metabolomics. PLoS ONE. 2015; 10:e0124219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huan T., Forsberg E.M., Rinehart D., Johnson C.H., Ivanisevic J., Benton H.P., Fang M., Aisporna A., Hilmers B., Poole F.L.. Systems biology guided by XCMS Online metabolomics. Nat. Methods. 2017; 14:461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wadi L., Meyer M., Weiser J., Stein L.D., Reimand J.. Impact of outdated gene annotations on pathway enrichment analysis. Nat. Methods. 2016; 13:705–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marco-Ramell A., Palau-Rodriguez M., Alay A., Tulipani S., Urpi-Sarda M., Sanchez-Pla A., Andres-Lacueva C.. Evaluation and comparison of bioinformatic tools for the enrichment analysis of metabolomics data. BMC Bioinformatics. 2018; 19:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K.. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017; 45:D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wishart D.S., Feunang Y.D., Marcu A., Guo A.C., Liang K., Vázquez-Fresno R., Sajed T., Johnson D., Li C., Karu N.. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2017; 46:D608–D617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hastings J., de Matos P., Dekker A., Ennis M., Harsha B., Kale N., Muthukrishnan V., Owen G., Turner S., Williams M.. The ChEBI reference database and ontology for biologically relevant chemistry: enhancements for 2013. Nucleic Acids Res. 2012; 41:D456–D463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Smith C.A., O’Maille G., Want E.J., Qin C., Trauger S.A., Brandon T.R., Custodio D.E., Abagyan R., Siuzdak G.. METLIN: a metabolite mass spectral database. Therapeut. Drug Monitor. 2005; 27:747–751. [DOI] [PubMed] [Google Scholar]
- 24. Kim S., Thiessen P.A., Bolton E.E., Chen J., Fu G., Gindulyte A., Han L., He J., He S., Shoemaker B.A.. PubChem substance and compound databases. Nucleic Acids Res. 2015; 44:D1202–D1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jewison T., Su Y., Disfany F.M., Liang Y., Knox C., Maciejewski A., Poelzer J., Huynh J., Zhou Y., Arndt D.. SMPDB 2.0: big improvements to the small molecule pathway database. Nucleic Acids Res. 2013; 42:D478–D484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pluskal T., Castillo S., Villar-Briones A., Orešič M.. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics. 2010; 11:395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lommen A., Kools H.J.. MetAlign 3.0: performance enhancement by efficient use of advances in computer hardware. Metabolomics. 2012; 8:719–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kind T., Fiehn O.. Metabolomic database annotations via query of elemental compositions: mass accuracy is insufficient even at less than 1 ppm. BMC Bioinformatics. 2006; 7:234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kind T., Fiehn O.. Seven Golden Rules for heuristic filtering of molecular formulas obtained by accurate mass spectrometry. BMC Bioinformatics. 2007; 8:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Johnson C.H., Ivanisevic J., Siuzdak G.. Metabolomics: beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016; 17:451–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hanash S.M., Pitteri S.J., Faca V.M.. Mining the plasma proteome for cancer biomarkers. Nature. 2008; 452:571–579. [DOI] [PubMed] [Google Scholar]
- 33. Goveia J., Pircher A., Conradi L.C., Kalucka J., Lagani V., Dewerchin M., Eelen G., DeBerardinis R.J., Wilson I.D., Carmeliet P.. Meta‐analysis of clinical metabolic profiling studies in cancer: challenges and opportunities. EMBO Mol. Med. 2016; 8:1134–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tzoulaki I., Ebbels T.M., Valdes A., Elliott P., Ioannidis J.P.. Design and analysis of metabolomics studies in epidemiologic research: a primer on-omic technologies. Am. J. Epidemiol. 2014; 180:129–139. [DOI] [PubMed] [Google Scholar]
- 35. Tseng G.C., Ghosh D., Feingold E.. Comprehensive literature review and statistical considerations for microarray meta-analysis. Nucleic Acids Res. 2012; 40:3785–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Patti G.J., Yanes O., Siuzdak G.. Innovation: Metabolomics: the apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012; 13:263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xia J., Fjell C.D., Mayer M.L., Pena O.M., Wishart D.S., Hancock R.E.. INMEX–a web-based tool for integrative meta-analysis of expression data. Nucleic Acids Res. 2013; 41:W63–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Walsh C.J., Hu P., Batt J., Santos C.C.D.. Microarray meta-analysis and cross-platform normalization: integrative genomics for robust biomarker discovery. Microarrays. 2015; 4:389–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cambiaghi A., Ferrario M., Masseroli M.. Analysis of metabolomic data: tools, current strategies and future challenges for omics data integration. Brief. Bioinformatics. 2016; 18:498–510. [DOI] [PubMed] [Google Scholar]
- 40. Ritchie M.D., Holzinger E.R., Li R., Pendergrass S.A., Kim D.. Methods of integrating data to uncover genotype-phenotype interactions. Nat. Rev. Genet. 2015; 16:85–97. [DOI] [PubMed] [Google Scholar]
- 41. Chong J., Xia J.. Computational approaches for integrative analysis of the metabolome and microbiome. Metabolites. 2017; 7:E62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xia J., Gill E.E., Hancock R.E.. NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat. Protoc. 2015; 10:823–844. [DOI] [PubMed] [Google Scholar]
- 43. Yao Q., Xu Y., Yang H., Shang D., Zhang C., Zhang Y., Sun Z., Shi X., Feng L., Han J.. Global prioritization of disease candidate metabolites based on a multi-omics composite network. Scientific Rep. 2015; 5:17201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Thevenot E.A., Roux A., Xu Y., Ezan E., Junot C.. Analysis of the human adult urinary metabolome variations with age, body mass index, and gender by implementing a comprehensive workflow for univariate and OPLS statistical analyses. J. Proteome Res. 2015; 14:3322–3335. [DOI] [PubMed] [Google Scholar]
- 45. Lê Cao K.-A., Boitard S., Besse P.. Sparse PLS discriminant analysis: biologically relevant feature selection and graphical displays for multiclass problems. BMC Bioinformatics. 2011; 12:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Giacomoni F., Le Corguillé G., Monsoor M., Landi M., Pericard P., Pétéra M., Duperier C., Tremblay-Franco M., Martin J.-F., Jacob D.. Workflow4Metabolomics: a collaborative research infrastructure for computational metabolomics. Bioinformatics. 2014; 31:1493–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Davidson R.L., Weber R.J., Liu H., Sharma-Oates A., Viant M.R.. Galaxy-M: a Galaxy workflow for processing and analyzing direct infusion and liquid chromatography mass spectrometry-based metabolomics data. Gigascience. 2016; 5:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wanichthanarak K., Fan S., Grapov D., Barupal D.K., Fiehn O.. Metabox: A toolbox for metabolomic data analysis, interpretation and integrative exploration. PLoS ONE. 2017; 12:e0171046. [DOI] [PMC free article] [PubMed] [Google Scholar]