Abstract
The success of widely used oligonucleotide-based experiments, ranging from PCR to microarray, strongly depends on an accurate design. The design process involves a number of steps, which use specific parameters to produce high quality oligonucleotides. Oli2go is an efficient, user friendly, fully automated multiplex oligonucleotide design tool, which performs primer and different hybridization probe designs as well as specificity and cross dimer checks in a single run. The main improvement to existing oligonucleotide design web-tools is that oli2go combines multiple steps in an all-in-one solution, where other web applications only accomplish parts of the whole design workflow. Especially, the oli2go specificity check is not only performed against a single species (e.g. mouse), but against bacteria, viruses, fungi, invertebrates, plants, protozoa, archaea and sequences from whole genome shotgun sequence projects and environmental samples, at once. This allows the design of highly specific oligonucleotides in multiplex applications, which is further assured by performing dimer checks not only on the primers themselves, but in an all-against-all fashion. The software is freely accessible to all users at http://oli2go.ait.ac.at/.
INTRODUCTION
Oligonucleotides are short, single-stranded nucleic acids used for an increasing variety of biological experiments. They are essential components in various applications such as polymerase chain reaction (PCR), microarrays, fluorescence in situ hybridization (FISH) and other DNA-based technologies. Independent of the use case, the success of oligonucleotide-based experiments strongly depends on an accurate design and appropriate parameter selection. The design process involves several steps, which use specific parameters to produce oligonucleotides for conducting high quality experiments. Combining the recommendations from different publications, the following factors need to be addressed during the design process: oligonucleotide length and melting temperature (Tm), primer pairing, amplification product size, secondary structure formation, sensitivity and specificity of subsequent reactions (1–4). The length of oligonucleotides is a critical factor for biological experiments because specificity and Tm depend on this physical parameter (2). It directly influences the prediction of Tm delivered by different calculation models. However, the model needs to be selected carefully, as errors in Tm estimation may negatively influence the experiment outcome, e.g. no or non-specific amplification of DNA duplexes (5). Bakhtiarizadeh et al. compared the most common Tm calculator tools for PCR oligonucleotide design showing that Primer3 Plus and Primer-BLAST offer the most precise algorithms for an accurate Tm prediction (6–8). The primer pairing parameter needs to consider the size of the PCR product, a minimal Tm difference between the primers, or matched ΔG values (1,4,9). However, it should be noted that the enthalpy change upon primer binding is sequence dependent. This means that primers with the same Tm can bind differently to the target due to different hybridization behaviors. Therefore, it is better to match ΔG values instead of Tm (9). As oligonucleotides are single stranded, they can form intramolecular secondary structures, such as hairpin loops, but also intermolecular structures, such as primer dimers (10). Both cases should be avoided, as they may lead to false-negative signals in the experiment (10).
In order to increase the possibility of achieving a specific amplification reaction, the sequence of the amplified product of the primers needs to be unique in comparison to other templates (2). Moreover, hybridization probes must bind exclusively to the target in order to prevent cross-hybridization to non-targets (11). In summary, the final set of primers or probes strongly depends on the selected models and parameters. To date, a high number of web-tools for oligonucleotide designing are available (7,8,12–15). However, none of them covers the whole workflow (Supplementary Table S1). Each tool differs in the used design pipeline and input parameters. Primer3 involves singleplex primer and probe design for one target sequence but does not offer a specificity check for primers and probes. PrecisePrimer performs primer design for PCR primers involving useful pre-set options for different polymerase buffers and batch design, but lack options for probe design, cross dimerization and specificity checks. MFEprimer performs a primer dimer and specificity check, which implements the nearest-neighbor model to evaluate the binding stability for multiplex reactions, but misses an oligonucleotide design step (16). Furthermore, the specificity can only be checked using one background database derived from a relatively small pool of species. In addition, the primer dimer formation calculation is limited to maximal 50 primers per run. Here we introduce oli2go, a web-based fully automated multiplex oligonucleotide design tool for the design of highly specific primers and hybridization or ligation probes targeting bacteria, viruses, fungi, invertebrates, plants, protozoa, archaea as well as sequences from whole genome shotgun sequence projects and environmental samples. Oli2go combines all essential steps for a high quality probe and primer design for a variety of biological experiments in an all-in-one solution. A 45-plex assay targeting antibiotic resistance genes was successfully designed using oli2go and experimentally tested to evaluate the performance of the software.
METHODS AND IMPLEMENTATION
The workflow of oli2go is illustrated in Figure 1. The following subsections describe the main features of each step in detail.
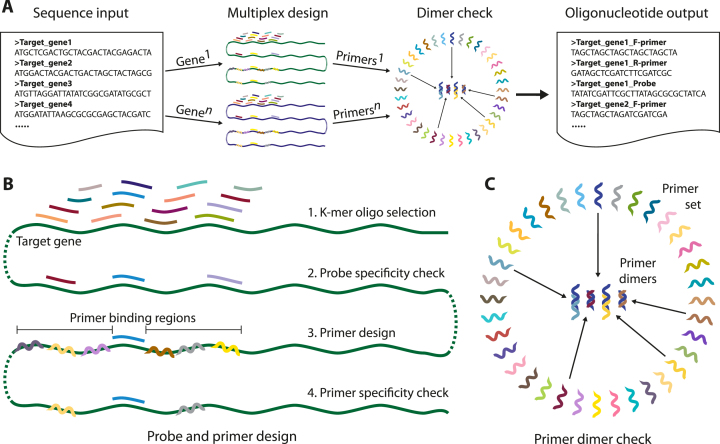
Figure 1.
An overview of the oli2go software. (A) Illustrates the workflow starting with the input of n DNA sequences, followed by the multiplex design, which is performed independently for each input sequence. Subsequently, a primer dimer check is performed using all primers produced in the multiplex design. The main output contains primers and probes for each input sequence in FASTA format. (B) Provides more details on the multiplex probe and primer design steps, which involve k-mer selections, Tm calculations, hairpin checks, probe and primer specificity checks as well as probe and primer pairing for each input sequence independently. (C) Visualizes the primer dimer check, where all primers targeting all input sequences, resulting from the preceding multiplex design, are checked for primer dimer formation.
Input
The home page of the web-based tool oli2go is used to upload the input sequences and to specify the design parameters. The sequences have to be provided in FASTA format, either by upload or by using a designated input box. The data should include a minimum of two sequences, as oli2go is designed to handle more than one sequence for multiplex reactions. Sequences containing ambiguous nucleotides are supported, but should be used carefully as each variable position within the sequence increases the number of computational steps. As specificity checks are performed for each possible variable position, an increase in run time will be the consequence. Designated input parameters are necessary for primer and probe design and dimerization checks. Dependent on the use case, the default parameters should be tuned meaningfully. Several papers describe in detail the selection of optimized parameters for primer and probe design (3,4,17,18). Additionally, oli2go supports the option to generate two-part hybridization probes used in ligation-based experiments.
File preparation
Input sequences are first aligned using the standalone version of the National Center for Biotechnology Information’s (NCBI) Basic Local Alignment Search Tool (BLAST) version 2.7.0+ and a comprehensive collection of databases (Table 1). These databases are a collection of sequence files covering >100 million sequences from bacteria, viruses, fungi, archaea, invertebrates, environmental samples, protozoa, plants and whole genome shotgun (WGS) projects, downloaded from the File Transfer Protocol (FTP) server of NCBI. The user selects databases for the file preparation and probe specificity checks. The BLAST results comprise all hits that show >90% sequence similarity to the query sequence and form the basis for the specificity check of the probes.
Table 1. NCBI database sources used for the probe specificity check.
| Source | Number of sequences | Database fraction |
|---|---|---|
| Bacteria | 7 658 345 | 7.55% |
| Environmental samples | 7 276 975 | 7.18% |
| Invertebrates | 27 651 271 | 27.27% |
| Patented sequences | 31 140 928 | 30.71% |
| Plants | 3 798 824 | 3.75% |
| Viruses | 1 837 439 | 1.81% |
| Archaea | 38 310 | 0.04% |
| Fungi | 3 889 143 | 3.84% |
| Protozoa | 3 880 518 | 3.83% |
| WGS project sequences | 14 220 046 | 14.02% |
| Total amount of sequences | 101 391 799 | 100.00% |
The number of sequences and their share of the entire data pool are listed.
Primer and probe selection
The selection of primers and probes starts with the creation of k-mers, ranging from the minimum user-defined primer and probe size to the maximum one, using a step size of 1. Afterward, the Tm is calculated for each k-mer (16,19). Candidates where the Tm is within the defined range are then checked for hairpin formation. The hairpin check is implemented using Primer3’s nucleotide thermodynamic alignment tool ntthal (12). This software uses the tables of thermodynamic parameters suggested by SantaLucia to calculate the secondary structure Tm and ΔG value of the most stable duplex (16). Oligonucleotides are accepted if their secondary structure Tm and ΔG value are below the user-defined thresholds.
Probe specificity check
The probe specificity check is one of the key features of oli2go. This step analyzes each possible probe candidate with BLAST against the user-defined databases (Table 1). The resulting alignment hits are compared to the target sequence hits generated in the file preparation workflow step. Only probes that bind to the same sequences as their target sequence, will be accepted.
Primer definition and specificity check
The specific probes resulting from the preceding specificity check are used to find possible forward and reverse primer candidates that flank the hybridization oligonucleotide. The detection capability of the probe is dependent on the specificity of the associated primers and the preceding DNA amplification reaction. Oli2go will output eligible primer pairs (each with one forward and reverse primer) that generate a product within the defined size range, do not form any secondary structures with each other, and show minimum difference in ΔG values. The primer specificity check is performed to minimize the risk of primer binding to human background DNA. Primer candidates are aligned using the Burrows-Wheeler Aligner (BWA) to the human reference genome downloaded from the NCBI FTP server (20).
Primer dimer check
The cross dimer or primer dimer check is an important design step to optimize primer performance in multiplex reactions. Oli2go uses Primer3’s ntthal and the user-defined ΔG and Tm values to check for cross dimerization. Specific forward and reverse primer pairs resulting from the preceding design task form the input for this last workflow step. It starts with the input sequence that has fewest specific primers. These primers are checked against all other possible primers of the other input sequences. The first results involve primer pairs which do not exceed the cross dimerization thresholds. If the results contain at least one primer pair for each sequence, each one is checked against the other primers in the results. Finally, for each input sequence one primer pair forming no cross dimerization with all other sequences is returned.
Output
The output is presented on a separate web-page and includes a table showing the resulting primers and probes, their Tm’s, product sizes, hairpin Tm’s, and ΔG values. The table also contains web links to NCBI’s online BLAST and Primer-BLAST to perform additional analysis. This table can also be downloaded as comma-separated values (CSV) file. Furthermore, primer and probe sequences as well as the initial input sequences are available in FASTA format. The used design parameters can be downloaded as text file.
Implementation
The software workflow runs on a Linux server (64 CPUs, 256GB RAM). The main software packages used for the implementation are BLAST 2.7.0+, ntthal (which is part of Primer3 2.3.7), BWA, and Python 2.7 together with the Biopython library (21). In order to maximize the utilization of the server resources, most of the workflow steps are running in parallel using multithreading. The highly responsive user interface is implemented using Bootstrap 3.3.7 and enables the user to use oli2go on almost any device capable of entering the internet via browser ranging from Laptops, Tablets to Smartphones. Oli2go is freely accessible to all users at http://oli2go.ait.ac.at/.
EVALUATION AND DISCUSSION
Oli2go represents an efficient, accurate, user friendly design tool for multiplex oligonucleotide projects. Consequently, researchers do no longer have to use multiple tools, each having their own requirements, input formats, and parameters, to find suitable primers and probes for their experiments. Furthermore, oli2go significantly reduces the number of manual, error prone steps.
Run time evaluation
Oli2go has been successfully tested for the design of an 45-plex assay targeting antibiotic resistance genes. The computationally most expensive steps include BLAST analysis for a high number of specificity checks that have to be computed per target sequence (>100). Alternative approaches to improve the performance of the specificity check have been evaluated without success. String searching methods and data structures, such as suffix trees and Burrows-Wheeler-Transformation have been tested, but failed due to the high data amount of the background databases and the related memory usage (20). Also applications for sequence assembly, such as Kraken, could not be used, as the oligonucleotide sizes are usually too small for the implemented algorithms (22). Although BLAST was not intended for specificity checks of oligonucleotides due to the lack of, e.g. thermodynamic models in the algorithm, it showed the best performance considering run time, memory usage, and result quality. More precise algorithms using thermodynamic models or molecular dynamics simulations cannot handle the large number of possible oligonucleotide interactions in multiplex applications within a realistic time frame (16,23). Comparing the run time of oli2go with an example of a conventional oligonucleotide design workflow, we can prove that oli2go is significantly faster (Figure 2). During a conventional primer design process, the user has to use several independent programs and sequence databases. Parsing the output and setting the input data between the used tools need to be done manually, resulting in an extremely inefficient work process. Also the manual examination of all specificity check results using online BLAST is impossible to do within in a reasonable time. It does not even involve as many sequence databases as oli2go for one run. The cross dimer check using MFEprimer is another case where the results should be handled with care, as it is not as accurate as the cross dimer check provided with oli2go (Supplementary Table S2). Even with these limitations in background data and thermodynamic alignments, the manual design workflow (Figure 2) still showed worse performance than oli2go.
Figure 2.
Schematic illustration of the run time evaluation. (A) shows the run times of oli2go compared to the conventional manual design workflow shown in B). The manual workflow was simulated by means of three different scenarios. The three scenarios differ by the number of required primer dimer checks. In the most unlikely of these three scenarios, no primer dimers were identified after the first run of MFEprimer. (B) illustrates the manual design workflow. It starts with the manual input of target sequences to Primer3. As Primer3 can only handle one sequence, this step has to be done for each target at least once. The resulting probes are merged and submitted to online BLAST where the probe specificity check is performed. The results of each probe need to be checked individually by the user. The exclamation mark indicates that certain input sequences result in more than 100 000 alignments making it almost impossible to examine all hits using online BLAST. The primer specificity check is performed running Primer-BLAST on each primer pair separately. After the visual evaluation of the results, specific primer candidates are merged in a single file and checked for cross dimerization using MFEprimer. The final exclamation mark indicates that the MFEprimer cross dimer check results deviate from oli2go because MFEprimer uses a downgraded thermodynamical model (Supplementary Table S2). The arrows in the figure highlight the iterative nature of the oligonucleotide design process which is responsible for the high work load in comparison to the fully automated oli2go workflow.
Experimental evaluation
A 45-plex assay targeting the 45 most common antibiotic resistance genes was designed with oli2go as proof of principle. Sequences from the Comprehensive Antibiotic Resistance Database (CARD) were used as input for the probe and primer design (24). The primer performance was evaluated using PCR and agarose gel analysis (Supplementary Figure S1). The probes were tested on a microarray platform resulting in highly specific signals (Supplementary Figure S2).
Outlook
Oli2go has high potential in improving assay design of oligonucleotide-based experiments. It is known, that computational automation and big data handling, as performed by oli2go, become more and more important for biologists (25). Oli2go has already proven its usability and service to the scientific community during a successful testing period of 6 months performed by an external project team (>10 people, >100 input sequences). Due to the highly positive outcome of this test and the increasing demand for automated multiplex oligonucleotide design solutions, we are consistently working on further improvements of oli2go.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Union’s Horizon 2020 research and innovation program [634137]. Funding for open access charge: H2020 [634137].
Conflict of interest statement. None declared.
REFERENCES
- 1. Schretter C., Milinkovitch M.C.. Oligonucleotide design by multilevel optimization. Unit Evol. Genet. 2005; 5:1–9. [Google Scholar]
- 2. Abd-Elsalam K.A. Bioinformatic tools and guideline for PCR primer design. Afr. Jo. Biotechnol. 2003; 2:91–95. [Google Scholar]
- 3. Rychlik W. Selection of primers for polymerase chain reaction. Methods Mol. Biol. 1993; 15:31–40. [DOI] [PubMed] [Google Scholar]
- 4. Dieffenbach C.W., Lowe T.M., Dveksler G.S.. General concepts for PCR primer design. PCR Methods Appl. 1993; 3:S30–S37. [DOI] [PubMed] [Google Scholar]
- 5. Steger G. Thermal denaturation of double-stranded nucleic acids: prediction of temperatures critical for gradient gel electrophoresis and polymerase chain reaction. Nucleic Acids Res. 1994; 22:2760–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bakhtiarizadeh M.R., Najaf-Panah M.J., Mousapour H., Salami S.A.. Versatility of different melting temperature (Tm) calculator software for robust PCR and real-time PCR oligonucleotide design: A practical guide. Gene Rep. 2016; 2:1–3. [DOI] [PubMed] [Google Scholar]
- 7. Untergasser A., Nijveen H., Rao X., Bisseling T., Geurts R., Leunissen J.A.M.. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007; 35:W71–W74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T.L.. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012; 13:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. SantaLucia J. Physical principles and visual-OMP software for optimal PCR design. PCR Primer Des. 2007; 402:3–33. [DOI] [PubMed] [Google Scholar]
- 10. SantaLucia J. Jr, Hicks D.. The thermodynamics of DNA structural motifs. Annu. Rev. Biophys. Biomol. Struct. 2004; 33:415–440. [DOI] [PubMed] [Google Scholar]
- 11. Ilie L., Mohamadi H., Golding Geoffrey B., Smyth W.F.. BOND: basic oligonucleotide design. BMC Bioinformatics. 2013; 14:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B.C, Remm M., Rozen S.G.. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012; 40:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pauthenier C., Faulon J.-L.. PrecisePrimer: an easy-to-use web server for designing PCR primers for DNA library cloning and DNA shuffling. Nucleic Acids Res. 2014; 42:W205–W209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Qu W., Zhou Y., Zhang Y., Lu Y., Wang X., Zhao D., Yang Y., Zhang C.. MFEprimer–2.0: a fast thermodynamics-based program for checking PCR primer specificity. Nucleic Acids Res. 2012; 40:W205–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pandey R.V., Pulverer W., Kallmeyer R., Beikircher G., Pabinger S., Kriegner A., Weinhäusel A.. MSP-HTPrimer: a high-throughput primer design tool to improve assay design for DNA methylation analysis in epigenetics. Clin. Epigenet. 2016; 8:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. SantaLucia J. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:1460–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bustin S., Huggett J.. qPCR primer design revisited. Biomol. Detect. Quant. 2017; 14:19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Robertson J.M., Walsh-Weller J.. An introduction to PCR primer design and optimization of amplification reactions. Forensic DNA Profil. Protoc. 1998; 98:121–154. [DOI] [PubMed] [Google Scholar]
- 19. Rychlik W., Spencer W.J., Rhoads R.E.. Optimization of the annealing temperature for DNA amplification in vitro. Nucleic Acids Res. 1990; 18:6409–6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li H., Durbin R.. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cock P.J.A., Antao T., Chang J.T., Chapman B.A., Cox C.J., Dalke A., Friedberg I., Hamelryck T., Kauff F., Wilczynski B. et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009; 25:1422–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wood D.E., Salzberg S.L.. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014; 15:R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Šulc P., Romano F., Ouldridge T.E., Rovigatti L., Doye J.P.K., Louis A.A.. Sequence–dependent thermodynamics of a coarse-grained DNA model. J. Chem. Phys. 2012; 137:135101. [DOI] [PubMed] [Google Scholar]
- 24. Jia B., Raphenya A.R., Alcock B., Waglechner N., Guo P., Tsang K.K., Lago B.A., Dave B.M., Pereira S., Sharma A.N. et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2016; 45:D566–D573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marx V. Biology: The big challenges of big data. Nature. 2013; 498:255–260. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.