Abstract
Background
Salvia splendens Ker-Gawler, scarlet or tropical sage, is a tender herbaceous perennial widely introduced and seen in public gardens all over the world. With few molecular resources, breeding is still restricted to traditional phenotypic selection, and the genetic mechanisms underlying phenotypic variation remain unknown. Hence, a high-quality reference genome will be very valuable for marker-assisted breeding, genome editing, and molecular genetics.
Findings
We generated 66 Gb and 37 Gb of raw DNA sequences, respectively, from whole-genome sequencing of a largely homozygous scarlet sage inbred line using Pacific Biosciences (PacBio) single-molecule real-time and Illumina HiSeq sequencing platforms. The PacBio de novo assembly yielded a final genome with a scaffold N50 size of 3.12 Mb and a total length of 808 Mb. The repetitive sequences identified accounted for 57.52% of the genome sequence, and 54,008 protein-coding genes were predicted collectively with ab initio and homology-based gene prediction from the masked genome. The divergence time between S. splendens and Salvia miltiorrhiza was estimated at 28.21 million years ago (Mya). Moreover, 3,797 species-specific genes and 1,187 expanded gene families were identified for the scarlet sage genome.
Conclusions
We provide the first genome sequence and gene annotation for the scarlet sage. The availability of these resources will be of great importance for further breeding strategies, genome editing, and comparative genomics among related species.
Keywords: annotation, evolution, reference genome, Salvia splendens, scarlet sage; single-molecule real-time sequencing
Data Description
Background information
Salvia L., with nearly 1,000 species of shrubs, herbaceous perennials, and annuals, is the largest genus in the mint family (Lamiaceae: Nepetoideae: Mentheae: Salviinae) [1–4]. The genus is widely distributed throughout the world. Many species of this genus are extensively used for culinary purposes, essential oil production, and Chinese herbal remedies, such as S. officinalis [3] and S. miltiorrhiza (Danshen). Additionally, they are used as ornamental plants valued for their flowers and for their aromatic foliage, such as S. splendens (Fig. 1a-k).
Figure 1:
Images of the scarlet sage, Salvia splendens. (a-b) Flowers of the sequenced cultivar of S. splendens, “Aoyunshenghuo (Olympic flame).” (c) The scarlet sage with different flower colors in bedding. (d-k) The scarlet sage with flowers of different pure colors, or bi-colors.
Salvia splendens (National Center for Biotechnology Information [NCBI] taxon ID:180675), scarlet or tropical sage, is a herbaceous perennial species that is native to Brazil. While it is a perennial in warmer climate zones, it grows as an annual in cooler areas. Salvia splendens,characterized by its dense flowers, wide variation of colors (e.g., scarlet, purple, pink, blue, lavender, salmon, yellow green, white, and bicolor), and long-lasting flowering (3–9 weeks or longer), is a very popular bedding plant that is widely cultivated in public gardens all over the world [3, 5]. Additionally, S. splendens can provide outstanding visual effects when grown in beds, borders, and containers; and its long life span ranges from late spring to the occurrence first frost. Furthermore, the flower is easy to maintain and fairly free of pests and diseases due to Lamiaceae's characteristic insect-repellent fragrance content [6]. The plant blends nicely with other annuals and perennial plants for the best visual effects in an ensemble setting. In addition, this plant requires little deadheading and it attracts various butterfly species. Salvia splendens is a prolific and durable bloomer, thrives in full sun, and survives in a large range of soil moisture regimes.
Traditional breeding activities using phenotypic selection as well as performing targeted variety hybridizations between elite cultivars have resulted in a large number of new cultivars with different performances regarding flowering characteristics (e.g., related to color, flowering time, and flowering period), individual growth performance, height, and tolerance to moisture and temperature extremes. However, little is known about the molecular mechanisms underlying such economically important characteristics for ornamental varieties. To date, few genetic markers [7] are available for marker-assistant breeding and genetic modification.
Here, we present the first high-quality genome assembly for S. splendens with a hybrid assembly strategy using Pacific Biosciences (PacBio) single-molecule real-time (SMRT) and Illumina HiSeq short-read sequencing platforms. The genome assembly, its structural and functional annotation, provide a valuable reference for the genomic dissection of the phenotypic variation in Salvia and new breeding strategies. This reference genome could also be used in comparative genomics with the recently released Salvia genome (S. miltiorrhiza) [8,9] and the mint genome (Mentha longifolia) [10] to study the biosynthesis of important fragrant and medicinal compounds.
Plant material
We chose the elite variety S. splendens, “Aoyunshenghuo (Olympic flame)” (Fig. 1a-b), for whole-genome sequencing. The variety was originally developed by multiple rounds of selection/selfing of one hybrid to obtain this inbred line. This cultivar is characterized by resistance to drought and high temperatures and by improved performance related to a longer flowering period. It is well adapted to climate conditions across North China and therefore grows well in Beijing. Because of the high homozygosity obtained due to advanced generation selfing, this cultivar shows no phenotypic segregation, a characteristic of important commercial value. Seeds of this cultivar were provided by the Beijing Institute of Landscape Architecture germplasm bank.
PacBio SMRT sequencing
High-quality high-molecular-weight genomic DNA was extracted from leaves of two soil-grown seedlings (huo1 and huo1_1) following ∼20 kb SMRTbell Libraries” protocol [11]. Plants for DNA extraction were placed in the dark for 48 hours before harvesting the leaf material. DNA was purified using the Mobio PowerClean Pro DNA Clean-Up Kit; quality was assessed using standard agarose gel electrophoresis and Thermo Fisher Scientific Qubit fluorometry. Genomic DNA was sheared to a size range of 15–40 kb using either AMPure beads (Beckman Coulte) or g-TUBE (Covaris) and enzymatically repaired and converted into SMRTbell template libraries as recommended by PacBio. Following this procedure, hairpin adapters were ligated following exonuclease-based digestion (of the remaining damaged DNA fragments and those fragments without adapters at both ends). Subsequently, the resulting SMRTbell templates were size selected using Blue Pippin electrophoresis (Sage Sciences). Templates ranging from 15 to 50 kb were sequenced on a PacBio RS II instrument using P6-C4 sequencing chemistry (25 SMRT cells for individual huo1) and on a PacBio Sequel instrument using S/P2-C2 sequencing chemistry (8 SMRT cells for the other individual, huo1_1). A total of 8,858 ,116 PacBio post-filtered reads were generated. This produced 65,962,079,028 bp (roughly 82x the assembled genome) of single-molecule sequencing data, with an average read length of 7,446 bp (Supplementary Fig. S1 and Table S1).
Illumina short-read sequencing
DNA was extracted from leaf tissue of the same soil-grown seedlings (huo1 and huo1_1) using the Qiagen DNeasy Plant Mini Kit. Two 500-bp paired-end (PE) libraries (huo1 and huo1_1) were prepared using the NEBNext Ultra DNA Library Prep Kit for Illumina sequencing with an Illumina HiSeq X Ten machine. Short reads were processed with Trimmomatic v0.33 (Trimmomatic, RRID:SCR_011848) [12,13] and Cutadapt v1.13 (cutadapt, RRID:SCR_011841) [14,15] to remove adapter sequences and leading and trailing bases with a quality score below 20 and reads with an average per-base-quality of 20 over a 4-bp sliding window. Reads <70 nucleotides in length after trimming were removed from further analysis. A total of 265.53 million reads were generated. This produced 36.83 Gb (roughly 40x the assembled genome) of raw sequencing data, with an average cleaned read length of 137 bp (Supplementary Table S1).
Estimation of genome size, heterozygosity, and repeat content
All generated PacBio reads were filtered and corrected with Canu v1.5 (Canu, RRID:SCR_015880) [16]; thereafter, Jellyfish (Jellyfish, RRID:SCR_005491) [17] was used to count the occurrence of k-mers based on the processed data. Finally, gce 1.0.0 [18] was used to estimate the overall characteristics of the genome, such as genome size, repeat contents, and heterozygous rate. In this study, 22,117,819,357 k-mers were generated, and the peak k-mer depth was 31 (Supplementary Fig. S2). The genome size was estimated to be approximately 711 Mb (Supplementary Table S2), and the final cleaned data corresponded to the coverage of about 33-fold. Repeat and error rates were estimated to be 47.99% and 0.27%, respectively, and the heterozygosity rate was 0.06%.
De novo genome assembly
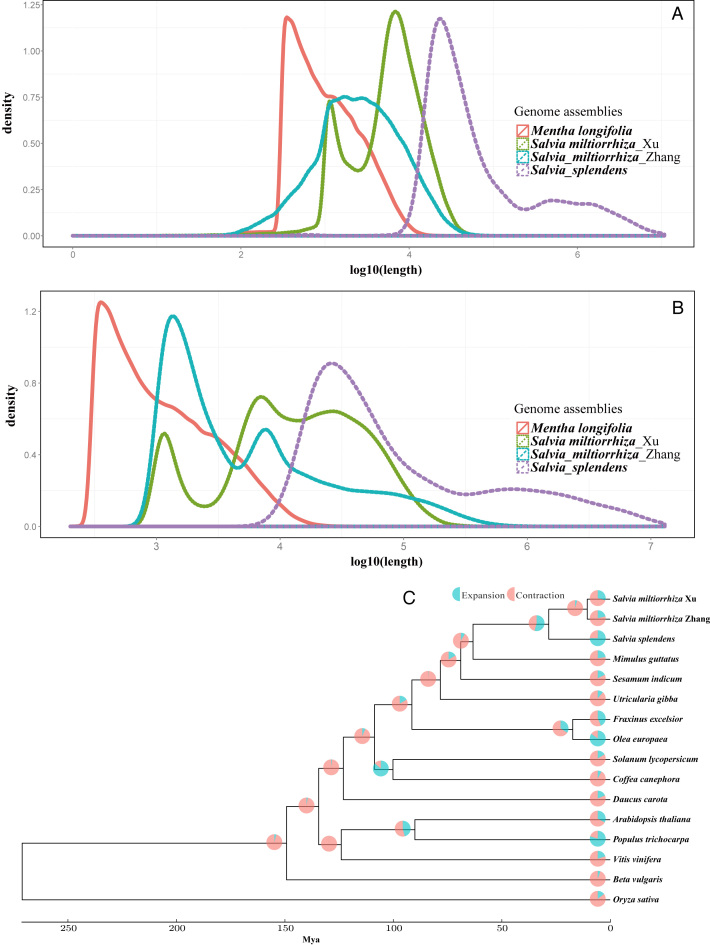
Thede novo assembly was conducted as follows in a progressive manner. First, primary assemblies were generated from PacBio long reads of the 31 Gb from the “huo1” sequenced individual by four overlap-layout-consensus–based assemblers, Canu (produced assembly v0.1), MECAT 1.1 (assembly v0.2) [19], FALCON v0.7 (Falcon, RRID:SCR_016089) [20,21] after Canu correction (v0.3), and SMARTdenovo 1.0.0 [22] after Canu correction (v0.4) (Supplementary Table S3). Based on the size of the assembled genome, the total number of assembled contigs, N50, the L50, maximum length of the contigs, and the completeness of the genome assembly as assessed by using Benchmarking Universal Single-Copy Orthologs (BUSCO) criteria v2.0.1 (BUSCO, RRID:SCR_015008) [23] (1,440 single-copy orthologs of the Viridiplantae database) with the BLAST E-value cutoff of 10–5, the assembly (v0.1) from Canu was chosen for further polishing and scaffolding. In this selected primary assembly, the assembled genome size was 808 Mb distributed across 2,306 contigs with N50 of 2.06 Mb, L50 of 109, and maximum contig length of 8.88 Mb. We also confirmed, on average, 92.1% gene completeness in this assembly (Supplementary Table S3). In the following steps, the arrow algorithm v2.2.1 [24] was used to further improve the assembly based on PacBio long reads (v1.0), after which SSPACE-LongRead 1.1 [25] and SSPACE-standard 3.0 (SSPACE, RRID:SCR_005056) [26] were used for subsequent scaffold assembly based on PacBio long reads of 35 Gb from the second sequenced individual “hou1_1” and Illumina short reads, respectively. Finally, after scaffold processing and subsequent gap filling with SOAPdenovo and GapCloser (GapCloser, RRID:SCR_015026) [27] (v1.1), arrow v2.2.1 algorithm (based on PacBio long reads) and Pilon (Pilon, RRID:SCR_014731) (based on Illumina short reads, and run two times, parameters for Pilon: –changes –diploid –dumpreads), we obtained the final genome assembly (v1.2). Mapping of Illumina reads was done using Bowtie2 v2.3.0 (Bowtie, RRID:SCR_005476) [28]. We detected 400,170 single-nucleotide polymorphisms (SNPs), 96,854 insertions, and 62,637 deletions, respectively, for the first pilon run. Subsequently, there was a greatly decreased number of variants for the second pilon run ( 40,465 SNPs, 6,935 insertions, and 9,976 deletions, respectively). In this final assembly, we gained an assembled genome size of 808 Mb characterized by 2,204 contigs and 1,525 scaffolds (with contig N50 of 2.27 Mb and scaffold N50 of 3.12 Mb) and by gene completeness of 92.2% (Table 1 and Supplementary Table S3). This assembly represents the highest continuity and completeness among the recently released genome assemblies for the Salvia genus [8,9] and for mint [10], as it was examined by length distribution plotting of contigs and scaffolds, as shown in Fig. 2A, B.
Table 1:
Statistics of the final genome assembly of the scarlet sage
| Contig | Scaffold | |||
|---|---|---|---|---|
| Size (bp) | Number | Size (bp) | Number | |
| Total size | 807,514,799 | - | 809,159,598 | - |
| Total number | - | 2,204 | - | 1,525 |
| N10 | 6,529,455 | 10 | 8,157,631 | 9 |
| N50 | 2,267,074 | 100 | 3,123,266 | 73 |
| N90 | 265,262 | 456 | 433,303 | 324 |
| Max. | 10,812,588 | - | 12 , 944,193 | - |
| Min. | 500 | - | 9,495 | - |
| Mean | 366,386 | - | 530,596 | - |
| Median | 38,049 | - | 48,557 | - |
| Gap | - | - | 1,644,799 (0.2%) | 679 |
| GC (Guanine Cytosine) content | 38.84% | - | 38.76% | - |
Figure 2:
Quality of scarlet sage genome assembly and the phylogenomic inferences. Quality was assessed by comparing the scarlet genome with the recently released genomes of related species. Length distribution of contigs (A) and scaffolds (B). (C) Phylogenetic tree, divergence time, and profiles of gene families that underwent expansion or contraction. Salvia miltiorrhiza Zhang [15] and S. miltiorrhiza Xu [15] are two genome assemblies reported for S. miltiorrhiza.
DNA repeats annotation
RepeatModeler v1.0.10 (RepeatModeler, RRID:SCR_015027) [29] was used to de novo identify and classify repeat families in the genome assembly. Subsequently, the outputs from the RepeatModeler and RepBase [30] library were combined and used as the repeat library for subsequent RepeatMasker (RepeatMasker, RRID:SCR_012954) (v4.0.7, rmblast-2.2.28) [31] analyses, which was used to fully discover and identify repeats within the assembled genome. In summary, 57.52% of the genome was annotated as repeats, among which we found 1.08% simple repeats and 40.35% known transposable elements (TE). Long terminal repeats (LTRs) constituted the greatest proportion (26.49%) of the genome, and DNA TE made up 11.91% of the genome. Gypsy (18.15% of the genome) and Copia (7.92%) TEs were the largest components of LTRs. The results of repeat annotations are summarized in Supplementary Table S4.
RNA sequencing, transcriptome assembly, and functional annotation
RNA was extracted from the two cultivated lines with different flower colors (red and purple) using tissue obtained from roots, shoots, leaves, calyxes, and corollas. Frozen tissue from all samples was ground manually using a mortar and pestle, and RNA was isolated using the NEBNext Poly(A) mRNA Magnetic Isolation Module. RNA quality was assessed using an Agilent 2100 BioAnalyzer. Sequencing libraries were prepared using the NEBNext Ultra RNA Library Prep Kit for Illumina; 150 bp PE sequencing was performed using an Illumina HiSeq X Ten.
A total of 1,344 million raw reads from RNA sequencing were processed by Trimmomatic and Cutadapt and aligned to the genome assembly with HiSat2 v2.1.0 (HiSat2, RRID:SCR_015530) [32]. Base quality was checked with FastQC (FastQC, RRID:SCR_014583) [33] before and after data cleaning. Respective statistics of RNA sequencing data are shown in Supplementary Table S1. Reference genome–guided transcriptome assemblies were independently prepared with Cufflinks v2.1.1 (Cufflinks, RRID:SCR_014597) [34], StringTie v1.3.3b (StringTie, RRID:SCR_016323) [35], and Trinity v2.0.6 (Trinity, RRID:SCR_013048) [36]. De novo assembly was generated using Trinity. Then, transcriptome assemblies were combined and further refined using CD-HIT v4.6 [37], and finally, 192,169 unique transcripts were obtained. The summary of the transcriptome assemblies is shown in Supplementary Table S5.
AUGUSTUS v3.2.3 (Augustus, RRID:SCR_008417) [38] was used for ab initio gene prediction, using model training based on coding sequences from Arabidopsis thaliana and S. miltiorrhiza (with two sets of proteins from independent genome annotation [8,9]). Then, transcripts from RNA sequencing were aligned to the repeat-masked reference genome assembly with BlastN and TblastX from BLAST v2.2.28+ (NCBI BLAST, RRID:SCR_004870) [39] (E-value cutoff of 10–5). Protein sequences from A. thaliana and S. miltiorrhiza were aligned to the repeat-masked reference genome assembly with BlastX (E-value cutoff of 10–5). After optimization with Exonerate v2.4.0 [40,41], gene model predictions were prepared using the MAKER package v2.31.9 (MAKER, RRID:SCR_005309) [42] provided within AUGUSTUS. To assess the quality of the gene prediction, annotation edit distance (AED) scores were generated for each of the predicted genes as part of the MAKER pipeline. The putative function for each identified gene was assessed by performing a BLAT (BLAST-like alignment tool) (BLAT, RRID:SCR_011919) [43] search of the peptide sequences against the UniProt database (UniProt, RRID:SCR_002380) [44]. Protein annotation against PFAM (Pfam, RRID:SCR_004726) [45] and InterProScan (InterProScan, RRID:SCR_005829) [46] ID were also conducted using the scripts provided in the MAKER package. The completeness of gene annotation was checked using BUSCO (1,440 single-copy orthologs of the Viridiplantae database) with a BLAST E-value cutoff of 10−5.
A total of 54,008 genes could be predicted, with average lengths of gene regions, genes (exons and introns), coding DNA sequence, and exons of 3,430.43 bp, 1,696.34 bp, 1,293.62 bp, and 265.94 bp, respectively (Supplementary Table S6). The comparisons among genomes from related species regarding lengths of genes, exons, and introns are shown in Fig. 2. The distribution of AED tagged by MAKER is shown in Supplementary Fig. S3, in which about 97% of the annotated genes ( 52,338 genes) had an AED <0.5 (Supplementary Table S6), indicating that the annotation is well supported. The result from BUSCO assessment of the quality of the genome assembly and annotation is shown in Supplementary Table S7. We identified 92.08% of the universal single-copy genes (1,326 genes of the total 1,440 genes), supporting the high quality of the genome assembly. Among the 1,326 BUSCO conserved single-copy genes detected in the scarlet genome, 466 genes were found to be single copies, while 860 genes were duplicated (Supplementary Table S7).
The predicted genes were annotated against several functional databases, including the NCBI nonredundant protein database (NR; [47]), the Swiss-Prot protein database [48, 44], the Translated EMBL-Bank (part of the International Nucleotide Sequence Database Collaboration, TrEMBL, [49]) [44], the protein families database (Pfam; [50]), the Cluster of Orthologous Groups for eukaryotic complete genomes (KOG) database [51], the KO (the Kyoto Encyclopedia of Genes and Genomes, Orthology) database [52,53], and Gene Ontology (GO) [54,55]. It was found that 94.67% of all predicted genes could be annotated with the following protein related databases: NR (94.60%), Swiss-Prot (63.40%), TrEMBL (93.50%), Pfam (82.10%), KOG (90.05%), KO (37.40%), and GO (78.80%) (Supplementary Table S8).
Identification of orthologous genes and phylogenetic inference
To analyze gene families, we downloaded the protein sequences of 15 genome assemblies of 14 additional species (Salvia miltiorrhiza [8,9], Fraxinus excelsior [56], Olea europaea [57], Mimulus guttatus [58], Utricularia gibba [59], Sesamum indicum [60], Coffea canephora [61], Solanum lycopersicum [62], Daucus carota [63], Vitis vinifera [64], Arabidopsis thaliana [65], Populus trichocarpa [66], Oryza sativa [67], and Beta vulgaris [68]) (Supplementary Table S9). Orthologous and paralogous gene clusters were identified among species using OrthoMCL v2.0.9 [69]. Recommended settings were used for all-against-all BLASTP comparisons (Blast+ v2.3.056) [39] and OrthoMCL [26] analyses.
A total of 35,808 OrthoMCL families were built based on effective database sizes of all vs all BLASTP with an E-value of 10–5 and a Markov chain clustering default inflation parameter. We identified 1,306 gene families (3,797 genes) that were specific to the scarlet sage genome when compared with the other 15 genomes (Supplementary Table S10), and we detected 10,770 gene families that have expanded in the scarlet sage lineage using CAFE v4.0 [70,71] (Fig. 2C). The expanded gene families were enriched for 60 significant (q<0.05) GO terms of three functional categories, i.e., BP (Biological Process), CC (Cellular Component), and MF (Molecular Function)(Supplementary Table S11), and one KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway (amino acid metabolism) (Supplementary Table S12) significant at q <0.05. Also, 3,579 genes and 78 gene families were detected to be contracted and found to have rapidly evolved within the scarlet sage genome (Fig. 2C). Subsequently, 134 orthologous proteins among the 16 analyzed genomes were acquired and aligned with MUSCLE v3.8.31 (MUSCLE, RRID:SCR_011812) [72] using default settings. A maximum likelihood phylogenetic tree was then generated using the concatenated amino acid sequences in PhyML v3.0 (PhyML, RRID:SCR_014629) [73] with the GTR+G+I model. The divergence time was estimated with r8s v1.81 [74] and calibrated against the timing of divergence between A. thaliana and V. vinifera (124 Mya) [75] as well as against the A. thaliana and P. trichocarpa divergence time (90 Mya) [76]. The phylogenetic analysis identified the close relationship among the three Salvia genomes; their divergence time was estimated to be about 28.21 Mya (Fig. 2C).
Secondary metabolic pathways: gene annotations, gene clusters, and comparative genomics
The mint family is recognized as providing promising sources of bioactive secondary metabolites [77]. In fact, a diverse variety of bioactive secondary metabolites can be found with a wide range of pharmacological activities including antimicrobial, antispasmodic, carminative, antioxidant, antiulcer, cytoprotective, heptoprotective, cholagogue, chemo-preventive, anti-inflammatory, and antidiabetogenic. Here, we obtained enzymatic annotations for coding genes by using the E2P2 package v3.1 [78]. Then, we mapped genes to flavonoid and menthol biosynthesis pathways by querying the Plant Metabolic Network (v12.5) [79,80]. Regarding the flavonoid biosynthesis pathway, we found an abundance of genes encoding annotated enzymes in this pathway, especially of note the 41 genes for flavanone synthase I (EC: 1.14.11.9) (Supplementary Fig. S5 and Supplementary File 1). With respect to menthol biosynthesis, certain genes are still lacking annotations for enzymes such as (+)-pulegone reductase (EC: 1.3.1.81), (-)-isopiperitenone reductase (EC: 1.3.1.82), and menthol-dehydrogenase (lacking EC number) (Supplementary Fig. S6 and Supplementary File 1). However, this pathway mapping analysis provides a highly valuable reference for the genetic dissection of key metabolic genes for the scarlet sage.
The presence of metabolic gene clusters for secondary metabolic pathways is common in bacteria and filamentous fungi and is also widely reported in plants [81–83]. Using the newly created and robust computational tool kit, plantSMASH [84], we identified 85 gene clusters potentially related to secondary metabolic biosynthesis in the scarlet sage genome, as reported here, and 23 gene clusters in the S. miltiorrhiza genome [8]. The genomic position, gene composition, and functional annotation of the identified gene clusters are summarized in Supplementary Table S13 and Supplementary Files 2 and 3. The gene clusters were found to be potentially related to the biosynthesis of alkaloids, saccharides, polyketides, terpenes, and lignans. It was previously reported that physical clustering of terpene synthase genes (TPS) and cytochrome P450 mono-oxygenase genes is frequently associated with consecutive enzymatic actions in terpenoid biosynthesis [85]. Interestingly, we detected eight such gene clusters within the scarlet sage genome but none in the S. miltiorrhiza genome, which could be due, in part, to the draft status of the genome assembly for S. miltiorrhiza. Furthermore, significant gene co-expression across different organs was detected for one TPS gene and two of four P450 genes located in a single gene cluster (i.e., cluster 63; Supplementary Table S13 and Supplementary File 2). Evidence for moderate or significant co-expression among clustered genes was revealed and is shown in Supplementary File 2.
Based on the collinearity elucidated by former OrthoMCL analyses, a comparative genomic study between the scarlet sage and S. miltiorrhiza genomes revealed six pairs of gene clusters that share synteny between these two congeneric plants, and two blocks from the scarlet sage share synteny with one block from S. miltiorrhiza (Supplementary Fig. S7). Among the shared synteny blocks, four could be related to saccharide, one to lignan, and another to polyketide biosynthesis. The smaller number of gene clusters detected for S. miltiorrhiza and, subsequently, fewer shared synteny blocks of metabolic gene cluster between these two species may be partially attributed to the present state of the S. miltiorrhiza genome assembly, which is 100 times more fragmented than that of the scarlet sage. Thus, here, we provide a starting point for comparative genomics among plant species within the mint family.
In summary, we presented the draft assembly for the scarlet sage genome using a PacBio long-read dominated strategy that was responsible for obtaining the high-quality sequence assembly. Also, the almost complete homozygosity within the sequenced inbred line's genome was a key factor for the high continuity gained in this study. The novel genome data generated in the present study will provide a valuable resource for studying the molecular underpinnings of the various phenotypic variations found within Salviasp. and sets the foundation for molecular-informed breeding strategies and genome editing approaches for this valued ornamental flowering plant. Moreover, this genome assembly is useful for comparative genomic studies among related species.
Supplementary Material
2/21/2018 Reviewed
2/22/2018 Reviewed
5/12/2018 Reviewed
2/26/2018 Reviewed
ACKNOWLEDGEMENTS
This study was funded by the Beijing Key Laboratory of Greening Plants Breeding (NO. Z201605) and Fundamental Research Funds for the Central Universities (NO. YX2013-41).
Availability of supporting data
The genome assembly, annotations, and other supporting data are available via the GigaScience database GigaDB [86]. The raw sequence data have been deposited in the Short Read Archive under NCBI BioProject ID PRJNA422035.
Additional files
Figure S1. Length distribution of PacBio subreads.
Figure S2. K-mer frequency distribution at k-mer size of 17. A k-mer refers to an artificial sequence division of K nucleotides. From k-mer frequency, genomic characteristics (genome size, repeat structure and heterozygous rate) could be estimated. Peaks at depths of 31 and 62 were annotated with dash lines.
Figure S3. Distribution of AED scores from gene prediction. AED, Annotation Edit Distance, AED = 0 indicates perfect agreement between annotation and the evidence; AED = 1 indicates no evidence support for annotation.
Figure S4. Length distribution of annotated genes, exons and introns. a-c for annotated genes, exons and introns from different genome assemblies.
Figure S5. Flavonoid biosynthesis pathway. Flavonoid biosynthesis pathways by querying the Plant Metabolic Network (https://www.plantcyc.org/), enzymatic coding genes of the scarlet sage were shown for key reactions.
Figure S6. Menthol biosynthesis pathway. Menthol biosynthesis pathways by querying the Plant Metabolic Network (https://www.plantcyc.org/), enzymatic coding genes of the scarlet sage were shown for key reactions.
Figure S7. Shared synteny addressed for metabolic gene clusters between Salvia genomes. a-f: display of the different pairs of synteny blocks. Genes are colored along the contigs/scaffolds to compare between scarlet sage and Salvia miltiorrhiza Zhang [8], with metabolic genes highlighted with olive drab color, other homologous genes are shown in grey.
Table S1. Summary of PacBio and Illumina sequencing data generated in the present study. IDs of the study, sample, library and accession in NCBI SRA and sequencing platform, material origins of the sequenced DNA or RNA, the statistics of the raw and cleaned data are shown.
Table S2. Estimation of genome characteristics based on 17-mer statistics.
Table S3. Statistics of the different versions of the genome assembly of the scarlet sage. NA: data not available; * statistics for contigs/scaffolds.
Table S4. Summary of the annotated interspersed repeats in the genome assembly of the scarlet sage.
Table S5. Summary of the transcriptome assemblies.
Table S6. Summary of the annotated genes. AED: Annotation Edit Distance; gene regions (including UTRs, exons and introns); genes (including 5’, 3’ UTRs, exons and introns).
Table S7. Summary of BUSCO evaluation of gene prediction.
Table S8. Summary of functional annotation of predicted genes.
Table S9. Genomic data used for gene families analyses. Origins, download links, assembly versions, genome properties and references of 15 analyzed genomes are shown.
Table S10. Summary of gene family analyses. Unique groups and genes, single-copy and duplicated groups and genes are summarized for the 16 analyzed genomes of 15 plant species.
Table S11. GO enrichment of expanded gene families. (A) “Category” is the Gene Ontology (GO) term ID; (B) “p_value” is the over represented P-value indicating the observed frequency of a given term among analyzed genes is equal to the expected frequency based on the null distribution; i.e., lower P-values indicate stronger evidence for overrepresentation; (C) “q_value” is the Benjamini and Hochberg adjusted P-value, (D) “numEPInCat” is the number of expanded gene families in the corresponding GO category; (E) “numInCat” is the number of detected gene families in the corresponding GO category; (F) “Term” is the GO term; (G) “Ontology” indicates which ontology the term comes from. 60 significant (q<0.05) GO-terms of three different functional categories are indicated in bold.
Table S12. KEGG enrichment of expanded gene families. (A) “KO category” is the KEGG Orthology (KO) category ID; (B) “p_value” is the over represented P-value indicating the observed frequency of a given term among analyzed genes is equal to the expected frequency based on the null distribution; i.e., lower P-values indicate stronger evidence for overrepresentation; (C) “q_value” is the Benjamini and Hochberg adjusted P-value, (D) “numEPInCat” is the number of expanded gene families in the corresponding KO category; (E) “numInCat” is the number of detected gene families in the corresponding KO category; (F) “Pathway” is the KEGG pathway; (G) “Class” indicates which KEGG class the pathway comes from. One significant (q<0.05) KEGG pathway is indicated in bold.
Table S13. Summary of metabolic gene clusters detected in genomes of Salvia miltiorrhiza and S. splendens. (A) “Genome” denotes the genome origination; (B) “Cluster” is the code for a certain gene cluster detected; (C) “Record” denotes the contig/scaffold ID from where the gene cluster was detected; (D) “Type” denotes the functional assignment for the gene cluster; (E) “From”, “To” and “Size” denote the genomic position and range of the gene cluster; (F) “Core domains” denote the domain annotation for the metabolic genes in the cluster; (G) “CD-HIT Cluster” indicate the number of genes in the cluster; (H) “Gene cluster genes” is showing the ID of genes in the cluster.
Supplementary File 1. Genes (Gene ID, name and EC number) mapped to flavonoid and menthol biosynthesis pathways.
Supplementary File 2. Structure of a metabolic gene cluster (polyketide synthesis) and gene expression patterns of Salvia splendens. Genomic position, gene composition, functional annotation of gene cluster are shown, also including a heatmap of tissue specific expression of the genes within the presented cluster is shown. HG: root of red flower (individual); HJ: stem of red flower (individual); HY: leave of red flower (individual); HE: calyx of red flower (individual); HHG: corolla of red flower (individual); ZG: root of purple flower (individual); ZJ: stem of purple flower (individual); ZY: leave of purple flower (individual); ZE: calyx of purple flower (individual); ZHG: corolla of purple flower (individual).
Supplementary File 3. Structure of a metabolic gene cluster (alkaloid synthesis). Genomic position, gene composition, functional annotation of gene cluster were shown.
Abbreviations
AED: annotation edit distance; BUSCO: Benchmarking Universal Single-Copy Orthologs; GO: Gene Ontology; LTR: long terminal repeat; Mya: million years ago; NCBI: National Center for Biotechnology Information; PE: paired end; SMRT: single-molecule real-time; SNP: single-nucleotide polymorphism; TE: transposable element; TSP: terpene synthase genes.
Competing interests
The authors declare that they have no competing interests.
Author contributions
A.X.D., H.B.X., R.C.C., J.F.M., F.M., and I.P. conceived and designed the study; A.X.D., H.B.X., Z.J.L., H.L., Y.Q.S., S.N., Z.N.Z., R.F.C., H.L.Z., R.G.Z., and Q.Z.Y. prepared the materials and conducted the experiments; and J.F.M., H.B.X., F.M., and I.P. wrote the manuscript.
References
- 1. Drew BT, González-Gallegos JG, Xiang C-L, et al. Salvia united: the greatest good for the greatest number. Taxon. 2017;66(1):133–45. [Google Scholar]
- 2. Sutton J. The Gardener's Guide to Growing Salvias. Portland: David & Charles; 1999. [Google Scholar]
- 3. Clebsch B, Barner CD. The New Book of Salvias: Sages for Every Garden. Portland: Timber Press; 2003. [Google Scholar]
- 4. Walker JB, Sytsma KJ, Treutlein J, et al. Salvia (Lamiaceae) is not monophyletic: implications for the systematics, radiation, and ecological specializations of Salvia and tribe Mentheae. Am J Bot. 2004;91(7):1115–25. [DOI] [PubMed] [Google Scholar]
- 5. Griffiths M, Society RH. Index of Garden Plants. London: Macmillan; 1994. [Google Scholar]
- 6. Regnault-Roger C. The potential of botanical essential oils for insect pest control. Integrated Pest Management Reviews. 1997;2(1):25–34. [Google Scholar]
- 7. Ge X, Chen H, Wang H, et al. De novo assembly and annotation of Salvia splendens transcriptome using the Illumina platform. PLoS One. 2014;9(3):e87693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang G, Tian Y, Zhang J, et al. Hybrid de novo genome assembly of the Chinese herbal plant danshen (Salvia miltiorrhiza Bunge). GigaScience. 2015;4(1):62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xu H, Song J, Luo H et al. Analysis of the genome sequence of the medicinal plant Salvia miltiorrhiza. Molecular Plant. 2016;9(6):949–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vining KJ, Johnson SR, Ahkami A, et al. Draft genome sequence of Mentha longifolia and development of resources for mint cultivar improvement. Molecular Plant. 2017;10(2):323–39. [DOI] [PubMed] [Google Scholar]
- 11. Preparing Arabidopsis Genomic DNA for Size-Selected ~20 kb SMRTbellTM Libraries http://www.pacb.com/wp-content/uploads/2015/09/Shared-Protocol-Preparing-Arabidopsis-DNA-for-20-kb-SMRTbell-Libraries.pdf. Accessed 20 Sept 2017.
- 12. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Alberto CM, Sanso AM, Xifreda CC. Chromosomal studies in species of Salvia (Lamiaceae) from Argentina. Botanical Journal of the Linnean Society. 2003;141(4):483–90. [Google Scholar]
- 14. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal. 2011;17(1):10–12. [Google Scholar]
- 15. The Gene Ontology (GO). Database and informatics resource. Nucleic Acids Res. 2004;32(database issue):D258–D61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Koren S, Walenz BP, Berlin K, et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27(5):722–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Marçais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27(6):764–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu B, Shi Y, Yuan J et al. Estimation of genomic characteristics by analyzing k-mer frequency in de novo genome projects. Quantitative Biology2013;35(s1-3):62-67.
- 19. Xiao C-L, Chen Y, Xie S-Q et al. MECAT: fast mapping, error correction, and de novo assembly for single-molecule sequencing reads. Nat Methods. 2017;14:1072. [DOI] [PubMed] [Google Scholar]
- 20. Chin CS, Peluso P, Sedlazeck FJ et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat Methods. 2016;13(12):1050–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. FALCON: experimental PacBio diploid assembler. https://github.com/PacificBiosciences/FALCON/. Accessed 01 Dec 2017.
- 22.Ultra-fast de novo assembler using long noisy reads. https://github.com/ruanjue/smartdenovo. Accessed 01 Dec 2017. [DOI] [PMC free article] [PubMed]
- 23. Simão FA, Waterhouse RM, Ioannidis P et al. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–2. [DOI] [PubMed] [Google Scholar]
- 24.PacBio variant and consensus caller. https://github.com/PacificBiosciences/GenomicConsensus. Accessed 01 Dec 2017.
- 25. Boetzer M, Pirovano W. SSPACE-LongRead: scaffolding bacterial draft genomes using long read sequence information. BMC Bioinformatics. 2014;15(1):211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Boetzer M, Henkel CV, Jansen HJ et al. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27(4):578–9. [DOI] [PubMed] [Google Scholar]
- 27. Luo R, Liu B, Xie Y, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012;1:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.RepeatModeler - 1.0.10. http://www.repeatmasker.org/RepeatModeler/. Accessed 01 Jul 2017.
- 30. Bao W, Kojima KK, Kohany O. Repbase update, a database of repetitive elements in eukaryotic genomes. Mobile DNA. 2015;6(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.RepeatMasker. http://www.repeatmasker.org/. Access 01 Jul 2017.
- 32. Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.FastQC. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed 10 Feb 2018.
- 34. Trapnell C, Williams BA, Pertea G et al. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pertea M, Pertea GM, Antonescu CM, et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33(3):290–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Grabherr MG, Haas BJ, Yassour M et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol. 2011;29:644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fu L, Niu B, Zhu Z, et al. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28(23):3150–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stanke M, Diekhans M, Baertsch R et al. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008;24(5):637–44. [DOI] [PubMed] [Google Scholar]
- 39. Boratyn GM, Schäffer AA, Agarwala R, et al. Domain enhanced lookup time accelerated BLAST. Biology Direct. 2012;7(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.A generic tool for sequence alignment. https://www.ebi.ac.uk/about/vertebrate-genomics/software/exonerate. Accessed 10 Jan 2018.
- 41. Slater GSC, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cantarel BL, Korf I, Robb SM et al. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008;18(1):188–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12(4):656–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bairoch A, Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000;28(1):45–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bateman A, Birney E, Cerruti L, et al. The Pfam protein families database. Nucleic Acids Res. 2002;30(1):276–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Quevillon E, Silventoinen V, Pillai S, et al. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33(web server issue):W116–W20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.National Center for Biotechnology Information. http://www.ncbi.nlm.nih.gov. Accessed 01 Dec 2017.
- 48.ExPASy Bioinformatics Resources Portal. http://www.expasy.ch/sprot. Accessed 01 Dec 2017.
- 49.UniProt. http://www.ebi.ac.uk/uniprot. Accessed 01 Dec 2017.
- 50.Pfam. http://pfam.xfam.org/. Accessed 01 Dec 2017.
- 51.The KOG Browser. http://genome.jgi-psf.org/help/kogbrowser.jsf. Accessed 01 Dec 2017.
- 52.KO (KEGG ORTHOLOGY) Database. http://www.genome.jp/kegg/ko.html. Accessed 01 Dec 2017.
- 53. Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28(1):27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gene Ontology Consortium. http://www.geneontology.org. Accessed 01 Dec 2017.
- 55. The Gene Ontology Consortium et al. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sollars ESA, Harper AL, Kelly LJ et al. Genome sequence and genetic diversity of European ash trees. Nature. 2016;541:212. [DOI] [PubMed] [Google Scholar]
- 57. Unver T, Wu Z, Sterck L, et al. Genome of wild olive and the evolution of oil biosynthesis. Proc Natl Acad Sci. 2017;114(44):E9413–E22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hellsten U, Wright KM, Jenkins J et al. Fine-scale variation in meiotic recombination in Mimulus inferred from population shotgun sequencing. Proc Natl Acad Sci. 2013;110(48):19478–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lan T, Renner T, Ibarra-Laclette E et al. Long-read sequencing uncovers the adaptive topography of a carnivorous plant genome. Proc Natl Acad Sci. 2017;114(22):E4435–E41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang L, Yu S, Tong C et al. Genome sequencing of the high oil crop sesame provides insight into oil biosynthesis. Genome Biol. 2014;15(2):R39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Denoeud F, Carretero-Paulet L, Dereeper A, et al. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science. 2014;345(6201):1181–4. [DOI] [PubMed] [Google Scholar]
- 62. Tomato Genome Consortium The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485:635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Iorizzo M, Ellison S, Senalik D, et al. A high-quality carrot genome assembly provides new insights into carotenoid accumulation and asterid genome evolution. Nat Genet. 2016;48:657. [DOI] [PubMed] [Google Scholar]
- 64. French-Italian Public Consortium for Grapevine Genome Consortium for Grapevine Genome Characterization The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463. [DOI] [PubMed] [Google Scholar]
- 65. Cheng C-Y, Krishnakumar V, Chan AP et al. Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 2017;89(4):789–804. [DOI] [PubMed] [Google Scholar]
- 66. Tuskan GA, DiFazio S, Jansson S et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science. 2006;313(5793):1596–604. [DOI] [PubMed] [Google Scholar]
- 67. Ouyang S, Zhu W, Hamilton J, et al. The TIGR rice genome annotation resource: improvements and new features. Nucleic Acids Res. 2007;35(suppl_1):D883–D7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Dohm JC, Minoche AE, Holtgräwe D, et al. The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature. 2013;505:546. [DOI] [PubMed] [Google Scholar]
- 69. Li L, Stoeckert CJ, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13(9):2178–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. De Bie T, Cristianini N, Demuth JP et al. CAFE: a computational tool for the study of gene family evolution. Bioinformatics. 2006;22(10):1269–71. [DOI] [PubMed] [Google Scholar]
- 71. Badouin H, Gouzy J, Grassa CJ, et al. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature. 2017;546:148. [DOI] [PubMed] [Google Scholar]
- 72. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Guindon S, Dufayard J-F, Lefort V, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–21. [DOI] [PubMed] [Google Scholar]
- 74. Sanderson MJ. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics. 2003;19(2):301–2. [DOI] [PubMed] [Google Scholar]
- 75. Doyle JA. Molecular and fossil evidence on the origin of angiosperms. Annu Rev Earth Planet Sci. 2012;40(1):301–26. [Google Scholar]
- 76. Wang H, Moore MJ, Soltis PS, et al. Rosid radiation and the rapid rise of angiosperm-dominated forests. Proc Natl Acad Sci. 2009;106(10):3853–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Mimica-Dukic N, Bozin B. Mentha L. species (Lamiaceae) as promising sources of bioactive secondary metabolites. Curr Pharm Des. 2008;14(29):3141–50. [DOI] [PubMed] [Google Scholar]
- 78.E2P2. https://gitlab.com/rhee-lab/E2P2/tree/master. Accessed 10 Feb 2018.
- 79.PLANT METABOLIC PATHWAY DATABASES. https://www.plantcyc.org/. Accessed 10 Feb 2018.
- 80. Schläpfer P, Zhang P, Wang C, et al. Genome-wide prediction of metabolic enzymes, pathways, and gene clusters in plants. Plant Physiol. 2017;173(4):2041–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Osbourn A. Secondary metabolic gene clusters: evolutionary toolkits for chemical innovation. Trends Genet. 2010;26(10):449–57. [DOI] [PubMed] [Google Scholar]
- 82. Nützmann H-W, Osbourn A. Gene clustering in plant specialized metabolism. Curr Opin Biotechnol. 2014;26:91–9. [DOI] [PubMed] [Google Scholar]
- 83. Hans-Wilhelm N, Ancheng H, Anne O. Plant metabolic clusters–from genetics to genomics. New Phytol. 2016;211(3):771–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kautsar SA, Suarez Duran HG, Blin K et al. plantiSMASH: automated identification, annotation and expression analysis of plant biosynthetic gene clusters. Nucleic Acids Res. 2017;45(W1):W55–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Boutanaev AM, Moses T, Zi J, et al. Investigation of terpene diversification across multiple sequenced plant genomes. Proc Natl Acad Sci. 2015;112(1):E81–E8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Dong A, Xin H, Li Z et al. Supporting data for “High quality assembly of the reference genome for scarlet sage, Salvia splendens, an economically important ornamental plant.”. GigaScience Database. 2018. 10.5524/100463. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
2/21/2018 Reviewed
2/22/2018 Reviewed
5/12/2018 Reviewed
2/26/2018 Reviewed