Abstract
Recently, advanced text-mining techniques have been shown to speed up manual data curation by providing human annotators with automated pre-annotations generated by rules or machine learning models. Due to the limited training data available, however, current annotation systems primarily focus only on common concept types such as genes or diseases. To support annotating a wide variety of biological concepts with or without pre-existing training data, we developed ezTag, a web-based annotation tool that allows curators to perform annotation and provide training data with humans in the loop. ezTag supports both abstracts in PubMed and full-text articles in PubMed Central. It also provides lexicon-based concept tagging as well as the state-of-the-art pre-trained taggers such as TaggerOne, GNormPlus and tmVar. ezTag is freely available at http://eztag.bioqrator.org.
INTRODUCTION
Efficient access to information contained in the biomedical literature plays several key roles in experiments, from the early stages of planning to the final interpretation of the results. This biological knowledge can be obtained effectively from expert-curated databases such as UniProt (1). However, the increasing number of new publications makes the cost of manual curation more challenging. Since manual curation alone is not sufficient to keep biological databases up to date (2), computer-assisted curation by text mining techniques has gained popularity in recent years (3,4).
While there are numerous web-based annotation tools available (5–12), they are mostly task-specific, tuned on certain gold standard sets and/or knowledge bases (13). Due to the limited resources available (14–17), computer-assisted annotation has normally focused on common biological concepts such as gene/protein, chemical and disease names. Certain annotation tools (7,8) support more entity types, however they are rule-based in general, i.e. assigning concept types is achieved by lexical pattern matching. Moreover, only few studies suggest the idea of adaptive bio-entity annotation via interactive learning (12,18).
Another critical issue when developing an annotation tool is whether it supports full text articles. Even though biocurators read full-text articles as well as abstracts for manual curation, full-text articles have not been well supported by existing annotation tools (13). This is partially due to the difficulty of parsing various XML formats, as well as complex copyright issues for certain journals. For example, the most common user requests for PubTator (11), a widely used annotation tool for biomedical concepts we introduced in 2013, have been to support PubMed Central (PMC) full-text articles and to provide more flexibility for text-mined annotation. The latter is essential for some users because annotation guidelines may differ even for common bio-entity types.
To address these problems, we introduce ezTag, a user-friendly annotation tool that allows biocurators to perform annotation and provide training data interactively. Compared to other bio-entity annotation tools, ezTag has several unique features. First, ezTag supports all PubMed abstracts and PMC open access articles. We achieved this by standardizing the text from both repositories into BioC format (19); it also supports any other document in BioC format. Second, ezTag users have multiple ways of annotating bio-entities: (i) the pre-trained state-of-the-art bio-entity taggers (20–22), (ii) the string pattern match tagger, which uses a user-provided lexicon and (iii) the customized tagger by training TaggerOne (20). Third, ezTag explicitly supports training and annotating text iteratively, hence it helps produce a set of annotated documents and a customized tagging module in any bio-entities efficiently. Other features include a user-friendly interface based on PubTator user feedback, automatic session ID-based login, i.e. no manual login required and RESTful API support for customized tagging modules. As a result, biocurators can annotate documents without much help from software developers. Also, software developers without text mining experience can benefit from our RESTful APIs.
Throughout this paper, concept tagging is used to describe bio-entity annotation and it may or may not include assigning concept IDs (i.e. normalization or grounding).
SYSTEM DESCRIPTION
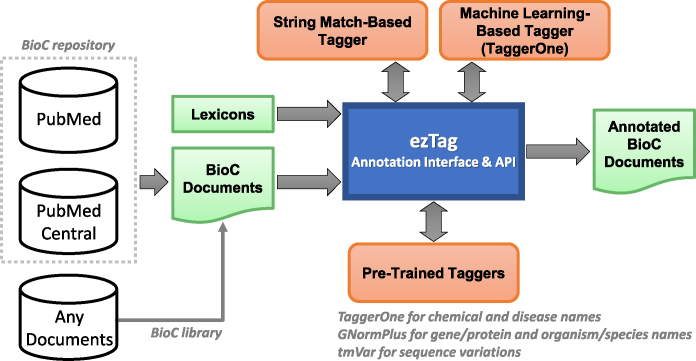
Figure 1 illustrates the system overview of ezTag. As shown in the figure, ezTag utilizes multiple resources to provide bio-entity annotation in biomedical text. For input, any documents in the BioC format (http://bioc.sourceforge.net) (19) can be uploaded to the interface. We chose BioC for input and output for better data interoperability. PubMed abstracts and PMC full-text articles are pre-processed in BioC and ready for upload using PubMed and PMC IDs. These BioC documents are also accessible through RESTful APIs (https://www.ncbi.nlm.nih.gov/research/bionlp/APIs), thus users can always process and share the same BioC documents. Lexicons (Figure 1) are used in two different scenarios. One is for the string match-based tagger, and the other is to assign concept IDs for the machine learning-based tagger (TaggerOne in the figure).
Figure 1.
System overview. ezTag connects multiple resources to provide efficient and effective biological concept tagging. Input and output documents are handled using the BioC format, and user-provided lexicons are used for string match and machine learning-based taggers. The options for automatic concept tagging in text are (i) the string match-based tagger using a lexicon, (ii) the machine learning-based tagger using TaggerOne for customized tagging modules and (iii) the pre-trained taggers.
The core functionalities of ezTag are manual annotation and automatic annotation. ezTag has three modules for automatic annotation: the string match-based tagger, the machine learning-based tagger and pre-trained taggers. The string match-based tagger uses a user-provided lexicon for identifying bio-entities and assigning concept IDs (i.e. normalization). Since this step may be used as a starting point for interactive learning (which will be explained later), we implemented a trie structure (23) for strict string match but also allowed small variations such as abbreviations, Greek letters, upper/lowercases, hyphens and other stopwords. The machine learning-based tagger system is TaggerOne (20). TaggerOne is a semi-Markov model for joint named entity recognition and normalization. Users can train TaggerOne for a set of annotated documents, then use the trained model to tag concepts in a new set of documents. Providing a lexicon is optional; however, if one is provided by the user, then TaggerOne also learns to assign concept IDs.
In addition to the customizable modules, string match and machine learning based taggers, ezTag provides annotations from pre-trained taggers. The common bio-entities we support here are chemical, disease, gene/protein, organism/species and sequence variations. We utilize three state-of-the-art performance tools for those common types. The pre-trained TaggerOne (20) is used for annotating chemical and disease names. GNormPlus (21) is used for annotating gene/protein and organism/species. tmVar (22) is for sequence variations. Table 1 lists all pre-trained concept taggers used in ezTag and the bio-entities and nomenclatures they use. The last column of the table also shows the normalization performance (F1 scores) of each concept tagger based on the gold standard sets reported in (20–22,24). Note that the F1 performance at the mention level (i.e. identifying bio-entities only) is typically higher than those at the normalization level.
Table 1. Pre-trained concept tagging tools used in ezTag.
| Pre-trained tagger | Bio-entity | Nomenclature | F1 score (normalization) |
|---|---|---|---|
| TaggerOne | Chemical | MeSH | 0.895 |
| Disease | MEDIC | 0.807 | |
| GNormPlus | Gene | NCBI Gene | 0.867 |
| Species | NCBI Taxonomy | 0.854 | |
| tmVar | Sequence variation | NCBI dbSNP | 0.903 |
MEDIC is a disease vocabulary created by Comparative Toxicogenomics Database. All other vocabularies are products of National Library Medicine. F1 scores are taken from their corresponding publications.
Implementation
We developed ezTag using Ruby on Rails and MySQL as a backend database. RESTful APIs were implemented in C++ and Perl. All the web pages in ezTag are HTML5/CSS compatible, thus it supports the latest version of popular web browsers such as Chrome, Safari, Firefox and Internet Explorer. On rare occasions, Internet Explorer may not correctly display some icons due to HTML5 compatibility issues. The source code of the ezTag web interface is available at https://github.com/ncbi-nlp/ezTag.
USAGE
User interface
ezTag was motivated by the feedback from PubTator users and designed to merge useful features of PubTator (11), TaggerOne (20) and BioC Viewer (25). The two primary approaches to providing assisted annotations for concept tagging are string match and machine learning. In ezTag, we support both approaches by implementing a lexicon-based string match tagger and integrating with multiple machine learning-based taggers. ezTag also allows users to choose a training set for a customized tagging module.
For a smooth annotation experience, users should first create a collection for an annotation project. An annotation task is then started by uploading documents in BioC format or using PubMed and PMC IDs in the collection. ezTag has top menus for lexicons and customized models (These are called ‘Lexicons’ and ‘Models’ in the web pages, respectively). In this way, lexicons and models can be used to annotate any collection present in the user’s repository.
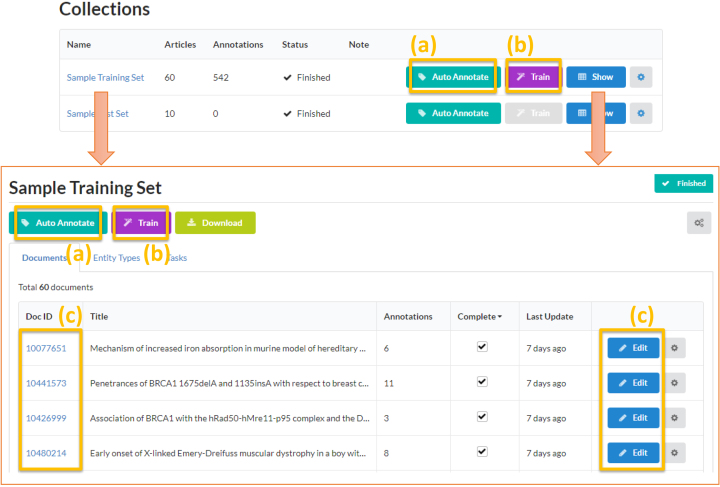
Figure 2 shows a screenshot of the ‘sample training set’ collection page. As described earlier, ezTag has two main functions, automatic annotation and manual annotation ((a) and (c) in the figure, respectively). Users can also create a customized module using the collection to train a model ((b) in the figure).
Figure 2.
ezTag user interface for the sample training set. An annotation project (e.g. the sample training set here) is called ‘collection’ in ezTag. Uploaded documents belong to a collection and these documents are used for (a) auto annotation (i.e. pre-trained, lexicon or machine learning based concept tagging), (b) training a machine learning-based tagger (i.e. TaggerOne) and (c) manual annotation.
Input and output
ezTag uses the BioC format for both input and output documents. Annotated or unannotated documents are used as input. The output is a set of documents annotated automatically or manually. If a tagging module was created by training a collection, the customized module is also an output of ezTag.
Manual annotation
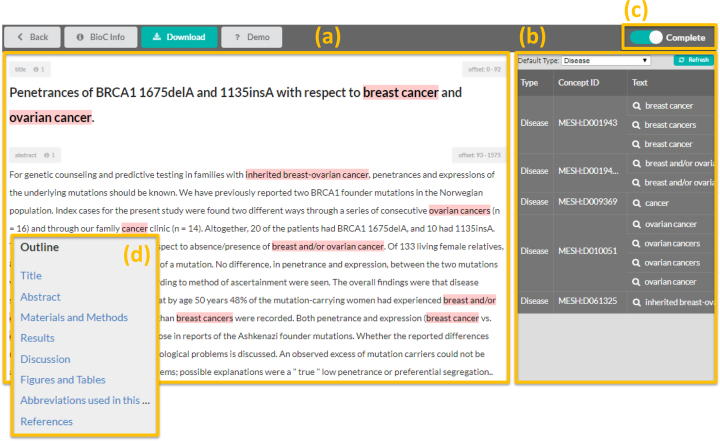
ezTag has a manual annotation tool supporting an arbitrary number of bio-entity types (Figure 3). Manual annotation can be used for adding annotations from scratch, or to refine existing annotations. For easy browsing in full-text articles, the annotation window has an outline view ((d) in the figure), which will appear in the left of the window when the document has multiple sections. Clicking a section in the outline view will move the mouse focus to where the section is in the main text column. Since ezTag aims to be a general annotation tool, only manual typing in is allowed for entering concept IDs in the current version. The last step of manual annotation is to toggle on the ‘Complete’ button ((c) in the figure), which indicates that the annotation is complete and the document may be used for training. When the ‘Complete’ button is on, the document will be used to train a customized tagging model. When it is off, the document will be used for tagging concepts. Note that, although ezTag does not fully support highlighting overlapping annotations, it keeps and displays all annotations in the annotation table.
Figure 3.
Manual annotation page in ezTag. There are two main windows: (a) main text and (b) annotation table. Users can add an annotation by a mouse drag on text (a), and tag the annotation by typing ID(s) (b). The complete button (c) is used to mark whether annotation of the document is done. Using this mark, ezTag decides how a document should be used, i.e. either for automatic annotation or for training TaggerOne. A browsable outline will appear in the left if a document has multiple sections (d).
Automatic annotation (‘Auto Annotate’)
Using lexicons
When no annotated set or pre-trained tagger is available for the desired entity type, but there is a dictionary of concept names available, this is a good option to start with. Our string match-based tagger uses a user-provided lexicon to tag text. When concept IDs are given in the lexicon, the tagger also assigns the IDs along with annotated text. The user can then review and refine the annotated text as a next step. This also can be followed by training our machine learning-based tagger, TaggerOne, for a new customized tagger.
Using pre-trained tagging models
This auto annotation can be used when user targeted bio-entities fall in one of types that our pre-trained models support. If users have different annotation guidelines in mind, annotated text from this step can be refined and the result can be fed into TaggerOne for a customized tagger.
Using customized tagging models
TaggerOne is a model that jointly predicts mentions and their normalized IDs. Since it does not require specific bio-entity types, one can use TaggerOne to train and predict a wide variety of entity types. After training TaggerOne (‘Train’ in the ezTag interface), one obtains a customized tagger, and this model will be available in the auto annotation menu (as a pre-trained tagging model). As in other auto annotate steps, the output from a customized tagging model for a new set can be used to obtain an improved customized tagger (see ‘Use case’ for more details).
Programmatic access via RESTful API
In addition to a user-friendly web interface, ezTag provides a RESTful API for accessing customized concept taggers. For a customized tagger, users can input text and get annotations programmatically via API. A help page of how to use the API is shown in each customized model web page. One can share a customized tagger with anyone using a RESTful API. RESTful APIs for pre-trained taggers are also available and the instructions can be found at https://www.ncbi.nlm.nih.gov/research/bionlp/APIs.
Session-based automatic logins
ezTag does not require a manual login, but allows users to continue their annotation processes by assigning session IDs. Once a user enters ezTag, a session ID is automatically created (if it does not exist already). A user session is recorded in web cookies, hence normally there is no need to re-login using the session ID. Users can also get a unique URL for each session ID via email to ensure their session remains accessible.
USE CASE: INTERACTIVE LEARNING FOR ADAPTIVE ANNOTATION
ezTag provides multiple ways of annotating text: manual annotation and auto annotation. Auto annotation has three options: string match tagger, machine learning tagger for customized models and pre-trained taggers for common bio-entity types. Because of the flexibility provided by these diverse options, users can perform interactive learning for adaptive entity tagging.
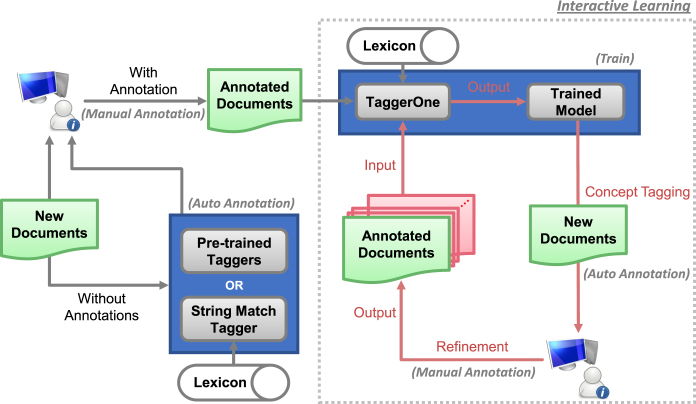
Figure 4 presents our interactive learning workflow using ezTag, with the interactive part highlighted in the gray box with the dotted line. As shown in the top left, if annotated training data already exists, ezTag allows the user to train a machine learning tagging model on-the-fly using TaggerOne, that in turn provides pre-annotations for human review. If no training data is initially available, pre-annotations can be obtained either through pre-trained methods or a string matching approach with a user-provided concept lexicon. In either case, when computer pre-annotations are reviewed and refined by the human annotator, they can be iteratively fed into TaggerOne to build a new and improved model that in turn provides higher quality pre-annotations. As a result, for input documents with/without annotations, users obtain a customized concept tagging model and/or higher quality annotated documents as output.
Figure 4.
Interactive learning workflow. New documents are first manually annotated or refined after applying the pre-trained or string match tagger. Interactive learning follows three steps interactively: (i) Training TaggerOne using annotated documents, (ii) tagging biological concepts in new documents using the trained model and (iii) refining the documents by correcting annotation mistakes.
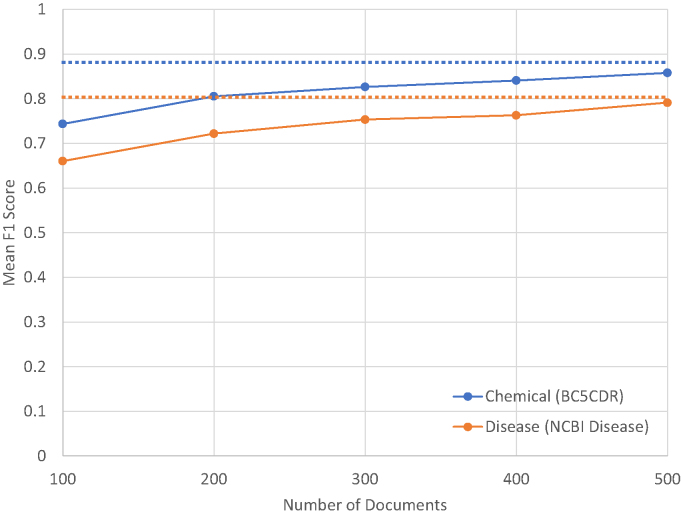
To check the effectiveness of interactive learning, we performed ablation tests using BioCreative V CDR (26) and NCBI Disease (17) datasets for chemical and diseases, respectively. We assume users annotate 100 documents in each iteration, and add them to the existing training set, i.e. starting from 100 documents, the number of annotated documents are accumulated after each iteration. The performance is measured by F1 scores on the (independent) test set. This experiment simulates the interactive process and shows how much improvement one would get through the process. We ran the experiments five times for each dataset, and averaged F1 scores in terms of chemical/disease name identification (see Figure 5). The 100 documents added each iteration were randomly chosen. We observe that the performance after five iterations approaches the upper bound (i.e. the performance when we could obtain using all training examples). Moreover, a tagger that is imperfect but useful for pre-annotation is likely obtainable from a relatively small number of documents. In practice, the performance realized will depend on the entity type and the quantity and variety of entity mentions that appear in the documents annotated, and should be evaluated periodically as annotation proceeds.
Figure 5.
Performance changes over accumulated training documents for identifying chemical and disease names. The dotted lines indicate the upper bound that TaggerOne can achieve, i.e. when all training documents are used.
CONCLUSION
ezTag is a versatile annotation tool that enables users to perform both simple annotation and complex interactive learning for adaptive concept tagging. With or without pre-existing training data, users can obtain annotated text for a wide variety of bio-entities. This made possible by combining the user-friendly web interface with a new string match based tagger and state-of-the-art annotation tools such as TaggerOne, GNormPlus and tmVar. Moreover, the interactive learning framework via ezTag will reduce the annotation burden for new or adaptive concept tagging. While PubTator, our earlier tool, pre-annotates common bio-entities in PubMed abstracts, ezTag annotates any documents including PubMed abstracts and PMC full-text articles on the fly. The popular PubTator tool still helps access high quality pre-annotated text, but ezTag will complement PubTator by supporting full-text articles and adaptive annotation for more bio-entities. Currently, we display only text from the body of PMC articles, however curators often use figures and tables for annotation. In the future, we plan to include figures and tables in BioC and display the graphics in ezTag for better annotation experience. Another limitation is the lack of disjoint annotation and PDF support, which also remain as future work.
DATA AVAILABILITY
ezTag is free and open to all users and there is no login requirement. ezTag can be accessed at http://eztag.bioqrator.org. The source code of the ezTag web interface is also available at https://github.com/ncbi-nlp/ezTag.
FUNDING
Ministry of Science and ICT (MSIT) [NRF-2014M3C9A3064706 to D.K.]; Ministry of Education [NRF-2012R1A1A2044389 to D.K.]; Intramural Research Program of the National Institutes of Health, National Library of Medicine (to S.K., C.W., R.L., Z.L.). Funding for open access charge: Intramural Research Program of the National Institutes of Health, National Library of Medicine.
Conflict of interest statement. None declared.
REFERENCES
- 1. The UniProt, C. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017; 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baumgartner W.A. Jr, Cohen K.B., Fox L.M., Acquaah-Mensah G., Hunter L.. Manual curation is not sufficient for annotation of genomic databases. Bioinformatics. 2007; 23:i41–i48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Singhal A., Simmons M., Lu Z.. Text mining genotype-phenotype relationships from biomedical literature for database curation and precision medicine. PLoS Comput. Biol. 2016; 12:e1005017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Poux S., Arighi C.N., Magrane M., Bateman A., Wei C.H., Lu Z., Boutet E., Bye A.J.H., Famiglietti M.L., Roechert B. et al. On expert curation and scalability: UniProtKB/Swiss-Prot as a case study. Bioinformatics. 2017; 33:3454–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rak R., Batista-Navarro R.T., Rowley A., Carter J., Ananiadou S.. Text-mining-assisted biocuration workflows in Argo. Database. 2014; 2014:bau070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kwon D., Kim S., Shin S.Y., Chatr-aryamontri A., Wilbur W.J.. Assisting manual literature curation for protein-protein interactions using BioQRator. Database. 2014; 2014:bau067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Campos D., Lourenco J., Matos S., Oliveira J.L.. Egas: a collaborative and interactive document curation platform. Database. 2014; 2014:bau048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pafilis E., Buttigieg P.L., Ferrell B., Pereira E., Schnetzer J., Arvanitidis C., Jensen L.J.. EXTRACT: interactive extraction of environment metadata and term suggestion for metagenomic sample annotation. Database. 2016; 2016:baw005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Salgado D., Krallinger M., Depaule M., Drula E., Tendulkar A.V., Leitner F., Valencia A., Marcelle C.. MyMiner: a web application for computer-assisted biocuration and text annotation. Bioinformatics. 2012; 28:2285–2287. [DOI] [PubMed] [Google Scholar]
- 10. Rinaldi F., Clematide S., Marques H., Ellendorff T., Romacker M., Rodriguez-Esteban R.. OntoGene web services for biomedical text mining. BMC Bioinformatics. 2014; 15(Suppl. 14):S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wei C.H., Kao H.Y., Lu Z.. PubTator: a web-based text mining tool for assisting biocuration. Nucleic Acids Res. 2013; 41:W518–W522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cejuela J.M., McQuilton P., Ponting L., Marygold S.J., Stefancsik R., Millburn G.H., Rost B., FlyBase C.. tagtog: interactive and text-mining-assisted annotation of gene mentions in PLOS full-text articles. Database. 2014; 2014:bau033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Neves M., Leser U.. A survey on annotation tools for the biomedical literature. Brief. Bioinform. 2014; 15:327–340. [DOI] [PubMed] [Google Scholar]
- 14. Leser U., Hakenberg J.. What makes a gene name? Named entity recognition in the biomedical literature. Brief. Bioinform. 2005; 6:357–369. [DOI] [PubMed] [Google Scholar]
- 15. Campos D., Matos S.R, Oliveira J.L.S.. Biomedical Named Entity Recognition: a Survey of Machine-Learning Tools. 2012; London: InTech. [Google Scholar]
- 16. Eltyeb S., Salim N.. Chemical named entities recognition: a review on approaches and applications. J. Cheminform. 2014; 6:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dogan R.I., Leaman R., Lu Z.. NCBI disease corpus: a resource for disease name recognition and concept normalization. J. Biomed. Inform. 2014; 47:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yimam S.M., Biemann C., Majnaric L., Sabanovic S., Holzinger A.. An adaptive annotation approach for biomedical entity and relation recognition. Brain Inform. 2016; 3:157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Comeau D.C., Islamaj Dogan R., Ciccarese P., Cohen K.B., Krallinger M., Leitner F., Lu Z., Peng Y., Rinaldi F., Torii M. et al. BioC: a minimalist approach to interoperability for biomedical text processing. Database. 2013; 2013:bat064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Leaman R., Lu Z.. TaggerOne: joint named entity recognition and normalization with semi-Markov Models. Bioinformatics. 2016; 32:2839–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wei C.H., Kao H.Y., Lu Z.. GNormPlus: an integrative approach for tagging genes, gene families, and protein domains. Biomed Res. Int. 2015; 2015:918710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wei C.H., Phan L., Feltz J., Maiti R., Hefferon T., Lu Z.. tmVar 2.0: integrating genomic variant information from literature with dbSNP and ClinVar for precision medicine. Bioinformatics. 2018; 34:80–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brass P. Advanced Data Structures. 2008; Cambridge: Cambridge University Press. [Google Scholar]
- 24. Wei C.H., Kao H.Y., Lu Z.. SR4GN: a species recognition software tool for gene normalization. PLoS One. 2012; 7:e38460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shin S.Y., Kim S., Wilbur W.J., Kwon D.. BioC viewer: a web-based tool for displaying and merging annotations in BioC. Database. 2016; 2016:baw106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li J., Sun Y., Johnson R.J., Sciaky D., Wei C.H., Leaman R., Davis A.P., Mattingly C.J., Wiegers T.C., Lu Z.. BioCreative V CDR task corpus: a resource for chemical disease relation extraction. Database. 2016; 2016:baw068. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
ezTag is free and open to all users and there is no login requirement. ezTag can be accessed at http://eztag.bioqrator.org. The source code of the ezTag web interface is also available at https://github.com/ncbi-nlp/ezTag.