Abstract
Familial dysautonomia (FD) is a rare genetic disease with no treatment, caused by an intronic point mutation (c.2204+6T>C) that negatively affects the definition of exon 20 in the elongator complex protein 1 gene (ELP1 also known as IKBKAP). This substitution modifies the 5′ splice site and, in combination with regulatory splicing factors, induces different levels of exon 20 skipping, in various tissues. Here, we evaluated the therapeutic potential of a novel class of U1 snRNA molecules, exon-specific U1s (ExSpeU1s), in correcting ELP1 exon 20 recognition. Lentivirus-mediated expression of ELP1-ExSpeU1 in FD fibroblasts improved ELP1 splicing and protein levels. We next focused on a transgenic mouse model that recapitulates the same tissue-specific mis-splicing seen in FD patients. Intraperitoneal delivery of ELP1-ExSpeU1s-adeno-associated virus particles successfully increased the production of full-length human ELP1 transcript and protein. This splice-switching class of molecules is the first to specifically correct the ELP1 exon 20 splicing defect. Our data provide proof of principle of ExSpeU1s-adeno-associated virus particles as a novel therapeutic strategy for FD.
Introduction
Familial dysautonomia (FD, OMIM 223900) is a rare autosomal recessive disorder characterized by progressive degeneration of the sensory and autonomic nervous system (1,2). This disease affects mainly the Ashkenazi Jew population with a carrier frequency of 1 in 27 and 1 in 18 in Ashkenazi Jewish of Polish descent (3). The severely compromised autonomic and sensory nervous system leads to multiple symptoms, such as cardiac instability, gastrointestinal dysfunction, pneumonia, postural hypotension, defective tear production and reduced pain sensitivity. There is also ataxia and progressive atrophy of the optic nerve that eventually leads to vision loss (2,4,5). More than 99% of FD patients are homozygous for the c.2204 + 6T > C intronic splice site mutation in the ELP1 gene (6,7). This gene, located on chromosome 9q31, is 66.60 kb long and has 37 exons (8,9). The ELP1 mRNA is ∼5.9 kb long and encodes a 150 kDa protein of 1332 amino acids, called elongator complex protein 1 (ELP1) (6,10). This protein, also known as the I kappa B kinase complex-associated protein (IKAP), has been implicated in various cellular processes including cell migration, Jun N-terminal kinases (JNK) signaling, exocytosis and t-RNA modification (11–14). FD patients have lowered ELP1 protein in several tissues, particularly in the central and peripheral nervous systems (15). It remains unclear how ELP1 loss leads to neurodegenerative disease.
The c.2204 + 6T > C intronic ELP1 mutation is located at position +6 of the 5′ splice site where it induces various degrees of skipping of exon 20. Transcripts without exon 20 have a premature termination codon in exon 21 and are targeted by nonsense-mediated decay, decreasing the total amount of ELP1 protein (16). The mutation reduces the recruitment of the U1 small nuclear ribonucleoprotein (snRNP) particle to the 5′ splice site to different degrees in different tissues (17), doing so most strongly in neuronal tissues (15).
The process of exon definition requires both the canonical sequences (splice sites, polypyrimidine tract and branch site) as well as exonic and intronic cis-acting regulatory elements with both enhancer or silencer functions, the exonic and intronic splicing enhancers and silencers (18–20). These sequences are bound by splicing factors and the ultimate inclusion or exclusion rates of the exon relies on the complex interaction between these positive and negative elements. In the context of ELP1 exon 20, the regulatory splicing factors are largely unknown (21), particularly those that induce exon 20 skipping.
Exon-specific U1s (ExSpeU1s) are a novel therapeutic strategy to promote recognition of defective exons (22). ExSpeU1s are U1 snRNP-like particles that bind by complementarity to intronic regions downstream of the 5′ splice site of skipped exons. Even if they bind to intronic sequences they do not seem to be acting as antisense molecules as they facilitate exon recognition by recruiting the spliceosomal components (22–28). ExSpeU1s can be used to correct exon skipping caused mutations at the consensus 5′ splice site (other than at the invariant GT dinucleotide), the polypyrimidine tract or at exonic regulatory elements. ExSpeU1s have successfully restored splicing in various genetic-disorder-genes including hemophilia B, cystic fibrosis, Netherton syndrome and spinal muscular atrophy (22–27). Recently, we have also demonstrated the efficacy and safety of ExSpeU1s in a severe spinal muscular atrophy mouse model where, the transgenic expression of the ExSpeU1 corrected the exon skipping defect and resulted in a strong phenotypic improvement (28). Moreover, consistent with the fact that ExSpeU1s bind at non-conserved intronic sequences, they resulted in very few off target events (28).
The currently available treatments for FD are only supportive and based on the attenuation of the symptoms (4,5). Furthermore, the large size of the ELP1 mRNA (∼5.9 kb) makes a classical gene therapy approach with adeno-associated virus (AAV) vectors difficult, where it has been successfully exploited for several other smaller-gene disorders (29–32). Several general splicing fidelity compounds can correct ELP1 splicing (33–38). The plant cytokinin, kinetin, rescues ELP1 splicing in FD cell lines (15,34,39) and in vivo in both a transgenic mouse model (16) and in FD patients (33). A kinetin analog, rectifier of aberrant splicing also improves ELP1 splicing in FD cells (38), as does the glycoside digoxin. The latter is thought to act by down-regulating the general splicing factor SRSF3 (35). As these compounds do not specifically target the ELP1 exon 20, it is clear that more specifically targeted approaches would be preferable. Here, we identify ExSpeU1s molecules that efficiently promote ELP1 exon 20 inclusion by counteracting the negative effect of inhibitory splicing factors.
Results
ExSpeU1s rescue the aberrant splicing of ELP1 C.2204 + 6T > C in a minigene assay
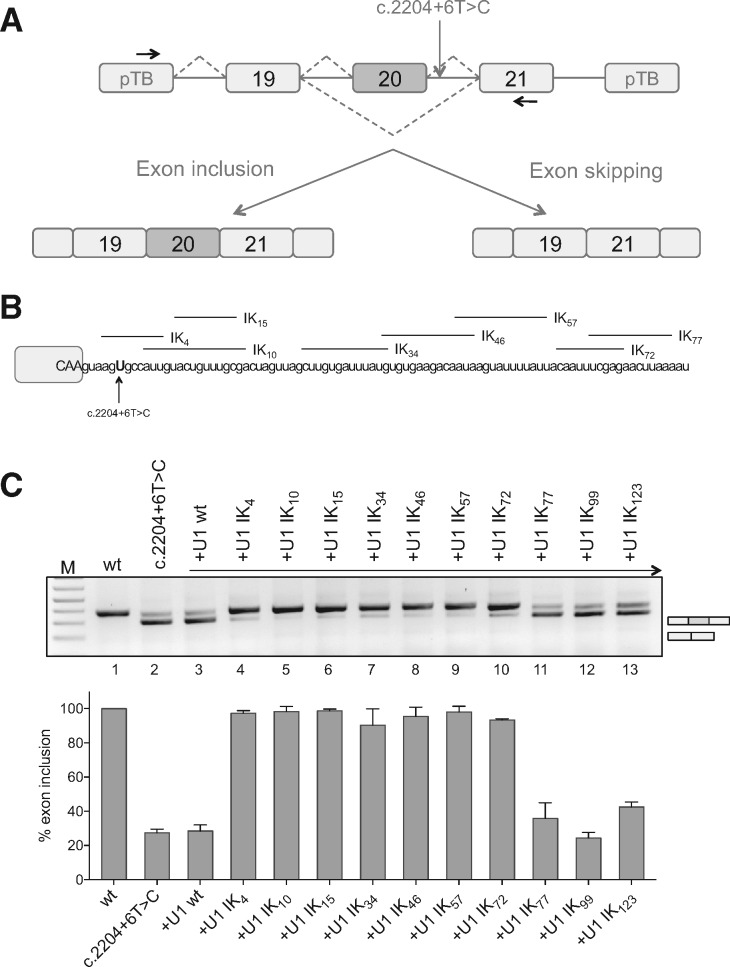
To identify a panel of ExSpeU1s that rescue ELP1 exon 20 splicing of the C.2204 + 6T > C mutation, we took advantage of a minigene system expressing the wild-type and the mutant sequences of the genomic ELP1 region spanning from intron 18 to 21 (Fig. 1A). We transfected the C.2204 + 6T > C construct transfected into neuronal SH-SY5Y cells and evaluated the splicing rescue activity of eleven ExSpeU1s that bind intronic regions between position +4 and position +123 downstream of the 5′ splice site of ELP1 exon 20 (Fig. 1B). The mutant RNA had the typical aberrant splicing caused by the intronic point mutation (∼25% exon 20 inclusion) whereas the wild-type minigene was correctly processed (Fig. 1C). To evaluate their effect on ELP1 splicing, we co-transfected the ExSpeU1-coding plasmids with the ELP1 mutant minigene. Seven of the ExSpeU1s spanning the intronic region from position +4 through +81 (ExSpeU1 Ik4–72) induced a significant rescue of the splicing defect, improving the percentage of exon inclusion to ∼90–100% (Fig. 1C, lanes 4–10). In contrast, ExSpeU1s that bind to further downstream sequences (Fig. 1C, lanes 11–13) and the wild-type U1 snRNA (Fig. 1C, lane 3) were ineffective. Finally we used different doses of four active ExSpeU1s (ik10, ik15, ik46 and ik57) to evaluate how ExSpeU1 efficacy depends on proximity of the targeted sequence to the 5′ splice site. As shown in Supplementary Material, Figure S1, increasing the distance from the 5′ splice site progressively reduces splicing rescue efficiency.
Figure 1.
ExSpeU1s correct the FD splicing defect in an in vitro splicing assay. (A) Schematic representation of the ELP1 minigenes. Boxes represent exons and lines introns. The intronic C.2204+6T>C mutation is indicated. Arrows indicate the locations of the primers used for analysis of splicing products. The two alternative transcripts are indicated below. (B) Schematic representation of the ELP1-ExSpeU1s binding regions in the intronic sequence downstream of the ELP1 exon 20 5′ splice site. Exonic and intronic sequences are in upper and lower cases, respectively. (C) Results of co-transfection, of ELP1 minigenes along with ELP1-ExSpeU1s, in SH-SY5Y cells. The upper band of 349 bp corresponds to transcripts including the exon 20; the lower band of 275 bp to exon 20 skipping. Wild-type ELP1 (wt), mutant (C.2204+6T>C) and ExSpeU1s (Ik4–123) are indicated. A schematic representation of the splicing pattern is on the right of the RT-PCR gel analysis. The graph represents the percentage of exon 20 inclusion and data are expressed as mean±SD of three independent experiments.
The most effective ExSpeU1s promote exon definition counteracting the activity of inhibitory splicing factors and do not act as antisense molecules
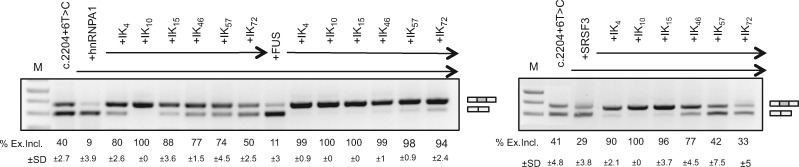
To select the most active ExSpeU1s for subsequent in vivo experiments, we tested their ability to promote exon 20 inclusion more in detail. We reasoned that the most active molecule should be able to counteract the activity of splicing factors that reduce the exon recognition by the spliceosome. In order to identify the inhibitory splicing factors, we evaluated various splicing factors in co-transfection experiments, including SR proteins, heterogeneous ribonuclear particles (hnRNPs), proteins involved in U2 recognition, and tissue-specific splicing factors. For a more sensitive readout of splicing inhibition, we overexpressed these splicing factors in Hek293T cells, in which ∼40% of the mutant exon 20 is included. Four splicing factors (SRSF3, hnRNP A1, U2AF1 and FUS) reduced exon 20 inclusion (Supplementary Material, Fig. S2). Consistently, siRNA against hnRNP A1, SRSF3 and FUS significantly increased exon inclusion (Supplementary Material, Fig. S3). We therefore tested if the previously identified ExSpeU1s could rescue the C.2204 + 6T > C ELP1 exon 20 inclusion in the presence of these three inhibitory splicing factors. The results shown in Figure 2 showed that Ik10 and Ik15 were the most effective.
Figure 2.
ExSpeU1s counteract the effect of inhibitory splicing factors. ELP1 mutant minigene was co-transfected with different ExSpeU1s (Ik4–72) along with plasmids coding for the indicated inhibitory splicing factors in Hek293T cells: hnRNP A1, FUS and SRSF3. The exon 20 inclusion and exon 20 skipping bands are indicated. The upper band of 349 bp corresponds to transcripts including exon 20; the lower band of 275 bp to exon 20 skipping. The PCR products were separated on a 2% agarose gel and the intensity of the bands were measured with ImageJ software. The percentage of exon 20 inclusion expressed as mean±SD of three independent experiments is indicated.
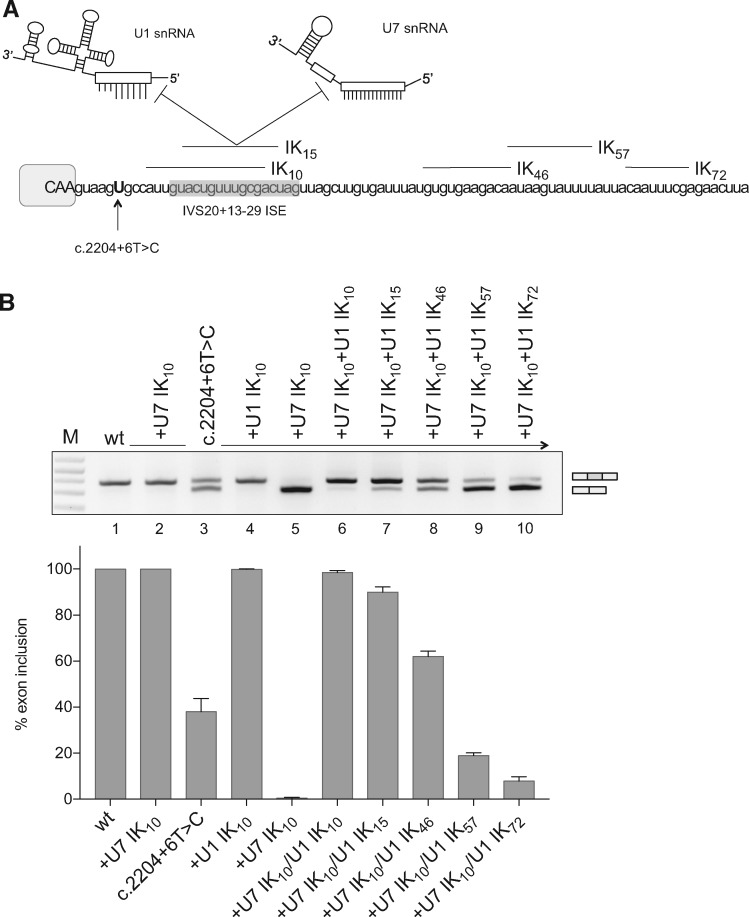
Next, we sought to clarify whether these ExSpeU1s work as antisense molecules masking a putative intronic splicing silencer. For this, we prepared an U7 snRNA (U7-Ik10) that binds to the same targeting sequence of the ExSpeU1 Ik10 (Fig. 3A). U7 snRNA derivatives have been extensively used to target RNA splicing regulatory elements with an antisense mechanism (40–42). In contrast to the positive effect of ExSpeU1 Ik10, transfection of the U7-Ik10 induced complete exon 20 skipping, suggesting that the intronic sequence between +10 and +24 mainly function as a splicing enhancer (Fig. 3B, lane 5). We then tested if the ExSpeU1s could rescue the inclusion of C.2204 + 6T > C ELP1 exon 20 in the presence of this U7 antisense molecule. Co-transfection experiments with equal amounts of plasmids showed that ExSpeU1s Ik10 and Ik15 counteracted the U7-Ik10 splicing inhibition (Fig. 3B, lanes 6 and 7). Consistent with a reduced splicing rescue activity of more distal ExSpeU1s (Supplementary Material, Fig. S1), ExSpeU1 lk46 had a small effect on U7-Ik10 splicing inhibition (Fig. 3B, lane 8) whereas Ik57 and Ik72 located more downstream were inactive (lanes 9 and 10). To better clarify the relationship between ExSpeU1s Ik10 and U7-Ik10 we also performed competition experiments by co-transfecting different amounts of plasmids. ExSpeU1 Ik10 was at least ∼10 times more efficient than the U7-Ik10 antisense in promoting exon inclusion (Supplementary Material, Fig. S4). This effect might be due to the fact that, unlike U7, ExSpeU1 is structurally similar to the endogenous U1snRNP (28), and thus probably directly recruited into the spliceosome on the defective exon. However, different expression levels of the ExSpeU1 versus the U7 might be the reason, as ExSpeU1 and U7 have polymerase II and III promoters, respectively. Overall these results indicate that Ik10 and Ik15 do not act as antisense molecules and that the region they target is mainly acting as an intronic splicing enhancer.
Figure 3.
ExSpeU1s do not act as antisense molecules. (A) Schematic representation of ExSpeU1 and U7 snRNAs along with their binding region in ELP1 intron 20. Exonic and intronic sequences are in upper and lower case, respectively. The recently described intronic splicing enhancer (48) is indicated. (B) Co-transfection experiment of ELP1 minigenes with ExSpeU1s (Ik10–72) and modified-U7 snRNA (U7-Ik10) in Hek293T cells. The exon 20 inclusion and exon 20 skipping bands are indicated. PCR products were separated on a 2% agarose gel. Exon 20 inclusion is quantified in the histograms. Data are expressed as mean±SD of three independent experiments.
Lentiviral transduction of FD fibroblasts with ExSpeU1 Ik10 restores ELP1 protein levels
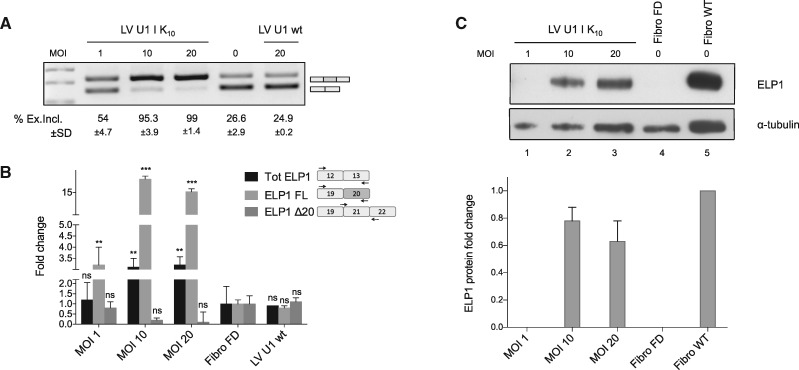
Next, we evaluated the effect of ExSpeU1s splicing of the endogenous ELP1 gene in primary fibroblasts from FD patients. We used lentivirus to transduce various concentrations of the most potent ExSpeU1 Ik10, using a lentivirus with wild-type U1 snRNA as a control (Fig. 4). Increasing multiplicities of infection of Lentivirus-Ik10 induced dose-dependent expression of the corresponding snRNA in the transduced fibroblasts, as detected by specific RT-PCR (Supplementary Material, Fig. S5) and Lentivirus-Ik10 significantly rescued ELP1 splicing in FD cells. Transduction of FD fibroblasts with multiplicities of infection 1, 10 and 20 increased the percentage of exon 20 inclusion to 54 ± 4.7, 95 ± 3.9 and 99 ± 1.4, respectively, and the control had no effect (Fig. 4A). Quantitative analysis by real-time PCR confirmed a respective increase in the full-length ELP1 transcript of ∼3, 16 and 15-fold, and, the levels of transcript lacking exon 20 (ELP1 Δ20) went down concomitant with increased lentiviral load (Fig. 4B). To evaluate whether the increase of ELP1 full-length transcripts was exclusively due to splicing correction, we also analyzed the total amount of ELP1 mRNAs by qPCR (Fig. 4B). There was an ∼3-fold increase of the total ELP1 mRNA in cells treated with ExSpeU1 Ik10 multiplicities of infection 10 and 20, whereas multiplicity of infection =1 and the control had no effect (Fig. 4B). Finally, we evaluated the effect of ExSpeU1 on ELP1 protein levels by western. Consistent with the RNA analysis we found ELP1 protein expression recovered in FD fibroblasts treated with just the two highest doses of ExSpeU1 Ik10 (Fig. 4C). Compared to normal fibroblasts, the ExSpeU1’s treatment of the FD fibroblasts recovers ∼80% of the protein level (Fig. 4C).
Figure 4.
Lentiviral delivery of ExSpeU1 rescues ELP1 splicing and protein expression in FD patient fibroblasts. (A) Endpoint PCR analysis of the ELP1 splicing pattern in FD fibroblasts treated with lentiviral particles expressing the ExSpeU1 Ik10 (Lentivirus U1 Ik10) and the wild-type U1 snRNA (Lentivirus U1 wt) at different multiplicities of infection. The upper band of 202 bp corresponds to transcripts including the exon 20, the lower band of 128 bp corresponds to exon 20 skipping. The percentage of exon 20 inclusion of three independent experiments is reported as mean±SD. (B) Quantitative analysis of ELP1 mRNAs’ isoforms by qPCR. The histograms represent the fold change of total (Tot ELP1), full-length (ELP1 FL) and exon 20 skipping (ELP1 Δ20) ELP1 mRNAs compared to the untreated cells (n.t.). A schematic representation of each amplicon is shown. Statistical analysis was performed using a two-ways ANOVA (***P<0.0001, **P < 0.001; ns, not significant). (C) ELP1 protein detection by western blot with human-specific primary antibody in FD (Fibro FD) and normal (Fibro WT) fibroblasts. α-tubulin was used for internal normalization and ELP1 fold change of FD-treated fibroblasts was referred to normal fibroblasts.
AAV9 delivery of ExSpeU1s rescues ELP1 splicing and protein levels in vivo
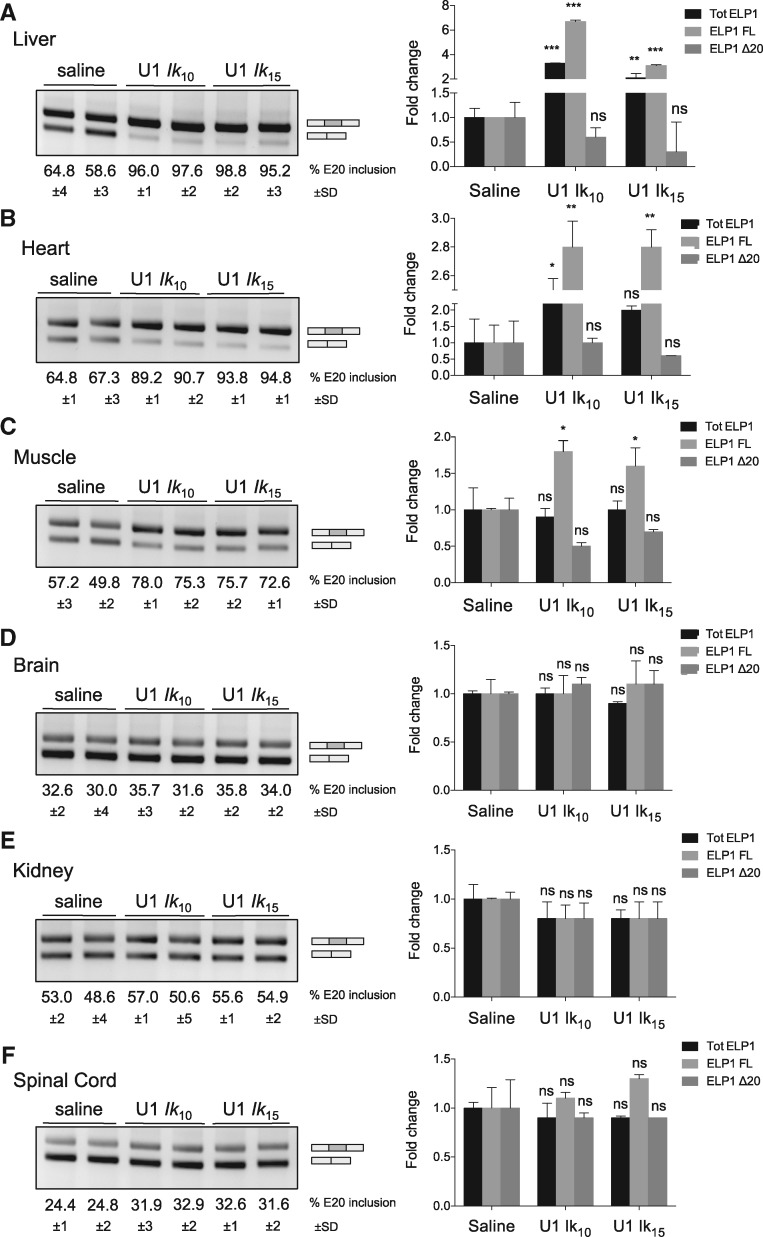
To evaluate ExSpeU1s in vivo, we used an AAV vector serotype 9 (AAV9) carrying the two most active ExSpeU1s, Ik10 and Ik15 in TgFD9 transgenic model mice that carry the entire human ELP1 C.2204 + 6T > C mutant gene (43). Importantly, this mouse recapitulates the same tissue-specific ELP1 splicing pattern observed in FD patients. We injected newborn FD mice at postnatal days 0 and 2 with Ik10 and Ik15 AAV9-ExSpeU1s or saline solution. Mice were sacrificed at 7 days after birth for splicing analysis in brain, liver, heart, muscle, brain, kidney and spinal cord. The splicing of the human transgene ELP1 was evaluated by quantitative and endpoint RT-PCR (Fig. 5). Strikingly, the AAV-ExSpeU1s improved ELP1 splicing in liver, heart and muscle (Fig. 5A–C). In liver, exon 20 inclusion increased from 60 to 97% and wild-type ELP1 transcripts increased around 5-fold (Fig. 5A). Similarly in the heart, exon inclusion increased from ∼65 to 90% and full-length ELP1 transcripts increased ∼3-fold and muscle splicing improved from ∼52 to 75% and transcripts increased ∼1.8-fold (Fig. 5B and C). ELP1 splicing was largely unaffected in brain, kidney and spinal cord (Fig. 5D–F). The slight increase in spinal cord samples was not statistically significant. Although our analysis includes male and female mice we do not expect any significant sex-dependent difference. In the previously described SMA transgenic mice that overexpress ExSpeU1s (28) we did not observe any sex-related difference in the ExSpeU1 rescue activity (unpublished data). In addition, there is no evidence of any sex-dependent splicing difference in FD mice (43) or in humans (2,4,5).
Figure 5.
AAV9-mediated delivery of ExSpeU1s Ik10 and Ik15 rescues the hELP1 splicing defect in FD mouse model. (A–F) Endpoint (gels) and quantitative (histogram) PCR analysis of hELP1 mRNA isoforms in tissues of transgenic FD mice treated with 3.55×1011 and 2.65×1011 viral genomes/mouse of AAV9-ExSpeU1 Ik10 (U1 Ik10) and AAV9-ExSpeU1 Ik15 (U1 Ik15), respectively. The upper band of 202 bp corresponds to exon 20 inclusion and the lower band of 128 bp corresponds to exon 20 skipping. The percentage of exon inclusion is expressed as mean±SD of groups of four animals per condition. The histograms on the right represent the fold change of ELP1 mRNAs’ isoforms (total ELP1, ELP1 full-length and ELP1 Δ20) compared to four control animals per condition. Statistical analysis was performed by two-way ANOVA (***P<0.0001, **P<0.001, *P<0.01; ns, not significant).
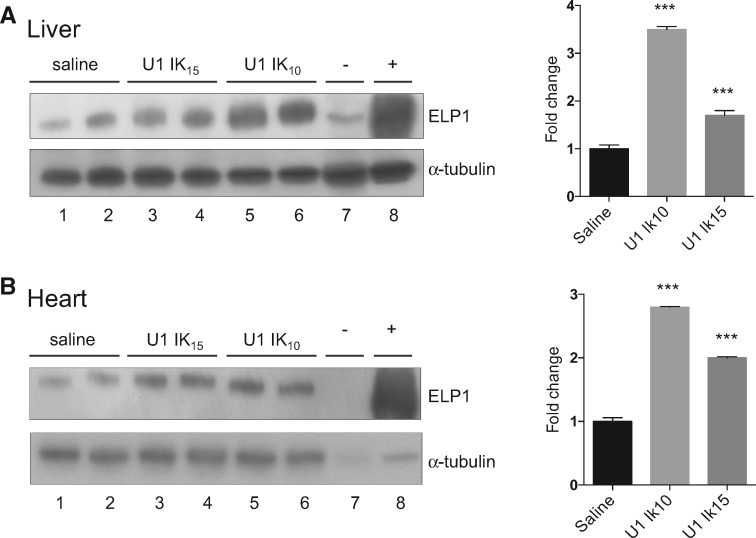
Finally, we analyzed total protein lysates from liver and heart by western blot. ExSpeU1 Ik10 increased ELP1 protein ∼3-fold in liver and heart, and ExSpeU1 Ik15 treatment nearly doubled the amount of ELP1 protein (Fig. 6A and B).
Figure 6.
ExSpeU1s Ik10 and Ik15 increase ELP1 protein levels in liver and heart of FD mice. (A and B) ELP1 western blots (left) and densitometric analysis (right) of two liver and heart samples of FD mice treated with saline, 3.55×1011 viral genomes/mouse of AAV9-ExSpeU1 Ik10 (U1 Ik10) and 2.65×1011 viral genomes/mouse of AAV9-ExSpeU1 Ik15 (U1 Ik15). α-tubulin was used as reference for internal normalization. Each group includes two animals and data are expressed as mean±SD. Statistical analysis was performed by two-way ANOVA (***P<0.0001).
Discussion
FD is a lethal disease with no available treatment that targets the underlying molecular defect. Here we propose for the first time a novel splice-switching strategy based on an ExSpeU1. These are small RNA molecules that specifically correct the splicing defect by targeting complementary intronic sequences downstream of ELP1 exon 20. In FD cellular and mouse models, the ExSpeU1-mediated splicing correction significantly rescues ELP1 protein levels. These results, coupled with the fact that AAV vectors are actively explored for efficient gene delivery in clinical trials, establish ELP1-ExSpeU1 as a highly promising strategy for FD.
To select the most potent splice-switching molecule for the in vivo studies we first evaluated splice-regulating protein factors that have a negative effect on exon 20 incorporation. As ExSpeU1s mainly act on exon definition, we reasoned that the most potent molecules for subsequent preclinical development should be able to overcome the negative effect of these splicing regulators. We found that hnRNPA1/A2, SRSF3 and FUS induce exon 20 skipping in mutant (Fig. 2B). SRSF3 has been previously implicated in exon 20 regulation as its digoxin-mediated reduction was shown to induce exon 20 inclusion (35). The other two factors have not been implicated in exon 20 regulation. hnRNPA1/A2 is a well-established negative splicing factor that inhibits many exons (44,45). FUS is a splicing factor that binds along the whole length of nascent RNA, with a preference for single stranded RNA just upstream of structured guanine-cytosine-rich regions, especially in introns near repressed exons (46). Cytoplasmic accumulation, aggregation and nuclear clearance of FUS, have been observed in a subset of neurodegenerative diseases, including amyotrophic lateral sclerosis and frontotemporal dementia (46,47). Future studies will establish exactly how these three inhibitory splicing factors affect the tissue-specificity of the FD splicing defect.
As the two most active ExSpeU1s bind to the intronic region 10–29 nucleotides downstream of the ELP1 exon 20 5′ splice site, we evaluated an antisense U7 RNA that binds to the same sequence. In contrast to ExSpeU1, the U7 RNA induced exon skipping. This evidence suggests that the ExSpeU1 does not have antisense activity and that the intronic sequence is mainly a splicing enhancer. Indeed, a very recent report identified a splicing enhancer in this region (from position 13 to 29) that binds RBM24 and promotes U1 snRNP recognition of the defective 5′ splice site (48). We speculate that binding of the ExSpeU1 particle in this critical position efficiently substitutes the missing U1 snRNP at the nearby 5′ splice site. However, more detailed studies are required to better define the composition of this intronic regulatory element and splicing mechanism.
Importantly, ExSpeU1s not only improve the FD splicing pattern but also increase the total amount of hELP1 mRNA transcript. In fibroblasts and in some AAV-treated tissues, IK-ExSpeU1s increase the total amount of hELP1 transcripts 3 and 2–3 fold, respectively (Figs 4B and 5A and B). As the exon-skipped transcript is frame-shifted and contains premature stop codons, the increase in hELP1 transcripts could be due to rescued full-length transcript avoiding nonsense-mediated decay. However, ExSpeU1s might also have a direct effect on hELP1 pre-mRNA stability, as we previously reported to be the case for SMN2 (24).
AAV vectors are a safe and effective delivery system used for classical substitutive gene replacement therapies actively explored in several diseases and clinical trials (29–32). The AAV strategy had not been previously considered for FD as the AAV particles cannot easily accommodate the large size of the ELP1 reading frame and associated regulatory elements. In contrast, the very short length of the ExSpeU1 coding gene (∼700 bp including regulatory elements) can be easily accommodated into AAV particles and in this paper, we show that the ExSpeU1 molecules can be delivered in vivo by these vectors. We evaluate the therapeutic potential of the ELP1-ExSpeU1s in vivo with the previously described TgFD9 transgenic mouse carrying the entire human ELP1 gene with the major FD splicing mutation (43). This FD mouse model, carrying the endogenous elp gene does not have any phenotypic abnormality, but displays the same tissue-specificity of exon skipping observed in FD patients. AAV ExSpeU1 corrects the ELP1 exon 20 skipping and increases the ELP1 protein in several mouse tissues. This finding strongly suggests that our approach will be an effective therapeutic option for FD.
FD symptoms are already visible in newborn babies and neurodegeneration worsens throughout life (49). Therefore, potential treatments should start as soon as possible and a small improvement in the amount of ELP1 might be sufficient to reduce the degenerative processes (5). AAV vectors are currently used to target different tissues and evidence from animal models and clinical trials indicate that they can potentially reach the most important target tissues affected in FD, including dorsal root ganglion, brain and retina (31,50,51). Experiments in symptomatic mouse models are required to evaluate whether the small and not significant improvement that we observed in neuronal tissues will be sufficient to improve the pathology. It will also likely be useful to evaluate a more specific delivery system (intracerebral or systemic) and different AAV serotypes (52). A mouse model that recapitulates the FD splicing defect and FD symptoms has been elusive due to embryonic lethality of the endogenous elp1 knockout (53). Very recently however, we have developed a humanized mouse that does have both mis-splicing and FD symptoms (54). This mouse model will be invaluable to test the efficacy and safety of the selected ExSpeU1s, providing the necessary preclinical evidence for therapeutic development.
FD is a neurodevelopmental and neurodegenerative disorder, whose main therapeutic targets are the afferent neurons in dorsal root ganglia, cranial nerve ganglia and retina (4,5). Recent studies have highlighted retinal dysfunction in the onset of blindness in FD (55,56) and FD patients’ retinas have thinner nerve fibers due to loss of retinal ganglion cells (55,56). ExSpeU1 will likely be efficiently delivered in the eye by AAV.
In conclusion, our results, demonstrating that ELP1-ExSpeU1s delivered by AAV are active on ELP1 transcripts and ELP1 protein in vivo, represent a proof of principle of the therapeutic potential of these molecules for FD treatment. Moreover, the ExSpeU1 platform can be applied for the correction of similar splicing defects in other human pathologies.
Material and Methods
Creation of expression vectors
ELP1 hybrid minigenes were created by cloning the genomic region spanning intron 18–21 into the previously described pTB-NdeI minigene (57). The intronic C.2204 + 6T > C mutation was inserted by site-directed mutagenesis using the QuikChange Site-directed Mutagenesis Kit II (Agilent, Santa Clara, USA). Constructs were validated by sequence analysis. ExSpeU1 snRNAs were created by replacing the sequence between the BclI and BglII sites with oligonucleotides as previously reported (22). The oligonucleotide sequences are in Supplementary Material, Table S1. Modified U7 snRNAs were created by PCR amplification of the U7SmOPT vector using specific primers, U7Ik10F and SP6R, as previously described (22). Lentiviral vectors were created by cloning the U1 expression cassettes into pRRL human immunodeficiency virus-derived backbone at the BamHI and PstI sites (Supplementary Material, Fig. S6A). Lentiviral vectors expressing the snRNAs (U1 wt, U1 Ik10) were prepared as previously described (58). Virus preps were titered as previously described (58), yielding 7.8×108 and 1.1×109 IG/ml infectious genomes per ml (IG/ml) for U1-Ik10 and U1 wt, respectively.
Cell cultures, transfections and lentiviral transduction
Hek293T cells were grown in Dulbecco’s modified eagle medium with Glutamax I (ThermoFisher, Waltham, MA USA) supplemented with nonessential amino acids, 125 U/ml penicillin, 125 µg/ml streptomycin and 10% fetal bovine serum. Cells were maintained at 37°C with 5% CO2. Transfection experiments were carried out in 6-well plates using Lipofectamine 2000 (ThermoFisher) according to manufacturer’s instructions using 1 µg of each plasmid. Human FD fibroblasts were purchased from the Coriell Institute (GM04959, GM04899) and maintained as suggested by the supplier. At day 0, FD fibroblasts were seeded at a density of 6×104 cells/well. At day 1, the cells were transduced with lentiviral ExSpeU1 Ik10 and lentiviral U1 wild-type with increasing concentrations (multiplicities of infection, 0–20). Polybrene (Sigma, St. Louis, MO USA) was used at a concentration of 10 µg/ml to reduce the electronegativity between cells and viruses’ surfaces. Samples were harvested at 72 h post-transduction for RNA and protein analysis.
RNA isolation and ELP1 splicing analysis
Total RNA was extracted with TRIzol (ThermoFisher) following the manufacturer’s instructions. Reverse-transcription was performed using 1 µg of total RNA and Superscript Vilo MasterMix (ThermoFisher). ELP1 minigene splicing was analyzed by endpoint PCR using specific primers, alpha 2, 3 F and ELP1E×21 R (Supplementary Material, Table S1). PCR conditions were 94° C for 5 min, 40 cycles at 94° C for 30 s, 56°C for 30 s, 72° for 40 sfollowed by 72°C for 10 min. PCR products were analyzed by electrophoresis on a 2% agarose gel and the intensity of the bands was quantified with ImageJ Software (NIH). Endpoint PCR analysis of human ELP1 splicing was performed with human-specific ELP1 primers ELP1E×19 F and ELP1E×21 R. Quantitative PCR was performed with iQ-SYBR Green Supermix using human-specific ELP1 primers: ELP1FLF and ELP1FLR for the full-length ELP1 transcripts, ELP1Δ20F and ELP1Δ20 R for transcripts lacking the exon 20. Total ELP1 mRNA was quantified with human-specific primers TotELP1-F and TotELP1-R. Quantitative PCR conditions were: 1 cycle at 95°C for 3 min; 40 cycles at 95°C for 10 s and at 59°C for 45 s. GAPDH was used as control in human and mouse samples (named hGAPDH-F/hGAPDH-R or mGAPDH-F/mGAPDH-R, respectively). Primers’ sequences are in supplementary material (Supplementary Material, Table S1).
Protein isolation and western blot analysis
Protein samples were obtained by tissue lysis with ice-cold RIPA buffer (Sigma) and Proteinase Inhibitor Cocktail (Roche). Only 50 µg of total protein was separated on NuPAGE 4–12% Bis-Tris precast gels (ThermoFisher) and transferred to nitrocellulose membranes. The membranes were blocked for 1 h in 5% non-fat milk and successively blotted overnight at +4°C in 5% milk with a primary antibody against ELP1 (1:500; Anaspec) and a primary antibody against α-tubulin (1:5000; Santa Cruz Biotechnology). Membranes were washed and incubated with specific secondary antibodies for 1 h at room temperature (anti-Rabbit 1:2000 and anti-mouse 1:5000). Protein bands were detected with ECL western blotting substrate (ThermoFisher) followed by exposure to X-ray film (Kodak).
AAV9 production and titration
AAV vectors were created by cloning the U1 expression cassettes into the AAV9 backbone using the XbaI site and prepared as previously described (59) (Supplementary Material, Fig. S6B). Genome-containing vectors were produced in roller bottles by a triple-transfection protocol including a cesium chloride gradient purification step (59). Titers of AAV vector stocks were determined by qPCR. The determined titers were of 7.10×1012 VG/ml for the AAV9-ExSpeU1 Ik10 and of 5.30×1012 VG/ml for the AAV9-ExSpeU1 Ik15.
In vivo analysis in FD mouse model
The transgenic FD mouse model TgFD9 has been previously described by Hims et al. (43). Mice were housed and handled according to institutional guidelines, and experimental procedures approved by the International Centre for Genetic Engineering and Biotechnology board. All in vivo experimental procedures were approved by the competent Italian competent authorities and Ethics Committees according to the European Directive 2010/63/EU.
Newborn mice were intraperitoneally injected at two different post-natal days (0 and 2). Four TgFD9 mice (two males and two females) were treated with AAV9-ExSpeU1 Ik10, four TgFD9 mice (one male and three females) with AAV9-ExSpeU1 Ik15 and four TgFD9 mice (two males and two females) with saline. Injections with a final volume of 25 µl were performed with a micro-syringe (Hamilton) with a 33-gauge removable needle. Mice were sacrificed at post-natal day 7 to collect tissues and organs (brain, liver, heart, kidney, muscle and spinal cord). RNA and protein extraction was performed as described in RNA isolation and protein isolation paragraphs.
Statistical analysis
Data are represented as mean±SD of triplicate experiments. Data were analyzed by two-way ANOVA followed by a Bonferroni correction for false discovery rate with Prism software (GraphPad Software, La Jolla, CA).
Supplementary Material
Supplementary Material is available at HMG online.
Conflict of Interest statement. S.A.S. has a financial interest in PTC Therapeutics, Inc., a company developing small-molecule drugs that target mRNA splicing. S.A.S.’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies. F.P. and M.P. are listed as inventors in the US patent n. 9 669 109 ‘A modified human U1snRNA molecule, a gene encoding for the modified human U1snRNA molecule, an expression vector including the gene, and the use thereof in gene therapy of familial dysautonomia and spinal muscular atrophy’. As such the inventors could potentially benefit from any future commercial exploitation of patent rights, including the use of ExSpeU1s in Familial D.
Supplementary Material
Funding
This work was supported by the Telethon Foundation (grant number GGP17006 to F.P.), the Association Francais contre les Myopathies (AFM; grant n. 19500 to F.P.) the Muscular Dystrophy Association (MDA 383229 to F.P.), the National Institute of Health (R01NS095640 to S.A.S.) and the Elizabeth G Riley and Dan E. Smith, Jr MGH Research Scholar Award to S.A.S. Funding to pay the Open Access publication charges for this article was provided by ICGEB institutional funds.
References
- 1. Riley C.M., Day R.L., David M.G., Langford W.S. (1949) Central autonomic dysfunction with defective lacrimation; report of five cases. Pediatrics, 3, 468–478. [PubMed] [Google Scholar]
- 2. Axelrod F.B. (2005) Familial dysautonomia: a review of the current pharmacological treatments. Expert. Opin. Pharmacother., 6, 561–567. [DOI] [PubMed] [Google Scholar]
- 3. Jackson M.Z., Gruner K.A., Qin C., Tourtellotte W.G. (2014) A neuron autonomous role for the familial dysautonomia gene ELP1 in sympathetic and sensory target tissue innervation. Development, 141, 2452–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Norcliffe-Kaufmann L., Slaugenhaupt S.A., Kaufmann H. (2017) Familial dysautonomia: history, genotype, phenotype and translational research. Prog. Neurobiol., 152, 131–148. [DOI] [PubMed] [Google Scholar]
- 5. Dietrich P., Dragatsis I. (2016) Familial dysautonomia: mechanisms and models. Genet. Mol. Biol., 39, 497–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Slaugenhaupt S.A., Blumenfeld A., Gill S.P., Leyne M., Mull J., Cuajungco M.P., Liebert C.B., Chadwick B., Idelson M., Reznik L.. et al. (2001) Tissue-specific expression of a splicing mutation in the IKBKAP gene causes familial dysautonomia. Am. J. Hum. Genet., 68, 598–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Axelrod F.B. (2002) Hereditary sensory and autonomic neuropathies. Familial dysautonomia and other HSANs. Clin. Auton. Res., 12 (Suppl. 1), I2–14. [DOI] [PubMed] [Google Scholar]
- 8. Axelrod F.B. (2006) A world without pain or tears. Clin. Auton. Res., 16, 90–97. [DOI] [PubMed] [Google Scholar]
- 9. Axelrod F.B., Gold-von Simson G. (2007) Hereditary sensory and autonomic neuropathies: types II, III, and IV. Orphanet. J. Rare Dis., 2, 39.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Anderson S.L., Coli R., Daly I.W., Kichula E.A., Rork M.J., Volpi S.A., Ekstein J., Rubin B.Y. (2001) Familial dysautonomia is caused by mutations of the IKAP gene. Am. J. Hum. Genet., 68, 753–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hawkes N.A., Otero G., Winkler G.S., Marshall N., Dahmus M.E., Krappmann D., Scheidereit C., Thomas C.L., Schiavo G., Erdjument-Bromage H.. et al. (2002) Purification and characterization of the human elongator complex. J. Biol. Chem., 277, 3047–3052. [DOI] [PubMed] [Google Scholar]
- 12. Krappmann D., Hatada E.N., Tegethoff S., Li J., Klippel A., Giese K., Baeuerle P.A., Scheidereit C. (2000) The I kappa B kinase (IKK) complex is tripartite and contains IKK gamma but not IKAP as a regular component. J. Biol. Chem., 275, 29779–29787. [DOI] [PubMed] [Google Scholar]
- 13. Holmberg C., Katz S., Lerdrup M., Herdegen T., Jaattela M., Aronheim A., Kallunki T. (2002) A novel specific role for I kappa B kinase complex-associated protein in cytosolic stress signaling. J. Biol. Chem., 277, 31918–31928. [DOI] [PubMed] [Google Scholar]
- 14. Close P., Hawkes N., Cornez I., Creppe C., Lambert C.A., Rogister B., Siebenlist U., Merville M.P., Slaugenhaupt S.A., Bours V.. et al. (2006) Transcription impairment and cell migration defects in elongator-depleted cells: implication for familial dysautonomia. Mol. Cell, 22, 521–531. [DOI] [PubMed] [Google Scholar]
- 15. Cuajungco M.P., Leyne M., Mull J., Gill S.P., Lu W., Zagzag D., Axelrod F.B., Maayan C., Gusella J.F., Slaugenhaupt S.A. (2003) Tissue-specific reduction in splicing efficiency of IKBKAP due to the major mutation associated with familial dysautonomia. Am. J. Hum. Genet., 72, 749–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hims M.M., Ibrahim E.C., Leyne M., Mull J., Liu L., Lazaro C., Shetty R.S., Gill S., Gusella J.F., Reed R.. et al. (2007) Therapeutic potential and mechanism of kinetin as a treatment for the human splicing disease familial dysautonomia. J. Mol. Med. (Berl.), 85, 149–161. [DOI] [PubMed] [Google Scholar]
- 17. Carmel I., Tal S., Vig I., Ast G. (2004) Comparative analysis detects dependencies among the 5' splice-site positions. RNA, 10, 828–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cartegni L., Chew S.L., Krainer A.R. (2002) Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet., 3, 285–298. [DOI] [PubMed] [Google Scholar]
- 19. Pagani F., Baralle F.E. (2004) Genomic variants in exons and introns: identifying the splicing spoilers. Nat. Rev. Genet., 5, 389–396. [DOI] [PubMed] [Google Scholar]
- 20. Chen M., Manley J.L. (2009) Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol., 10, 741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Daguenet E., Dujardin G., Valcarcel J. (2015) The pathogenicity of splicing defects: mechanistic insights into pre-mRNA processing inform novel therapeutic approaches. EMBO Rep., 16, 1640–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fernandez Alanis E., Pinotti M., Dal Mas A., Balestra D., Cavallari N., Rogalska M.E., Bernardi F., Pagani F. (2012) An exon-specific U1 small nuclear RNA (snRNA) strategy to correct splicing defects. Hum. Mol. Genet., 21, 2389–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tajnik M., Rogalska M.E., Bussani E., Barbon E., Balestra D., Pinotti M., Pagani F. (2016) Molecular basis and therapeutic strategies to rescue factor IX variants that affect splicing and protein function. PLoS Genet., 12, e1006082.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dal Mas A., Rogalska M.E., Bussani E., Pagani F. (2015) Improvement of SMN2 pre-mRNA processing mediated by exon-specific U1 small nuclear RNA. Am. J. Hum. Genet., 96, 93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dal Mas A., Fortugno P., Donadon I., Levati L., Castiglia D., Pagani F. (2015) Exon-specific U1s correct SPINK5 exon 11 skipping caused by a synonymous substitution that affects a bifunctional splicing regulatory element. Hum. Mutat., 36, 504–512. [DOI] [PubMed] [Google Scholar]
- 26. Pinotti M., Rizzotto L., Balestra D., Lewandowska M.A., Cavallari N., Marchetti G., Bernardi F., Pagani F. (2008) U1-snRNA-mediated rescue of mRNA processing in severe factor VII deficiency. Blood, 111, 2681–2684. [DOI] [PubMed] [Google Scholar]
- 27. Balestra D., Scalet D., Pagani F., Rogalska M.E., Mari R., Bernardi F., Pinotti M. (2016) An exon-specific U1snRNA induces a robust factor IX activity in mice expressing multiple human FIX splicing mutants. Mol. Ther. Nucleic Acids, 5, e370.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rogalska M.E., Tajnik M., Licastro D., Bussani E., Camparini L., Mattioli C., Pagani F. (2016) Therapeutic activity of modified U1 core spliceosomal particles. Nat. Commun., 7, 11168.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nathwani A.C., Reiss U.M., Tuddenham E.G., Rosales C., Chowdary P., McIntosh J., Della Peruta M., Lheriteau E., Patel N., Raj D.. et al. (2014) Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N. Engl. J. Med., 371, 1994–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Scoto M., Finkel R.S., Mercuri E., Muntoni F. (2017) Therapeutic approaches for spinal muscular atrophy (SMA). Gene Ther., 24, 514–519. [DOI] [PubMed] [Google Scholar]
- 31. Bennett J., Wellman J., Marshall K.A., McCague S., Ashtari M., DiStefano-Pappas J., Elci O.U., Chung D.C., Sun J., Wright J.F.. et al. (2016) Safety and durability of effect of contralateral-eye administration of AAV2 gene therapy in patients with childhood-onset blindness caused by RPE65 mutations: a follow-on phase 1 trial. Lancet, 388, 661–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mingozzi F., High K.A. (2011) Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat. Rev. Genet., 12, 341–355. [DOI] [PubMed] [Google Scholar]
- 33. Axelrod F.B., Liebes L., Gold-Von Simson G., Mendoza S., Mull J., Leyne M., Norcliffe-Kaufmann L., Kaufmann H., Slaugenhaupt S.A. (2011) Kinetin improves IKBKAP mRNA splicing in patients with familial dysautonomia. Pediatr. Res., 70, 480–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Slaugenhaupt S.A., Mull J., Leyne M., Cuajungco M.P., Gill S.P., Hims M.M., Quintero F., Axelrod F.B., Gusella J.F. (2003) Rescue of a human mRNA splicing defect by the plant cytokinin kinetin. Hum. Mol. Genet., 13, 429–436. [DOI] [PubMed] [Google Scholar]
- 35. Liu B., Anderson S.L., Qiu J., Rubin B.Y. (2013) Cardiac glycosides correct aberrant splicing of IKBKAP-encoded mRNA in familial dysautonomia derived cells by suppressing expression of SRSF3. Febs. J., 280, 3632–3646. [DOI] [PubMed] [Google Scholar]
- 36. Bochner R., Ziv Y., Zeevi D., Donyo M., Abraham L., Ashery-Padan R., Ast G. (2013) Phosphatidylserine increases IKBKAP levels in a humanized knock-in IKBKAP mouse model. Hum. Mol. Genet., 22, 2785–2794. [DOI] [PubMed] [Google Scholar]
- 37. Keren H., Donyo M., Zeevi D., Maayan C., Pupko T., Ast G. (2010) Phosphatidylserine increases IKBKAP levels in familial dysautonomia cells. PLoS One, 5, e15884.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yoshida M., Kataoka N., Miyauchi K., Ohe K., Iida K., Yoshida S., Nojima T., Okuno Y., Onogi H., Usui T.. et al. (2015) Rectifier of aberrant mRNA splicing recovers tRNA modification in familial dysautonomia. Proc. Natl. Acad. Sci. U S A, 112, 2764–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lee G., Papapetrou E.P., Kim H., Chambers S.M., Tomishima M.J., Fasano C.A., Ganat Y.M., Menon J., Shimizu F., Viale A.. et al. (2009) Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature, 461, 402–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Biferi M.G., Cohen-Tannoudji M., Cappelletto A., Giroux B., Roda M., Astord S., Marais T., Bos C., Voit T., Ferry A.. et al. (2017) A new AAV10-U7-mediated gene therapy prolongs survival and restores function in an ALS mouse model. Mol. Ther., 25, 2038–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Odermatt P., Trub J., Furrer L., Fricker R., Marti A., Schumperli D. (2016) Somatic therapy of a mouse SMA model with a U7 snRNA gene correcting SMN2 splicing. Mol. Ther., 24, 1797–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nuzzo F., Radu C., Baralle M., Spiezia L., Hackeng T.M., Simioni P., Castoldi E. (2013) Antisense-based RNA therapy of factor V deficiency: in vitro and ex vivo rescue of a F5 deep-intronic splicing mutation. Blood, 122, 3825–3831. [DOI] [PubMed] [Google Scholar]
- 43. Hims M.M., Shetty R.S., Pickel J., Mull J., Leyne M., Liu L., Gusella J.F., Slaugenhaupt S.A. (2007) A humanized IKBKAP transgenic mouse models a tissue-specific human splicing defect. Genomics, 90, 389–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kashima T., Manley J.L. (2003) A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat. Genet., 34, 460–463. [DOI] [PubMed] [Google Scholar]
- 45. Hua Y., Vickers T.A., Okunola H.L., Bennett C.F., Krainer A.R. (2008) Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am. J. Hum. Genet., 82, 834–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rogelj B., Easton L.E., Bogu G.K., Stanton L.W., Rot G., Curk T., Zupan B., Sugimoto Y., Modic M., Haberman N.. et al. (2012) Widespread binding of FUS along nascent RNA regulates alternative splicing in the brain. Sci. Rep., 2, 603.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vance C., Rogelj B., Hortobagyi T., De Vos K.J., Nishimura A.L., Sreedharan J., Hu X., Smith B., Ruddy D., Wright P.. et al. (2009) Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science, 323, 1208–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ohe K., Yoshida M., Nakano-Kobayashi A., Hosokawa M., Sako Y., Sakuma M., Okuno Y., Usui T., Ninomiya K., Nojima T.. et al. (2017) RBM24 promotes U1 snRNP recognition of the mutated 5' splice site in the IKBKAP gene of familial dysautonomia. RNA, 23, 1393–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dietrich P., Alli S., Shanmugasundaram R., Dragatsis I. (2012) IKAP expression levels modulate disease severity in a mouse model of familial dysautonomia. Hum. Mol. Genet., 21, 5078–5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Machida A., Kuwahara H., Mayra A., Kubodera T., Hirai T., Sunaga F., Tajiri M., Hirai Y., Shimada T., Mizusawa H.. et al. (2013) Intraperitoneal administration of AAV9-shRNA inhibits target gene expression in the dorsal root ganglia of neonatal mice. Mol. Pain, 9, 36.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Foust K.D., Poirier A., Pacak C.A., Mandel R.J., Flotte T.R. (2008) Neonatal intraperitoneal or intravenous injections of recombinant adeno-associated virus type 8 transduce dorsal root ganglia and lower motor neurons. Hum. Gene Ther., 19, 61–70. [DOI] [PubMed] [Google Scholar]
- 52. Tanguy Y., Biferi M.G., Besse A., Astord S., Cohen-Tannoudji M., Marais T., Barkats M. (2015) Systemic AAVrh10 provides higher transgene expression than AAV9 in the brain and the spinal cord of neonatal mice. Front. Mol. Neurosci., 8, 36.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chen Y.T., Hims M.M., Shetty R.S., Mull J., Liu L., Leyne M., Slaugenhaupt S.A. (2009) Loss of mouse Ikbkap, a subunit of elongator, leads to transcriptional deficits and embryonic lethality that can be rescued by human IKBKAP. Mol. Cell Biol., 29, 736–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Morini E., Dietrich P., Salani M., Downs H.M., Wojtkiewicz G.R., Alli S., Brenner A., Nilbratt M., LeClair J.W., Oaklander A.L.. et al. (2016) Sensory and autonomic deficits in a new humanized mouse model of familial dysautonomia. Hum. Mol. Genet., 25, 1116–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mendoza-Santiesteban C.E., Hedges T.R. III, Norcliffe-Kaufmann L., Warren F., Reddy S., Axelrod F.B., Kaufmann H. (2012) Clinical neuro-ophthalmic findings in familial dysautonomia. J. Neuroophthalmol., 32, 23–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mendoza-Santiesteban C.E., Hedges Iii T.R., Norcliffe-Kaufmann L., Axelrod F., Kaufmann H. (2014) Selective retinal ganglion cell loss in familial dysautonomia. J. Neurol., 261, 702–709. [DOI] [PubMed] [Google Scholar]
- 57. Pagani F., Buratti E., Stuani C., Baralle F.E. (2003) Missense, nonsense and neutral mutations define juxtaposed regulatory elements of splicing in CFTR exon 9. J. Biol. Chem., 278, 26580–26588. [DOI] [PubMed] [Google Scholar]
- 58. Charrier S., Ferrand M., Zerbato M., Precigout G., Viornery A., Bucher-Laurent S., Benkhelifa-Ziyyat S., Merten O.W., Perea J., Galy A. (2011) Quantification of lentiviral vector copy numbers in individual hematopoietic colony-forming cells shows vector dose-dependent effects on the frequency and level of transduction. Gene Ther., 18, 479–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ayuso E., Mingozzi F., Montane J., Leon X., Anguela X.M., Haurigot V., Edmonson S.A., Africa L., Zhou S., High K.A.. et al. (2010) High AAV vector purity results in serotype- and tissue-independent enhancement of transduction efficiency. Gene Ther., 17, 503–510. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.