Abstract
HotSpot Wizard is a web server used for the automated identification of hotspots in semi-rational protein design to give improved protein stability, catalytic activity, substrate specificity and enantioselectivity. Since there are three orders of magnitude fewer protein structures than sequences in bioinformatic databases, the major limitation to the usability of previous versions was the requirement for the protein structure to be a compulsory input for the calculation. HotSpot Wizard 3.0 now accepts the protein sequence as input data. The protein structure for the query sequence is obtained either from eight repositories of homology models or is modeled using Modeller and I-Tasser. The quality of the models is then evaluated using three quality assessment tools—WHAT_CHECK, PROCHECK and MolProbity. During follow-up analyses, the system automatically warns the users whenever they attempt to redesign poorly predicted parts of their homology models. The second main limitation of HotSpot Wizard’s predictions is that it identifies suitable positions for mutagenesis, but does not provide any reliable advice on particular substitutions. A new module for the estimation of thermodynamic stabilities using the Rosetta and FoldX suites has been introduced which prevents destabilizing mutations among pre-selected variants entering experimental testing. HotSpot Wizard is freely available at http://loschmidt.chemi.muni.cz/hotspotwizard.
INTRODUCTION
Proteins are macromolecules with many biological functions. Apart from their irreplaceable role in all living organisms, they are also widely used in many fields, including medicine (1), enzymology (2), synthetic biology (3) and material science (4). Naturally occurring proteins often do not meet the specifications for practical applications. Therefore, protein engineers modify sequences to obtain enhanced properties or completely new functions. Directed evolution, which has been an extremely successful protein engineering technology, does not require a molecular understanding of the impact of mutation on the protein structure (5). Modified proteins are generated in iterative rounds of mutation and screening or selection of the best hits that possess the required property (6). The obvious disadvantage to this method is that only a tiny fraction of all protein variants contain the desired property. Analysis of libraries containing millions of mutants is costly and time-consuming. Semi-rational protein engineering is an approach that implements in silico identification of important regions of the protein so that mutagenesis is better located, resulting in smaller high-quality libraries (7). The key step to semi-rational protein engineering is the selection of hotspot residues whose mutations will bring the largest improvement to the target protein properties (8).
HotSpot Wizard 2.0 (9) is an interactive web server used for the identification of hotspots in proteins by automated multi-step calculation and a comprehensive presentation of results. The tool makes protein design accessible to researchers with no prior knowledge of bioinformatics. After entering an input protein structure, 19 prediction tools and 3 databases are used for protein annotation. HotSpot Wizard then provides four different strategies for selecting hotspots: (i) functional hotspots corresponding to highly mutable residues located in the active site pocket or access tunnels, (ii) stability hotspots corresponding to flexible residues, (iii) stability hotspots from back-to-consensus analysis and (iv) correlated hotspots corresponding to pairs of co-evolving residues. The users can design a smart library based on naturally accepted substitutions from phylogenetic analysis. HotSpot Wizard 2.0 (9) has been used for over 10 000 protein structures by more than 1000 unique users since its release. For example, HotSpot Wizard has been used for the design of smart libraries of oxyhaemoglobin protein (10), for analysis leading to thermostabilization of a xylanase (11) and for identification of hotspots in a mutagenesis study of the transcription factor DREB1A (12). Previous implementations of HotSpot Wizard had two major drawbacks: (i) a requirement for the tertiary structure as essential input information and (ii) identification of positions for mutagenesis without quantification of the effects of individual substitutions on protein stability. HotSpot Wizard 3.0 shows dramatically enhanced usability by overcoming both these key limitations.
There are about 135 000 protein structures available in the RCSB Protein Data Bank (13), but there are more than 98 000 000 known protein sequences (14). Usage of HotSpot Wizard 2.0 is limited to the proteins with an available 3D structure. A solution to this problem is the prediction of the protein structure from its sequence by comparative (homology) modeling or threading (15). Homology modeling is based on the fact that members of a protein family with similar sequences also have similar tertiary structures (16,17). In HotSpot Wizard 3.0, it is possible to enter a sequence for a protein and have its tertiary structure retrieved from the repositories of models or constructed ad hoc. As the quality of the protein structure is critical for further structure analyses carried out by HotSpot Wizard, a robust quality assessment of the protein structure is provided using three well-established tools. The current implementation of our web server predicts hot-spots for mutagenesis and designs smart libraries based on phylogeny, but does not provide any quantitative analysis of individual substitutions, which is important, for example, in studies analyzing structure–function relationships. Moreover, screening or selection for multiple mutations at several different positions can still be time-consuming and so pre-selection of the most appropriate mutations is desirable. To help our users rationally decrease the number of variants for experimental testing, protein stability prediction has been introduced to discard potentially destabilizing mutations.
MATERIALS AND METHODS
Searches of structural databases and model depositories
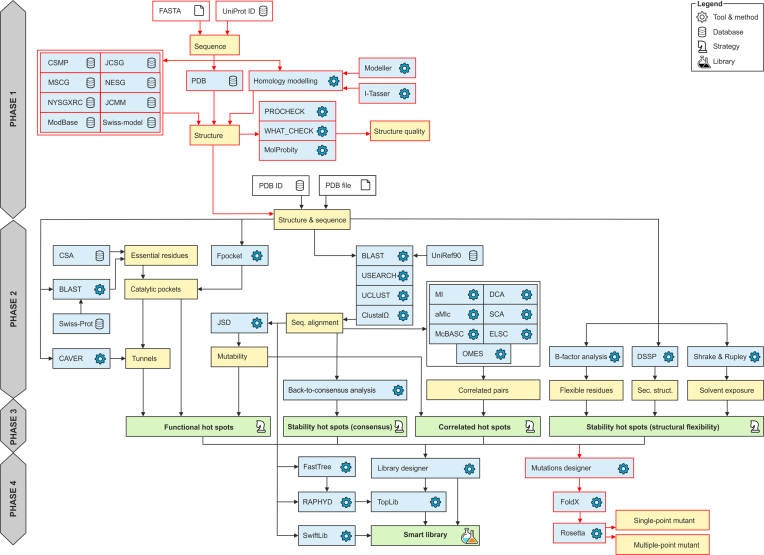
The overall workflow of HotSpot Wizard 3.0 is outlined in Figure 1. When a protein sequence is used as an input, HotSpot Wizard: (i) searches experimentally determined structures, (ii) searches computationally modeled structures and (iii) constructs a homology model. The first step in this workflow is searching the RCSB Protein Data Bank (13). In this phase, only protein structures with a 100% sequence identity match (or part of the sequence matching the input with 100% sequence identity) are provided as a starting structure for the analysis. If no such structure is found, the Protein Model Portal (18) is searched. The Protein Model Portal collates models of protein structures from eight different resources: Center for Structures of Membrane Proteins, CSMP (19), Joint Center for Structural Genomics, JCSG (20), Midwest Center for Structural Genomics, MCSG (21), Northeast Structural Genomics Consortium, NESG (22), New York SGX Research Center for Structural Genomics, NYSGXRC (23), Joint Center for Molecular Modeling, JCMM (24), ModBase (25) and SWISS-MODEL Repository (26). HotSpot Wizard queries the Protein Model Portal and then lists all available hits. After selection of one of these models, the structure is downloaded directly to Hotspot Wizard from the repository.
Figure 1.
Workflow diagram of HotSpot Wizard 3.0. The workflow consists of four phases: (1) construction of a model of a structure, (2) annotation of a protein, (3) identification of mutagenesis hot spots and (4) design of mutations and a smart library. Phase 1 is applied only when a sequence is submitted as the input information. The new modules in version 3.0 are highlighted in red.
Homology modeling
Whenever a homology model is not found or the user is not satisfied with the quality of the models available in public depositories, HotSpot Wizard carries out the homology modeling during the phase 1 (Figure 1). There is a wide range of homology modeling tools available. Twelve tools were initially considered for our workflow: SWISS-MODEL (27), Rosetta (28), Robetta (29), PHYRE2 (30), Pcons (31), Modeller (32), I-Tasser (33), IntFold (34), IMP (35), HHPred (36), RaptorX (37) and Sparks-X (38). These tools were analyzed for their availability as well as performance using Continuous Automated Model Evaluation, CAMEO (18) and Critical Assessment of Protein Structure Prediction, CASP (39). These community-wide comparisons evaluate structure predictions with available experimental data. Based on results from CASP and CAMEO, six tools were selected for further consideration, installed locally and tested (Modeller, Sparks-X, RaptorX, Rosetta, I-Tasser and SWISS-MODEL). RaptorX is very accurate with good coverage (i.e. percentage of submitted models, which could be successfully modeled), but it uses the less accurate Modeller for comparative modeling in its standalone version. Sparks-X is very fast with good coverage, but the version available for download does not provide modeling, only template identification. I-Tasser is the slowest of all the tools considered, but it is very accurate and is ranked the best by CASP. Rosetta has good accuracy and coverage, but it requires a template protein and an alignment as an input defined by user. SWISS-MODEL is fast with good coverage, but it is not available as a standalone version. Modeller is one of the fastest and the most robust tools with reasonable accuracy for modeling cases with good templates. We selected two tools for implementation with HotSpot Wizard: (i) I-Tasser, which is ranked the most accurate of all the tools considered, but also very slow (∼3 days for an average-sized protein) and (ii) Modeller, which is less accurate, but very fast (∼5 min for an average-sized protein). Both tools can be run in a fully automatic mode, or the template protein and/or the pairwise alignment can be entered as an input information.
Quality assessment of the model
It is essential to assess the quality of the homology model prior to its further use for identification of hotspots or for the design of libraries. It is important to identify low quality models and the parts of the protein structure which were not modeled well. The results of today’s modeling tools are far from perfect due to many difficulties with accurate protein structure prediction. Quality assessment is therefore an essential part of the phase 1 of the HotSpot Wizard workflow (Figure 1). Several quality assessment tools were considered and three of them, providing diverse quality metrics, were implemented. PROCHECK (40) is used for analysis of protein backbone torsion angles using Ramachandran diagrams and identification of the outliers from the allowed values. MolProbity (41) provides several parameters representing the quality of the whole structure as well as individual residues (number of poor rotamers, Ramachandran outliers, favored Ramachandran conformations, bad bonds and bad angles in the protein). WHAT_CHECK (42) generates a detailed report about structure quality (checks on secondary structure, coordinate problems, unexpected atoms, B-factor, occupancy checks, nomenclature related problems, geometric checks, torsion-related checks, bump checks, packing, accessibility, threading, water, ion and hydrogen bond-related checks).
Mutation design based on thermodynamic stability
Mutation design is part of the phase 4 of the HotSpot Wizard computation (Figure 1). Force field calculations are used for quantifying the change in protein thermodynamic stability after mutation. Rosetta (43) is used to evaluate ΔΔG between the wild-type and the mutant structures. Either single-point or multiple-point mutants can be evaluated. If the single-point mutations are pre-selected, multiple mutant structures are evaluated according to the user’s selected positions and intended amino acid substitutions. The user can also select several mutations in a single round and calculate the energy of combined multiple-point mutants. For stability evaluation, FoldX (44) is first used for repairing protein structure by filling in the missing atoms and patching the structure. Then, minimalization of the structure using Rosetta is carried out using default settings. After that, a Rosetta stability calculation according to protocol 3 (45) is carried out, which results in the prediction of ΔΔG value for each mutation.
DESCRIPTION OF THE WEB SERVER
Sequence input and homology modeling
Initially, the user selects one of two types of input data: a structure or a sequence (Figure 2A). If a sequence is selected, there are three types of input. The user can either manually enter the protein sequence, specify the UniProt ID or upload the FASTA file. After entering the sequence, the user is provided with the results from searching the Protein Data Bank or the Protein Model Portal. This result is displayed in the form of a table (Figure 2B). In the case of the Protein Data Bank results, PDB ID, resolution and the link to the Protein Data Bank are provided. The user can then pick one of the proteins and continue with the HotSpot Wizard workflow. In the case of the results from the Protein Model Portal model provider, following information is listed: (i) used template, (ii) sequence identity with a template, (iii) range of the alignment, (iv) coverage and (v) reliability of the model. Links to a model in the Protein Model Portal and the template structure in the Protein Data Bank are provided in the table. Coverage and reliability of the models are represented by a color ranging from green to red (Figure 2C). If the user selects a model with unsatisfactory coverage (<80%) or insufficient reliability (low reliability value), a warning is displayed. When a protein model is selected which cannot be downloaded automatically, the user is asked to download it manually and then upload it as a structure for further analysis. The user can then select one of the models provided and continue with the HotSpot Wizard workflow or, if none of the models is satisfactory, carry out homology modeling and construct their own model. If the user carries out homology modeling, several parameters must be set first (Figure 2D). The user can select between Modeller, which is faster but less accurate, or I-Tasser, which is more accurate but slow. The second important parameter that must be specified prior to calculation is either automatic or manual identification of the template structure and alignment. The template can be provided either by entering the PDB ID or by uploading a PDB file. In the case of the user entering the alignment, pairwise alignment of the template and an input sequence in FASTA format must be provided. The process of hotspot identification can then begin after all these essential inputs have been defined.
Figure 2.
Graphic user interface of the sequence input in the HotSpot Wizard 3.0. (A) Selection between structure and sequence input. (B) After entering of the sequence, searching for existing structures in PDB database is performed. (C) If no existing structure is found, search in homology model databases is performed. (D) Setting of homology modeling parameters—user can choose between Modeller and I-Tasser and eventually enter his own template or sequence alignment.
Quality assessment of the model
Results of the quality assessment are shown in separate windows consisting of three tabs containing various quality assessment analyses. The first tab shows the MolProbity overall quality assessment table (Supplementary Figure S1A). In this table, the number and percentage of poor rotamers, Ramachandran outliers, favored Ramachandran conformers, bad bonds and bad angles are shown. Colored highlights are used to distinguish between good and unsatisfactory models. The second tab shows the MolProbity quality assessment results for each residue, displayed in the form of plots (Supplementary Figure S1B). A plot of MolProbity Ramachandran scores and MolProbity rotamer scores is given. In the last tab, there is a Ramachandran plot for the protein created by PROCHECK with outlier residues highlighted (Supplementary Figure S1C). The contents of all these tabs can be downloaded in PDF format together with a full quality assessment report created by WHAT_CHECK.
Mutations design based on stability
The stability changes introduced by specific mutations can be accessed through a newly introduced Mutations design module (Supplementary Figure S2A). There are three tabs in the Mutation design window—the first for definition of single-point mutants, the second for multiple-point mutants and the third summarizing the status of submitted jobs. In the case of single-point mutations, the user can select particular amino acids for each of the selected hotspots. The amino acid residues for mutagenesis can be selected based on: (i) amino acid frequency, (ii) mutational landscape, (iii) physico-chemical properties or (iv) user selection (Supplementary Figure S2B). After selection of the mutations, the stability of each single-point mutation is evaluated by the Rosetta software suite. The results are shown in the table—stabilizing mutations are highlighted in green, destabilizing mutations are highlighted in red (Supplementary Figure S2C). There are two options for setting multiple-point mutants. Either a particular amino acid can be selected for each position in the multiple-point tab or the results table from a previous single-point calculation can be used for recombination with the most promising substitutions. In both cases, only a single substitution for each position can be selected (Supplementary Figure S2D). After the calculation is finished, Hotspot Wizard reports the overall stability change as well as the decomposition of energy terms, both of which provide excellent assistance for mutagenesis experiments (Supplementary Figure S2E). The stability prediction can be downloaded in CSV format with the sequence of designed mutants being provided in FASTA format. These reports can also be generated in PDF or HTML formats. The third tab shows a table with the history of previously evaluated stabilities for the job. For each calculation, the job id, date and time of computation, status of the job (failed or finished), mutation type (single-point or multiple-point), selected positions and mutations are shown (Supplementary Figure S2F). The results page from any previous calculations can be revisited at any time.
EXPERIMENTAL VALIDATION
We have carried out validation of individual steps of the workflow as well as thoroughly tested the final version of the web server. The homology modeling tools were selected for implementation based on the results of CAMEO comparison (Supplementary Data 1). The reliability, coverage and availability of a standalone version of all the software code were considered during the selection process. The reliability of the Rosetta protocol 3 employed in the Design module was benchmarked against experimental stability data previously collected for multiple-point mutants in our laboratory (46) as well as 1573 single-point mutants available in the ProTherm and HotMuSiC databases (Supplementary Data 2). These tests confirmed a significant correlation between half-lives and calculated changes in free energy ΔΔG, as well as an ability of the fast protocol 3 to correctly classify stabilizing and destabilizing mutations. Functionality of the Mutation design module was validated by saturation mutagenesis at the hotspot position L177 located at the tunnel mouth of the haloalkane dehalogenase LinB (47). Theoretical predictions correctly identified the variant L177W, which was found to be the most stable also experimentally (Supplementary Data 3). At last, we used the HotSpot Wizard 3.0 workflow for computational mutagenesis of six residues lining the active site cavity and the access tunnel of the haloalkane dehalogenases from non-pathogenic and pathogenic bacteria Sphingobium japonicum UT26 and Mycobacterium tuberculosis Rv2579, respectively (48). Single-point mutations and combined sixfold mutants were predicted using the automated protocols with crystal structures and homology models (Supplementary Data 4).
CONCLUSIONS AND OUTLOOK
HotSpot Wizard 3.0 is a new version of a popular web server used for the automated prediction of hotspots and the design of smart libraries in semi-rational protein design. In this version, homology modeling of the protein structure dramatically increases the usability of the platform by increasing the number of possible inputs and solves the limitation imposed by the number of available experimental structures. For homology modeling, Modeller and I-Tasser are used. The quality of the models created is evaluated using three different tools to identify wrongly modeled regions, which should be used for further computational design only with extreme care. The users are automatically warned whenever they attempt to redesign poorly resolved regions, for example the residues lying outside allowed regions of the Ramachandran plot. Rational design is further supported by the novel Mutation design module employing force field calculations for estimating the effect of substitution on protein thermodynamic stability. This new module can dramatically reduce the number of variants selected for experimental testing and can also help to pre-select mutations for identified positions during construction of smart libraries. In the future, we want to focus on more systematic use of multiple structural data from the Protein Data Bank, and on development of a novel engineering strategy for the design of biocatalysts that catalyze specific chemical reactions. Extensive databases searches will be coupled with the computational design module for identification of the best starting protein template for such an engineering exercise.
Supplementary Material
ACKNOWLEDGEMENTS
Computational resources were supplied by the Ministry of Education, Youth and Sports of the Czech Republic under the Projects CESNET (Project No. LM2015042) and CERIT-Scientific Cloud (Project No. LM2015085).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Education, Youth and Sports of the Czech Republic from the National Programme of Sustainability II [LQ1602, LQ1605, LO1214]; European Regional Development Fund [LM2015051, LM2015047, LM2015055]; Grant Agency of the Czech Republic [16–06096S]; European Union [720776, 722610]; Brno University Technology [FIT-S-17–3994 to L.S.]. Funding for open access charge: Czech Ministry of Education.
Conflict of interest statement. None declared.
REFERENCES
- 1. Hawkins M.J., Soon-Shiong P., Desai N.. Protein nanoparticles as drug carriers in clinical medicine. Adv. Drug Deliv. Rev. 2008; 60:876–885. [DOI] [PubMed] [Google Scholar]
- 2. Godfrey T., Reichelt J.. Industrial applications. Industrial Enzymology: The Application of Enzymes in Industry. 1982; London: Macmillan, The Nature Press; 582. [Google Scholar]
- 3. Bromley E.H., Channon K., Moutevelis E., Woolfson D.N.. Peptide and protein building blocks for synthetic biology: from programming biomolecules to self-organized biomolecular systems. ACS Chem. Biol. 2008; 3:38–50. [DOI] [PubMed] [Google Scholar]
- 4. De La Rica R., Matsui H.. Applications of peptide and protein-based materials in bionanotechnology. Chem. Soc. Rev. 2010; 39:3499–3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cheng F., Zhu L., Schwaneberg U.. Directed evolution 2.0: improving and deciphering enzyme properties. Chem. Commun. 2015; 51:9760–9772. [DOI] [PubMed] [Google Scholar]
- 6. Romero P.A., Arnold F.H.. Exploring protein fitness landscapes by directed evolution. Nat. Rev. Mol. Cell Biol. 2009; 10:866–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lutz S. Beyond directed evolution—semi-rational protein engineering and design. Curr. Opin. Biotechnol. 2010; 21:734–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cheng Z., Peplowski L., Cui W., Xia Y., Liu Z., Zhang J., Kobayashi M., Zhou Z.. Identification of key residues modulating the stereoselectivity of nitrile hydratase towards rac‐Mandelonitrile by Semi‐rational engineering. Biotechnol. Bioeng. 2017; 115:1–12. [DOI] [PubMed] [Google Scholar]
- 9. Bendl J., Stourac J., Sebestova E., Vavra O., Musil M., Brezovsky J., Damborsky J.. HotSpot Wizard 2.0: automated design of site-specific mutations and smart libraries in protein engineering. Nucleic Acids Res. 2016; 44:W479–W487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Talukdar P., Talapatra S.N.. Oxy-haemoglobin protein engineering: an automated design for hotspots stability, site-specific mutations and smart libraries by using HotSpot Wizard 2.0 software. Int. J. Adv. Res. Comput. Sci. 2017; 8:220–228. [Google Scholar]
- 11. Wang X., Ma R., Xie X., Liu W., Tu T., Zheng F., You S., Ge J., Xie H., Yao B. et al. Thermostability improvement of a Talaromyces leycettanus xylanase by rational protein engineering. Sci. Rep. 2017; 7:15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vatansever R., Uras M.E., Sen U., Ozyigit I.I., Filiz E.. Isolation of a transcription factor DREB1A gene from Phaseolus vulgaris and computational insights into its characterization: protein modeling, docking and mutagenesis. J. Biomol. Struct. Dyn. 2016; 35:1–12. [DOI] [PubMed] [Google Scholar]
- 13. Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E.. The Protein Data Bank. Nucleic Acids Res. 2000; 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017; 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baker D., Sali A.. Protein structure prediction and structural genomics. Science. 2001; 294:93–96. [DOI] [PubMed] [Google Scholar]
- 16. Cavasotto C.N., Phatak S.S.. Homology modeling in drug discovery: current trends and applications. Drug Discov. Today. 2009; 14:676–683. [DOI] [PubMed] [Google Scholar]
- 17. Schwede T. Protein modeling: what happened to the ‘protein structure gap’. Structure. 2013; 21:1531–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Haas J., Roth S., Arnold K., Kiefer F., Schmidt T., Bordoli L., Schwede T.. The Protein Model Portal—a comprehensive resource for protein structure and model information. Database. 2013; 2013:bat031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Csmp.ucsf.edu CSMP | Home. 2017; (20 December 2017, date last accessed)http://csmp.ucsf.edu/index.htm. [Google Scholar]
- 20. Jcsg.org The Joint Center for Structural Genomics (JCSG) Homepage. 2017; (20 December 2017, date last accessed)http://www.jcsg.org/. [Google Scholar]
- 21. Mcsg.anl.gov 2017; (20 December 2017, date last accessed)http://www.mcsg.anl.gov/. [Google Scholar]
- 22. Nesg.org NESG - NorthEast Structural Genomics consortium. 2017; (20 December 2017, date last accessed)http://www.nesg.org/. [Google Scholar]
- 23. Venkatagiriyappa V. NYSGRC. 2017; (20 December 2017, date last accessed)http://www.nysgxrc.org/psi3-cgi/index.cgi. [Google Scholar]
- 24. Jcmm.burnham.org Joint Center for Molecular Modeling (JCMM). 2017; (20 December 2017, date last accessed)http://jcmm.burnham.org/. [Google Scholar]
- 25. Pieper U., Webb B.M., Dong G.Q., Schneidman-Duhovny D., Fan H., Kim S.J., Tainer J.A.. ModBase, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2013; 42:D336–D346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kiefer F., Arnold K., Künzli M., Bordoli L., Schwede T.. The SWISS-MODEL repository and associated resources. Nucleic Acids Res. 2008; 37:D387–D392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Schwede T.. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014; 42:W252–W258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Song Y., DiMaio F., Wang R.Y.R., Kim D., Miles C., Brunette T.J., Baker D.. High-resolution comparative modeling with RosettaCM. Structure. 2013; 21:1735–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim D.E., Chivian D., Baker D.. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004; 32:W526–W531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015; 10:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Larsson P., Skwark M.J., Wallner B., Elofsson A.. Improved predictions by Pcons. net using multiple templates. Bioinformatics. 2010; 27:426–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Webb B., Sali A.. Protein structure modeling with MODELLER. Methods Mol. Biol. 2014; 1137:151–115. [DOI] [PubMed] [Google Scholar]
- 33. Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y.. The I-TASSER Suite: protein structure and function prediction. Nat. Methods. 2015; 12:7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McGuffin L.J., Atkins J.D., Salehe B.R., Shuid A.N., Roche D.B.. IntFOLD: an integrated server for modelling protein structures and functions from amino acid sequences. Nucleic Acids Res. 2015; 43:W169–W173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Russel D., Lasker K., Webb B., Velázquez-Muriel J., Tjioe E., Schneidman-Duhovny D., Sali A.. Putting the pieces together: integrative modeling platform software for structure determination of macromolecular assemblies. PLoS Biol. 2012; 10:e1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hildebrand A., Remmert M., Biegert A., Söding J.. Fast and accurate automatic structure prediction with HHpred. Proteins. 2009; 77:128–132. [DOI] [PubMed] [Google Scholar]
- 37. Källberg M., Wang H., Wang S., Peng J., Wang Z., Lu H., Xu J.. Template-based protein structure modeling using the RaptorX web server. Nat. Protoc. 2012; 7:1511–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang Y., Faraggi E., Zhao H., Zhou Y.. Improving protein fold recognition and template-based modeling by employing probabilistic-based matching between predicted one-dimensional structural properties of query and corresponding native properties of templates. Bioinformatics. 2011; 27:2076–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kryshtafovych A., Fidelis K., Moult J.. CASP10 results compared to those of previous CASP experiments. Proteins. 2014; 82:164–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M.. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993; 26:283–291. [Google Scholar]
- 41. Chen V.B., Arendall W.B., Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Richardson D.C.. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hooft R W., Vriend G., Sander C., Abola E.E.. Errors in protein structures. Nature. 1996; 381:272–272. [DOI] [PubMed] [Google Scholar]
- 43. Kellogg E.H., Leaver‐Fay A., Baker D.. Role of conformational sampling in computing mutation‐induced changes in protein structure and stability. Proteins. 2011; 79:830–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schymkowitz J., Borg J., Stricher F., Nys R., Rousseau F., Serrano L.. The FoldX web server: an online force field. Nucleic Acids Res. 2005; 33:W382–W388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kellogg E.H., Leaver‐Fay A., Baker D.. Role of conformational sampling in computing mutation‐induced changes in protein structure and stability. Proteins. 2011; 79:830–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bednar D., Beerens K., Sebestova E., Bendl J., Khare S., Chaloupkova R., Prokop Z., Brezovsky J., Baker D., Damborsky J.. FireProt: energy-and evolution-based computational design of thermostable multiple-point mutants. PLoS Comput. Biol. 2015; 11:e1004556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chaloupková R., Sykorova J., Prokop Z., Jesenska A., Monincova M., Pavlova M., Tsuda M., Nagata Y., Damborsky J.. Modification of activity and specificity of haloalkane dehalogenase from Sphingomonas paucimobilis UT26 by engineering of its entrance tunnel. J. Biol. Chem. 2003; 278:52622–52628. [DOI] [PubMed] [Google Scholar]
- 48. Nagata Y., Prokop Z., Marvanova S., Sykorova J., Monincova M., Tsuda M., Damborsky J.. Reconstruction of mycobacterial dehalogenase Rv2579 by cumulative mutagenesis of haloalkane dehalogenase LinB. Appl. Environ. Microbiol. 2003; 69:2349–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.